Abstract
A screen for genes required in Drosophila eye development identified an UNC-104/Kif1 related kinesin-3 microtubule motor. Analysis of mutants suggested that Drosophila Unc-104 has neuronal functions that are distinct from those of the classic anterograde axonal motor, kinesin-1. In particular, unc-104 mutations did not cause the distal paralysis and focal axonal swellings characteristic of kinesin-1 (Khc) mutations. However, like Khc mutations, unc-104 mutations caused motoneuron terminal atrophy. The distributions and transport behaviors of green fluorescent protein-tagged organelles in motor axons indicate that Unc-104 is a major contributor to the anterograde fast transport of neuropeptide-filled vesicles, that it also contributes to anterograde transport of synaptotagmin-bearing vesicles, and that it contributes little or nothing to anterograde transport of mitochondria, which are transported primarily by Khc. Remarkably, unc-104 mutations inhibited retrograde runs by neurosecretory vesicles but not by the other two organelles. This suggests that Unc-104, a member of an anterograde kinesin subfamily, contributes to an organelle-specific dynein-driven retrograde transport mechanism.
INTRODUCTION
To maintain the structural and biochemical order required for proper function, cells of higher organisms use cytoskeleton-based force-generating machinery to carry RNAs, proteins, and organelles to specific destinations. Neurons are especially dependent on such transport processes, because although most synthesis of new components occurs in the soma (cell body), often more than 99% of the cell's volume is in its axon. To support an axon, a neuron must continuously transport new cytoplasmic materials out of the cell body toward the terminal (anterograde transport) and old, unused, or endosome-associated materials back to the cell body (retrograde transport). Disruption of this cycle causes a decline in neurotransmission at the terminal, poor retrograde neurotrophic signaling, and atrophy or “dying back” of the axon (Chevalier-Larsen and Holzbaur, 2006; Duncan and Goldstein, 2006).
The general mechanistic principles of axonal transport center around cytoskeletal filaments (microtubules and F-actin) and three families of force-generating motor proteins (myosins, dyneins and kinesins). A motor links to an axonal cargo and pulls it stepwise along a filament track, using ATP as an energy source (Vale and Milligan, 2000). Long-distance transport in axons is accomplished by members of the kinesin and dynein families, which use microtubules as tracks. Composed of head-to-tail polymers of α- and β-tubulin dimers, microtubules in axons are organized with their β ends (plus-ends) toward the axon terminal and their α ends (minus-ends) toward the cell body (Heidemann et al., 1981). Cytoplasmic dynein, for which there seems to be just one variety of force-producing heavy chain subunit, is minus-end directed, and it is the primary motor for retrograde axonal transport. The cytoplasmic dynein heavy chain has many associated nonforce-producing subunits whose functions are not well understood. Some are regulatory subunits, and some may serve as specific adaptors to link the motor to its different retrograde cargoes (Mallik and Gross, 2004; Chevalier-Larsen and Holzbaur, 2006).
There are many different subfamilies of kinesins with members whose amino-terminal ATPase and microtubule binding “motor domain” sequences suggest that they might contribute to anterograde transport (Wickstead and Gull, 2006). Function tests in model systems and human disease genetics currently indicate that members of the kinesin-1 and kinesin-3 subfamilies are especially critical for anterograde axonal transport. Mutations in human KIF5A (a kinesin-1) and KIF1B (a kinesin-3) can cause, respectively, hereditary spastic paraplegia (Reid et al., 2002) and Charcot-Marie-Tooth disease (Zhao et al., 2001). Although the motor regions of kinesins-1 and -3 have similar sequences, there are differences that endow them with distinct biophysical capabilities (e.g., velocity and processivity) when tested in vitro (e.g., Tomishige et al., 2002). This and the fact that the “stalk-tail” cargo binding regions of kinesin-1 and kinesin-3 motor subunits are not conserved suggests that kinesins-1 and -3 carry different sets of anterograde cargoes at different rates. However, the identities of those cargo sets and how defects in their axonal transport relate to mechanisms of neurodegeneration are not well understood.
To gain insight into axonal transport mechanisms and more specifically into the functions of a new UNC-104/KIF1A-like kinesin-3 that we and Pack-Chung et al. (2007) have identified in Drosophila, we applied genetics, immunolocalization, time-lapse microscopy, and digital tracking to study the distributions and movements of organelles in motor axons. Tests of mutants show that Drosophila Unc-104 is critical for normal axon terminal development. It is a key anterograde motor for large neuropeptide-filled vesicles and small transport vesicles, but not for mitochondria. Comparison of the two vesicle types indicates that Unc-104–driven motion is strongly influenced by the identity but not by the size of its cargo. This suggests that organelle-specific components are more important for defining transport behavior than cytoplasmic drag. Analysis of unc-104 mutants revealed an unexpected inhibition of neurosecretory vesicle retrograde runs, but no detectable inhibition of retrograde runs by vesicles or mitochondria, suggesting that Unc-104, an anterograde microtubule motor, is required for a specific cytoplasmic dynein-mediated retrograde transport mechanism.
MATERIALS AND METHODS
Fly Culture and Genetics
Flies were cultured at 25°C in a 12-h light/dark cycle on standard soft medium (0.5% agar, 7% molasses, 6% cornmeal, and 0.8% killed yeast) seeded with live yeast. Descriptions of the previously characterized mutant alleles and balancers used can be found in FlyBase (http://flybase.bio.indiana.edu/) and elsewhere (Pilling et al., 2006).
Amorphic alleles of unc-104 were isolated in a screen for mutations that disrupt photoreceptor connectivity (Newsome et al., 2000) (T. Suzuki and B. Dickson, unpublished). Hypomorphic alleles were isolated in a standard F2 lethal screen for ethyl methanesulfonate-mutagenized chromosomes (Saxton et al., 1991) that failed to complement amorphic unc-104 alleles (Supplemental Table S1).
To generate animals with fluorescently labeled organelles, meiotic recombination was used to generate third chromosomes with a neuronal Gal4 driver P{GawB}D42, which expresses Gal4 in cells of the optic lopes, the ventral ganglion and motoneurons, but not sensory neurons, and a Gal4-UAS GFP-organelle responder (Pilling et al., 2006). The responders used were as follows: 1) P{w+mC = UAS-ANFGFP}3, which expresses a fusion protein that concentrates in large dense core vesicles (Rao et al., 2001), referred to here as atrial natriuretic factor::green fluorescent protein (ANF::GFP); 2) P{w+mC = UAS-mitoGFP.AP}3, which is 13.1-cM distant from P{GawB}D42 and expresses a fusion protein that concentrates in the matrix of mitochondria (Pilling et al., 2006), referred to here as mitoGFP; and 3) P{w+mC = UAS-syt.eGFP}3, which expresses a fusion protein targeted to small clear core transport vesicles (Zhang et al., 2002), referred to here as synaptotagmin (syt)::GFP. Those recombinant driver-responder chromosomes were used to construct strains with unc-104 alleles on chromosome 2 balanced by a translocation (T(2;3)CyO, TM6B Hu Tb e) that allowed recognition of unc-104/unc-104 larvae by body shape. The unc-104 alleles used were unc-104P350, unc-10O1.2, and unc-104O3.1 (Supplemental Table S1).
Transgenic Unc-104-GFP Construct
A full-length Unc-104 cDNA, isolated from PgR7 (Senti et al., 2003) by digestion with Kpn1 and Xba1 was ligated into the Drosophila transformation vector pUAST, fused in-frame with a gene for enhanced GFP (S65T) downstream of a GAL4-UAS that allowed tissue-specific expression of the Unc-104::GFP fusion gene (Brand and Perrimon, 1993). The final transposable element, P{w+mC = UAS-unc-104.GFP.RVB}, was transformed into flies by using a helper plasmid containing a transposase gene. The transgene is referred to here as unc-104::GFP.
