Abstract
Under certain conditions of nutrient stress, the budding yeast Saccharomyces cerevisiae initiates a striking developmental transition to a filamentous form of growth, resembling developmental transitions required for virulence in closely related pathogenic fungi. In yeast, filamentous growth involves known mitogen-activated protein kinase and protein kinase A signaling modules, but the full scope of this extensive filamentous response has not been delineated. Accordingly, we have undertaken the first systematic gene disruption and overexpression analysis of yeast filamentous growth. Standard laboratory strains of yeast are nonfilamentous; thus, we constructed a unique set of reagents in the filamentous Σ1278b strain, encompassing 3627 integrated transposon insertion alleles and 2043 overexpression constructs. Collectively, we analyzed 4528 yeast genes with these reagents and identified 487 genes conferring mutant filamentous phenotypes upon transposon insertion and/or gene overexpression. Using a fluorescent protein reporter integrated at the MUC1 locus, we further assayed each filamentous growth mutant for aberrant protein levels of the key flocculence factor Muc1p. Our results indicate a variety of genes and pathways affecting filamentous growth. In total, this filamentous growth gene set represents a wealth of yeast biology, highlighting 84 genes of uncharacterized function and an underappreciated role for the mitochondrial retrograde signaling pathway as an inhibitor of filamentous growth.
INTRODUCTION
In its most familiar growth form, the baker's yeast Saccharomyces cerevisiae divides mitotically by budding, forming two independent and separate daughter cells from a single mother cell. In response to specific environmental cues, however, some strains of S. cerevisiae are capable of forming multicellular filaments—chains of cells that remain physically connected after cytokinesis (Gimeno et al., 1992; Kron, 1997; Madhani and Fink, 1998). In yeast, this form of filamentous growth is thought to constitute a foraging mechanism initiated under conditions of limited nutrient availability (Gimeno et al., 1992; Liu et al., 1993; Cullen and Sprague, 2000). Similar filamentous growth transitions are evident in many fungal species; in particular, many fungal pathogens transition between unicellular and filamentous growth forms, and, in fact, this transition is required for virulence in most of these organisms (Alspaugh et al., 1997; Lo et al., 1997b). For example, in the opportunistic human pathogen Candida albicans, environmental cues of temperature, pH, and serum source have been found to trigger a distinct morphogenetic program resulting in the transition from a cellular yeast form to a filamentous growth form (Liu et al., 1994; Singh et al., 1997). Furthermore, a mutant strain of C. albicans impaired in its ability to undergo filamentous growth is avirulent in a mouse model of disseminated candidiasis (Lo et al., 1997a). Thus, filamentous growth is relevant to our understanding of fungal pathogenesis, and the budding yeast has emerged as an informative model for studies of filamentous growth.
In S. cerevisiae, filamentous growth is induced under conditions of nitrogen stress or by growth in media containing short-chain alcohols (Gimeno et al., 1992; Lorenz et al., 2000; Miled et al., 2001). In each case, this environmental stress elicits substantive changes in yeast cell morphology and growth: the yeast cells delay in G2/M, exhibit an elongated morphology, display an altered budding pattern, remain physically attached, and invade their growth substrate (Kron et al., 1994). Collectively, this results in the formation of filamentous chains of elongated cells, called pseudohyphae. Specifically, a diploid strain of yeast extends pseudohyphal filaments over the surface of its growth substrate and downward into the agar medium; classically, this form of growth is referred to as diploid pseudohyphal growth. A haploid strain of yeast under conditions of nutrient sufficiency extends invasive filaments downward, but it does not extend surface-spread filaments; classically, this is referred to as haploid invasive growth (Gancedo, 2001). Alternative growth conditions, however, can induce surface filamentation in haploid yeast as well (Lorenz et al., 2000). Thus, to avoid confusion, we use the term filamentous growth and simply refer to the growth as occurring in a haploid or diploid strain of yeast.
Yeast filamentous growth is regulated by at least two known signaling pathways: 1) the nutrient-sensing cyclic AMP–protein kinase A (PKA) pathway and 2) a mitogen-activated protein kinase (MAPK) pathway (Buehrer and Errede, 1997; Madhani and Fink, 1997; Lengeler et al., 2000; Cullen et al., 2004). During filamentous growth, the GTP-binding protein Ras2p is activated through a sensor system that is poorly understood at present; Ras2p, in turn, stimulates the synthesis of cAMP, resulting in the activation of PKA (Robertson and Fink, 1998). PKA promotes filamentous growth, in part, by activating the key filamentous growth transcription factor Flo8p, a pseudogene in nonfilamentous lab strains of yeast (Pan and Heitman, 1999). The filamentous growth MAPK pathway encompasses the p21-activated kinase (PAK) Ste20p (Peter et al., 1996) and the MAPK cascade itself of Ste11p, Ste7p, and Kss1p (Cook et al., 1997). Kss1p activates a presumably large set of targets, including the transcription factor Ste12p, which heterodimerizes with the transcription factor Tec1p to regulate expression of genes contributing to filamentous growth (Liu et al., 1993; Madhani et al., 1997; Borneman et al., 2006).
At present, the cell-surface glycoprotein Muc1p (formerly Flo11p) is the most well-characterized downstream effector of yeast filamentous growth (Gagiano et al., 1999; Guo et al., 2000). Muc1p is required for cell–cell adhesion, or flocculation, and filamentous growth in budding yeast, and it is the only known target of both the PKA and MAPK pathways described above (Lo and Dranginis, 1996, 1998). MUC1 contains an unusually large 2.8-kb promoter with experimentally verified recognition sequences for the MAPK pathway-regulated transcription factor complex Ste12/Tec1p and for the PKA pathway-regulated transcription factor Flo8p (Rupp et al., 1999; Borneman et al., 2006). Accordingly, MUC1 is expressed at very low levels under normal conditions of vegetative growth, and it is induced during filamentous growth (Caro et al., 1997; Strittmatter et al., 2006).
The MAPK and PKA pathways described above are incompletely defined, and they likely represent only a fraction of the signaling and metabolic pathways mediating yeast filamentous growth. Through a number of independent observations, reviewed in Rua et al. (2001), the filamentous growth PKA and MAPK modules have been linked with genes functioning in the establishment of cell polarity, bud site selection, and cell cycle progression. The extensive genetic and morphological changes underlying the transition to yeast filamentous growth suggest a very broad network of associated signaling pathways—a network that may be best investigated through systematic and large-scale approaches. To this end, we present the first systematic gene disruption and overexpression analysis of yeast filamentous growth. Using transposon-based directed allele replacement (Kumar et al., 2000, 2002c), we have generated 3627 transposon insertion mutants for subsequent phenotypic analysis; we also have individually introduced 2043 gene overexpression constructs into the same filamentous strain background to assess overexpression-induced filamentous growth phenotypes. Collectively, this study encompasses 4528 yeast genes (∼78% of the total gene complement in S. cerevisiae), identifying 487 genes contributing to filamentous growth. Among other interesting points, this gene set highlights the importance of mitochondrial function during filamentous growth, and through follow-up studies using targeted gene deletions, we identify the mitochondrial retrograde signaling pathway as a key negative regulator of yeast filamentous growth.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
All mutants in this study were constructed in strain Y825 derived from the filamentous Σ1278b genetic background. The genotype of Y825 is as follows: MATa ura3-52 leu2Δ0. Transposon insertion and gene overexpression mutants were generated as described below. Precise start codon-stop codon gene deletions were generated using the one-step gene replacement strategy of Baudin et al. (1993) with the KanMX6 disruption cassette from plasmid pFA6a-KanMX6 (Longtine et al., 1998) unless otherwise noted. Strains lacking mitochondrial DNA (rho0 strains) were generated by treatment with ethidium bromide (Sherman et al., 1994); the loss of mitochondrial DNA was confirmed by staining with 4,6-diamidino-2-phenylindole.
