Abstract
We previously revealed a novel signal pathway involving S100A11 for inhibition of the growth of normal human keratinocytes (NHK) caused by high Ca++ or transforming growth factor β. Exposure to either agent resulted in transfer of S100A11 to nuclei, where it induced p21WAF1. In contrast, S100A11 has been shown to be overexpressed in many human cancers. To address this apparent discrepancy, we analyzed possible new functions of S100A11, and we provide herein evidence that 1) S100A11 is actively secreted by NHK; 2) extracellular S100A11 acts on NHK to enhance the production of epidermal growth factor family proteins, resulting in growth stimulation; 3) receptor for advanced glycation end products, nuclear factor-κB, Akt, and cAMP response element-binding protein are involved in the S100A11-triggered signal transduction; and 4) production and secretion of S100A11 are markedly enhanced in human squamous cancer cells. These findings indicate that S100A11 plays a dual role in growth regulation of epithelial cells.
INTRODUCTION
S100 proteins are small molecules (10∼12 kDa) with two EF-hands, a canonical hand at the C terminus and a modified, S100-specific hand at the N terminus (Donato 2001; Heizmann et al., 2002; Marenholz et al., 2004). Binding of Ca++ to EF-hand motifs changes the conformation and hence the function of S100 proteins (Rety et al., 2000; Dempsey et al., 2003). Although S100 family proteins share such structural features, they show different cell type- and functional state-specific expression profiles in different tissues, and they exhibit distinct functions in diverse physiological and pathological contexts (Donato 2001; Heizmann et al., 2002; Emberley et al., 2004; Marenholz et al., 2004).
S100A11 (S100C) was cloned from the chicken gizzard in 1991 (Todoroki et al., 1991), and it was later shown to bind to annexin A1 (Mailliard et al., 1996), but its function has remained largely unknown. Because S100A11 is encoded in epidermal differentiation complex on chromosome 1 in humans, we studied the biological role of S100A11 in normal human keratinocytes (NHK), and we found that it is an essential element in the signal transduction for high Ca++- and transforming growth factor (TGF)β-induced suppression of the growth of NHK (Sakaguchi et al., 2003, 2004, 2005). On exposure of NHK to either agent, S100A11 was transferred to nuclei, where it induces p21WAF1. Blocking the S100A11-mediated pathway abrogated the growth suppression by high Ca++ and TGFβ, even though the canonical nuclear factor of activated T cells (NFAT)1-mediated pathway for high Ca++ and the Smad-mediated pathway for TGFβ remained intact. Thus, high Ca++ and TGFβ have a common S100A11-mediated pathway in addition to unique pathways for exhibiting an inhibitory effect on the growth of NHK, and both pathways are indispensable (Sakaguchi et al., 2005).
Conversely, S100A11 has been shown to be overexpressed in many different types of human cancer, including those derived from the esophagus (Ji et al., 2004), stomach (Oue et al., 2004), colorectum (Tanaka et al., 1995; Stulik et al., 1999), pancreas (Ohuchida et al., 2006), thyroid (Torres-Cabala et al., 2004), uterus (Kanamori et al., 2004), and soft tissues (Schaefer et al., 2004). Although the widely observed up-regulation of S100A11 indicates that S100A11 may be involved in growth enhancement, malignant progression of cancer cells, or both, no mechanistic interpretation has been provided. If S100A11 plays a role solely in a growth-suppressive pathway (Sakaguchi et al., 2005), its up-regulation in cancer cells might simply be a compensatory response to enhanced growth of malignant cells. It is more likely, however, that S100A11 is involved in yet-unknown mechanisms featured in cancer cells.
Some S100 family members are known not only to exert their function intracellularly but also to be secreted and act on cell surface receptors. S100A8 and S100A9 are secreted by inflammatory cells, such as neutrophils, activated monocytes, and macrophages, and they act as chemotactic molecules for those cells (Roth et al., 2003; Ryckman et al., 2003; Foell et al., 2004). Serum levels of S100A8 and S100A9 have been shown to be increased in chronic inflammatory diseases, including rheumatoid arthritis (Liao et al., 2004), multiple sclerosis (Bogumil et al., 1998), Crohn's disease (Lugering et al., 1995), and connective tissue diseases (Kuruto et al., 1990). S100B is abundantly produced and secreted by astrocytes, and it functions as a trophic factor for neurons and glial cells in autocrine and paracrine manners (Adami et al., 2001). Increased serum level of S100B has been observed in various inflammatory, ischemic, and psychiatric diseases of the human brain (Rothermundt et al., 2003). Recently, Cecil et al. (2005) reported that S100A11 was secreted from and induced hypertrophic change in human chondrocytes.
In the present study, we examined possible new roles of S100A11 in an alternative pathway of growth regulation of NHK. We found that S100A11 is secreted from NHK and acts on the cells to promote cell growth via induction of epidermal growth factor (EGF). Thus, S100A11 functions as a dual mediator for growth regulation of NHK.
MATERIALS AND METHODS
Cells and Chemicals
Neonatal human epidermal keratinocytes (KURABO, Osaka, Japan) were cultured in Epilife medium with HKGS Growth Supplement (Cascade Biologics, Portland, OR). A431, BSCC-93, and HSC-5 were purchased from American Type Culture Collection (Manassas, VA) and cultured in DMEM with 10% fetal bovine serum (FBS). For A431, the medium was supplemented with 4 mM l-glutamate and 4.5 g/l glucose. Inhibitors for EGF receptor (AG1478), nuclear factor (NF)-κB (NF-κB activation inhibitor IV) and Akt (protein kinase B) were purchased from Calbiochem (San Diego, CA). Histamine, substance P, and recombinant human TGFβ were purchased from Sigma-Aldrich (St. Louis, MO). Recombinant human EGF, interferon (IFN)α, tumor necrosis factor (TNF)-α, and interleukin (IL)-6 were provided by Peprotech (London, England). Recombinant proteins of human IL-1F9, IL-8/CXCL8, CXCL1, and soluble receptor for advanced glycation end products (RAGE) (sRAGE/Fc chimera), neutralizing mouse antibodies against human EGF and RAGE, and control mouse immunoglobulin (Ig)G (R&D Systems, Minneapolis, MN) were obtained from the designated sources.