Immunostaining
Wandering third instar larvae were dissected and fixed as described previously (Hurd and Saxton, 1996). After 20 min of fixation, larvae were washed four times with phosphate-buffered saline containing 3% Triton X-100. The primary antibodies used were mouse monoclonal anti-cysteine string protein at 1:500 (Zinsmaier et al., 1994), rabbit anti-synaptotagmin at 1:500 (Littleton et al., 1993), and rabbit anti-syntaxin at 1:500 (Hata et al., 1993). The fluorescent secondary antibodies used were affinity-purified Alexa 488-conjugated goat anti-mouse and Alexa 594-conjugated goat anti-rabbit immunoglobinin G (H+L) at 1:1000 (Invitrogen, Carlsbad, CA).
Imaging of fixed/stained tissues was done with a PerkinElmer UltraVIEW LCI Spinning Disk confocal fluorescence system on a Nikon Eclipse TE200 microscope equipped with a Nikon 40× objective, except for D in Supplemental Figure S1, which was collected with a 60× objective. Images were processed in NIH Image version 1.62b7 (National Institutes of Health, Bethesda, MD) and Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Segmental Nerve Ligation
To generate physical blockades of axonal transport in segmental nerves, a fine nylon fiber (Henry and Raff, 1990) was tied with an overhand knot to tightly constrict wandering third instar larvae midway between head and tail (Horiuchi et al., 2005). After 2 h, ligated larvae were pinned to a Sylgard-lined dish, submersed in Schneider's insect medium and incised along the dorsal midline, except for the immediate area of the fiber. After removal of fat body, gut, and salivary glands, larvae were fixed (Horiuchi et al., 2005). The fiber was then cut, dissection was completed, and specimens were immunostained as described above. Constrictions in individual segmental nerves varied in width, probably due to variable tissue surroundings and ligation tightness.
Live Imaging of GFP-tagged Organelle Behavior in Larval Axons
Wandering third instar larvae with GFP fusions driven by D42-Gal4 were dissected quickly (<5 min) in Schneider's medium, and the resulting preparations were laid on microscope slides with the cuticle side against the glass. After adding fresh medium and coverslip fragments as spacers, coverslips were placed over the specimens and anchored with Valap (petroleum jelly:lanolin:paraffin [1:1:1]) at the corners. Imaging was initiated at 10–15 min and terminated at 25–30 min after the start of dissection (Pilling et al., 2006).
Imaging protocols were selected that allowed the most efficient analysis of the transport behavior of each organelle class. GFP-Syt was imaged continuously with a frame collected every 0.7–0.9 s on a Nikon widefield E800 fluorescence microscope with an Orca ER charge-coupled device (CCD) camera controlled by MetaMorph software. Because of larger brighter organelles and the abundance of stationary organelles, mitoGFP in a 30–50-μm-wide region of a nerve was partially photobleached and imaged with an MRC600 scanning confocal fluorescence microscope (Bio-Rad, Hercules, CA) at one frame per second (Pilling et al., 2006). ANF::GFP also produced bright organelle signals, but transport was relatively fast. To prevent streaking distortion of organelle images, a high-speed spinning disk confocal system (UltraVIEW LIC; PerkinElmer Life and Analytical Sciences, Boston, MA) with an Orca ER CCD camera (Hamamatsu, Bridgewater, NJ) was used to collect frames at two per second after initial photobleaching of a 30–50-μm-wide section of nerve.
Tracking and Statistics
Digital tracking of organelles in time-lapse image series was done using NIH Image and a tracking protocol described previously (Pilling et al., 2006). For each of the three microscopes, a stage micrometer was used to calculate X and Y pixel dimensions, which were then used to calculate real distances between organelle positions in succeeding video frames. The elapsed time between each succeeding frame was recorded and used to calculate velocities.
Data from organelle tracking were used to define anterograde runs, retrograde runs, and pauses as described previously (Pilling et al., 2006). To determine whether the means of these transport parameters were significantly different between wild-type and mutant larvae for each type of organelle (Figure 7 and Table 1), linear contrast statistical analyses of aggregated means for each organelle were done as detailed previously (Pilling et al., 2006). The significance of differences in flux, net transport velocity (Figure 5 and Table 1), and comparisons of organelles and Unc-104::GFP run velocities (Table 2) were analyzed by F-tests to determine variance and with t tests with unequal or equal variance at 95% confidence intervals.
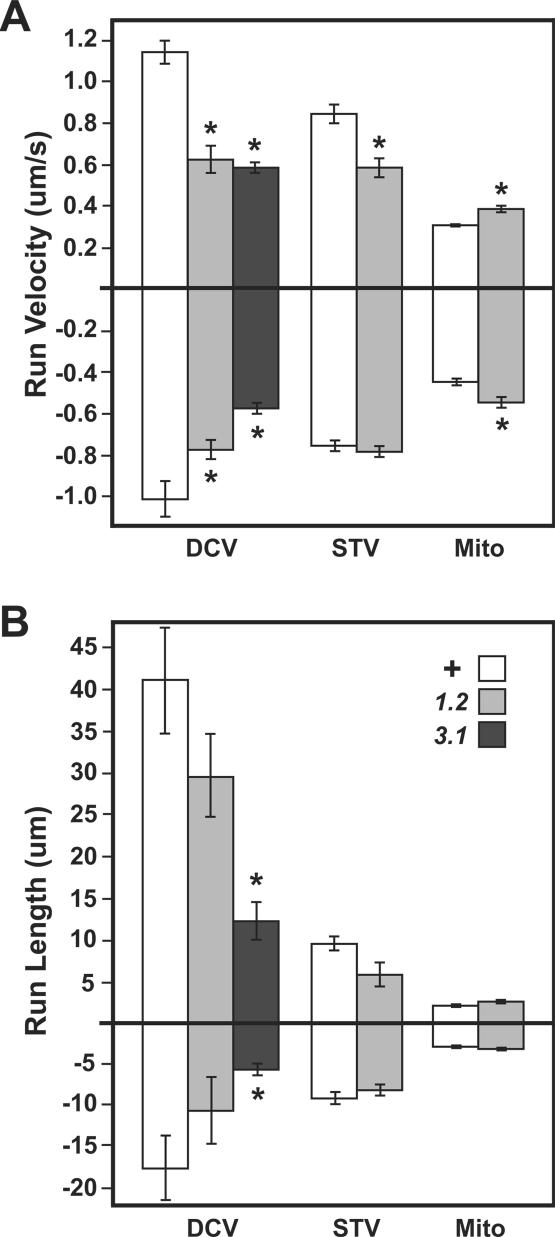
Figure 7.
Changes in run behaviors for different organelles in unc-104 mutant axons. Runs were defined as periods of uninterrupted organelle motion in one direction bounded by pauses or reverse runs. Means (±SEMs) are shown for run velocities (A) and run lengths (B) for DCVs, STVs, and mitochondria (Mito). Sample sizes and additional data can be found in Table 1. Wild-type (unshaded) and unc-104 mutant (shaded) values were compared using linear contrast. Significant differences are noted by an asterisk (p < 0.05). The genotypes tested were wild-type (+), unc-104O1.2/unc-104P350 (1.2), and unc-104O3.1/unc-104P350 (3.1).
Table 1.
