Abstract
Nuclear export of mRNAs is mediated by the Tap/Nxt1 pathway. Tap moves its RNA cargo through the nuclear pore complex by direct interaction with nucleoporin phenylalanine-glycine repeats. This interaction is strengthened by the formation of a Tap/Nxt1 heterodimer. We now present evidence that Tap can form a multimeric complex with itself and with other members of the NXF family. We also show that the homotypic Tap complex can interact with both Nxt1 and nucleoporins in vitro. The region mediating this oligomerization is localized to the first 187 amino acids of Tap, which overlaps with its RNA-binding domain. Removal of this domain greatly reduces the ability of Tap to bind nucleoporins in vitro and in vivo. This is the first report showing that the Tap amino terminus modulates the interaction of Tap with nucleoporins. We speculate that this mechanism has a regulatory role for RNA export independent of RNA binding.
INTRODUCTION
Eukaryotes depend on soluble transport receptors to move proteins and RNAs across the nuclear envelope (Pemberton and Paschal, 2005; Tran and Wente, 2006). These receptors shuttle through the nuclear pore complex (NPC) by transient interactions with specific phenylalanine-glycine (FG) repeats lining the central channel. Members of the karyopherin-β family of transport receptors are responsible for moving proteins, ribosomal subunits, tRNAs, snRNAs, and some mRNAs across the pores and rely on the small GTPase Ran to confer directionality through the NPC (Cullen, 2003; Rodriguez et al., 2004). Tap/NXF1, a member of the NXF family of transport receptors, and its cofactor Nxt1/p15 are thought to mediate export for the majority of cellular mRNAs (Tan et al., 2000; Wilkie et al., 2001; Izaurralde, 2002). Unlike transport mediated by the karyopherin-β family, mRNA export does not require Ran (Clouse et al., 2001; Schmitt and Gerace, 2001). It is proposed that directionality for the Tap/RNA transport is orchestrated by the DEAD-box helicase Dbp5 bound to cytoplasmic fibrils (for recent reviews, see Cole and Scarcelli, 2006; Tran and Wente, 2006; Stewart, 2007).
Tap is a ubiquitously expressed protein that was first identified for its ability to export viral RNA bearing the constitutive transport element (CTE) (Bray et al., 1994; Ernst et al., 1997a,b; Grüter et al., 1998). Until recently, the cis-acting CTE had only been identified on transcripts of simple type D retroviruses such as the Mason–Pfizer monkey virus and the simian retrovirus type 1 (Bray et al., 1994; Zolotukhin et al., 1994). However, a study by Li et al. (2006) has identified a functional CTE motif within intron 10 of Tap pre-mRNA, which is sometimes retained upon alternate splicing. Interestingly, this alternate intron-retaining Tap mRNA is exported to the cytoplasm by Tap whereupon it is translated into a shorter Tap variant. Three functional domains of Tap have been well characterized: the RNA-binding domain, the Nuclear Transport Factor 2 (NTF2)-like domain, and the ubiquitin-associated (UBA) domain (Liker et al., 2000; Suyama et al., 2000; Fribourg et al., 2001; Grant et al., 2002, 2003). These last two domains mediate the interaction of Tap with the NPC (Katahira et al., 1999; Bachi et al., 2000; Fribourg et al., 2001; Lévesque et al., 2001; Schmitt and Gerace, 2001; Thakurta et al., 2004). Nxt1 heterodimerizes with the NTF2-like domain, and this association imparts Tap with greater binding efficiency for nucleoporins (Suyama et al., 2000; Fribourg et al., 2001; Lévesque et al., 2001, 2006; Wiegand et al., 2002).
Structural studies have revealed that the RNA-interacting domain of Tap is located at its amino terminus and that it is composed of two RNA recognition motifs (RRMs) and one leucine-rich region (LRR; Liker et al., 2000; Ho et al., 2002). The RRMs bear the typical βαββαβ fold found on many other RNA-binding proteins, such as polyA-binding protein (PAB), the Sex-lethal (Sxl) protein, and the U1A and U2B“ splicing factors (Liker et al., 2000). Whereas the RRM region is sufficient for binding RNAs in vitro, both RRM and LRR are required for RNA export in vivo (Katahira et al., 1999; Liker et al., 2000). Both domains are also necessary to bind CTE (Liker et al., 2000). The Tap LRR is composed of parallel β-pleated sheets arranged on the inner surface of a concave structure overlaid with α-helices on the convex side, homologous to the U2A' LRR (Liker et al., 2000; Ho et al., 2002). Although Tap can bind to RNA directly in vitro, the current paradigm is that recruitment of Tap to export-ready RNAs in vivo is through adaptor proteins (Cullen, 2003; Rodriguez et al., 2004; Huang and Steitz, 2005). Such putative adaptor proteins are members of the transport/transcription complex and serine/arginine-rich family of splicing factors (Huang and Steitz, 2005; Reed and Cheng, 2005).
We now report a new mechanism regulating Tap function based on the ability of Tap to self-oligomerize and form a multimeric complex with other members of the NXF family. We show that the first 187 residues of Tap are required for self-assembly and that Tap oligomers bind both nucleoporins and Nxt1. We also observed that a Tap mutant unable to assemble into the oligomerized form is able to bind nucleoporins. However, compared with wild-type (WT) Tap, this binding was significantly reduced, and it was further aggravated when Nxt1 was included in the binding reaction. We speculate that the formation of oligomeric Tap complexes may be necessary for the Tap RNA export function.
MATERIALS AND METHODS
Plasmids
FLAG-Tap WT, glutathione transferase (GST)-Tap WT, pSVK3-Nxt1, and HA-NTF2 constructs have all been described previously (Guzik et al., 2001; Lévesque et al., 2001). Hemagglutinin (HA)-Tap was generated by cleaving Tap cDNA from the FLAG-Tap vector with BamH1 and EcoR1 and ligating it into the respective sites on the pCDNA3.1 vector (Invitrogen, Carlsbad, CA) that was modified to contain an amino-terminal HA epitope (MAYPYDVPDYA; pCDNA3.1-HA). Point mutations within the FLAG-Tap and GST-Tap vectors were introduced using the QuikChange site-directed mutagenesis protocol (Stratagene, La Jolla, CA). Tap carboxy-terminal deletion mutants of FLAG-Tap and GST-Tap were made by replacing two adjacent and in-frame amino acids with two stop codons within the full-length sequence. Amino-terminal deletion mutants of Tap were amplified using a standard polymerase chain reaction (PCR) protocol and ligated into the BamH1 and EcoR1 sites of pCDNA3-FLAG vector or pGEX-4T3. The pCDNA3-FLAG plasmid was modified from the original pCDNA3 vector (Invitrogen) by addition of an amino-terminal FLAG sequence (MDYKDDDDK). GST-p62 (Hu et al., 1996), GST-Nup98 (GLFG domain residues 221-504), and pEGFP-NXF5 (Jun et al., 2002) were a generous gift from Drs. L. Gerace (Scripps Research Institute, La Jolla, CA), M. A. Powers (Emory University School of Medicine, Atlanta, GA), and G. Froyen (Flanders Interuniversity Institute for Biotechnology, Leuven, Belgium), respectively. An NXF5 cDNA insert was obtained by cleaving a GFP-NXF5 construct with BglII and ApaI. The obtained fragment was then ligated into the BamHI and Apa1 sites of the pCDNA3-FLAG or pGEX-4T3 vectors to generate FLAG-NXF5 and GST-NXF5, respectively. All DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and cloned vectors verified by DNA sequencing at the University of Illinois at Urbana-Champaign core facility. Detailed maps of these oligonucleotides and vectors are available upon request. Human NXF2 (clone ID 3921074) and NXF3 (clone ID 5170537) cDNAs inserted into the pCMV-SPORT6 were purchased from Open Biosystems (Huntsville, AL).
