Abstract
Secreted modular calcium-binding protein-2 (SMOC-2) is a recently-identified SPARC-related protein of unknown function. In mRNA profiling experiments we, found that SMOC-2 expression was elevated in quiescent (G0) mouse fibroblasts and repressed after mitogenic stimulation with serum. The G0-specific expression of SMOC-2 was similar to that of platelet-derived growth factor-β receptor (PDGFβR), a major mitogenic receptor. Therefore, we tested a possible role for SMOC-2 in growth factor-induced cell cycle progression. SMOC-2 overexpression augmented DNA synthesis induced by serum and fibroblast mitogens (including PDGF-BB and basic fibroblast growth factor). Conversely, SMOC-2 ablation by using small interfering RNA attenuated DNA synthesis in response to PDGF-BB and other growth factors. Mitogen-induced expression of cyclin D1 was attenuated in SMOC-2–ablated cells, and cyclin D1-overexpressing cells were resistant to inhibition of mitogenesis after SMOC-2 ablation. Therefore, cyclin D1 is limiting for G1 progression in SMOC-2–deficient cells. SMOC-2 ablation did not inhibit PDGF-induced PDGFβR autophosphorylation or PDGF-BB–dependent activation of mitogen-activated protein kinase and Akt kinases, suggesting that SMOC-2 is dispensable for growth factor receptor activation. However, integrin-linked kinase (ILK) activity was reduced in SMOC-2–ablated cells. Ectopic expression of hyperactive ILK corrected the defective mitogenic response of SMOC-2–deficient cells. Therefore, SMOC-2 contributes to cell cycle progression by maintaining ILK activity during G1. These results identify a novel role for SMOC-2 in cell cycle control.
INTRODUCTION
The mammalian cell cycle comprises successive phases of DNA synthesis termed S phase and mitosis (M phase) that are separated by intervening Gap phases (G1 and G2). Many cells can exit the cell cycle to enter a state reversible growth arrest termed quiescence (G0) (Pardee, 1989). Most cultured cells are grown in medium supplemented with exogenous growth factors supplied in the form of 10% serum. In many cell lines, serum-deprivation of confluent cultures (typically a reduction to 0.5% serum) results in quiescence. Treatment of quiescent cells with serum stimulates synchronous entry of cells into G1 and promotes a round of cell cycle progression. Certain purified growth factors present in serum, such as platelet-derived growth factor (PDGF) can promote G1 entry and DNA synthesis and may account for much of the mitogenic activity of serum. Cells become independent of the requirement for growth factors at a point termed the R point that occurs ∼1 h before the onset of S phase. After entry into S phase, cells are committed to completion of the cell cycle in the absence of additional growth factor stimuli (Pardee, 1989).
Growth factors (GFs) such as PDGF act on specific cell surface receptor tyrosine kinases (RTKs) that are autophosphorylated as a result of ligand binding. RTK autophosphorylation results in recruitment and activation of various signaling proteins (including phosphatidylinositol 3-kinase, Raf, GTPase activation protein, phospholipase Cγ). The ensuing second messenger production and protein kinase cascades trigger transcriptional activation of genes that are termed immediate early (IE), delayed, or late depending on their kinetics of activation by active RTKs (Cochran et al., 1983; Cochran et al., 1984; Lau and Nathans, 1985, 1987; Dean et al., 1986; Almendral et al., 1988; Bravo et al., 1988; Ryseck et al., 1988; reviewed in Nathans et al., 1988; Herschman, 1991). The products of GF-induced genes enable cells to overcome restriction points that are otherwise imposed upon cell cycle progression (Pardee, 1989). Cyclin D1 is a target of many mitogenic signaling events and its mitogen-induced expression is crucial for G1 progression (Sherr, 1993, 1994, 1995). When expressed, cyclin D1 contributes to activation of cyclin-dependent kinases (CDKs) 4 and 6, which phosphorylate and inactivate the retinoblastoma protein (Rb) tumor suppressor protein (Sherr, 1993, 1994, 1995). Rb inactivation results in derepression of genes required for the G1/S transition (Nevins et al., 1997; Nevins, 1998).
It is becoming increasingly apparent that growth factor receptor-induced pathways cooperate with integrin signaling events to control cell proliferation (Assoian and Schwartz, 2001; Schwartz and Assoian, 2001; Giancotti and Tarone, 2003). Integrins are heterodimeric transmembrane receptors made up of α and β subunits that mediate interactions between cells and the extracellular matrix (ECM) (Arnaout et al., 2005). Ligation of integrin αβ heterodimers by components of the ECM enables transduction of extracellular signals to the cell interior. Similar to growth factor-activated RTKs, integrins interact with intracellular proteins that regulate the cell cycle. Effectors of integrin signaling that contribute to the mitogenic response include integrin-linked kinase (ILK) and focal adhesion kinase. Many RTK and integrin-induced signals contribute to cell cycle progression by stimulating cyclin D1 expression via transcriptional (Assoian and Schwartz, 2001) and posttranscriptional (Huang et al., 1998) mechanisms.
Clearly, both RTK and integrin-mediated signaling pathways are influenced by the extracellular environment. Potentially, the ECM can regulate proliferation indirectly by controlling availability or presentation of RTK ligands (such as PDGF) to cells. Additionally, direct interactions of ECM components with cell surface RTKs or integrins may modulate mitogenic signal transduction events. Matricellular proteins are secreted factors that mediate interactions between cells and the ECM (Bornstein and Sage, 2002). In general matricellular factors are hypothesized to act as extracellular modulators of signaling events between the ECM, adhesion molecules, growth factors, and growth factor receptors (Bornstein and Sage, 2002). Thus, matricellular proteins have been implicated in regulation of cell–matrix interactions, cell adhesion, spreading, migration, wound repair, and angiogenesis during development and in response to injury (Brekken and Sage, 2000, 2001; Bornstein and Sage, 2002). Secreted protein acidic and rich in cysteine (SPARC), also termed BM40 and osteonectin, is one of the best-studied matricellular proteins (Brekken and Sage, 2000). SPARC binds directly to several growth factors, including PDGF (Motamed et al., 2002), vascular endothelial growth factor (VEGF) (Kupprion et al., 1998; Francki et al., 1999), fibroblast growth factor (FGF)2 (Motamed et al., 2003), transforming growth factor (TGF) β (Francki et al., 2004), and insulin-like growth factor-1 (Francki et al., 2003) to negatively regulate receptor-mediated signaling by these ligands. SPARC also associates in a complex with ILK and modifies ILK-mediated signaling (Barker et al., 2005). SPARC-null cells have altered proliferation (Bradshaw et al., 1999), and SPARC levels are often elevated in cancer (Paley et al., 2000). Therefore, SPARC likely plays important roles in cell growth control during normal development and disease (Framson and Sage, 2004). However, putative mechanisms by which most matricellular factors might regulate mitogenic events initiated at the cell surface have not been elucidated.
This study is concerned with a putative matricellular factor termed secreted modular calcium-binding protein-2 (SMOC-2), whose precise biological roles are unclear, but influences angiogenic factor signaling (Rocnik et al., 2006). SMOC-2 (Vannahme et al., 2002) and a related protein termed SMOC-1, also of unknown function (Vannahme et al., 2003), share considerable sequence similarity with SPARC, possibly suggesting similar biological roles for SMOC-1, SMOC-2, and SPARC in regulation of growth factor signaling and mitogenesis. In RNA profiling experiments described here, we found that SMOC-2 mRNA levels are elevated in G0 mouse fibroblasts and subsequently down-regulated concomitant with serum-induced reentry into the cell cycle. The quiescence-specific pattern of SMOC-2 expression is similar to that of PDGFβR (Vaziri and Faller, 1995), a major mitogen receptor in fibroblasts. Therefore, we have investigated a role for SMOC-2 in growth control.
MATERIALS AND METHODS
Chemicals and Antibodies
Anti-Myc rabbit polyclonal antibody (2272) and anti-phospho-Akt (Ser 473) polyclonal antibody (9271) were purchased from Cell Signaling Technology (Danvers, MA); Anti-cyclin D1 monoclonal antibody (sc-8396) and anti-cyclin A polyclonal antibody (sc-751) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibodies against fibronectin (610077), ILK (611802), and Rb (554136) were purchased from BD Biosciences Transduction Laboratories (Lexington, KY). A rabbit polyclonal anti-SMOC-2 antibody was raised against a synthetic peptide (YPTLWTEQVKSRQNK) corresponding to residues 193–207 of murine and human SMOC-2. Peptide synthesis, immunizations, and affinity purification of antibody were performed by Genemed Synthesis (San Francisco, CA). The SMOC-1 antibody was described previously by Vannahme et al. (2002).
Cells and Culture
Swiss 3T3 cells and Rat1 fibroblasts were obtained from American Type Culture Collection (Manassas, VA). Cells were grown in DMEM (supplemented with penicillin and streptomycin) containing 10% heat-inactivated fetal bovine serum. MCF10A normal human breast epithelial cells, MCF10CA1h human breast carcinoma cells (low grade, well differentiated), and MCF10CA1d (undifferentiated, metastatic to the lung) were kindly provided by Dr. Thiagalingam (Boston University School of Medicine). MCF10A cells were grown in DMEM/F-12 (1:1) containing 10 mM HEPES, 0.029 M NaHCO3, 10 μg/ml insulin, 20 ng/ml epidermal growth factor (EGF), 0.5 μg/ml hydrocortisone, and 5% horse serum (Invitrogen, Carlsbad, CA). MCF10CA1h and MCF10CA1d cells were grown in DMEM/F-12 (1:1) containing 10 mM HEPES, 0.029 M NaHCO3, and 5% horse serum. For synchronization studies, cells were grown to 90% confluence, and then they were placed in medium containing 0.5% serum for 24 h to induce quiescence. Quiescent cells were then stimulated with 10% serum or purified factors for various times before harvest.
Adenovirus and Infection
cDNAs encoding green fluorescent protein (GFP), SMOC-2, and Myc-SMOC-2 were subcloned into the shuttle vector pAC-cytomegalovirus (CMV). The resulting shuttle vectors were cotransfected into 293T cells with the pJM17 plasmid to generate recombinant adenovirus as described previously (Guo et al., 2002). Adonovirus vectors were by centrifugation on CsCl gradients and gel filtration chromatography as described previously (Guo et al., 2002).
Microarray Analysis
Swiss 3T3 cells were synchronized by serum-deprivation and restimulation as described above. RNA samples isolated using the single step method of Chomczynski and Sacchi (1987), and they were submitted to Genome Systems (www.genomesystems.com; St. Louis, MO) for labeling and hybridization to DNA chips containing 10,000 arrayed mouse expressed sequence tags (ESTs). Genome Systems provided a list of transcripts corresponding to arrayed EST clones that were differentially expressed between the samples. EST clones corresponding to some of the differentially expressed RNAs were purchased from Genome Systems and used as probes in RNA blotting experiments as described below.
RNA Blot Analysis
For each sample, 20 μg of total RNA was electrophoresed on a 1% agarose gel in the presence of formaldehyde. After separation, RNA was transferred to nitrocellulose filters using 20X standard saline citrate (SSC) (1X SSC is 0.15 M NaCl and 0.015 M sodium citrate). RNA was UV cross-linked to the filters using a Stratalinker device (Stratagene, La Jolla, CA). Filters were hybridized with random-primed 32P-labeled cDNA probes in 50% formamide-5X SSC-1% SDS-5X Denhardt's solution-100 μg of salmon sperm DNA per milliliter. Washes were carried out at high stringency, and autoradiography was performed at −70°C with enhancing screens.