Organelle transport in wild-type and unc-104 mutant larval motor axons
| Organellea | Genotypeb | Fluxc (org./min) | Net velocityd,e (μm/s) | Forward runsd,f |
Reverse runsd,f |
Pauses% Time | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| % Time | Velocity (μm/s) | Length (μm) | % Time | Velocity (μm/s) | Length (μm) | |||||
| Anterograde organelles |
||||||||||
| DCV(ANF) | +/+ | 104.0 ± 7.4 | 1.05 ± 0.10 | 85.14 ± 3.87 | 1.14 ± 0.06 | 41.75 ± 6.54 | 1.24 ± 0.89 | −0.41 ± 0.06 | 1.21 ± 0.50 | 13.6 ± 3.59 |
| O1.2/P350 | 34.4 ± 4.3* | 0.72 ± 0.06* | 82.01 ± 3.14 | 0.62 ± 0.07* | 29.65 ± 5.12 | 3.07 ± 1.11 | −0.42 ± 0.04 | 1.33 ± 0.32 | 14.9 ± 2.91 | |
| O3.1/P350 | 22.2 ± 6.4* | 0.49 ± 0.05* | 82.73 ± 3.23 | 0.58 ± 0.03* | 12.31 ± 2.42* | 2.20 ± 0.97 | −0.39 ± 0.05 | 0.88 ± 0.13* | 15.1 ± 2.69 | |
| STV(Syt) | +/+ | 8.4 ± 1.2 | 0.69 ± 0.09 | 78.48 ± 2.20 | 0.84 ± 0.05 | 9.63 ± 0.93 | 3.47 ± 0.80 | −0.36 ± 0.03 | 1.06 ± 0.13 | 18.1 ± 1.92 |
| O1.2/P350 | 1.6 ± 0.5* | 0.51 ± 0.06 | 54.20 ± 6.30* | 0.58 ± 0.05* | 5.70 ± 1.54 | 11.16 ± 5.23 | −0.40 ± 0.06 | 1.35 ± 0.39 | 34.7 ± 0.05* | |
| Mitochondria | +/+ | 4.1 ± 0.8 | 0.19 ± 0.02 | 59.09 ± 2.35 | 0.30 ± 0.01 | 2.06 ± 0.18 | 1.14 ± 0.36 | −0.25 ± 0.02 | 0.58 ± 0.54 | 39.8 ± 2.24 |
| O1.2/P350 | 1.9 ± 0.2* | 0.26 ± 0.04 | 57.24 ± 3.41 | 0.38 ± 0.02* | 2.52 ± 0.31 | 1.08 ± 0.36 | −0.28 ± 0.03 | 0.67 ± 0.15 | 41.7 ± 3.44 | |
| Retrograde organelles |
||||||||||
| DCV(ANF) | +/+ | 38.8 ± 1.9 | −0.62 ± 0.09 | 67.80 ± 4.26 | −1.02 ± 0.09 | −17.42 ± 4.06 | 7.27 ± 1.93 | 0.36 ± 0.02 | 1.20 ± 0.22 | 24.9 ± 3.70 |
| O1.2/P350 | 31.4 ± 2.4* | −0.56 ± 0.06 | 64.77 ± 3.33 | −0.78 ± 0.05* | −10.60 ± 4.09 | 8.60 ± 1.99 | 0.33 ± 0.03 | 1.15 ± 0.19 | 26.6 ± 3.31 | |
| O3.1/P350 | 13.0 ± 2.5* | −0.36 ± 0.10 | 60.41 ± 4.3 | −0.58 ± 0.03* | −5.39 ± 0.81* | 9.03 ± 1.71 | 0.32 ± 0.04 | 0.90 ± 0.11 | 30.6 ± 3.80 | |
| STV(Syt) | +/+ | 10.0 ± 1.1 | −0.62 ± 0.09 | 76.83 ± 2.24 | −0.76 ± 0.03 | −9.01 ± 0.81 | 3.51 ± 0.62 | 0.38 ± 0.04 | 1.16 ± 0.13 | 19.7 ± 1.94 |
| O1.2/P350 | 11.2 ± 1.0 | −0.60 ± 0.08 | 73.08 ± 2.07 | −0.79 ± 0.03 | −7.98 ± 0.75 | 7.33 ± 1.00* | 0.32 ± 0.02 | 0.92 ± 0.07 | 19.6 ± 1.54 | |
| Mitochondria | +/+ | 2.4 ± 0.7 | −0.21 ± 0.02 | 45.07 ± 4.12 | −0.45 ± 0.02 | −2.77 ± 0.22 | 4.25 ± 0.60 | 0.23 ± 0.01 | 0.54 ± 0.04 | 50.7 ± 4.22 |
| O1.2//P350 | 1.8 ± 0.4 | −0.20 ± 0.02 | 37.53 ± 3.22 | −0.55 ± 0.03* | −2.90 ± 0.24 | 7.49 ± 2.08 | 0.24 ± 0.01 | 0.60 ± 0.05 | 55.0 ± 3.40 | |
a Organelle-targeted GFPs were imaged in motor axons of third instar segmental nerves by time-lapse fluorescence microscopy.
b Wild-type (+), unc-104O1.2 (O1.2), unc-104O3.1 (O3.1), and unc-104P350 (P350) alleles were used.
c Flux for DCVs and mitochondria represents the mean number (±SEM) of organelles per minute that entered the photobleached zone (1 nerve/animal, 5 animals/genotype). For Syt vesicles, flux represents the total number of vesicles that could be tracked in one nerve of each animal divided by total observation time. F-tests were used to determine variance, and t tests were done with unequal or equal variance at 95% confidence intervals. Significant differences for mutant relative to wild type means are noted by an asterisk (p < 0.05).
d All values other than flux were determined by measuring the position of the center of each organelle as a function of time in each video frame (1 nerve/animal, 5 animals/genotype). For DCVs and mitochondria, five organelles were tracked in each direction for each animal. For STVs, because all observable transported organelles were tracked, sample sizes from five larvae were 70 anterograde and 85 retrograde for wild type and 13 anterograde and 93 retrograde for mutant larvae. The significance of differences between wild type and mutant means were determined by linear contrast (asterisk indicates p < 0.05).
e Net velocity was determined by summing all velocities for each organelle over all time intervals, including forward runs (positive), reverse runs (negative), and pauses. Note that mean values for forward run velocity can be less than net velocity if short duration runs are numerous and slow, whereas long duration runs are few and fast. F-tests were used to determine variance, and t tests were done with unequal or equal variance at 95% confidence intervals. Significant differences for mutant relative to wild type means are noted by an asterisk (p < 0.05).
f Most organelles showed a strong directional bias with frequent long runs in a forward direction interrupted by pauses and infrequent short runs in the opposite or reverse direction. Thus, organelles were classed as either “anterograde” or “retrograde” and runs as either “forward” or “reverse.”
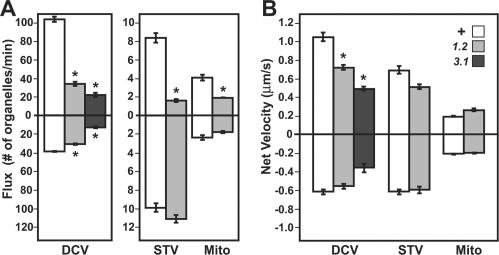
Figure 5.
Influence of unc-104 mutations on axonal organelle flux and net transport rates. (A) Mean flux values (±SEM) for DCVs (ANF::GFP) and mitochondria (mitoGFP) were estimated by counting in one segmental nerve per larva (n = 5 larvae per genotype) the number of clearly defined organelles per unit time that entered the field of view moving in either the anterograde (charted above the origin) or retrograde (below the origin) directions. Because of the low abundance of distinct syt::GFP punctae, flux approximations were made by dividing the total number of directionally transported organelles seen anywhere in the nerve by the total time of imaging. Genotypes were wild-type (+), unc-104O1.2/unc-104P350 (1.2), and unc-104O3.1/unc-104P350 (3.1). (B) Net velocity for a single organelle is a summation of all its position changes divided by total time. Means (±SEM) were determined for five organelles in each direction from five larvae for each genotype, except for STVs in which all distinct organelles were tracked (see Table 1 for STV sample sizes). For both A and B, differences between wild-type (unshaded bars) and unc-104 mutant (shaded bars) means were assessed using F-tests for variance followed by two tailed t tests at either equal or unequal variance with 95% confidence intervals. Significant differences for a given organelle type are indicated by asterisks (p < 0.05).
Table 2.
