Abstract
Signaling by the nonreceptor tyrosine kinase Abelson (Abl) plays key roles in normal development, whereas its inappropriate activation helps trigger the development of several forms of leukemia. Abl is best known for its roles in axon guidance, but Abl and its relatives also help regulate embryonic morphogenesis in epithelial tissues. Here, we explore the role of regulation of Abl kinase activity during development. We first compare the subcellular localization of Abl protein and of active Abl, by using a phosphospecific antibody, providing a catalog of places where Abl is activated. Next, we explore the consequences for morphogenesis of overexpressing wild-type Abl or expressing the activated form found in leukemia, Bcr-Abl. We find dose-dependent effects of elevating Abl activity on morphogenetic movements such as head involution and dorsal closure, on cell shape changes, on cell protrusive behavior, and on the organization of the actin cytoskeleton. Most of the effects of Abl activation parallel those caused by reduction in function of its target Enabled. Abl activation leads to changes in Enabled phosphorylation and localization, suggesting a mechanism of action. These data provide new insight into how regulated Abl activity helps direct normal development and into possible biological functions of Bcr-Abl.
INTRODUCTION
A significant insight into cancer was provided by chronic myelogenous leukemia (CML). The identification of a characteristic chromosome translocation, the Philadelphia chromosome (Ph) (Nowell and Hungerford, 1961; Rowley, 1973), ultimately led to discovery of the c-abl gene, and the revelation that it encodes a nonreceptor tyrosine kinase. This culminated in development of an Abelson (Abl) kinase-specific inhibitor, imatinib, leading to major advances in treatment of CML (for review, see Deininger et al., 2005). In parallel, scientists pursued normal functions of Abl family kinases in cultured cells and during animal development, revealing key roles in regulating many cellular processes, including cytoskeletal function. However, many questions remain about the normal roles of Abl and how the translocation alters its activity and thus alters cell behavior.
Abl normally exists in an inactive conformation, characterized by intramolecular interactions between the kinase, Src homology (SH)2 and SH3 domains, and locked into place by insertion of the myristoylated N terminus into a binding pocket on the kinase domain (Nagar et al., 2003). The mutant kinase found in CML is constitutively active, due to at least two events (for review, see Advani and Pendergast, 2002). The fusion protein retains most abl coding sequences, but deletes the myristoylation site from the extreme N terminus of Abl, thus “unlocking” the closed, inactive conformation. Sequences from Bcr also contribute to fusion protein activity. Different forms of Ph+ leukemia are associated with slightly different translocations; all have the same region of Abl, but they differ in the amount of Bcr. The shortest fusion protein, p185, is usually associated with acute lymphocytic leukemia, whereas CML patients typically express a longer fusion protein, p210. Most studies agree that the N-terminal Bcr dimerization domain, present in both fusion proteins, disrupts the autoinhibited “closed” conformation (Smith et al., 2003) and promotes transautophosphorylation and kinase activation.
Abl has a long C-terminal tail that mediates cytoskeletal interactions. Near the C terminus is a site that binds and bundles actin (Van Etten et al., 1994; Hantschel et al., 2005). This region is retained in the Bcr-Abl fusion protein, which localizes to the cytoplasm and colocalizes with actin. Cytoskeletal interactions are thought to localize Abl to sites of action. However, Abl binding to actin inhibits the kinase activity of Abl (Woodring et al., 2001), raising the possibility that Abl docks at actin in an inactive form for action in the neighborhood. The mammalian Abl paralogue, Arg, has a second actin-binding site (Wang et al., 2001) and also carries a microtubule-binding site, allowing it to cross-link actin and microtubules (Miller et al., 2004).
Studies in cultured mammalian cells and model animals both suggest that Abl plays a key role at the interface between signal transduction and cytoskeletal regulation. In cultured cells, Abl regulates cytoskeletal and adhesive responses to extracellular stimuli (for review, see Hernandez et al., 2004). For example, Abl is activated by platelet-derived growth factor stimulation, and is required for the cell ruffling and chemotaxis that occur in response (Plattner et al., 1999; 2004). Loss of both Abl and Arg or expression of kinase-dead Abl leads to increased cell motility, whereas overexpression of Abl inhibits migration (Kain and Klemke, 2001). This may suggest a role for Abl in modulating cell–matrix adhesion (for review, see Hernandez et al., 2004). Like Abl, Bcr-Abl also influences cell behavior and the cytoskeleton. In vivo, Bcr-Abl reduces adhesion to bone marrow stroma (Gordon et al., 1987), whereas in vitro it has complex, cell-type–dependent effects on adhesion to fibronectin and cell migration (Salesse and Verfaillie, 2002). Bcr-Abl also has other cytoplasmic functions (for review, see Advani and Pendergast, 2002), allowing it to stimulate proliferation and inhibit apoptosis, rendering cells independent of cytokines. Localization of Bcr-Abl exclusively to the cytoplasm suggests that these cytoskeletal and signaling events are critical.
Parallel studies of Abl function in whole animals also revealed roles connecting signaling and the cytoskeleton. In Drosophila, loss-of-function mutations (Henkemeyer et al., 1987) lead to defects in axon outgrowth in the central and peripheral nervous systems (CNS and PNS, respectively; for review, see Moresco and Koleske, 2003). Abl acts downstream of several axon guidance receptors, including the Robo family and the receptor tyrosine phosphatase Lar, to regulate both the actin and microtubule cytoskeletons in growth cones, thus mediating growth cone guidance. Drosophila Abl also plays important roles in epithelial tissues (Baum and Perrimon, 2001; Grevengoed et al., 2001), regulating the actin cytoskeleton and thus morphogenetic movements in follicle cells and the embryonic epidermis. In the epidermis, Abl works with DE-cadherin, regulating cytoskeletal responses required for cell shape changes and migration.
In mice the two Abl paralogues complicate analysis. Both single mutant mice are relatively normal—abl mutants have defects in lymphocyte and osteoblast development, whereas arg mutants have behavioral problems correlated with changes in synaptic function (for review, see Hernandez et al., 2004). However, abl; arg double mutants die before E11, and they have defects in neural tube closure (Koleske et al., 1998), probably due to defects in the actin cytoskeleton. A recent brain-specific knockout also revealed roles for Abl and Arg in dendrite branch maintenance (Moresco et al., 2005). Surprisingly, the single Caenorhabditis elegans abl gene is dispensable for viability and morphogenesis (Deng et al., 2004).
Several potential effectors may allow Abl family kinases to modulate cytoskeletal activity, including p190RhoGAP, Crk, Abi proteins, and neural Wiskott Aldrich syndrome protein (N-WASP) (for review, see Hernandez et al., 2004). Abl/Arg also can directly affect actin assembly and bundling. In Drosophila, Enabled (Ena), a member of the Ena/VASP family of actin regulators, is an essential Abl target in both the CNS (Gertler et al., 1990) and epithelia (Grevengoed et al., 2001; 2003). Ena/VASP proteins regulate actin polymerization, promoting production of unbranched actin filaments by acting as anticapping proteins (Bear et al., 2002; Barzik et al., 2005). Abl negatively regulates Ena (Gertler et al., 1990), at least in part by regulating its intracellular localization (Grevengoed et al., 2003). Whereas Abl phosphorylates Ena (Comer et al., 1998), mutations of the phosphorylation sites suggest this is not essential for negative regulation. The mammalian Ena relatives Mena and VASP are also Abl targets (Howe et al., 2002; Tani et al., 2003).
Studies of Bcr-Abl in model organisms offer the opportunity to assess the effects of Abl activation on cell behavior in vivo. Fogerty et al. (1999) generated transgenic Drosophila expressing p185 or p210 Bcr-Abl. In these, Bcr and the N-terminal Abl sequences are derived from the human oncogenes (these regions of Abl are highly similar in fly and human), whereas the C-terminal Abl tail is derived from Drosophila, maximizing the likelihood that the Bcr-Abl proteins would effectively interact with endogenous Drosophila regulators and targets. Each transgene is under the control of a GAL4-activated promotor, allowing it to be introduced into the genome in a silent state. Expression of Bcr-Abl can be activated in a tissue and temporally specific manner by crossing to lines expressing GAL4 in particular tissues or times. Interestingly, each Bcr-Abl isoform can rescue abl mutants, suggesting that their signaling properties are not grossly altered. However, neural-specific expression of these oncogenes in a wild-type background disrupts CNS development, whereas neural-specific overexpression of wild-type Abl does not, suggesting that regulated kinase activity is important (Fogerty et al., 1999). Interestingly, some neuronal Bcr-Abl phenotypes resembled those of ena loss-of-function, and Ena phosphorylation was increased in Bcr-Abl expressing flies, consistent with Ena being a Bcr-Abl target.
We used this model system to extend our analysis of Abl function during morphogenesis, examining in parallel the effects of overexpressing wild-type Abl and misexpressing Bcr-Abl. The embryonic epithelium, which undergoes many types of morphogenetic change, provides an excellent model to study the effects of inappropriate Abl activation on cell adhesion and cytoskeletal regulation, revealing insight into the mechanisms of action of Abl during morphogenesis.
MATERIALS AND METHODS
Fly Stocks and Cuticle Analysis
Flies carrying UAS-Bcr-Abl and UAS-Abl transgenes were provided by F. Fogerty (University of Wisconsin, Madison, WI). In all experiments, females carrying the UAS transgene were crossed to males with a GAL4 driver (engrailed [en]-GAL4, e22-GAL4, and arm-GAL4:VP16). Sources of fly stocks are as follows: moesin-green fluorescent protein (GFP), Dan Kiehart (Duke University, Durham, NC); UAS-GFP-actin, Paul Martin (University of Bristol, Bristol, United Kingdom); UAS-PKNG58AeGFP, A. Jacinto (Lisbon, Portugal). Wild-type was Canton S. All other fly stocks were obtained from the Bloomington Stock Center (Department of Biology, Indiana University, Bloomington, IN), and they are described at FlyBase (flybase.bio.indiana.edu). Flies were grown at 25°C. Cuticle preparations were as in Wieschaus and Nüsslein-Volhard (1986).
Immunoprecipitation and Immunoblotting
Live embryos were visually selected at the cellular blastoderm stage in halocarbon oil (series 700; Halocarbon Products, River Edge, NJ) and aged defined periods of time. For immunoblotting, embryos were homogenized in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (62.5 mM Tris-HCl, pH 8.0, 40% glycerol, 8% β-mercaptoethanol, 0.06% bromophenol blue, and 1.3% SDS) with 0.5 mM sodium orthovanadate and Complete EDTA-free protease inhibitors (Roche Molecular Biochemicals, Lewes, United Kingdom). Samples were analyzed by 6% SDS-PAGE and transferred to nitrocellulose. Membranes were blocked and incubated with primary and secondary antibodies in 5% nonfat dry milk (or 5% bovine serum albumin for phosphospecific antibodies) in Tris-buffered saline/Tween 20. Signal was detected using the ECL kit (GE Healthcare Bio-Sciences, Little Chalfont, Buckinghamshire, United Kingdom). For immunoprecipitations, embryos were homogenized in NET buffer (50 mM Tris, pH 7.5, 400 mM NaCl, 5 mM EDTA, and 1% NP-40) on ice. Homogenate was clarified by centrifugation at 3500 × g for 6 min, and incubated with primary antibodies for 1 h at 4°C. Protein A-Sepharose beads were added, incubated 2 h at 4°C, washed six times with NET buffer and boiled in SDS-PAGE sample buffer. Antibodies used were as follows: mouse monoclonals anti-phosphotyrosine (clone 4G10, 1:250; Upstate Biotechnology, Lake Placid, NY), anti-phosphotyrosine-RC20:HRP0 (1:500; BD Biosciences, San Jose, CA), anti-actin (1:500; Chemicon International, Temecula, CA), anti-armadillo (Arm) (1:250), anti-Pnut (1:30), anti-BicD (1:500), and anti-Ena (1:250; all Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); rat monoclonals anti-α-catenin (1:250) and anti-DCAD1 (1:500; both Developmental Studies Hybridoma Bank); rabbit anti-c-Abl (phospho Y412) (1:250; Abcam, Cambridge, MA) and anti-Bcr (N-20) (1:250; Santa Cruz Biotechnology, Santa Cruz, CA); and guinea pig anti-dAbl (1:50; Grevengoed et al., 2001).