Isolation of the SMOC-2 cDNA
I.M.A.G.E. EST clone ID #482198 was selected for further analysis based on the interesting pattern of expression of the corresponding mRNA. EST 482198 contains a partial 3′ untranslated region (UTR), but it lacks an open reading frame. To obtain a full-length cDNA corresponding to EST 482198, we generated a cDNA library from Swiss 3T3 cells in the pSPORT1 vector (Stratagene). The resulting cDNA library was used as a template in a polymerase chain reaction (PCR) using a forward primer corresponding to the pSPORT1 vector (5′-TAATACGACTCACTATAGGGAAAGCTGGTACG-3′) and a reverse primer corresponding to the 3′ UTR of EST 482198 and containing a BamHI restriction site (5′-CGGGATCCAATATCTCATTTCTACAAAAGTGAATT-3′). Products of the PCR reaction were analyzed by electrophoresis on agarose gels. The largest PCR product obtained (1878 base pairs) was gel purified, digested with EcoRI and BamHI, and ligated into pcDNA3.1. The insert was sequenced, and it was found to contain an open reading frame encoding a putative protein of 420 amino acids. The same predicted protein was later identified independently by Hartmann and colleagues (Vannahme et al., 2003) and designated SMOC-2 based on its sequence similarity to SMOC-1 (Vannahme et al., 2002).
DNA Synthesis Assay
DNA synthesis was measured by [3H]thymidine incorporation assays as described previously (Vaziri and Faller, 1995). In brief, cells were seeded in 12-well culture plates. To measure rates of DNA synthesis, 1μCi/ml [3H]thymidine (PerkinElmer Life and Analytical Sciences, Boston, MA) was added to each well for 1 h. The resulting cells were fixed with 2 ml of 5% trichloroacetic acid (TCA). The fixed monolayers were washed three times with TCA to remove unincorporated [3H]thymidine. The plates were dried overnight, and then 1 ml of 0.3 M NaOH was added to each well to solubilize the fixed nuclei. Three hundred microliters of NaOH-solubilized material from each well was transferred to a scintillation vial and neutralized by adding 100 μl of acetic acid. Three milliliters of Ecoscint scintillation fluid was added to the neutralized mixture, and radioactivity was measured by scintillation counting.
Reverse Transcriptase (RT)-PCR Analysis of SMOC-2 and Cyclin D1 mRNA
Total cellular RNA was extracted from 3T3 cells by using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. The concentration and purity of the RNA were determined by measurements of absorbance at 260 and 280 nm by using a spectrophotometer. One microgram of RNA was used for each RT-PCR reaction. RT-PCR was performed using the ThermoScript RT-PCR system (Invitrogen) according to the manufacturer's instructions. The following primers were used: 5′-CAGGTCCAGTGTCACAGCTACAC-3′(mouse SMOC-2 forward), 5′-GGTCTTGTTCTGCCGACTCTTAAC-3′ (mouse SMOC-2 reverse), 5′-GACTCCCCACGATTTCATCGAACAC-3′(mouse cyclinD1 forward), 5′-GAAGGGCTTCAATCTGTTCCTGGCAG-3′(mouse cyclinD1 reverse), 5′-GGCTACAGCTTCACCACCACAGC-3′(mouse β-actin forward), and 5′-CCACAGGATTCCATACCCAAGAAGG-3′ (mouse β-actin reverse). The amplified products were separated on 1.0% agarose gels and visualized under an UV transilluminator.
Immunofluorescence Microscopy
Rat1 cells grown on chamber slides were fixed with 4% paraformaldehyde solution for 10 min, and then they were permeabilized with 0.2% Triton X-100 for 10 min. Slides were blocked in 5% bovine serum albumin (BSA) for 1 h, followed by overnight incubation at 4°C with primary antibodies. After extensive washing with phosphate-buffered saline (PBS), cells were incubated with fluorescein isothiocyanate (FITC)- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). After washing, the slides were 4,6-diamidino-2-phenylindole (DAPI) stained and mounted with VECTASHIELD solution (Vector Laboratories, Burlingame, CA). Slides were imaged using a Nikon Eclipse E800 fluorescent microscope. For F-actin staining, Rat1 cells were grown on chamber slides, rinsed twice with PBS, and fixed with 3.7% formaldehyde solution in PBS. The fixed cells were washed twice with PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 min. The permeabilized cells were incubated with 1% BSA in PBS for 30 min at room temperature. After blocking with BSA, the cells were labeled with 5 U/ml Alexa Fluor 488-labeled phalloidin (Invitrogen) in PBS containing 1% BSA for 20 min at room temperature. The labeled cells were washed three times with PBS, and then they were mounted with VECTASHIELD solution and imaged as described for immunofluorescence staining.
Preparation of Whole Cell Extracts and Immunoblotting
Plates of cells were rinsed three times with PBS to remove the growth medium. Washed monolayers were lysed in 0.5 ml of ice-cold lysis buffer containing 50 mM HEPES, pH 7.4, 0.1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 80 mM β-glycerophosphate, and protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The lysates were transferred to microcentrifuge tubes and centrifuged at 10,000 × g for 5 min to remove nuclei and other insoluble material. The cleared extracts were normalized for protein content, boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, resolved by electrophoresis on 10% acrylamide gels, and transferred to nitrocellulose. Membranes were probed with various antibodies and signals were detected by chemiluminescence (PerkinElmer Life and Analytical Sciences).
Isolation of Sodium Deoxycholate-insoluble ECM
Exponentially growing cells in 10-cm culture dishes (for immunoblotting) or chamber slides (for immunofluorescence staining) were rinsed twice with PBS, followed by three 5-min incubations with sodium deoxycholate (DOC)-EDTA buffer (0.1% DOC, 1 mM EDTA, and 10 mM Tris-Cl, pH 8.0) at 4°C. For immunofluorescence staining, the DOC-resistant material remaining on the chamber slides was fixed with ice-cold methanol for 10 min, rinsed with PBS, and then probed with primary and secondary antibodies as described above. For immunoblotting, DOC-resistant extracellular matrix fractions were lysed with ice-cold lysis buffer and analyzed exactly as described for whole cell extracts above.
ILK Protein Kinase Assays
ILK activity assays were performed exactly as described by Delcommenne et al. (1998). In brief, cells were lysed in kinase lysis buffer containing 50 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% NP-40, 1 mM sodium-orthovanadate, and 1 mM β-glycerophosphate, supplemented with protease inhibitor cocktail (Roche Applied Science). Lysates were clarified by centrifugation at 10,000 × g for 10 min. Soluble supernatants were normalized for protein content, and they were precleared for 1 h at 4°C with protein A/G-Sepharose beads (Santa Cruz Biotechnology). Precleared supernatants containing ∼500 μg of protein were incubated for 2 h at 4°C with 1 μg of anti-ILK antibody (BD Biosciences, San Jose, CA) or with a nonspecific immunoglobulin G (for negative controls) on a rotating platform. Twenty-five microliters of packed protein A/G-Sepharose beads was added to each tube, and the incubations were continued for another hour. Immune complexes were recovered by centrifugation at 10,000 × g for 10 s. The resulting beads were washed twice with kinase lysis buffer and then twice with kinase wash buffer containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 0.5% NP-40. After the final wash, beads were suspended in kinase assay buffer containing 10 μg of myelin basic protein (MBP), 1 mM unlabeled ATP, 10 μCi of [γ-32P]ATP, and 1 mM DTT. Kinase reactions were incubated for 30 min at 30°C. Reactions were terminated by the addition of SDS-PAGE reducing buffer. Phosphorylated proteins were electrophoresed on 12% SDS-PAGE gels. The resulting gels were fixed and stained with Coomassie Blue. The gels were dried, and phosphorylated proteins were visualized by autoradiography.
Fluorescence-activated Cell Sorting (FACS) Analysis
After various treatments, cells were trypsinized, recovered by centrifugation (5000 × g; 1 min), and fixed with 35% ethanol in PBS overnight. The fixed cells were recovered by centrifugation (1000 × g; 5 min), rinsed with PBS, and then resuspended in 1 ml of PBS containing 8 μg of RNAse A and 50 μg of propidium iodide (PI). The nuclear suspensions were incubated in the dark at room temperature for 30 min before analysis by flow cytometry. FACS analysis was performed on a BD Biosciences flow cytometer with CellQuest software.
Ablation of SMOC-2 and ILK Expression by Using Small Interfering RNA (siRNA)
3T3 cells were grown to 50% confluence and then transfected with siRNA duplexes against SMOC-2 (siSMOC-2), ILK (siGENOME SMARTpool M-040115-00-0020) or with control (siCy3, Cy3-Luciferase-GL2 Duplex) RNA duplex oligonucleotides (Dharmacon RNA Technologies, Lafayette, CO) by using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. The SMOC-2 siRNA target sequence used is TTAAGAGGTTCCTGCGAAA.
Whole Mount In Situ Hybridization
We assessed distribution of SMOC-2 mRNA in embryonic day (E)12–13 mouse embryos as described by Malpel et al. (2000). Briefly, digoxigenin (DIG)-labeled riboprobes (sense and antisense) (MaxiScript kit; Ambion, Austin, TX) were generated from a plasmid carrying a mouse SMOC-2 cDNA. Embryos were rehydrated, digested with proteinase K, prehybridized for 1 h at 70°C in solution containing 50% formamide, 5X SSC buffer, 1% SDS, 50 mg/ml yeast RNA, and heparin, followed by overnight hybridization with DIG-labeled RNA probes. Samples were then incubated with anti-DIG alkaline phosphatase conjugate overnight at 4°C. Signal was visualized with BM Purple substrate solution (Roche Applied Science), as described in the manufacturer's protocol.
Reproducibility
All results are representative of experiments that were repeated at least three times with similar results on each occasion. Statistical analyses were performed using a paired t test.
RESULTS
Identification of SMOC-2 as a Mitogen-regulated mRNA
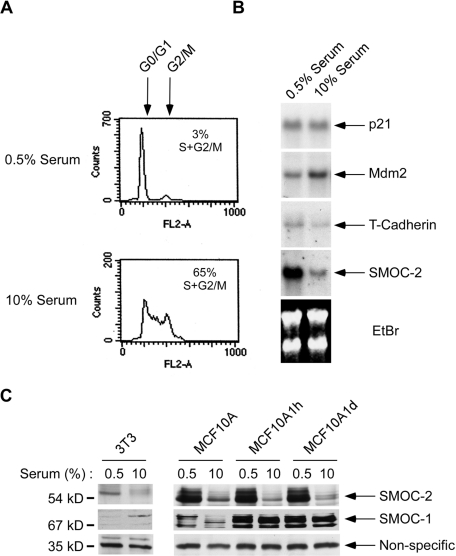
During microarray profiling experiments, we noticed that the mRNA corresponding to I.M.A.G.E. EST clone ID #482198 was abundant in quiescent Swiss 3T3 cells, but it was repressed after serum stimulation (unpublished observations). As described in Materials and Methods, we identified clone 482198 as SMOC-2. Our Northern blot analyses confirmed that SMOC-2 mRNA levels were elevated during quiescence and then repressed in response to serum concomitant with G1/S phase progression (Figure 1, A and B). The effects of serum on SMOC-2 mRNA levels were relatively specific as shown by RNA blot analysis of other mRNAs, including p21 and Mdm2 (Figure 1B). Therefore, SMOC-2 mRNA levels are specifically increased during quiescence.