Comparison of anterograde run velocities; Unc-104-GFP versus GFP-organelles
| Run velocity (μm/s)a | n | p value | |
|---|---|---|---|
| Unc-104-GFP | 1.05 ± 0.08 | 15 | |
| ANF::GFP (DCVs) | 1.14 ± 0.06 | 25 | <0.38 |
| Syt::GFP (STVs) | 0.84 ± 0.05* | 25 | <0.03 |
| MitoGFP (mitochondria) | 0.30 ± 0.01* | 25 | <0.006 |
a Unc-104::GFP particles were tracked as described for Table 1. Mean anterograde run velocities (±SEM) were compared using F-tests for variance and then t tests with unequal or equal variance. Velocities that are significantly different from that of Unc-104::GFP at >95% confidence are noted with an asterisk.
Organelle flux was calculated for mitoGFP and ANF::GFP by counting the number of fluorescent organelles that moved past the anterior and posterior bleach boundaries per minute. MitoGFP flux was measured from 300 frames of each image series. ANF::GFP organelles were sufficiently abundant that accurate counts could be obtained from 200 frames. Because of lower numbers of clearly distinguishable syt::GFP organelles the total number of directionally transported punctae were counted and divided by the total elapsed time; thus, syt::GFP flux values are not directly comparable with those for the other two organelle types.
RESULTS
A Drosophila Kinesin-3 with Essential Functions in Neurons
Using a whole eye mosaic screen for mutations that disrupt photoreceptor connectivity in the Drosophila visual system (Newsome et al., 2000), we isolated eight alleles of a gene that encode a 1671 amino-acid kinesin-3. One of four Drosophila kinesin-3 family members, phylogenetic comparisons of motor domain sequences places it in a clade that includes Caenorhabditis elegans UNC-104 and human Kif-1A, B, and C (Miki et al., 2005; Wickstead and Gull, 2006). A cDNA sequence for the gene was submitted to GenBank as dunc-104 (accession no. AF247761). The gene was subsequently renamed unc-104 by Flybase in accordance with standard Drosophila nomenclature rules (Flybase report FBrf0129389). Sequencing revealed that four of the eight unc-104 alleles isolated had nonsense mutations (Supplemental Table S1). All heteroallelic combinations of the eight original alleles tested caused similar late embryonic lethality, suggesting that all were amorphic (functionally equivalent to nulls).
To facilitate study of the functions of Drosophila Unc-104 in a mature nervous system with minimal developmental or pleiotropic defects that arise from the amorphic genotypes, we conducted an F2 lethal screen for hypomorphic (partial-loss-of-function) alleles. Two were isolated and characterized (unc-104O1.2 and unc-104O3.1). When combined with a nonsense allele (unc-104P350), unc-104O3.1 caused lethality in the larval stages such that late third instars were rare; however, unc-104O1.2 allowed development through the third instar and into the pupal stages (Supplemental Table S1). Observation of both types of hypomorphic mutant larvae revealed sluggish, somewhat uncoordinated crawling movements, consistent with neuronal defects. However, there was no sign of the dystonic posterior paralysis (tail flipping) phenotype that is characteristic of even nonlethal hypomorphic genotypes for Drosophila Khc, which encodes the central subunit of the classic axonal transport motor kinesin-1 (Saxton et al., 1988; Yang et al., 1988; Saxton et al., 1991; Brendza et al., 1999).
To test the possibility that Unc-104 has essential functions in neurons, we determined if Gal4-UAS controlled neuron-specific expression of a GFP-tagged wild-type unc-104 cDNA transgene could prevent the lethality caused by unc-104O1.2 or unc-104O3.1/unc-104P350. Microscopy of larvae in which Unc-104::GFP expression was induced by the “motoneuron driver” D42-Gal4 (Rao et al., 2001; Pilling et al., 2006) showed fluorescence in the larval brain, in segmental nerves that contain motor axons, and at motor axon terminals on bodywall muscles. Some of the fluorescence in nerves was in large immobile inclusions, perhaps representing Unc-104::GFP aggregates. The remainder was diffuse, with occasional small fluorescent particles that moved in both anterograde and retrograde directions. Their mean anterograde velocity was 1.05 ± 0.08 μm/s (SEM), consistent with previous studies of kinesin-3 motors in vivo (Zhou et al., 2001; Lee et al., 2003) and in vitro (Nangaku et al., 1994; Okada et al., 1995). Unc-104-GFP expression in unc-104O1.2/unc-104P350 and unc-104O3.1/unc-104P350 mutants showed fluorescence distributions similar to wild type and rescued mutant lethality, allowing the development of adults. The rescue was partial, however, because some animals died as pharate adults, and mature adults were behaviorally depressed. However, the marked suppression of lethality by neuronal expression of the GFP-tagged wild-type protein indicates both that the Unc-104::GFP is functional and that Unc-104 is critical in neurons. This and its identity as an anterograde kinesin-3 support the premise that Drosophila Unc-104 makes essential contributions to anterograde axonal transport.
Terminal Atrophy, but No Focal Axonal Organelle Accumulations in unc-104 Mutants
Mutations that inhibit kinesin-1 and cytoplasmic dynein, the known major axonal transport motors in Drosophila, cause motoneuron terminal atrophy and focal axonal swellings that are filled with organelles (Hurd and Saxton, 1996; Gindhart et al., 1998; Bowman et al., 1999; Martin et al., 1999; Pilling et al., 2006). The terminal atrophy is likely due to impaired trafficking of structural and trophic factors needed to build and sustain terminals. The focal organelle accumulations were originally termed organelle jams or clogs, reflecting the idea that they form because of general steric hindrance of transport caused by stalled axonal organelles.
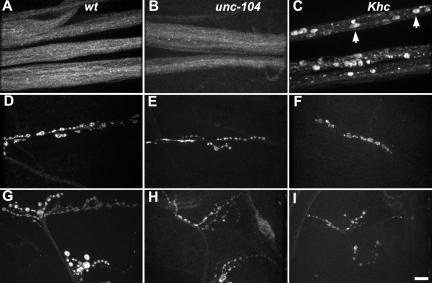
To test for similar axonal transport phenotypes in unc-104 mutants, we compared the distribution of two proteins that associate with small transport vesicles (STVs), cysteine string protein (CSP) and synaptotagmin, in wild-type, unc-104 mutant and Khc mutant larvae, by using immunofluorescence microscopy (Figure 1 and Supplemental Figure S1). In segmental nerves of unc-104 mutants, as in wild type, staining was diffuse, finely punctate, and evenly distributed. This was in marked contrast to Khc mutant nerves, in which CSP and synaptotagmin were concentrated in the large accumulations typical of focal axonal swellings (Figure 1C and Supplemental Figure S1D). Because even nonlethal Khc mutant genotypes cause swellings (Martin et al., 1999), whereas relatively severe unc-104 lethal genotypes do not, it is unlikely that the phenotypic difference is due to differences in allele severity. This argues that kinesin-1 and Unc-104 have important functional differences in Drosophila neurons and that focal swellings may reflect specific rather than general defects in axonal organelle transport mechanisms.
Figure 1.
Influence of unc-104 mutations on CSP distribution in larval axons. CSP, a vesicle associated protein, was immunolocalized in wild-type (A, D, and G), unc-104O1.2/unc-104P350 mutant (B, E, and H), and Khc6/Khc27 mutant (C, F, and I) third instars. (A–C) Segmental nerves. (D–F) Motor axon terminals on muscle 6/7. (G–I) Motor axon terminals on muscles 12/13. Note that unc-104 mutations did not cause CSP accumulation in the sort of focal axonal swellings that are caused by Khc mutations (short arrows in C). However, unc-104 mutations did seem to cause a reduction in terminal size that was particularly noticeable for the terminals on muscles 12/13 (G–I). Bar, 10 μm.