Immunofluorescence and Microscopy
Embryos were dechorionated in bleach and fixed in 1:1 3.7% formaldehyde in phosphate-buffered saline:heptane for 20 min at room temperature. Vitelline membranes were removed in methanol. For actin visualization, embryos were fixed in equal amounts of 37% formaldehyde and heptane for 5 min at room temperature; vitelline membranes then were removed manually. Embryos were mounted in Aqua PolyMount (Polysciences, Warrington, PA), and digital images were obtained on a confocal laser-scanning microscope (LSM510; Carl Zeiss MicroImaging, Jena, Germany) by using 40× (Plan-NeoFluar; numerical aperture [NA] 1.3) and 63× (Plan-Apochromat; NA 1.4) objectives, and LSM software. Antibodies used were mouse monoclonals anti-Ena (1:200), anti-Rho1 (1:100), anti-Crb (1:25), anti-Dlg (1:100; all Developmental Studies Hybridoma Bank) and anti-phosphotyrosine (4G10) (1:1000; Upstate Biotechnology); rabbit anti-c-Abl (phospho Y412) (1:250) and anti-Bcr (N-20) (1:250); and guinea pig anti-dAbl (1:100). Actin was visualized with Alexa 488 and Alexa 568 phalloidin (Invitrogen, Carlsbad, CA). Secondary antibodies conjugated to cyanine (Cy)3, Cy5, Alexa 488, and Alexa 568 (Invitrogen) were used. Adobe Photoshop 7.0 (Abobe Systems, Mountain View, CA) was used to adjust input levels so the main range of signals spanned the entire output gray scale, and it was used to adjust brightness and contrast. When protein levels were compared, compared images were equally adjusted.
Time-Lapse Microscopy
Embryos were dechorionated in bleach and mounted in halocarbon oil (series 700; Halocarbon Products Corporation) on a gas permeable membrane (petriPERM 50, hydrophobic; Sartorius). A Perkin Elmer-Cetus Ultraview spinning-disk confocal, ORCA-ER digital camera (Hamamatsu), and Metamorph software were used. Images were acquired every 10 s, except video 2, in which images were acquired every 15 s. Image J was used for quantitation of lamellipodial area and filopodial length.
RESULTS
Abl and Bcr-Abl as Tools to Explore the Effects of Abl Activity during Morphogenesis
To explore the mechanisms by which Abl acts during morphogenesis, we activated Abl by overexpression or by expressing constitutively active Bcr-Abl isoforms. We used transgenic lines expressing wild-type Abl or a Bcr-Abl isoform under the control of the GAL4-UAS system. Different GAL4 drivers allow us to express these lines at different levels and in different cells, thus providing a broad range of Abl hyperactivity. To begin our analysis, we examined expression levels in embryos of several independent lines of each construct, to control for effects of insertion site. We used the e22c-GAL4 driver, which drives expression ubiquitously in both the embryonic epidermis and amnioserosa. We examined UAS-Bcr-Abl lines by immunoblotting with anti-Bcr; most lines expressed Bcr-Abl at roughly similar levels (Supplemental Figure 1A; we used lines with high-level expression for further experiments). We examined the UAS-Abl lines using our antibody raised against Drosophila Abl (Grevengoed et al., 2001). Once again, expression levels in most lines were equivalent (Supplemental Figure 1B). Expression of UAS-Abl driven by e22c-GAL4 is roughly 4–9 times the level of endogenous Abl (range of two different lines each assayed twice). We could not directly compare expression levels of UAS-Abl and UAS-Bcr-Abl, because our fly Abl antibody does not recognize Bcr-Abl as well as it recognizes fly Abl (Supplemental Figure 1C; contrast reactivity with anti-phosphotyrosine and anti-Abl), likely because the region used as an epitope for our anti-fly Abl antibody is derived from human Abl in the Bcr-Abl transgene. However, given the uniformity in expression levels of different lines of the same construct, we suspect that transcription of Abl and Bcr-Abl lines driven by the same driver is likely to be similar, although we cannot rule out differences in protein stability.
Both Abl and Bcr-Abl Are Enriched at the Cortex When Overexpressed
One key question is where within the cell Abl acts during morphogenesis. As a first step to assess this, we examined the localization of overexpressed Abl and misexpressed Bcr-Abl. In wild-type embryos, Abl is cortically localized in early embryogenesis, but after germband extension cortical enrichment is reduced, with primarily diffuse cytoplasmic staining during germband retraction or dorsal closure (Bennett and Hoffmann, 1992; Fox and Peifer, 2007). At later stages, contrast enhancement of the signal revealed weak cortical enrichment in cells expressing normal levels of Abl (Figure 1D), but it was subtle at best. However, when we overexpressed wild-type Abl in stripes of cells using the en-GAL4 driver, we saw cortical enrichment in addition to significant cytoplasmic staining (Figure 1, A and B″, arrow). Cortical enrichment increased during dorsal closure (Figure 1C″, arrow). When Abl was expressed at even higher levels, using arm-GAL4:VP16, the degree of cortical enrichment was increased (Figure 1, E–H). Interestingly, kinase activity was not essential for cortical enrichment, because the kinase-dead mutant exhibited a similar localization (Figure 1J″). We also examined the effect of Abl overexpression on the levels of tyrosine (Tyr)-phosphorylated proteins, using anti-phosphotyrosine (PTyr) antibodies. Normally, PTyr is significantly enriched at the cortex in adherens junctions (AJs) (e.g., Cox et al., 1996). Overexpression of wild-type Abl led to a significant increase in the levels of PTyr signal in both the cytoplasm and at the cortex, which increased in parallel with Abl accumulation as development proceeded (Figure 1, B′ and C′). This depended on Abl kinase-activity (Figure 1J′). We also examined the localization of the Bcr-Abl fusion proteins, using antibodies against Bcr. Like overexpressed Abl, both p185 Bcr-Abl (Figure 1, L″ and N″, arrows) and p210 Bcr-Abl (Figure 1P″, arrow) accumulated in the cytoplasm but were enriched at the cell cortex. This localization also did not require kinase activity of the fusion-protein (Figure 1R″, arrow). Both activated forms of Bcr-Abl stimulated very high levels of accumulation of PTyr both at the cortex and in the cytoplasm (Figure 1L′, N′, and P′), and once again this was dependent on kinase activity (Figure 1R′). Because Abl can exist in an autoinhibited conformation predicted to be cytoplasmic, and because Abl overexpression or Bcr-fusion is likely to trigger activation, these data are consistent with the hypothesis that active Abl is enriched at the cell cortex.
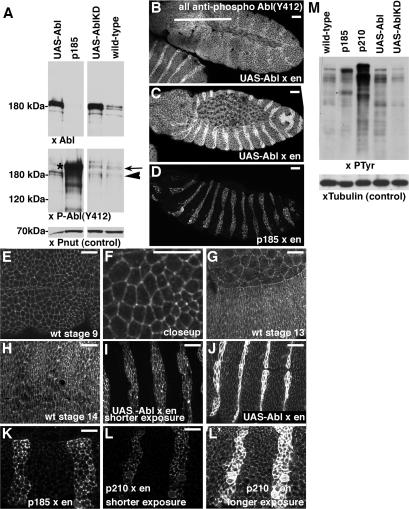
Figure 1.
Abl and Bcr-Abl localization. Embryos, anterior to the left, lateral view unless indicated. Antigens are indicated. (A–D) Wild-type UAS-Abl x en-GAL4. A and B, stage 11. C and D, stage 14. Arrows, cells overexpressing Abl. Arrowheads, cells not overexpressing Abl. (E–H) Wild-type UAS-Abl x arm-GAL4:VP16. E and F, stage 10. G and H, stage 14, ventral views. I and J, UAS-Abl-kinase dead x en-GAL4, stage 13. J, arrow, cortical Abl-kinase dead. (K–N) UAS-p185Bcr-Abl x en-GAL4. K and L, stage 10. M and N, stage 13. O and R, UAS-p210Bcr-Abl x en-GAL4. O and P, stage 12. Q and R, UAS-p210Bcr-Abl-kinase dead x en-GAL4, stage 14, ventral view. L″, N″, P″, R″, arrows, increasing levels of cortical Bcr-Abl. Bars, 20 μm.
Activated Abl Is Strongly Enriched at the Cell Cortex
We next tested this hypothesis directly. Abl activity is regulated, in part, by the phosphorylation of tyrosine residues 245, in the SH3-SH2 domain linker, and 412, in the activation loop of the kinase domain (Brasher and Van Etten, 2000). Antibodies have been generated to the regions surrounding each of these phosphorylated residues that thus specifically recognize activated Abl. In Drosophila, the tyrosine residue corresponding to Tyr412 (Tyr 539/522 in Drosophila) and its surrounding residues are well conserved (13/15 identical amino acids centered on the Y).
We thus examined whether an antibody against this phosphorylated tyrosine recognizes activated fly Abl. On immunoblots, our anti-Abl antibody recognizes a doublet at ∼180 kDa (Figure 2A, xAbl, lane 4). The activated Abl antibody recognizes this doublet (Figure 2A, xPAbl, lane 4, arrowhead), and it also recognizes bands at >200 kDa (Figure 2A, xPAbl, lane 4, arrow) which may be cross-reacting proteins. Overexpression of either wild-type Abl (line UA22) or kinase-dead Abl (line UL6) elevated the intensity of the ∼180-kDa bands recognized by anti-Abl antibody (Figure 2A, xAbl, lanes 1, 3). Overexpression of wild-type Abl leads to an increase in the signal detected by the activated Abl antibody (Figure 2A, xPAbl, lane 1 vs. 4, arrowhead). In contrast, overexpression of kinase-dead Abl does not lead to a corresponding increase in the signal detected by activated Abl antibody (Figure 2A, xPAbl, lane 3 vs. 4, arrowhead); active-Abl signal remains in the kinase-dead Abl lane due to the endogenous wild-type Abl present in these embryos. The activated Abl antibody also recognizes the Bcr-Abl fusion proteins (Figure 2A, xPAbl, lane 2 asterisk; data not shown). This signal is very strong, consistent with the expectation that a larger fraction of Bcr-Abl should be in the activated state (the region of the fusion protein around Tyr 412/539 is derived from fly Abl, and thus this difference is not due to a difference in cross-reactivity between species). As a separate test of whether the activated Abl antibody recognized activated Abl in Drosophila, we overexpressed Abl or Bcr-Abl in stripes in the embryonic epidermis, and stained the embryos with anti-PAbl (Figure 2, B–D; the signal in the Bcr-Abl panel is turned down relative to the others to avoid saturating the signal in the stripes). Both Abl overexpression and Bcr-Abl expression led to elevated PAbl staining, with the elevation most pronounced after Bcr-Abl overexpression, consistent with its expected high level of activity. These data suggest that activated Abl antibody is a good tool to examine activated Abl in Drosophila, although caution is warranted by the presence of cross-reacting bands.
