Abstract
The fragile X mental retardation protein (FMRP) is a selective RNA-binding protein that regulates translation and plays essential roles in synaptic function. FMRP is bound to specific mRNA ligands, actively transported into neuronal processes in a microtubule-dependent manner, and associated with polyribosomes engaged in translation elongation. However, the biochemical relationship between FMRP–microtubule association and FMRP–polyribosome association remains elusive. Here, we report that although the majority of FMRP is incorporated into elongating polyribosomes in the soluble cytoplasm, microtubule-associated FMRP is predominantly retained in translationally dormant, polyribosome-free messenger ribonucleoprotein (mRNP) complexes. Interestingly, FMRP–microtubule association is increased when mRNPs are dynamically released from polyribosomes as a result of inhibiting translation initiation. Furthermore, the I304N mutant FMRP that fails to be incorporated into polyribosomes is associated with microtubules in mRNP particles and transported into neuronal dendrites in a microtubule-dependent, 3,5-dihydroxyphenylglycine-stimulated manner with similar kinetics to that of wild-type FMRP. Hence, polyribosome-free FMRP–mRNP complexes travel on microtubules and wait for activity-dependent translational derepression at the site of function. The dual participation of FMRP in dormant mRNPs and polyribosomes suggests distinct roles of FMRP in dendritic transport and translational regulation, two distinct phases that control local protein production to accommodate synaptic plasticity.
INTRODUCTION
The fragile X mental retardation protein (FMRP) is a critical factor for neuronal function and development. The lack of FMRP causes fragile X syndrome, the most frequent form of familial mental retardation in humans (O'Donnell and Warren, 2002; Bear et al., 2004; Bagni and Greenough, 2005; Penagarikano et al., 2007). FMRP is a selective RNA-binding protein abundantly expressed in brain neurons, where it is incorporated into polyribosomes that are already engaged in translation elongation (Feng et al., 1997a; Ceman et al., 2003; Stefani et al., 2004), and it is implicated in controlling translation efficiency of its mRNA ligands with various potential mechanisms (Zalfa et al., 2003; Feng, 2002; Antar and Bassell, 2003; Jin and Warren, 2003; Garber et al., 2006). Consistent with this view, FMRP has been shown to suppress translation of its bound mRNAs in vitro (Laggerbauer et al., 2001; Li et al., 2001), in transfected cells (Mazroui et al., 2002; Wang et al., 2004), and in the brain (Lu et al., 2004; Qin et al., 2005). In addition, FMRP and its mRNA ligands are transported to dendritic shafts and spines as well as developing growth cones (Miyashiro et al., 2003; Antar et al., 2004, 2005; Ferrari et al., 2007), providing a mechanism for FMRP to regulate local translation to accommodate synaptic development and plasticity (Bagni and Greenough, 2005). Indeed, dendritic transport of FMRP and Fmr1 mRNA, as well as localized translation of FMRP at the synaptic terminals, can be stimulated by the activation of group I metabotropic glutamate receptors (GP1-mGluRs) (Weiler et al., 1997; Antar et al., 2004; Ferrari et al., 2007). Moreover, the absence of FMRP results in deregulation of GP1-mGluR–stimulated protein synthesis in synaptoneurosomes (Weiler et al., 2004; Muddashetty et al., 2007). Hence, the absence of FMRP-regulated local translation in response to synaptic activity is proposed as an underlying mechanism for the mental impairment in fragile X syndrome (Willemsen et al., 2004; Penagarikano et al., 2007).
Several microscopic studies indicate that FMRP is transported into neuronal processes in a microtubule-dependent manner (De Diego Otero et al., 2002; Antar et al., 2005). However, despite the fact that FMRP colocalizes with ribosome proteins and microtubule motors (De Diego Otero et al., 2002; Ling et al., 2004) and can be detected in biochemically isolated large granules containing clustered ribosomes (Aschrafi et al., 2005; Elvira et al., 2006), the biochemical relationship between FMRP-associated polyribosomes and microtubule-dependent FMRP transport remains undefined. It is not understood whether FMRP travels on microtubules after it is incorporated into polyribosomes through translation initiation on its mRNA targets, or alternatively whether dendritic transport of FMRP is in the form of polyribosome-free messenger ribonucleoprotein (mRNP) complexes that are sequestered from translation initiation. How FMRP controls transport and translation of its mRNA targets, which in turn governs neuronal function and development, is an important question in understanding FMRP function and pathogenesis of fragile X mental retardation.
The goal of this study is to dissect the biochemical relationship between the dynamic association of FMRP with polyribosomes and microtubules. We found that in contrast to the microtubule-dependent dendritic transport of FMRP (De Diego Otero et al., 2002; Antar et al., 2005), polyribosome association of FMRP is not affected by microtubule disruption. In fact, microtubule-bound FMRP is not associated with polyribosomes, but it largely cosediments with mRNP complexes and ribosomal subunits on a linear sucrose gradient. Releasing FMRP into polyribosome-free mRNP complexes, but not short polyribosomes, as a result of pharmacological inhibition of translation, increased the amount of microtubule-associated FMRP. Moreover, the I304N mutant FMRP that fails to associate with polyribosomes (Feng et al., 1997a) binds microtubules, colocalizes with RNA on microtubule polymers, and is transported into the dendrites of hippocampal neurons in a microtubule-dependent manner with similar kinetics as that of wild-type FMRP. Together, these results suggest that microtubule-dependent transport of FMRP and its mRNA ligands is largely in the form of translationally dormant free mRNP complexes, waiting to be activated for polyribosome association and local translation upon synaptic stimulation.
MATERIALS AND METHODS
Cell Cultures
The CAD cell line was propagated and maintained at 60–85% confluence in DMEM/F-12 containing 8–10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) as described previously (Qi et al., 1997). The immortalized fragile X fibroblast cell line was maintained at 60–85% confluence in DMEM containing 10% FBS. Transfection of the fragile X fibroblast cell line was performed using the Nucleofector technology following manufacturer's protocol (Amaxa Biosystems, Gaithersburg, MD). Hippocampal neurons from day 18 rat embryos or from P0 FVB mice were cultured on glass coverslips (18 mm for immunofluorescence and 40 mm for live imaging; Bioptechs, Butler, PA) and cocultured with astrocytes using the Goslin and Banker method (Goslin and Banker, 1989). After 9–10 days in vitro (DIV), cells were transfected with enhanced green fluorescent protein (EGFP)-FMRP-I304N driven by the human FMR1 promoter (Darnell et al., 2005b) by using calcium phosphate precipitation or Lipofectamine 2000 (Invitrogen).