To determine if Unc-104 influences axon terminal organization, CSP was imaged at neuromuscular junctions on muscles 6/7 and 12/13 (Figure 1, D–I). The axons that innervate 12/13 produce a highly branched terminal structure with boutons of diverse sizes. Those that innervate 6/7 are less branched with less diverse bouton sizes (Jia et al., 1993). In unc-104 mutants, both terminal types seemed reduced in size relative to wild-type. To quantify this, we compared the number of boutons per 12/13 terminal. The mean for wild-type was 86.6 ± 16.6 (SEM). The large variance was due to substantial differences in the lengths of thin neurites that produce small boutons (type II). The means for unc-104O1.2 or unc-104O3.1/unc-104P350 were significantly reduced to 41.0 ± 2.1 and 40.2 ± 1.8, respectively (p = 0.05, n = 5 larvae per genotype). The small variances were in part due to the fact that few type II boutons were visible. These terminal defects in larvae corroborate a recent detailed study of terminal development in unc-104 (imac) mutant embryos that defines essential functions for Unc-104/imac in embryonic bouton formation and synapse development (Pack-Chung et al., 2007). In summary, these observations suggest that despite lack of the classic tail flipping and focal swelling phenotypes caused by kinesin-1 inhibition, Drosophila Unc-104, like kinesin-1 is required for normal axon terminal development. This is consistent with both motors functioning in the transport of structural and/or signaling materials to the axon terminal.
Unc-104 Specifically Affects the Distribution and Movement of Neuropeptide Vesicles
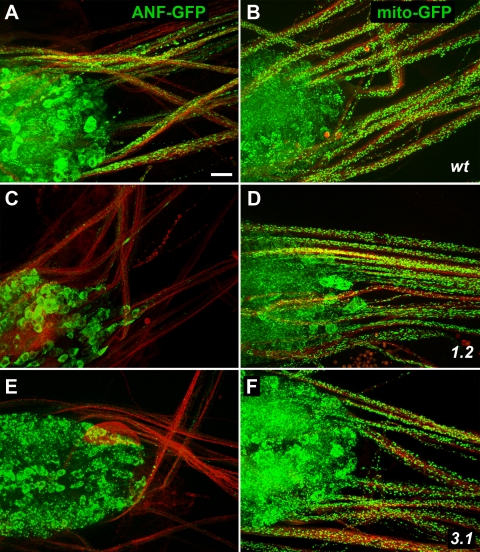
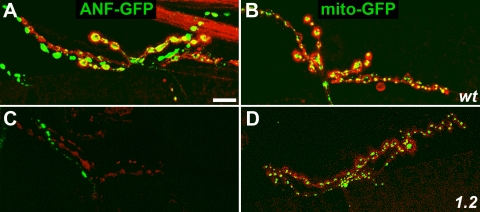
To test the influence of Unc-104 on specific axonal organelle transport, the expression of mito::GFP and ANF::GFP, targeted to the matrix of mitochondria and the lumens of neuropeptide-bearing large dense-core vesicles (DCVs), respectively, was driven by D42-Gal4. The genetic load associated with the driver-responder chromosome hindered growth of unc-104O3.1/unc-104P350 third instars, so most experiments focused on unc-104O1.2/unc-104P350 mutants. Larvae were fixed and immunostained with anti-syntaxin to allow imaging of axonal membranes, and then they were examined by fluorescence microscopy (Figures 2 and 3). In wild type, GFP fluorescence was intense in ventral ganglia, bright punctae were scattered along axons, and motoneuron terminals were strongly fluorescent. In unc-104 mutants, the distribution of mitochondria in ventral ganglia and axons was not distinguishable from the wild-type pattern. Mutant terminals were small, but mitochondria were clearly present. Although DCVs in unc-104 mutants were abundant in ventral ganglia, they were greatly reduced in axons and terminals. Interestingly, however, DCV fluorescence remained bright in one (occasionally two) thin 12/13 neurites in unc-104 mutants (Figure 3C). One explanation of this is that multiple neurons likely contribute to the 12/13 neuromuscular junction and that they have cell-specific variations in transport mechanisms such that terminal accumulation of DCVs in one neuron is relatively insensitive to Unc-104 inhibition.
Figure 2.
Effects of unc-104 mutations on organelle distribution in the ventral ganglion and motor axons. Wild-type (wt) (A and B), unc-104O1.2/unc-104P350 (1.2) (C and D), and unc-104O3.1/unc-104P350 (3.1) (E and F) larvae, in which D42-GAL4 induced expression of either a DCV tag (ANF::GFP; green in A, C, and E) or a mitochondrial tag (mitoGFP; green in B, D, and F). To allow imaging of axons independent of GFP presence, neuromuscular systems were fixed and immunostained with antibodies to syntaxin (red). Motoneuron cell bodies are located in the ventral ganglion (left side of each panel) and axons are in segmental nerves (extending to the right). Bar, 10 μm.
Figure 3.
Effects of unc-104 mutations on GFP-tagged organelle distribution in motor axon terminals. Wild-type (wt) (A and B) and unc-104O1.2/unc-104P350 mutant (1.2) (C and D) larvae with D42-GAL4–driven expression of ANF::GFP (green in A and C) or mitoGFP (green in B and D). Immunostaining with anti-syntaxin highlights axon plasma membranes (red). Portions of muscle 12/13 axon terminals in segment A4 are shown. Note in unc-104 mutants the absence of DCVs (ANF::GFP) and the aberrant terminal structure. Mitochondria remained abundant. Bar, 10 μm.
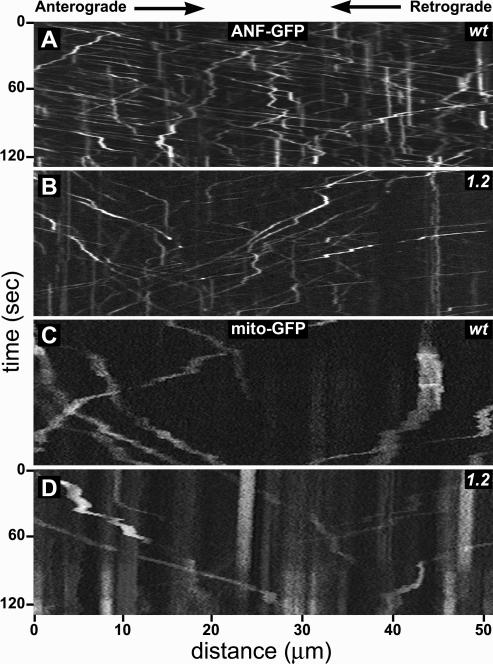
To identify Unc-104–dependent transport mechanisms more directly, wild-type and mutant larvae expressing GFP-organelle tags in motoneurons were dissected to expose segmental nerves, mounted in culture medium, and imaged at one to two frames per second (Supplemental Movies S1–S6). Kymographs from the movies (Figure 4) show that in wild-type axons, most DCVs were mobile and made long runs in one primary direction (diagonal line segments) interrupted by pauses (vertical line segments) and infrequent short runs in the opposite direction. Some DCVs, however, moved in an erratic manner, switching between anterograde and retrograde movement such that no primary direction of travel was evident. Kymographs of DCVs in unc-104 mutant nerves suggested a distinct decrease in both organelle abundance and velocity (Figure 4B). In wild-type axons, GFP-mitochondria were much less abundant than DCVs (Figure 4C). About one-half were stationary or oscillated slightly, consistent with previous studies (Hollenbeck and Saxton, 2005; Pilling et al., 2006). Mobile mitochondria showed slower overall transport rates (steeper line slopes) than DCVs and seemed to have shorter runs. Transport of mitochondria in unc-104 mutants and wild-type were not distinguishable in kymographs (Figure 4, C and D). These results agree with the organelle distribution studies shown in Figures 2 and 3, suggesting that Drosophila Unc-104 is important for the transport of DCVs but not for the transport of mitochondria.
Figure 4.
