Abstract
Tim23p is an essential channel-forming component of the multisubunit TIM23 complex of the mitochondrial inner membrane that mediates protein import. Radiolabeled Tim23p monocysteine mutants were imported in vitro, incorporated into functional TIM23 complexes, and subjected to chemical cross-linking. Three regions of proximity between Tim23p and other subunits of the TIM23 complex were identified: Tim17p and the first transmembrane segment of Tim23p; Tim50p and the C-terminal end of the Tim23p hydrophilic region; and the entire hydrophilic domains of Tim23p molecules. These regions of proximity reversibly change in response to changes in membrane potential across the inner membrane and also when a translocating substrate is trapped in the TIM23 complex. These structural changes reveal that the macromolecular arrangement within the TIM23 complex is dynamic and varies with the physiological state of the mitochondrion.
INTRODUCTION
Mitochondria are morphologically complex organelles containing four subcompartments: the outer membrane (OM), the intermembrane space (IMS), the inner membrane (IM), and the matrix. Because most proteins that reside in mitochondria are nuclear-encoded and synthesized on cytosolic ribosomes, several multisubunit mitochondrial translocases are required to direct mitochondrial proteins to their correct locations (Bohnert et al., 2007; Neupert and Herrmann, 2007). Proteins targeted to mitochondria initially engage the TOM (translocase of the outer membrane) complex that mediates both translocation across, and integration into, the OM (Bohnert et al., 2007; Neupert and Herrmann, 2007). Two major translocases of the inner membrane (TIM) mediate the translocation and integration of specific sets of precursor proteins at the IM. The carrier translocase (TIM22 complex) mediates the integration of polytopic membrane proteins (MPs) into the IM in a manner that is strictly dependent on the membrane potential (Δψ) across the membrane (Rehling et al., 2004). The presequence translocase (TIM23 complex) mediates the Δψ-dependent translocation of soluble proteins with N-terminal presequences into the matrix, as well as the integration of single spanning MPs into the IM (Neupert and Brunner, 2002). Translocation of soluble substrates across the IM into the matrix requires the ATP-powered PAM (presequence translocase-associated motor) complex (Bohnert et al., 2007; Neupert and Herrmann, 2007).
The TIM23 complex of Saccharomyces cerevisiae consists of four integral membrane proteins: Tim23p, Tim17p, Tim50p, and Tim21p. Tim23p consists of a C-terminal membrane-bound region with four predicted transmembrane (TM) helices and a hydrophilic N-terminal region in the IMS. Electrophysiological studies have shown that Tim23p forms a voltage-gated, cation-selective channel (Lohret et al., 1997; Truscott et al., 2001); therefore Tim23p is thought to comprise at least part of the protein-conducting pathway through the IM. The multifunctional IMS region of Tim23p can be divided into two parts: the N-terminal half has been shown to span the OM and may help position the TIM23 complex relative to the OM (Donzeau et al., 2000), and the C-terminal half binds presequences and mediates the dimerization of Tim23p molecules through a heptad leucine repeat motif (Bauer et al., 1996). In addition, the Tim23p IMS region regulates channel activity (Truscott et al., 2001). The Δψ plays two major roles in Tim23p function: it regulates Tim23p dimerization (Bauer et al., 1996), and it provides an electrophoretic force necessary for pulling the basic presequence of matrix-targeted substrates across the IM (Neupert and Brunner, 2002). Tim17p is homologous to the C-terminal region of Tim23p, and although its function is less clear, recent studies indicate that it is involved in substrate-induced voltage gating of the TIM23 channel (Meier et al., 2005; Martinez-Caballero et al., 2007), and in recruitment of the PAM complex (Chacinska et al., 2005). Tim50p, which contains a single TM segment and a large C-terminal domain in the IMS region, is essential for the recognition of substrates emerging from the TOM complex and in their transfer to the TIM23 channel (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003) and has been implicated in maintaining the permeability barrier across the IM (Meinecke et al., 2006). Tim21p is a single-spanning IM protein with a C-terminal region that directly tethers the TIM23 and TOM complexes and is involved in the substrate-directed switching of the TIM23 complex between matrix translocation and inner membrane sorting modes (Chacinska et al., 2005; Mokranjac et al., 2005). The associated PAM complex, responsible for mediating the ATP-driven translocation of soluble substrates across the IM into the matrix, consists of mtHsp70 (matrix heat-shock protein), the nucleotide exchange factor Mge1, Pam16p/Tim16p, Pam18p/Tim14p, Pam17, and Tim44 (van der Laan et al., 2005; Neupert and Herrmann, 2007).
Several studies have addressed the quaternary structure of the TIM23 complex, particularly with respect to the essential membrane proteins Tim23p, Tim17p, and Tim50p. Tim23p and Tim17p appear to be present in the complex in equimolar amounts (Bömer et al., 1997; Ryan et al., 1998; Moro et al., 1999) and together comprise the core complex of the presequence translocase (Dekker et al., 1997). Tim23p monomers associate through their IMS regions (Bauer et al., 1996) and not through their hydrophobic regions (Ryan et al., 1998). However, the hydrophobic domain of Tim23p has been shown to physically interact with Tim17p (Ryan et al., 1998). In addition, the IMS domain of Tim50p interacts with the C-terminal part of the IMS region of Tim23p (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003).
This complex is also highly dynamic and versatile, showing marked changes in its activity and subunit composition under different conditions. For example, early work demonstrated that Tim23p existed in separate, dynamically interacting subcomplexes (Bömer et al., 1997) and that newly imported Tim subunits could be exchanged for endogenous subunits (Dekker et al., 1997). Recent work has shown that the subunit interactions of the TIM23 complex vary in response to polypeptide substrates. Transiting precursor proteins can stably connect the TIM23-PAM and TOM translocases to form a supercomplex (Chacinska et al., 2003; Frazier et al., 2003) and interaction with the TOM complex is mediated by Tim21p (Chacinska et al., 2005; Mokranjac et al., 2005). The TIM23 complex switches between different modular states depending on sorting information in the polypeptide substrate: an inner membrane sorting form of TIM23 associates with Tim21p but lacks the PAM complex, whereas a matrix import form of TIM23 associates with the PAM complex but loses its association with Tim21p (Chacinska et al., 2005).
To extend our understanding of TIM23 complex quaternary structure, regions in Tim23p adjacent to Tim17p, Tim50p, and other Tim23p molecules have been identified by chemical cross-linking. These regions of proximity reversibly change in response to changes in the Δψ and also to the presence of substrate. This high-resolution analysis refines our understanding of Tim23p subunit interactions and defines structural rearrangements within an assembled TIM23 complex.
MATERIALS AND METHODS
Plasmids and Yeast Strains
The SP6-containing plasmid pJE29 (Ryan et al., 1998) was used to generate the library of Tim23p variants. The substitution of all endogenous Cys residues to Ala to create Tim23pΔCys, and the subsequent introduction of a Cys codon (5′-TGC-3′) at each selected location was done using the Quikchange protocol (Stratagene, La Jolla, CA). The primary sequence of each construct was confirmed by DNA sequencing. mRNA was transcribed in vitro using SP6 RNA polymerase and PCR-generated DNA fragments encoding the full-length Tim23p as described (Flanagan et al., 2003).
Mitochondria were isolated as described previously (Daum et al., 1982) from either wild-type strain D273-10B or from strain RJ474, a strain expressing Tim23p-HA (Tim23p with a triple hemagglutinin [HA] epitope at its carboxy terminus; Davis et al., 1998).