Recombinant Protein Preparation
S100A11 cDNA was obtained by RT-PCR and integrated into pET3a (Novagen, Madison, WI). S100A11 protein was purified by differential precipitation using successively polyethylenimine and ammonium sulfate followed by ion exchange column chromatography. Dimer S100A11 was prepared by incubating S100A11 in a HEPES buffer, pH 7.5, at 4°C for 2 wk, and the resulting dimer was fractionated by gel filtration. For biotinylation of S100A11, 3-fold excess moles of Biotin-(AC5)2sulfo-OSu (Dojindo, Gaithersburg, MD) was reacted with S100A11 at room temperature for 2 h, and the nonreacted reagent was inactivated with Nap-5 (GE Healthcare Bio-Sciences, Little Chalfont, Buckinghamshire, United Kingdom). About 0.95 molecules of biotin was conjugated per S100A11 molecule as assayed by the HABA method (Pierce Chemical, Rockford, IL). Labeling of S100A11 by Cy3 was performed using a Cy3 antibody labeling kit (GE Healthcare Bio-Sciences).
cDNAs of S100A2 and S100A6 and cDNA of S100A10 were cloned into pGEX2T and pGEX6P1 vectors, respectively (GE Healthcare). The glutathione transferase (GST)-fused recombinant proteins were purified using a Sephadex 4B column after cleaving with Thrombin or PreScission protease (GE Healthcare Bio-Sciences) under conventional conditions.
Western Blot Analysis
Western blot analysis was performed under conventional conditions. Antibodies used are listed in Supplemental Material. The membranes were then treated with horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (Cell Signaling Technology, Danvers, MA), and positive signals were finally visualized by a chemiluminescence system (ECL Plus; GE Healthcare Bio-Sciences).
Rabbit anti-human S100A11 antibody was raised in our laboratory using S100A11 dimer as an immunogen. The purified antibody was confirmed to react with monomer S100A11 and dimer S100A11 and also to neutralize S100A11 (Supplemental Figure S9).
Subfractionation of cells was performed using a subfractionation kit (NE-PER nuclear and cytoplasmic extraction reagent kit; Pierce Chemical). The plasma membrane fraction was prepared under conditions similar to those described previously (Sarkadi et al., 1992).
Northern Blot Analysis
Northern blot analysis was performed under conventional conditions. Twenty micrograms of total RNAs was electrophoresed, transferred to Nytran Plus nylon membranes (GE Healthcare Bio-Sciences), and probed with a 502-base pair fragment of EGF cDNA amplified using forward (5′-atgctgctcactcttatcat-3′) and reverse (3′-ctggatcaactgctacattt-5′) primers.
Arrays
Extracts prepared from NHK cultured with or without S100A11 were analyzed using a proteome profiler array (Human Phospho-MAPK Array; R&D Systems) and a cytokine antibody array (Human Cytokine Antibody Array IV; RayBiotech, Norcross GA).
A Method to Fish Promoter-binding Proteins
We newly developed a method to identify proteins binding to a long region of a given promoter (Supplemental Figure S5). Nuclear extracts preabsorbed with an excess amount of streptavidin agarose were incubated in a buffer [0.1% Triton X-100, 60 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 50 μg/ml poly(dI-dC), 1 mM dithiothreitol (DTT), 5% glycerol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 200 μg/ml phenylmethanesulfonyl fluoride, and 20 mM HEPES, pH 7.4] at 4°C for 120 min with a biotinylated 2081-base pair genomic fragment of the human EGF promoter that was prepared using forward (5′-biotin-atctgggcaactgtagatga-3′) and reverse (3′-aaggtgagtatgattgaccc-biotin-5′) primers. The resulting protein–primer complex was isolated with streptavidin agarose (Invitrogen, Carlsbad, CA) and then subjected to Western blot analysis. Oligomers used for competitive experiments for which results were shown in Figure 4C were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
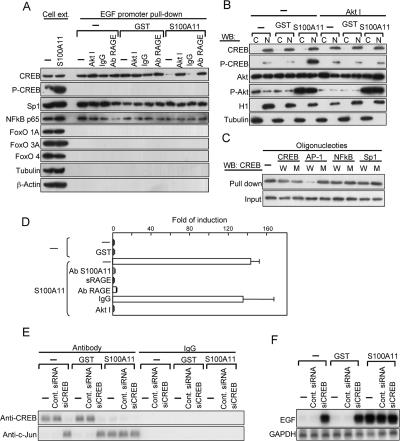
Figure 4.
Transcriptional activation of EGF gene by S100A11. (A) Transcription factors bound to the EGF promoter identified by a newly developed method. NHK were precultured in HKGS-free EpiLife for 24 h. An Akt inhibitor and anti-RAGE antibody were added 1 h before the addition of S100A11 or GST and harvested 6 h later. Cell extracts were incubated with a biotinylated EGF promoter fragment (2085 base pairs), and proteins pulled down with streptavidin beads were analyzed by Western blot analysis. (B) Copresence of activated Akt and phosphorylated CREB in nuclei of NHK exposed to S100A11. Histone H1 (H1) and tubulin were used as indicators for nuclei and cytoplasm, respectively. (C) Competitive inhibition of binding of CREB to the EGF promoter by transcription factor-binding elements. Wild-type (W) or mutant (M) oligomers were added to the incubation mixture under conditions similar to those described in A. (D) A luciferase assay for the seventh AP-1 site of the EGF promoter (see Supplemental Figure S7). The seventh AP-1 site was responsive to S100A11 (10 ng/ml), and this was abrogated by blocking of the function of S100A11 (with an antibody, 20 μg/ml), RAGE (with soluble RAGE at 1 μg/ml), and Akt (with an inhibitor at 10 μM). (E) Chromatin immunoprecipitation assay for proteins bound to the seventh AP-1 site described in D. (F) Effect of down-regulation of CREB by siRNA on induction of the EGF gene examined by Northern blot analysis.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed under conditions similar to those described previously (Sakaguchi et al., 2005). Rabbit antibodies against human cAMP response element-binding protein (CREB) and c-Jun were obtained from Santa Cruz Biotechnology. Forward (5′-agcgagttatctcctctttggcagt-3′) and reverse (3′-acagagcaaggcaaaggcttagaga-5′) primers were used to amplify a 290-base pair fragment covering an AP1 site of the EGF promoter (−167 to +122).
Small Interfering RNAs (siRNAs)
siRNAs against human CREB (predesigned siRNA for CREB) and control siRNA (Silencer Negative control 2 siRNA) were purchased from Ambion (Austin, TX). siRNAs were transfected using Lipofectamine 2000 (Invitrogen).
