Abstract
The 21 nucleotide RNA trafficking signal (RTS), originally identified in myelin basic protein mRNA, but also found in a variety of other localized RNAs, is necessary and sufficient for transport of RNA along microtubules in oligodendrocytes. The RTS binds specifically to the RNA binding protein, hnRNP A2. Together, the RTS and hnRNP A2 comprise cis/trans determinants for several steps in the RNA trafficking pathway. Here we show that insertion of the RTS into green fluorescent protein (GFP) RNA enhances translation without affecting stability of microinjected RNA. In dicistronic RNA, the RTS enhances cap-dependent translation without affecting internal ribosome entry site (IRES)-dependent translation. The translation enhancer function of the RTS is position, copy number, and cell type independent, hnRNP A2 dependent, and saturable with increasing amounts of injected RNA. This represents one of the first specific translation enhancer elements identified in a mammalian system.
Keywords: translation enhancer, hnRNP A2, RNA trafficking signal, dicistronic RNA, eIF4E
Eucaryotic mRNAs follow defined intracellular trafficking pathways from their sites of transcription to their sites of translation (Dreyfuss et al. 1996). The pathway for each mRNA is determined by specific cis-acting sequence elements within the RNA and by cognate trans-acting factors in the cell (McCarthy and Kollmus 1995; Gavis 1997). Specific cis/trans determinants for RNA trafficking have been identified for RNAs encoding actin (Ross et al. 1997), bicoid (Macdonald and Struhl 1988; Macdonald et al. 1993), nanos (Gavis et al. 1996b), oskar (Ephrussi and Lehmann 1992; Kim-Ha et al. 1993), Vg-1 (Mowry and Melton 1992; Deshler et al. 1997), and myelin basic protein (MBP)1 (Ainger et al. 1997; Hoek et al. 1998).
Translation regulation plays an integral role in the RNA trafficking pathway. Translation is thought to be repressed while the RNA is in transit and activated once the RNA is localized. This steers expression of specific gene products to spatially restricted regions of the cell where the RNA is localized, and minimizes ectopic expression elsewhere in the cell. A variety of general mRNA structural features such as the 5′ cap (Svitkin et al. 1996), 3′ poly A tail (Hentze 1995), Kozak consensus sequence (Kozak 1989), internal ribosome entry sites (IRES) (Sachs et al. 1997), and AU rich elements (ARE) (Zhou et al. 1997) affect translational efficiency. In Drosophila, multiple factors regulating translation during development have been identified genetically (Wilson et al. 1996; Seydoux 1996), and specific cis/trans determinants for translation suppression have been identified biochemically (Simbert et al. 1996; Dahanukar and Wharton 1996; Gavis et al. 1996a). Recently, motifs with translation enhancer activity have been identified in the 5′UTR of Arabidopsis ferredoxin gene, fed A, and a photosystem I gene, psaDb, of Nicotiana sylvestris (Yamamoto et al. 1995). However, few specific cis/trans determinants for translation regulation have been identified in mammalian systems. The work described here provides evidence that a specific cis-acting RNA trafficking sequence (RTS) and its cognate trans-acting ligand (hnRNP A2), originally identified as cis/trans determinants for transport of MBP mRNA in oligodendrocytes, also function to enhance translation in mammalian systems.
The oligodendrocyte is the cell that elaborates myelin in the CNS (Bunge et al. 1962). MBP is a major structural component of the myelin membrane (Mikoshiba et al. 1991) and is required for major dense line formation during myelin compaction. The unique topology of the oligodendrocyte has facilitated elucidation of intracellular RNA trafficking because the different steps in the pathway are confined to spatially resolvable subcellular compartments (Ainger et al. 1993). Nuclear export occurs at the nuclear envelope, assembly of RNA into granules occurs in the perikaryon, RNA transport occurs along microtubules in the processes and veins, and RNA localization and translation activation occur in the myelin compartment. Deletion mapping and microinjection experiments with chimeric RNAs have delineated a 21-nucleotide RNA trafficking sequence (RTS) in the 3′UTR of MBP mRNA that is necessary and sufficient for RNA transport in oligodendrocytes (Ainger et al. 1997). RTS-like sequences are also found in a variety of other RNAs that are localized in other cell types, suggesting that the RTS is a general RNA trafficking signal. In vitro binding experiments indicate that the RTS binds with sequence specificity and high affinity to hnRNP A2, a ubiquitously expressed RNA binding protein that is believed to play a role in intracellular RNA trafficking (Hoek et al. 1998). The RTS and hnRNP A2 comprise cis/trans determinants for multiple steps in trafficking of MBP mRNA in oligodendrocytes and perhaps of RTS-containing RNAs in other cell types as well (Carson et al. 1998). The work described here was undertaken to determine if RTS/A2 determinants also function to regulate translation.