Linear Sucrose Gradient Fractionation
The fractionation experiments were carried out essentially as described previously (Feng et al., 1997a). Briefly, CAD cells, with or without treatment of microtubule disruption reagents as described in the corresponding experiments, were incubated with 100 μg/ml cycloheximide for 15 min to arrest polyribosome migration, unless treated by other translation inhibitors as indicated in the corresponding figure legends. Cells were then lysed to isolate postmitochondrial extracts, followed by fractionation on 15–45% sucrose gradient. EDTA-treated lysate was fractionated on a parallel gradient lacking MgCl2 but containing 1 mM EDTA to dissociate ribosomes into subunits. Fractions were collected from each gradient tube by up-ward replacement with monitored absorption at OD254 by using a fractionator (Isco, Lincoln, NE). Two percent of each fraction was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) immunoblot as described previously (Wang et al., 2004).
Cytoskeleton Isolation and Analysis of FMRP–Microtubule Association
To separate microtubule polymers from free tubulin, CAD cells were treated with 10 nM paclitaxel (Taxol; Sigma-Aldrich, St. Louis, MO) for 1 h before lysed in the gradient lysis buffer containing 150 mM KCl, 2 mM MgCl2, 50 mM Tris, pH 7.5, 2 mM EGTA, 2%glycerol, 10 nM paclitaxel, 0.125% Triton X-100, protease inhibitor cocktail (EDTA-free; Roche Diagnostics, Indianapolis, IN), and 10 U of RNasin (Promega, Madison, WI) at room temperature for 5 min. For RNase treatment, the lysate buffer contains 1.2 μg/μl RNase A1 and 30 units of RNase T1 but no RNasin. Nuclei were pelleted at 700 g for 1 min at room temperature, and the cytoplasmic supernatant was centrifuged for 10 min at 16,000 g at room temperature to pellet microtubule polymers. The microtubule pellet and the postmicrotubule supernatant were brought to 1X Laemmli buffer followed by SDS-PAGE analysis. For analyzing microtubule-associated polyribosomes, the microtubule pellet was resuspended in the gradient buffer in the absence of paclitaxel on ice. The released polyribosomes and mRNPs were subjected to standard linear sucrose gradient fractionation as described above.
To analyze microtubule association of the Flag-tagged wild-type and mutant FMRP in the fragile X fibroblast, cells were transfected with the plasmids Flag-2.17 (Wang et al., 2004) and Flag-2.17-BOPM that carries the I304N mutation (Eberhart et al., 1996) by using the Nucleofector kit for fibroblasts. The EGFP-N3 plasmid was cotransfected to monitor transfection efficiency. Twenty-four hours after transfection, cells were treated with 100 nM paclitaxel for 30 min before lysed in the gradient buffer containing 100 nM paclitaxel and 0.5% Igepal CA-630 at room temperature. After removing nuclei and unlysed cells, the supernatant was centrifuged at 16,000 g for 15 min at room temperature to pellet microtubule polymers. An aliquot of the input lysate and the resuspended microtubule pellet were subjected to SDS-PAGE immunoblot.
Antibodies and Immunoblot Detection
For immunoblot analysis, the protein quantity of each sample was estimated by Bradford assay following manufacture's protocol (Bio-Rad, Hercules, CA) before subjected to SDS-PAGE. After an overnight transfer, the blots were subjected to Ponceau S staining (Sigma-Aldrich, St. Louis, MO) to confirm equal protein loading before carrying out immunoblot analysis. The primary antibodies were diluted as follows: FMRP (IC3), 1:1000; α-tubulin, 1:4000; and eukaryotic initiation factor (eIF)5a, 1:5000 (Santa Cruz Biotechnology, Santa Cruz, CA).
For detection of microtubule colocalization of fluorescently tagged wild-type and I304N mutant FMRP in the fragile X fibroblast, cells were fixed in 3% paraformaldehyde in PHEMO buffer [0.068 M piperazine-N,N′-bis(2-ethanesulfonic acid), 0.025M HEPES acid, 0.015 M EGTA Na2, 0.03 M MgCl2, and 10% dimethyl sulfoxide) at room temperature. After washing, the slides were blocked in phosphate-buffered saline (PBS) containing 5% normal goat serum, followed by incubation with the rat monoclonal antibody against α-tubulin (1:1000; Chemicon International, Temecula, CA) for 1 h at room temperature. Fluorescein isothiocyanate or Texas Red-conjugated anti-rat immunoglobulin (Ig)G were incubated with the corresponding slides for 30 min at room temperature. After washes, fluorescence was detected at room temperature by using the Zeiss LMS510 confocal microscopic imaging system.
For immunofluorescence analysis of hippocampal neurons, transfected cells (8–24 h after transfection) were exposed to 50 μM DHPG for 15–30 min in media and then rinsed four times in 20 mM HEPES-buffered Hank's modified salt solution and then fixed in 4% formaldehyde/4% sucrose in PBS for 20–30 min. Cells were then blocked in PBS with 2% bovine serum albumin and 1% Triton X-100 (PBSAT) for 1 h, followed by incubation with primary antibodies in the same buffer for 1 h. Cells were then washed 10 times in PBSAT, and Cy5 anti-mouse secondary antibodies were added (along with propidium iodide for RNA, as indicated in the corresponding figure legend) for 15 min, washed again 10 times in PBSAT, two times in PBS, and then briefly in distilled water just before mounting in antifade mounting buffer. Primary antibodies used are as follows: mouse anti-tubulin (DM1a clone; Sigma-Aldrich), 1:2000; and monoclonal anti-microtubule-associated protein MAP2 (Chemicon International), 1:2000. Total dendritic and cell body fluorescence was quantified by tracing the cell compartments using the ROI tools in IPLab software (BD Biosciences Bioimaging, Rockville, MD), normalized to area in each image, and background was subtracted using a random area on the coverslip adjacent to each cell.