Luciferase Assay
Genomic fragments of 2081 and 779 base pairs of the human EGF promoter were inserted into pGL4.14-Basic (Promega, Madison, WI) at Kpn1 and Xho1 sites. For more detailed conditions, see Supplemental Material. Nucleotide sequences of the constructs were confirmed by sequencing.
The plasmids were cotransfected with a pTracer-EF/lacZ vector (Invitrogen) into NHK by using Trans IT-keratinocyte transfection reagent (Mirus, Madison, WI). Luciferase activity of cell extracts was measured with a Luciferase reporter assay system (LucLite; Packard Bioscience, Groningen, The Netherlands). The luciferase activities were normalized to β-galactosidase activity derived from a cotransfected plasmid.
RESULTS
Secretion of S100A11 by NHK
At first, we examined possible secretion of S100A11 by NHK. NHK were incubated in EpiLife without HKGS. Addition of FBS, EGF, and IL-1F9 dose-dependently enhanced production and secretion of S100A11 (Figure 1). Similar amounts of S100A11 protein were recovered from the medium with those from the cells when analyzed 24 h after replacement of the medium. Calcium, IL-8, CXCL1, and TGFβ enhanced production but not secretion of S100A11 (Supplemental Figure S1A). IL-6, TNF-α, IFNα, substance P, and histamine showed no appreciable effects (Supplemental Figure S1B). Secretion of S100A11 was not due to decomposition of NHK, because no appreciable increase in the release of lactate dehydrogenase from treated NHK was observed (data not shown).
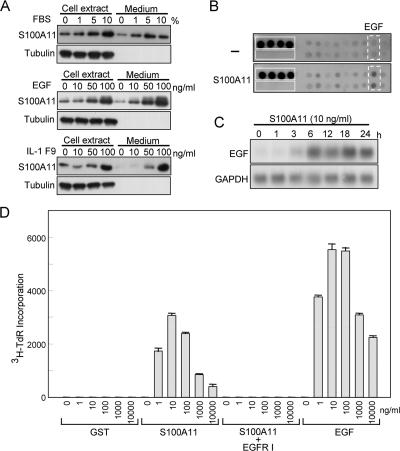
Figure 1.
Secretion of and growth stimulation by S100A11 in NHK. (A) Production and secretion of S100A11. NHK were cultured in EpiLife medium without HKGS but with designated additives for 24 h. Equivalent aliquots of whole protein preparations from the cell extract, and the culture medium was applied for Western blot analysis. Tubulin was used as a control for the applied amount of protein preparations. (B) Screening for cytokines induced by S100A11 (10 ng/ml; 24 h) by using a human cytokine antibody array. Dotted squares, EGF; solid squares, positive and negative controls. (C) Induction of EGF by S100A11 in NHK determined by Northern blot analysis. NHK were cultured in HKGS-free EpiLife 24 h before addition of S100A11. (D) Growth stimulation of NHK by exogenous S100A11. NHK were cultured in HKGS-free EpiLife 24 h before addition of S100A11 and incubated for a further 24 h with the designated cytokines. [3H]Thymidine (1 μCi/ml) was added to the medium 1 h before harvest, and radioactivity in an insoluble fraction was counted. GST was used as a control. Epidermal growth factor receptor (EGFR) I, 10 μg/ml an inhibitor for the EGF receptor AG1478.
Enhancement of the Growth of NHK by Exogenous S100A11
To study functions of secreted S100A11, we prepared extremely pure recombinant S100A11 protein mainly using ion-exchange column chromatography (Supplemental Figure S2). Addition of the purified S100A11 to the HKGS-free culture of NHK remarkably stimulated DNA synthesis even at a concentration as low as 1 ng/ml (Figure 1D). GST produced in the same Escherichia coli system and EGF were used as negative and positive controls, respectively.
Screening for cytokines induced by exogenous S100A11 by using a cytokine antibody array resulted in identification of EGF (Figure 1B). Induction of the EGF gene was confirmed by Northern blot analysis (Figure 1C). The amount of EGF protein secreted over a period of 48 h by NHK exposed to 10 ng/ml S100A11 was determined by enzyme-linked immunosorbent assay to be 17.5 pg/ml in 10 ml of HKGS-free EpiLife medium, whereas EGF level in the medium of untreated NHK was below the detection limit (< 5 pg/ml). An inhibitor of the EGF receptor abrogated the growth stimulation of NHK exerted by S100A11 (Figure 1D). In addition to EGF, betacellulin, epiregulin, heparin binding (HB)-EGF, and amphiregulin (although delayed in comparison with the other ligands) but not TGFα were induced by S100A11 among known ligands to the EGR receptor (Supplemental Figure S3). These results indicate that exogenous S100A11 stimulates the growth of NHK mainly through activation of the EGF receptor. We hence studied the intracellular signal transduction pathway for the induction of EGF, a representative mediator for growth stimulation by exogenous S100A11.
Akt, an Essential Element for EGF Induction by S100A11
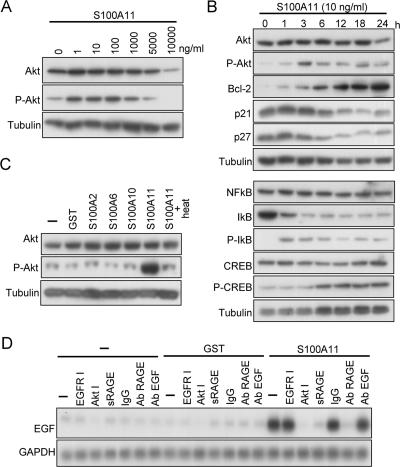
To gain insight into intracellular signaling of the EGF induction by S100A11, we screened protein kinases that were activated in NHK exposed to S100A11 by using a phosphoprotein antibody array. We found that phosphorylation of Akt was induced by S100A11 (Supplemental Figure S4), and this was confirmed by Western blot analysis (Figure 2A). Even 1 ng/ml S100A11 increased the phosphorylation level of Akt, and concentrations higher than 5 μg/ml showed a suboptimal effect. The phosphorylation level of Akt was elevated at 1 h and further elevated at 3 h after exposure of NHK to S100A11 (Figure 2B). The enhanced phosphorylation state of Akt was sustained up to 24 h. It should also be noted that antiapoptotic Bcl-2 was substantially up-regulated, whereas negative growth regulators p21WAF1 and p27KIP1 were down-regulated. Induction of phosphorylation of Akt seemed specific to S100A11 among S100 family members, because S100A2, S100A6, and S100A10 showed no effect (Figure 2C). Heating inactivated S100A11 and resulted in loss of its capacity to induce phosphorylation of Akt, excluding the possibility of involvement of contaminated LPS that is heat-stable in nature. Activation of Akt is essential for the EGF induction by S100A11 because an Akt inhibitor abrogated it (Figure 2D).