Filamentous growth was induced on solid medium according to standard protocols using low-nitrogen growth medium (2% glucose, 50 μm ammonium sulfate, 0.17% yeast nitrogen base without amino acid and ammonium sulfate, with essential amino acids for nutritional auxotrophies) supplemented with 1% (vol/vol) butanol (Lorenz et al., 2000). Filamentous growth was induced in liquid culture by using YPD rich medium (1% bacto yeast extract, 2% bacto peptone, and 2% glucose) supplemented with 1% butanol as described in Lorenz et al. (2000). Low-autofluorescence medium was prepared according to standard protocols for appropriate synthetic complete (SC) medium, substituting yeast nitrogen base (YNB) without ammonium sulfate, folic acid, and riboflavin for YNB without ammonium sulfate (Guthrie and Fink, 1991).
Construction of the Yeast Transposon Insertion and Gene Overexpression Collections in a Filamentous Strain of Yeast
Transposon insertion alleles were generated in a previous study (Ross-Macdonald et al., 1999; Kumar et al., 2002b, 2004). Insertion alleles likely to represent null alleles were determined manually by inspection of the insertion site relative to the open reading frame coding sequence. A representative and nonredundant collection of transposon insertion alleles was generated by cherry-picking individual bacterial cultures into 96-well plates; we prepared plasmid DNA from these cultures by using standard alkaline lysis protocols adapted for application in 96-well format. Plasmid DNA was digested with NotI to release inserts of yeast genomic DNA, each carrying a single transposon insertion; this DNA was introduced individually in 96-well format into strain Y825 by standard methods of lithium acetate-mediated transformation adapted for high throughput as described previously (Kumar et al., 2000, 2004). Transformants were selected on SC −Ura, and three independent transformants were saved per transformation.
Gene overexpression plasmids were constructed as reported in Kumar et al. (2002a), and they represent the set of genes successfully cloned into pYES2/GS by topoisomerase I-mediated ligation. In this study, overexpression plasmids were introduced individually into Y825 by the high-throughput lithium acetate-mediated transformation protocol referenced above, and transformants were selected on SC −Ura.
Phenotypic Profiling of Filamentous Growth Mutants
In yeast, filamentous growth phenotypes may be identified through a number of assays. Surface-spread filaments are readily apparent from visual inspection of colony morphology (Gimeno et al., 1992). Invasive filaments can be identified through a plate-washing assay, in which a gentle stream of water is run over a Petri plate streaked with yeast, such that surface-spread cells are washed away and invasive cells left behind (Gimeno et al., 1992). Filamentous growth also may be assessed in liquid culture, because filamentous cells exhibit an elongated morphology and cluster to a greater degree than nonfilamentous cells (Lorenz et al., 2000; Zeitlinger et al., 2003). Filamentation in haploid yeast is stimulated by growth in fusel alcohols; the short-chain alcohols are thought to mimic the end products of amino acid catabolism in nitrogen-poor conditions (Lorenz et al., 2000). The resulting filamentous growth form exhibits both surface-spread and invasive filaments. Thus, all of the assays described above may be used to determine alcohol-induced filamentous growth in haploid strains of yeast, and our screening approach used each of these methods. Here, phenotypes were assessed qualitatively and were binned in the indicated categories.
For this analysis, haploid yeast strains (Y825 background) carrying integrated transposon insertion alleles were arrayed in 96-well format for inoculation in liquid YPD supplemented with 1% (vol/vol) butanol. Mutants were grown in 96-well format with shaking at 30°C for ∼2–3 d, and they were subsequently screened for cell morphology and cell clustering by standard microscopy. Liquid cultures were spotted onto multiwell microscope slides, and they were imaged individually at 10× magnification. Each mutant strain was assayed for increased or decreased filamentous growth, as indicated by the observed degree of cell clustering and cell elongation; in our hands, this assay yielded the most obvious and easily distinguished filamentous growth phenotypes. Only insertion alleles for which at least two of three independent transformants scored positive were considered further. These mutants exhibiting altered filamentous growth phenotypes were examined on solid medium to define colony morphology and cell invasiveness. For this analysis, mutants were grown in liquid Sc −Ura medium overnight, and cultures were spotted in 96-well format on solid SC −Ura low nitrogen medium (50 μM ammonium sulfate) supplemented with 1% (vol/vol) butanol. Spotted cultures were grown at 30°C 3–5 d before analysis of surface-spread filamentation by standard microscopy. Only insertional mutants exhibiting filamentous growth phenotypes in both liquid and solid media were scored as filamentous growth mutants in this screen. Filamentous growth mutants also were assayed on solid medium for invasive phenotypes by the plate-washing assay described in Gimeno et al. (1992).
Gene overexpression mutants were analyzed similarly, except that transformants were sequentially grown in standard liquid SC −Ura medium for 1 d, in liquid SC −Ura medium with raffinose as the carbon source overnight, and in liquid SC −Ura medium with 1% (vol/vol) butanol and galactose as the carbon source for 2 d. Mutants were subsequently analyzed as described above. Overexpression strains exhibiting filamentous growth mutants in liquid culture were analyzed on solid growth medium for colony morphology and invasiveness. Mutants were initially grown in liquid SC −Ura medium overnight, and then they were spotted onto solid low-nitrogen medium (50 μM ammonium sulfate) with galactose as the carbon source and 1% (vol/vol) butanol. Cultures were grown 2–3 d at 30°C before examination by microscopy and by the plate-washing assay referenced above.
Determining Muc1p and Yef3p Abundance
To determine the abundance of Muc1p (as an indicator of filamentous growth) and Yef3p (as a general indicator of protein levels), we established a dual reporter system in which Muc1p is fused to green fluorescent protein (GFP) at its carboxy terminus, and Yef3p is fused to Discosoma sp. fluorescent protein (DsRed) at its carboxy terminus. Muc1p-GFP was constructed by integrating the GFP-KanMX6 cassette from pFA6a-KanMX6 (Longtine et al., 1998) at the 3′ end of MUC1 in Y825. To generate the Yef3p–DsRed fusion, we constructed a DsRed-URA3 cassette and integrated this cassette at the 3′ end of YEF3.
Transposon insertion mutants containing the dual reporters were incubated in liquid SC −Ura medium supplemented with 1% (vol/vol) butanol overnight, and they were then incubated in fresh medium at an optical density (650 nm) of 0.3 (∼3.9 × 106 haploid cells) for an additional 3 h of growth. Insertion mutants were analyzed for GFP and DsRed fluorescence in black 96-well assay plates by using the Victor2 multilabel counter (PerkinElmer-Cetus, Waltham, MA); cell density was determined using the ThermoMax microplate reader (Molecular Devices, Vienna, VA). Gene overexpression mutants were cultured in liquid as described above (phenotypic profiling), with 3 h of galactose induction before analysis. Insertion mutants and overexpression mutants of interest were also imaged by fluorescence microscopy to consider cell–cell variability in Muc1p and Yef3p abundance. To minimize autofluorescence, the media described above were made with YNB lacking folic acid and riboflavin (Guthrie and Fink, 1991).
RESULTS
Generating Transposon Insertion Alleles and Overexpression Constructs in a Filamentous Strain of Budding Yeast
Many large-scale phenotypic studies have been performed in S. cerevisiae by using the genome-wide deletion collection (Winzeler et al., 1999); however, this collection has been constructed in a nonfilamentous strain of yeast and is, therefore, inappropriate for studies of filamentous growth. Ideally, filamentous growth studies should be performed in the yeast strain Σ1278b, because this strain is known to undergo an extensive and easily controlled transition to filamentous growth (Grenson, 1966; Gimeno et al., 1992).