Figure 1.
Identification of SMOC-2 as a serum-repressed mRNA. Replicate plates of serum-starved cultures of Swiss 3T3 cells were treated with 10% serum or were left untreated for controls. After 17 h, cells were analyzed for cell cycle distribution by flow cytometry (A). Alternatively, RNA was extracted from the cells, separated on agarose gels, and analyzed by Northern blotting with cDNA probes for p21, Mdm2, T-cadherin, and SMOC-2 (B). (C) Immunoblot analysis of SMOC-2 and SMOC-1 levels in Swiss 3T3, MCF10A, MCF10A1h, and MCF10A1d cells after culture in 0.5 or 10% serum. A nonspecific band recognized by the secondary antibody is shown as a loading control.
We generated anti-SMOC-2 polyclonal antibodies by using an immunogenic peptide corresponding to a conserved region of mouse and human SMOC-2. Immunoblotting experiments with anti-SMOC-2 antibodies showed that SMOC-2 protein levels were elevated after serum starvation in Swiss 3T3 fibroblasts and in several other cells tested, including MCF10A, MCF10CA1h, and MCF10CA1d (Figure 1C). Therefore, the serum-dependent changes in SMOC-2 mRNA levels correlated with expression of SMOC-2 protein. For comparison, we also performed immunoblot analysis of the SMOC-2–related protein SMOC-1 in lysates from quiescent and serum-stimulated Swiss 3T3 cells. In contrast to SMOC-2, expression of SMOC-1 was not repressed by serum. Note that the diffuse nature of SMOC-1 and SMOC-2 bands on immunoblots has been observed by previous workers and seems to represent extensive glycosylation of these factors (Vannahme et al., 2002, 2003). Together, the results of Figure 1 show that SMOC-2 expression is induced during quiescence and down-regulated in response to serum stimulation concomitant with cell cycle reentry.
SMOC-2 Is Expressed in Distinct Developing Structures of Mouse Embryos
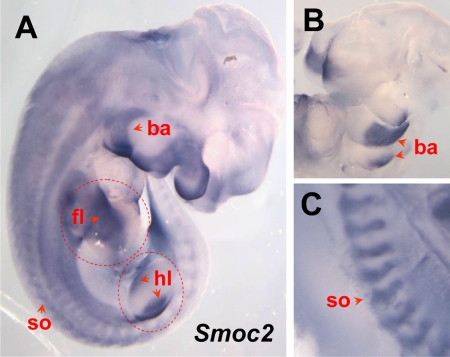
To investigate the potential role of Smoc2 in development, we performed a preliminary analysis of its expression pattern in midgestation mouse embryos. Whole mount in situ hybridization at E12.5–E13 revealed a well-defined distribution of SMOC-2 transcripts in structures, such as the developing limb, branchial arches, and somites (Figure 2). In both forelimbs and hindlimbs, transcripts were restricted to proximal regions, and they were clearly excluded from distal sites, including the apical ectodermal ridge (Figure 2A). Signals were also present in a number of internal organs, such as the lung, stomach, and reproductive tract (data not shown). A comprehensive analysis of the Smoc2 expression pattern pre- and postnatally is underway and will be reported elsewhere. Nevertheless, the results of Figure 2 demonstrate a broad pattern of Smoc2 expression, and they suggest that SMOC-2-dependent events might serve important roles in multiple tissues during development. Furthermore, mouse embryo-derived cells such as the fibroblasts used in our study are widely used in the fields of growth factor and integrin signaling. Therefore, it is likely that the role we have discovered for SMOC-2 in mitogenic signaling and cell cycle control (described below) is relevant to cell culture model systems studied commonly by other laboratories.
Figure 2.
Smoc2 is expressed in distinct structures of the developing mouse embryo. (A–C) Whole mount in situ hybridization of Smoc2 in the mouse embryo at E12.5–E13, showing strong signals in forelimb (fl) and hindlimb (hl), somites (so), and branchial arches (ba).
Subcellular Distribution of SMOC-2
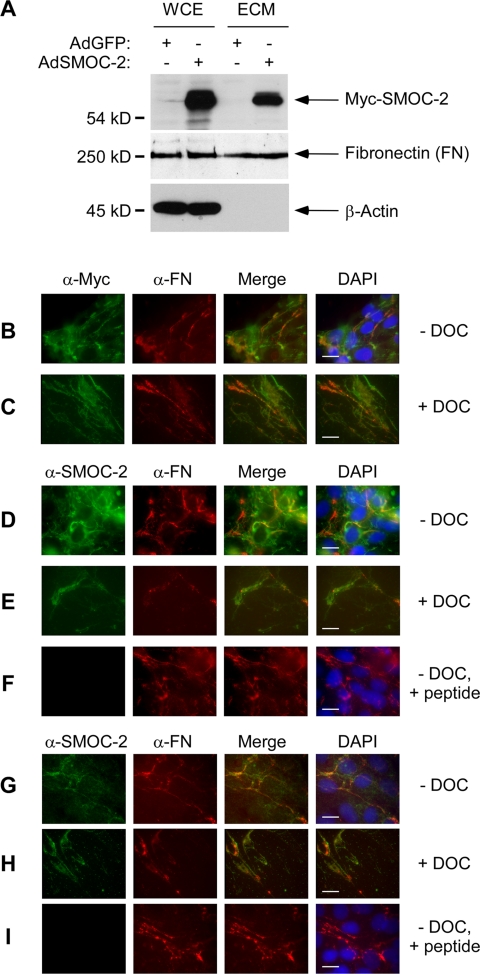
To gain insight into possible biological roles of SMOC-2, we investigated its subcellular distribution. We constructed a recombinant adenovirus vector for ectopic expression of Myc-tagged SMOC-2 (designated AdSMOC-2). We infected Rat1 fibroblasts with AdSMOC-2 (or with a GFP control adenovirus vector designated AdGFP). Extracts from AdSMOC-2– and AdGFP-infected cells were analyzed for expression of epitope-tagged SMOC-2 by immunoblotting. As shown in Figure 3A, we detected a 54-kDa protein (corresponding to the expected molecular weight for Myc-tagged SMOC-2) in lysates from AdSMOC-2–infected (but not AdGFP-infected) cells. Because SMOC-2 contains a putative N-terminal signal peptide and it has sequence similarity to matricellular proteins (Vannahme et al., 2003), we asked whether SMOC-2 was present in the ECM. We performed DOC fractionations (Midwood and Schwarzbauer, 2002) to isolate ECM components from AdGFP- and AdSMOC-2–infected cells. ECM-containing fractions from AdGFP- and AdSMOC-2–infected cultures were analyzed for SMOC-2 expression. As shown in Figure 3A, we detected Myc-SMOC-2 in ECM preparations from AdSMOC-2–infected (but not control) cells.
Figure 3.
Subcellular distribution of SMOC-2. (A) Whole cell lysates and ECM preparations from AdCon- and Ad-SMOC-2–infected Rat1 cells were resolved by SDS-PAGE, transferred to nitrocellulose and probed with an anti-Myc antibody. The same samples were probed with antibodies against FN and β-actin. AdSMOC-2–infected (B–F) or uninfected (G–I) Rat1 cells were fixed and stained with anti-Myc and anti-fibronectin (B and C) or anti-SMOC-2 and anti-fibronectin (D–I) antibodies. Bound primary antibodies were detected using a FITC-conjugated antibody (for detecting anti-Myc and anti-SMOC-2) and a Cy3-conjugated antibody (for detecting anti-fibronectin). In some experiments, cells were extracted with DOC buffer before fixing and antibody staining (C, E, and H). Some antibody incubations with were carried out in the presence of a competitor peptide corresponding to the epitope recognized by the SMOC-2 antisera (F and I). Bars, 10 μm.
We also performed immunofluorescence microscopy to determine the distribution of ectopically expressed SMOC-2 in Rat1 cells. AdCon (“empty” adenovirus control vector) or AdSMOC-2–infected cells were fixed and stained directly with anti-Myc antibodies as described under Materials and Methods. Alternatively, the cells were extracted with DOC, and the resulting detergent-insoluble remnant ECM was fixed and analyzed for SMOC-2 expression by immunofluorescence microscopy.
In AdSMOC-2–infected cells probed with anti-Myc, we observed a broad expression pattern of SMOC-2 with a slight increase in anti-Myc staining intensity at the cellular periphery (Figure 3B). For comparison, we also probed the cells with an antibody against fibronectin (FN, a known ECM component). As shown in Figure 3B, there was some similarity between the distribution patterns of fibronectin and SMOC-2. Interestingly, when we analyzed the DOC-insoluble ECM preparations by immunofluorescence microscopy, SMOC-2 was present (Figure 3C). As expected, the ECM preparations retained fibronectin, but they were DAPI negative and β-actin free (indicating efficient extraction of the nuclei and non-ECM compartments).
To complement the immunofluorescence microscopy experiments with the anti-Myc antibody, we performed similar studies using a polyclonal antibody raised against a peptide corresponding to an internal domain of SMOC-2 (see Materials and Methods). As shown in Figure 3, we observed similar patterns of staining of ectopically expressed Myc-SMOC-2 with anti-SMOC-2 and anti-Myc antibodies (compare Figure 3, B and C, with D and E). As expected, the staining of SMOC-2 was inhibited when the antibody incubations were performed in the presence of competitor SMOC-2 antigenic peptide. Fibronectin staining was unaffected by the SMOC-2 competitor peptide. The peptide competition experiments demonstrate specificity of the anti-SMOC-2 antibody.
Because the SMOC-2 localization experiments in Figure 3, B–F, were performed with ectopically expressed SMOC-2, it was of interest to determine the subcellular distribution of endogenous SMOC-2. Therefore, uninfected Rat1 cells were also probed with antibodies against SMOC-2 and fibronectin. The distribution of endogenous SMOC-2 was similar to that of ectopically overexpressed Myc-SMOC-2 in fixed cells and DOC-extracted ECM (Figure 3, G and H). As expected, detection of endogenous SMOC-2 (but not of fibronectin) was prevented when we performed antibody staining in the presence of competitor SMOC-2 peptide (Figure 3I). Together, our data show that ectopically expressed and endogenous SMOC-2 are present in the ECM. The ECM localization of SMOC-2 is similar to the localization reported for other matricellular proteins such as SMOC-1 and SPARC (Brekken and Sage, 2000; Vannahme et al., 2002). However, our observation that SMOC-2 assembles fibrils that partly colocalize with fibronectin is not a typical matricellular characteristic, and it may suggest roles for SMOC-2 other than those reported for other matricellular factors.
Mitogenic Role of SMOC-2
Because of the similar growth arrest-specific expression patterns of SMOC-2 and other cell cycle regulators such as PDGFβR (Vaziri and Faller, 1995), an important regulator of G0-S phase transition (Pardee, 1989), we hypothesized that SMOC-2 might play a role in growth control. Furthermore, we previously showed that SMOC-2 overexpression promotes VEGF-induced angiogenic responses in endothelial cells (Rocnik et al., 2006), also consistent with a role for SMOC-2 in cell growth. Therefore, in experiments described below, we investigated a possible role for SMOC-2 in mitogenic signaling and cell cycle control.