Figure 2.
Activation of Akt by S100A11 in NHK. (A) Induction of phosphorylation of Akt. NHK were cultured in HKGS-free EpiLife 24 h before addition of S100A11 protein and incubated for a further 24 h before harvest. Cell extracts were analyzed by Western blot analysis. (B) Time course of activation of Akt and related proteins by S100A11. The experiment was performed under conditions similar to those described in A except for incubation times after addition of S100A11. (C) Specificity in Akt activation among S100 family members. The experiment was performed under conditions similar to those described in A. The recombinant proteins were added at 10 ng/ml. S100A11 was heated at 100°C for 20 min. (D) Inhibition of EGF induction by S100A11 by various agents demonstrated by Northern blot analysis. The experiment was performed under conditions similar to those described in A except for incubation time of 6 h after addition of S100A11 or GST at 10 ng/ml.
Function of RAGE as a Receptor for S100A11
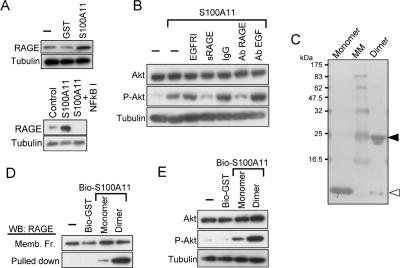
RAGE is a multiligand receptor binding to at least some S100 family members, including S100A11 (Marenholz et al., 2004; Cecil et al., 2005). RAGE was expressed in NHK as demonstrated by Western blot analysis, and the protein level was enhanced by S100A11 (Figure 3A). Addition of a NF-κB inhibitor resulted in an undetectable level of RAGE, indicating that NF-κB is needed not only for induction of RAGE by S100A11 but also for constitutive expression of RAGE. Blocking of RAGE function by either soluble RAGE or an antibody against RAGE completely suppressed the induction of EGF (Figure 2D) and phosphorylation of Akt (Figure 3B) by S100A11. Neither an EGF receptor inhibitor nor an antibody against EGF showed any effect on phosphorylation of Akt, indicating that activation of Akt is upstream to the action of EGF. S100A11 is known to form covalently linked dimers or multimers, and this process facilitates the binding to RAGE (Marenholz et al., 2004; Moroz et al., 2002). We prepared monomer and dimer preparations of S100A11 as described in Materials and Methods (Figure 3C). The S100A11 dimers bound to RAGE more readily and enhanced phosphorylation of Akt to a higher level than did the monomers (Figure 3, D and E).
Figure 3.
Involvement of RAGE in signal transduction for S100A11. (A) Expression of RAGE in NHK as detected by Western blot analysis. Top, NHK were precultured in HKGS-free EpiLife for 24 h. Cells were harvested 6 h after addition of S100A11 or GST (10 ng/ml). Bottom, after being precultured as described above, NHK were incubated with an inhibitor of NF-κB (0.5 μg/ml) followed by exposure to S100A11 at 10 ng/ml for 16 h. (B) Abrogation of S100A11-induced Akt activation by functional blocking of RAGE. EGFR I, 10 μg/ml AG1478; sRAGE, 1 μg/ml soluble RAGE; antibody (Ab), 20 μg/ml antibody. NHK were pre-cultured in HKGS-free EpiLife for 24 h. The agents were added 1 h before the addition of S100A11 and harvested 6 h later. (C) Preparation of monomer S100A11 and dimer S100A11 (see Materials and Methods). The membrane was stained with Coomassie Brilliant Blue. MM, molecular markers. White arrowhead, monomer; black arrowhead, dimer. (D) Binding of S100A11 monomer and S100A11 dimer to RAGE. Biotinylated proteins (0.1 nmol) were incubated with 1 mg of membrane preparation of NHK, pulled down with a streptavidin agarose, and analyzed for RAGE by Western blot analysis. (E) Phosphorylation of Akt by S100A11 monomer and S100A11 dimer (10 μg/ml). The experiment was performed under conditions similar to those described in B.
Mechanisms of Transcriptional Activation of EGF by S100A11
To explore mechanisms of transcriptional activation of the EGF gene, we first pulled down proteins bound to the EGF promoter by a newly developed method that enables fishing for proteins binding to a long region covering 2085 base pairs (see Materials and Methods and Supplemental Figure S5). CREB, simian virus 40 promoter factor 1, and NF-κB were detected among candidate transcription factors examined (Figure 4A). NF-κB was probably activated by S100A11 through phosphorylation and subsequent degradation of inhibitor of nuclear factor-κB (IκB) (Figure 2B). Exposure of NHK to S100A11 changed the binding mode of only CREB, i.e., liberated from the EGF promoter, and this was associated with phosphorylation of CREB (Figures 2B and 4A). CREB is one of the direct substrates of phosphorylation by Akt (Du and Montminy, 1998). In NHK, activated Akt was partly present together with CREB in nuclei, and the Akt inhibitor blocked phosphorylation of CREB induced by S100A11 (Figure 4B and Supplemental Figure S6). Binding of CREB to the EGF promoter region was inhibited by an activator protein (AP)-1 element (Figure 4C), indicating that CREB binds to AP-1 sites of the promoter. The EGF promoter contains 7 AP-1 sites. The proximate seventh site was shown by a luciferase assay to be critical for the induction by S100A11 (Supplemental Figure S7). Transcriptional activation via the 7th AP-1 site by S100A11 was completely suppressed by application to NHK of an antibody against S100A11, soluble RAGE, an antibody against RAGE, or an Akt inhibitor (Figure 4D). A chromatin immunoprecipitation assay revealed that on exposure to S100A11 CREB was liberated from the default binding site of the EGF promoter in NHK (Figure 4E). c-Jun showed a reciprocal binding behavior to the site. Down-regulation of CREB by siRNA (Supplemental Figure S8) induced transcription of the EGF gene by itself (Figure 4F). These results indicate that liberation of CREB from the 7th AP-1 site through phosphorylation triggers transcriptional activation of the EGF gene by AP-1.