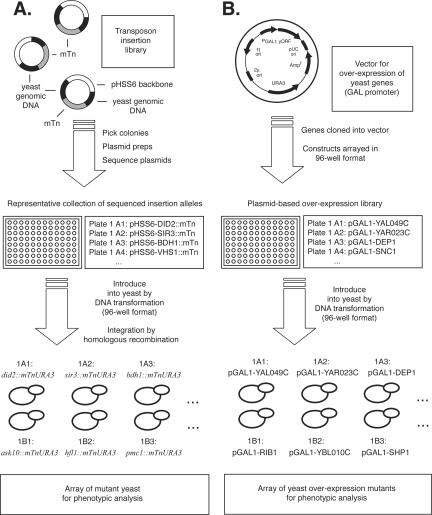
To construct a genome-wide set of gene disruption mutants in the Σ1278b genetic background, we used a plasmid-based collection of gene disruption alleles generated previously by large-scale mutagenesis of the yeast genome (Figure 1A) (Ross-Macdonald et al., 1999; Kumar et al., 2002c). The specifics of these insertion alleles are presented in Kumar et al. (2004). The assembled collection encompasses 3627 plasmid-based transposon insertion alleles (representing 63% of all annotated protein coding genes in yeast), in 38 96-well plates. Transposon insertion alleles were introduced individually into a haploid strain of the Σ1278b genetic background by using protocols described previously (Kumar et al., 2000). Yeast strains for overexpression analysis were generated using a collection of high-copy plasmids carrying genes cloned under transcriptional control of a galactose-inducible promoter (Figure 1B) (Kumar et al., 2002). In total, this plasmid-based collection encompasses 2043 yeast genes (35% of the yeast genome), and the identity of each gene has been verified by DNA sequencing. Overexpression constructs from this collection also were introduced into the filamentous Σ1278b strain for phenotypic analysis as outlined in Figure 1B.
Figure 1.
Construction of yeast gene disruption and overexpression arrays for phenotypic analysis of filamentous growth. (A) Diagrammatic representation of the plasmid-based mini-transposon (mTn) insertion alleles and the approach by which these insertion alleles were introduced into a filamentous strain of yeast. The transposon-mutagenized yeast DNA was introduced into the filamentous Σ1278b strain in 96-well format; thus, the final yeast gene disruption array reflects the same order as the arrayed transposon insertion plasmids. As indicated, the mTn carries the URA3 marker. (B) Overview of the gene overexpression plasmids and the construction of a corresponding yeast gene overexpression array in the Σ1278b strain. The overexpression plasmid carries the galactose-inducible GAL1 promoter; each yeast gene is maintained on this plasmid in the filamentous yeast strain.
Collectively, the yeast gene disruption and overexpression collections encompass 4528 genes (Table 1), representing 78% of the predicted gene complement in S. cerevisiae. These collections include 816 essential genes, 632 of which are represented in the transposon insertion collection. On introduction into haploid yeast, the majority (90%) of transposon insertions in essential genes failed to yield colonies, indicating that these insertion represented null alleles. Overexpression constructs of essential genes (184) yielded viable colonies upon introduction into yeast. Thus, 247 essential genes were analyzed in this study, largely by gene overexpression.
Table 1.
Summary of gene disruptions and overexpression analysis
| Results | No. (%)a |
|---|---|
| Gene disruption analysis | |
| Plasmid preps/yeast transformations | 3635 |
| Genes screened | 3627 |
| Identified filamentous growth genes | 309 (8.5) |
| Mutants with decreased filamentous growth | 240 (6.6) |
| Mutants with increased filamentous growth | 69 (1.9) |
| Noninvasive mutants | 243 (6.7) |
| Gene overexpression analysis | |
| Plasmid preps/yeast transformations | 2361 |
| Genes screened | 2043 |
| Identified filamentous growth genes | 199 (9.7) |
| Mutants with decreased filamentous growth | 43 (2.1) |
| Mutants with increased filamentous growth | 156 (7.6) |
| Noninvasive mutants | 38 (1.9) |
| Cumulative (nonredundant) | |
| Genes screened | 4528 |
| Identified filamentous growth genes | 487 (10.8) |
a Percentage of identified genes relative to total genes screened per study.
A Systematic Screen for Filamentous Growth Mutants
Haploid gene disruption and overexpression mutants were screened for filamentous growth phenotypes in the presence of short-chain alcohols to enable analysis of surface-spread filamentation, cell elongation/clustering, and cell invasiveness (see Materials and Methods). By this approach, we identified 487 genes (∼11% of all genes examined in this study) conferring filamentous growth phenotypes upon gene disruption and/or overexpression (Table 1). This gene set encompasses 309 genes, yielding mutant filamentous growth phenotypes by transposon-based gene disruption and 199 genes exhibiting filamentous growth phenotypes by overexpression. By plate-washing assays, 243 transposon insertion mutants and 38 gene overexpression mutants are noninvasive. Thus, surface-spread filamentation phenotypes do not always correlate with invasive growth phenotypes. Disruption mutants with filamentous growth phenotypes typically exhibited decreased filamentous growth (240 of 309 disruption mutants), and the mutants were generally noninvasive. Conversely, the majority of gene overexpression mutants with filamentous growth phenotypes exhibited exaggerated filamentation (156 of 199 overexpression mutants) and were invasive (Table 1). Few genes, however, exhibited both gene disruption and overexpression phenotypes, consistent with previous observations from large-scale deletion and overexpression studies in yeast (Sopko et al., 2006). Specifically, 1142 genes are redundant between the transposon insertion and overexpression mutant collections. Of these genes, 217 yielded a mutant phenotype in either screen, and 20 exhibited filamentous growth phenotypes as both disruption and overexpression alleles (e.g., the transcription factor PHD1 indicated in Figure 2, A and B). Six of these 20 genes are involved in the regulation of nitrogen metabolism, including the known filamentous growth genes PHD1, TEC1, and RAS2. It is also noteworthy that four of 20 genes (MGM101, MGR1, MSK1, and MSW1) are involved in mitochondrial functions; the importance of mitochondrial function during filamentous growth is addressed later in this text. Aside from PHD1, TEC1, and RAS2, these genes do not, however, exhibit any enrichment for genetic or physical interactions with each other or with known filamentous growth signaling modules or pathway components. The results from this analysis are summarized further in Table 1, and they are available in full as Supplemental Tables S1 and S2).
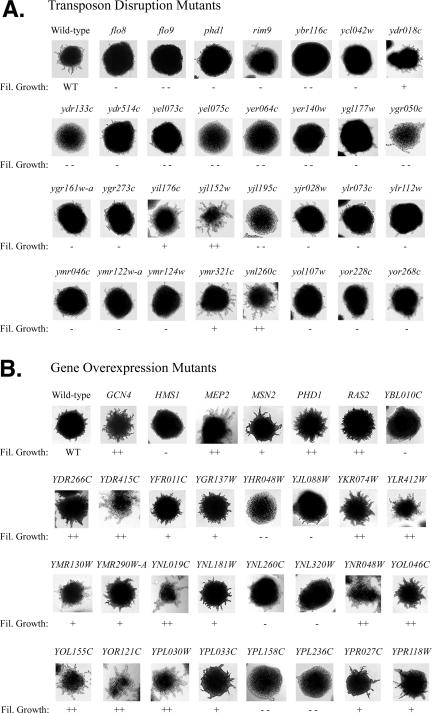
Figure 2.