Live Cell Imaging with Fluorescence Recovery after Photobleaching (FRAP) Analysis
After 8–24 h of green fluorescent protein (GFP)-FMRP-I304N expression, transfected coverslips were transferred to the Bioptechs live imaging chamber and imaging media (minimal essential medium with B27 supplements, 5 mM l-glutamine, and 20 mM HEPES, pH 7.2; Invitrogen) was perfused containing 50 μM DHPG. Cells were imaged between 15 and 45 min after exposure to DHPG. For microtubule depolymerization experiments with FRAP analysis, the cells were preincubated with 2 μg/ml nocodazole for 15 min before being transferred to the imaging chamber, at which time the cells were perfused with media containing nocodazole in the presence of DHPG. GFP-expressing cells were visualized and subjected to FRAP by using a FluoView 500 confocal laser scanning microscope (Olympus, Melville, NY) equipped with an argon blue laser (488 nm) as described previously (Antar et al., 2004). Briefly, a dendritic region was chosen and an image captured. The region was then bleached (100% power) for <1 min and allowed to recover over 5 min, imaging during this time at 10-s intervals at low power (10%). Recovery rates calculated for five control cells (11 dendrites) and six nocodazole-treated cells (14 dendrites) were averaged from each 1-min time point (1–5 min) and graphed using Excel (Microsoft, Redmond, WA). Rates of recovery were calculated from the equation of the best-fit curve of the graph for y = 0.5 in Excel (50% recovery).
RESULTS
FMRP–Polyribosome Association Is Independent of Microtubules
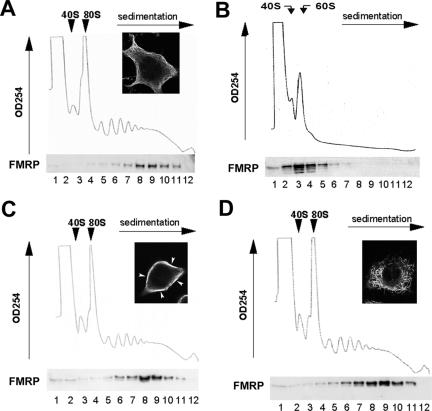
Several lines of evidence indicate that microtubules are essential for dendritic transport of FMRP (De Diego Otero et al., 2002; Antar et al., 2005). However, whether FMRP–polyribosome association also depends on microtubule integrity has not been studied previously. To address this question, we analyzed FMRP-polyribosome association in the immortalized brain catecholaminergic neuronal cell line CAD using linear sucrose gradient fractionation. As shown in Figure 1A, the majority of cytoplasmic FMRP (∼80%) cofractionated with large polyribosome complexes in cytoplasmic extracts (fractions 6–11). EDTA treatment resulted in dissociation of polyribosomes into ribosomal subunits and released FMRP into complexes that cosediment with mRNPs and ribosomal subunits (Figure 1B, fractions 1–5). Interestingly, when microtubules are either robustly bundled by paclitaxel-mediated stabilization (Figure 1C) or severely depolymerized by nocodazole treatment (Figure 1D), FMRP-polyribosome association was unaffected. A nearly identical sedimentation profile of FMRP was observed in untreated cells (Figure 1A) and in cells with severe microtubule perturbation (Figure 1, C and D). In addition, disruption of actin dynamics by cytochalasin B treatment did not affect FMRP–polyribosome association (data not shown). Hence, unlike dendritic transport of FMRP, polyribosome association of FMRP is independent of microtubule and actin cytoskeletal integrity.
Figure 1.
Polyribosome association of FMRP is independent of microtubules. Cytoplasmic lysates derived from cells mock treated (A), incubated with 100 nM paclitaxel (C), or with 30 nM nocodazole (D) overnight are fractionated on 15–45% (wt/vol) linear sucrose gradient containing 5 mM MgCl2. In addition, lysates are treated with 10 mM EDTA to dissociate polyribosomes into subunits (B). In each panel, the sedimentation of ribosomal subunits (40S and 60S), the monoribosome (80S) and the translating polyribosomes are shown on the top. Representative images of nicely organized microtubule network in mock-treated cells (A), the robust microtubule bundling caused by paclitaxel (C), and the disruption of the microtubule network caused by nocodazole (D) are shown in the corresponding insets. Each gradient was fractionated into 12 1-ml fractions, and 2% of each fraction was subjected to immunoblot for FMRP. The image of the blot is shown in relation to the corresponding sedimentation profile with fraction number marked at the bottom of the corresponding lanes.
FMRP–Microtubule Association Is Sensitive to RNAse Treatment, Predominantly in the Form of Polyribosome-free mRNP Complexes
To determine how much cytoplasmic FMRP is associated with the microtubule cytoskeleton, we isolated soluble cytoplasm and cytoskeletal pellet from the postnuclear extracts derived from CAD cells that were mock treated or exposed to drugs that alter assembly of either the microtubule or the actin cytoskeleton (Figure 2). Immunoblot analysis revealed that ∼10% of total FMRP was associated with the cytoskeleton pellet (P), whereas the majority of FMRP was present in the postcytoskeletal supernatant (S). Increased microtubule assembly, as a result of paclitaxel pretreatment, doubled the amount of FMRP associated with the cytoskeletal pellet (Figure 2A). In addition, RNAse treatment of the paclitaxel-stabilized microtubule pellet significantly reduced the amount of FMRP detected by immunoblot (Figure 2B), suggesting that microtubule-associated FMRP requires RNA. Reciprocally, destabilization of microtubules by nocodazole reduced the amount of FMRP in the cytoskeletal pellet (Figure 2C). In contrast, disruption of the actin cytoskeleton by pretreatment with cytochalasin B did not alter the amount of cytoskeleton-associated FMRP (Figure 2D). These results suggest that FMRP-containing RNA complexes associate with microtubules but not with actin polymers in total cytoskeleton.