Recombinant Protein
GST and GST-fusion proteins were expressed in the BL21 Escherichia coli strain, induced with 1 mM isopropyl fl-d-thiogalactoside and bound to glutathione-agarose beads (Pierce Chemical, Rockford, IL) as described previously (Lévesque et al., 2001, 2006). Untagged recombinant Tap was prepared by first binding GST-Tap to glutathione-agarose beads and then cleaved by ∼13 units/ml thrombin (Sigma-Aldrich, St. Louis, MO) in cleavage buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 2.5 mM CaCl2) for 2 h at room temperature. Cleaved protein was recovered from the unbound fraction. GST-NXF5 was resolublized from inclusion bodies with 6 M guanidine-HCl and subsequently diluted down to 3 M guanidine with Tris-sodium-EDTA buffer (50 mM Tris, pH 8, 50 mM NaCl, and 1 mM EDTA) for binding to glutathione beads. Recombinant Nxt1 was prepared as described previously (Black et al., 2001).
In vitro-translated proteins were prepared using the coupled transcription/translation rabbit reticulocyte system (Promega, Madison, WI) and radiolabeled by adding Redivue l-[35S]methionine (1000 Ci/mmol; GE Healthcare, Piscataway, NJ). FLAG-Tap (WT and mutants), HA-Tap, FLAG-NXF5, HA-NTF2, and pSVK-Nxt1 constructs were transcribed using T7 polymerase, whereas NXF2 and NXF3 were transcribed using Sp6 polymerase (Promega).
GST Pull-Down
Each pull-down reaction contained 5 μg of GST-tagged protein bound to 20 μl of packed glutathione beads. In vitro-synthesized protein (10 μl) was diluted in phosphate-buffered saline (PBS) composed of 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT), and 0.1% (vol/vol) Tween 20 to a final volume of 500 μl. Reactions were incubated overnight at 4°C, rotating and unbound were proteins removed by washing beads 6 times with PBS plus 0.1% (vol/vol) NP-40. Bound proteins were eluted in Laemmli sample buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE). After gels were fixed and stained by Coomassie Blue R250, they were treated with Autofluor (National Diagnostics, Atlanta, GA) and dried. 35S-proteins were detected by autoradiography. In some experiments, 1.5 μg of untagged recombinant Tap was used instead of 35S-proteins and bound protein detected by Western blotting (WB) using a rabbit polyclonal antibody (pAb) raised against amino acids 522-619 of Tap. More details about the production and characterization of the Tap pAb are provided in Supplemental Material.
Densitometry analysis was performed using NIH Image software, version 1.63 (http://rsb.info.nih.gov/nih-image/), or Adobe Photoshop CS3 extended (Adobe Systems, Mountain View, CA). Total binding for each band was calculated by adding the level of signal for each pixel within the band. Total binding was then compared with the binding of 35S-FLAG-Tap WT arbitrarily assigned a value of 100% binding.
Solid-Phase Binding Assay
GST-tagged proteins or GST diluted in buffer A (20 mM HEPES, 110 mM potassium acetate, 2 mM magnesium acetate, and 0.5 μM EGTA, pH 7.4) were adsorbed to high-binding capacity 96-well plates (Corning, Corning, NY) for 24 h at 4°C. Unbound proteins were removed and replaced with 3% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich) in buffer A for 24 h. Equal volumes of binding reaction composed of in vitro-translated 35S-FLAG-Tap (WT or mutant) in buffer A supplemented with 5 μg/μl BSA, 0.2 mM DTT, and 0.2% (vol/vol) Tween 20 were added to each well and incubated for 18–24 h. In some experiments, 66 nM recombinant Nxt1 was added to the binding reaction. After washing of the unbound fraction with 0.5X buffer A plus 0.2% (vol/vol) Tween 20, bound protein was eluted with 5% (wt/vol) SDS and quantified by liquid scintillation counting.
Gel Filtration Chromatography
35S-Tap containing rabbit reticulocyte lysate was treated with DNase (12.9 μM) and RNase A (24.3 μM) for 30 min at room temperature. The lysate (100 μl) was then applied to a Superdex 200 gel filtration column (GE Healthcare) in buffer A supplemented with 2 mM DTT at a flow rate of 0.5 ml/min using the ÁKTA system (GE Healthcare). Fractions were collected in 0.5 ml increments, precipitated with 20% (wt/vol) trichloroacetic acid, resuspended in Laemmli sample buffer, and separated by SDS-PAGE. 35S-Tap containing fractions were detected by autoradiography. Column retention times were calibrated using thyroglobulin (667 kDa), ferritin (440 kDa), catalase (215 kDa), aldolase (191 kDa), BSA (64 kDa), ovalbumin (45.8 kDa), chymotrypsinogen A (19.9 kDa), and RNase A (15.4 kDa).
Mammalian Cell Culture and Transfection
Suspension HeLa S3 cells were grown at 37°C and 2% CO2 in Joklik media supplemented with 5% newborn calf serum (HyClone Laboratories, Logan, UT), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen). HeLa cell extracts were prepared in buffer B (50 mM NaCl, 120 mM Tris, pH 8.0, 1 mM EDTA, and 0.5% [vol/vol] NP-40) supplemented with 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1 mM DTT. Protein concentration was determined with a bicinchonic acid protein assay kit (Sigma-Aldrich).
COS-7 cells were grown in DMEM (Invitrogen) supplemented with 10% newborn calf serum at 37°C in 5% CO2. Before transient transfection by electroporation, cells were trypsinized and resuspended at 107 cells/ml in Opti-MEM (Invitrogen). For each transfection, 0.8 ml of cells and 10 μg of each plasmid were added to a 0.4-mm electroporation cuvette, and the solution was subjected to 180 mV with 960 μFd.
Protein Immunoprecipitation (IP) and Western Blot
COS-7 cells expressing HA-Tap and FLAG-Tap constructs were harvested 36 h after transfection. Cells were lysed in buffer B with protease inhibitors (1 μg/ml aprotinin, leupeptin, and pepstatin A and 1 mM phenylmethylsulfonyl fluoride) and 1 mM DTT by pipetting up and down 40 times. Cell extract was clarified by centrifugation at 18,000 × g at 4°C for 15 min. In some cases, extracts were treated with 100 μg/ml RNase A before IP. Cleared cell extracts were incubated with protein A/G-agarose beads (Calbiochem, San Diego, CA) preloaded with mouse monoclonal HA.11 antibody (Covance, Berkeley, CA). Immunoprecipitations were incubated overnight at 4°C, rotating. After incubation, unbound proteins were removed, and the beads were washed four times with cold buffer B. Bound protein was eluted in Laemmli sample buffer, separated by SDS-PAGE, and transferred to an Immobilon-P membrane (Millipore, Billerica, MA). The membrane was blocked with 5% (wt/vol) powdered milk dissolved in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) for 2 h at room temperature. Primary antibodies were diluted in TBS-T and incubated with the blot overnight at 4°C. The HA.11 monoclonal antibody (mAb) (∼5 μg/ml; Covance) was used to detect HA-Tap and the M2 mAb (0.49 μg/ml; Sigma-Aldrich) was used to detect FLAG-Tap. Detection of endogenous and recombinant Tap was done with the Tap pAb antibody (1:1000). Following three washes in TBS-T, the membrane was incubated with donkey anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibody (20 ng/ml; Jackson ImmunoResearch Laboratories, West Grove, PA) diluted in TBS-T for 1 h at room temperature and washed with TBS-T. The proteins were visualized using enhanced chemiluminescence (GE Healthcare).
Immunofluorescence (IF)
Detection of FLAG-Tap WT and mutant by indirect IF was done as described previously in COS-7 cells 24 h after transfection by electroporation (Lévesque et al., 2001). FLAG-tagged proteins were detected using M2 antibodies followed by anti-mouse Cy3 conjugated antibody (Jackson ImmunoResearch Laboratories). All images were captured with an LSM 510 confocal microscope by using apochromat an 63/1.4 differential interference contrast (DIC) lens at 1640 × 1640 resolution. All images were acquired using the same settings unless otherwise noted and processed with Adobe Photoshop 7.0.