A visual library of yeast filamentous growth mutants. (A) Colony morphologies of transposon-based gene disruption mutants. This sampling of images highlights a subset of previously uncharacterized genes exhibiting disruption phenotypes related to filamentous growth; for comparison, we have also included the wild-type background and mutants with transposon insertions in known filamentous growth genes (FLO8, FLO9, PHD1, and RIM9). All gene disruption mutants were spotted in 96-well format on solid SC – Ura low nitrogen medium (50 μM ammonium sulfate) supplemented with 1% (vol/vol) butanol, and mutants of interest were streaked for single colonies (shown here). The severity of the phenotype is indicated below each image (+, exaggerated filamentous growth; ++, strongly exaggerated filamentous growth; −, decreased filamentous growth; −−, strongly decreased filamentous growth). (B) Colony morphologies of gene overexpression mutants. To drive gene overexpression under conditions inducing filamentous growth, the mutants were grown in low-nitrogen SC −Ura with galactose and 1% butanol. Again, the images depict single colonies, and the severity of the phenotype is indicated below each image.
A Visual Library of Filamentous Growth Phenotypes
The filamentous growth mutants identified in this study present a striking array of colony morphologies; in particular, we highlight here a complement of previously uncharacterized genes presenting filamentous growth phenotypes upon transposon insertion or gene overexpression (Figure 2). By convention, these uncharacterized genes are listed by their systematic names—a standardized nomenclature applied to all yeast genes derived from the physical location of each open reading frame in the yeast genome (e.g., YBR116c). In total, 59 previously uncharacterized genes (with no common name or functional description available in the Saccharomyces Genome Database at www.yeastgenome.org as of April 2007) were identified in the disruption screen, and 27 uncharacterized genes were identified by gene overexpression; two genes (YKR074w and YNL260c) were identified in both screens. For comparison, we also include here several examples of functionally characterized genes previously known to affect filamentous growth upon gene disruption (FLO8, FLO9, PHD1, and RIM9) or gene overexpression (GCN4, HMS1, MEP2, MSN2, PHD1, and RAS2). The filamentous growth phenotypes resulting from disruption or overexpression of these known filamentous growth genes are comparable in severity to the observed phenotypes of the uncharacterized genes; thus, we expect these previously uncharacterized genes play significant roles in enabling wild-type filamentous growth.
Collectively, the images gathered in this study constitute a unique visual library of yeast filamentous growth phenotypes. To maximize the widespread utility of these data, we have captured an image of each filamentous growth mutant, and we have assembled these images into an archived file that can be freely downloaded. Information for downloading this file is presented as Supplemental Material. With this resource, users can inspect any specific mutants of interest in great detail. We have also assigned a broad score to each mutant, indicating the extent of its filamentous growth relative to wild type; in this way, mutants can be easily grouped by the severity of the phenotype. This data set is available in Supplemental Table S1.
A Genome-Wide Set of Genes Affecting Filamentous Growth
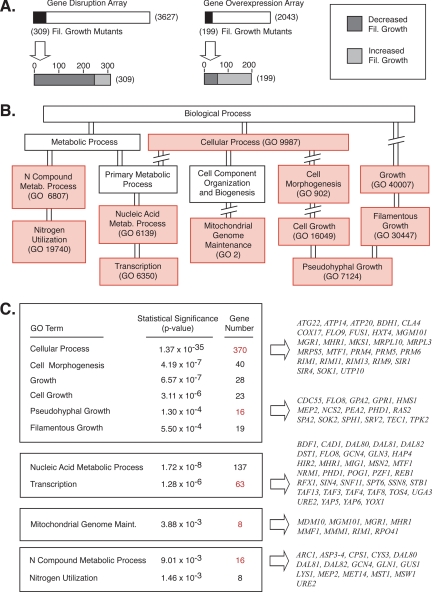
To characterize this filamentous growth gene set, we have annotated each gene identified in our screen with information from the Gene Ontology (GO) project, and we have searched the gene set for any enriched GO terms (Consortium, 2006). GO descriptors are available for three information categories: the cellular compartment to which the protein localizes, the molecular function for the gene product, and the biological process to which the gene contributes. In our gene set, we find no enrichment for proteins localized to a particular subcellular compartment and relatively little enrichment for proteins of a particular molecular function. In contrast, however, we see significant enrichment for gene products associated with a number of biological processes. Figure 3B presents a tree diagram highlighting GO biological process terms statistically enriched among the 487 nonredundant genes identified in this study. Specific biological processes are indicated in Figure 3C; genes from this screen associated with the indicated processes are listed here, along with p values describing the statistical significance of each enriched gene subset. Complete gene lists are presented in Supplemental Table S3.
Figure 3.
Statistically significant enrichment for gene functions in the filamentous growth mutants. (A) Overview of the gene disruption and overexpression screen results. (B) Tree diagram indicating related GO functional categories enriched in the filamentous growth gene set. GO biological process terms are indicated in boxes; biological processes enriched in the filamentous growth mutant data set are shaded in light red. The GO numerical identifier for each of these biological processes is indicated in parentheses. (C) The statistical significance of each enriched GO biological process term is indicated along with the actual number of identified genes belonging to this GO process category. As the GO categories can overlap significantly, we have indicated four general groupings (boxes), representing closely related processes. Sample gene subsets for at least one enriched GO biological process in each grouping are presented; the corresponding gene number for each of these listed gene sets is highlighted in red. Complete gene lists are available in Supplemental Table S3.
Not surprisingly, this data set is significantly enriched for genes known to affect filamentous growth (a subcategory of cell growth and morphology). By prior GO annotation, 65 genes have been associated with yeast pseudohyphal growth (Consortium, 2006). We identified 16 of these genes here (p value 1.3 × 10−4) (Figure 3C); 14 genes were not represented in either the transposon insertion or overexpression collection; thus, they were unavailable for analysis. To ensure that our scoring criteria are effective in identifying genes affecting filamentous growth, we generated gene disruption or overexpression constructs for 17 previously known filamentous growth genes absent from our plasmid-based allele collections: BEM1, BUD1, BUD5, BUD9, CDK1, CLB1, CLB2, CLN2, CLN3, ELM1, GRR1, MIH1, MSB3, MSS11, MUC1, and NDD1. Analysis of each mutant yielded observable filamentous growth phenotypes, suggesting that our scoring strategy is appropriate, although our screen is not saturated.
In addition to known filamentous growth genes, we also identified a statistically significant set of genes contributing to cell growth and morphogenesis, nitrogen metabolism, nucleic acid metabolism (including transcription-related processes), and mitochondrial function. In particular, these latter two processes are not conceptually intuitive contributors to the filamentous growth response. From previous studies, however, we know that filamentous differentiation does require an extensive transcriptional program regulated by a sizable cohort of transcription factors (Rupp et al., 1999; Prinz et al., 2004; Borneman et al., 2006). It is further interesting to note that the gene overexpression screen, but not the gene disruption screen, yielded an enriched set of transcription-associated genes, including the transcription factors FLO8, GCN4, MSN2, and PHD1. Compared with other protein classes, overexpression of a transcription factor may be more likely to yield exaggerated filamentous growth. The requirement for mitochondrial function during filamentous growth is also consistent with the prior observation that a rho0 strain of yeast impaired in mitochondrial function is unable to undergo filamentous growth (Kang and Jiang, 2005).
Although GO annotations can be limited in consistency and detail, this analysis, nonetheless, raises two points. First, we identified a set of genes extending well beyond the scope of previously known filamentous growth genes. Second, our data set is enriched for genes functioning in cell processes associated with filamentous growth, encompassing known filamentous growth genes, and genes involved in the regulation of nitrogen metabolism, cell morphogenesis, and growth.