Figure 2.
Microtubule dynamics, but not actin dynamics, influences the association of FMRP with cytoskeleton, which is sensitive to RNAse treatment. Cells were preincubated with 100 nM paclitaxel, 100 nM nocodazole, 10 μg/ml cytochalasin B, or mock treated for 40 min. Cytoskeleton was isolated from postnuclear extracts by centrifugation in three independent experiments. Proportional amounts of the S containing soluble cytosol and the P were subjected to immunoblot in the top panels. The ratio of densitometer reading of FMRP in P and S was calculated, normalized to the mock-treated control, and it is graphically displayed in the bottom panels. (A) Paclitaxel pretreatment increases association of FMRP with cytoskeleton pellet. *p < 0.05 in t test. (B) FMRP association with paclitaxel-treated cytoskeleton pellet is abrogated upon treatment by a combination of 1.2 μg/μl RNase A and 30 U of T1. **p < 0.01 in t test. (C) Nocodazole pretreatment decreases association of FMRP with cytoskeleton pellet. *p < 0.05 in t test. (D) Cytochalasin B pretreatment does not alter association of FMRP with cytoskeleton pellet.
To delineate the biochemical nature of the microtubule-associated FMRP-containing complexes, we performed parallel linear sucrose gradient fractionation experiments using extracts derived from the microtubule pellet (MT) and that from the nonmicrotubule supernatant (non-MT). We observed that FMRP cofractionated with large polyribosomes in the non-MT cytoplasm (Figure 3A, middle), which was shifted to fractions containing mRNPs and ribosomal subunits upon EDTA treatment (Figure 3B, middle). In contrast, microtubule-associated FMRP complexes were not detected in polyribosomes, but they cofractionated with EDTA-released mRNPs and ribosomal subunits (Figure 3, A and B, bottom). These results suggest that unlike the majority of FMRP in the soluble cytoplasm, microtubule-associated FMRP is predominantly in the form of translationally dormant, polyribosome-free mRNP complexes.
Figure 3.
Microtubule-associated FMRP cosediment with mRNPs and ribosomal subunits but not large polyribosomes. (A) Microtubule polymers (MT) were separated from nonmicrotubule soluble cytoplasm (non-MT), resuspended and followed by linear sucrose gradient fractionation in parallel with the non-MT extracts as described in the legend of Figure 1. Two percent of each fraction was subjected to immunoblot analysis for detection of FMRP and P0, a structural protein on the 60S large ribosomal subunit, as indicated on the left of each blot. The fraction numbers are indicated at the bottom of each lane. (B) In a parallel experiment, MT and non-MT lysates were treated with EDTA and fractionated on an EDTA-containing gradient to dissociate polyribosomes into subunits, as indicated by the loss of polyribosome peaks in the OD254 profile and the shift of the P0 signals to the top fractions of the gradient.
FMRP–Microtubule Association Is Increased upon Releasing FMRP into Ribosome-free mRNPs from Elongating Polyribosomes
Because FMRP associates with microtubules predominantly as polyribosome-free mRNP particles, we wondered whether releasing FMRP from polyribosomes might increase the association of FMRP with microtubules. We inhibited translation initiation in CAD cells by preincubation with NaF, which caused ribosome run-off and released FMRP into polyribosome-free mRNP complexes that sediment in top fractions on linear sucrose gradient (Figure 4A, middle). Alternatively, we inhibited translation with homoharringtonine (Hrt), which still retained FMRP on short polyribosomes (Figure 4A, right). Cells that were briefly treated with cycloheximide to freeze translating polyribosomes on their mRNA template were analyzed in parallel experiments as a control (Figure 4A, left). Under the aforementioned conditions, we analyzed the amount of FMRP associated with the microtubule cytoskeleton. As shown in Figure 4B, releasing FMRP into polyribosome-free mRNP by NaF treatment significantly increased the amount of FMRP in the microtubule pellet (Figure 4B, left). In contrast, releasing FMRP into short polyribosome complexes by hyringtonine-treatment did not alter the amount of FMRP in the microtubule pellet. This result supports our conclusion that FMRP associates with microtubules in the form of translationally repressed mRNP complexes that are not incorporated into polyribosomes.
Figure 4.
Influence of FMRP–microtubule association due to polyribosome runoff. Cells were treated with 40 μM NaF to inhibit translation initiation and release mRNPs, or with 2 μM Hrt to generate short polyribosomes, or with 100 μg/ml cycloheximide (Cxh) to freeze polyribosomes on the translation template mRNAs as a control. (A) Linear sucrose gradient fractionation of cells subjected to the aforementioned treatment and immunoblot of FMRP as described in the legend of Figure 1. Note the release of FMRP from polyribosomes (right, fractions 4–9) into mRNPs upon NaF treatment (middle, fractions 1–3), and the release of FMRP into short polyribosomes upon Hrt treatment (left, fractions 4–6). (B) NaF treatment releases FMRP into mRNPs and increases FMRP in paclitaxel-stabilized MT pellet. The ratio of densitometer reading of FMRP in the MT pellet (P) to that in the soluble cytosol (S) was calculated and normalized to that for control cells. Results from three independent experiments were subjected to t test. **p < 0.01. (C) Hrt treatment releases FMRP into short polyribosomes without altering the amount of FMRP in paclitaxel-stabilized MT pellet. Three independent experiments were conducted and the P/S ratio was subjected to t test.
The I304N Mutation Does Not Affect FMRP–Microtubule Association, Despite the Inability for Incorporation into Polyribosomes
The I304N mutation in the second KH domain of FMRP was identified from a patient with extremely severe clinical expression of fragile X syndrome (De Boulle et al., 1993). This mutation ablates FMRP–polyribosome association (Feng et al., 1997a) and affects FMRP binding to the kissing complex RNA ligand (Darnell et al., 2005a), but it does not affect binding to the G-quartet FMRP-binding RNA motif (Darnell et al., 2001) and cellular poly(A) RNA (Feng et al., 1997a). To test whether the I304N mutation may affect FMRP–microtubule association, we transfected fluorescent-tagged wild-type and I304N mutant FMRP individually into an immortalized fibroblast cell line derived from a fragile X patient that lacks endogenous FMRP (Figure 5). Confocal microscopy revealed that both the wild-type and the I304N mutant FMRP formed particles that were clearly aligned on individual microtubules in the cell body and at the leading edge (Figure 5A). In addition, in the transfected fragile X cells, comparable levels of the wild-type and I304N-FMRP were detected in the paclitaxel-stabilized microtubule pellets (Figure 5B), suggesting that I304N-FMRP and wild-type FMRP associates with microtubules with similar efficiency, despite the fact that the I304N mutation is incapable for polyribosome association (Feng et al., 1997a).