The recombinant protein pSu9-DHFR (consisting of the first 69 residues of subunit 9 of the FO-ATPase fused to full length dihydrofolate reductase with a C-terminal hexahistidine tag) was subcloned into the pET30a(+) vector (Novagen, Madison, WI) and expressed in BL21 Star (DE3) cells (Invitrogen, Carlsbad, CA). Variants of this protein containing single Cys sites in the presequence were created by site-directed mutagenesis as above. After growth to an A600 of ∼0.8, cells were induced with 1 mM IPTG for 2.5 h, pelleted cells were disrupted by sonication, and protein was purified from cell lysate on a Ni-NTA matrix using an imidazole gradient (Stan et al., 2000).
Translation, Import, and Chemical Cross-Linking
Tim23p mRNA was translated (26°C, 40 min) in a wheat germ–based extract in the presence of [35S]Met (0.25 μCi/μl) as described (Flanagan et al., 2003). Mitochondrial import reactions (26°C, 20 min) were carried out in import buffer (0.25 M sucrose, 20 mM HEPES-KOH, pH 7.5, 80 mM KCl, 5 mM MgCl2, 2 mM potassium phosphate, pH 7.5, 3 mg/ml bovine serum albumin [BSA; fatty acid-free], 3 mM NADH, 1.5 mM ATP) containing mitochondria (0.25 mg/ml) and translation reaction (8% vol/vol). After import, mitochondria were sedimented (12,500 × g, 5 min) and resuspended in cross-linking buffer (CB, Import buffer lacking BSA). For dissipation of the IM potential after Tim23p import, ionophores and uncouplers were added as indicated from ethanol stocks (final ethanol concentration = 0.06% vol/vol). Where indicated, purified pSu9-DHFR was added in the indicated concentrations in the presence of 2 μM methotrexate (MTX). Osmotic shock to disrupt the OM was performed as before (Davis et al., 1998). For proteolysis experiments, samples were treated with 50 μg/ml proteinase K for 20 min on ice followed by the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF).
For chemical cross-linking, the homobifunctional thiol reactive reagent Bis-maleimidoethane (BMOE) was added from a dimethyl sulfoxide (DMSO) stock to a final concentration of 50 μM (final DMSO concentration = 2.5% vol/vol) to mitochondria (0.13 mg/ml) in CB. The pH of the incubation was maintained within the proper range to ensure that the maleimide group will react specifically with sulfhydryls (Smyth et al., 1964). For cysteine labeling, the reagent 4-acetamido-4′-[(iodoacetyl)amino] stilbene-2,2′-disulfonic acid (IASD) was added to a final concentration of 4.5 mM to mitochondria as above in CB with 1 mM dithiothreitol (DTT). BMOE cross-linking and IASD labeling reactions were incubated at 26°C for 20 and 10 min, respectively, and quenched by the addition of 40 mM DTT (cross-linking reactions) or 200 mM DTT (labeling reactions).
Immunoprecipitation and Coimmunoprecipitation
For immunoprecipitation after cross-linking reactions, mitochondria (25 μg) were sedimented and the pellets were resuspended in 100 μl of 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1% (vol/vol) SDS. Solubilized mitochondria were diluted 10-fold with lysis buffer (10 mM Tris-HCl, pH 7.5, 10–150 mM NaCl, depending on antiserum used, 5 mM EDTA, 1 mM PMSF, and 1% Triton X-100, vol/vol), and a clarifying spin (18,000 × g, 10 min) was used to remove undissolved material. Supernatants were transferred to new tubes, and protein A-Sepharose beads precoupled with antibody were added. After overnight rocking at 4°C, beads were washed three times in 1 ml of the respective lysis buffer, once in 1 ml of 10 mM Tris-HCl (pH 7.5), and eluted with 30 μl of SDS-sample buffer.
For coimmunoprecipitation, mitochondria (125 μg) were sedimented and solubilized in 250 μl ice-cold solubilization buffer (30 mM HEPES-KOH [pH 7.5],10 mM NaCl, 0.5 mM EDTA, 10% [vol/vol] glycerol, 1% [vol/vol] digitonin [2× recrystallized in ethanol]), 1 mM PMSF, and standard protease inhibitors]. After incubation (4°C, 30 min), the extract was cleared by centrifugation (12,500 × g for 10 min), the supernatant was rocked with 8 μl of respective antibody (4°C, 2.5 h), and incubation with precoupled beads and elution was performed as above.
RESULTS
Experimental Design
The objective of this work was to identify TIM23 complex subunits that are proximal to specific regions of Tim23p and to determine how their proximity may vary with changes in membrane potential and with the presence of substrate. To this end, we used cysteine-scanning mutagenesis, positioning single Cys residues at each of 54 different sites of a 222-residue Tim23p construct devoid of native Cys residues (Figure 1A). In vitro–translated [35S]Tim23p monocysteine mutants were integrated into the IM of isolated mitochondria and assembled into functional TIM23 complexes. Endogenous proteins adjacent to specific regions of Tim23p were then identified by chemical cross-linking and immunoprecipitation. Specifically, a homobifunctional thiol-reactive reagent may create a covalent adduct (cross-link) by reacting with the free sulfhydryl group on a given Tim23p monocysteine variant and with a Cys residue on a nearby native protein. Immunoprecipitation of the radiolabeled adduct with antibodies to different TIM23 complex subunits can then be used to identify the cross-linked protein. This approach relies upon the fortuitous proximity and reactivity of Cys groups on the imported [35S]Tim23p mutant and on the endogenous target proteins. However, a positive result, the formation of a covalent linkage, has the advantage of unambiguously identifying the region of proximity between the native subunit and the imported [35S]Tim23p molecule. In this respect, this approach provides the same high-resolution positional information as techniques using site-specific incorporation of photocrosslinking reagents (e.g., (Kanamori et al., 1999; McCormick et al., 2003; Saksena et al., 2004; Davis et al., 2007).
Figure 1.
Membrane integration and assembly of Tim23p mutants. (A) Linear representations of the essential membrane proteins in the TIM23 complex. Large boxes denote proposed Tim23p TMS regions (TMS1-4); thin boxes denote the hydrophilic N-terminal region and loops (L1-3) in the IMS (shifted upward) and in the matrix (shifted downward). Residue numbers of Tim23p are shown at the top. Open ovals show native Cys positions in S. cerevisiae Tim23p (Cys98, Cys209, and Cys213) that were changed to Ala in the Tim23pΔCys construct; closed ovals show native Cys positions in Tim17p and Tim50p. Gray rectangles in Tim23p show positions of individual monocysteine mutations used in this study. Slash bars in the Tim50p cartoon are breaks between positions 100 and 140 and between positions 240 and 450. (B) Tim23p monocysteine mutants are imported into mitochondria and integrated into the IM with efficiency similar to wild-type. Wild-type, Cys-less (ΔCys), and the indicated monocysteine mutants of in vitro–translated [35S]Tim23p were incubated with energized mitochondria or with mitochondria in which the membrane potential had been dissipated (−Δψ, lanes 5, 10, 15, 20, and 25) with 1 μM valinomycin. Samples were then subjected to the treatments after import: addition of protease directly to intact mitochondria (lanes 2, 7, 12, 17, and 22 for energized mitochondria, and lanes 5, 10, 15, 20, and 25 for −Δψ mitochondria); hypotonic swelling to produce mitoplasts and then addition of protease to produce the ∼14-kDa fragment (marked by the arrowhead) characteristic of membrane integration (“swelling”, lanes 3, 8, 13, 18, and 23); addition of the detergent Triton X-100 followed by addition of protease (“TX-100”, lanes 4, 9, 14, 19, and 24). Lanes 1, 6, 11, 16, and 21 show 20% of the total in vitro translation product added to each import reaction. (C) Tim23p monocysteine mutants are assembled into TIM23 complexes with efficiency similar to wild type. Wild-type (WT), G186D (corresponding to the Tim23-1 mutant; Lohret et al., 1997), ΔCys, and the indicated monocysteine mutants of [35S]Tim23p were incubated with energized mitochondria or mitochondria with a dissipated membrane potential (WT −Δψ), then subjected to wash in a buffer containing 1% digitonin to maintain the TIM23 complex, and coimmunoprecipitated with antibodies directed against Tim17p (CoIP, lanes 2, 4, 6, 8, 10, 12, and 14). Lanes 1, 3, 5, 7, 9, 11, and 13 show 5% of total in vitro translation product added to each import reaction before membrane solubilization.