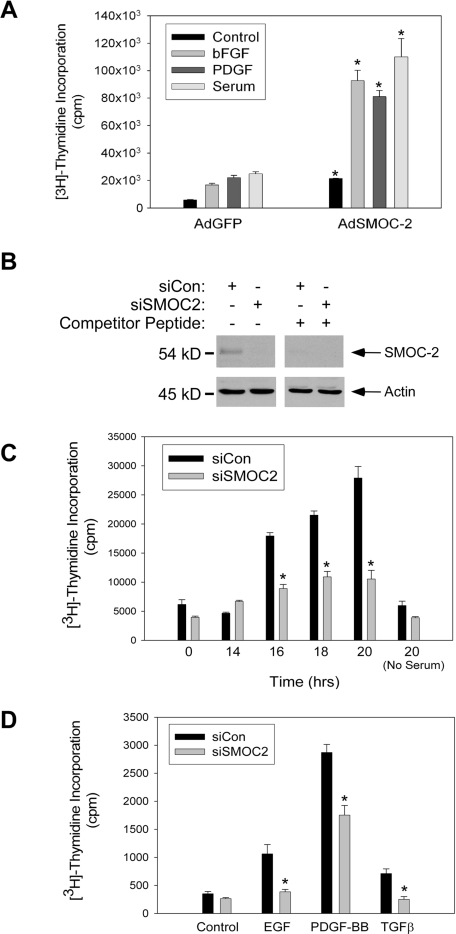
First, we determined the effect of SMOC-2 overexpression on DNA synthesis responses to serum, basic (b)FGF, and PDGF-BB. As shown in Figure 4A, cells overexpressing SMOC-2 showed high basal levels of DNA synthesis relative to control cultures. Moreover, DNA synthesis in response to serum, bFGF, and PDGF were increased in SMOC-2–overexpressing cells (Figure 4A). Both the -fold induction of DNA synthesis, and absolute levels of DNA synthesis attained in response to mitogens were increased as a consequence of SMOC-2 overexpression.
Figure 4.
Role of SMOC-2 in growth factor-induced DNA synthesis. (A) Quiescent Rat1 cells were infected with AdGFP or AdSMOC-2 for 24 h. The resulting cultures were stimulated with serum (10%), bFGF (3 ng/ml), or PDGF-BB (10 ng/ml) for 20 h. Rates of DNA synthesis in the resulting cultures were determined using measurements of [3H]thymidine incorporation. Data points represent the mean for two samples, with error bars representing the range. *p < 0.005 compared with AdGFP-infected cells. (B) Quiescent Swiss 3T3 cells were transfected with siRNA duplexes targeting SMOC-2 (siSMOC-2) or with control Cy3 oligonucleotide duplexes (siCon). Duplicate samples of extracts were from the resulting cells were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. One half of the nitrocellulose membrane was probed sequentially with anti-SMOC-2 and anti-β-actin (left). The other half of the membrane was processed similarly except that primary antibody incubations were performed in the presence of 100 μg/ml SMOC-2 competitor peptide (right). (C) Swiss 3T3 cells were transfected with siSMOC-2 or siCon oligonucleotide duplexes. The resulting cells were made quiescent by serum deprivation. Quiescent siSMOC-2 or Cy3-transfected cells were stimulated to enter the cell cycle by the addition of 10% serum. At different time points after serum stimulation, rates of DNA synthesis were determined using measurements of [3H]thymidine incorporation. Data points represent the mean for two samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells. (D) Swiss 3T3 cells were transfected with siSMOC-2 or control Cy3 oligonucleotide duplexes and made quiescent by serum deprivation as described for C. Quiescent cultures were treated with EGF (10 ng/ml), PDGF-BB (5 ng/ml), or TGFβ (5 ng/ml), or they were left untreated for controls. Twenty hours after growth factor treatment, rates of DNA synthesis were determined using measurements of [3H]thymidine incorporation. Note that in the experiment shown here, cells received 10-fold less [3H]thymidine than was used for the DNA synthesis assay in Figure 4C. Data points represent the mean for two samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells.
Because overexpressed SMOC-2 stimulated DNA synthesis (Figure 4A), we asked whether endogenous SMOC-2 plays any role in serum or growth factor-induced DNA synthesis. Therefore, we designed siRNA duplexes to ablate SMOC-2 expression. We transfected Swiss 3T3 cells with a SMOC-2-directed siRNA duplex (designated siSMOC-2) or with a control RNA oligonucleotide (designated siCon). Extracts from the resulting cells were analyzed by immunoblotting with our anti-SMOC-2 antibody. As shown in Figure 4B (left), expression levels of a 54-kDa protein (corresponding to the expected size for SMOC-2) were reduced by >90% in cells transfected with siSMOC-2 relative to controls. In a parallel immunoblotting experiment, we performed the primary antibody incubation in the presence of the antigenic peptide used to generate our anti-SMOC-2 antibody. As expected, the competitor peptide specifically prevented detection of the 54-kDa protein recognized by our anti-SMOC-2 antibody. Therefore, our siRNA strategy was effective for reducing SMOC-2 protein levels in 3T3 cells.
Having validated our siRNA protocol, we determined the effect of SMOC-2 ablation on serum-induced DNA synthesis. Figure 4C shows rates of DNA synthesis in control and siSMOC-2–transfected Swiss 3T3 cells at different time points after serum stimulation. As expected, rates of DNA synthesis in control cells increased ∼16 h after serum treatment, and they continued to rise over the next 8 h. Interestingly, in siSMOC-2–transfected cells, the levels of serum-induced DNA synthesis attained at 16, 18, and 20 h were attenuated by 54.8 ± 4.068 (p = 0.0004) (Figure 4C). Therefore, SMOC-2 is important for serum-induced entry into S phase.
Serum comprises a mixture of fibroblast mitogens that includes PDGF, EGF, insulin, and other factors (Pardee, 1989). It was of interest to determine whether SMOC-2 is necessary for DNA synthesis in response to specific factors, or whether SMOC-2 expression is a general requirement for mitogen-induced DNA synthesis. Therefore, we tested the effect of SMOC-2 ablation on induction of DNA synthesis by several known fibroblast mitogens, including PDGF-BB, EGF, and TGFβ. As expected, PDGF-BB induced a strong DNA synthesis response, EGF induced a moderate response, and the response to TGFβ was weak in control cultures (Figure 4D). Interestingly, SMOC-2 ablation attenuated DNA synthesis induced by all three growth factors (Figure 4D). We have not yet performed an exhaustive analysis of the requirement of SMOC-2 for DNA synthesis in response to known fibroblast mitogens. However, the results presented here suggest a general requirement for SMOC-2 in growth factor-induced DNA synthesis.
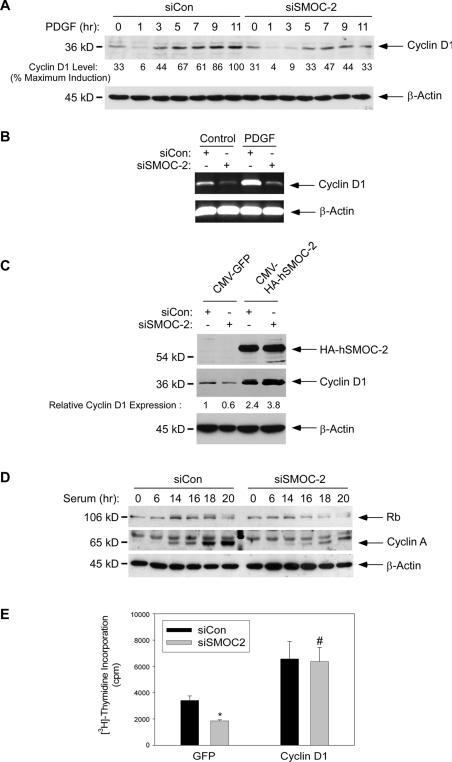
SMOC-2 Is Required for Cyclin D1 Expression
Mitogen-induced cyclin D1 expression is a key rate-limiting step for G1 progression in many cell types (Sherr, 1993), including rodent fibroblasts (Quelle et al., 1993). To investigate the role of SMOC-2 in G1 progression, we determined the effect of SMOC-2 ablation on PDGF-induced cyclin D1 expression. As shown in Figure 5A, PDGF-induced expression of cyclin D1 protein was detectable after 3–5 h, and maximal cyclin D1 levels were attained 9–11 h after PDGF treatment in control (siCon) cells. Interestingly, in SMOC-2-depleted cells (siSMOC-2), the PDGF-induced levels of cyclin D1 expression were reduced relative to controls at every time point tested (Figure 5A). The maximal level of cyclin D1 expression in SMOC-2–depleted cells was 47% of that induced by PDGF in control cultures (Figure 5A). In four different experiments (shown in Figures 5C, 7C, 8A, and 8C), PDGF-induced cyclin D1 expression was inhibited by 51 ± 5.68% (p < 0.001). Therefore, PDGF-induced DNA synthesis and cyclin D1 expression are inhibited to a similar extent as a result of SMOC-2-ablation. We also determined the effect of SMOC-2-ablation on cyclin D1 mRNA levels. As shown by the RT-PCR analyses in Figure 5B, PDGF-induced (and basal) expression levels of cyclin D1 mRNA were specifically reduced by SMOC-2 ablation.
Figure 5.
Role of SMOC-2 in mitogen-induced cyclin D1 expression. (A) Swiss 3T3 cells were transfected with siCon or siSMOC-2 oligonucleotide duplexes. The resulting cells were made quiescent, then treated with PDGF-BB (3 ng/ml) or left untreated for controls. At different time points after PDGF-BB treatment, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting by using antibodies against cyclin D1 and β-actin. (B) Swiss 3T3 cells were transfected with siCon or siSMOC-2 oligonucleotide duplexes. The resulting cells were made quiescent, then treated with PDGF-BB (3 ng/ml) for 6 h (or left untreated for controls). RNA extracted from the resulting cells was analyzed for cyclin D1 and β-actin levels by using RT-PCR as described in Materials and Methods. (C) Swiss 3T3 cells were cotransfected with siCon or siSMOC-2 together with expression vectors for hemagglutinin (HA)-hSMOC-2 or GFP. The resulting cells were made quiescent, then treated with PDGF-BB (3 ng/ml) for 9 h. Extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with anti-HA, anti-cyclin D1, and anti-β-actin antibodies as described under Materials and Methods. (D) Swiss 3T3 cells were transfected with siCon or siSMOC-2 oligonucleotide duplexes. The resulting cells were made quiescent by serum deprivation, and then they were restimulated with 10% serum. At different time points after serum treatment, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting by using antibodies against Rb, cyclin A, and β-actin. (E) Swiss 3T3 cells were cotransfected with siCon or siSMOC-2 together with expression vectors for cyclin D1 (Quelle et al., 1993) or GFP. The resulting cells were made quiescent and then treated with PDGF-BB (3 ng/ml) and [3H]thymidine (1μCi/ml) for 17 h. Rates of DNA synthesis were determined using measurements of [3H]thymidine incorporation as described under Materials and Methods. Data points represent the mean for two samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells. #p = 0.7354 and is not statistically significant compared with siCon-transfected cells.
Figure 7.