Figure 2.
Localization of active Abl. (A) Anti-PAbl(Y412) recognizes activated Abl in Drosophila. Immunoblots of embryo extracts from wild-type embryos or embryos expressing the indicated transgenes with e22c-GAL4, blotted with the antibodies indicated. UAS-Abl was line UA22, p185 was line 17, and UAS-Abl-kinase-dead (KD) was line UL6. Arrowhead, band at position of wild-type Abl. Arrow, presumed cross-reacting band. Asterisk, band at position of Bcr-Abl. Peanut (Pnut) is a loading control. (B–L) Embryos, anterior to the right. All dorsal up except E, F, H, and J, which are ventral views. Staining with anti-PAbl(Y412). B and C, wild-type UAS-Abl x en-GAL4, stages 11 and 13, respectively. D, UAS-p185Bcr-Abl x en-GAL4, stage 13. E–H, wild-type, stages indicated. I and J, wild-type UAS-Abl x en-GAL4, stage 12, shorter and longer exposures. K, UAS-p185Bcr-Abl x en-GAL4, stage 11. L and L′, UAS-p210Bcr-Abl x en-GAL4, stage 11, shorter and longer exposures. (M) Levels of phosphotyrosine induced by transgenes. Embryo extracts from wild-type embryos or embryos expressing the indicated transgenes with e22c-GAL4, blotted with the antibodies indicated. Lines as in A, plus UAS-p210 was line 31. Bars, 20 μm.
We thus examined subcellular localization of activated Abl, both in wild-type and in cells overexpressing wild-type Abl. In contrast to total Abl (Figure 1, B″ and D, arrowheads), the activated Abl epitope remains significantly enriched at the apicolateral cell cortex of both epidermal and amnioserosal cells in the extended germband (Figure 2, B, E, and F), with elevated levels at tricellular junctions where three cells meet. This cortical enrichment is also seen in both epidermal and amnioserosal cells during dorsal closure (Figure 2G), and it remains in the epidermis after closure (Figure 2H). This is consistent with a role for Abl activation during germband retraction and dorsal closure, and, in fact, both processes are affected by Abl loss-of-function (Grevengoed et al., 2001). When we overexpressed wild-type Abl in stripes using en-GAL4, the activated-Abl signal was substantially elevated (Figure 2, C, I, and J), although the pattern of cortical accumulation remained largely unchanged, consistent with this reflecting localization of activated Abl and not a cross-reacting protein. Misexpression of either p185 or p210 also led to substantially elevated activated-Abl signal in cells expressing the construct, with substantial cortical enrichment (Figure 2, D, K, and L). Together, these data suggest that although Abl localizes to the cytoplasm and cortex, active Abl is highly enriched at the cell cortex.
Expression of Abl or Bcr-Abl Leads to Embryonic Lethality and Disrupts Morphogenesis
Loss of Abl disrupts a number of processes during embryonic morphogenesis (Grevengoed et al., 2001). We hypothesized that regulated Abl kinase activity also plays a key role in regulating these processes. To test this hypothesis, we examined the biological consequences of activating Abl in epithelial cells, by using three GAL4 drivers to express the transgenes in different spatial patterns and/or levels of expression. en-GAL4 drives expression in the posterior-most two to three cells of each epidermal segment, with little or no expression in the amnioserosa. e22c-GAL4 drives expression ubiquitously and at fairly uniform levels in both the embryonic epidermis and amnioserosa. arm-GAL4:VP16 is the strongest driver, and it also leads to ubiquitous expression in the epidermis and amnioserosa. With all three GAL4-drivers, the onset of transgene expression is during the midextended germband stage, with increased accumulation during germband retraction and subsequent stages.
We first examined whether Abl activation affects embryonic viability. When we expressed wild-type Abl ubiquitously, using either e22c-GAL4 or arm-GAL4:VP16, it led to embryonic lethality, although the penetrance of this lethality was lower with e22c-GAL4 (Table 1), suggesting that embryos can tolerate reasonably high levels of wild-type Abl. Embryonic lethality requires kinase activity, because expression of kinase-dead Abl was not embryonic lethal (Table 1). We also expressed the two isoforms of Bcr-Abl by using the same two GAL4-drivers. Expression of either p185 or p210 resulted in highly penetrant embryonic lethality (Table 1), which once again required kinase activity. We also expressed a version of p185 carrying a mutation altering the putative Grb2 binding site in the Bcr portion of Bcr-Abl (Y177F). Expression of this construct was also embryonic lethal (Table 1). In contrast, expression of Abl or Bcr-Abl using the segmentally restricted en-GAL4 driver did not, in general, lead to embryonic lethality (Table 1; the background lethality of different wild-type stocks is 2–10%). These data suggest that widespread unregulated Abl kinase activity is deleterious, whereas embryos can tolerate reasonably high levels of wild-type Abl.
Table 1.
Abl overexpression and Bcr-Abl misexpression cause embryonic lethality
| Driver | Transgene | % embryonic lethalitya | nb |
|---|---|---|---|
| e22-GAL4 | p185 (17) | 100 | 389 |
| p185 (6a1) | 78 | 343 | |
| p185-Y177F (14b) | 80 | 255 | |
| p210 (31) | 96 | 350 | |
| p210 (11a) | 56 | 355 | |
| p210 (6) | 64 | 348 | |
| P210 kinase dead (11a) | 8 | 347 | |
| P210 kinase dead (15a) | 2 | 361 | |
| Wild-type Abl (UA3) | 68 | 698 | |
| Kinase-dead Abl (UL6) | 8 | 366 | |
| arm-GAL4:VP16 | p185 (17) | 100 | 240 |
| p185 (6a1) | 100 | 319 | |
| P185-Y177F (14b) | 100 | 257 | |
| p210 (11a) | 96 | 309 | |
| p210 (11) | 100 | 298 | |
| P210 kinase dead (15a) | 8 | 333 | |
| Wild-type Abl (UA22) | 82 | 319 | |
| Wild-type Abl (UA3) | 100 | 328 | |
| Kinase-dead Abl (UL1) | 4 | 174 | |
| en-GAL4 | p185 (17) | 12 | 242 |
| p185 (6a1) | 12 | 238 | |
| p210 (31) | 12 | 464 | |
| Wild-type Abl (UA22) | 4 | 167 |
a Corrected for fraction of embryos of correct genotype. Both e22-GAL4 and arm-GAL4:VP16 are heterozygous, so only 50% of the embryos receive the driver and thus could be affected. Some of the transgenes were also maintained as balanced stocks and thus are also heterozygous.
b Number of embryos scored.
Previous work demonstrated that regulated Abl activity is necessary for the correct development of the CNS and that CNS-specific expression of Bcr-Abl disrupts the CNS and results in embryonic lethality (Fogerty et al., 1999). Therefore, we assessed whether the embryonic lethality we observed using our GAL4 drivers was due to CNS disruption. Expression of the strongest p210 lines by using e22c-GAL4 led to CNS defects (Figure 3, A vs. D)—longitudinal axons were reduced (arrowheads), whereas commissures were thickened (arrows). However, e22c-GAL4 is not expressed at high levels in the CNS (data not shown), and overexpression of Abl (Figure 3B) or p185 Bcr-Abl (Figure 3C) by using this driver did not substantially disrupt the CNS, suggesting that alterations in the CNS are not the sole cause of embryonic lethality.
Figure 3.
Lethality often occurs without severe effects on the CNS. Axons of the embryonic CNS, revealed by anti-BP102 antibody. (A) Wild type. (B–D) Indicated transgenes crossed to e22c-GAL4. Arrows, commissures. Arrowheads, longitudinal axons. Bar, 20 μm.
We next assessed how Abl activation affects the epithelial epidermis and the morphogenetic movements that shape it, by examining the embryonic cuticle. Cuticle is secreted by the epidermis late in embryogenesis, and thus it provides a sensitive assay for the integrity of the epidermal epithelium and the events of morphogenesis. The wild-type cuticle completely encloses the embryo (Figure 4, A and B), as a result of proper completion of dorsal closure during mid-late embryogenesis. A second complex set of morphogenetic movements, termed head involution, internalizes tissues that secrete the “head skeleton” (Figure 4B, arrow), the structures at the front end of the digestive tract. Disruption of head involution leads to disruptions of the head skeleton or, in more extreme cases, to loss of head cuticle. When we drove wild-type Abl (Figure 4C and Supplemental Table 1) or p185 Bcr-Abl (Figure 4D and Supplemental Table 1) lines with e22c-GAL4, two phenotypic consequences were seen. These were mild effects on the head skeleton (Figure 4, C and D, arrows), suggesting partial disruption of head involution, and disruption of the final stages of dorsal closure, resulting in a failure of the two epidermal sheets to properly pair with their neighbors, leading to puckering of the dorsal cuticle (data not shown). We also assessed a version of p185 with a mutation in the Grb2 binding site that is important for some Bcr-Abl functions in mammalian cells—this had consequences similar to normal p185 (Supplemental Table 1). Expression of p210 Bcr-Abl by using e22c-GAL4 often had more severe consequences, with defects ranging from modest (Figure 4E, arrow) to total disruption of head involution (Figure 4F, arrow, and Supplemental Table 1); some p210 lines also exhibited defects in dorsal closure, including holes in the dorsal cuticle (Figure 4G, arrowhead). When we used the stronger arm-GAL4:VP16 driver, effects of Abl activation were more dramatic (Supplemental Table 1). Overexpression of wild-type Abl (Figure 4H) or misexpression of p185 Bcr-Abl (Figure 4I) now led to severe disruption of head involution (arrows) and partially penetrant dorsal closure defects (arrowheads). Misexpression of p210 Bcr-Abl led to even more dramatic consequences, which at their most severe included disruption of epidermal integrity (Figure 4J). These data suggested that p210 exhibits more biological activity in this context.
Figure 4.
Abl overexpression and Bcr-Abl expression cause defects in morphogenesis. Embryonic cuticles, anterior to the top. (A and B) Wild-type, ventral and dorsal views. Arrow, head skeleton. (C–J) Average cuticle phenotypes of indicated transgenes crossed to indicated drivers; C–I, arrows, defects in head involution: arrows in C–E indicate mild defects in head skeleton, whereas arrows in F–I indicate more severe head holes. F–I, arrowheads, defects in dorsal closure indicated by dorsal holes (G and I) or dorsal scars (F and H). (K and L) Embryos overexpressing wild-type Abl by using e22c-GAL4, which are either wild type (K) or homozygous mutant (L) for ena. An ena mutant cuticle is essentially wild type. Arrow, head hole. Arrowhead, dorsal hole. Bar, 20 μm.