Import and Assembly of Tim23p Mutants
A cysteine-less variant of S. cerevisiae Tim23p was prepared by replacement of the native Cys98, Cys209, and Cys213 using site-directed mutagenesis (Figure 1A). As judged by proteolysis experiments with in vitro imported protein (see below), replacement of Cys98 with Ser rendered Tim23p unable to integrate into the IM (data not shown); however, all three positions tolerated replacement with Ala to create a functional Tim23pΔCys, and this construct was used as a background for preparation of all subsequent monocysteine variants.
We first tested whether Tim23pΔCys and Tim23p monocysteine mutants underwent biogenesis similar to that of wild-type Tim23p in isolated mitochondria. Tim23p that is integrated into the IM of intact mitochondria will be protected from added protease. When incubated with intact energized mitochondria, wild-type [35S]Tim23p was integrated into the IM and protected from externally added proteinase K (Figure 1B, lane 2). After osmotic swelling and hypotonic rupture of the OM to produce mitoplasts, the protease degraded the N-terminal region of Tim23p in the IMS and thereby reduced the [35S]Tim23p to the ∼14-kDa proteolysis fragment that is diagnostic (Bömer et al., 1997; Davis et al., 1998) for proper integration (Figure 1B, lane 3, arrow). As controls, incubation of the wild-type construct with de-energized mitochondria prevented integration into the IM (Figure 1B, lane 5), and addition of detergent to solubilize membranes before protease addition rendered the [35S]Tim23p completely sensitive to protease (Figure 1B, lane 4). All mutants were examined with these protease assays, and representative results are shown (Figure 1B) for Tim23pΔCys and mutants with Cys substitutions in TMS1 (F114C), L1 (K131C), and TMS2 (I161C). For all mutants used in this study, the percentage of protein protected from external protease in intact mitochondria was similar (within ∼20%) to wild-type, indicating that the Cys substitutions used did not interfere with translocation across the OM. Moreover, the amount of proteolytic fragment after hypotonic swelling was similar in all cases, indicating that none of the Cys substitutions interfered with integration into the IM or with topogenesis.
To determine whether Tim23pΔCys and each monocysteine mutant were properly assembled into TIM23 complexes of isolated mitochondria, we performed coimmunoprecipitation experiments (Bömer et al., 1997; Ryan et al., 1998). After import into mitochondria and membrane solubilization under nondenaturing conditions that keep the TIM23 complex intact, wild-type [35S]Tim23p was coimmunoprecipitated with antiserum to Tim17p (Figure 1C, lane 2). As negative controls, wild-type [35S]Tim23p incubated with de-energized mitochondria (Figure 1C, lane 4) and the [35S]Tim23p G186D mutant (Lohret et al., 1997; Ryan et al., 1998; Figure 1C, lane 6) were not precipitated by Tim17p antiserum, thereby demonstrating the requirement of proper integration and assembly for coimmunoprecipitation. Tim23pΔCys and all monocysteine variants exhibited assembly similar to wild type, as demonstrated by representative mutants (Figure 1C, lanes 8, 10, 12, and 14). We conclude that the monocysteine mutants used in this study were able to assemble into TIM23 complexes with efficiency similar to wild type. Notably, the coimmunoprecipitation efficiencies shown here are similar to those obtained previously with in vitro–translated and imported Tim23p (Bömer et al., 1997). Although the exact steady-state amount of imported [35S]Tim23p that assembles into a complex cannot be determined by this assay, these results indicate that the monocysteine mutations do not significantly affect Tim23p assembly, and that proximity to other subunits obtained by cross-linking will reflect the native situation.
Tim23p Cross-Linking to TIM23 Complex Subunits
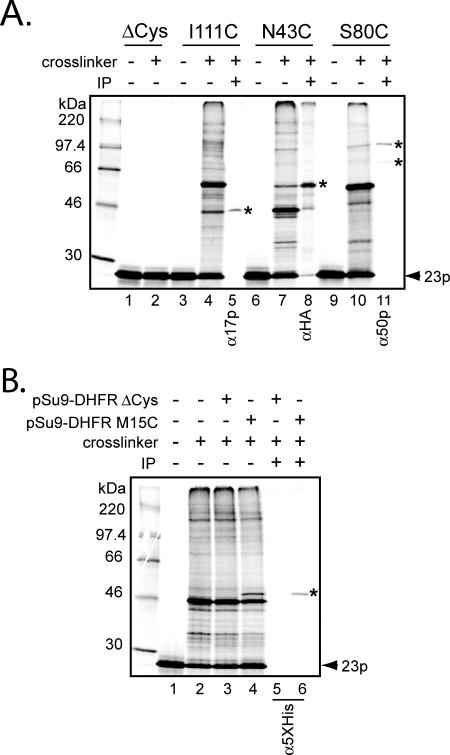
Having established that the Tim23p monocysteine mutants properly integrated into the IM and assembled into TIM23 complexes, we used these constructs in chemical cross-linking experiments to detect subunits of the endogenous complex that are proximal to different regions of Tim23p. Our primary interest was in identifying cross-linked adducts between the imported Tim23p monocysteine mutants and the essential membrane-bound components of the TIM23 complex (Tim17p, Tim23p, and Tim50p). Figure 2 shows typical results of experiments using the thiol-reactive homobifunctional cross-linking reagent BMOE, which has a spacer arm of 8 Å. As a negative control, imported [35S]Tim23pΔCys did not show any high-molecular-weight adducts after incubation with the chemical cross-linker (Figure 2A, compare lanes 1 and 2), indicating that under the reaction conditions used, there was no spurious cross-linking to non-Cys residues on the imported [35S]Tim32p molecule.
Figure 2.
Cross-linking and immunoprecipitation of Tim23p monocysteine mutants. (A) After [35S]Tim23p monocysteine mutants were imported into isolated mitochondria (either D273-10B, or mitochondria from the strain expressing Tim23p-HA [lanes 6–8]), samples were treated either with the cross-linking reagent BMOE (+cross-linker) or with solvent only (−cross-linker), as indicated. Cross-linked samples were then used for immunoprecipitation (lanes 5, 8, and 11) using antiserum against Tim17p, HA, and Tim50p (α17p, αHA, and α50p, respectively). Cross-linked adducts identified by immunoprecipitation are indicated by asterisks. Other cross-linked bands were not identified. (B) Imported Tim23p S34C forms a cross-linked adduct with the pSu9-DHFR translocation intermediate. After import of [35S]Tim23p S34C into isolated mitochondria, samples were incubated with either 1 μM pSu9-DHFR ΔCys (lanes 3 and 5), 1 μM pSu9-DHFR M15C (lanes 4 and 6), or protein buffer only (lanes 1 and 2). Samples were then treated with BMOE (lanes 2–6) or solvent only as shown. Immunoprecipitation of selected samples (lanes 5 and 6) was done using antibodies against the pentahistidine epitope, which recognizes the affinity tag on the pSu9-DHFR proteins. The cross-link between Tim23p and substrate is identified by the asterisk.