SMOC-2 is necessary for ILK activation. (A) Swiss 3T3 cells were transfected with siCon or siSMOC-2 oligonucleotide duplexes. The resulting cells were made quiescent, and then they were treated with PDGF-BB (3 ng/ml) or left untreated for controls. Cells were lysed and ILK was immunoprecipitated as described under Materials and Methods. Immune complexes were assayed for in vitro ILK kinase activity (top), or they were analyzed by immunoblotting with anti-ILK antibodies (bottom). (B) Swiss 3T3 cells were cotransfected with siCon or siSMOC-2 oligonucleotide duplexes together with an expression vector for hyperactive ILK (CMV-ILK343D) or empty vector (for control). After transfection, cells were serum-deprived for 24 h. Then, some cultures were treated with PDGF-BB (10 ng/ml) for 20 min. After PDGF treatment, the cells were lysed, ILK was immunoprecipitated, and the resulting immune complexes were assayed for MBP-directed kinase activity in vitro (as described under Materials and Methods). (C) Quiescent Swiss 3T3 cells were cotransfected with siCon or siSMOC-2 oligonucleotide duplexes and an expression vector for hyperactive ILK or empty vector for control. Cells were stimulated with PDGF-BB (10 ng/ml) to induce cell cycle reentry. After 0, 5, and 10 h of PDGF-BB treatment, cells were lysed and the resulting samples were tested for ILK, cyclin D1, and β-actin levels by immunoblotting. The lanes indicated by the dashed box show the inhibitory effect on SMOC-2 RNAi on cyclin D1 expression, which is most evident 10 h after PDGF treatment. (D) Quiescent Swiss 3T3 cells were cotransfected with siCon or siSMOC-2 oligonucleotide duplexes and an expression vector for hyperactive ILK or empty vector for control. Cells were stimulated with PDGF-BB (10 ng/ml) to induce cell cycle reentry. After 18 h of PDGF treatment, rates of DNA synthesis were determined as described under Materials and Methods. Data points represent the mean for two samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells. #p equals 0.156 and is not statistically significant compared with siCon-transfected cells.
Figure 8.
ILK is necessary for cyclin D1 induction and DNA synthesis. (A) Quiescent Swiss 3T3 cells were transfected with siCon or siILK oligonucleotide duplexes. The resulting cells were treated with PDGF-BB (10 ng/ml). At 0, 5, and 9 h after PDGF treatment, cells were lysed and extracts were analyzed for ILK, cyclin D1, and β-actin expression by SDS-PAGE and immunoblotting. For the 0- and 9-h time points, ILK was immunoprecipitated from cell extracts, and the resulting immune complexes were assayed for in vitro ILK kinase activity. (B) Quiescent 3T3 cells were cotransfected with siCon or siILK oligonucleotide duplexes. The resulting cells were stimulated for 22 h with PDGF-BB (10 ng/ml), or they were left untreated for controls. Rates of [3H]thymidine incorporation were determined as described under Materials and Methods. Data points represent the mean for three samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells. (C) Quiescent 3T3 cells were cotransfected siSMOC-2 (or siCon) oligonucleotides and a plasmid encoding dominant-negative ILK (designated ILK R211A) or a CMV-GFP control vector. Transfected cultures were treated with PDGF-BB (10 ng/ml) for 10 h, and lysates from the resulting cells were analyzed for ILK and cyclin D1 expression by SDS-PAGE and immunoblotting. (D) Triplicate cultures of quiescent 3T3 cells were cotransfected siSMOC-2 (or siCon) oligonucleotides and a plasmid encoding dominant-negative ILK (designated ILK R211A) or a CMV-GFP control vector, as described for C. The resulting cells were stimulated for 22 h with PDGF-BB (10 ng/ml), or they were left untreated for controls. Rates of [3H]thymidine incorporation were determined as described under Materials and Methods. Data points represent the mean for three samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells.
It was important to demonstrate that the effect of siSMOC-2 on cyclin D1 expression was not due to “off-target” effects of the RNA oligonucleotides. Therefore, we performed experiments to complement the defective cyclin D1 expression of siSMOC-2 cultures by using a human SMOC-2 cDNA (hSMOC-2), which differs from mouse SMOC-2 with respect to nucleotide sequence and is therefore resistant to siSMOC-2. Quiescent 3T3 cells were transfected with siCon or siSMOC-2 oligonucleotides. The resulting cells were retransfected with CMV-hSMOC-2 or CMV-GFP (for control) and treated with PDGF. After 9 h of PDGF treatment, cells were harvested and analyzed for cyclin D1 expression by immunoblotting. As expected, siSMOC-2 elicited a 40% decrease in cyclin D1 protein levels in control CMV-GFP–expressing cells (Figure 5C). Interestingly, hSMOC-2 expression induced cyclin D1 expression 2.4-fold relative to CMV-GFP–transfected controls. Moreover, the elevated cyclin D1 levels resulting from CMV-hSMOC-2 transfection were not reduced by the siSMOC-2 RNA interference (RNAi) (Figure 5C). Therefore, the effect of ablating endogenous SMOC-2 on cyclin D expression can be overcome by ectopically expressed hSMOC-2. We conclude that SMOC-2 is specifically required for efficient cyclin D1 expression during G1.
Because SMOC-2 is important for cyclin D1 expression, we predicted that SMOC-2-deficiency would also affect distal cyclin D1-CDK4/6–dependent mitogenic signaling events during G1.
Therefore, we determined the effect of SMOC-2 ablation on mitogen-induced RB and cyclin A expression. As shown in Figure 5D, the serum-induced expression of Rb and its phosphorylation (evident as an electrophoretic mobility shift) were reduced after SMOC-2 ablation. Moreover, the mitogen-induced expression of cyclin A was dramatically attenuated in SMOC-2–ablated cells. Therefore, SMOC-2 is required for growth factor-induced expression of G1 and S phase cyclins.
Our analyses of cell cycle regulators (Figure 5, A–D) indicated that SMOC-2 contributes to G1 progression by promoting expression of cyclin D1. If cyclin D1 is rate limiting for G1 progression in SMOC-2–ablated cells, we predicted that ectopic expression of cyclin D1 would complement the defective DNA synthesis resulting from SMOC-2 deficiency. Other investigators have also used cyclin D reconstitution experiments to bypass cell cycle blocks specifically associated with reduced expression of endogenous cyclin D1 (Roussel et al., 1995; Brewer et al., 1999; Agami and Bernards, 2000). Therefore, we determined the effect of SMOC-2-ablation on DNA synthesis in cyclin D1-overexpressing fibroblasts. A CMV-driven cyclin D1 expression vector (Quelle et al., 1993), or a CMV-GFP construct for control, were transfected into SMOC-2 ablated (or siRNA control) 3T3 cells. As expected, SMOC-2 ablation inhibited PDGF-induced DNA synthesis response by 46% in control (CMV-GFP–transfected) cells (Figure 5E). In contrast, PDGF-induced DNA synthesis was only inhibited by 4% in cyclin D1-overexpressing cells (Figure 5E). Therefore, cyclin D1-overexpressing cells were insensitive to SMOC-2 ablation.
Similar to any reconstitution study, a caveat of the experiment is that the high-level expression of cyclin D1 achieved by a CMV-driven vector might nonspecifically override the G1 block resulting from SMOC-2 deficiency. Therefore, our cyclin D1 reconstitution data alone do not prove that cyclin D1 deficiency is the cause of the cell cycle arrest in SMOC-2–ablated cells. However, when viewed together with our other data showing effects of reduced SMOC-2 expression on D1 induction, and the stimulation of D1 expression by ectopically expressed SMOC-2, our data are most consistent with the idea that cyclin D1 mediates mitogenic effects of SMOC-2. Together, our results suggest that a key role of SMOC-2 in growth factor-induced G1 progression is mediated via induction of cyclin D1.
SMOC-2 Is Dispensable for PDGFβR Signaling
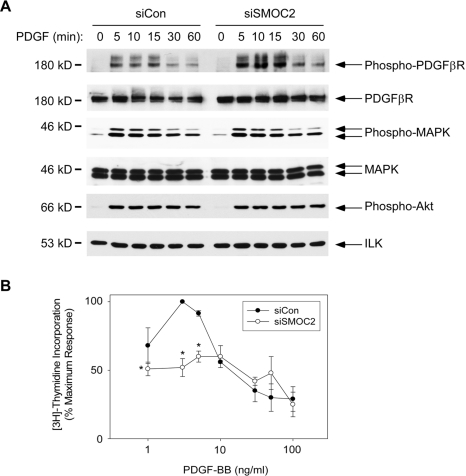
The requirement of SMOC-2 for PDGF-induced DNA synthesis suggested the possibility that SMOC-2 is necessary for efficient activation and signaling by the PDGFβR (the major PDGF receptor isoform expressed by 3T3 cells). Therefore, we determined the effect of SMOC-2-deficiency on ligand induced PDGFβR phosphorylation. As shown in Figure 6A, PDGFβR was phosphorylated with similar kinetics in response to PDGF-BB in both control and SMOC-2–ablated cultures. Moreover, SMOC-2 deficiency did not reduce the levels of PDGFβR phosphorylation induced by ligand treatment. In fact, levels of phosphorylated and unphosphorylated PDGFβR were typically slightly higher in SMOC-2-ablated cells, most likely reflecting the decrease in proliferation-induced PDGFR down-regulation and desensitization (Vaziri and Faller, 1995) resulting from SMOC-2 deficiency.
Figure 6.
SMOC-2 is dispensable for ligand-induced PDGFβR autophosphorylation and activation of MAPK and Akt. (A) Swiss 3T3 cells were transfected with siCon or siSMOC-2 oligonucleotide duplexes. The resulting cells were made quiescent by serum deprivation, and then they were stimulated with recombinant PDGF-BB (3 ng/ml) or were left untreated for controls. At different time points after PDGF-BB treatment, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting using antibodies against phospho-PDGFβR, PDGFR, phospho-MAPK, MAPK, phospho-Akt, and ILK. (B) Swiss 3T3 cells were transfected siCon or siSMOC-2 oligonucleotide duplexes. The resulting cells were made quiescent, and then they were treated with different concentrations of PDGFBB (0–100 ng/ml) and [3H]thymidine (1μCi/ml). Eighteen hours after stimulation, rates of DNA synthesis in the resulting cultures were determined using measurements of [3H]thymidine incorporation. Data points represent the mean for two samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells.
Mitogen-activated protein kinase (MAPK) and Akt are important mediators of PDGFR-induced DNA synthesis. PDGF-BB–induced activation of MAPK and Akt (as determined using phospho-specific antibodies against the active forms of these kinases) was also unaffected by SMOC-2 ablation (Figure 6A). The lack of effect of SMOC-2 deficiency on PDGF-induced MAPK and Akt activation further indicates that proximal PDGF signaling events do not require SMOC-2.
We also determined whether the inhibitory effect of SMOC-2 deficiency could be overcome by high concentrations of PDGF. As shown in Figure 6B, PDGF-BB concentrations of 0–10 ng/ml induced a dose-dependent increase in DNA synthesis in control and SMOC-2–deficient cells. PDGF-BB concentrations of >10 ng/ml were less effective at promoting DNA synthesis. The decreased DNA synthesis responses to PDGF-BB concentrations above 10 ng/ml results is expected and results from a 1:1 stoichiometry of PDGF-BB:PDGFR that precludes receptor dimerization and transphosphorylation. Nevertheless, as shown in Figure 6B, high concentrations of PDGF-BB did not overcome the reduced DNA synthesis resulting from SMOC-2 deficiency. Therefore, the reduced DNA synthesis of SMOC-2–deficient cells cannot be compensated for by increased PDGFβR signaling. Together, our data demonstrate that the defective cell cycle progression of SMOC-2–ablated cells is not due to reduced PDGFβR activation.