To examine the mechanism underlying the differential activity of Abl, p185 and p210 in Drosophila embryos, we examined overall levels of phosphorylated tyrosine triggered by misexpression of each transgene. We found that the levels of phosphotyrosine correlated perfectly with biological activity, with Abl < p185 < p210 (Figure 2M). Thus, activation of Abl by overexpression or Bcr-fusion disrupts morphogenesis, with the severity of these disruptions correlating with the level of Abl activation.
Abl Activation Affects Cell Shape Changes during Dorsal Closure and Segmental Groove Retraction
These data demonstrate that activated Abl disrupts the end product of morphogenesis, the embryonic cuticle. To get a more detailed mechanistic understanding of how activated Abl affects morphogenesis, we examined the biological and cell biological consequences of Abl activation. We first examined fixed embryos, looking for alterations in the cytoskeleton or cell adhesion that might help explain the defects in morphogenesis. We followed the expression of Bcr-Abl by using an antibody directed against Bcr. To follow Abl overexpression, we used either an antibody against Drosophila Abl or an antibody directed against the activated form of mammalian Abl (Figure 2). We used phalloidin to reveal cell shapes and to mark the actin cytoskeleton.
We examined embryos beginning with the onset of germband retraction, soon after initiation of transgene expression, and continuing through dorsal closure. During wild-type gastrulation, embryos elongate in the anterior-posterior body-axis and narrow in the dorsal-ventral axis, such that the posterior end is now tucked behind its head. During germband retraction the tail end moves back to the posterior (Figure 5A), leaving a sheet of epidermal cells covering the ventral and lateral surfaces of the embryo, and the aminoserosal cells spread over the dorsal surface (Figure 5A, arrowhead). Before the end of germband retraction, there were only subtle effects on morphogenesis. Germband extension was not affected, but it may occur before significant expression of the transgenes.
Figure 5.
Cellular and cytoskeletal consequences of ubiquitous Abl overexpression and Bcr-Abl expression. Embryos, anterior left and, unless noted, lateral view, stained with phalloidin to visualize F-actin. Transgenes and GAL4 drivers are indicated [e22c-GAL4 (e22), arm-GAL4:VP16 (VP16)]. (A–G) Stage 12. (H–P) Stage 13. (Q–W) Stage 14 (R–V, dorsal views). Arrows, segmental grooves. Arrowheads in A and B, amnioserosa. Arrowheads in C, D, and G, leading edge cells. Arrowheads in H–W, leading edge actin cable. Brackets in L–N, elongation of leading edge cells. Long arrows in H, K, L, and O–Q, normal amnioserosa (H and L) or holes in amnioserosa (K and O–Q). X–CC, ventral views. X and CC, stage 12. Y–BB, stage 14. Arrows, segmental groove. Arrowheads, apical cell surface. Bars, 20 μm.
We also followed development of the segmental grooves. Wild-type grooves form dorsally toward the end of stage 11 (just preceding germband retraction), and they extend into the ventral and lateral epidermis as germband retraction begins (Martinez-Arias, 1993; Larsen et al., 2003). Groove formation initiates by apical constriction of the “bottle” cells (the most posterior cells of each segment), followed by inward folding of two-three neighbors on each side (Larsen et al., 2003). Grooves deepen during retraction (Figure 5A, arrow), and then they begin to regress at the onset of dorsal closure (stage 13; Figure 5H, arrow). They disappear first in the dorsal and ventral epidermis (Figure 5L), and disappear later laterally. Overexpression of wild-type Abl or misexpression of p185 Bcr-Abl using e22-GAL4 (Figure 5, A vs. B and C vs. D; data not shown) did not have significant consequences on segmental grooves. However, high level expression of p185 (by using arm-GAL4VP16; Figure 5, E–G) or expression of p210 by using e22-GAL4 (data not shown) led to substantial deepening of segmental grooves by the end of germband retraction (Figure 5C vs. E–G, arrows); whereas segmental grooves are a normal feature at this stage, they do not extend all the way to the leading edge in wild-type (Figure 5C, arrow).
We next examined dorsal closure. This is one of the most dramatic events of morphogenesis, involving a complex, coordinated series of cell shape changes, migration events, and cytoskeletal alterations. Before dorsal closure, columnar epidermal cells cover the ventral and lateral sides of the embryo (Figure 5, A and C), whereas squamous amnioserosal cells (Figure 5A, arrowhead) cover the dorsal surface. During germband retraction, cells at the leading edge of each lateral epidermal sheet first change shape (Figure 5C, arrowhead) and then elongate along the dorsal-ventral axis. A wave of elongation then spreads ventrally, until all epidermal cells are elongated along this axis (Figure 5L, bracket). As they elongate, leading edge cells assemble an actomyosin cable along their leading edge (Figure 5, H and L, arrowheads), which is anchored at cell–cell junctions, forming a supracellular contractile purse-string (Kiehart et al., 2000; Jacinto et al., 2002). Meanwhile, amnioserosal cells (Figure 5, H and L, long arrows) begin to constrict their apical ends—this occurs slowly in all cells, with a subset of cells initiating rapid constriction and disappearing from the epithelial sheet. The combined forces of elongation/migration of epithelial cells, actin cable contraction and amnioserosal cell apical constriction drive dorsal closure, culminating in the complete encapsulation of the embryo in the epidermis and the internalization of the amnioserosa (Kiehart et al., 2000; Hutson et al., 2003). The final stages of the process involve zipping together of the leading edge (Figure 5T, arrowhead) and precise alignment of cells from the two sides.
As dorsal closure was initiated, effects of Abl activation became more pronounced, altering many but not all of the cytoskeletal and morphogenetic movements occurring during this process. The effects of overexpression of wild-type Abl were largely confined to deepening of segmental grooves (Figure 5, I and W, arrows); these effects were most pronounced in embryos overexpressing Abl at the highest levels (by using arm-GAL4VP16; data not shown). However, wild-type Abl overexpression did not disrupt other events of dorsal closure, including cell shape changes and actin cable formation (Figure 5W and Y vs. Z).
Ubiquitous misexpression of p185 or p210 Bcr-Abl had more severe effects on dorsal closure, with p210 the most severe. The normally uniform dorsal-ventral elongation of epidermal cells (Figure 5L, bracket) was disrupted, with some cells elongating less than their neighbors (Figure 5, M and N, bracket). Moderate activation (p185 or p210 x e22c-GAL4) led to persistence of very deep segmental grooves throughout dorsal closure (Figure 5, J, K, M–O, and U, arrows), in contrast to wild-type (Figure 5, H, L, and T). This is likely due to defects in the cell shape changes of segmental groove cells, but the dramatic groove depth in mutants made direct observation of cell shape impossible. These two defects combined to cause significant defects in the completion of dorsal closure and in alignment of segments at the dorsal midline (Figure 5R vs. S and T vs. V, arrowheads; the actin projections on epidermal cells in Figure 5, U–W are dorsal hairs; because mutants close more slowly, dorsal hairs form before closure is completed). However, despite these defects, cell shape changes in the ventral epidermis were not dramatically disrupted (Figure 5Y vs. AA). Strong activation of Bcr-Abl (p185 x arm-GAL4:VP16, data not shown; p210 x e22-GAL4, Figure 5, H and L vs. N and O) led to even deeper segmental grooves (Figure 5O, arrow) and affected amnioserosal cell shapes (Figure 5O), although the actin cable remained intact (Figure 5, N and O, arrowheads). In these genotypes, dorsal closure often failed completely, likely due in part to disruption of integrity of the amnioserosal epithelium (Figure 5, K and O, long arrows). High-level expression of p210 also led to abnormal apical actin accumulation in epidermal cells (Figure 5X vs. CC, arrowheads). The very strongest defects were seen when p210 was expressed with arm-GAL4:VP16. This led to extreme deepening and persistence of segmental grooves, dorsally and even midventrally (Figure 5Y vs. Q and BB; ventrally, these grooves engulf the forming denticle belts), and to dramatic disruption of the amnioserosa (Figure 5, P and Q, long arrows). Thus, inappropriate Abl activation can disrupt many but not all of the cell shape changes and movements of morphogenesis, and the strength of these defects is graded based on level of expression and of activation.
We also examined the effects of localized expression of Abl or Bcr-Abl in the posterior cells of each segment, by using en-GAL4. This allowed us to directly compare neighboring cells with and without Abl activation, looking for more subtle changes in actin and cell architecture. Little effect on morphogenesis was seen through germband retraction, even in embryos expressing the strongest p210 lines (Figure 6, A–D). Dorsal closure was essentially wild type in embryos expressing increased wild-type Abl or expressing p185 in en-stripes. As dorsal closure was completed, there was a subtle deepening of segmental grooves dorsally (Figure 6, J′ and L′), when they should have disappeared (Figure 6J′). However, there was little effect on cell shape changes (Figure 6N′) or the actin cable, and dorsal closure proceeded normally (Figure 6, K′ and L′, yellow arrowheads, and N′). Expression of p210 triggered deeper, more persistent segmental grooves (Figure 6E′ vs. F′ and G). In addition, misexpressing leading edge cells were slightly less elongated along the dorsal-ventral axis than their wild-type counterparts, and they had slightly elevated apical actin (Figure 6H′ vs. I′, arrows), but the actin cable remained unaltered (Figure 6I′, arrowhead). As dorsal closure was completed, expression of p210 in en-stripes led to a drastic deepening and persistence of the segmental grooves dorsally (Figure 6M′), with the p210-expressing cells invaginated deep into the embryos (Figure 6O′, arrowheads).
Figure 6.
The cellular and cytoskeletal consequences of localized Abl overexpression and Bcr-Abl expression. Embryos, anterior left, and, unless noted, lateral view. Antigens indicated. Transgenes indicated were driven by en-GAL4. (A–D) Stage 12. Brackets in C and D, en-GAL4 expression domain. (E–I) Stage 13 (G is a ventral view). E–G, blue arrowheads, en-GAL4–expressing cells. Yellow arrowheads, segmental grooves. H and I, arrows, en-GAL4–expressing cells. White arrowheads, leading edge actin cable. (J–O) Stage 14. White arrowheads, segmental grooves. Yellow arrowheads, leading edge actin cable. (N′) Red arrowhead, unaltered cell shapes. (O) Red arrowheads, hyperconstricted Bcr-Abl–expressing cells. Bars, 20 μm.
One potential mechanism by which activated Abl might alter cell shapes is by perturbing apical-basal cell polarity. To test this possibility, we examined localization of a number of cell polarity markers in embryos expressing Bcr-Abl p210 (Supplemental Figure 2, A–C), p185 (data not shown), or overexpressing wild-type Abl (Supplemental Figure 2, D–F) in stripes in epidermis. This allowed us to directly compare protein localization in adjacent expressing and nonexpressing cells. We examined the AJ protein DE-cadherin (Supplemental Figure 2, A and D), the apical determinant Crumbs (Crb; Supplemental Figure 2, B and E), and the basolateral determinant Dlg (Supplemental Figure 2, C and F). In all cases, the apical-basal localization of these polarity markers was unaltered. Thus, it is unlikely that the changes in cell shape we observe are primarily due to effects on cell polarity. We also assessed whether Bcr-Abl might alter microtubule localization, because elongating epidermal cells and stretched amnioserosal cells both have interesting microtubule arrangements in wild-type. We saw no dramatic differences in microtubule localization in cells expressing Bcr-Abl (Supplemental Figure 2, G and H).