RESULTS
Tap Can Self-Associate and Form Oligomers with Other Human NXF Family Members
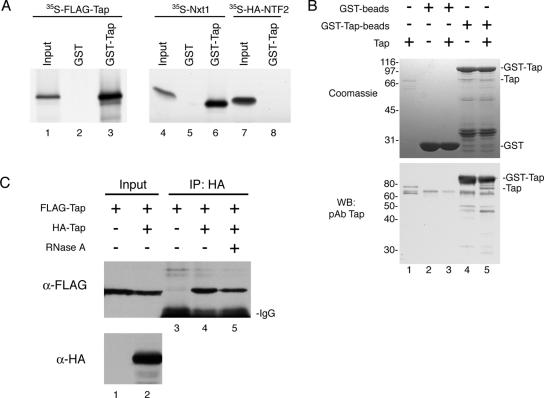
An initial GST pull-down assay demonstrated that in vitro translated 35S-FLAG-Tap can bind to GST-Tap but not to GST coupled to glutathione-Sepharose beads (Figure 1A, lanes 3 and 2, respectively). Treatment of the lysate with RNase A before incubation with GST-Tap did not abolish the interaction (data not shown). The same experiment was carried out in parallel using in vitro translated 35S-Nxt1. Nxt1 bound GST-Tap as expected (lane 6), whereas its homologue 35S-HA-NTF2 did not (lane 8). To rule out the possibility that the binding was bridged by other proteins present in the reticulocyte lysate, we repeated the experiment by incubating recombinant Tap with GST-Tap (Figure 1B). Because of a contaminating band in the GST-Tap beads (top, lanes 4 and 5) that comigrates with untagged Tap (top, lane 1) on SDS-gels, bound Tap was detected instead by Western blot (bottom) by using the Tap pAb. Once again, we found that Tap could bind GST-Tap specifically in the absence of any RNAs or adaptor proteins (Figure 1B, compare lanes 4 and 5).
Figure 1.
Complex formation of human Tap in vitro and in vivo. (A) In vitro translated 35S-FLAG-Tap (lanes 1–3), 35S-Nxt1 (lanes 4–6), and 35S-HA-NTF2 (lanes 7 and 8) were incubated with GST alone (lanes 2 and 5) or with GST-Tap (lanes 3, 6, and 8) immobilized on glutathione-coated agarose beads. Bound proteins were detected by autoradiography. Input lanes represent 5% of total 35S-labeled protein added to each reaction. (B) In a similar assay, 2 μg of recombinant Tap (input; lane 1) was assayed for binding to GST (input; lane 2) or GST-Tap beads (input; lane 4). Bound protein (lanes 3 and 5) was loaded onto an SDS-PAGE gel and detected by Coomassie Blue R250 staining (top) or by Western blotting by using the Tap pAb (bottom). (C) Co-IP of FLAG-Tap from COS-7 cells expressing FLAG-Tap alone (lane 1) or FLAG-Tap with HA-Tap (lane 2). Protein complexes were immunoprecipitated using the HA.11 mAb, and the presence of FLAG-Tap bound to HA-Tap was detected by Western blotting (lanes 3–5). Some of the FLAG-Tap + HA-Tap cell extract was pretreated with RNase A before IP (lane 5).
We also examined the formation of the Tap homotypic complex in cells by co-IP to verify the biological relevance of this novel association. The HA.11 mAb antibody was used to IP HA–Tap complexes from extracts of COS-7 cells expressing HA-Tap and/or FLAG-Tap as indicated (Figure 1C). Although all cells expressed FLAG-Tap, FLAG-Tap was only coimmunoprecipitated from cell extracts containing HA-Tap (compare lanes 3 and 4). Treatment of the lysate with RNase A before immunoprecipitation reduced the amount of FLAG-Tap in the bound fraction by ∼50%, an effect that we found to be reproducible. However, a significant amount of FLAG-Tap remained associated with HA-Tap in RNase A-treated samples (lane 5), consistent with the notion that Tap can self-associate independently of RNAs in vivo. The decrease in FLAG-TAP associated with HA-Tap observed after RNase treatment could represent the loss of Tap monomers or oligomers loaded onto the same RNA molecule. A second explanation would be that although Tap can self-oligomerize independently of RNA, its assembly into a homotypic complex may be stimulated by RNA binding. Our current experiment cannot distinguish between these two possibilities.
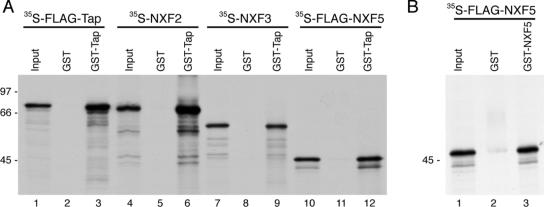
Because of sequence homologies between Tap and other human NXFs, we also tested the potential interaction of Tap with its homologues. In vitro-translated 35S-labeled FLAG-Tap, NXF2, NXF3, and FLAG-NXF5 were each incubated with either GST- or GST-Tap bound to glutathione beads. We found that all of the human NXF family members bound GST-Tap (Figure 2A). Using the same GST pull-down assay, we also found that 35S-FLAG-NXF5 can bind GST-NXF5 (Figure 2B).
Figure 2.
Tap interacts with other human NXF family members. (A) The ability of the other NXF family members to interact with Tap was tested using a GST pull-down assay. In vitro synthesized 35S-FLAG-Tap (lanes 1–3), 35S-NXF2 (lanes 4–6), 35S-NXF3 (lanes 7–9), and 35S-FLAG-NXF5 (lanes 10–12) were incubated with glutathione-agarose beads coated with GST or GST-Tap. Bound proteins were resolved by SDS-PAGE followed by autoradiography. Input lanes correspond to 5% of radioactive protein used for each condition. (B) Using the same assay, 35S-FLAG-NXF5 was tested for binding GST-NXF5 (lane 3) or GST alone (lane 2).
Characterization of the Multi-Tap Complex
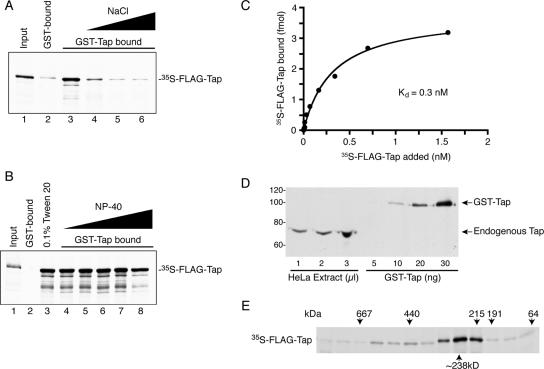
To characterize the nature of the interaction, we tested the effect of increasing salt and detergent concentration on Tap–Tap complex formation (Figure 3, A and B, respectively). We found that the self-oligomerization of Tap was contingent on the salt concentration in the binding buffer. The presence of 500 mM NaCl greatly reduced Tap binding and 1 M NaCl reduced the binding to background levels (Figure 3A, lanes 4 and 5, respectively). Conversely, 0.l% Tween 20 (Figure 3B, lane 3) and as high as 2% NP-40 (Figure 3B, lane 7) had no effect on the assembly of the complex. A small decrease in bound 35S-FLAG-Tap was observed when 5% NP-40 was included in the binding reaction (Figure 3B, lane 8), but this decrease also corresponded to an equivalent loss of GST-Tap protein bound to glutathione beads (data not shown). These results suggest that electrostatic rather than hydrophobic interactions are primarily responsible for the formation of the Tap complex.
Figure 3.