The [35S]Tim23p I111C mutant, with a Cys substitution in TMS1, produced a number of high-molecular-weight adducts after incubation with the cross-linker (Figure 2A, compare lanes 3 and 4). Immunoprecipitation revealed that the adduct with a molecular weight of ∼45 kDa was a cross-link between the imported [35S]Tim23p I111C and endogenous Tim17p (Figure 2A, lane 5).
To test for proximity between imported [35S]Tim23p variants and endogenous Tim23p, we used mitochondria isolated from cells expressing Tim23p-HA, a fusion Tim23p protein in which the influenza HA epitope is inserted at the carboxy terminus (Davis et al., 1998). Import of [35S]Tim23p monocysteine mutants into these mitochondria, followed by cross-linking and immunoprecipitation with antiserum to the HA tag, was then used to detect proximity to endogenous Tim23p molecules. The [35S]Tim23p N43C mutant, with a Cys substitution in the IMS region, produced a cross-linking adduct of ∼60 kDa (Figure 2A, lane 7) that was identified as Tim23p-HA by immunoprecipitation (Figure 2A, lane 8).
The [35S]Tim23p S80C mutant, another construct with an IMS Cys substitution, produced one cross-linking adduct of ∼95 kDa and a less intense adduct of ∼70 kDa (Figure 2A, lane 10), both of which were identified as cross-links to Tim50p by immunoprecipitation (Figure 2A, lane 11). The apparent molecular mass of the major Tim23p-Tim50p adduct based on this SDS-PAGE analysis is significantly higher than the ∼79 kDa expected for a covalent Tim23p-Tim50p adduct. This variation in electrophoretic mobility could be due to the site of covalent linkage between a [35S]Tim23p derivative and endogenous Tim50p, a phenomenon observed in other systems (Plath et al., 1998; McCormick et al., 2003, but see below). Indeed, multiple photoadduct bands have been observed previously between Tim50p and imported Tim23p (Davis et al., 2007).
Tim23p Cross-Linking to Translocation Intermediates
To determine whether imported Tim23p monocysteine mutants were assembled into functional TIM23 complexes, we examined whether the mutants were adjacent to substrate proteins that had been trapped in the TIM23 complex during import. When pSu9-DHFR, a fusion protein between the first 69 amino acids of subunit 9 of the mitochondrial FO-ATPase and mouse dihydrofolate reductase (Rapaport et al., 1998), was imported in the presence of its specific ligand MTX, the resultant tightly folded DHFR-MTX domain could not cross the TOM complex channel and therefore formed a translocation intermediate in which the substrate presequence was engaged with the TIM23 complex (Eilers and Schatz, 1986). Thus, [35S]Tim23p S34C, a mutant with a Cys substitution in the N-terminal IMS region, was first imported into mitochondria. Purified pSu9-DHFR was then incubated with those mitochondria in the presence of MTX, after which cross-linker was added (Figure 2B). Although several proteins were cross-linked to imported [35S]Tim23p S34C, an adduct of ∼45 kDa was observed with pSu9-DHFR M15C (bearing a single Cys residue at position 15 of the presquence; Figure 2B, lane 4), but not with pSu9-DHFR ΔCys (a version of the fusion protein lacking Cys residues in the presequence; Figure 2B, lane 3). This band was identified as a cross-link between pSu9-DHFR and newly imported [35S]Tim23p because the radioactive species was immunoprecipitated with pentahistidine antiserum that recognizes the hexahistidine affinity tag on the pSu9-DHFR protein (Figure 2B, lanes 5 and 6). Identical results were obtained when the sample was immunoprecipiated with DHFR antiserum (data not shown). Taken together, these data show that the trapped substrate cross-links to the Tim23p mutant via its Cys residue, and hence confirm that the imported Tim23p monocysteine mutants are assembled into functional, translocation-competent TIM23 complexes.
Mapping Tim23p Proximity to TIM23 Complex Components
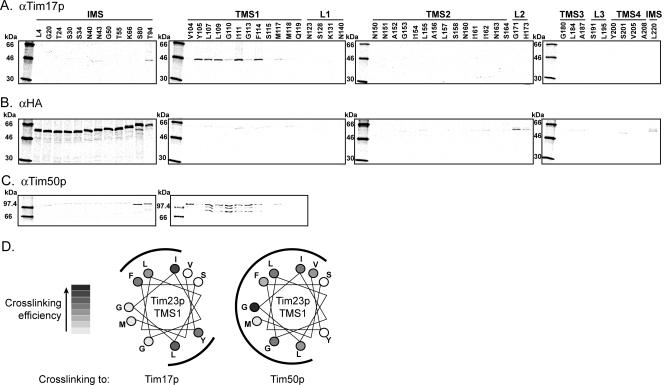
We expanded our cross-linking analysis to explore regions of proximity between our library of Tim23p monocysteine mutants and endogenous Tim17p, Tim23p-HA, and Tim50p. We found robust cross-linking adducts between endogenous Tim17p and imported [35S]Tim23p mutants that had Cys substitutions in TMS1 (Figure 3A). Because S. cerevisiae Tim17p contains two Cys residues in IMS-facing loops and two Cys residues predicted to be within the membrane (Tim17p TMS4, Figure 1A), Cys residues in Tim23p TMS1 and Tim17p TMS4 are likely to be proximal, provided that the local environment is conducive to the cross-linking reaction (see Discussion). These cross-linking results are also consistent with the previously observed coimmunoprecipitation of Tim17p with the C-terminal (membrane-embedded) domain of Tim23p (Ryan et al., 1998). A helical wheel projection showing Tim23p TMS1 sites that cross-link to Tim17p (Figure 3D) indicates that cross-linking to Tim17p occurs on opposite sides of the Tim23p TMS1 helix. These data suggest that multiple copies of Tim17p associate with Tim23p on opposite faces of TMS1 and/or that TMS1 of Tim23p has significant rotational flexibility, but exists stably in only a few orientations. Whatever the case, these results demonstrate both the high resolution of this cross-linking approach and also the specificity of the quaternary arrangement of proteins in the TIM23 complex.
Figure 3.
Proximity of Tim23p sites to TIM23 complex subunits. Multiple monocysteine [35S]Tim23p mutants were imported into isolated mitochondria, subjected to cross-linking with BMOE, and then immunoprecipitated with antibodies directed against either Tim17p (A), HA (B), or Tim50p (C). (D) Helical wheel representation of Tim23p TMS1 showing all positions with Cys substitutions from Val104 to Met117. Cross-linking efficiency to Tim17p and Tim50p is ranked as indicated with regions of strong cross-linking indicated by outer arcs.
Cross-links to endogenous Tim23p-HA were primarily detected with imported [35S]Tim23p mutants having Cys substitutions in the IMS region. In addition, significant adduct formation occurred at the IMS-facing L2 and at position 220 near the C-terminus, along with a few weak adducts detected at other sites throughout the protein (Figure 3B). Because the native Cys residues in endogenous Tim23p of S. cerevisiae include a single Cys located in the IMS just N-terminal to TMS1 and two Cys in TMS4 (Figure 1A), the most likely cross-linking partner for the N-terminal [35S]Tim23p mutants is the IMS-facing Cys residue in Tim23p-HA, especially given the short span of the BMOE cross-linker and the extensive range over which cross-linking occurred. Proximity between the IMS regions of Tim23p assembled into a TIM23 complex is consistent with previous work showing that dimerization of Tim23p molecules occurs via the second half of the N-terminal domains (Bauer et al., 1996). But the similarity in cross-linking yields for all sites tested in the mutant Tim23p IMS domain strongly indicates that the N-terminal half of Tim23p does not occupy a fixed position in the TIM23 complex. Instead, the IMS domain appears flexible and free to move such that residues from 4 to 94 on the imported mutants are able to contact Cys98 of the endogenous Tim23p during the course of the cross-linking reaction.