Figure 5.
The I304N mutation in FMRP that abrogates FMRP–polyribosome association does not affect the association of FMRP with microtubules. (A) An immortalized fragile X fibroblast cell line was transfected to express RFP-FMRP (a) or GFP-I304N-FMRP (b) for 20 h. Cells were stained for α-tubulin to visualize individual microtubule polymers. Enlarged image indicating the alignment of RFP-FMRP and GFP-I304N granules (white rectangles in a and b) with individual microtubule bundles are shown in c and d. (B) Immunoblot analysis of paclitaxel-stabilized MT isolated from postnuclear lysates (Inp) from a fragile X fibroblast cell line expressing Flag-tagged wild-type FMRP or Flag-I304N FMRP. The same blot was reprobed by antibodies for eIF5a and total tubulin (Tub). (C) Colocalization of the GFP-I304N-FMRP with RNA and microtubules in the dendrites. Primary rat hippocampal neurons (10 DIV) were transfected with GPF-FMRP-I304N for 8 h before fixed and stained for total RNA and tubulin. Top panels show transfected neuron expressing the GFP-I304N-FMRP, in which RNA granules stained by propidium iodide and tubulin polymers stained by Cy5 fluorescence of anti-tubulin antibodies are visualized by fluorescent microscopy. A proximal dendritic segment of a transfected neuron (white outlined) is shown with higher magnification in the bottom panels. Arrows highlight areas where FMRP-I304N granules (green) colocalize with RNA (red) on microtubule bundles (blue) within the dendrite.
The I304N Mutant FMRP Is Transported to Neuronal Dendrites in a Microtubule-dependent Manner, Which Is Stimulated upon Activation of GP1-mGluR
To examine whether I304N-FMRP can form RNP complexes on microtubules in primary neurons, the GFP-I304N-FMRP fusion construct was transfected into hippocampal cultures (10 DIV) and processed for immunofluorescence analysis (Figure 5C). The GFP-I304N-FMRP fusion protein clearly formed granules typical for mRNPs. However, these complexes seemed to be somewhat diffuse in dendrites, with apparently fewer dendritic particles and smaller sizes than those seen in neurons stained for endogenous FMRP and the wild-type FMRP-GFP fusion protein (Antar et al., 2004). Nonetheless, the GFP-I304N-FMRP granules (green) contained RNA as marked by propidium iodide (red), and they were aligned on individual microtubule bundles (blue). Furthermore, time-lapse recording revealed active movement of these complexes into dendrites (see below; also see Supplemental Movies). When transfected neurons were treated with the GP1-mGluR agonist DHPG, we observed a significant increase in the dendritic levels of the GFP-I304N-FMRP compared with that in mock-treated cells (Figure 6, A–D). Quantitative analysis revealed an approximately twofold increase of dendritic localization in response to DHPG treatment (Figure 6E), similar to the DHPG-stimulated localization reported previously for the GFP-tagged wild-type FMRP (Antar et al., 2004).
Figure 6.
DHPG stimulates dendritic transport of FMRP-I304N in hippocampal neurons. Primary hippocampal neurons (10 DIV) derived from P0 FVB mice were transfected with GFP-I304N-FMRP for 12–16 h before subjected to DHPG treatment. (A and B) Representative images of GFP-I304N-FMRP granules in an unstimulated control cell and a DHPG-stimulated cell. (C and D) Immunofluorescent staining of MAP2 marks the dendrites in the corresponding cells in A and B. (E) Average ratios for dendrite/cell body fluorescence in DHPG-stimulated (hatched bar) and unstimulated cells (white bar) were calculated using at least 34 dendrites from more than 12 cells within three separate experiments are expressed graphically. *p < 0.01 as determined by Student's t test. Bar 20 μm (A).
To further determine whether the GFP-I304N-FMRP is transported into dendrites in a microtubule-dependent manner, we performed FRAP in live neurons. Transfected hippocampal neurons were stimulated with DHPG, and the transport kinetics of the I304N-FMRP was measured in the absence and presence of nocodazole (Figure 7). A robust transport of the I304N-FMRP into dendrites was observed after photobleaching (Figure 7, B–D). In contrast, a brief treatment (30–60 min) by nocodazole markedly impaired the fluorescence recovery after photobleaching (Figure 7, F–H). Quantification of recovery rates (Figure 7I) revealed that nocodazole treatment caused a 32-fold decrease in transport kinetics for the I304N-FMRP, similar to that for wild-type FMRP (Antar et al., 2005). Together, these data suggest that the I304N mutation does not affect the ability of FMRP to form transport-competent mRNP complexes or binding microtubules in neurons, despite the fact that this mutation prevents FMRP from association with polyribosomes (Feng et al., 1997a), dimerization (Laggerbauer et al., 2001), and binding to specific RNA ligands (Darnell et al., 2005a).
Figure 7.