Characterization of the Tap complex. (A) The formation of the Tap complex was tested against increasing salt concentration in the binding buffer. These binding reactions were done in the absence of any detergent. 35S-FLAG-Tap was incubated with GST-coated beads in normal binding buffer containing 137 mM NaCl (lane 2), GST-Tap–coated beads in normal binding buffer (lane 3), or GST-Tap–coated beads in binding buffer containing 500 mM (lane 4), 1 M (lane 5), or 2 M (lane 6) NaCl. Bound proteins were resolved by SDS-PAGE followed by autoradiography. Input lanes correspond to 5% of radioactive protein used for each condition. (B) In a parallel experiment, the effect of nonionic detergent on the formation of the Tap complex was tested. 35S-FLAG-Tap was incubated with GST (lane 2) or GST-Tap (lane 3) in normal binding buffer supplemented with 0.1% Tween 20. In addition, the binding of 35S-FLAG-Tap was assessed in the presence of 0.1% (lane 4), 0.5% (lane 5), 1% (lane 6), 2% (lane 7), and 5% (lane 8) NP-40 in the binding reaction. (C) The affinity of Tap for itself was tested using a solid-phase assay. Increasing concentrations of 35S-FLAG-Tap as indicated was added to immobilized recombinant GST or GST-Tap. The level of bound radioactive protein was measured by scintillation counting. Total binding values for GST-Tap were corrected for nonspecific binding by subtracting corresponding GST-bound values. Each data point represents the mean specific binding of three replicates (±SD). The Kd was determined by nonlinear regression fit analysis using the Prism 4 software (GraphPad Software, San Diego, CA). (D) The level of endogenous Tap from HeLa cell extract was compared with that of known quantities of recombinant Tap by Western blotting. Tap protein was visualized using the Tap pAb. (E) 35S-FLAG–Tap complexes were separated by gel filtration chromatography. Fractions were resolved by SDS-PAGE and visualized by autoradiography. The elution points of protein standards are indicated above their corresponding fractions.
We also tested the affinity of Tap for itself using a solid-phase binding assay. Briefly, 25 ng of GST or GST-Tap protein was immobilized to a multiwell plate. Equal volumes of binding reactions composed of increasing amounts of in vitro translated 35S-FLAG-Tap were added to GST- or GST-Tap–containing wells and incubated overnight. We found that the binding of 35S-FLAG-Tap to GST-Tap was saturable, and we calculated the Kd of the interaction to be 0.3 nM (Figure 3C). To determine whether the observed Kd was compatible with the formation of such a complex in vivo, we estimated the average cellular concentration of endogenous Tap in HeLa S3 cells. Whole cell extract was made from a predetermined number of cells (1.17 × 105 cells/μl). Western blot analysis was used to compare the amount of Tap in the extract to a range of recombinant Tap standards of known concentration (Figure 3D). We then calculated the amount of Tap per cell and using the published volume for HeLa S3 cells, 1.8 pl for the cytoplasm and 1.2 pl for the nucleus (Görlich et al., 2003). We determined the cellular Tap concentration to be ∼1 amol of Tap per cell. Because most Tap is present in the nucleus, we can estimate the concentration of nuclear Tap per cell to be 0.8 μM. This level is well within the range of Tap concentration that can oligomerize in vitro, and it further validates the physiological relevance of this interaction.
We next assessed the number of Tap molecules incorporating into this Tap multimer by gel filtration. In vitro-translated 35S-FLAG-Tap treated with RNase and DNase was separated on a S200 Superdex gel filtration column and the presence of Tap in each fraction detected by autoradiography (Figure 3E). No significant amount of FLAG-Tap was observed in fractions corresponding to the ∼70-kDa monomeric form of Tap. Instead, the majority of Tap eluted with an apparent molecular weight of ∼238 kDa, suggesting that the majority of Tap exists as a trimeric or tetrameric complex. Less abundant higher molecular weight species of Tap complexes also could be detected ranging from 440 to 570 kDa.
The Amino Terminus of Tap Is Responsible for Its Self-Oligomerization
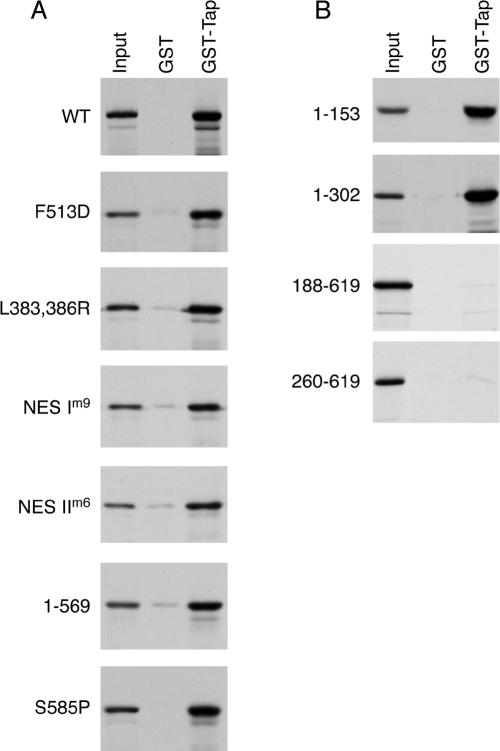
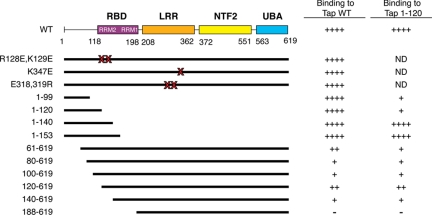
The central portion of Tap includes an NTF2-like domain, which heterodimerizes with the respective NTF2-like domain of Nxt1. NTF2 itself exists as a dimer. Therefore, we reasoned that this NTF2-like domain may also mediate formation of the multimeric Tap complex. We tested Tap constructs with mutations in the NTF2-like domain for their ability to interact with Tap WT by GST pull-down assays (Figure 4A). The F513D mutation, known to disrupt the interaction of Tap with Nxt1, had no effect on the Tap oligomer formation (Lévesque et al., 2001, 2006). The L383,386R, NES Im9, or NES IIm6 mutants, all of which are defective for NPC binding, did not abrogate Tap binding either (Fribourg et al., 2001; Thakurta et al., 2004; Lévesque et al., 2006). We next tested the effect of mutating additional residues located on the same side as the binding interface with Nxt1 (I518A, V480A+D482A, and D438A+D457A) or on the opposite surface (R440A). None of these mutations prevented Tap complex assembly (data not shown). Two UBA mutants defective for NPC binding (Tap S585P or Tap 1-569) could also oligomerize with Tap WT (Bear et al., 1999; Lévesque et al., 2001, 2006).
Figure 4.
The amino terminus of Tap is required for the Tap complex formation. In vitro translated Tap mutants were compared for binding to GST or GST-Tap WT immobilized onto glutathione-coated beads. Bound proteins were separated by SDS-PAGE and visualized by autoradiography. Input lanes correspond to 5% of radioactive protein loads. (A) Tap constructs with mutations in the NTF2-like or UBA domains. (B) Tap deletion constructs.
We then tested Tap constructs spanning the amino terminus (fragments 1-153 and 1-302) or carboxy terminus (residues 188-619 and 260-619) for their ability to bind full length GST-Tap, and we determined that the interaction domain is located at the amino terminus of Tap (Figure 4B). We found that the first 153 amino acids of Tap were sufficient to mediate the interaction and that both Tap 1-153 and 1-302 could bind Tap WT at comparable levels (Figure 4B). Tap 1-153 still retains the RRM2-fold and the region known to interact with other RNA-binding proteins. That all NXF family members display high conservation within this region suggests that oligomerization represents an important aspect of the function of the NXF family (Izaurralde, 2002).