Abl Activation Alters Cell Behavior during Dorsal Closure
These data suggested that Abl activation affects cell shape changes and morphogenetic movements, especially during dorsal closure. To address the effect on Abl activation on cell behavior directly, we examined morphogenesis in living embryos expressing GFP-tagged cytoskeletal proteins. This reveals a wider range of cell behaviors than can be seen in fixed embryos, and it also adds dynamic information. We, initially examined the effects of ubiquitous overexpression of Abl and misexpression of Bcr-Abl, using e22c-GAL4.
We first assessed dorsal closure in living embryos ubiquitously expressing Moe-GFP (F-actin–binding domain of moesin fused to GFP). This reports F-actin localization in live embryos (Figure 7A and Supplemental Video 1; Edwards et al., 1997). It is a superb reporter for cytoskeletal rearrangements, cell shape changes, and other events during complex morphogenetic processes. In wild-type embryos, when dorsal closure begins, epidermal cells have elongated along the dorsal-ventral axis and cover the dorsal and lateral surfaces of the embryo (Figure 7A, 0:00, white arrow), whereas squamous amnioserosal cells (Figure 7A, black arrowheads) cover the dorsal surface. Epidermal leading edge cells assemble the actomyosin cable (Figure 7A, white arrowheads), the contraction of which helps drive closure (Kiehart et al., 2000; Jacinto et al., 2002). Leading edge cells extend filopodia and lamellipodia over the surface of the adjacent amnioserosal cells (Jacinto et al., 2000), whereas amnioserosal cells apically constrict (Figure 7A, black arrowheads), also helping drive closure (Kiehart et al., 2000; Hutson et al., 2003). Apical ends of the amnioserosal cells are covered by filopodia (Figures 7A, 0:00, black arrowhead, 8, A and C, and Supplemental Video 2). As the epithelial sheets approach one another, they zipper together (Figure 7A, black arrows).
Figure 7.
Effect of Abl overexpression and Bcr-Abl misexpression on morphogenesis and cell behavior. Stills, movies of dorsal closure in living embryos expressing moesin-GFP. Times in hours and minutes. (A) Wild-type. Black arrowheads, amnioserosal cells. White arrowheads, actin cable at leading edge. White arrow, leading edge cells elongated in dorsal-ventral axis. Black arrows, zippering together as two sheets meet. (B) Embryo expressing Bcr-Abl p185(6a1) by using e22c-GAL4. Note delay in closure. Black arrowheads, abnormally shaped amnioserosal cells, often with ectopic lamellipodia (e.g., 0:27). White arrowheads, uneven leading edge. Black arrow, puckering during zippering. White arrow, persistent segmental groove. (C) Embryo expressing Bcr-Abl p210(11a) by using e22c-GAL4. Black arrowhead, abnormally shaped amnioserosal cells. White arrowheads, uneven leading edge. Black arrows, rip in aminoserosa. White arrows, persistent segmental groove. (D) Embryo overexpressing Abl (UA22) by using e22c-GAL4. Black arrowheads, amnioserosal cells. White arrowheads, uneven leading edge. White arrows, persistent segmental grooves. Bar, 20 μm.
Figure 8.
Abl overexpression and Bcr-Abl misexpression alter filopodia and lamellipodia. (A–E) Stills, movies of amnioserosa in living embryos expressing moesin-GFP. Misexpression driven by e22c-GAL4. A and C, wild-type (WT). Amnioserosal cells surrounded by filopodia (arrows). B, p210(11a). D, p185(6a1). Note reduction of filopodia and replacement with lamellipodia (arrows). E, Wild-type Abl overexpression (UA22). Filopodia are reduced (arrow) without appearance of lamellipodia. (F–H and K and L) Stills, movies of dorsal closure in living embryos expressing actin-GFP (F–H) or GFP-Ena (K and L) and the indicated transgene by using the en-GAL4 driver. Time = minutes:seconds. F, wild-type. Cell projections are highly dynamic lamellipodia with filopodia extensions. G, p185. Lamellipodia remain but filopodia are reduced substantially (arrows). H, overexpressed Abl. All protrusions are reduced in size and number. Most remaining protrusions are short and blunt (arrows). (I) Quantitation of filopodial number, with standard deviations. Filopodia were defined as any protrusion <1.25 μm wide extending beyond the lamellipodium or leading edge, and they were quantitated within en-GAL4 stripes in three to five embryos of each genotype as the leading edges moved from 26.9 to 7.9 μm apart. (J) Quantitation of average lamellipodial area, with standard deviations. Lamellipodia were defined as any projection in the direction of migration >1.3 μm in width and >1.3 μm in length, and area was calculated in ImageJ in frames 1 min apart during the same part of dorsal closure using for filopodial measurements. p values are in text. K, Co-overexpression of GFP-Ena and p185 restores filopodia. L, expression of GFP-Ena alone. Bars, 5 μm.
Misexpression of Bcr-Abl had dramatic consequences on cell behavior during dorsal closure, despite the fact that in most animals it went to completion. In embryos misexpressing p185 Bcr-Abl ubiquitously using e22c-GAL4, several defects were apparent (Figure 7B and Supplemental Video 3). Epidermal cells elongated along the dorsal-ventral axis, but the leading edge was very uneven (Figure 7B, white arrowheads), making it difficult to assess integrity of the actin cable in live embryos. We suspect this is due in part to an increase in depth and persistence of segmental grooves. Although amnioserosal cells (Figure 7B, black arrowheads) constricted apically, their cell shapes were abnormal, and some cells were significantly delayed in constriction. Further, the filopodia that normally project from amnioserosal cells were lost, and replaced with broad lamellipodia extending over neighboring cells (Figure 7B, 0:27, black arrowhead; Figure 8D; and Supplemental Video 4). The opening in the dorsal epidermis was oval rather than almond-shaped (Figure 7B, 0:27 vs. A, 0:27), and the amnioserosa was in a deeper focal plane than the epidermis during late closure, suggesting defects in zipping together the epidermal sheets. As closure was completed, cells in the two sheets did not precisely align with their counterparts from the other side of the embryo, causing puckering of the dorsal surface (Figure 7B, 3:09, black arrow). Despite these defects, however, closure was completed, but it took considerably longer than in wild-type (average time to close the last 48 μm = 159 min (n = 3) for p185 vs. 79 min (n = 5) for wild-type; p = 0.003).
Expression of strong p210 lines using the same driver had similar but more severe consequences (Figure 7C and Supplemental Video 5). The leading edge was uneven (Figure 7C, white arrowheads), segmental grooves were deep and persistent (Figure 7C, white arrowheads, the dorsal opening oval-shaped (Figure 7C, 0:51), and amnioserosal cells were abnormal in shape and extended lamellipodia rather than filopodia (Figures 7C, 0:00, black arrowhead; and 8B). One additional defect was seen in embryos misexpressing p210—the amnioserosal cell sheet often ruptured (Figure 7C, 1:21–2:10, black arrows), thus contributing to the dorsal holes in cuticles of this genotype. Because rupture ended dorsal closure, it was impossible to calculate time to closure.
Misexpression of wild-type Abl affected dorsal closure in largely similar ways (Figure 7D and Supplemental Video 6), but the effects were less severe than those of Bcr-Abl. Abl overexpression led to an uneven leading edge (Figure 7D, 0:51, white arrowheads) and deepened segmental grooves (Figure 7D, 2:10 and 2:37, arrows). The shape of the dorsal opening was also more oval (Figure 7D, 0:51), as was seen in embryos expressing Bcr-Abl, and in the later stages of dorsal closure the amnioserosa was in a deeper focal plane than the epidermis; both suggest that zippering together of the two epidermal sheets is slowed. Abl overexpression had distinct effects on the cell protrusions produced by amnioserosal cells from Bcr-Abl; filopodia were reduced but lamellipodial formation was not triggered by Abl overexpression (Figures 7D, black arrowheads; and 8E). Abl overexpression also significantly slowed dorsal closure (average time for Abl-overexpressing embryos to close 128 min [n = 5] vs. 79 min in wild type [n = 5]; p = 0.001). Thus, live analysis revealed dramatic effects of Abl activation on cell behavior during dorsal closure, even in genotypes where closure was successful.
Bcr-Abl Reduces Filopodial Number in Leading Edge Cells
We next zoomed in on behavior of individual cells. During dorsal closure, epidermal cells at the leading edge send out dynamic, actin-based cell processes, which can be visualized by expressing actin-GFP in stripes of epidermal cells (Jacinto et al., 2002). In wild-type embryos, these actin-based cell processes consist of broad lamellipodia from which emerge filopodia, thus, resembling neuronal growth cones (Supplemental Video 7 and Figure 8F, 5:00, arrows). These structures are highly dynamic: new processes constantly emerge, evolve, and retract. We thus assessed whether Bcr-Abl expression affected the cell processes formed by leading edge cells.
When we expressed both Bcr-Abl and actin-GFP by using en-GAL4, we saw a striking change in the nature of actin processes formed. Cells still produced broad lamellipodia, but there were far fewer filopodia extending from them (Supplemental Video 8 and Figure 8G, arrows). To quantitate this, we measured the number and maximum length of each filopodium (Figure 8I; see figure legend for methodology). Wild-type leading edges made an average of 21.9 filopodia per en-stripe (n = 9). In contrast, embryos misexpressing p185 made 6.4 filopodia per en-stripe (n = 14), whereas embryos misexpressing p210 made 6.2 filopodia (n = 12) and 8.8 filopodia (n = 20) per en-stripe in the two lines quantitated. All were significantly different from wild-type (p < 7 × 10−7). In embryos misexpressing p185, the remaining filopodia were also somewhat shorter than in wild-type (2.14 μm [n = 90] in p185 vs. 2.54 μm [n = 197] in wild-type; p = 0.008). To our surprise, the filopodia in embryos expressing p210 were not shorter than those in wild-type—in fact, in they were somewhat longer (2.91 μm [n = 74] and 2.94 μm [n = 175] in p210 vs. 2.54 μm in wild type). We also compared total area of the lamellipodial cell projections made by wild-type and Bcr-Abl misexpressing embryos (Figure 8J; see figure legend for methodology). No dramatic differences were seen—lamellipodial area in p185-expressing embryos slightly increased (from 36 μm2 in wild-type [n = 262] to 42 μm2 [n = 478]; Figure 8J) and lamellipodial area in p210-expressing embryos was statistically unchanged or slightly decreased in the two lines we assessed (34 μm2 [n = 391] and 29 μm2 [n = 634]; Figure 8J).
Overexpression of wild-type Abl had distinct effects from Bcr-Abl. Leading edge cells overexpressing Abl produced fewer protrusions overall (Supplemental Video 9 and Figure 8H). Many of the protrusions produced were short “filopodia” that emerged directly from the cell, rather than from a lamellipodium. Filopodia were substantially reduced in number (11.6 filopodia per en-stripe [n = 16] vs. 21.9 per en-stripe in wild-type; p = 7 × 10−5; Figure 8I) and in length (2.07 μm [n = 186] in Abl-overexpressers vs. 2.54 μm in wild-type; p = 6 × 10−5). In addition, total lamellipodial area was substantially reduced from wild-type (Figure 8J; from 36 μm2 in wild-type [n = 262] to 11 μm2 in Abl overexpressers [n = 390]; p < 3 × 10−56), contrasting with Bcr-Abl misexpression. Thus, Bcr-Abl and Abl overexpression both affect cell behavior and the actin cytoskeleton during morphogenesis, and their effects on cell behavior are similar but not identical.