Muc1p Abundance in Filamentous Growth Mutants
Muc1p is considered a principal downstream effector of yeast filamentous growth, and it is the only known gene whose expression integrates signals from both the filamentous growth MAPK and PKA pathways (Gagiano et al., 1999; Rupp et al., 1999; Guo et al., 2000); accordingly, we examined each identified filamentous growth mutant to determine whether the observed phenotype corresponded with altered levels of Muc1p. For this analysis, we constructed a Muc1p-fluorescent protein reporter. In each mutant, we integrated a cassette containing GFP and a selectable marker at the 3′ end of MUC1 as a readout of MUC1 promoter activity. This reporter strain is competent for filamentous growth, and, upon translation, the GFP fusion results in fluorescence at the cell periphery, consistent with results from previous studies (Lo and Dranginis, 1996). Fluorescence was assessed in 96-well format by using a plate reader, and individual mutants of interest were examined at the single cell level by microscopy to account for cell–cell heterogeneity in MUC1 expression. To confirm the accuracy of the fluorescence measurements, we assessed Muc1p-GFP levels by Western blotting in five selected mutants, and we observed corroborating results in each case (Supplemental Figure SF1).
To consider whether these effects are specific to MUC1 and filamentous growth pathways, we constructed a second reporter consisting of red fluorescent protein (DsRed) fused to the carboxy terminus of the translation elongation factor Yef3p. YEF3 is an essential gene encoding translation factor (EF)-3; Yef3p stimulates EF-1α in binding aminoacyl-tRNA to the ribosome; therefore, it functions outside of pathways specific to filamentous growth. A cassette carrying DsRed and an appropriate selectable marker were fused at the 3′ end of YEF3 in each identified filamentous growth mutant exhibiting significantly modified levels of Muc1p. As with Muc1-GFP, we assessed Yef3p–DsRed levels in each mutant by using a fluorescence plate reader and normalized the resulting values by cell density.
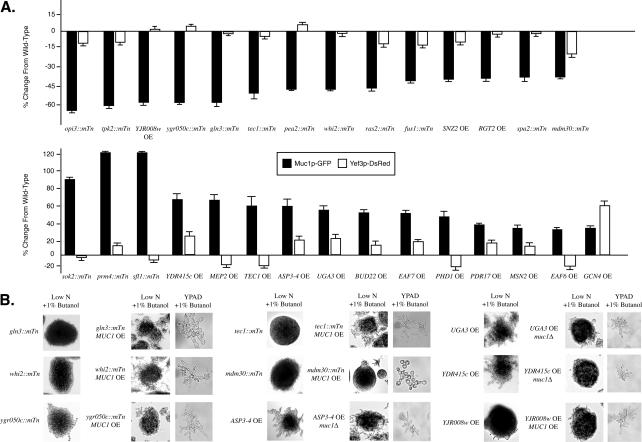
Figure 4A indicates Muc1-GFP and Yef3p-DsRed levels in gene disruption and overexpression mutants relative to levels observed in a wild-type strain; we highlight here all mutants exhibiting significant differences in Muc1p abundance without corresponding differences in Yef3p expression. In total, we report 27 genes conferring filamentous growth phenotypes upon disruption and/or overexpression through a molecular mechanism controlling Muc1p abundance distinct from any general effect on protein expression over the proteome. These mutants identify candidate filamentous growth genes in which the filamentous growth response may be mediated, in part, through direct or indirect regulation of Muc1p abundance. For example, Tec1p is a transcription factor that heterodimerizes with Ste12p; the Ste12/Tec1p complex is required for filamentous growth, and it is known to activate expression of MUC1. We found that disruption of TEC1 resulted in decreased levels of Muc1p but approximately wild-type levels of Yef3p. Conversely, overexpression of TEC1 resulted in increased Muc1p abundance but slightly decreased levels of Yef3p. This assay correctly identified numerous bona fide filamentous growth genes (including TEC1, TPK2, SOK2, PHD1, RAS2, and FUS1). Interestingly, we found that disruption or overexpression of the previously uncharacterized genes YGR050c, YJR008w, and YDR415c resulted in mutant filamentous growth phenotypes exhibiting modified levels of Muc1p relative to Yef3p; these genes, therefore, very likely represent new filamentous growth genes.
Figure 4.
Analysis of Muc1p abundance in filamentous growth mutants. (A) Dual reporter system describing the change in Muc1p-GFP abundance from wild type and the corresponding change in Yef3p-DsRed abundance. Data are presented here for those genes exhibiting changes in Muc1-GFP levels consistent with the observed filamentous growth phenotype without comparable changes in Yef3p-DsRed abundance. For comparison, the mdm30::mTn mutant and GCN4 overexpression mutant are included; Yef3p-DsRed levels differ significantly from wild-type in these mutants, suggesting that, in these cases, the observed changes in Muc1p-GFP levels likely represent general effects on protein production. For this analysis, error bars are generated from three biological replicates. (B) Studies of epistasis using MUC1 deletion and overexpression alleles in mutants disrupted for GLN3, WHI2, YGR050c, TEC1, and MDM30, and in mutants overexpressing ASP3-4, UGA3, YDR415c, and YJR008w. Filamentous growth phenotypes at the colony and cell levels are indicated, along with growth conditions for each analysis.
To define further whether these genes function in MUC1-dependent pathways, we performed a series of epistasis studies in which MUC1 was either ectopically expressed or deleted in selected gene disruption and overexpression mutants from Figure 4A. Specifically, MUC1 was expressed from a galactose-inducible promoter in mutants disrupted for GLN3, WHI2, YGR050c, TEC1, and MDM30, and in a mutant overexpressing YJR008w. Conversely, MUC1 was deleted in hyperfilamentous mutants overexpressing ASP3-4, UGA3, and YDR415c. Filamentous growth phenotypes of the mutants are indicated in Figure 4B. The analysis of tec1::mTn serves as a positive control, and, as expected, overexpression of MUC1 rescues filamentous growth in this mutant. Analysis of mdm30::mTn serves as a negative control: disruption of MDM30 does not produce an effect specific to MUC1, and overexpression of MUC1 in this background does not restore filamentous growth. MUC1 overexpression restored filamentation in mutants deleted for GLN3, a transcriptional activator of genes subject to nitrogen catabolite repression (Mitchell and Magasanik, 1984), and in mutants deleted for WHI2, a phosphatase activator involved in the general stress response in yeast (Kaida et al., 2002). Overexpression of MUC1 also restored filamentous growth in a mutant deleted for YGR050c; however, MUC1 overexpression did not complement the loss of filamentation observed upon overexpression of YJR008w. Deletion of MUC1 decreased filamentous growth in a mutant overexpressing the cell wall asparaginase ASP3-4 (Kim et al., 1988), and even more strongly in mutants overexpressing the transcriptional activator UGA3 and the uncharacterized gene YDR415c. Thus, GLN3, WHI2, ASP3-4, UGA3, YGR050c, and YDR415c function in pathways regulating MUC1.
Mitochondrial Function during Filamentous Growth and the Retrograde Signaling Pathway
By gene disruption and overexpression analysis, we identify a data set statistically enriched (p value 3.9 × 10−3) in genes contributing to mitochondrial function (Figure 3C). Independent studies by Kang and Jiang (2005) identified two genes, IML1 and UGO1, required for both mitochondrial function and filamentous growth in haploid yeast; here, we report eight genes from our screens (Figure 3C) contributing to mitochondrial genome maintenance as annotated by GO. Furthermore, a rho0 strain containing a deleted version of the mitochondrial genome is unable to undergo filamentous growth even in the filamentous Σ1278b genetic background (Figure 5B) (Kang and Jiang, 2005). Interestingly, mitochondrial function is not required for normal vegetative growth on a fermentable carbon source, because rho0 cells are viable under standard growth conditions. Thus, mitochondrial function is required for filamentous growth, but the mechanism by which mitochondrial function is transduced into a signal affecting filamentous growth is unknown.
Figure 5.