Live transport behavior of organelles in unc-104 mutant axons. Each panel, extracted from a time-lapse movie of GFP-organelles in motor axons of a larval segmental nerve, shows a kymograph representation of fluorescent organelle positions as a function of time. Anterograde movements have negative slopes, whereas retrograde movements have positive slopes. Stationary organelles appear as vertical streaks. Before each movie, the field of view was photobleached, which reduced signal from stationary organelles, allowing better contrast for organelles that subsequently moved into the bleached area. (A and B) ANF::GFP shows DCV behavior in wild-type (wt) and unc-104O1.2/unc-104P350 (1.2) axons. Note the lower abundance of anterograde DCVs and their slower movements (larger negative slopes) compared with wild type. (C and D) MitoGFP shows mitochondrial behavior. Intact time-lapse movies of organelle transport can been seen in Supplemental Movies S1–S6.
Unc-104 Has Direct Influences on DCV Flux in Axons
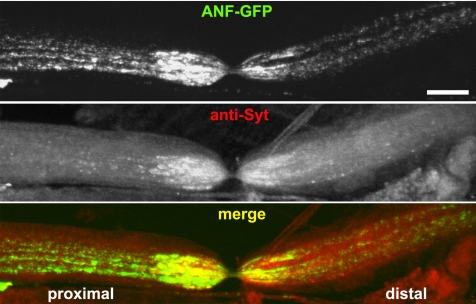
To better assess Unc-104 influences on general transport behaviors, flux and net transport rates of GFP-tagged DCVs, mitochondria, and STVs were quantified in wild-type and mutant motor axons (Figure 5 and Table 1). Consistent with the kymograph portrayal in Figure 4, flux for DCVs in wild-type axons was high, with a strong anterograde bias (3-fold). Flux for mitochondria was relatively low (∼20-fold fewer moving organelles), with a 1.7-fold anterograde bias. The flux of STVs tagged with syt::GFP was also low, and it seemed to be balanced with no evident anterograde bias. A previous study of ligated mouse sciatic nerves suggested that synaptotagmin is transported only anterograde (Yonekawa et al., 1998). To determine whether the retrograde syt::GFP transport we observed reflected normal behavior for endogenous synaptotagmin, we ligated wild-type larvae that expressed ANF::GFP in motoneurons. After fixation and immunostaining, anti-synaptotagmin signal was relatively even on proximal and distal sides of the ligation, consistent with retrograde and anterograde transport (Figure 6). In contrast, ANF::GFP signal showed a greater accumulation on the proximal sides of ligations, consistent with a substantial anterograde transport bias for DCVs. This suggests that syt::GFP does report accurately on the bidirectionality of endogenous synaptotagmin-bearing organelles in Drosophila. The contrast with the mouse sciatic nerve results may be due to differences in experimental approach, or it may reflect real differences in axonal transport mechanisms for flies and mice.
Figure 6.
Accumulation of axonal transport cargoes at a physical blockade. Live wild-type larvae with expression of ANF::GFP driven in motoneurons by D42-Gal4 were ligated with a fine thread for 2 h and subsequently dissected, fixed, and stained with antibodies to endogenous synaptotagmin (anti-Syt). Motoneuron cell bodies were to the left (proximal) and terminals were to the right (distal). Note that both proteins accumulated on both sides of the ligation-induced segmental nerve constriction, but although synaptotagmin seemed relatively balanced, ANF::GFP accumulated more heavily on the proximal side. Bar, 10 μm.
In unc-104 mutants, anterograde flux was significantly reduced for all three organelles (Figure 5A and Table 1). Relative to wild type, anterograde values for DCVs, STVs, and mitochondria were inhibited 3-fold, 5.2-fold, and 2-fold, respectively. This does not mean that Unc-104 is an anterograde motor for all three organelles. Flux is a complex parameter that encompasses both the abundance of moving organelles and their net velocity. To help distinguish abundance versus velocity effects, we quantified the movements of organelles that remained visible for long periods and that did not undergo conversions in primary transport direction (Pilling et al., 2006). For each organelle, the net distance traveled was divided by total tracking time, ignoring underlying run-pause behaviors. In unc-104 mutants, net velocity was reduced by 32% for anterograde DCVs, supporting a function for Unc-104 in anterograde DCV transport. This is consistent with a previous report of anterograde DCV transport by UNC-104 in C. elegans (Zahn et al., 2004), suggesting that the two proteins are orthologues. Mean net velocity for anterograde STVs in unc-104 mutants was reduced by 25% relative to wild type, but that difference was not significant. Net velocities for mitochondria in unc-104 mutants also were not different from wild type. These results suggest that the most of the reduction in anterograde STV and mitochondrial flux was due to reduced abundance of anterograde organelles. Relatively little is known about control of STV movement, but mitochondrial movement is known to be controlled by local axon metabolic needs and by other signals (Chada and Hollenbeck, 2004; Hollenbeck and Saxton, 2005). General changes in physiology caused by depressed Unc-104 driven anterograde vesicle transport may alter such signals and thus reduce the recruitment of STVs and mitochondria from cell bodies into axons. Alternatively, Unc-104 might function within the cell body to help initiate anterograde transport of STVs and mitochondria.
Unc-104 Has Specific Influences on Both Anterograde and Retrograde DCV Runs
To gain further insight into the transport functions of Unc-104, we compared run-pause parameters in wild-type and mutant motor axons. In unc-104O1.2 and unc-104O3.1/unc-104P350 axons, anterograde DCVs had 41 and 44% slower forward run velocities and 30 and 70% shorter run lengths than in wild type, respectively (Figure 7 and Table 1). Thus, the identity of Unc-104 as a member of an anterograde kinesin family, and the observation that mean anterograde DCV run velocity was similar to that of Unc-104::GFP particles (Table 2) agree that Drosophila Unc-104 serves as a major motor for anterograde DCV transport. Tracking of syt::GFP and mito::GFP in unc-104O3.1/unc-104P350 animals was not feasible due to the scarcity and small size of late third instars of the correct genotypes, so analysis was focused on unc-104O1.2/unc-104P350 animals. Anterograde STV run velocity was significantly reduced, and there was a shift from time spent in anterograde runs to time spent paused. This is consistent with anterograde transport of some STVs by Unc-104 as suggested by studies of homologues in C. elegans and mouse (Hall and Hedgecock, 1991; Yonekawa et al., 1998). There was no reduction in anterograde run parameters for mitochondria; in fact, there was a slight increase in run velocity, suggesting again that mitochondrial transport is sensitive to axon physiology changes.
Surprisingly, unc-104 mutations compromised retrograde DCV run parameters, causing significant reductions in run velocity and length (Figure 7 and Table 1). This suggests that Unc-104 has an important positive influence on retrograde transport by cytoplasmic dynein. Previous studies in larval motor axons, by using the same approaches used here, showed that Khc mutations inhibited retrograde flux of mitochondria. Part of the retrograde effect was suggested to be due to reduced anterograde delivery of dynein into distal parts of the axon, which should decrease axonal dynein concentration and thus hinder retrograde transport of all organelles (Pilling et al., 2006). Our data indicate that unc-104 mutations caused no reductions in retrograde flux or specific retrograde run parameters for STVs and mitochondria. This suggests that the negative effects of unc-104 mutations on retrograde DCV run velocity and length were not due to general effects, such as decreased axonal dynein concentration. Rather, Unc-104 may specifically enhance the transport performance of a DCV-associated dynein motor complex.
DISCUSSION
To gain insight into mechanisms of axonal transport, we studied the consequences of inhibition of a kinesin-3 in Drosophila. The founder of the kinesin-3 subfamily, UNC-104, was discovered as a C. elegans protein required for coordinated crawling behavior (Otsuka et al., 1991). Subsequent studies of C. elegans UNC-104, mammalian Kif1A, B, and other kinesin-3 family members have revealed that different kinesin-3 motors, which move relatively fast toward microtubule plus-ends, can transport a variety of different cargoes, including endosomes, mitochondria, and various vesicles (Hall and Hedgecock, 1991; Nangaku et al., 1994; Okada et al., 1995; Wedlich-Soldner et al., 2002; Zahn et al., 2004). Our results indicate that Drosophila Unc-104 can carry at least two anterograde vesicle types in motor axons, ANF neuropeptide DCVs and synaptotagmin STVs. Unc-104 may also transport other types of organelles, but we found no evidence for transport of axonal mitochondria.