Ena Is a Key Target of Activated Abl
These dramatic cell biological effects suggest that deregulated Abl alters cell behavior and are consistent with an effect on actin dynamics. To understand the mechanisms of Abl action, we need to identify its key targets. One known target of Abl is the actin modulator Ena, which Abl negatively regulates (Gertler et al., 1990). In parallel work, we examined the effects of loss of maternal and zygotic Ena function on morphogenesis (Gates et al., 2007). Strikingly, many of the phenotypes induced by Ena depletion were very similar to those caused by Abl activation; both caused defects in head involution, resulted in substantially deepened segmental grooves, slowed dorsal closure, and substantially reduced the numbers of filopodia produced by leading edge cells. These data suggest that down-regulation of Ena activity may be a key part of the mechanism by which deregulated Abl kinase disrupts morphogenesis.
To test whether Ena regulation is important for effects of activated Abl on morphogenesis, we first tested whether reducing Ena levels modifies the consequences of Abl activation. We overexpressed Abl in a background zygotically mutant for ena (reducing but not eliminating Ena, due to its maternal contribution). Reduction of the Ena dose substantially enhanced the cuticle phenotype of Abl overexpression (Figure 4, K vs. L, and Supplemental Table 1), consistent with Ena down-regulation playing an important role in the effects of Abl activation.
If Ena is a key Abl target, the negative regulation triggered by Abl activation might be alleviated by Ena overexpression. To test this mechanistic hypothesis, we examined whether Ena overexpression would alleviate the effects of activated Abl on cell behavior, by co-overexpressing Bcr-Abl with a GFP-tagged form of Ena (Gates et al., 2007). As described above, Bcr-Abl expression substantially reduced the production of filopodia. GFP-Ena localizes to the ends of filopodia, allowing us to visualize them in living embryos (Gates et al., 2007). We found that co-overexpression of GFP-Ena restored filopodia to leading edge cells expressing p185 Bcr-Abl (Figure 8K; co-overexpressing embryos were selected using a GFP-marked Balancer chromosome); the filopodia resembled those seen in embryos expressing GFP-Ena alone (Figure 8L). These data further support the hypothesis that Ena is a key target of Abl action during morphogenesis.
Abl Activation Disrupts Ena Localization
These data raise the question of the mechanism by which activated Abl regulates the actin regulator Ena. Activated Abl and Ena both localize to AJs, especially at tricellular junctions (Figure 9, A vs. B). When Abl is inactivated in early embryos, Ena accumulates at ectopic locations (Grevengoed et al., 2003). Our genetic interaction experiments described above (Figure 4, K vs. L) suggest that Ena misregulation accounts, at least in part, for the morphogenesis defects seen upon Bcr-Abl misexpression. We thus examined Ena localization in embryos misexpressing wild-type or activated Abl to determine whether alterations in its localization explain some of the cell biological effects we observed. In wild-type embryos, Ena localizes to AJs of epidermal epithelial cells, with enrichment at tricellular junctions where three cells meet (Figure 9, B and D′, white arrowheads). During dorsal closure Ena also localizes to prominent dots at AJs of leading edge cells, where they meet the amnioserosa (Figure 9D′, arrow), and Ena also accumulates at higher levels at the dorsal and ventral cell interfaces of the single row of epidermal cells that initiates the segmental groove (Figure 9D′, blue arrowhead). In the amnioserosa, Ena outlines the apical ends of the cells (Figure 9, G, I, and L, red arrowheads).
Figure 9.
Abl overexpression and Bcr-Abl misexpression alter Ena localization. Embryos, anterior left and, unless noted, lateral view. Antigens and transgenes indicated. (A–F) Wild-type or en-GAL4 x indicated transgene. A, stage 12. B, C, and F, stage 14. D and E, stage 13. Brackets, en-GAL4 expression domain. White arrowheads, tricellular junctions. Blue arrowheads, segmental groove cells. Blue arrows, ectopic cortical P-Abl and Ena. (G–N) Wild-type or e22c-GAL4 x indicated transgene. G and H, stage 12. G, arrowhead, tricellular junction. H, arrowhead, ectopic cortical Ena. I–N, stage 14 (K–M are dorsal views). Red arrowheads, amnioserosal cell cortex. Blue arrowheads, tricellular junctions or ectopic cortical Ena. Arrows, Ena at leading edge. Bars, 10 μm.
Activation of Abl by expression of Bcr-Abl significantly alters subcellular localization of Ena. Localized expression in en-stripes of either p185 (Figure 9, C and C′, brackets) or p210 (data not shown) led to reduction or loss of Ena enrichment at tricellular junctions and at the cortex. Ena localization to AJs of leading edge cells was more resistant to disruption (data not shown). The effect on Ena localization is dependent on kinase activity, because it did not occur in a kinase-dead p210 mutant (Figure 9, E′ vs. D′, brackets). Ubiquitous expression of Bcr-Abl had similar effects on Ena localization. This was first apparent during germband retraction, when Ena was reduced at tricellular junctions in embryos expressing p210, and cells exhibited more uniform cortical Ena staining (Figure 9, G vs. H, arrowheads). At the onset of dorsal closure, Ena enrichment at tricellular junctions was even more reduced (Figure 9, I vs. J, K, blue arrowheads), as was cortical Ena in amnioserosal cells (Figure 9, I and L vs. J and M, red arrowheads). Epidermal cells retained punctate cortical Ena, which was strongest at anterior and posterior cell interfaces (Figure 9, J and M), and cytoplasmic Ena staining seemed elevated. The elevated Ena levels at leading edge AJs (Figure 9I, arrow) were relatively resistant to disruption (Figure 9K, arrows), but Ena became less restricted to AJs (Figure 9, J and M, arrows). Abl overexpression in en-stripes had similar, but more subtle effects; in overexpressing cells, Ena was more uniformly cortical and less enriched at tricellular junctions (Figure 9, F and F′). Uniform overexpression of Abl (Figure 9N) had effects on Ena reminiscent of but somewhat weaker than those of Bcr-Abl expression. These data are consistent with a model in which altered Ena localization plays an important role in the effects of Abl activation.
To determine whether the changes we observed were due to an alteration of Ena accumulation or simply a change in its localization, we examined the total levels of Ena in embryos ubiquitously expressing Bcr-Abl. No effects on Ena levels were seen (Figure 10A; data not shown). However, consistent with Ena being a target of Bcr-Abl, tyrosine-phosphorylation of Ena significantly increased (Figure 10B). Interestingly, a second tyrosine-phosphorylated protein of ∼120 kDa coimmunoprecipitates with Ena in extracts from embryos misexpressing p185 (Figure 10B); the identity of this protein is not known. In our previous work on embryos deficient for maternal and zygotic Abl, we saw reduced recruitment of the AJ proteins Arm and α-catenin into junctions and reduced levels of their accumulation (Grevengoed et al., 2001). We thus also examined the levels and Tyr-phosphorylation of Arm in embryos overexpressing or misexpressing Bcr-Abl—no consistent differences were seen (Figure 10C). We also observed no consistent changes in the association of Arm with its junctional partner DE-cadherin, as assessed by coimmunoprecipitation (Figure 10C). In embryos misexpressing Bcr-Abl, levels of DE-cadherin or α-catenin were also unchanged (Figure 10A).
Figure 10.
Abl overexpression and Bcr-Abl expression alter Ena phosphorylation but not levels. In all cases the transgenes were ubiquitously misexpressed by using e22c-GAL4. (A) Embryonic extracts from wild-type embryos or embryos ubiquitously misexpressing the indicated transgene, immunoblotted with the antibodies indicated (Pnut is a loading control). P210K>R is a kinase-dead transgene, and p185Y>F has an alteration in the Grb2-binding site. (B) Ena was immunoprecipitated from wild-type or p185-expressing embryos (e22c-GAL4) and immunoblotted with antibodies to PTyr or Ena. (C) Arm was immunoprecipitated from wild-type, Abl-overexpressing, or p185-expressing embryos (e22c-GAL4) and immunoblotted with antibodies to PTyr, Arm, or DE-cad.
Abl Activation and Rho Regulation
Another key set of effectors regulating cell protrusiveness and cell behavior are Rho family GTPases. Drosophila Rho1 is encoded by a single gene and good reagents are available to examine Rho1 protein localization (Magie et al., 1999) and the localization of active Rho1 in vivo (Simoes et al., 2006). We thus examined whether Abl activation alters Rho1 activation or localization. Neither Abl overexpression nor Bcr-Abl expression altered levels or localization of an in vivo probe that detects Rho1 activity (UAS-PKNG58AeGFP = RBD-GFP; Figure 11, A–C). Likewise, neither altered accumulation of Rho1 protein or its recruitment to the membrane (Figure 11, D and E; its overall levels were also not changed; data not shown). Of course, this does not rule out subtle changes in Rho1 activity in discrete places or times, nor does it rule out effects on other Rho family members. It will be particularly important to examine the possible involvement of Rac as tools become available for studying its activation in Drosophila.
Figure 11.
Abl activation and Rho. Embryos, anterior to the left, and, unless noted, lateral view. Antigens and transgenes indicated. (A–C) en-GAL4 x the Rho-activity probe-GFP (UAS-PKNG58AeGFP) in wild-type or embryos expressing the indicated transgene. A and B, stage 15. C, stage 13. (D and E) Rho localization, stage 13. (F–I) DE-cad and Ena (F and G) or active Abl (H and I) localization in embryos expressing DN-Rho x en-GAL4. Bars, 10 μm.
The localization of both active Abl and Ena to AJs, and especially tricellular junctions, raised the possibility that Rho might regulate their localization, through its effects on AJs. Expression of dominant-negative Rho disrupts the integrity of AJs and the localization of DE-cadherin to the plasma membrane (Bloor and Kiehart, 2002; Figure 11, F and G). We found that expression of dominant-negative Rho resulted in the loss of both active Abl (Figure 11, H and I) and Ena (Figure 11, F and G) from the plasma membrane, whereas the cortical localization of a control protein, Diaphanous, was unaffected. We suspect that the primary effect of Rho is on AJs, with secondary effects on localization of active Abl and Ena.
DISCUSSION
Deregulation of Abl Kinase Activity Disrupts Morphogenesis
Loss-of-function mutations in abl disrupt many morphogenetic events (see Introduction). To further our mechanistic understanding of the roles of Abl, we explored whether deregulated kinase activity disrupts morphogenesis. Inappropriate activation of Abl affects many of the same morphogenetic events disrupted by loss of Abl. Normally, Abl is likely to exist primarily in an inactive form (Nagar et al., 2003). Docking with ligands for the SH2 or SH3 domains may help trigger the active conformation. Embryos are relatively resistant to overexpression of wild-type Abl. We suspect that increasing protein levels are largely accommodated by normal regulatory mechanisms until levels become extremely high. Consistent with this, we found that active Abl has a more restricted localization than total Abl, suggesting that Abl activation is normally restricted to the apical cell cortex and in particular to tricellular junctions, with a pool of inactive Abl in the cytoplasm. Increasing wild-type Abl levels may drive formation of more active Abl, or it may titrate negative regulators.