Nxt1 Can Bind Tap Oligomers
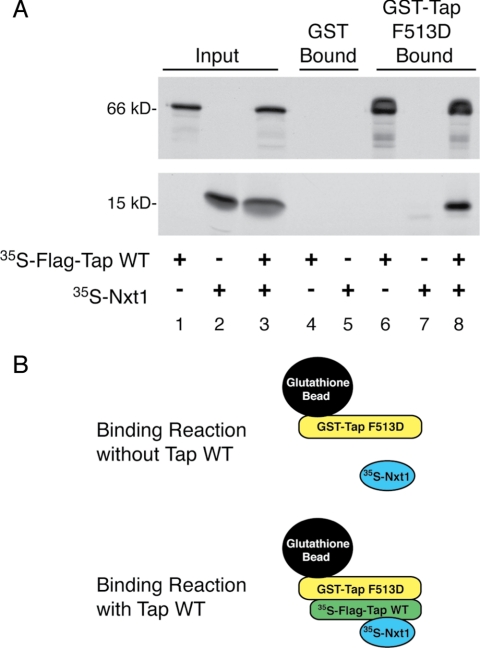
The interaction of Tap with Nxt1 significantly enhances its RNA export activity (Braun et al., 2001; Guzik et al., 2001; Wiegand et al., 2002). We knew from our previous work that Tap 260-619 and Tap WT can both bind Nxt1 equally well (Guzik et al., 2001). Because our results in Figure 4B showed that the Tap 260-619 mutant cannot assemble into Tap oligomers, we can then infer that Nxt1 can bind the monomeric form of Tap. To try to understand the significance of the oligomerization of Tap in terms of export function, we tested whether Nxt1 could also bind the Tap multimeric complex. We reasoned that failure of the oligomeric Tap to associate with Nxt1 would suggest that the Tap oligomer was less efficient at exporting RNA cargo through the NPC than the Tap monomer. To test this idea, we used a GST-Tap construct bearing the Tap F513D mutation, which is deficient for Nxt1 binding but can still associate with Tap WT (Lévesque et al., 2006; Figure 4A). As expected, 35S-FLAG-Tap WT alone bound to the GST-Tap F513D mutant whereas 35S-Nxt1 by itself did not (Figure 5, lanes 6 and 7, respectively). When the 35S-Nxt1 was added to the binding reaction together with 35S-FLAG-Tap WT, Nxt1 was recruited to the GST-Tap F513D bound fraction via its interaction with 35S-FLAG-Tap WT (lane 8). Therefore, Nxt1 can bind both monomeric and oligomeric forms of Tap.
Figure 5.
Oligomerized Tap interacts with Nxt1. (A) In vitro-translated 35S-FLAG-Tap (lanes 1, 4, and 6), 35S-Nxt1 (lanes 2, 5, and 7), or 35S-FLAG-Tap together with 35S-Nxt1 (lanes 3 and 8) were incubated with GST or GST-Tap F513D beads. Bound proteins were separated by SDS-PAGE and visualized by autoradiography. Input lanes correspond to 5% of radioactive protein loads. (B) Graphic representation of the above-mentioned experiment showing that the GST-Tap F513D mutant, unable to bind Nxt1 directly, can recruit 35S-Nxt1 indirectly when in a multimeric complex with 35S-FLAG-Tap WT.
Monomeric and Oligomeric Tap Binds Nucleoporins Differently
The export function of Tap depends on its direct association with nucleoporins. We have previously reported that Nxt1 increases the affinity of Tap for nucleoporins and increases its ability to recruit CTE RNA to nucleoporins (Lévesque et al., 2001, 2006). Indeed Tap mutants unable to bind Nxt1 do not export RNA efficiently (Guzik et al., 2001; Lévesque et al., 2006). Although the oligomerization domain of Tap was mapped to the amino terminus, away from the nucleoporin-binding regions, it is still plausible that while in an oligomeric complex, the NPC-binding domains could be occluded or sterically hindered from interacting with nucleoporins; conversely, oligomerization may induce an arrangement of NPC-binding domains more conducive to nucleoporin interactions.
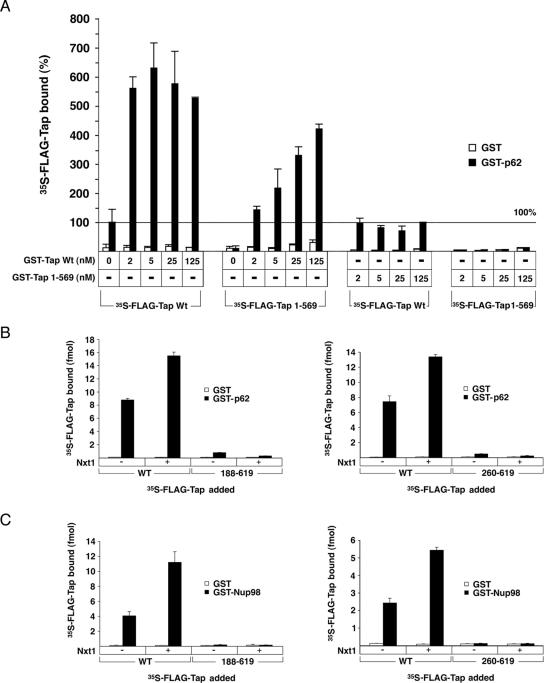
The ability of multimeric Tap to associate with nucleoporins was tested using a solid-phase binding assay. We exploited the fact that the Tap 1-569 mutant is unable to associate with the nucleoporin p62, but it could still form a mutlimeric complex with Tap WT (Lévesque et al., 2006; Figure 4A). In vitro synthesized 35S-FLAG-Tap 1-569 and 35S-FLAG-Tap WT were mixed with increasing concentrations of purified GST-Tap WT. Each protein mixture was then incubated with GST or GST-p62 immobilized to ELISA plates. The molar amount of 35S-FLAG Tap (WT or mutant) bound to GST-p62 or GST for each condition was converted to a percent binding, with 35S-FLAG-Tap WT alone set to 100%. Figure 6A shows that the addition of only 2 nM recombinant GST-Tap WT stimulated the binding of 35S-FLAG-Tap WT to p62 by fivefold. As expected, the 35S-FLAG-Tap 1-569 mutant was unable to bind GST-p62 on its own, but it was recruited to the nucleoporin when GST-Tap WT was also included in the binding reaction. To rule out the possibility that this observation was simply the result of increasing the overall Tap concentration in the binding reaction, we also tested the effect of adding recombinant GST-Tap 1-569. No binding of FLAG-Tap 1-569 could be obtained with the addition of mutant GST-Tap. The GST-Tap 1-569 also failed to stimulate the binding of FLAG-Tap WT to p62. These results support the idea that the heterotypic Tap WT/Tap 1-569 complex can associate with p62. The maximum level of 35S-FLAG-Tap 1-569 bound to p62 never reached the same level as that of 35S-FLAG-Tap WT bound, suggesting that the binding ability of the complex is linked to the presence of multiple functional NPC binding domains. We have also performed a similar experiment with immobilized His-p62 and obtained a similar stimulation of both 35S-FLAG-Tap WT and 35S-FLAG-Tap 1-569 binding to His-p62 in the presence of GST-Tap (data not shown).
Figure 6.
Tap oligomers interact with nucleoporins by solid-phase binding assay. (A) Immobilized GST (□) or GST-p62 (■) were incubated with 35S-FLAG-Tap WT or 1-569 deletion mutant. Increasing amounts of recombinant GST-Tap WT or 1-569 were also added to the binding reactions, as indicated. The level of bound radioactive protein was measured by liquid scintillation counting. Binding of 35S-FLAG-Tap WT alone when bound to GST-p62 was arbitrarily assigned a value of 100% and used to normalize the binding values obtained for all other conditions. Each data point represents the mean of three replicates (±SD). (B) Binding of 35S-FLAG-Tap 188-619 and 35S-FLAG-Tap 260-619 to immobilized GST (□) and GST-p62 (■) were compared with that of 35S-FLAG-Tap WT in the presence or absence of recombinant Nxt1. The level of bound radioactive protein was measured by liquid scintillation counting. Each data point represents the mean of three replicates (±SD). (C) Binding of 35S-FLAG-Tap mutant and WT to GST (□) or GST-Nup98-GLFG (■) in the presence or presence of Nxt1 as described in B.