SMOC-2 Is Necessary for ILK Activation
Our analyses indicated that SMOC-2 is required for PDGF-induced cyclin D1 expression, yet it is dispensable for PDGFβR activation. Efficient cyclin D1 expression and G1 progression require the concerted actions of RTK (e.g., PDGFβR) and integrin-mediated signals. We considered the possibility that the cell cycle defect of SMOC-2–ablated cells results from changes in integrin signaling. Therefore, we sought to determine the effects of SMOC-2 deficiency on integrin-dependent signal transduction pathways. We investigated ILK as a candidate SMOC-2–dependent effector of integrin signaling because similar to SMOC-2, ILK is necessary for cyclin D1 expression and G1 progression (Tan et al., 2004). Moreover, Sage and colleagues recently showed that the SMOC-2–related matricellular factor SPARC is involved in ILK activation (Barker et al., 2005).
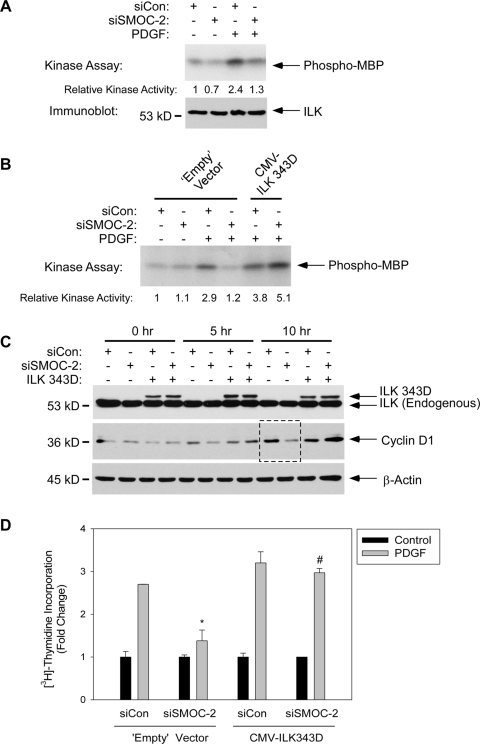
Therefore, we measured the kinase activity of ILK immunoprecipitated from control and SMOC-2–ablated cells. As shown in Figure 7A, the MBP-directed kinase activity of anti-ILK immune complexes from SMOC-2–depleted cells was reduced by ∼42% in both quiescent and PDGF-treated cultures (Figure 7A). In four separate experiments (shown in Figures 7, A and B, and 8A), ILK activity as determined by inositol phosphate kinase assays was inhibited by 50.2 ± 4.4% (n = 4, p < 0.0001). Levels of ILK protein were unaffected by SMOC-2 status or by PDGF (Figures 7A and 8A), demonstrating that the effects of SMOC-2 ablation reflected changes in the activation status of ILK, not ILK protein levels. Therefore, SMOC-2 is necessary for efficient ILK activation.
The results shown in Figure 7A suggested that the cell cycle defect of SMOC-2–deficient cells might result from reduced ILK activity. Therefore, we predicted that the defective mitogenic response of SMOC-2–ablated cells might be corrected by the hyperactive ILK S343D mutant (Cordes, 2004). We introduced a plasmid encoding hyperactive ILK (or an empty vector control plasmid) into replicate plates of siCon and siSMOC-2–transfected cells. As expected, ILK-associated kinase activity in the ILK S343D-expressing cells was constitutively high and insensitive to inhibition after SMOC-2 ablation (Figure 7B). In fact, we routinely observed slight increases in ILK 343D-dependent kinase activity after SMOC-2 ablation, although the significance of this effect is unclear. We also determined the effect of ectopically expressed ILK343D on PDGF-induced cyclin D1 expression and DNA synthesis in SMOC-2–ablated cells. Similar to the results of Figure 5, SMOC-2 depletion inhibited PDGF-induced cyclin D1 expression (Figure 7C) and DNA synthesis (Figure 7D) in control (empty vector-transected) cells. PDGF-induced cyclin D1 expression was reduced by 60% in siSMOC-2–transfected cells relative to siCon controls (Figure 7C, see region of blot indicated by the dashed line). PDGF-induced DNA synthesis was inhibited to a similar extent (48% inhibition) as a result of SMOC-2 ablation. Interestingly, ectopic expression of hyperactive ILK prevented the siSMOC-2–induced inhibition of cyclin D1 expression (Figure 7C, see last two lanes). Ectopically expressed hyperactive ILK also conferred normal levels of PDGF-induced DNA synthesis (Figure 7D) in SMOC-2–deficient cells. Together, our results suggest that SMOC-2 is necessary for ILK activation and that hyperactive ILK can correct the cell cycle defect of SMOC-2–deficient cells. These data suggest that reduced ILK activity underlies the defective G1/S progression of SMOC-2–deficient cells.
ILK Is Necessary for PDGF-induced Cyclin D1 Expression and DNA Synthesis in 3T3 Cells
The results of Figure 7 showed that SMOC-2 is required for efficient ILK activation, suggesting that defective ILK signaling is responsible for the cell cycle retardation of SMOC-2–deficient cells. It was important to test whether ILK is necessary for cyclin D1 expression and DNA synthesis in our experimental system, as has been reported for other cell types (Tan et al., 2004).
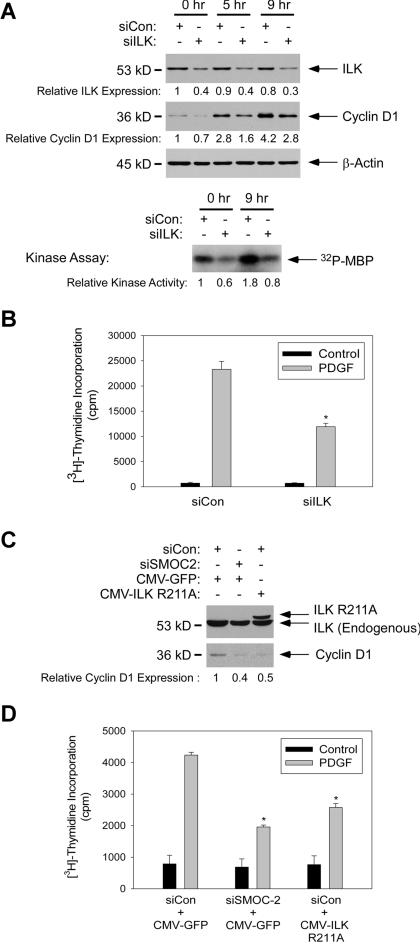
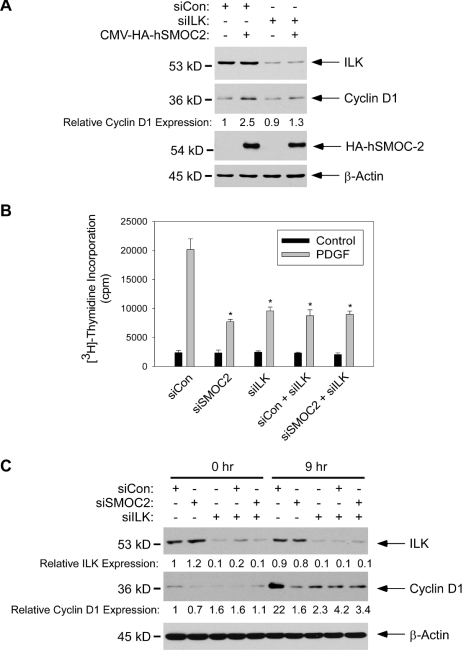
Therefore, we used ILK-directed siRNA (designated siILK) to ablate ILK expression in 3T3 cells. As shown in Figure 8A, we typically achieved ∼60% knockdown of ILK protein by using siRNA. The siRNA-induced decrease in ILK protein expression resulted in a 56% decrease in activity measured using anti-ILK IP kinase assay (Figure 8A). We determined the effect of ILK ablation on PDGF-induced cyclin D1 expression. As shown in Figure 8A, ILK ablation inhibited the PDGF-induced expression of cyclin D1 by 43 and 44% at 5- and 9-h time points after PDGF treatment, respectively. In [3H]thymidine incorporation assays conducted in parallel, PDGF-induced DNA synthesis was inhibited by 50% as a result of ILK ablation. In a complementary approach to test the role of ILK in cell cycle progression of 3T3 cells we determined the effects of ectopically expressed “dominant-negative” R211A mutant ILK (designated ILK R211A). As shown in Figure 8, C and D, transiently transfected ILK R221 inhibited PDGF-induced cyclin D1 expression and DNA synthesis by 50 and 40%, respectively, in 3T3 cells. Therefore, similar to results reported by other workers, perturbation of ILK signaling inhibits cyclin D1 expression and DNA synthesis. Importantly, the results of Figure 8 indicate that the ∼50% decrease in ILK-associated kinase activity resulting from SMOC-2 depletion (Figure 7) is sufficient to account for the inhibitory effects of SMOC-2 on cyclin D1 expression and DNA synthesis.
If SMOC-2 induces cyclin D1 via ILK, we predicted that SMOC-2–induced cyclin D1 expression should be attenuated in ILK-ablated cells. Therefore, we overexpressed SMOC-2 in control and ILK-ablated cells, and then we determined cyclin D1 expression by immunoblot analysis of extracts from the resulting cells. Consistent with the results of Figure 5C, SMOC-2 induced cyclin D1 protein levels by 2.5-fold in siCon-transfected cells (Figure 9A). However, in the ILK-ablated cells, cyclin D1 expression was only increased by 1.3-fold as a result of SMOC-2 overexpression (Figure 9A).
Figure 9.
SMOC-2 and ILK regulate a common signaling pathway leading to cyclin D1 expression. (A) Quiescent Swiss 3T3 cells were cotransfected with siCon or siILK together with expression vectors for HA-hSMOC-2 or GFP. The transfected cells were treated with PDGF-BB (3 ng/ml) for 9 h. Extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with anti-HA, anti-cyclin D1, and anti-β-actin antibodies as described under Materials and Methods. (B) Triplicate cultures of quiescent 3T3 cells were cotransfected with siCon, siSMOC-2, and siILK individually or in combination as described in A. The resulting cells were treated with PDGF-BB (10 ng/ml) for 22 h (or were left untreated for controls). Rates of [3H]thymidine incorporation were then determined as described under Materials and Methods. Data points represent the mean for three samples, with error bars representing the range. *p < 0.001 compared with siCon-transfected cells. (C) Quiescent 3T3 cells were cotransfected with siCon, siSMOC-2, and siILK individually or in combination. The total amount of siRNA used for each transfection was kept constant. The transfected cells were treated with PDGF-BB (10 ng/ml). At 0 and 9 h after PDGF treatment, cells were lysed, and extracts were analyzed for ILK, cyclin D1, and β-actin expression by SDS-PAGE and immunoblotting.
Moreover, if SMOC-2 and ILK participate in a common pathway, we predicted that the inhibitory effects of SMOC-2 depletion and ILK depletion on DNA synthesis should not be additive. Therefore, we used siRNA to silence ILK and SMOC-2 individually, and in combination. As shown in Figure 9, B and C, individual and combined knockdown of SMOC-2 and ILK inhibited DNA synthesis and cyclin D1 expression to a similar extent. These data are consistent with an epistatic relationship between ILK and SMOC-2. Together, our experiments suggest that ILK mediates the stimulatory effects of SMOC-2 on cyclin D1 expression and DNA synthesis.