Misexpression of Bcr-Abl has more drastic consequences on morphogenesis, consistent with the constitutive activation of Bcr-Abl. In some cases, effects were simply quantitatively stronger, e.g., both Abl and Bcr-Abl affected head involution and segment grooves. However, other processes such as dorsal closure were only affected by Bcr-Abl. These processes may simply be less sensitive, affected only by very high level Abl activity. Alternately, Bcr-Abl may have cell biological effects in Drosophila distinct from those of Abl.
Many Effects of Abl Activation Occur via Ena Regulation
The best-known target of Drosophila Abl is Ena. Abl negatively regulates Ena (Gertler et al., 1995; Comer et al., 1998), in part by restricting its localization (Grevengoed et al., 2003). In parallel work, we examined effects on embryogenesis of depleting maternal and zygotic Ena (enaM; Z; Gates et al., 2007). This allows us to evaluate which effects of Abl activation result from negative regulation of Ena.
Ena loss-of-function and Abl activation share striking similarities. Both disrupt head involution. In both segmental grooves are substantially deepened and persist long after they normally retract. Finally, both alter cell behavior during dorsal closure in similar ways: dorsal closure is significantly slowed, leading edge cells produce fewer filopodia, and epithelial cell matching and zippering are disrupted. These data are consistent with the idea that Ena is the major target of both Abl and Bcr-Abl during Drosophila morphogenesis. This mechanism of action is further supported by other data. First, reduction in Ena levels enhances effects of Bcr-Abl overexpression. Second, overexpression of GFP-Ena partially rescues the effects of Abl activation on filopodial behavior. Finally, Ena localization is regulated by Abl. In abl loss-of-function mutants, Ena accumulates inappropriately at the apical cortex of early embryos and at the leading edge during dorsal closure (Grevengoed et al., 2001, 2003). In contrast, in embryos overexpressing wild-type Abl, Ena is lost from places it normally accumulates (e.g., tricellular junctions), and it localizes instead at lower levels all around the cell cortex and in the cytoplasm. These data are consistent with Ena being a key Abl target.
Current models of Ena function provide good insight into some of the biological and cell biological effects of Abl activation. Both Ena inactivation (Gates et al., 2007) and Abl activation (this study) led to a reduction in filopodia produced by leading edge cells and defects in the last stages of dorsal closure. These roles fit well with the role of Ena as an anti-capping protein that may also mediate filament bundling into filopodia (Bear et al., 2002; Barzik et al., 2005). Reduction in Abl function leads to the formation of excess filopodia with elevated levels of Ena at the tips (Gates et al., 2007), further supporting this regulatory mechanism. Abl activation and Ena loss of function also have parallel effects on head involution and segmental groove formation. In both biological events a row of cells adopts an unusual localization of Ena, with elevated Ena levels and Ena planar polarized at the dorsal-ventral cell boundaries (Gates et al., 2007; Figure 9E′, blue arrowhead). It seems reasonable that the substantial alterations of Ena subcellular localization caused by Abl activation could interfere with Ena function in these key subsets of cells. However, it remains unclear precisely how localized Ena activity contributes to the distinctive cell shape changes of cells of the segmental grooves or head fold.
One key question is the mechanism(s) by which Abl regulates Ena. The data above and our earlier loss-of-function experiments (Grevengoed et al., 2001; 2003) are consistent with a model in which Abl regulates Ena localization, restricting its activity to places it is essential. Abl may form a complex with Ena, keeping it in an inactive state. Consistent with this, Abl can bind Ena (Ahern-Djamali et al., 1999), Ena and active Abl colocalize to tricellular junctions and leading edge cell AJs, and Abl overexpression or Bcr-Abl misexpression leads to elevated Abl activity all around the cell cortex, disrupting normal Ena localization. Perhaps Abl docks inactive Ena at sites near where its activity will be needed. For example, Ena at leading edge cell AJs could be the source of Ena needed at the leading edge to make filopodia. Although this model is attractive, some data cannot be easily accommodated by it, e.g., Abl and active Abl both localize to the cortex of syncytial embryos (Fox and Peifer, 2007), but Ena is not normally localized there, and Ena localizes there in the absence of Abl (Grevengoed et al., 2003), suggesting that there may be alternate mechanisms by which Abl regulates Ena. Further experiments are needed to test these hypotheses.
One way Abl may regulate Ena is by phosphorylation (Fogerty et al., 1999). The effects on embryogenesis of Abl and Bcr-Abl require kinase activity. However, mutating all the Ena phosphorylation sites does not lead to the “activated” phenotype expected if this is the sole mechanism of negative regulation (i.e., mimicking abl loss-of-function); instead, it has a weak ena loss-of-function phenotype (Comer et al., 1998). Thus, Abl regulation of Ena involves more than direct phosphorylation. Abl may phosphorylate itself and other partners, creating or disrupting protein complexes. Consistent with this, although Ena phosphorylation sites are not conserved in its mouse homologues, mouse Abl promotes Mena phosphorylation (Tani et al., 2003) and binds VASP (Howe et al., 2002). Given the clear ability of Bcr-Abl to alter Ena localization/activity in Drosophila, further exploration of Ena/VASP proteins as possible targets in mammalian cells seems warranted.
One Kinase, Diverse Responses
Although many effects of Abl activation can be explained by negative regulation of Ena, a subset of the effects suggest alternate targets. For example, effects of Abl overexpression on leading edge cell behavior are more drastic than those seen in enaM/Z mutants, e.g., reduced lamellipodial activity was not seen after Ena was inactivated (Gates et al., 2007), and high-level Bcr-Abl expression during embryogenesis is more detrimental than Ena loss. Thus, both Abl and Bcr-Abl likely have Ena-independent effects on actin and cell behavior in Drosophila.
One critical issue in interpreting our results is whether Bcr-Abl acts in Drosophila simply as a deregulated form of Abl, or whether it has additional effects on cell behavior. In most of our assays, Bcr-Abl expression had effects similar to but stronger than those of Abl overexpression. In some cases, high-level Bcr-Abl expression affected processes not affected by high-level Abl overexpression (e.g., amnioserosa integrity), but we may not have achieved sufficient levels of wild-type Abl overexpression to mimic them. However, one striking effect of Bcr-Abl was not seen either with Abl overexpression or Ena loss-of-function: the explosive production of lamellipodia by amnioserosal cells, which normally only produce filopodia. Perhaps the Bcr part of the fusion recruits additional proteins that influence its abilities. Alternately Bcr-Abl may stimulate signaling pathways such as those of c-Jun NH2-terminal kinase or mitogen-activated protein kinase, targets of mammalian Bcr-Abl (Advani and Pendergast, 2002); both affect Drosophila epidermal cell shape or fates (Szüts et al., 1997; Xia and Karin, 2004). Further studies of the mechanisms of action of Bcr-Abl in Drosophila may offer clues into additional targets of Abl.
Although both Abl and Bcr-Abl modulate actin dynamics, the cytoskeletal response they program is complex. In fibroblasts, loss of Abl prevents ruffling in response to PDGF (Plattner et al., 1999), whereas loss of Arg reduces lamellipodial dynamics (Miller et al., 2004). These data suggest that Abl regulates formation of branched actin filaments involved in lamellipodia, consistent with its ability to speed migration. Many of our observations are consistent with this, including reduced filopodial number after Abl activation, and Bcr-Abl–triggered lamellipodia. Likewise, Drosophila Abl inhibits dendrite branching (Li et al., 2005). However, in other contexts, Abl modulates actin differently. Mouse Abl and Arg maintain dendrite branching (Moresco et al., 2005), and Abl promotes actin microspikes in fibroblasts plated on fibronectin (Woodring et al., 2002). Both are consistent with promoting unbranched actin. Bcr-Abl expression also has distinct effects in different cell types, triggering ruffling and filopodial extension in BaF3 cells (Salgia et al., 1997), while preventing spreading and polarization on fibronectin in dendritic cells (Dong et al., 2003).
Bcr-Abl adds additional complexity. We saw distinct effects of Bcr-Abl expressed at different levels. This dose sensitivity mimics that seen in myeloid cells expressing different levels of Bcr-Abl, which differ in adhesion to fibronectin and ability to induce tumors (Barnes et al., 2005). A second complexity involves differences between p210 and p185. In Drosophila, p210 produced consistently stronger phenotypes and also triggered higher levels of tyrosine-phosphorylated proteins. p185 and p210 differ in their biochemical and biological activities in mammals as well (Advani and Pendergast, 2002), and p185 and p210 cause distinct diseases in patients, and induce different pathways of differentiation in primary bone marrow cells. However, in human cells, p185 is the more active kinase. Further exploration of these functional distinctions will help illuminate the different pathways activated by Abl and Bcr-Abl during normal development and oncogenesis.
Thus, both Abl and Bcr-Abl have distinct and at times seemingly opposite cytoskeletal effects in different cells. Perhaps this is not surprising, given the array of cytoskeletal regulators Abl can target, including those promoting unbranched actin filaments, such as Ena/VASP, and those regulating Arp2/3 and branching, such as WASP and WASP family Verprolin-homologous protein (Hernandez et al., 2004). The choice of target may be dictated by upstream inputs regulating Abl, and the consequences for actin dynamics will depend on the suite of other regulators active in the same cell. Understanding how individual cells integrate these inputs and outputs is one challenge for the future.
Supplementary Material
ACKNOWLEDGMENTS
We are especially grateful to Fran Fogerty, without whose insights and reagents this work would have been impossible. We also thank the Bloomington Drosophila Stock Center, D. Kiehart, P. Martin, A. Jacinto, and the Developmental Studies Hybridoma Bank for reagents; and J. Gates for help with live imaging and data analysis. This work was supported by National Institutes of Health (NIH) grant GM-47857 (to M.P.) T.L.S. was supported by NIH grants 5T3CA09156, 1F32GM20797, and the Thomas F. and Kate Miller Jeffress Memorial Trust; L.M.K. was supported in part by a Smallwood Fellowship; E.M.R. is a Leukemia and Lymphoma Society Fellow; C.C.F.H. is a student in the Gulbenkian Ph.D. Program in Biomedicine, Portugal; and M.P. is the Hooker Professor of Biology.
Abbreviations used:
- Abl
Abelson
- AJ
adherens junction
- Arm
armadillo
- CML
chronic myelogenous leukemia
- CNS
central nervous system
- en
engrailed
- Ena
Enabled
- KD
kinase-dead
- Ph
Philadelphia chromosome
- PNS
peripheral nervous system
- PTyr
phosphotyrosine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0008) on October 24, 2007.