We also compared the nucleoporin-binding ability of the monomeric Tap 188-619 or Tap 260-619 to that of Tap WT by using the same solid-phase binding assay. Because all of the characterized nucleoporin interaction domains are still intact in these deletion mutants, we assume that any binding differences observed between the mutants and WT Tap are attributable to the oligomeric state of the protein. Unexpectedly, by removing the oligomerization domain of Tap, we affected its association with nucleoporins in two ways. First, in the absence of Nxt1, binding to p62 was 11-fold lower for Tap 188-619 and 15-fold lower for Tap 260-619 compared with Tap WT (Figure 6B). Binding of these same mutants to Nup98 could not be detected above background (Figure 6C). The discrepancy in the level of WT versus mutant Tap being recruited to the nucleoporins was more than the fourfold disparity expected to distinguish recruitment of a Tap tetramer versus monomer to each binding site. This difference suggests that the formation of the Tap oligomer has a synergistic effect on its binding to nucleoporins. The effect may be the consequence of a local enrichment of FG-binding domains near the nucleoporin substrate as a result of the multimeric Tap complex binding nucleoporins, thereby causing an apparent increase in avidity. Alternatively, a change in protein conformation during Tap–Tap complex assembly could modify the affinity of each Tap molecule for nucleoporins.
The second difference observed between WT and mutant protein was in how Nxt1 modified their interaction with nucleoporins. The addition of recombinant Nxt1 stimulates the binding of FLAG-Tap WT to both nucleoporins as reported previously (Lévesque et al., 2001, 2006; Figure 6, A and B). Surprisingly, the addition of Nxt1 had the opposite effect on the binding of the monomeric form, even though Tap WT and Tap 260-619 seem to bind Nxt1 equally well (Guzik et al., 2001). Nxt1 actually decreased the binding of Tap 188-619 and Tap 260-619 to p62 by ∼40% (Figure 6, B and C, respectively). Our results suggest that oligomerization of Tap has a significant impact on the way Tap interacts with nucleoporins and suggest that the multimeric form may be more efficient at exporting RNA.
Cellular Distribution of Tap Mutants
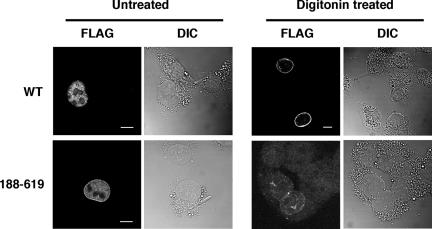
FLAG-Tap WT or 188-619 mutant were expressed in COS-7 cells and their cellular distribution examined by IF (Figure 7). Just like FLAG-Tap WT, the monomeric FLAG-Tap 188-619 could still localize to the nucleus ruling out the possibility that Tap oligomerization is necessary for its nuclear import. Whereas the WT FLAG-Tap seemed mostly nucleoplasmic, a larger pool of FLAG-Tap 188-619 could be found at the rim than throughout the nucleoplasm. We cannot determine from these experiments whether the difference in distribution between WT and mutant Tap is due to an increased accumulation of mutant Tap at the rim or decreased association of Tap 188-619 within the nucleoplasm. However, a decrease in nucleoplasmic staining would be consistent with the fact that the Tap 188-619 mutant is missing its RNA-binding domain. When cells expressing FLAG-Tap WT are treated with 0.005% digitonin before fixation, a large pool of the protein is released from the nucleoplasm but a significant fraction of WT FLAG-Tap remained associated with the NPC. Under the same conditions, FLAG-Tap 188-619 could not be detected either in the nucleoplasm or at the pores when using the same microscopy settings (data not shown). However, to verify that we were looking at fields that contained digitonin-treated cells expressing the Tap 188-619 mutant, images were also captured by increasing the detector gain. Using these settings, some trace of mutant protein in association with the nuclear rim could still be detected as seen in Figure 7. The loss of mutant protein from the NPC when cells are permeabilized correlates with the observed decreased affinity obtained with this mutant in our in vitro binding assay.
Figure 7.
Tap WT and monomer Tap 188-619 are both nuclear. IF of COS-7 cells expressing FLAG-Tap WT or FLAG-Tap 188-619. FLAG-Tap was detected using M2 mAb. Some cells were treated with 0.005% digitonin before fixation as indicated. Bar, 10 μm.
Mapping of the Residues Involved in Tap Multimeric Complex Formation
The structural domains of the RNA binding region have been well characterized. The region spanning amino acids 120-198 have the typical βαββαβ topology consistent with that of other RMM proteins such as the U2B“ and PAB (Liker et al., 2000). The Tap LRR domain, structurally homologous to the U2A' protein, has the standard concave β-pleated sheet structure, which is usually involved in protein–protein or protein–RNA interactions. Both the U2B” and U2A' proteins form a heterodimeric complex through their respective RRM and LRR domains, which regulates their interaction with the U2 snRNA (Price et al., 1998). Because of the similarities between the spliceosomal U2B“ and U2A' heterodimerizing domains and Tap, we speculated that corresponding domains of Tap may also interact in trans to form a multimer. To test our hypothesis, we selected residues for mutagenesis within these two regions for further experiments. We used point mutants within the LRR and RRM regions previously shown to be important for CTE binding (Liker et al., 2000); combined mutations of residues R128E and K129E were first selected, because structural analysis positioned these residues on the exposed surface of the RRM fold. These mutations did not prevent association with Tap WT (data not shown). We also chose to test the combined E318R and E319R mutations or a single K347E mutation within the LRR domain for the same reasons. These residues are found on the concave β-pleated sheet surface but neither mutation interfered with Tap–Tap interaction.
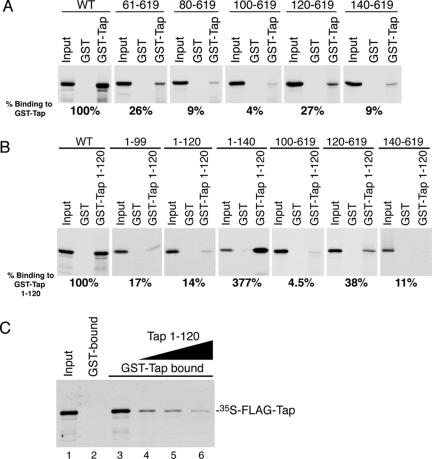
Because of the likelihood that oligomeric Tap formation involves the interaction of more than one set of residues, we decided to pursue our domain mapping by making overlapping deletion constructs within amino acids 1–140. Mutants were transcribed in vitro and tested for binding full-length GST-Tap. All of the carboxy-terminal deletion mutants tested (Tap 1-99, Tap 1-120, and Tap 1-140) bound Tap WT and full-length Tap (data not shown). In contrast, amino terminus deletions impeded the formation of a Tap complex. Tap 61-619 bound 26% of the level of Tap WT. Removal of the next 40 amino acids (Tap 80-619 and 100-619) decreased binding to below 10% of WT levels. An even shorter deletion mutant, Tap 120-619, which still retains its RNA binding domain, bound Tap WT to the same extent as the Tap 61-619 mutant. However, binding of Tap 140-619, which is missing the RRM2-fold, only bound to ∼9% of the level of Tap WT (Figure 8A). Results are summarized in Figure 9.
Figure 8.
Mapping of the Tap oligomerization domain. In vitro-translated 35S-FLAG-Tap deletion mutants were compared for binding to GST and GST-Tap WT (A) or GST-Tap 1-120 (B) immobilized onto glutathione-coated beads. Bound proteins were separated by SDS-PAGE and visualized by autoradiography. Input lanes correspond to 5% of radioactive protein loads. The level of binding for each mutant was determined by densitometry analysis and compared with the binding levels of 35S-FLAG-Tap WT. Values are expressed as percent binding as a function of 35S-FLAG-Tap WT binding. (C) In vitro-translated 35S-FLAG-Tap WT was incubated with 1 μg of GST-Tap WT immobilized onto glutathione-coated beads in the absence (lane 3) or presence of 10 μg (lane 4), 20 μg (lane 5), or 40 μg (lane 6) of untagged recombinant Tap 1-120 peptide. Bound fractions were processed and analyzed as described above.