Growth Regulation via Endogenous S100A11 in NHK and Squamous Cell Carcinomas (SSCs)
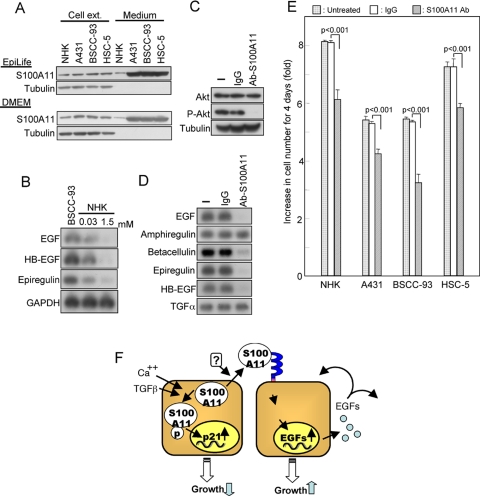
Finally, we examined involvement of endogenous S100A11 in growth regulation of NHK and squamous carcinoma cell lines A431, BSCC-93, and HSC-5. Growing squamous carcinoma cell lines produced and secreted larger amounts of S100A11 than did NHK regardless of the culture medium, i.e., EpiLife or DMEM (Figure 5A). EGF, HB-EGF, and epiregulin were constitutively expressed at higher levels in BSCC-93 cells than in NHK (Figure 5B). Higher expression levels of the EGF family members were also observed in A431 and HSC-5 cells (data not shown). In BSCC-93 cells, the constitutively active state of endogenous Akt was suppressed by addition of an anti-S100A11 antibody (Figure 5C). The antibody was confirmed to efficiently block the binding of S100A11 to the cell surface receptor (Supplemental Figure S9). In accordance with the reduced phosphorylation level of endogenous Akt, expression of EGF, betacellulin, epiregulin, and HB-EGF, which were induced by S100A11 in NHK (Supplemental Figure S3), was suppressed by a neutralizing anti-S100A11 antibody (Figure 5D). Growth of the three squamous cancer cell lines and NHK was partially but significantly compromised by addition of the neutralizing antibody against S100A11 (Figure 5E). These results indicate that secreted endogenous S100A11 is partly involved in sustaining growth of the cells.
Figure 5.
S100A11 in SSC cell lines. (A) Production and secretion of S100A11 in SCCs (A431, BSCC-93, and HSC-5) determined under conditions similar to those described in the legend to Figure 1A. (B) Higher constitutive expression levels of EGF, HB-EGF, and epiregulin in BSCC-93 cells than in NHK (cultivated with Ca++ concentration of 0.03 or 1.5 mM) as determined by Northern blot analysis. (C) Down-regulation of constitutively active Akt in BSCC-93 cells by a neutralizing antibody against S100A11 (20 μg/ml; 24 h) as shown by Western blot analysis. (D) Down-regulation of constitutive EGF family mRNA levels by an anti-S100A11 antibody (20 μg/ml; 24 h). (E) Compromise of the growth of NHK and SCCs by an anti-S100A11 antibody. Increase in cell number over a 4-d culture period with or without the antibody (20 μg/ml added at 0 and 2 culture days) was determined. (F) Schematic presentation of the ambivalent role of S100A11 in growth regulation of NHK.
Ambivalent Nature of S100A11 as a Mediator for Growth Regulation
As described previously (Sakaguchi et al., 2003, 2004, 2005), S100A11 functions as an essential mediator of high calcium and TGFβ for growth suppression of NHK. Conversely, our results demonstrated a growth enhancing function of secreted S100A11. The ambivalent pathways are schematically summarized in Figure 5F.
DISCUSSION
Our previous studies clearly demonstrated that S100A11 functions as an essential mediator for growth suppression of NHK triggered by representative growth inhibitors, high Ca++ and TGFβ, in cells (Sakaguchi et al., 2003, 2004, 2005). Alternatively, the present study provides evidence that S100A11 enhances growth of NHK through induction of EGF and some other ligands to the EGF receptor when acting on the cells from the outside. Thus, S100A11 plays an ambivalent role with respect to growth regulation of NHK.
Involvement of S100A11 in both growth-suppressive and growth-stimulatory signaling in NHK seems to be paradoxical. We have been learning, however, more and more complex functions of a given protein in multiple biological contexts with recent progress in molecular and cellular biology, and S100A11 is not an exception. S100A11 is constitutively expressed in NHK. Under ordinary culture conditions for NHK, the medium contains EGF. EGF enhanced the production and secretion of S100A11 (Figure 1A), which in turn induced EGF in an autocrine/paracrine manner (Figure 1, B and C). Exposure of growing NHK to either Ca++ or TGFβ overrides growth of NHK supported by EGF, and S100A11 cell autonomously functions as an indispensable mediator of the growth suppression via phosphorylation and translocation into nuclei (Sakaguchi et al., 2003, 2004, 2005). The mechanistic link between EGF and ambivalent S100A11 demonstrated in the present study may contribute to more exquisite growth regulation and fine tunings.
Secretion of S100A11 was first described in chondrocytes by Cecil et al. (2005). S100A11 was markedly overexpressed and secreted into the surrounding matrix by human chondrocytes in osteoarthritis. In the present study, we showed that S100A11 was also secreted from epithelial cells. S100A11 lacks a classical signal peptide sequence for secretion via the endoplasmic reticulum-Golgi pathway. Although several possible mechanisms have been proposed and partly verified for unconventional secretion routes (Nickel, 2005), the mechanisms by which of S100A11 and other secretory S100 family members are secreted remain to be clarified. In this respect, it is noteworthy that S100A13 was shown to be cosecreted with fibroblast growth factor (FGF)1, both of which lack the conventional signal sequence (Landriscina et al., 2001). An inhibitor of actin stress fiber formation suppressed secretion of FGF1 and hence of S100A13, which is known to bind to the actin fiber (Prudovsky et al., 2002). S100A11 also has a capacity to bind to the actin fiber (Sakaguchi et al., 2000). Phosphorylation of S100A11 does not seem to be involved in the secretion. High Ca++ induced phosphorylation of S100A11 (Sakaguchi et al., 2003), but it did not affect its secretion (Supplemental Figure S1A), whereas EGF enhanced the secretion (Figure 1A), but it showed no effect on phosphorylation of S100A11 (data not shown). S100A11 protein was identified in human serum by condensation using affinity chromatography and detection by Western blotting (data not shown), indicating that secretion of S100A11 takes place in vivo and is not merely an in vitro phenomenon.