It is known that axonal transport involves the energetic motion of individual organelles, each pulled along cytoskeletal filaments by motor proteins (Chevalier-Larsen and Holzbaur, 2006). The time-lapse analysis reported here emphasizes how distinct the transport behaviors of different organelles can be, and it raises questions about what the mechanistic underpinnings of those differences are. One possibility is that velocity varies inversely with organelle size (Allen et al., 1982), implying that cytoplasmic resistance to movement (viscous drag) is a key determinant of transport behavior and thus of cargo distribution dynamics. Mitochondria in Drosophila larval axons range widely in length, up to several micrometers, and they have an average diameter of 150 nm (Hurd and Saxton, 1996; Pilling et al., 2006). DCVs are mostly spherical with diameters of about 100 nm (Renden et al., 2001). Mean DCV run velocity and length were, respectively, 4-fold and 20-fold greater than those of mitochondria, consistent with an inverse size–velocity relationship. However, although DCV diameter is two- to three-fold greater than that of STVs (∼30 nm) (Hurd and Saxton, 1996; Renden et al., 2001), means for DCV run velocity and length were, respectively, 1.5- and 4-fold greater than those of STVs. Furthermore, it was previously reported that run velocities for mitochondria in larval motor axons were independent of mitochondria lengths (Pilling et al., 2006). These observations argue that transport behavior is determined mainly by organelle identity and organelle-specific differences in transport mechanisms, rather than by differences in size-dependent viscous drag.
One likely source of transport mechanism differences is the intrinsic mechanochemical capabilities of different motors. The results presented here indicate that many anterograde DCVs in Drosophila motor axons use Unc-104 (kinesin-3). Previous work in the same system showed that anterograde mitochondria use Khc (kinesin-1). DCV runs have higher velocity and longer anterograde runs than mitochondria, consistent with in vitro tests showing that dimeric Unc-104 constructs move with higher velocity and processivity than dimeric Khc constructs (Tomishige et al., 2002). This sort of straightforward mechanochemical difference, however, fails to explain why synaptotagmin-tagged STVs, which also use Unc-104, have slower, shorter runs than DCVs. Furthermore, retrograde run velocities and lengths that we measured for the three organelle types were quite different, despite the fact that cytoplasmic dynein heavy chain (Dhc64C) is the only known fast retrograde microtubule motor available in Drosophila (Walker et al., 1990; Rasmusson et al., 1994; Goldstein and Gunawardena, 2000). Thus, although differences in the mechanochemical properties of motors are important to differential organelle transport behavior, it seems clear that motor performance can be influenced by cargo identity.
Cargo-specific factors that might alter the output of a motor include posttranslational motor modification, motor-cargo linkage proteins, and the presence of other motors on the same organelle (Schnapp, 2003; Mallik and Gross, 2004). Kinesin-3s are reported to be monomeric in vitro (Okada et al., 1995), and individual monomers move slowly on microtubules. However, artificially induced dimerization allows faster more processive motion, supporting the hypothesis that clustering of motors on an organelle may be an important determinant of transport behavior (Okada and Hirokawa, 1999; Tomishige et al., 2002). Because Unc-104 may link directly to vesicle membranes via an FH lipid anchor domain, a variation in clustering controlled by lipid raft dynamics could produce variation in velocity and processivity (Klopfenstein et al., 2002; Klopfenstein and Vale, 2004). In addition, some cargoes are known to use multiple types of anterograde motors. Recent studies have shown that two different kinesins with distinct velocities, when active on the same dendritic cargo, generate motion at an intermediate velocity (Pan et al., 2006). Thus, the slower velocities of the STVs reported here might reflect mixed use of fast Unc-104 and slower Khc, whereas faster DCV velocities could reflect clusters of Unc-104 alone.
Our organelle tracking results suggest a specific positive influence of anterograde Unc-104 on retrograde DCV run velocity and length. A previous study of mitochondrial transport in Drosophila axons showed that kineisn-1 is critical for the dynein-driven retrograde flux of mitochondria (Pilling et al., 2006). Although that sort of positive influence of an opposing motor might reflect a direct physical interaction between kinesin-1 and the dynein complex (Ligon et al., 2004), it could also reflect simple logistical dependence. First, for normal numbers of mitochondria to move retrograde, normal numbers must be transported anterograde. Because kineisn-1 is the anterograde motor, Khc mutations result in low numbers of mitochondria in distal axons (Pilling et al., 2006). Second, dynein itself must be transported to the distal axon, before it can function in retrograde transport, and kinesin-1 is likely responsible for some of that anterograde dynein movement (Brendza et al., 2002; Lenz et al., 2006; Pilling et al., 2006). In contrast, the retrograde DCV run velocity and length decreases we observed in unc-104 mutant axons were not general, i.e., for STVs or mitochondria, statistically significant decreases in retrograde run velocity or length were not seen. This suggests that Unc-104 has an organelle-specific positive influence on the function of DCV-bound dynein.
How could Unc-104 contribute to DCV retrograde transport? First, it might be responsible for delivering DCV-specific dynein regulatory factors into the axon that enhance retrograde run velocity and length. This would require no specific association of Unc-104 with retrograde organelles. However, the fact that retrograde movement of Unc-104::GFP has been observed in axons of C. elegans (Zhou et al., 2001) and Drosophila (this report), along with a report that C. elegans UNC-104 is a retrograde cargo of dynein (Koushika et al., 2004) suggest more direct possibilities. First, DCV-specific motor docking complexes might juxtapose anterograde and retrograde motors such that Unc-104 itself acts as an allosteric activator for dynein. Second, Unc-104 on DCVs might facilitate their retrograde transport biophysically, for example, intermittently generating reverse strain and motion that helps dynein-DCV complexes get past steric barriers in the axon.
It is apparent that neurons use a diverse array of microtubule-based transport mechanisms to support long axons. Each type of organelle, RNP, and protein complex should have an ideal distribution and replacement rate for maintaining proper axon physiology and function. Thus, although it seems that only a few basic force-generating motors are used, diversity in their transport output via cargo-specific motor-motor influences and other regulatory schemes is likely important for optimizing nervous system function (Wong et al., 2002). Because motor proteins have complex effects on multiple processes in neurons and other cells, identifying cargo-specific motor control factors will be important, both for understanding the basic mechanisms of cytoplasmic organization and for providing new potential targets for drugs that can slow the progress of axonal transport-related neurodegenerative diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joe Duffy for advice and reagents, Aaron Pilling and Curt Lively for help with statistics, James Powers for microscope development, and Daniel S. Saxton for meiotic recombination mapping of P{w+mC = UAS-mitoGFP.AP}3 and P{GawB}D42. This work was supported by National Institutes of Health GM-46295 (to W.M.S.), Boehringer Ingelheim (to B.J.D.), and predoctoral fellowships from the American Heart Association, Midwest Affiliate (to R.V.B. and D.H.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0261) on November 7, 2007.