Only Tim23p monocysteine mutants with Cys substitutions in the IMS and in TMS1 were used to test for proximity to native Tim50p. Of the IMS sites, only Cys residues located in the C-terminal part of this domain (positions 80 and 94) yielded robust cross-linked adducts (Figure 3C). This result is consistent with previous observations that the IMS domain of Tim50p directly interacts with the C-terminal half of the IMS domain of Tim23p (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). In addition, strong cross-linking was detected between TMS1 of Tim23p and Tim50p (Figure 3C), and a helical projection indicates that this cross-linking occurs over a broad arc of the TMS1 helix (Figure 3D). Given that S. cerevisiae Tim50p has only a single Cys (Figure 1A), the cross-linking partners can be unambiguously identified: Cys sites in the extreme C-terminal IMS region and in TMS1 of Tim23p are proximal to Cys286 of Tim50p. Because each protein bears a single Cys, a single band representing the Tim23p-Tim50p adduct would be expected to be immunoprecipitated with antiserum to Tim50p. The presence of multiple bands seen here in certain cases may, for example, suggest partial proteolysis of the sample during immunoprecipitation. Notably, the extensive and overlapping cross-linking of Tim17p and Tim50p with Tim23p TMS1 suggests that an imported Tim23p exists in multiple different states within the TIM23 complex and/or that Tim23p has sufficient conformational flexibility to position TMS1 proximal to either Tim17p or Tim50p (see Discussion).
It is important to note that using homobifunctional reagents with the same maleimide reactive groups, but with longer spacer arm lengths (up to 16.1 Å), did not yield any qualitative differences in the cross-linking profiles reported here.
Membrane Potential-dependent Cross-Linking of Tim23p
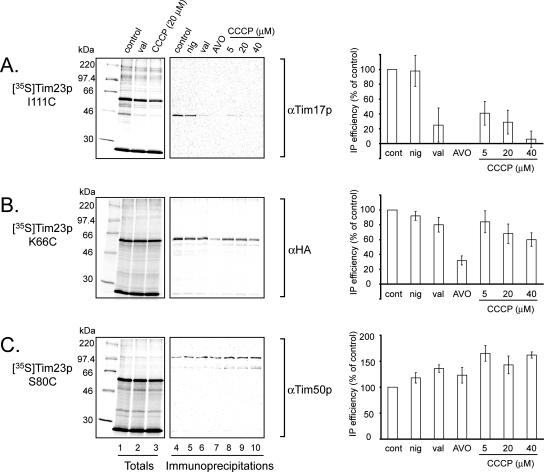
Isolated Tim23p and the TIM23 complex display voltage gating behavior (Lohret et al., 1997; Truscott et al., 2001). Other channels, such as voltage-gated potassium channels, undergo conformational changes and transmembrane movements to varying extents in response to changing electric field across the membrane (Tombola et al., 2005). We therefore examined whether our cross-linking strategy would detect changes in TIM23 complex quaternary structure if the Δψ across the IM changed. In this approach, the energized import of a given [35S]Tim23p monocysteine mutant was followed by the addition of different uncoupling agents and ionophores to dissipate the Δψ before cross-linking.
Collapse of the Δψ by the K+ ionophore valinomycin or by carbonyl cyanide 3-chlorophenylhydrazone (CCCP) specifically blocked cross-linking between [35S]Tim23p I111C and Tim17p (Figure 4A, compare lane 1 with lanes 2 and 3 [totals] and lane 4 with lanes 6 through 10 [immunoprecipitations]). Indeed, dissipation of the Δψ by CCCP completely blocked cross-linking between all TMS1 Cys sites and native Tim17p (data not shown). In contrast, nigericin, which catalyzes the electroneutral exchange of H+ and K+ and thus collapses only the pH gradient (believed to constitute a minor fraction of the protonmotive force across the IM; Nicholls and Ferguson, 1992), caused only a small change in [35S]Tim23p I111C-Tim17p cross-linking (Figure 4A, lane 5). These results therefore show that the proximity of residues in Tim23p TMS1 and Tim17p is strongly dependent on the Δψ across the IM. It should be noted, however, that the Tim23p-Tim17p core complex has been shown to remain stable after dissipation of the Δψ by blue native PAGE (Dekker et al., 1997). Therefore, any interactive surface(s) between these two subunits that remain(s) after Δψ dissipation likely exists between Tim23p and a site on Tim17p that lacks a nearby Cys (and therefore any potential cross-linking site in our experiments).
Figure 4.
Sensitivity of Tim23p cross-linking to membrane potential. Effect of reduction in electrochemical gradient across the IM on cross-linking between endogenous subunits and [35S]Tim23p monocysteine mutants I111C (A), K66C (B), and S80C (C). Samples were treated subsequent to [35S]Tim23p import, but before the cross-linking reaction, with the following reagents as indicated: control (EtOH only, lanes 1 and 4), val (1 μM valinomycin, lanes 2 and 6), nig (1 μM nigericin, lane 5), AVO (4 μM antimycin A, 1 μM valinolycin, 8 μM oligomycin; lane 7), and CCCP at the indicated concentrations (lanes 8–10). For all treatments, [EtOH] = 0.06% (vol/vol). Totals: complete cross-linked sample was analyzed by SDS-PAGE. Immunoprecipitation: parallel samples were subjected to immunoprecipitation with antiserum against the indicated protein before SDS-PAGE. Bar graphs to the right show average band intensities from immunoprecipitations, normalized to control, with standard deviations from three independent experiments.
Cross-linking between [35S]Tim23p mutants and endogenous Tim23p-HA varied with the electrochemical gradient in a different way (Figure 4B). [35S]Tim23p K66C, bearing a Cys substitution in the C-terminal half of the IMS region, shows only a slight valinomycin-dependent decrease in cross-linking to Tim23p-HA (Figure 4B, compare lanes 4 and 6) and only a slight decrease upon titration with CCCP (Figure 4B, compare lanes 4 and lanes 8–10). However, cross-linking between [35S]Tim23p K66C and endogenous Tim23p-HA was significantly reduced with the inclusion of antimycin A (an inhibitor of complex III of the respiratory chain) and oligomycin (an inhibitor of the FO/F1-ATPase; Figure 4B, lane 7). A separate series of experiments revealed that the presence of 4 μM antimycin A alone reduced the extent of cross-linking between [35S]Tim23p K66C and Tim23p-HA to 43% of the control value. Qualitatively similar patterns were observed when [35S]Tim23p S30C and T94C cross-linking to Tim23p-HA was analyzed (data not shown). Interestingly, the presence of potassium cyanide (an inhibitor of complex IV) did not reduce the efficiency of cross-linking between Tim23p IMS regions (data not shown), indicating a specific effect of antimycin A that is not simply attributable to reduced Δψ nor to a general block of respiratory electron flow. We note also that these drugs can affect matrix ATP levels and thus our observed effects may be due in part to altered ATP concentrations.
In dramatic contrast to the above results, the extent of adduct formation between [35S]Tim23p S80C and native Tim50p unexpectedly increased in the presence of the valinomycin and nigericin ionophores (Figure 4C, lanes 5 and 6) and with increasing concentrations of the uncoupler CCCP (Figure 4C, lanes 8–10), consistent with a greater degree of proximity between the two subunits as Δψ decreases. The presence of 4 μM antimycin A alone increased cross-linking efficiency between [35S]Tim23p S80C and Tim50p by 35% (data not shown). Thus, the structural arrangement of Tim23p within the TIM23 complex is significantly altered with respect to Tim17p, Tim50p, and other Tim23p molecules when the Δψ is dissipated.