Figure 9.
Summary diagram of ability of various Tap mutants to oligomerize with GST-Tap WT or GST-Tap 1-120 as determined by GST pull-down experiment. ND, not determined.
The same deletion mutants were also tested for binding GST-Tap 1-120 (Figure 8B). Although FLAG-Tap 1-99 and 1-120 behaved as well as FLAG-Tap WT when tested for binding to full-length GST-Tap (Figure 9), the binding of these same deletion mutants to GST-Tap 1-120 was greatly reduced (17 and 14% of FLAG-Tap WT, respectively). Conversely, FLAG-Tap 1-140 bound to GST-Tap 1-120 about threefold better than FLAG-Tap WT. In addition, we observed that FLAG-Tap 120-619 could bind GST-Tap 1-120, although at a lower extent than FLAG-Tap WT (38%). These results suggest that residues located between amino acids 120–140, which include the RRM2 domain, associate with residues located within the first 120 amino acids. They provide additional evidence that more than one domain of Tap is required for oligomerization and that Tap does not bind as head-to-head but rather as a head-to-tail oligomer.
To further demonstrate the role of the amino terminal domain in Tap oligomerization, we performed a standard GST pull-down reaction and showed that a Tap 1-120 peptide could compete with the binding of 35S-FLAG-Tap WT to GST-Tap WT (Figure 8C). The addition of 40 μg of recombinant Tap 1-120 inhibited the binding of 35S-FLAG-Tap WT to GST-Tap WT by 91% (lane 6).
DISCUSSION
This study demonstrates for the first time that Tap can assemble into a homotypic complex both in vitro and in vivo. The gel filtration data suggests that the Tap complex contains three to four Tap molecules. Our mutational analysis has delimited the Tap oligomerization region to the amino terminus, which includes part of the RNA-binding domain. More specifically, we found that two nonoverlapping deletion constructs, Tap 1-120 and Tap 120-619, could bind Tap WT providing evidence that more than one set of residues mediates this interaction. This was further supported by our finding that the self-association of Tap 1-120 was threefold lower than its binding to the Tap 120-619 fragment. Although we cannot rule out the possibility that some of these interactions are potentiated as a result of the deletion, we think that there is sufficient evidence to propose that each domain associates with the alternate domain in trans on Tap WT.
Oligomerization of RNA-binding proteins through their RNA-binding regions seems to be a recurring theme (Samuels et al., 1998; Kasashima et al., 2002; Yuan et al., 2002; Adinolfi et al., 2003). The initial structural analysis of Tap RRMs and LRR regions were found analogous to the U2B“ and U2A' splicing factors, respectively. U2B” and U2A' form a heteromeric complex that regulates binding to their cognate U2 RNA (Liker et al., 2000). It would thus be probable that the binding in trans of Tap RRM and LRR regions could also mediate the assembly of the Tap homotypic complex, Although, this seems unlikely because an amino deletion construct with an intact LRR region (Tap 188-619) failed to bind Tap WT in the present study.
Reintroduction of the RRM domain to the carboxy terminus (Tap 120-619) restored the ability of Tap to self-assemble, supporting its role for Tap–Tap binding. The RRM fold is the most common type of RNA-binding motifs found in as many as 500 different human proteins (Maris et al., 2005). These domains are thought to have primarily originated as protein–RNA interaction motifs, but they have more recently evolved to also mediate protein–protein interactions, either dependently or independently of RNAs. Interestingly, RRM-bearing proteins are implicated with all steps of RNA processing from splicing, nonsense-mediated decay, and transport (Maris et al., 2005). Some RRM proteins, such as HuD, Aly/Ref, and Sxl, can also form homotypic complexes through their RRM domains (Samuels et al., 1998; Kasashima et al., 2002; Golovanov et al., 2006). Therefore, our finding that the Tap RRM can mediate self-assembly is not unprecedented. We do not yet know how the formation of the multimeric Tap complex impacts RNA binding, but we do know that the oligomerization can occur in the absence of RNAs. Our findings that the oligomerization domain also overlaps with the region known to interact with adaptor proteins considered essential to the recruitment of export-ready mRNAs to Tap, implies an additional regulatory function for the multimeric Tap complex. It is probable that the monomeric and heteromeric Tap species interact with these adaptor proteins differently. Interestingly, many of these proteins also have RRM domains such as Aly/Ref, U2AF35, 9G8, ASF/SF2, and Srp20, and they bind the amino terminus of Tap directly (Stutz et al., 2000; Zolotukhin et al., 2002; Huang et al., 2003).
When comparing the binding abilities of Tap WT to the binding of Tap 188-619 and Tap 260-619, two mutant proteins unable to assemble into a multimeric complex, we found that these mutants bound nucleoporins with reduced efficiency. This discrepancy could be explained the binding of FG repeats by multimeric Tap being of higher avidity compared with monomeric Tap due to the local recruitment of multiple FG-binding domains in the vicinity of the nucleoporin substrate. However, we also observed that the nucleoporin-binding ability of Tap 188-619 and Tap 260-619 was further decreased when Nxt1 was included in the assay. We and others have previously reported that the association of Tap WT with Nxt1 stimulates its nucleoporin-binding affinity (Lévesque et al., 2001, 2006; Wiegand et al., 2002). Therefore, the oligomerization state of Tap seems to also regulate the effect of Nxt1. These results demonstrate for the first time that the Tap amino terminus can modulate NPC binding. Because we know that Tap 260-619 can bind Nxt1 and Tap WT (Guzik et al., 2001), it is unclear how the amino terminus dictates whether Nxt1 impedes or stimulates the interaction of Tap with nucleoporins. We speculate that the Tap multimeric assembly alters the conformation of the NPC-binding domain. However, the structural analyses of Tap and FG repeat interaction used protein fragments lacking the oligomerization domain (Fribourg et al., 2001; Grant et al., 2002, 2003). It is thus difficult to envision how the multimeric state of Tap may influence its interaction with FG repeats. Because formation of oligomers at the amino terminus can modulate NPC binding at the carboxy terminus, perhaps association of Tap with adaptor proteins through its amino terminus also modulates the association of Tap with nucleoporins.
We report that Tap is not only able to complex with itself but can also do so with all of the other human NXF family members in vitro, all of which have highly conserved RRM domains (Izaurralde, 2002). We suggest that NXFs may be capable of interacting with each other and perhaps modulate each other's functions. This possibility also brings into question the role of novel Tap variant identified by Li et al. (2006). This Tap protein, derived from an intron-retaining transcript, encodes a shorter Tap protein lacking the nucleoporin-binding domains, but it retains the first 356 amino acids of Tap. Although we have not yet tested whether the small Tap variant can bind Tap WT, we think that it should because the oligomerization domain is conserved.
The current study cannot determine whether monomeric or multimeric Tap species predominates in vivo or which of these is more proficient at moving across the NPC. It has been postulated that low affinity interactions between nucleoporins and transport receptors allow for more efficient movement through the pores, at least in the karyopherin-β family members (Ribbeck and Görlich, 2001). If the same principle holds true for Tap, one could reason that in the presence of Nxt1, the monomeric form of Tap would be a better transporter than Tap WT. However, this hypothesis was disputed in a study by Guzik et al. (2001), which compared the ability of various Tap constructs to export RNAs by using a transfection assay. In their assay, Tap fragments were tethered to the mutant RevM10 protein and expressed in 293 cells along with a reporter transcript containing a Rev-recognition element (RRE). Because the RevM10 mutant is unable to export its RRE transcript via the Crm1 pathway, export of the RRE RNA has to be mediated by the Tap moiety of the chimeric protein. This assay has the advantage of being able to assess the export efficiency of various Tap constructs independently of their RNA-binding abilities. They observed that the chimeric protein containing residues 262-619 of Tap exported RNA with a sevenfold lower efficiency than the RevM10-Tap 61-619 construct, even though both constructs still retained their complete NPC-binding domains. The RNA export efficiency obtained between the different Tap constructs correlated well with the NPC-binding affinity of similar constructs in our assays.