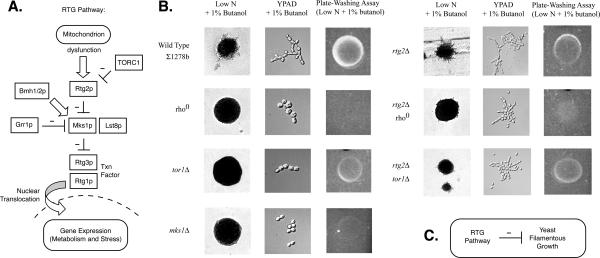
The mitochondrial retrograde signaling pathway and filamentous growth. (A) Overview of the pathway and relevant proteins. (B) Phenotypic analysis of RTG pathway mutants. Gene deletion and rho0 mutants were analyzed for filamentous growth phenotypes as indicated. Colony morphology was assessed on low-nitrogen solid growth medium supplemented with 1% butanol; cell morphology was assessed in rich liquid medium supplemented with 1% butanol. (C) The RTG pathway inhibits filamentous growth. Retrograde signaling acts as part of the mechanism by which mitochondrial dysfunction is transduced into a signal affecting filamentous growth.
To consider the extent to which mitochondrial dysfunction is responsible for filamentous growth defects, we screened each gene disruption mutant exhibiting decreased filamentous growth for impaired respiratory function. Each loss-of-function filamentous growth mutant was grown on medium with glycerol substituted for glucose. Glycerol is a nonfermentable carbon source; yeast strains grown on glycerol must meet their energy needs through aerobic respiration, and mitochondrial mutants are, therefore, unable to grow (Guthrie and Fink, 1991; Ross-Macdonald et al., 1999). Of 309 mutants screened for growth on glycerol, 23 mutants were inviable (Table 2). Muc1p abundance is typically decreased in these mutants, but the effect is also frequently evident in Yef3p levels (e.g., as in mdm30::mTn indicated in Figure 4A); therefore, mitochondrial function likely affects many pathways, acting upstream of the filamentous growth MAPK and PKA signaling modules.
Table 2.
Disrupted genes conferring filamentous growth and glycerol growth phenotypes
| Disrupted gene | Filamentous growtha | Growth on glycerol | Description |
|---|---|---|---|
| ATP14 | −− | Inviable | Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase |
| COA1 | − | Inviable | Putative protein of unknown function; the authentic, nontagged protein is detected in highly purified mitochondria |
| FMC1 | −− | Inviable | Mitochondrial matrix protein, required for assembly or stability at high temperature of the F1 sector of mitochondrial F1F0 ATP synthase |
| FMP25 | − | Inviable | Putative protein of unknown function; the authentic, nontagged protein is detected in highly purified mitochondria |
| FMP38 | − | Inviable | Putative protein of unknown function; the authentic, nontagged protein is detected in highly purified mitochondria |
| IFM1 | −− | Inviable | Mitochondrial translation initiation factor 2 |
| MDJ1 | − | Inviable | Protein involved in folding of mitochondrially synthesized proteins in the mitochondrial matrix |
| MDM30 | − | Inviable | F-box protein; physically associates with mitochondria and is required for normal mitochondrial fusion in rich medium, during sporulation, and in mating cells |
| MEF2 | −− | Inviable | Mitochondrial elongation factor involved in translational elongation |
| MGM101 | − | Inviable | Protein involved in mitochondrial genome maintenance; component of the mitochondrial nucleoid |
| MHR1 | − | Inviable | Protein involved in homologous recombination in mitochondria and in transcription regulation in nucleus |
| MMF1 | − | Inviable | Mitochondrial protein involved in maintenance of the mitochondrial genome |
| MMM1 | −− | Inviable | Mitochondrial outer membrane protein required for normal mitochondrial morphology and mitochondrial DNA stability |
| MST1 | −− | Inviable | Mitochondrial threonyl-tRNA synthetase |
| MSW1 | −− | Inviable | Mitochondrial tryptophanyl-tRNA synthetase |
| QCR9 | − | Inviable | Subunit 9 of the ubiquinol cytochrome c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain |
| RIM1 | −− | Inviable | Single-stranded DNA-binding protein essential for mitochondrial genome maintenance; involved in mitochondrial DNA replication |
| RPO41 | − | Inviable | Mitochondrial RNA polymerase; single subunit enzyme similar to those of T3 and T7 bacteriophages |
| SUV3 | −− | Inviable | ATP-dependent RNA helicase, component of the mitochondrial degradosome along with the RNase Dss1p |
| WHI2 | −− | Inviable | Protein required, with binding partner Psr1p, for full activation of the general stress response, possibly through Msn2p dephosphorylation |
| YOX1 | −− | Inviable | Homeodomain-containing transcriptional repressor, binds to Mcm1p and to early cell cycle boxes in the promoters of cell cycle-regulated genes expressed in M/G1 phase |
| YDR133c | −− | Inviable | Uncharacterized open reading frame |
a −−, severe filamentous growth defect; −, intermediate filamentous growth defect.
With this in mind, the retrograde signaling (RTG) pathway represents a possible avenue by which mitochondrial signals may affect filamentous growth. The yeast retrograde signaling pathway, illustrated in Figure 5A, is a mitochondria-to-nucleus pathway transducing changes in mitochondrial function to specific adaptive changes in nuclear gene expression (Butow and Avadhani, 2004; Liu and Butow, 2006). The pathway encompasses one positive regulator, Rtg2p, four negative regulators, Lst8p, Mks1p, Bmh1p, and Bmh2p, and two terminal transcription factors, Rtg1p and Rtg3p (Liao and Butow, 1993; Jia et al., 1997; Liu et al., 2001). The mitochondrial retrograde response pathway is also known to engage in cross talk with the target of rapamycin (TOR) pathway (Komeili et al., 2000), likely through Mks1p, a negative regulator of the Ras2p signaling pathway (Edskes et al., 1999).
Because the majority of these genes were not represented in our transposon insertion collection, we generated a series of gene deletions targeting RTG pathway regulatory components in the filamentous Σ1278b genetic background. The results of this analysis are indicated in Figure 5B. Deletion of the positive RTG pathway regulator RTG2 results in exaggerated alcohol-induced filamentous growth, whereas deletion of the RTG pathway negative regulator MKS1 results in a loss of filamentous growth. Deletion of MKS1 has been shown to diminish filamentous growth induced under conditions of nitrogen starvation (Edskes et al., 1999); we see a similar phenotype in mks1Δ upon conditions of alcohol induction. Loss-of-filamentation phenotypes have also been observed in BMH1 and BMH2 mutants (Gelperin et al., 1995; Roberts et al., 1997). Consistent with these observations, deletion of TOR1 and transposon-mediated disruption of RAS2 yields loss-of-filamentation phenotypes. From these results, the retrograde signaling pathway negatively regulates filamentous growth. It is interesting to note, however, that deletion of RTG2 in a rho0 background restores filamentous growth slightly: in liquid medium, cell clustering is comparable with wild type, but cell morphology seems less elongated, whereas, on solid medium, limited filamentation can be observed. Deletion of RTG2 in a tor1Δ mutant also restores filamentous growth to an intermediate level; again, cell morphology is less elongated in liquid medium, and only limited filamentation is evident on solid medium. The TORC1 complex is known to regulate many pathways, and this intermediate phenotype reflects the fact that the RTG pathway is not its sole downstream effector. Thus, retrograde signaling is a significant pathway by which mitochondrial dysfunction inhibits filamentous growth, but it is likely not the only avenue by which that signal is transduced.