RAGE is a multiligand receptor that belongs to the immunoglobulin superfamily. RAGE was first identified as a receptor for glycation end products of proteins (Schmidt et al., 1992), but it was soon found to bind diverse ligands, including amyloid β (Yan et al., 1996), HMGB1 (amphoterin) (Hori et al., 1995), and some S100 family members (Donato, 2003; Hofmann et al., 1999). Cecil et al. (2005) showed that exogenous S100A11 acted on articular chondrocytes to increase the production of type X collagen and CXCL8/IL8 in culture and that addition of an anti-RAGE antibody abrogated the action of S100A11. Abrogation of the effect of exogenous S100A11 by a neutralizing antibody was also observed in the present study (Figures 3B and 4A). In addition, it was shown that RAGE was coprecipitated with S100A11 from extracts of NHK with a higher efficiency for dimerized S100A11, a functionally more active form than monomeric S100A11 (Figure 3D). Although binding to the common receptor RAGE, different S100 family members seem to exert distinct biological effects. Heterodimeric S100A8 and S100A9, candidate ligands for RAGE (Ehlermann et al., 2006), neither induced EGF nor activated Akt, but they induced cytokines such as IL-8, IL-1F9, and TNF-α in NHK (data not shown). Hsieh et al. (2004) showed that intracellular translocation of S100A4, S100A12, S100A13, and S100B was brought about by application of the respective proteins onto endothelial cells. Although those S100 proteins seemed to share a common receptor, RAGE, intracellular translocation was observed in an individual protein-specific manner. The underlying mechanisms of the differential signal transduction by RAGE remain to be elucidated.
NF-κB is a well-known immediate mediator for RAGE-triggered signaling (Huttunen et al., 1999). In accordance with this, phosphorylation and subsequent degradation of IκB were observed within 1 h in NHK exposed to S100A11 (Figure 2B). Activation of Akt by phosphorylation was essential for the induction of EGF (Figures 2D and 4A). The phosphorylation level of Akt was appreciably elevated at 1 h and further elevated at 3 h after exposure of NHK to S100A11 (Figure 2B). The enhanced phosphorylation state of Akt was sustained up to 24 h. Transient and sustained modes of Akt activation have been shown to often lead to distinct outcomes in various biological contexts (Foulstone et al., 1999). The products of phosphatidylinositol 3-kinase (PI3K) are absolutely necessary for activation of Akt (Alessi et al., 1996), and PI3K is activated by RAGE via generation of reactive oxygen species (Alessi et al., 1996; Xu and Kyriakis, 2003).
To identify transcription factors involved in induction of the EGF gene, we developed a new method to fish proteins binding in vitro to a long region of the EGF promoter (Supplemental Figure S5). The method is facile and costless, and it can be applied for any promoters. Although the length of the target promoter region is restricted by size amplifiable by PCR, usually 2∼3 kb because of the necessity of a substantial amount of DNA, longer regions (>10 kb) may be analyzed by mixing flanking segments amplified separately. It is also possible to screen unknown promoter-binding proteins by combination with mass spectrometry. The electrophoretic mobility shift assay, another analytical method for in vitro protein-promoter binding, is usually used for a specific short segment of the promoter. The chromatin immunoprecipitation assay is a strong tool for analyzing in vivo binding of a given protein to a defined region of the promoter. It is also possible to screen unknown binding regions in genomic DNA when combined with a microarray (chromatin immunoprecipitation on chip; Wu et al., 2006).
S100A11 was produced and secreted at higher levels in squamous cell carcinoma cells than in NHK (Figure 5A). As expected, higher mRNA levels of EGF, HB-EGF, and epiregulin were observed in BSCC-93, A431, and HSC-5 cells than in NHK (Figure 5B; data not shown). If the S100A11-EGF family member axis constitutively functions in squamous carcinoma cells, sequestering of endogenous S100A11 would result in compromise of the pathway. When a neutralizing anti-S100A11 antibody was added to the culture medium, the constitutively higher phosphorylation state of Akt in BSCC-93 cells was almost completely suppressed (Figure 5C), and mRNA levels of EGF, betacellulin, epiregulin and HB-EGF were markedly reduced (Figure 5D). Addition of the anti-S100A11 antibody resulted in a decrease in the number of NHK and squamous carcinoma cells after 4 d of culture by 30∼40% (Figure 5E). The observed partial growth suppression was probably due to the fact that, among the EGF family members, TGFα and amphiregulin were hardly affected by S100A11 (Figure 5D) and the fact that some growth-stimulating pathways other than the S100A11-EGF family member axis also play significant roles. The advantageous effect for cancer cells of overexpressed S100A11 and hence activation of RAGE, however, is not necessarily restricted to growth stimulation. S100A11 induced expression of Bcl-2, a representative antiapoptotic protein (Figure 2B). RAGE-mediated signaling has been shown to confer cells resistance to induction of apoptosis in different biological systems (Huttunen et al., 2000; Arumugam et al., 2004). S100P, another ligand for RAGE, was shown to be overexpressed in pancreatic cancer (Crnogorac-Jurcevic et al., 2003; Sato et al., 2004) and to promote growth, survival, and invasion of pancreatic cancer cells (Arumugam et al., 2005). Angiogenesis, which is essential for growth and metastasis of cancer cells, has been shown to be promoted by ligands for the EGF receptor directly and/or via induction of vascular endothelial growth factor and its receptor (Goldman et al., 1993; Tsai et al., 1995). Recently, Gupta et al. (2007) showed that epiregulin is one of the genes that facilitate the assembly of new tumor blood vessels and thus promote lung metastasis. Together, the results suggest that overexpression of S100A11 and eventual activation of EGF family members contribute to facilitation of growth, survival, invasion, and metastasis because S100A11 and EGF family members compose a positive feedback loop. This process could be a target for exploitation of new therapeutic measures.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Department of Radiation Research Shikata Laboratory, Advanced Science Research Center (Okayama University) for experiments using isotopes and luciferase assay. This work was supported by Ministry of Education, Culture, Sports, Science and Technology of Japan grants 18013035 (to N.H.) and 7790150 (to M.S.).