Our observations from both published material and the present study lend support to our hypothesis that the Tap amino terminus plays a potentially important role for RNA export that is independent of the RNA-binding function of this protein domain. We propose that the multimeric assembly of Tap facilitates its movement across pores. All of the current popular models of nuclear trafficking allow for a correlation of efficient transport with multiple FG-binding sites, such as the many FG-interaction sites known on karyopherin-β (Macara, 2001; Ribbeck and Görlich, 2002; Rout et al., 2003; Isgro and Schulten, 2005; Peters, 2005; Isgro and Schulten, 2007). Therefore, oligomerized Tap would result in a local enrichment of FG-binding sites that would more efficiently transport the RNA-cargo across pores by either diffusing along FG-tracts or “dissolving” hydrophobic FG meshwork (Macara, 2001; Ribbeck and Görlich, 2001; Rout et al., 2003; Peters, 2005; Frey et al., 2006).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Craig Mizzen for helpful comments on the manuscript. This investigation was supported by American Heart Scientist Development grant 0335362Z.
Abbreviations used:
- CTE
constitutive transport element
- LRR
leucine-rich region
- NTF2
nuclear transport factor 2
- RRM
RNA-recognition motif
- UBA
ubiquitin-associated.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0255) on October 31, 2007.
REFERENCES
- Adinolfi S., Ramos A., Martin S. R., Dal Piaz F., Pucci P., Bardoni B., Mandel J. L., Pastore A. The N-terminus of the Fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry. 2003;42:10437–10444. doi: 10.1021/bi034909g. [DOI] [PubMed] [Google Scholar]
- Bachi A., et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J., Tan W., Zolotukhin A. S., Tabernero C., Hudson E. A., Felber B. K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Holaska J. M., Lévesque L., Ossareh-Nazari B., Gwizdek C., Dargemont C., Paschal B.M. NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J. Cell Biol. 2001;152:141–155. doi: 10.1083/jcb.152.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I. C., Herold A., Rode M., Conti E., Izaurralde E. Overexpression of Tap/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- Bray M., Prasad S., Dubay J. W., Hunter E. K., Jeang T., Rekosh D., Hammarskjöld M.-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse K. N., Luo M. J., Zhou Z., Reed R. A Ran-independent pathway for export of spliced mRNA. Nat. Cell Biol. 2001;3:97–109. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Scarcelli J. J. Transport of messenger RNA from the nucleus to the cytoplasm. Curr. Opin. Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Nuclear RNA export. J. Cell Sci. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- Ernst R. K., Bray M., Rekosh D., Hammarskjöld M.-L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997a;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- Ernst R. K., Bray M., Rekosh D., Hammarskjöld M.-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 1997b;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Richter R. P., Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Fribourg S., Braun I. C., Izaurralde E., Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Golovanov A. P., Hautbergue G. M., Tintaru A. M., Lian L.-Y., Wilson S. A. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA. 2006;12:1933–1948. doi: 10.1261/rna.212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Seewald M. J., Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 2003;22:1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R. P., Hurt E., Neuhaus D., Stewart M. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat. Struct. Biol. 2002;9:247–251. doi: 10.1038/nsb773. [DOI] [PubMed] [Google Scholar]
- Grant R. P., Neuhaus D., Stewart M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1Å resolution. J. Mol. Biol. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- Grüter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B. K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Guzik B. W., Lévesque L., Prasad S., Bor Y.-C., Black B. E., Paschal B. M., Rekosh D., Hammarskjöld M. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell Biol. 2001;21:2545–2554. doi: 10.1128/MCB.21.7.2545-2554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D. N., Coburn G. A., Kang Y., Cullen B. R., Georgiadis M. M. The crystal structure and mutational analysis of a novel RNA-binding domain found in the human Tap nuclear mRNA export factor. Proc. Natl. Acad. Sci. USA. 2002;99:1888–1893. doi: 10.1073/pnas.042698599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Guan T., Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Gattoni R., Stevenin J., Steitz J. A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang Y., Steitz J. A. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Isgro T. A., Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importin-β. Structure. 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Isgro T. A., Schulten K. Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J. Mol. Biol. 2007;366:330–345. doi: 10.1016/j.jmb.2006.11.048. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur. J. Cell Biol. 2002;81:577–584. doi: 10.1078/0171-9335-00273. [DOI] [PubMed] [Google Scholar]
- Jun L., Frints S., Duhamel H., Herold A., Abad-Rodrigues J., Dotti C., Izaurralde E., Maryen P., Froyen G. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr. Biol. 2002;11:1381–1391. doi: 10.1016/s0960-9822(01)00419-5. [DOI] [PubMed] [Google Scholar]
- Kasashima K., Sakashita E., Saito K., Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 2002;30:4519–4526. doi: 10.1093/nar/gkf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Sträßer K., Podtelejnikov A., Mann M., Jung J. U., Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque L., Bor Y.-C., Matzat L. H., Jin L., Berberoglu S., Rekosh D., Hammarskjöld M.-L., Paschal B. M. Mutations in Tap uncouple RNA export activity from translocation through the nuclear pore complex. Mol. Biol. Cell. 2006;17:931–943. doi: 10.1091/mbc.E04-07-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque L., Guzik B., Guan T., Coyle J., Black B. E., Rekosh D., Hammarskjöld M.-L., Paschal B. M. RNA export mediated by Tap involves NXT1-dependent interactions with the nuclear pore complex. J. Biol. Chem. 2001;276:44953–44962. doi: 10.1074/jbc.M106558200. [DOI] [PubMed] [Google Scholar]
- Li Y., Bor Y.-C., Misawa Y., Xue Y., Rekosh D., Hammarsjköld M.-L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- Liker E., Fernandez E., Izaurralde E., Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I. G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F. H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- Price S. R., Evans P. R., Nagai K. Crystal structure of the spliceosomal U2B“-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Reed R., Cheng H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C., Stutz F. Nuclear export of RNA. Biol. Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchinson J. D., Magnasco M. O., Chait B. T. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Samuels M., Deshpande G., Schedl P. Activities of the Sex-lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res. 1998;26:2625–2637. doi: 10.1093/nar/26.11.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I., Gerace L. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J. Biol. Chem. 2001;276:42355–42363. doi: 10.1074/jbc.M103916200. [DOI] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol. Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Stutz F., Bachi A., Doerks T., Braun I. C., Seraphin B., Wilm M., Bork P., Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with Tap/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Doerks T., Braun I. C., Sattler M., Izaurralde E., Bork P. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 2000;1:53–58. doi: 10.1093/embo-reports/kvd009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Zolotukhin A. S., Bear J., Patenaude D. J., Felber B. K. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurta A. G., Gopal G., Yoon J. H., Saha T., Dhar R. Conserved nuclear export sequences in Schizosaccharomyces pombe Mex67 and human TAP function in mRNA export by direct nuclear pore interactions. J. Biol. Chem. 2004;279:17434–17442. doi: 10.1074/jbc.M309731200. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125(6):1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Wiegand H. L., Coburn G. A., Zeng Y., Kang Y., Bogerd H. P., Cullen B. R. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie G. S., Zimyanin V., Kirby R., Korey C., Francis-Lang H., Van Vactor D., Davis I. Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA. 2001;7:1781–1792. [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Davydova N., Conte M. R., Curry S., Matthews S. Chemical shift mapping of RNA interactions with the polypyrimidine tract binding protein. Nucleic Acids Res. 2002;30:456–462. doi: 10.1093/nar/30.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin A. S., Tan W., Bear J., Smulevitch S., Felber B. K. U2AF participates in the binding of TAP (NXF1) to mRNA.J. Biol. Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
- Zolotukhin A. S., Valentin A., Pavlakis G. N., Felber B. K. Continuous propagation of RRE(-) and Rev(-)RRE(-) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J. Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.