Reversibility of Membrane Potential Effects on Cross-Linking
To test whether the observed Δψ-dependence of cross-linking efficiency was reversible, we counteracted the CCCP-induced Δψ collapse by washing the mitochondria with DTT and respiratory substrate (Yaffe, 1991). This same strategy was used previously to show that Δψ-dependent import of polytopic IM proteins can be restored after CCCP incubation and that such precursors are in fact productive on-pathway intermediates (Ryan et al., 1999; Davis et al., 2000, 2007). Thus, mitochondria were incubated with CCCP and then given a second wash, either with CCCP to keep the Δψ dissipated or with NADH and DTT to reestablish the Δψ before the cross-linking reaction (Figure 5). As shown above, incubation with CCCP reduced the cross-linking of [35S]Tim23p mutants I111C and K66C to Tim17p and to Tim23p-HA, respectively, and enhanced cross-linking between [35S]Tim23p S80C and Tim50p all to extents similar to those seen in Figure 4 (Figure 5, compare A and B samples). But when CCCP-treated mitochondria were incubated with NADH and DTT before cross-linking, the extents of the cross-linking were very similar to those of non-CCCP–treated samples for all mutants (Figure 5, compare A and C samples). The Δψ-dependent changes in cross-linking observed in this study are therefore reversible and likely reflect bona fide changes in subunit proximity that occur with changes in the energetic state of the IM.
Figure 5.
Reversibility of membrane potential effects on cross-linking. (A) Protocol schematic. After import of the [35S]Tim23p mutants shown, reactions were split into three aliquots, sedimented, and washed as follows. Sample A (positive control) was resuspended in a buffer containing ATP and NADH; samples B and C were resuspended in a buffer containing ATP and CCCP to collapse the Δψ. After a 10-min incubation, membranes were resedimented and resuspended as follows: Samples A and B were resuspended in the same buffers used for the first wash, and sample C was resuspended in buffer containing ATP, NADH, and DTT to inactivate the CCCP. After another 10 min incubation, mitochondria were resedimented and used for cross-linking reactions. (B) Reversibility tests for [35S]Tim23p mutants I111C, K66C, and S80C immunoprecipitated with antiserum to Tim17p, HA, and Tim50p, respectively. Bar graphs show corresponding average band intensities, normalized to control with standard deviations from three independent experiments.
Aqueous Access of Tim23p TMS1 Is Dependent on Membrane Potential
Because changes in Δψ effected such profound changes in subunit proximity, particularly for the Tim23p IMS and TMS1 sites, we examined whether changes in the Δψ could also alter the accessibility of these sites to a large hydrophilic reagent. Both energized and valinomycin-treated mitochondria containing an imported [35S]Tim23p monocysteine mutant were incubated with the large membrane-impermeant sulfhydryl reactive reagent IASD (Figure 6). IASD labeling can be detected by a shift in the electrophoretic mobility of the protein in SDS-PAGE (Krishnasastry et al., 1994). Under the reaction conditions used, the [35S]Tim23p S80C mutant, with a Cys residue in the IMS, was accessible to IASD in both fully energized and valinomycin-treated mitochondria, whereas the [35S]Tim23p L157C mutant, with a Cys in TMS2, was not labeled by IASD under either condition (Figure 6, compare lanes 2 and 3 with 1). In contrast, [35S]Tim23p mutants I111C and F114C, with Cys residues in TMS1, displayed Δψ-dependent IASD labeling. These TMS1 sites were partially accessible to IASD in fully energized mitochondria (Figure 6, lane 2) but the extent of IASD labeling increased significantly after valinomycin treatment (Figure 6, lane 3). Therefore, collapse of the Δψ increased the exposure of these TMS1 sites to an aqueous compartment contiguous with the IMS.
Figure 6.
Membrane potential-dependent IASD labeling of Tim23p. After import of the [35S]Tim23p mutants indicated, mitochondria were incubated with either 1 μM valinomycin (lane 3) or EtOH only (all other lanes) and then incubated in the absence (lanes 1 and 5) or presence (lanes 2–4) of IASD. IASD labeling is detected by a decrease in protein mobility in SDS-PAGE (arrowhead) relative to nontreated samples. The absence of labeling in samples in which DTT was added before IASD (lane 4) shows that no IASD labeling occurred after quenching of the reaction.
Substrate-dependent Cross-Linking of Tim23p to TIM23 Subunits
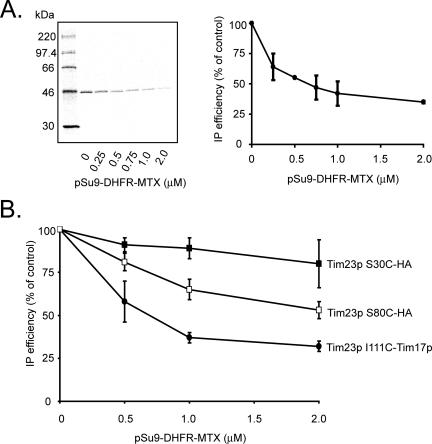
Finally, we tested whether the presence of substrate in the TIM23 complex might alter the cross-linking of its subunits. After the import of [35S]Tim23p monocysteine mutants, mitochondria were washed and supplied with pSu9-DHFR at different concentrations (up to 2 μM) in the presence of MTX to trap the substrate in the TIM23 complex. By using a Cys-less pSu9-DHFR substrate, no cross-linking between the overexpressed substrate and assembled [35S]Tim23p molecules could occur (Figure 2B). Given the estimated Tim23p concentration of 20 pmol per mg of mitochondrial protein (Dekker et al., 1997; Moro et al., 1999), substrate was added up to a roughly 200-fold molar excess over Tim23p molecules.
The extent of cross-linking between [35S]Tim23p I111C and endogenous Tim17p decreased dramatically when increasing amounts of pSu9-DHFR engaged the TIM23 complex (Figure 7A). Interestingly, cross-linking between Cys residues in the [35S]Tim23p IMS region and Tim23p-HA varied with pSu9-DHFR-MTX concentration in a position-dependent manner. Cross-linking between Tim23p-HA and [35S]Tim23p S30C, with a Cys in the N-terminal half of the IMS, was not strongly dependent on the presence of substrate (Figure 7B, ■). In contrast, cross-linking between Tim23p-HA and [35S]Tim23p S80C, with a Cys in the C-terminal half of the IMS, decreased significantly with increasing substrate concentration (Figure 7B, □). Thus, the cross-linking efficiencies between Tim23p TMS1 and Tim17p and between the C-terminal half of the Tim23p IMS region and other Tim23p subunits were significantly reduced by the presence of a translocation intermediate.
Figure 7.
Sensitivity of Tim23p cross-linking to the presence of substrate. (A) Cross-linking between [35S]Tim23p I111C and Tim17p with pSu9-DHFR-MTX titration. After import of [35S]Tim23p I111C, mitochondria were incubated with the indicated concentrations of pSu9-DHFR-MTX before cross-linking and immunoprecipitation with antiserum to Tim17p. (B) Cross-linking between [35S]Tim23p S30C and Tim23-HA (■) and between [35S]Tim23p S80C and Tim23-HA (□) with pSu9-DHFR-MTX titration as above. Parallel cross-linking between [35S]Tim23p I111C and Tim17p (•) are shown for comparison. The mean values and standard deviations of relative band intensities for three separate experiments are shown.