Kinetic analysis of GFP-I304N-FMRP granules by using FRAP with or without disruption of microtubule dynamics. Primary rat hippocampal neurons (9 DIV) transfected with GPF-FMRP-I304N for 8 h were treated with DHPG before subjected to FRAP analysis in the presence or absence of nocodazole. The image colors correspond to fluorescence intensities of GFP signal according to standard heat map format (Antar et al., 2005). A and E show a representative neuron expressing GFP-FMRP-I304N granules in the absence and presence of nocodazole, respectively. The white box outlines the dendritic area subjected to FRAP analysis in subsequent frames. The time-lapse images of GFP signal in mock-treated (B–D) and nocodazole-treated (F and G) illustrate the effect of nocodazole in the efficiency of fluorescence recovery. B and F, prebleach. C and G, postbleach. D and H, fluorescence recovery at 5 min after bleaching. Note that the dendritic GFP signal in the nocodazole-treated cell (E and F) seems more diffuse, less granular and has a much higher somatic signal than in nontreated cells (A and B), and the less robust recovery of fluorescence in the nocodazole-treated cell at 5 min after bleaching (H) compared with the untreated cell (D). (I) Graphical analysis of the average percentage of recovery of GFP signal after photobleaching from neurons treated with DHPG alone (red squares; see A–D) or DHPG and nocodazole (blue circles; see E–H), each shown with a best-fit curve. Bar, 20 μm.
DISCUSSION
In this study, we show for the first time that FMRP is associated with microtubules predominantly as translationally dormant, polyribosome-free mRNP. In contrast, the majority of cellular FMRP is incorporated into polyribosomes in the soluble cytoplasm, independently of microtubule integrity. FMRP–polyribosome association is neither required for microtubule association nor for dendritic transport of FMRP. Hence, neuronal FMRP are subjected to the following alternative cellular pathways: 1) to be incorporated into polyribosomes and govern translation elongation efficiency of its bound mRNA ligands, but perhaps lose the ability to associate with microtubules; or 2) to be sequestered from translation initiation and travel on microtubules toward neuronal dendrites. On arrival at the functional destination, these FMRP–mRNP complexes will be subjected to activity-dependent translational derepression, which controls localized production of critical proteins that are required for synaptic plasticity.
FMRP–polyribosome association is observed in various cell lines and in the mouse brain (Eberhart et al., 1996; Corbin et al., 1997; Ceman et al., 2003; Khandjian et al., 2004; Stefani et al., 2004; Wang et al., 2004). The FMRP-containing polyribosomes are engaged in translation elongation, but they display delayed ribosome runoff (Feng et al., 1997a; Ceman et al., 2003). Hence, suppression of translation elongation is one of the proposed models for FMRP to inhibit protein synthesis, which may be derepressed upon serine-phosphorylation of FMRP (Ceman et al., 2003). In addition to its presence in polyribosomes, FMRP is also known to associate with microtubules and motor proteins (De Diego Otero et al., 2002; Kanai et al., 2004) to be transported into neuronal processes in a microtubule-dependent manner (Antar et al., 2005). Unlike dendritic transport of FMRP, polyribosome-association of FMRP is not affected by microtubule disruption (Figure 1). Nonetheless, whether and how FMRP–polyribosome association is related to FMRP–microtubule association and dendritic transport of FMRP was not addressed by previous studies.
Association of mRNA and polyribosomes with microtubules has been well documented (Bassell et al., 1994; Hamill et al., 1994). In addition, a number of RNA-binding proteins, including FMRP, are capable of association with both ribosomes and microtubules and can be detected in neuronal processes (Bolognani et al., 2004; Brendel et al., 2004; Huang and Richter, 2004; Huttelmaier et al., 2005; Kosturko et al., 2005). A major paradigm for dendritic mRNA transport is the presence of translocating mRNAs in large granules that contain various RNA-binding proteins and densely packed ribosome clusters arrested from translation (for review, see Krichevsky and Kosik, 2001; Kiebler and Bassell, 2006). Although FMRP was detected in ribosome-containing large granules isolated from mouse brain (Kanai et al., 2004;Aschrafi et al., 2005; Elvira et al., 2006), how much FMRP is present in these granules and whether these granules are in the process to be transported into dendrites are not determined. Moreover, no evidence indicates that these clustered ribosomes are polyribosomes engaged in translation elongation. In fact, a recent proteomic study by Elvira et al. (2006) suggests that these large granules contain “an amorphous collection of ribosomes” unlikely representing polyribosomes. Because these large granules are not stimulated for dendritic localization by neuronal activation, whereas dendritic transport of FMRP is vigorously stimulated by neuronal depolarization and activation of mGluRs (Antar et al., 2004), FMRP must be present in other types of RNA granules for dendritic transport, in addition to the large granules that contain ribosome clusters. This is not surprising considering the diverse size and transport kinetics of RNA-containing granules (Kiebler and Bassell, 2006).
Indeed, several independent lines of evidence indicate that specific RNA-binding proteins that inhibit translation initiation also facilitate dendritic localization (Huang et al., 2003; Huttelmaier et al., 2005). This argues that some mRNPs must be transported in the form of dormant particles sequestered from polyribosomes (Huang and Richter, 2004; Dahm and Kiebler, 2005). However, it is not clear how this model pertains with the ribosome clusters in the large granules detected in the neuronal dendrites. Moreover, direct evidence for this model is not provided previously. Our results clearly show that in the CAD neuronal cell line, FMRP-containing complexes isolated from microtubules cofractionate with free mRNPs and ribosomal subunits (Figure 3), but not with large polyribosomes. In addition, these microtubule-associated FMRP complexes contain RNA (Figure 5C), and they can be removed from microtubules by RNase treatment (Figure 2B). Furthermore, releasing FMRP from polyribosomes into mRNPs by inhibiting translation initiation increased the FMRP–microtubule association (Figure 4A). In contrast, releasing FMRP from large polyribosomes into short polyribosomes did not increase FMRP–microtubule association (Figure 4B). Together, these results demonstrate FMRP as the first example among RNA-binding proteins for microtubule-dependent transport in dormant mRNP complexes sequestered from translation, some may contain ribosomal subunits. The observation that majority of the microtubule-associated FMRP is translationally quiescent, uncoupled from translating polyribosomes (Figure 3), is consistent with the activity of FMRP in suppressing ribosome subunits joining during translation initiation (Laggerbauer et al., 2001).