Abbreviations used:
- NHK
normal human keratinocytes
- RAGE
receptor for advanced glycation end products.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0682) on October 31, 2007.
REFERENCES
- Adami C., Sorci G., Blasi E., Agneletti A. L., Bistoni F., Donato R. S100B expression in and effects on microglia. Glia. 2001;33:131–142. [PubMed] [Google Scholar]
- Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Arumugam T., Simeone D. M., Schmidt A. M., Logsdon C. D. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J. Biol. Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- Arumugam T., Simeone D. M., Van Golen K., Logsdon C. D. S100P promotes pancreatic cancer growth, survival, and invasion. Clin. Cancer Res. 2005;11:5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- Bogumil T., Rieckmann P., Kubuschok B., Felgenhauer K., Bruck W. Serum levels of macrophage-derived protein MRP-8/14 are elevated in active multiple sclerosis. Neurosci. Lett. 1998;247:195–197. doi: 10.1016/s0304-3940(98)00263-8. [DOI] [PubMed] [Google Scholar]
- Cecil D. L., Johnson K., Rediske J., Lotz M., Schmidt A. M., Terkeltaub R. Inflammation-induced chondrocyte hypertrophy is driven by receptor for advanced glycation end products. J. Immunol. 2005;175:8296–8302. doi: 10.4049/jimmunol.175.12.8296. [DOI] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T., Missiaglia E., Blaveri E., Gangeswaran R., Jones M., Terris B., Costello E., Neoptolemos J. P., Lemoine N. R. Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J. Pathol. 2003;201:63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- Dempsey A. C., Walsh M. P., Shaw G. S. Unmasking the annexin I interaction from the structure of Apo-S100A11. Structure. 2003;11:887–897. doi: 10.1016/s0969-2126(03)00126-6. [DOI] [PubMed] [Google Scholar]
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Donato R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- Du K., Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- Ehlermann P., Eggers K., Bierhaus A., Most P., Weichenhan D., Greten J., Nawroth P. P., Katus H. A., Remppis A. Increased proinflammatory endothelial response to S100A8/A9 after preactivation through advanced glycation end products. Cardiovasc. Diabetol. 2006;5:6. doi: 10.1186/1475-2840-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley E. D., Murphy L. C., Watson P. H. S100 proteins and their influence on pro-survival pathways in cancer. Biochem. Cell Biol. 2004;82:508–515. doi: 10.1139/o04-052. [DOI] [PubMed] [Google Scholar]
- Foell D., Frosch M., Sorg C., Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clin. Chim. Acta. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- Foulstone E. J., Tavare J. M., Gunn-Moore F. J. Sustained phosphorylation and activation of protein kinase B correlates with brain-derived neurotrophic factor and insulin stimulated survival of cerebellar granule cells. Neurosci. Lett. 1999;264:125–128. doi: 10.1016/s0304-3940(99)00166-4. [DOI] [PubMed] [Google Scholar]
- Goldman C. K., Kim J., Wong W. L., King V., Brock T., Gillespie G. Y. Epidermal growth factor stimulates vascular endothelial growth factor production by human malignant glioma cells: a model of glioblastoma multiforme pathophysiology. Mol. Biol. Cell. 1993;4:121–133. doi: 10.1091/mbc.4.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G. P., Nguyen D. X., Chiang A. C., Bos P. D., Kim J. Y., Nadal C., Gomis R. R., Manova-Todorova K., Massague J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- Heizmann C. W., Fritz G., Schafer B. W. S100 proteins: structure, functions and pathology. Front. Biosci. 2002;7:d1356–d1368. doi: 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- Hofmann M. A., et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- Hori O., et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J. Biol. Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Hsieh H. L., Schafer B. W., Weigle B., Heizmann C. W. S100 protein translocation in response to extracellular S100 is mediated by receptor for advanced glycation endproducts in human endothelial cells. Biochem. Biophys. Res. Commun. 2004;316:949–959. doi: 10.1016/j.bbrc.2004.02.135. [DOI] [PubMed] [Google Scholar]
- Huttunen H. J., Fages C., Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- Huttunen H. J., Kuja-Panula J., Sorci G., Agneletti A. L., Donato R., Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J. Biol. Chem. 2000;275:40096–40105. doi: 10.1074/jbc.M006993200. [DOI] [PubMed] [Google Scholar]
- Ji J., Zhao L., Wang X., Zhou C., Ding F., Su L., Zhang C., Mao X., Wu M., Liu Z. Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2004;130:480–486. doi: 10.1007/s00432-004-0555-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T., et al. Increased expression of calcium-binding protein S100 in human uterine smooth muscle tumours. Mol. Hum. Reprod. 2004;10:735–742. doi: 10.1093/molehr/gah100. [DOI] [PubMed] [Google Scholar]
- Kuruto R., Nozawa R., Takeishi K., Arai K., Yokota T., Takasaki Y. Myeloid calcium binding proteins: expression in the differentiated HL-60 cells and detection in sera of patients with connective tissue diseases. J. Biochem. 1990;108:650–653. doi: 10.1093/oxfordjournals.jbchem.a123257. [DOI] [PubMed] [Google Scholar]
- Landriscina M., Soldi R., Bagala C., Micucci I., Bellum S., Tarantini F., Prudovsky I., Maciag T. S100A13 participates in the release of fibroblast growth factor 1 in response to heat shock in vitro. J. Biol. Chem. 2001;276:22544–22552. doi: 10.1074/jbc.M100546200. [DOI] [PubMed] [Google Scholar]
- Liao H., et al. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis. Rheum. 2004;50:3792–3803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- Lugering N., Stoll R., Kucharzik T., Schmid K. W., Rohlmann G., Burmeister G., Sorg C., Domschke W. Immunohistochemical distribution and serum levels of the Ca(2+)-binding proteins MRP8, MRP14 and their heterodimeric form MRP8/14 in Crohn's disease. Digestion. 1995;56:406–414. doi: 10.1159/000201267. [DOI] [PubMed] [Google Scholar]
- Mailliard W. S., Haigler H. T., Schlaepfer D. D. Calcium-dependent binding of S100C to the N-terminal domain of annexin I. J. Biol. Chem. 1996;271:719–725. doi: 10.1074/jbc.271.2.719. [DOI] [PubMed] [Google Scholar]