DISCUSSION
We present here a systematic and large-scale study of yeast filamentous growth, introducing 3627 sequenced transposon insertion alleles and 2043 gene overexpression constructs into the filamentous Σ1278b strain of yeast for subsequent phenotypic analysis. By this approach, we identify 487 genes affecting filamentous growth. This gene set encompasses components of the previously known filamentous growth MAPK and PKA pathways, as well as genes contributing to cellular morphogenesis, nitrogen metabolism, mitochondrial function, and transcription-related processes. This analysis offers the first phenotypic characterization of 84 yeast genes, illustrating the need to investigate gene function in nonstandard genetic backgrounds as a means of ascribing functions to previously uncharacterized genes. In total, we report 27 genes that directly or indirectly affect Muc1p abundance as part of the filamentous growth response, and, in particular, we identify GLN3, WHI2, ASP3-4, UGA3, YGR050c, and YDR415c as components of pathways that regulate MUC1. This study also highlights the importance of mitochondrial function during filamentous growth and identifies the mitochondrial retrograde signaling pathway as a negative regulator of yeast filamentous growth.
To support the broad dissemination of our results, we make all data from this study publicly available at https://lsinwfs03.lsi.umich.edu/NetStorage/(instructions for access are available as supplementary information to this text). In addition, any transposon insertion mutant or gene overexpression strain generated in this study can be requested free of charge from the authors.
Regulation of Muc1p Abundance as a Downstream Control Point During Filamentous Growth
Muc1p has garnered significant attention as a key downstream effector of yeast filamentous growth. As indicated by approaches coupling chromatin immunoprecipitation with DNA microarray analysis, the unusually large MUC1 promoter is directly targeted by at least six filamentous growth transcriptional regulators (Ste12p, Tec1p, Sok2p, Phd1p, Mga1p, and Flo8p) (Borneman et al., 2006). Previous studies have revealed that transcription of MUC1 is induced during filamentous growth (Caro et al., 1997; Strittmatter et al., 2006), and we observe this induction at the protein level as well. We, therefore, hypothesized that the genes identified in this study may affect filamentous growth through a mechanism involving the regulation of Muc1p abundance. To consider this possibility, we constructed a Muc1-GFP/Yef3p-DsRed dual reporter system, enabling us to identify changes in Muc1p abundance relative to general changes in protein expression over the proteome as a whole. By this method, we find 27 transposon insertion or gene overexpression mutants exhibiting 1) altered Muc1p abundance relative to Yef3p and 2) a corresponding filamentous growth phenotype. The gene products identified by this approach do not necessarily affect Muc1p abundance directly; in most cases, the encoded gene products are not obvious regulatory elements, and the effect is presumably indirect. Tec1p and Sok2p are notable exceptions, likely affecting MUC1 expression directly, considering the chromatin immunoprecipitation data referenced above.
Many of these genes had not been implicated previously in pathways regulating MUC1. To investigate a possible role for some of these genes in MUC1-dependent pathways, we performed a series of epistasis experiments with MUC1 deletion and overexpression alleles. We specifically selected the genes GLN3, WHI2, ASP3-4, UGA3, YDR415c, YGR050c, and YJR008w as putative MUC1 regulators, and we confirmed all but YJR008w as components of pathways regulating MUC1. Again, the effects may not be direct, and additional studies will be necessary to define the specific pathways in which these genes function.
On a related point, it is interesting that the change in Muc1p abundance did not correlate strongly with the filamentous growth phenotype in many mutants. Several factors may explain this observation. First, MUC1 expression and filamentous growth are heterogeneous even within a clonal population of cells (Halme et al., 2004). Although fluorescence readings for mutants of interest were confirmed at the single cell level by microscopy, Muc1p–GFP levels were initially surveyed using a plate reader, which would only provide a composite reading for the population as a whole. Plate reader data may, in fact, mask changes in Muc1p abundance if the observed difference is only evident in a relatively small subpopulation of cells. That stated, in our hands, we found plate reader results to be very comparable with results obtained by microscopy of single cells, and we do not consider this to wholly explain discrepancies between Muc1p abundance and filamentous growth phenotypes. Second, Muc1p activity may indeed differ from wild type in these mutants, but this modified activity may not result from altered Muc1p abundance. MUC1 possesses a long region of internal tandem repeats, and variability in the length of this region is known to correlate with MUC1 function (Voynov et al., 2006); thus, Muc1p activity may reflect variability in the length of this repeat region. Alternatively, Muc1p may be controlled by regulatory mechanisms affecting the subcellular localization of this protein, although regulated localization of Muc1p was not readily apparent in our microscopy analysis. Finally, and perhaps most importantly, Muc1p may not be the sole downstream effector of filamentous growth. Our data indicate that a subset of genes affect filamentous growth through a mechanism involving MUC1, but these mechanisms may not target MUC1 exclusively. In support of this possibility, filamentous growth can be induced in the absence of MUC1 on low-nitrogen medium supplemented with a fusel alcohol (Lorenz et al., 2000). Thus, despite the significant attention paid to MUC1, other genes may play equally important roles in enabling filamentous growth.
Previously Uncharacterized Genes in Filamentous Growth
The functionally uncharacterized genes identified in this study provide a particularly interesting starting point for further exploration of the yeast filamentous growth response. In particular, YDR415c and YGR050c function in pathways impinging upon MUC1. YGR050c is a dubious open reading frame, as designated by the Saccharomyces Genome Database, flanked by the uncharacterized open reading frame YGR051c and the putative CDC4 regulator SCM4. Transposon insertion mutants of these flanking genes failed to yield mutant filamentous growth phenotypes; so, the aberrant MUC1 regulation is derived from disruption of YGR050c. Collectively, however, it is difficult to draw obvious conclusions regarding the molecular functions of the 84 uncharacterized genes presented here. Two hybrid data sets (Uetz et al., 2000; Ito et al., 2001) and synthetic lethal interaction studies (Tong et al., 2004) do not reveal a statistically significant enrichment for known filamentous growth genes among putative partners of these genes. Additional individual studies may be necessary to place this gene set among regulatory and metabolic pathways affecting filamentous growth.
The Mitochondrial Retrograde Signaling Pathway Inhibits Filamentous Growth
Our transposon mutagenesis and overexpression analysis highlights the importance of mitochondrial function during filamentous growth, and, through additional follow-up studies, we identify the RTG pathway as one route by which mitochondrial signals may affect filamentous growth. The RTG pathway is known to activate expression of genes involved in glutamate biosynthesis (e.g., CIT1, CIT2, ACO1, IDH1, and IDH2) (Liao and Butow, 1993; Liu and Butow, 2006), and glutamate, together with its downstream metabolite glutamine, represents the principal nitrogen source used in biosynthetic reactions (Chen and Kaiser, 2002). The RTG pathway, therefore, ensures a sufficient level of glutamate to meet cellular demands for nitrogen, especially in respiration-deficient cells. This function of the RTG pathway is conceptually consistent with its observed role as an inhibitor of filamentous growth. Activation of the RTG pathway results in an increased concentration of total cellular amino acids (Chen and Kaiser, 2002), and we speculate this ameliorates nitrogen stress, thereby inhibiting filamentous growth. We do not, as of yet, understand the exact molecular mechanism by which nitrogen stress is sensed/transduced into signals affecting filamentous growth; nor do we understand the molecular link between filamentous growth and the RTG pathway. The TORC1 complex is the most obvious candidate, and the filamentous growth phenotypes we observe in relevant mutants are consistent with previous studies suggesting that TORC1 activates filamentous growth and inhibits the RTG pathway (Komeili et al., 2000; Gancedo, 2001). Interestingly, deletion of RTG2 in a rho0 mutant background results in intermediate filamentation; it does not restore wild-type levels of filamentous growth, suggesting that mitochobndrial dysfunction may activate additional pathways affecting the filamentous growth response. Identification of these pathways will require extensive investigation, and the gene set identified in this study provides a first step toward that end.