It is important to note that the detection of microtubule-associated FMRP as polyribosome-free mRNP in CAD cells does not contradict the presence of FMRP in large granules that contain ribosome clusters (Aschrafi et al., 2005; Elvira et al., 2006). However, how much FMRP is present in the ribosome-containing large granules and what percentage of these biochemically isolated granules associates with microtubules is not understood. In addition, whether these large granules are formed in dendrites from smaller FMRP– mRNP complexes after they left the soma remains unknown. Because many of the large granules are immobile in dendrites (Elvira et al., 2006), whether the large granules function as a format for dendritic transport, or alternatively as a local storage compartment for mRNAs under translational arrest and are poised for release for active translation in dendrites (Krichevsky and Kosik, 2001), still remains as an interesting question to be addressed by future studies.
Consistent with the results that the microtubule-associated FMRP–mRNP complexes are not incorporated into elongating polyribosomes, the I304N mutation that abolishes FMRP–polyribosome association (Feng et al., 1997a) does not affect the efficiency of FMRP–microtubule association (Figure 5) or dendritic transport of FMRP (Figures 6 and 7). This is in agreement with a previous study showing microtubule-dependent localization of GFP-I304N-FMRP in the neurites of PC12 cells (Schrier et al., 2004). However, this previous report could not detect I304N-FMRP in visible RNA-containing granules to resolve the incorporation of I304N-FMRP into RNPs. With our high-resolution imaging approach, the GFP-I304N-FMRP can be clearly visualized in granules, colocalized with RNA on microtubules in hippocampal neurons (Figure 6C), and actively transported into dendrites (Figure 7). Although the I304N mutation partially abrogates the activity of FMRP in binding a subclass of mRNAs, represented by the kissing complex (Darnell et al., 2005a), it does not prevent FMRP from binding the G-quartet and poly(A) RNA (Feng et al., 1997a; Darnell et al., 2001). Interestingly, the GFP-I304N-FMRP granules are smaller than those formed by the wild-type GFP-FMRP (Antar et al., 2004), possibly due to the oligomerization defects caused by the I304N mutation (Laggerbauer et al., 2001). The inability for I304N to associate with ribosomes may also contribute to the small size of these granules. Nonetheless, quantitative FRAP analysis demonstrated microtubule-dependent dendritic transport of I304N-FMRP granules (Figure 7), which is stimulated upon activation of GP1-mGluR (Figure 6) with similar kinetics of transport as that for the wild-type FMRP (Antar et al., 2005). These results provide a parallel line of evidence suggesting that dendritic transport of FMRP is independent and can be uncoupled from its association with polyribosomes.
Our result that FMRP can be detected in translationally repressed particles despite its predominant polyribosome association (Figure 3) is consistent with previous reports (Mazroui et al., 2002; Kim et al., 2006). An important finding in our study is the association of translationally repressed, polyribosome-free FMRP–mRNP complexes with microtubules (Figures 3 and 4). In the absence of FMRP, these mRNAs may be actively translated in the soma rather than repressed and transported into the dendrite. Because FMRP associates with polyribosomes in the synapse (Feng et al., 1997b), the translationally repressed FMRP–mRNP particles must be derepressed for local translation upon synaptic activation. The FMRP–polyribosome association may further control translation elongation/termination based on the phosphorylation status of FMRP (Ceman et al., 2003). Hence, the presence of FMRP in translationally dormant mRNPs for microtubule-dependent transport and in elongating polyribosomes in dendrites/synapses may represent two distinct phases for FMRP to control local protein synthesis. Although the I304N-FMRP-mRNPs are efficiently transported into dendrites (Figure 7), the mRNAs in these complexes may never be activated for translation at the synapse, or they may be misregulated for translation due to the lack of functional FMRP on polyribosomes. This may explain why the I304N mutant FMRP fails to rescue the synaptic defects when introduced into Fmr1 postsynaptic neurons (Pfeiffer and Huber, 2007).
At this point, the functional requirement of FMRP in mRNA transport has not been directly demonstrated. Previous studies examining dendritic mRNA localization in Fmr1 knockout mouse neurons have given conflicting results (Steward et al., 1998; Miyashiro et al., 2003), although neither study examined the stimulus-induced localization of FMRP ligands. Considering the fact that FMRP associates with microRNAs and the microRNA machinery (Jin et al., 2004), whether microRNA pathways may contribute to microtubule-dependent transport of FMRP–mRNPs is an intriguing possibility to be addressed by future studies. Delineating the role of FMRP in these scenarios will provide important insight regarding the molecular mechanisms for the pathogenesis of fragile X mental retardation, and for the fundamental rules that govern local protein synthesis required for synaptic plasticity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. S. T. Warren (Emory University) for providing the immortalized fragile X fibroblast cell line, Dr. J. Mandel (Université Louis Pasteur) for the IC3-producing hybridoma cells, and Dr. J. Darnell (Rockefeller University) for the plasmid encoding the GFP-I304N-FMRP driven by the human Fmr1 promoter. This work is supported by National Institutes of Health grant 5 P01 HD-35576 (Project III) and Fraxa foundation (to Y.F.), National Institutes of Health grant NS-051127 and the Dana Foundation grant (to G.B.), and a National Fragile X Foundation Basic Sciences Grant (to J.B.D.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0583) on October 31, 2007.