REFERENCES
- Advani A. S., Pendergast A. M. Bcr-Abl variants: biological and clinical aspects. Leuk. Res. 2002;26:713–720. doi: 10.1016/s0145-2126(01)00197-7. [DOI] [PubMed] [Google Scholar]
- Ahern-Djamali S. M., Bachmann C., Hua P., Reddy S. K., Kastenmeier A. S., Walter U., Hoffmann F. M. Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc. Natl. Acad. Sci. USA. 1999;96:4977–4982. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. J., Palaiologou D., Panousopoulou E., Schultheis B., Yong A. S., Wong A., Pattacini L., Goldman J. M., Melo J. V. Bcr-Abl expression levels determine the rate of development of resistance to imatinib mesylate in chronic myeloid leukemia. Cancer Res. 2005;65:8912–8919. doi: 10.1158/0008-5472.CAN-05-0076. [DOI] [PubMed] [Google Scholar]
- Barzik M., Kotova T. I., Higgs H. N., Hazelwood L., Hanein D., Gertler F. B., Schafer D. A. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Perrimon N. Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat. Cell Biol. 2001;3:883–890. doi: 10.1038/ncb1001-883. [DOI] [PubMed] [Google Scholar]
- Bear J. E., et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bennett R. L., Hoffmann F. M. Increased levels of the Drosophila Abelson tyrosine kinase in nerves and muscles: subcellular localization and mutant phenotypes imply a role in cell-cell interactions. Development. 1992;116:953–966. doi: 10.1242/dev.116.4.953. [DOI] [PubMed] [Google Scholar]
- Bloor J. W., Kiehart D. P. Drosophila RhoA regulates the cytoskeleton and cell-cell adhesion in the developing epidermis. Development. 2002;129:3173–83. doi: 10.1242/dev.129.13.3173. [DOI] [PubMed] [Google Scholar]
- Brasher B. B., Van Etten R. A. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J. Biol. Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- Comer A. R., Ahern-Djamali S. M., Juang J.-L., Jackson P. D., Hoffmann F. M. Phosphorylation of enabled by the Drosophila Abelson tyrosine kinase regulates the in vivo function and protein-protein interactions of Enabled. Mol. Cell. Biol. 1998;18:152–160. doi: 10.1128/mcb.18.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. T., Kirkpatrick C., Peifer M. Armadillo is required for adherens junction assembly, cell polarity, and morphogenesis during Drosophila embryogenesis. J. Cell Biol. 1996;134:133–148. doi: 10.1083/jcb.134.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger M., Buchdunger E., Druker B. J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- Deng X., et al. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat. Genet. 2004;36:906–912. doi: 10.1038/ng1396. [DOI] [PubMed] [Google Scholar]
- Dong R., et al. Dendritic cells from CML patients have altered actin organization, reduced antigen processing, and impaired migration. Blood. 2003;101:3560–3567. doi: 10.1182/blood-2002-06-1841. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Demsky M., Montague R. A., Weymouth N., Kiehart D. P. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev. Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Fogerty F. J., Juang J. L., Petersen J., Clark M. J., Hoffmann F. M., Mosher D. F. Dominant effects of the bcr-abl oncogene on Drosophila morphogenesis. Oncogene. 1999;18:219–232. doi: 10.1038/sj.onc.1202239. [DOI] [PubMed] [Google Scholar]
- Fox D. T., Peifer M. Abelson kinase and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 2007;134:567–578. doi: 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- Gates J., Mahaffey J. P., Rogers S. L., Emerson M., Rogers E. M., Sottile S. L., Van Vactor D., Gertler F. B., Peifer M. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–2039. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- Gertler F., Doctor J., Hoffman F. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 1990;248:857–860. doi: 10.1126/science.2188361. [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Comer A. R., Juang J., Ahern S. M., Clark M. J., Liebl E. C., Hoffmann F. M. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gordon M. Y., Dowding C. R., Riley G. P., Goldman J. M., Greaves M. F. Altered adhesive interactions with marrow stroma of haematopoietic progenitor cells in chronic myeloid leukaemia. Nature. 1987;328:342–344. doi: 10.1038/328342a0. [DOI] [PubMed] [Google Scholar]
- Grevengoed E. E., Fox D. T., Gates J., Peifer M. Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J. Cell Biol. 2003;163:1267–1279. doi: 10.1083/jcb.200307026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed E. E., Loureiro J. J., Jesse T. L., Peifer M. Abelson kinase regulates epithelial morphogenesis in Drosophila. J. Cell Biol. 2001;155:1185–1198. doi: 10.1083/jcb.200105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O., Wiesner S., Guttler T., Mackereth C. D., Rix L. L., Mikes Z., Dehne J., Gorlich D., Sattler M., Superti-Furga G. Structural basis for the cytoskeletal association of Bcr-Abl/c-Abl. Mol. Cell. 2005;19:461–473. doi: 10.1016/j.molcel.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Henkemeyer M., Gertler F., Goodman W., Hoffmann F. The Drosophila Abelson proto-oncogene homolog: identification of mutant alleles that have pleiotropic effects late in development. Cell. 1987;51:821–828. doi: 10.1016/0092-8674(87)90105-x. [DOI] [PubMed] [Google Scholar]
- Hernandez S. E., Krishnaswami M., Miller A. L., Koleske A. J. How do Abl family kinases regulate cell shape and movement? Trends Cell Biol. 2004;14:36–44. doi: 10.1016/j.tcb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Howe A. K., Hogan B. P., Juliano R. L. Regulation of vasodilator-stimulated phosphoprotein phosphorylation and interaction with Abl by protein kinase A and cell adhesion. J. Biol. Chem. 2002;277:38121–38126. doi: 10.1074/jbc.M205379200. [DOI] [PubMed] [Google Scholar]
- Hutson M. S., Tokutake Y., Chang M. S., Bloor J. W., Venakides S., Kiehart D. P., Edwards G. S. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- Jacinto A., Wood W., Balayo T., Turmaine M., Martinez-Arias A., Martin P. Dynamic actin-based epithelial adhesion and cell matching during Drosophila dorsal closure. Curr. Biol. 2000;10:1420–1426. doi: 10.1016/s0960-9822(00)00796-x. [DOI] [PubMed] [Google Scholar]
- Jacinto A., Wood W., Woolner S., Hiley C., Turner L., Wilson C., Martinez-Arias A., Martin P. Dynamic analysis of actin cable function during Drosophila dorsal closure. Curr. Biol. 2002;12:1245–1250. doi: 10.1016/s0960-9822(02)00955-7. [DOI] [PubMed] [Google Scholar]
- Kain K. H., Klemke R. L. Inhibition of cell migration by Abl family tyrosine kinases through uncoupling of Crk-CAS complexes. J. Biol. Chem. 2001;276:16185–16192. doi: 10.1074/jbc.M100095200. [DOI] [PubMed] [Google Scholar]
- Kiehart D. P., Galbraith C. G., Edwards K. A., Rickoll W. L., Montague R. A. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- Larsen C. W., Hirst E., Alexandre C., Vincent J. P. Segment boundary formation in Drosophila embryos. Development. 2003;130:5625–5635. doi: 10.1242/dev.00867. [DOI] [PubMed] [Google Scholar]
- Li W., Li Y., Gao F. B. Abelson, enabled, and p120 catenin exert distinct effects on dendritic morphogenesis in Drosophila. Dev. Dyn. 2005;234:512–522. doi: 10.1002/dvdy.20496. [DOI] [PubMed] [Google Scholar]
- Magie C. R., Meyer M. R., Gorsuch M. S., Parkhurst S. M. Mutations in the Rho1 small GTPase disrupt morphogenesis and segmentation during early Drosophila development. Development. 1999;126:5353–5364. doi: 10.1242/dev.126.23.5353. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A. Development and patterning of the larval epidermis of Drosophila. In: Bate M., Martinez-Arias A., editors. The development of Drosophila melanogaster. vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 517–608. [Google Scholar]
- Miller A. L., Wang Y., Mooseker M. S., Koleske A. J. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 2004;165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco E. M., Donaldson S., Williamson A., Koleske A. J. Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J. Neurosci. 2005;25:6105–6118. doi: 10.1523/JNEUROSCI.1432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco E. M., Koleske A. J. Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr. Opin. Neurobiol. 2003;13:535–544. doi: 10.1016/j.conb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Nagar B., Hantschel O., Young M. A., Scheffzek K., Veach D., Bornmann W., Clarkson B., Superti-Furga G., Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Nowell P. C., Hungerford D. A. Chromosome studies in human leukemia. II. Chronic granulocytic leukemia. J. Natl. Cancer Inst. 1961;27:1013–1035. [PubMed] [Google Scholar]
- Plattner R., Kadlec L., DeMali K. A., Kazlauskas A., Pendergast A. M. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner R., Koleske A. J., Kazlauskas A., Pendergast A. M. Bidirectional signaling links the Abelson kinases to the platelet-derived growth factor receptor. Mol. Cell Biol. 2004;24:2573–2583. doi: 10.1128/MCB.24.6.2573-2583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J. D. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Salesse S., Verfaillie C. M. Mechanisms underlying abnormal trafficking and expansion of malignant progenitors in CML: BCR/ABL-induced defects in integrin function in CML. Oncogene. 2002;21:8605–8611. doi: 10.1038/sj.onc.1206088. [DOI] [PubMed] [Google Scholar]
- Salgia R., Li J. L., Ewaniuk D. S., Pear W., Pisick E., Burky S. A., Ernst T., Sattler M., Chen L. B., Griffin J. D. BCR/ABL induces multiple abnormalities of cytoskeletal function. J. Clin. Invest. 1997;100:46–57. doi: 10.1172/JCI119520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes S., Denholm B., Azevedo D., Sotillos S., Martin P., Skaer H., Hombria J. C., Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- Smith K. M., Yacobi R., Van Etten R. A. Autoinhibition of Bcr-Abl through its SH3 domain. Mol. Cell. 2003;12:27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szüts D., Freeman M., Bienz M. Antagonism between EGFR and Wingless signaling in the larval cuticle of Drosophila. Development. 1997;124:3209–3219. doi: 10.1242/dev.124.16.3209. [DOI] [PubMed] [Google Scholar]
- Tani K., Sato S., Sukezane T., Kojima H., Hirose H., Hanafusa H., Shishido T. Abl interactor 1 promotes tyrosine 296 phosphorylation of mammalian Enabled (Mena) by c-Abl kinase. J. Biol. Chem. 2003;278:21685–21692. doi: 10.1074/jbc.M301447200. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Jackson P. K., Baltimore D., Sanders M. C., Matsudeira P. T., Janmey P. The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J. Cell Biol. 1994;124:325–340. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Miller A. L., Mooseker M. S., Koleske A. J. The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc. Natl. Acad. Sci. USA. 2001;98:14865–14870. doi: 10.1073/pnas.251249298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C. Looking at embryos. In: Roberts D. B., editor. Drosophila, A Practical Approach. Oxford, England: IRL Press; 1986. pp. 199–228. [Google Scholar]
- Woodring P. J., Hunter T., Wang J. Y. Inhibition of c-Abl tyrosine kinase activity by filamentous actin. J. Biol. Chem. 2001;276:27104–27110. doi: 10.1074/jbc.M100559200. [DOI] [PubMed] [Google Scholar]
- Woodring P. J., Litwack E. D., O'Leary D. D., Lucero G. R., Wang J. Y., Hunter T. Modulation of the F-actin cytoskeleton by c-Abl tyrosine kinase in cell spreading and neurite extension. J. Cell Biol. 2002;156:879–892. doi: 10.1083/jcb.200110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Karin M. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 2004;14:94–101. doi: 10.1016/j.tcb.2003.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.