REFERENCES
- Allen R. D., Metuzals J., Tasaki I., Brady S. T., Gilbert S. P. Fast axonal transport in squid giant axon. Science. 1982;218:1127–1129. doi: 10.1126/science.6183744. [DOI] [PubMed] [Google Scholar]
- Bowman A. B., Patel-King R. S., Benashski S. E., McCaffery J. M., Goldstein L. S., King S. M. Drosophila roadblock and Chlamydomonas LC 7, a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 1999;146:165–180. [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brendza K. M., Rose D. J., Gilbert S. P., Saxton W. M. Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J. Biol. Chem. 1999;274:31506–31514. doi: 10.1074/jbc.274.44.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza R. P., Serbus L. R., Saxton W. M., Duffy J. B. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 2002;12:1541–1545. doi: 10.1016/s0960-9822(02)01108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chada S. R., Hollenbeck P. J. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr. Biol. 2004;14:1272–1276. doi: 10.1016/j.cub.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Chevalier-Larsen E., Holzbaur E. L. Axonal transport and neurodegenerative disease. Biochim. Biophys. Acta. 2006;1762:1094–1108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Duncan J. E., Goldstein L. S. The genetics of axonal transport and axonal transport disorders. PLoS Genet. 2006;2:e124. doi: 10.1371/journal.pgen.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindhart J. G., Jr, Desai C. J., Beushausen S., Zinn K., Goldstein L. S. Kinesin light chains are essential for axonal transport in Drosophila. J. Cell Biol. 1998;141:443–454. doi: 10.1083/jcb.141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. S., Gunawardena S. Flying through the Drosophila cytoskeletal genome. J. Cell Biol. 2000;150:F63–F68. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Hedgecock E. M. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Hata Y., Slaughter C. A., Sudhof T. C. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Heidemann S. R., Landers J. M., Hamborg M. A. Polarity orientation of axonal microtubules. J. Cell Biol. 1981;91:661–665. doi: 10.1083/jcb.91.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J. J., Raff R. A. Evolutionary change in the process of dorsoventral axis determination in the direct developing sea urchin, Heliocidaris erythrogramma. Dev. Biol. 1990;141:55–69. doi: 10.1016/0012-1606(90)90101-n. [DOI] [PubMed] [Google Scholar]
- Hollenbeck P. J., Saxton W. M. The axonal transport of mitochondria. J. Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi D., Barkus R. V., Pilling A. D., Gassman A., Saxton W. M. APLIP1, a kinesin binding JIP-1/JNK scaffold protein, influences the axonal transport of both vesicles and mitochondria in Drosophila. Curr. Biol. 2005;15:2137–2141. doi: 10.1016/j.cub.2005.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd D. D., Saxton W. M. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. X., Gorczyca M., Budnik V. Ultrastructure of neuromuscular junctions in Drosophila: comparison of wild type and mutants with increased excitability. J. Neurobiol. 1993;24:1025–1044. doi: 10.1002/neu.480240804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein D. R., Tomishige M., Stuurman N., Vale R. D. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein D. R., Vale R. D. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell. 2004;15:3729–3739. doi: 10.1091/mbc.E04-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S. P., Schaefer A. M., Vincent R., Willis J. H., Bowerman B., Nonet M. L. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J. Neurosci. 2004;24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. R., Shin H., Ko J., Choi J., Lee H., Kim E. Characterization of the movement of the kinesin motor KIF1A in living cultured neurons. J. Biol. Chem. 2003;278:2624–2629. doi: 10.1074/jbc.M211152200. [DOI] [PubMed] [Google Scholar]
- Lenz J. H., Schuchardt I., Straube A., Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon L. A., Tokito M., Finklestein J. M., Grossman F. E., Holzbaur E. L. A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J. Biol. Chem. 2004;279:19201–19208. doi: 10.1074/jbc.M313472200. [DOI] [PubMed] [Google Scholar]
- Littleton J. T., Bellen H. J., Perin M. S. Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development. 1993;118:1077–1088. doi: 10.1242/dev.118.4.1077. [DOI] [PubMed] [Google Scholar]
- Mallik R., Gross S. P. Molecular motors: strategies to get along. Curr. Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- Martin M., Iyadurai S. J., Gassman A., Gindhart J. G., Jr, Hays T. S., Saxton W. M. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Okada Y., Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Nangaku M., Sato-Yoshitake R., Okada Y., Noda Y., Takemura R., Yamazaki H., Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Newsome T. P., Asling B., Dickson B. J. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Okada Y., Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- Okada Y., Yamazaki H., Sekine-Aizawa Y., Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Otsuka A. J., Jeyaprakash A., Garcia-Anoveros J., Tang L. Z., Fisk G., Hartshorne T., Franco R., Born T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- Pack-Chung E., Kurshan P. T., Dickman D. K., Schwarz T. L. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat. Neurosci. 2007;10:980–989. doi: 10.1038/nn1936. [DOI] [PubMed] [Google Scholar]
- Pan X., Ou G., Civelekoglu-Scholey G., Blacque O. E., Endres N. F., Tao L., Mogilner A., Leroux M. R., Vale R. D., Scholey J. M. Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J. Cell Biol. 2006;174:1035–1045. doi: 10.1083/jcb.200606003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling A. D., Horiuchi D., Lively C. M., Saxton W. M. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Lang C., Levitan E. S., Deitcher D. L. Visualization of neuropeptide expression, transport, and exocytosis in Drosophila melanogaster. J. Neurobiol. 2001;49:159–172. doi: 10.1002/neu.1072. [DOI] [PubMed] [Google Scholar]
- Rasmusson K., Serr M., Gepner J., Gibbons I., Hays T. S. A family of dynein genes in Drosophila melanogaster. Mol. Biol. Cell. 1994;5:45–55. doi: 10.1091/mbc.5.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E., et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am. J Hum. Genet. 2002;71:1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renden R., Berwin B., Davis W., Ann K., Chin C. T., Kreber R., Ganetzky B., Martin T. F., Broadie K. Drosophila CAPS is an essential gene that regulates dense-core vesicle release and synaptic vesicle fusion. Neuron. 2001;31:421–437. doi: 10.1016/s0896-6273(01)00382-8. [DOI] [PubMed] [Google Scholar]
- Saxton W. M., Hicks J., Goldstein L. S., Raff E. C. Kinesin heavy chain is essential for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell. 1991;64:1093–1102. doi: 10.1016/0092-8674(91)90264-y. [DOI] [PubMed] [Google Scholar]
- Saxton W. M., Porter M. E., Cohn S. A., Scholey J. M., Raff E. C., McIntosh J. R. Drosophila kinesin: characterization of microtubule motility and ATPase. Proc. Natl. Acad. Sci. USA. 1988;85:1109–1113. doi: 10.1073/pnas.85.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp B. J. Trafficking of signaling modules by kinesin motors. J. Cell Sci. 2003;116:2125–2135. doi: 10.1242/jcs.00488. [DOI] [PubMed] [Google Scholar]
- Senti K. A., Usui T., Boucke K., Greber U., Uemura T., Dickson B. J. Flamingo regulates R8 axon-axon and axon-target interactions in the Drosophila visual system. Curr. Biol. 2003;13:828–832. doi: 10.1016/s0960-9822(03)00291-4. [DOI] [PubMed] [Google Scholar]
- Tomishige M., Klopfenstein D. R., Vale R. D. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- Vale R. D., Milligan R. A. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Walker R. A., Salmon E. D., Endow S. A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Straube A., Friedrich M. W., Steinberg G. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 2002;21:2946–2957. doi: 10.1093/emboj/cdf296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol. Biol. Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R. W., Setou M., Teng J., Takei Y., Hirokawa N. Overexpression of motor protein KIF17 enhances spatial and working memory in transgenic mice. Proc. Natl. Acad. Sci. USA. 2002;99:14500–14505. doi: 10.1073/pnas.222371099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. T., Saxton W. M., Goldstein L. S. Isolation and characterization of the gene encoding the heavy chain of Drosophila kinesin. Proc. Natl. Acad. Sci. USA. 1988;85:1864–1868. doi: 10.1073/pnas.85.6.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa Y., Harada A., Okada Y., Funakoshi T., Kanai Y., Takei Y., Terada S., Noda T., Hirokawa N. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J. Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn T. R., Angleson J. K., MacMorris M. A., Domke E., Hutton J. F., Schwartz C., Hutton J. C. Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic. 2004;5:544–559. doi: 10.1111/j.1600-0854.2004.00195.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Rodesch C. K., Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- Zhao C., et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- Zhou H. M., Brust-Mascher I., Scholey J. M. Direct visualization of the movement of the monomeric axonal transport motor UNC-104 along neuronal processes in living Caenorhabditis elegans. J. Neurosci. 2001;21:3749–3755. doi: 10.1523/JNEUROSCI.21-11-03749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier K. E., Eberle K. K., Buchner E., Walter N., Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.