DISCUSSION
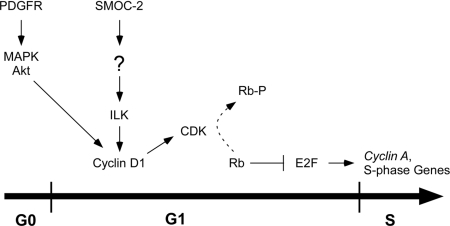
In this study, we have analyzed the expression and biological function of the putative matricellular factor SMOC-2. We have demonstrated that SMOC-2 is required for efficient DNA synthesis in response to PDGF and other growth factors. Our results indicate that SMOC-2 is dispensable for RTK activation. Instead, we suggest that the critical function of SMOC-2 during G1/S progression is maintenance of ILK activity and cyclin D1 expression (Figure 10).
Figure 10.
Hypothetical model describing requirement of SMOC-2 for cell cycle progression. See text for details.
Although we have shown that SMOC-2 is necessary for ILK activation and DNA synthesis, details of signaling events initiated by SMOC-2 (presumably at the cell surface) remain unknown. In contrast, mechanisms of signaling by the SMOC-2–related factor SPARC have been studied extensively and multiple biochemical activities have now been attributed to SPARC. For example, SPARC binds various mitogens (including VEGF, PDGF, and FGF) and inhibits receptor-mediated signaling by these growth factors (Brekken and Sage, 2000, 2001; Framson and Sage, 2004). In contrast with results reported for SPARC (which is antimitogenic), we have shown here that SMOC-2 promotes growth factor-induced DNA synthesis. Nevertheless, we considered the possibility that SMOC-2 might interact with growth factors, perhaps modulating their recruitment of to the cell surface or augmenting their interactions with receptors. However, SMOC-2 ablation does not affect PDGF-induced PDGFβR autophosphorylation or activation of PDGFβR effector kinases (MAPK and Akt), yet significantly attenuates PDGF-induced mitogenesis. Therefore, we consider it unlikely that a PDGF-binding activity is required for SMOC-2–dependent DNA synthesis after PDGF treatment. Instead, our analyses suggest that the major contribution of SMOC-2 to PDGF-induced DNA synthesis occurs via ILK, an integrin-activated protein kinase.
ILK is implicated in transcriptional activation of cyclin D1 (D'Amico et al., 2000), and our data also identify cyclin D1 as a distal effector of SMOC-2–dependent DNA synthesis. Induction of cyclin D1 is critical for G1 progression and requires cooperation between integrin and growth factor-mediated signaling pathways (Zhu et al., 1996; Danen and Yamada, 2001). Therefore, ILK provides a link between SMOC-2 and cyclin D1. Pestell and Li (2006) recently reported that cyclin D1 is necessary for VEGF-induced growth of vascular endothelial cells (Pestell and Li, 2006). We have shown that SMOC-2 is necessary for VEGF-induced mitogenesis and tube formation (hallmarks of the angiogenic response) in endothelial cells (Rocnik et al., 2006). Based on findings presented in this report, it is likely that SMOC-2 also contributes to angiogenesis via ILK-mediated induction of cyclin D1.
Clearly, further experiments are necessary to elucidate the mechanisms that mediate SMOC-2–dependent ILK activation. ILK activity is rapidly induced in response to engagement of integrins via adhesion to the ECM or Arg-Gly-Asp peptide (Wu et al., 1998; Barker et al., 2005). ILK catalytic activity is also subject to negative regulation by the phosphatase ILK-associated protein (ILKAP) (Wu and Dedhar, 2001; Wu, 2004, 2005). Potentially, SMOC-2 might facilitate integrin engagement by the ECM, thereby promoting integrin-dependent ILK activity. Alternatively, putative SMOC-2–binding partners/receptors might interact directly or indirectly with integrins to promote ILK activation. It is also possible that putative SMOC-2 effectors activate ILK independently of integrins, for example, via modulation of ILKAP. These hypothetical models of SMOC-2–mediated ILK activation are not necessarily mutually exclusive.
Interestingly, in phage display experiments to detect SPARC-interacting proteins, Sage and colleagues recently identified an interaction (presumably direct) between ILK and SPARC (Barker et al., 2005). Those workers also demonstrated that SPARC and ILK could be coimmunoprecipitated from cell extracts and that ILK fails to be activated in Sparc−/− cells (Barker et al., 2005). These findings are superficially similar to our results and might suggest that SPARC and SMOC-2 activate ILK via a common mechanism. However, it is unclear how matricellular proteins such as SPARC and SMOC-2 (which reside in the ECM) might interact directly with ILK (an intracellular signaling molecule) in a physiologically meaningful manner. It is possible that the interaction between ILK and SPARC occurs indirectly, perhaps via a transmembrane protein (although this would not explain the phage display interaction between SPARC and ILK). Alternatively, matricellular factors might activate ILK upon internalization. In contrast with results reported for SPARC, we have not detected any interactions between ILK and SMOC-2, even when both proteins are ectopically overexpressed (Liu and Vaziri, unpublished results). Moreover, ILK is generally proangiogenic and promitogenic. Therefore, it is unclear how ILK activation can account for the well-documented antiangiogenic and antimitogenic activities of SPARC. Nevertheless, a role for SPARC in ILK activation was also reported more recently by Shi et al. (2007). Therefore, ILK may represent a general effector of SMOC-2, SPARC, and possibly other matricellular proteins. It should be noted however, that the kinase domain of ILK differs from the catalytic loops and DGX motifs of other Ser/Thr kinases. Moreover, the catalytic domain of ILK is very divergent across species, possibly suggesting that ILK functions mainly as an adaptor rather than as a physiologically relevant protein kinase in vivo (Legate et al., 2006). Therefore, the protein kinase activity that we and other workers have detected in anti-ILK immune complexes might represent an ILK-associated kinase. Nevertheless, our data provides a novel link between SMOC-2 and ILK signaling.
Given the strong similarity between SPARC and SMOC-2, it is interesting that these factors have opposing activities with respect to angiogenesis. The opposing effects of SPARC and SMOC-2 on angiogenesis are reminiscent of the relationship between matricellular factors CCN1 and CCN3 which interact with integrins and HSPG to regulate growth (Brigstock, 2003). Whereas CCN1 acts as a tumor-promoting factor, CCN3 exhibits tumor-suppressive capabilities (Lombet et al., 2003; Bleau et al., 2005). It is possible that SPARC and SMOC-1/SMOC-2 have similar antagonistic relationships and that the balance between SPARC and SMOC activities dictates net proliferation, angiogenesis or other cellular outcomes.
Our work demonstrates that SMOC-2 expression levels can have a profound influence on mitogenesis. Therefore, regulated changes in SMOC-2 expression are likely to impact cell growth in vitro and in vivo. The quiescence-specific expression of SMOC-2 in cultured cells is similar to the expression pattern of PDGF receptors. PDGFαR and PDGFβR are highly expressed in G0 cells and are transcriptionally down-regulated in response to mitogens (Vaziri and Faller, 1995; Lih et al., 1996). Down-regulation of PDGFβR concomitant with cell cycle entry most likely represents a negative feedback mechanism that desensitizes cells to PDGF after cell cycle entry (Vaziri and Faller, 1995; Lih et al., 1996). Because SMOC-2 is required for mitogenesis, its down-regulation after cell cycle entry could help desensitize cells to mitogens.
The mechanism underlying cell cycle-dependent changes in SMOC-2 expression is unclear. We have isolated the 5′ region of the mouse Smoc2 gene and generated heterologous luciferase reporter constructs containing up to ∼1.2 kb of its promoter (Liu and Vaziri, unpublished). In transient transfection experiments, Smoc2 promoter-driven luciferase expression was insensitive to cell cycle state and mitogen treatments. Therefore, it is possible that nontranscriptional mechanisms regulate SMOC-2 levels during the cell cycle. Alternatively, it is possible that the large Smoc2 gene (>100 kb) is transcriptionally regulated via putative elements residing outside the 1.2-kb promoter region we have isolated.
Regardless of the precise mechanisms that determine SMOC-2 levels, it will be interesting to determine whether SMOC-2 expression is deregulated in diseases involving aberrant cell proliferation (such as cancer). Potentially, failure to appropriately down-regulate SMOC-2 and its resulting aberrant overexpression might contribute to malignancy. Interestingly, inappropriate expression of the SMOC-2-related matricellular factor SPARC has been implicated in tumorigenesis. SPARC is expressed at high levels in many cancers, particularly in stromal cells and in the vasculature (Framson and Sage, 2004). Massague and colleagues recently showed that SPARC and other extracellular factors are important for metastasis of breast cancer cells to the lung (Minn et al., 2005). Although a role for SMOC-2 in tumorigenesis has not been tested, it is possible that aberrant SMOC-2 overexpression could promote growth of tumor cells, particularly those that are dependent on mitogenic growth factors such as PDGF.
The distinct pattern of SMOC-2 we observed in various developing structures is intriguing, and suggests a role in control of specific events during organogenesis. Whether SMOC-2 controls rates of cell proliferation in the limbs or in somites, and whether it participates in some aspect of cell differentiation, remains to be investigated. Together, our results indicate that SMOC-2 is an important regulator of mitogenesis. Studies are underway to define the essential role(s) of SMOC-2 in growth factor signaling and cell cycle control during development and in disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Environmental Health Sciences grant ES09558 and by a research grant from Philip Morris USA, and Philip Morris International (to C.V.) and by National Institutes of Health/National Heart, Lung, and Blood Institute (P01) HL47049 (to W.V.C.). We thank Drs. Nils Cordes, Stephanie Hehglans, Charles Sherr, and Greg Hannigan for expression constructs. We thank Dr. Alex Toker, Dr. Kenn Albrecht, and Dorothy Pazin for valuable discussions and helpful advice with the manuscript.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0510) on November 7, 2007.