- Marenholz I., Heizmann C. W., Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem. Biophys. Res. Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- Moroz O. V., et al. The structure of S100A12 in a hexameric form and its proposed role in receptor signalling. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:407–413. doi: 10.1107/s0907444901021278. [DOI] [PubMed] [Google Scholar]
- Nickel W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic. 2005;6:607–614. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- Ohuchida K., Mizumoto K., Ohhashi S., Yamaguchi H., Konomi H., Nagai E., Yamaguchi K., Tsuneyoshi M., Tanaka M. S100A11, a putative tumor suppressor gene, is overexpressed in pancreatic carcinogenesis. Clin. Cancer Res. 2006;12:5417–5422. doi: 10.1158/1078-0432.CCR-06-0222. [DOI] [PubMed] [Google Scholar]
- Oue N., Hamai Y., Mitani Y., Matsumura S., Oshimo Y., Aung P. P., Kuraoka K., Nakayama H., Yasui W. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64:2397–2405. doi: 10.1158/0008-5472.can-03-3514. [DOI] [PubMed] [Google Scholar]
- Prudovsky I., Bagala C., Tarantini F., Mandinova A., Soldi R., Bellum S., Maciag T. The intracellular translocation of the components of the fibroblast growth factor 1 release complex precedes their assembly prior to export. J. Cell Biol. 2002;158:201–208. doi: 10.1083/jcb.200203084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rety S., Osterloh D., Arie J. P., Tabaries S., Seeman J., Russo-Marie F., Gerke V., Lewit-Bentley A. Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- Roth J., Vogl T., Sorg C., Sunderkotter C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 2003;24:155–158. doi: 10.1016/s1471-4906(03)00062-0. [DOI] [PubMed] [Google Scholar]
- Rothermundt M., Peters M., Prehn J. H., Arolt V. S100B in brain damage and neurodegeneration. Microsc. Res. Tech. 2003;60:614–632. doi: 10.1002/jemt.10303. [DOI] [PubMed] [Google Scholar]
- Ryckman C., Vandal K., Rouleau P., Talbot M., Tessier P. A. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M., Miyazaki M., Inoue Y., Tsuji T., Kouchi H., Tanaka T., Yamada H., Namba M. Relationship between contact inhibition and intranuclear S100C of normal human fibroblasts. J. Cell Biol. 2000;149:1193–1206. doi: 10.1083/jcb.149.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Miyazaki M., Takaishi M., Sakaguchi Y., Makino E., Kataoka N., Yamada H., Namba M., Huh N. H. S100C/A11 is a key mediator of Ca(2+)-induced growth inhibition of human epidermal keratinocytes. J. Cell Biol. 2003;163:825–835. doi: 10.1083/jcb.200304017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Miyazaki M., Sonegawa H., Kashiwagi M., Ohba M., Kuroki T., Namba M., Huh N. H. PKCalpha mediates TGFbeta-induced growth inhibition of human keratinocytes via phosphorylation of S100C/A11. J. Cell Biol. 2004;164:979–984. doi: 10.1083/jcb.200312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Sonegawa H., Nukui T., Sakaguchi Y., Miyazaki M., Namba M., Huh N. H. Bifurcated converging pathways for high Ca2+- and TGFbeta-induced inhibition of growth of normal human keratinocytes. Proc. Natl. Acad. Sci. USA. 2005;102:13921–13926. doi: 10.1073/pnas.0500630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkadi B., Price E. M., Boucher R. C., Germann U. A., Scarborough G. A. Expression of the human multidrug resistance cDNA in insect cells generates a high activity drug-stimulated membrane ATPase. J. Biol. Chem. 1992;267:4854–4858. [PubMed] [Google Scholar]
- Sato N., Fukushima N., Matsubayashi H., Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- Schaefer K. L., et al. Expression profiling of t(12;22) positive clear cell sarcoma of soft tissue cell lines reveals characteristic up-regulation of potential new marker genes including ERBB3. Cancer Res. 2004;64:3395–3405. doi: 10.1158/0008-5472.CAN-03-0809. [DOI] [PubMed] [Google Scholar]
- Schmidt A. M., et al. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J. Biol. Chem. 1992;267:14987–14997. [PubMed] [Google Scholar]
- Stulik J., Koupilova K., Osterreicher J., Knizek J., Macela A., Bures J., Jandik P., Langr F., Dedic K., Jungblut P. R. Protein abundance alterations in matched sets of macroscopically normal colon mucosa and colorectal carcinoma. Electrophoresis. 1999;20:3638–3646. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3638::AID-ELPS3638>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Adzuma K., Iwami M., Yoshimoto K., Monden Y., Itakura M. Human calgizzarin; one colorectal cancer-related gene selected by a large scale random cDNA sequencing and northern blot analysis. Cancer Lett. 1995;89:195–200. doi: 10.1016/0304-3835(94)03687-e. [DOI] [PubMed] [Google Scholar]
- Todoroki H., Kobayashi R., Watanabe M., Minami H., Hidaka H. Purification, characterization, and partial sequence analysis of a newly identified EF-hand type 13-kDa Ca(2+)-binding protein from smooth muscle and non-muscle tissues. J. Biol. Chem. 1991;266:18668–18673. [PubMed] [Google Scholar]
- Torres-Cabala C., Panizo-Santos A., Krutzsch H. C., Barazi H., Namba M., Sakaguchi M., Roberts D. D., Merino M. J. Differential expression of S100C in thyroid lesions. Int. J. Surg. Pathol. 2004;12:107–115. doi: 10.1177/106689690401200203. [DOI] [PubMed] [Google Scholar]
- Tsai J. C., Goldman C. K., Gillespie G. Y. Vascular endothelial growth factor in human glioma cell lines: induced secretion by EGF, PDGF-BB, and bFGF. J. Neurosurg. 1995;82:864–873. doi: 10.3171/jns.1995.82.5.0864. [DOI] [PubMed] [Google Scholar]
- Wu J., Smith L. T., Plass C., Huang T. H. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res. 2006;66:6899–6902. doi: 10.1158/0008-5472.CAN-06-0276. [DOI] [PubMed] [Google Scholar]
- Xu D., Kyriakis J. M. Phosphatidylinositol 3′-kinase-dependent activation of renal mesangial cell Ki-Ras and ERK by advanced glycation end products. J. Biol. Chem. 2003;278:39349–39355. doi: 10.1074/jbc.M302771200. [DOI] [PubMed] [Google Scholar]
- Yan S. D., et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer's disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.