REFERENCES
- Antar L. N., Afroz R., Dictenberg J. B., Carroll R. C., Bassell G. J. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar L. N., Bassell G. J. Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron. 2003;37:555–558. doi: 10.1016/s0896-6273(03)00090-4. [DOI] [PubMed] [Google Scholar]
- Antar L. N., Dictenberg J. B., Plociniak M., Afroz R., Bassell G. J. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Aschrafi A., Cunningham B. A., Edelman G. M., Vanderklish P. W. The fragile X mental retardation protein and group I metabotropic glutamate receptors regulate levels of mRNA granules in brain. Proc. Natl. Acad. Sci. USA. 2005;102:2180–2185. doi: 10.1073/pnas.0409803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C., Greenough W. T. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Bassell G. J., Singer R. H., Kosik K. S. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Bear M. F., Huber K. M., Warren S. T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Bolognani F., Merhege M. A., Twiss J., Perrone-Bizzozero N. I. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci. Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- Brendel C., Rehbein M., Kreienkamp H. J., Buck F., Richter D., Kindler S. Characterization of Staufen 1 ribonucleoprotein complexes. Biochem. J. 2004;384:239–246. doi: 10.1042/BJ20040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S., O'Donnell W. T., Reed M., Patton S., Pohl J., Warren S. T. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Corbin F., Bouillon M., Fortin A., Morin S., Rousseau F., Khandjian E. W. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- Dahm R., Kiebler M. Cell biology: silenced RNA on the move. Nature. 2005;438:432–435. doi: 10.1038/438432a. [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Fraser C. E., Mostovetsky O., Stefani G., Jones T. A., Eddy S. R., Darnell R. B. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005a;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Mostovetsky O., Darnell R. B. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005b;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- De Boulle K., Verkerk A.J.M.H., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van den Bos F., de Graaff E., Oostra B. A., Willems P. J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- De Diego Otero Y., Severijnen L. A., van Cappellen G., Schrier M., Oostra B., Willemsen R. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol. Cell Biol. 2002;22:8332–8341. doi: 10.1128/MCB.22.23.8332-8341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart D. E., Malter H. E., Feng Y., Warren S. T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Elvira G., et al. Characterization of an RNA granule from developing brain. Mol. Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Feng Y. Fragile X mental retardation: misregulation of protein synthesis in the developing brain? Microsc. Res. Tech. 2002;57:145–147. doi: 10.1002/jemt.10063. [DOI] [PubMed] [Google Scholar]
- Feng Y., Absher D., Eberhart D. E., Brown V., Malter H. E., Warren S. T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997a;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y., Gutekunst C. A., Eberhart D. E., Yi H., Warren S. T., Hersch S. M. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 1997b;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F., Mercaldo V., Piccoli G., Sala C., Cannata S., Achsel T., Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol. Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Garber K., Smith K. T., Reines D., Warren S. T. Transcription, translation and fragile X syndrome. Curr. Opin. Genet Dev. 2006;16:270–275. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Goslin K., Banker G. Experimental observations on the development of polarity by hippocampal neurons in culture. J. Cell Biol. 1989;108:1507–1516. doi: 10.1083/jcb.108.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill D., Davis J., Drawbridge J., Suprenant K. A. Polyribosome targeting to microtubules: enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. J. Cell Biol. 1994;127:973–984. doi: 10.1083/jcb.127.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Carson J. H., Barbarese E., Richter J. D. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Richter J. D. Regulation of local mRNA translation. Curr. Opin. Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., Singer R. H. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Jin P., Warren S. T. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem. Sci. 2003;28:152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- Jin P., Zarnescu D. C., Ceman S., Nakamoto M., Mowrey J., Jongens T. A., Nelson D. L., Moses K., Warren S. T. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Khandjian E. W., Huot M. E., Tremblay S., Davidovic L., Mazroui R., Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M. A., Bassell G. J. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Dong W. K., Weiler I. J., Greenough W. T. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J. Neurosci. 2006;26:2413–2418. doi: 10.1523/JNEUROSCI.3680-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosturko L. D., Maggipinto M. J., D'Sa C., Carson J. H., Barbarese E. The microtubule-associated protein tumor overexpressed gene binds to the RNA trafficking protein heterogeneous nuclear ribonucleoprotein A2. Mol. Biol. Cell. 2005;16:1938–1947. doi: 10.1091/mbc.E04-08-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A. M., Kosik K. S. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B., Ostareck D., Keidel E. M., Ostareck-Lederer A., Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Ku L., Wilkinson K. D., Warren S. T., Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S. C., Fahrner P. S., Greenough W. T., Gelfand V. I. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 2004;101:17428–17433. doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Wang H., Liang Z., Ku L., O'Donnell W. T., Li W., Warren S. T., Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl. Acad. Sci. USA. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Huot M. E., Tremblay S., Filion C., Labelle Y., Khandjian E. W. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- Miyashiro K. Y., Beckel-Mitchener A., Purk T. P., Becker K. G., Barret T., Liu L., Carbonetto S., Weiler I. J., Greenough W. T., Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Muddashetty R. S., Kelic S., Gross C., Xu M., Bassell G. J. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell W. T., Warren S. T. A decade of molecular studies of fragile x syndrome. Annu. Rev. Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Penagarikano O., Mulle J. G., Warren S. T. The pathophysiology of fragile X syndrome. Annu. Rev. Genomics Hum. Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Pfeiffer B. E., Huber K. M. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J. Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Wang J. K., McMillian M., Chikaraishi D. M. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M., Kang J., Burlin T. V., Jiang C., Smith C. B. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J. Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrier M., Severijnen L. A., Reis S., Rife M., van't Padje S., van Cappellen G., Oostra B. A., Willemsen R. Transport kinetics of FMRP containing the I304N mutation of severe fragile X syndrome in neurites of living rat PC12 cells. Exp. Neurol. 2004;189:343–353. doi: 10.1016/j.expneurol.2004.05.039. [DOI] [PubMed] [Google Scholar]
- Stefani G., Fraser C. E., Darnell J. C., Darnell R. B. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O., Bakker C. E., Willems P. J., Oostra B. A. No evidence for disruption of normal patterns of mRNA localization in dendrites or dendritic transport of recently synthesized mRNA in FMR1 knockout mice, a model for human fragile-X mental retardation syndrome. Neuroreport. 1998;9:477–481. doi: 10.1097/00001756-199802160-00022. [DOI] [PubMed] [Google Scholar]
- Wang H., Ku L., Osterhout D. J., Li W., Ahmadian A., Liang Z., Feng Y. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum. Mol. Genet. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- Weiler I. J., Irwin S. A., Klintsova A. Y., Spencer C. M., Brazelton A. D., Miyashiro K., Comery T. A., Patel B., Eberwine J., Greenough W. T. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler I. J., et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc. Natl. Acad. Sci. USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R., Oostra B. A., Bassell G. J., Dictenberg J. The fragile X syndrome: from molecular genetics to neurobiology. Ment. Retard Dev. Disabil. Res. Rev. 2004;10:60–67. doi: 10.1002/mrdd.20010. [DOI] [PubMed] [Google Scholar]
- Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Reis S., Oostra B., Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.