DISCUSSION
This comprehensive and high resolution study provides five main insights into the quaternary structure of the TIM23 complex. First, after importing each of 54 different Tim23p monocysteine mutants into intact and energized mitochondria, specific regions of a Tim23p molecule were shown to be adjacent to Tim17p, Tim50p, and at least one other Tim23p protein in a fully assembled and functional TIM23 complex (see below). Second, the structural proximity and hence arrangement of proteins within the TIM23 complex is dependent on the Δψ. Third, the Δψ-dependent conformational rearrangements are reversible, suggesting that they represent a true physiological response to the energized state of the IM. Fourth, different protein–protein proximities in the TIM23 complex are detected when a substrate is trapped inside the translocase, and these changes reflect Δψ-dependent changes. Fifth, these combined results show that an assembled TIM23 complex exists in a minimum of three states (energized, de-energized, and energized with substrate) within the IM that differ both structurally and functionally. The TIM23 complex is therefore a dynamic multicomponent assembly whose functional state is dictated, and presumably regulated, by changes in quaternary structure in response to cellular conditions.
Tim23p TMS1 is adjacent to Tim17p (Figures 3A and 8A) because nine residues of TMS1 (from Tyr105 to Phe114) cross-linked to Tim17p. Interestingly, these sites are found on two oppositely oriented faces of the TMS1 helix when mapped onto a helical projection (Figure 3D). Although rotational degeneracy of a transmembrane helix in the TIM23 complex could explain such differential cross-linking, the specificity with which cross-linked adducts were formed on the two faces strongly suggests that TMS1 of Tim23p contacts two separate Tim17p molecules on opposite sides. Given the topological placement of native Cys residues within Tim17p (Figure 8A), its two Cys residues in the N-terminal part of TMS4 are the most likely cross-linking partners for Tim23p TMS1 monocysteine mutants.
Figure 8.
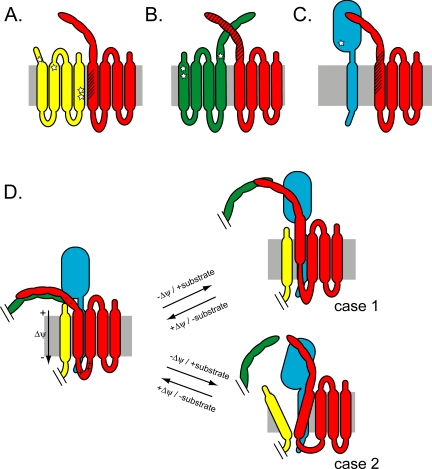
Summary of Tim23p cross-linking results. Schematic representation of imported [35S]Tim23p monocysteine mutants (red) and (A) Tim17p (yellow), (B) endogenous Tim23p (green), and (C) Tim50p (cyan) in the TIM23 complex. Stars represent approximate sites of native Cys residues in endogenous proteins, and hatch marks indicate regions on imported Tim23p that cross-link and hence are proximal to each subunit. (D) The data summarized in A–C are combined in the left panel to yield one (of many) potential quaternary structure arrangement of the TIM23 complex in energized mitochondria in the absence of substrate. Under these conditions, Tim23p TMS1 and the IMS region form extensive contacts with Tim17p and Tim23p, respectively, and Tim23p TMS1 shows low accessibility to IASD. The possibility that Tim23p TMS1 contacts two separate Tim17p molecules is not shown here. Charged residues flanking Tim23p TMS1 are indicated. Dissipation of the Δψ or the addition of substrate causes conformational changes that are similar, but differ in magnitude because the cross-linking efficiency changes are not equivalent. The right panels show two of the possible domain rearrangements that could simultaneously result in reduced cross-linking between Tim23p TMS1 and Tim17p, reduced cross-linking between the Tim23p IMS region and neighboring Tim23p molecules, enhanced cross-linking between the Tim23p IMS region and Tim50p, and increased Tim23p TMS1 accessibility to IASD. Case 1, Tim23p TMS1 translocation toward the IMS; case 2, formation of an aqueous crevice near the Tim23-Tim17 interface.
Among the TIM23 complex structural relationships characterized, the juxtaposition of Tim23p TMS1 and Tim17p was the most sensitive both to changes in Δψ (Figure 4A) and to the presence of substrate (Figure 7). It is not clear whether the Δψ- and substrate-dependent changes in Tim23p TMS1-Tim17p cross-linking have the same molecular basis (i.e., whether the collapse of the Δψ or the addition of substrate elicit the same conformational changes in the TIM23 complex). But whatever the origin, this dynamic Δψ- and substrate-dependent proximity may very well be related to the recently identified role of Tim17p in regulating the substrate-dependent voltage gating of Tim23p (Meier et al., 2005; Martinez-Caballero et al., 2007).
The Tim23p IMS region is proximal to other Tim23p subunits in the TIM23 complex (Figures 3B and 8B). The wide range of sites in the [35S]Tim23p mutants that cross-link to native Tim23p presumably through its single Cys in the IMS indicates a high degree of Tim23p conformational flexibility in the IMS. A decrease in the Δψ leads to a modest reduction in cross-linking between Tim23p IMS sites and other Tim23p subunits (Figure 4B), whereas the presence of a translocation intermediate significantly reduces cross-linking between Tim23p sites in the C-terminal half of the IMS region and other Tim23p subunits (Figure 7B). These results are consistent with previous work, indicating that Tim23p dimerizes via a putative leucine zipper structure in the C-terminal half of its IMS domain in a manner dependent on the functional state of the channel: dimer formation requires the Δψ, and matrix targeting presequences stimulate dimer dissociation (Bauer et al., 1996). Finally, the strong effect of antimycin A on Tim23p IMS cross-linking (Figure 4B) could result in part from the recently described coupling between Tim21p of the TIM23 complex and respiratory complexes III-IV (van der Laan et al., 2006). Alternatively, the effect could arise from the binding of antimycin A to a site other than complex III (e.g., Tzung et al., 2001).
The extreme C-terminal end of the IMS region of Tim23p is proximal to Tim50p (Figures 3C and 8C), consistent with previous work showing an interaction between the IMS region of Tim50p and the C-terminal half of the Tim23p IMS domain (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). In addition, we unexpectedly found cross-links between Tim50p and TMS1 of Tim23p (Figures 3C and 8C). Given that the putative cross-linking site on Tim50p (Cys268 in the mature protein) is in the C-terminal IMS domain, this suggests that either this region of Tim50p protrudes into the bilayer and/or that Tim23p has the conformational freedom to move TMS1 into the aqueous IMS to some degree. In any case, a helical projection of the sites that cross-link Tim23p TMS1 to Tim50p (Figure 3D) suggests that, in contrast to well-defined sites that cross-link Tim23p TMS1 to Tim17p (Figure 3D), there is significant rotational freedom between TMS1 and Tim50p such that cross-linking occurs over most of the helical circumference.
The cross-linking observed between Tim23p and Tim50p may also provide some insight into the functional role of Tim50p. The reactive Cys in Tim50p falls within a region of the IMS domain that is homologous to NIF/CDCc domains of CTD phosphatases (residues 165-344; Geissler et al., 2002; Mokranjac et al., 2003). Although the possible role of this domain in Tim50p function is not known, the potential specific interaction between Tim23p TMS1 and this region may provide clues to its function. Our unexpected observation that cross-linking between the IMS region of Tim23p and Tim50p increased when the Δψ collapsed suggests that Tim23p-Tim50p proximity and/or affinity increases upon loss of the Δψ. When coupled with the recent observation that the IMS region of Tim50p maintains the permeability barrier across the IM by promoting the closed state of the Tim23p channel (Meinecke et al., 2006), a regulatory mechanism is suggested. Specifically, the closer interaction between Tim23p and Tim50p that we detected upon lowering the Δψ may have evolved to maintain the IM permeability barrier and prevent unregulated collapse of IM gradients.
Our comprehensive cross-linking study has therefore provided unprecedented resolution in characterizing the proximity between TIM23 complex subunits, thereby significantly extending our understanding of the TIM23 complex quaternary structure. Although studies describing protein interactions based on coimmunoprecipitation cannot distinguish between direct contact and association through an intermediary subunit, a covalent bond that directly cross-links two macromolecules shows unambiguously that residues from each protein were adjacent. Moreover, the formation of covalent adducts usually identifies protein surfaces that interact for functional or structural reasons. But even though a lack of cross-linking cannot be taken as evidence for lack of proximity, the cross-linking approach used here maps the proximity of specific helical faces to other proteins at high resolution.