A Broad Spectrum of Genes and Pathways Contribute to Yeast Filamentous Growth
In budding yeast, the morphological and genetic changes associated with the transition to filamentous growth are extensive, and, perhaps not surprisingly, the genes and pathways underlying these changes seem to be equally extensive. The MAPK and PKA signaling modules initially identified as key components of the filamentous growth response are undeniably important in enabling filamentation, but the intense emphasis on these signaling modules does not adequately characterize the broad molecular scope of filamentous differentiation. From the results presented here, literally hundreds of other genes confer filamentous growth phenotypes of equal severity to those observed upon disruption of the MAPK and/or PKA pathway components. Although efforts to place these genes within specific pathways or networks will require sizable investments in labor and resources, our resulting knowledge base regarding yeast filamentous growth will expand tremendously as a result of such research, with concomitant hope of improved clinical antifungal therapeutics to follow.
Supplementary Material
ACKNOWLEDGMENTS
We thank Damian Krysan, Robert Fuller, Anthony Borneman, and Michael Snyder for providing filamentous yeast strains. This work was supported by grant RSG-06-179-01-MBC from the American Cancer Society (to A.K.), grant DBI 0543017 from the National Science Foundation (to A.K.), and Basil O'Connor Award 5-FY05-1224 from the March of Dimes (to A.K.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0519) on November 7, 2007.
REFERENCES
- Alspaugh J. A., Perfect J. R., Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer B. M., Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow R. A., Avadhani N. G. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Caro L. H., Tettelin H., Vossen J. H., Ram A. F., van den Ende H., Klis F. M. In silico identification of glycosyl-phosphatidyl-inositol-anchored plasma membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chen E. J., Kaiser C. A. Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:14837–14842. doi: 10.1073/pnas.232591899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G. O. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. G., Bardwell L., Thorner J. Inhibitory and activating functions forMAPK Kss1 in the S. cerevisiae filamentous growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- Cullen P. J., Sabbagh W., Graham E., Irick M. M., van Olden E. K., Neal C., Delrow J., Bardwell L., Sprague G. F. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sprague G. F. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:13461–13463. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H. K., Hanover J. A., Wickner R. B. Mks1p is a regulator of nitrogen catabolism upstream of Ure2p in Saccharomyces cerevisiae. Genetics. 1999;153:585–594. doi: 10.1093/genetics/153.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano M., van Dyk D., Bauer F. F., Lambrechts M. G., Pretorius I. S. Msn1p/Mss10p, Mss11p and Muc1p/Flo11p are part of a signal transduction pathway downstream of Mep2p regulating invasive growth and pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Microbiol. 1999;31:103–116. doi: 10.1046/j.1365-2958.1999.01151.x. [DOI] [PubMed] [Google Scholar]
- Gancedo J. M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Gelperin D., Weigle J., Nelson K., Roseboom P., Irie K., Matsumoto K., Lemmon S. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1995;92:11539–11543. doi: 10.1073/pnas.92.25.11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Grenson M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. II. Evidence for a specific lysine-transporting system. Biochim. Biophys. Acta. 1966;127:339–346. doi: 10.1016/0304-4165(66)90388-6. [DOI] [PubMed] [Google Scholar]
- Guo B., Styles C. A., Feng Q., Fink G. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Halme A., Bumgarner S., Styles C. A., Fink G. R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Rothermel B., Thornton J., Butow R. A. A basic helix-loop-helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell Biol. 1997;21:2506–2520. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Yashiroda H., Toh-e A., Kikuchi Y. Yeast Whi2 and Psr1-phosphatase form a complex and regulate STRE-mediated gene expression. Genes Cells. 2002;7:543–552. doi: 10.1046/j.1365-2443.2002.00538.x. [DOI] [PubMed] [Google Scholar]
- Kang C. M., Jiang Y. W. Genome-wide survey of non-essential genes required for slowed DNA synthesis-induced filamentous growth in yeast. Yeast. 2005;22:79–90. doi: 10.1002/yea.1195. [DOI] [PubMed] [Google Scholar]
- Kim K. W., Kamerud J. Q., Livingston D. M., Roon R. J. Asparaginase II of Saccharomyces cerevisiae: characterization of the ASP3 gene. J. Biol. Chem. 1988;263:11948–11953. [PubMed] [Google Scholar]
- Komeili A., Wedaman K. P., O'Shea E. K., Powers T. Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron S. J. Filamentous growth in budding yeast. Trends Microbiol. 1997;5:450–454. doi: 10.1016/S0966-842X(97)01131-1. [DOI] [PubMed] [Google Scholar]
- Kron S. J., Styles C. A., Fink G. R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., et al. Subcellular localization of the yeast proteome. Genes Dev. 2002a;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., DesEtages S., Coelho P., Roeder G., Snyder M. High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol. 2000;328:550–574. doi: 10.1016/s0076-6879(00)28418-8. [DOI] [PubMed] [Google Scholar]
- Kumar A., et al. Large-scale mutagenesis of the yeast genome using a Tn7-derived multipurpose transposon. Genome Res. 2004;14:1975–1986. doi: 10.1101/gr.2875304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Vidan S., Snyder M. Insertional mutagenesis: transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 2002b;350:219–229. doi: 10.1016/s0076-6879(02)50965-4. [DOI] [PubMed] [Google Scholar]
- Kumar A., Vidana S., Snyder M. Insertional mutagenesis: transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 2002c;350:219–229. doi: 10.1016/s0076-6879(02)50965-4. [DOI] [PubMed] [Google Scholar]
- Lengeler K. B., Davidson R. C., D'Souza C., Harashima T., Shen W. C., Wang P., Pan X., Waugh M., Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Butow R. A. RTG1 and RTG 2, two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G. R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Liu Z., Butow R. A. Mitochondrial Retrograde Signaling. Annu. Rev. Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Liu Z., Sekito T., Epstein C. B., Butow R. A. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 2001;20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. J., Kohler J., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997a;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 1996;178:7144–7151. doi: 10.1128/jb.178.24.7144-7151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Raitses E. I., Dranginis A. M. Development of pseudohyphae by embedded haploid and diploid yeast. Curr. Genet. 1997b;32:197–202. doi: 10.1007/s002940050266. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Cutler N. S., Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Styles C. A., Fink G. R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Miled C., Mann C., Faye G. Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth. Mol. Cell Biol. 2001;21:3714–3724. doi: 10.1128/MCB.21.11.3714-3724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol. Cell Biol. 1984;4:2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Neiman A. M., Park H. O., Lohuizen M. v., Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Avila-Campillo I., Aldridge C., Srinivasan A., Dimitrov K., Siegel A. F., Galitski T. Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res. 2004;14:380–390. doi: 10.1101/gr.2020604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. L., Mosch H. U., Fink G. R. 14–3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- Robertson L. S., Fink G. R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P., et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Rua D., Tobe B. T., Kron S. J. Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 2001;4:720–727. doi: 10.1016/s1369-5274(01)00274-0. [DOI] [PubMed] [Google Scholar]
- Rupp S., Summers E., Lo H. J., Madhani H., Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Singh P., Ghosh S., Datta A. A novel MAP-kinase kinase from Candida albicans. Gene. 1997;190:99–104. doi: 10.1016/s0378-1119(96)00758-5. [DOI] [PubMed] [Google Scholar]
- Sopko R., et al. mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Strittmatter A. W., Fischer C., Kleinschmidt M., Braus G. H. FLO11 mediated filamentous growth of the yeast Saccharomyces cerevisiae depends on the expression of the ribosomal RPS26 genes. Mol. Genet. Genomics. 2006;276:113–125. doi: 10.1007/s00438-006-0127-7. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Uetz P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Voynov V., Verstrepen K. J., Jansen A., Runner V. M., Buratowski S., Fink G. R. Genes with internal repeats require the THO complex for transcription. Proc. Natl. Acad. Sci. USA. 2006;103:14423–14428. doi: 10.1073/pnas.0606546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Fuctional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., Fink G. R., Young R. A. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.