REFERENCES
- Agami R., Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol. Cell. Biol. 1988;8:2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout M. A., Mahalingam B., Xiong J. P. Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- Assoian R. K., Schwartz M. A. Coordinate signaling by integrins and receptor tyrosine kinases in the regulation of G1 phase cell-cycle progression. Curr. Opin. Genet Dev. 2001;11:48–53. doi: 10.1016/s0959-437x(00)00155-6. [DOI] [PubMed] [Google Scholar]
- Barker T. H., et al. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J. Biol. Chem. 2005;280:36483–36493. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- Bleau A. M., Planque N., Perbal B. CCN proteins and cancer: two to tango. Front. Biosci. 2005;10:998–1009. doi: 10.2741/1594. [DOI] [PubMed] [Google Scholar]
- Bornstein P., Sage E. H. Matricellular proteins: extracellular modulators of cell function. Curr. Opin. Cell Biol. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- Bradshaw A. D., Francki A., Motamed K., Howe C., Sage E. H. Primary mesenchymal cells isolated from SPARC-null mice exhibit altered morphology and rates of proliferation. Mol. Biol. Cell. 1999;10:1569–1579. doi: 10.1091/mbc.10.5.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., Zerial M., Toschi L., Schurmann M., Muller R., Hirai S. I., Yaniv M., Almendral J. M., Ryseck R. P. Identification of growth-factor-inducible genes in mouse fibroblasts. Cold Spring Harb. Symp. Quant Biol. 1988;53:901–905. doi: 10.1101/sqb.1988.053.01.103. [DOI] [PubMed] [Google Scholar]
- Brekken R. A., Sage E. H. SPARC, a matricellular protein: at the crossroads of cell-matrix. Matrix Biol. 2000;19:569–580. doi: 10.1016/s0945-053x(00)00105-0. [DOI] [PubMed] [Google Scholar]
- Brekken R. A., Sage E. H. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- Brewer J. W., Hendershot L. M., Sherr C. J., Diehl J. A. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA. 1999;96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock D. R. The CCN family: a new stimulus package. J. Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cochran B. H., Reffel A. C., Stiles C. D. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- Cochran B. H., Zullo J., Verma I. M., Stiles C. D. Expression of the c-fos gene and of an fos-related gene is stimulated by platelet-derived growth factor. Science. 1984;226:1080–1082. doi: 10.1126/science.6093261. [DOI] [PubMed] [Google Scholar]
- Cordes N. Overexpression of hyperactive integrin-linked kinase leads to increased cellular radiosensitivity. Cancer Res. 2004;64:5683–5692. doi: 10.1158/0008-5472.CAN-04-1056. [DOI] [PubMed] [Google Scholar]
- D'Amico M., et al. The integrin-linked kinase regulates the cyclin D1 gene through glycogen synthase kinase 3beta and cAMP-responsive element-binding protein-dependent pathways. J. Biol. Chem. 2000;275:32649–32657. doi: 10.1074/jbc.M000643200. [DOI] [PubMed] [Google Scholar]
- Danen E. H., Yamada K. M. Fibronectin, integrins, and growth control. J. Cell Physiol. 2001;189:1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Ran W., Kindy M. S., Sonenshein G. E., Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J. Biol. Chem. 1986;261:9161–9166. [PubMed] [Google Scholar]
- Delcommenne M., Tan C., Gray V., Rue L., Woodgett J., Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc. Natl. Acad. Sci. USA. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Framson P. E., Sage E. H. SPARC and tumor growth: where the seed meets the soil? J. Cell. Biochem. 2004;92:679–690. doi: 10.1002/jcb.20091. [DOI] [PubMed] [Google Scholar]
- Francki A., Bradshaw A. D., Bassuk J. A., Howe C. C., Couser W. G., Sage E. H. SPARC regulates the expression of collagen type I and transforming growth factor-beta1 in mesangial cells. J. Biol. Chem. 1999;274:32145–32152. doi: 10.1074/jbc.274.45.32145. [DOI] [PubMed] [Google Scholar]
- Francki A., McClure T. D., Brekken R. A., Motamed K., Murri C., Wang T., Sage E. H. SPARC regulates TGF-beta1-dependent signaling in primary glomerular mesangial cells. J. Cell. Biochem. 2004;91:915–925. doi: 10.1002/jcb.20008. [DOI] [PubMed] [Google Scholar]
- Francki A., Motamed K., McClure T. D., Kaya M., Murri C., Blake D. J., Carbon J. G., Sage E. H. SPARC regulates cell cycle progression in mesangial cells via its inhibition of IGF-dependent signaling. J. Cell. Biochem. 2003;88:802–811. doi: 10.1002/jcb.10424. [DOI] [PubMed] [Google Scholar]
- Giancotti F. G., Tarone G. Positional control of cell fate through joint integrin/receptor protein kinase signaling. Annu. Rev. Cell Dev. Biol. 2003;19:173–206. doi: 10.1146/annurev.cellbio.19.031103.133334. [DOI] [PubMed] [Google Scholar]
- Guo N., Faller D. V., Vaziri C. Carcinogen-induced S-phase arrest is Chk1 mediated and caffeine sensitive. Cell Growth Differ. 2002;13:77–86. [PubMed] [Google Scholar]
- Herschman H. R. Primary response genes induced by growth factors and tumor promoters. Annu. Rev. Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Huang S., Chen C. S., Ingber D. E. Control of cyclin D1, p27(Kip1), and cell cycle progression in human capillary endothelial cells by cell shape and cytoskeletal tension. Mol. Biol. Cell. 1998;9:3179–3193. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupprion C., Motamed K., Sage E. H. SPARC (BM-40, osteonectin) inhibits the mitogenic effect of vascular endothelial growth factor on microvascular endothelial cells. J. Biol. Chem. 1998;273:29635–29640. doi: 10.1074/jbc.273.45.29635. [DOI] [PubMed] [Google Scholar]
- Lau L. F., Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985;4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc. Natl. Acad. Sci. USA. 1987;84:1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K. R., Montanez E., Kudlacek O., Fassler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- Lih C. J., Cohen S. N., Wang C., Lin-Chao S. The platelet-derived growth factor alpha-receptor is encoded by a growth-arrest-specific (gas) gene. Proc. Natl. Acad. Sci. USA. 1996;93:4617–4622. doi: 10.1073/pnas.93.10.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombet A., Planque N., Bleau A. M., Li C. L., Perbal B. CCN3 and calcium signaling. Cell Commun. Signal. 2003;1:1. doi: 10.1186/1478-811X-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpel S., Mendelsohn C., Cardoso W. V. Regulation of retinoic acid signaling during lung morphogenesis. Development. 2000;127:3057–3067. doi: 10.1242/dev.127.14.3057. [DOI] [PubMed] [Google Scholar]
- Midwood K. S., Schwarzbauer J. E. Tenascin-C modulates matrix contraction via focal adhesion kinase- and Rho-mediated signaling pathways. Mol. Biol. Cell. 2002;13:3601–3613. doi: 10.1091/mbc.E02-05-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamed K., Blake D. J., Angello J. C., Allen B. L., Rapraeger A. C., Hauschka S. D., Sage E. H. Fibroblast growth factor receptor-1 mediates the inhibition of endothelial cell proliferation and the promotion of skeletal myoblast differentiation by SPARC: a role for protein kinase A. J. Cell. Biochem. 2003;90:408–423. doi: 10.1002/jcb.10645. [DOI] [PubMed] [Google Scholar]
- Motamed K., Funk S. E., Koyama H., Ross R., Raines E. W., Sage E. H. Inhibition of PDGF-stimulated and matrix-mediated proliferation of human vascular smooth muscle cells by SPARC is independent of changes in cell shape or cyclin-dependent kinase inhibitors. J. Cell. Biochem. 2002;84:759–771. doi: 10.1002/jcb.10095. [DOI] [PubMed] [Google Scholar]
- Nathans D., Lau L. F., Christy B., Hartzell S., Nakabeppu Y., Ryder K. Genomic response to growth factors. Cold Spring Harb. Symp. Quant. Biol. 1988;53:893–900. doi: 10.1101/sqb.1988.053.01.102. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- Nevins J. R., Leone G., DeGregori J., Jakoi L. Role of the Rb/E2F pathway in cell growth control. J. Cell Physiol. 1997;173:233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Paley P. J., Goff B. A., Gown A. M., Greer B. E., Sage E. H. Alterations in SPARC and VEGF immunoreactivity in epithelial ovarian cancer. Gynecol. Oncol. 2000;78:336–341. doi: 10.1006/gyno.2000.5894. [DOI] [PubMed] [Google Scholar]
- Pardee A. B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Pestell R. G., Li Z. Antisense to cyclin D1 inhibits VEGF-stimulated growth of vascular endothelial cells: implication of tumor vascularization. Clin. Cancer Res. 2006;12:4459–4462. doi: 10.1158/1078-0432.CCR-06-0614. [DOI] [PubMed] [Google Scholar]
- Quelle D. E., Ashmun R. A., Shurtleff S. A., Kato J. Y., Bar-Sagi D., Roussel M. F., Sherr C. J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Rocnik E. F., Liu P., Sato K., Walsh K., Vaziri C. The novel SPARC family member SMOC-2 potentiates angiogenic growth factor activity. J. Biol. Chem. 2006;281:22855–22864. doi: 10.1074/jbc.M513463200. [DOI] [PubMed] [Google Scholar]
- Roussel M. F., Theodoras A. M., Pagano M., Sherr C. J. Rescue of defective mitogenic signaling by D-type cyclins. Proc. Natl. Acad. Sci. USA. 1995;92:6837–6841. doi: 10.1073/pnas.92.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryseck R. P., Hirai S. I., Yaniv M., Bravo R. Transcriptional activation of c-jun during the G0/G1 transition in mouse fibroblasts. Nature. 1988;334:535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- Schwartz M. A., Assoian R. K. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 2001;114:2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. D-type cyclins. Trends Biochem. Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- Shi Q., Bao S., Song L., Wu Q., Bigner D. D., Hjelmeland A. B., Rich J. N. Targeting SPARC expression decreases glioma cellular survival and invasion associated with reduced activities of FAK and ILK kinases. Oncogene. 2007;26:4084–4094. doi: 10.1038/sj.onc.1210181. [DOI] [PubMed] [Google Scholar]
- Tan C., Cruet-Hennequart S., Troussard A., Fazli L., Costello P., Sutton K., Wheeler J., Gleave M., Sanghera J., Dedhar S. Regulation of tumor angiogenesis by integrin-linked kinase (ILK) Cancer Cell. 2004;5:79–90. doi: 10.1016/s1535-6108(03)00281-2. [DOI] [PubMed] [Google Scholar]
- Vannahme C., Gosling S., Paulsson M., Maurer P., Hartmann U. Characterization of SMOC-2, a modular extracellular calcium-binding protein. Biochem. J. 2003;373:805–814. doi: 10.1042/BJ20030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannahme C., Smyth N., Miosge N., Gosling S., Frie C., Paulsson M., Maurer P., Hartmann U. Characterization of SMOC-1, a novel modular calcium-binding protein in basement membranes. J. Biol. Chem. 2002;277:37977–37986. doi: 10.1074/jbc.M203830200. [DOI] [PubMed] [Google Scholar]
- Vaziri C., Faller D. V. Repression of platelet-derived growth factor beta-receptor expression by mitogenic growth factors and transforming oncogenes in murine 3T3 fibroblasts. Mol. Cell. Biol. 1995;15:1244–1253. doi: 10.1128/mcb.15.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The PINCH-ILK-parvin complexes: assembly, functions and regulation. Biochim. Biophys. Acta. 2004;1692:55–62. doi: 10.1016/j.bbamcr.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Wu C. PINCH, N(i)ck and the ILK: network wiring at cell-matrix adhesions. Trends Cell Biol. 2005;15:460–466. doi: 10.1016/j.tcb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wu C., Dedhar S. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J. Cell Biol. 2001;155:505–510. doi: 10.1083/jcb.200108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Keightley S. Y., Leung-Hagesteijn C., Radeva G., Coppolino M., Goicoechea S., McDonald J. A., Dedhar S. Integrin-linked protein kinase regulates fibronectin matrix assembly, E-cadherin expression, and tumorigenicity. J. Biol. Chem. 1998;273:528–536. doi: 10.1074/jbc.273.1.528. [DOI] [PubMed] [Google Scholar]
- Zhu X., Ohtsubo M., Bohmer R. M., Roberts J. M., Assoian R. K. Adhesion-dependent cell cycle progression linked to the expression of cyclin D1, activation of cyclin E-cdk2, and phosphorylation of the retinoblastoma protein. J. Cell Biol. 1996;133:391–403. doi: 10.1083/jcb.133.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.