Several models could account for the multiple Δψ- and substrate-induced conformational changes in quaternary structure characterized here. Two possibilities are depicted in Figure 8D. Partial de-insertion of TMS1 from the membrane (case 1) would require that the energetic penalties of moving nonpolar residues into the aqueous phase and disrupting any associative energies with other TMSs in the membrane be offset by some compensatory binding energy (e.g., binding to Tim50p). Alternatively, loss of membrane potential and/or substrate addition may, for example, induce helix tilting and expansion of an aqueous crevice in this region (case 2). Future work will determine to what extent either model, or some combination thereof, is correct. But the conformational changes characterized by this study have provided insight into the Δψ- and substrate-dependent dynamics of the TIM23 complex and hence will direct further investigations.
ACKNOWLEDGMENTS
We are grateful to Yiwei Miao and Yuanlong Shao for outstanding technical assistance and to the members of the Johnson lab for helpful discussions. This work was supported by National Institutes of Health Grant GM26494 (A.E.J.), National Research Service Award GM70266 (N.N.A.), and by the Robert A. Welch Foundation (Chair Grant BE-0017).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0669) on October 24, 2007.
REFERENCES
- Bauer M. F., Sirrenberg C., Neupert W., Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- Bohnert M., Pfanner N., van der Laan M. A dynamic machinery for import of mitochondrial precursor proteins. FEBS Lett. 2007;581:2802–2810. doi: 10.1016/j.febslet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Bömer U., Meijer M., Maarse A. C., Hönlinger A., Dekker P.J.T., Pfanner N., Rassow J. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO J. 1997;16:2205–2216. doi: 10.1093/emboj/16.9.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Chacinska A., Rehling P., Guiard B., Frazier A. E., Schulze-Specking A., Pfanner N., Voos W., Meisinger C. Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 2003;22:5370–5381. doi: 10.1093/emboj/cdg532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Davis A. J., Alder N. N., Jensen R. E., Johnson A. E. The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochdondrial protein import. Mol. Biol. Cell. 2007;18:475–486. doi: 10.1091/mbc.E06-06-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. J., Ryan K. R., Jensen R. E. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol. Biol. Cell. 1998;9:2577–2593. doi: 10.1091/mbc.9.9.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. J., Sepuri N. B., Holder J., Johnson A. E., Jensen R. E. Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 2000;150:1271–1282. doi: 10.1083/jcb.150.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P.J.T., Martin F., Maarse A. C., Bömer U., Müller H., Guiard B., Meijer M., Rassow J., Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzeau M., Káldi K., Adam A., Paschen S., Wanner G., Guiard B., Bauer M. F., Neupert W., Brunner M. Tim23 links the inner and outer mitochondrial membranes. Cell. 2000;101:401–412. doi: 10.1016/s0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- Eilers M., Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- Flanagan J. J., Chen J.-C., Miao Y., Shao Y., Lin J., Bock P. E., Johnson A. E. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- Frazier A. E., Chacinska A., Truscott K. N., Guiard B., Pfanner N., Rehling P. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell. Biol. 2003;23:7818–7828. doi: 10.1128/MCB.23.21.7818-7828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A., Chacinska A., Truscott K. N., Wiedemann N., Brandner K., Sickmann A., Meyer H. E., Meisinger C., Pfanner N., Rehling P. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa S.-I., Nakai M., Shin I., Schultz P. G., Endo T. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. USA. 1999;96:3634–3639. doi: 10.1073/pnas.96.7.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnasastry M., Walker B., Braha O., Bayley H. Surface labeling of key residues during assembly of the transmembrane pore formed by staphylococcal α-hemolysin. FEBS Lett. 1994;356:66–71. doi: 10.1016/0014-5793(94)01240-7. [DOI] [PubMed] [Google Scholar]
- Lohret T. A., Jensen R. E., Kinnally K. W. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J. Cell Biol. 1997;137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Caballero S., Grigoriev S. M., Herrmann J. M., Campo M. L., Kinnally K. W. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 2007;282:3584–3593. doi: 10.1074/jbc.M607551200. [DOI] [PubMed] [Google Scholar]
- McCormick P. J., Miao Y., Shao Y., Lin J., Johnson A. E. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- Meier S., Neupert W., Herrmann J. M. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J. Biol. Chem. 2005;280:7777–7785. doi: 10.1074/jbc.M412158200. [DOI] [PubMed] [Google Scholar]
- Meinecke M., et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 2006;312:1523–1526. doi: 10.1126/science.1127628. [DOI] [PubMed] [Google Scholar]
- Mokranjac D., Paschen S. A., Kozany C., Prokisch H., Hoppins S. C., Nargang F. E., Neupert W., Hell K. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003;22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D., Popov-Čeleketić D., Hell K., Neupert W. Role of Tim21 in mitochondrial contact sites. J. Biol. Chem. 2005;280:23437–23440. doi: 10.1074/jbc.C500135200. [DOI] [PubMed] [Google Scholar]
- Moro F., Sirrenberg C., Schneider H.-C., Neupert W., Brunner M. The TIM17·23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J. 1999;18:3667–3675. doi: 10.1093/emboj/18.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W., Brunner M. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nicholls D. G., Ferguson S. J. Bioenergetics 2. London: Academic Press; 1992. [Google Scholar]
- Plath K., Mothes W., Wilkinson B. M., Stirling C. J., Rapoport T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Künkele K.-P., Dembowski M., Ahting U., Nargang F. E., Neupert W., Lill R. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Mol. Cell. Biol. 1998;18:5256–5262. doi: 10.1128/mcb.18.9.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Ryan K. R., Leung R. S., Jensen R. E. Characterization of the mitochondrial inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not with other Tim23p molecules. Mol. Cell. Biol. 1998;18:178–187. doi: 10.1128/mcb.18.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. T., Müller H., Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- Saksena S., Shao Y., Braunagel S. C., Summers M. D., Johnson A. E. Cotranslational integration and initial sorting at the endoplasmic reticulum translocon of proteins destined for the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 2004;101:12537–12542. doi: 10.1073/pnas.0404934101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. G., Blumenfeld O. O., Konigsberg W. Reactions of N-ethylmaleimide with peptides and amino acids. Biochem. J. 1964;91:589–595. doi: 10.1042/bj0910589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan T., Ahting U., Dembowski M., Künkele K.-P., Nussberger S., Neupert W., Rapaport D. Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J. 2000;19:4895–4902. doi: 10.1093/emboj/19.18.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F., Pathak M. M., Isacoff E. Y. How far will you go to sense voltage? Neuron. 2005;48:719–725. doi: 10.1016/j.neuron.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Truscott K. N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J.M., Rassow J., Pfanner N., Wagner R. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 2001;8:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- Tzung S.-P., Kim K. M., Basañez G., Giedt C. D., Simon J., Zimmerberg J., Zhang K.Y.J., Hockenbery D. M. Antimycin A mimics a cell-death-inducing Bcl-2 homology domain 3. Nat. Cell Biol. 2001;3:183–191. doi: 10.1038/35055095. [DOI] [PubMed] [Google Scholar]
- van der Laan M., Chacinska A., Lind M., Perschil I., Sickmann A., Meyer H. E., Guiard B., Meisinger C., Pfanner N., Rehling P. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 2005;25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Wiedemann N., Mick D. U., Guiard B., Rehling P., Pfanner N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Esaki M., Kanamori T., Tamura Y., Nishikawa S.-i., Endo T. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]