In eucaryotic cells, translation is often regulated at the initiation stage (Hentze 1995; Jackson and Wickens 1997). There are two mechanisms of translation initiation, cap dependent and internal ribosome entry site (IRES) dependent (Sachs et al. 1997). The two mechanisms use exactly the same molecular machinery (Sachs et al. 1997), except that cap-dependent initiation requires a 5′ cap on the mRNA (Svitkin et al. 1996) and cap-binding protein (eIF4E) in the cell (Jackson and Wickens 1997), whereas IRES-dependent initiation requires an IRES within the mRNA and an IRES-binding protein (possibly La autoantigen) in the cell (Svitkin et al. 1994). In this work, we examined the effect of the RTS on both cap-dependent and IRES-dependent translation. The RTS enhanced cap-dependent translation specifically and the effect was position, copy number, and cell type independent and hnRNP A2 dependent and saturable with increasing amounts of RNA.
Materials and Methods
Cell Culture
Neuroblastoma B104 cells and CHO cells were grown in 6% newborn calf serum in DME. Oligodendrocytes from shiverer mice were isolated after 12–14 d of growth in culture as described previously (Ainger et al. 1993).
Reagents
Restriction enzymes and RNA polymerase were obtained from New England BioLabs, Promega, Stratagene, and Epicentre Technologies. RNA and protein molecular markers were obtained from GIBCO BRL. Texas red–conjugated dextran (TRD) was obtained from Molecular Probes, Inc. Rabbit polyclonal antibody to ribophorin I (9R1) was obtained from R. Gould (New York State Institute for Basic Research and Developmental Disabilities, Staten Island, NY). Characterization of antibody 9R1 is described in Kreibich et al. 1983. Mouse monoclonal antibody to hnRNP A2 (EF-67) was obtained from W. Rigby (Dartmouth Medical School, Lebanon, NH). Recombinant hnRNP A2 was obtained from R. Smith (University of Queensland, Queensland, AU).
Recombinant DNA and In Vitro Transcription
Full-length cDNA for S65T mutant GFP cloned in pRSETB, was obtained from Dr. R.Y. Tsien (University of California, San Diego, CA). pGEM1A, containing the SP6 promoter and poly A, was obtained from Dr. G. Carmichael (University of Connecticut Health Center, Farmington, CT). The entire S65T mutant GFP cDNA was subcloned into the BamH1 site of pGEM1A to create pGFP′A. The RTS sequence (GCCAAGGAGCCAGAGAGCATG) was inserted into AvaI/SacI cut pGFP′A and HindIII-XbaI cut pGFP′A to create pGFP′3RTS and pGFP′5RTS, respectively. To create pGFP′53RTS, the RTS was inserted into both AvaI-SacI and HindIII-XbaI cut pGFP′A. To create dicistronic constructs, full-length cDNA for BFP, cloned in pQBI50 purchased from Quantum Biotechnologies Inc., was amplified by PCR using oligonucleotides that placed XbaI and XmaI cloning sites at the 5′ and 3′ ends, respectively, and then was inserted into XbaI-XmaI cut pGFP′A and pGFP3′RTS to replace the GFP ORF with the BFP ORF, creating pBFP and pBFP3′RTS. The IRES from EMCV and S65T GFP ORF cloned in pTR5-DC/GFP (a kind gift from D.D. Moser, Biotechnology Research Institute, Montreal, Canada) were amplified by PCR using oligonucleotides that placed XmaI cloning sites at both 5′ and 3′ ends, and then was inserted into XmaI cut pBFP and pBFP3′RTS to create pBDCG and pBDCG3R, respectively.
GFP mRNA and GFP/RTS mRNA, containing the RTS in the 3′UTR, 5′UTR, or both, were prepared by in vitro transcription using AmpliScribe SP6 Transcription Kits (Epicentre Technologies) using pGFP′A, pGFP′3RTS, pGFP′5RTS, and pGFP′53RTS, linearized with SapI as template DNA, and m7G(5′)ppp(5′)G added to the transcription mixture to produce capped mRNAs.
Translation Assays
To assay translation in vivo, capped GFP′A mRNA and GFP′3RTS mRNA at equivalent concentrations were microinjected into neuroblastoma B104 cells, CHO cells, or shiverer oligodendrocytes as described previously (Ainger et al. 1993). TRD was coinjected to provide a measure of the injected volume. Injected cells were incubated at 37°C for 20–24 h. The expressed GFP/TRD fluorescence was visualized by dual channel confocal microscopy, using a Zeiss LSM 410 confocal laser scanning system (Zeiss) with an infinity-corrected 60× 1.4 NA objective. Unless otherwise indicated, images were collected within the dynamic range of the frame store such that pixel values outside the cell were >0 and pixel values inside the cell were <255. GFP and TRD pixel values were integrated over the nucleus to avoid autofluorescent signal in the cytoplasm. The concentration of injected RNA in each cell was calculated based on the initial concentration of RNA and TRD in the pipet and the integrated TRD pixel values in the nucleus. Translation efficiencies were determined by calculating the GFP/RNA ratio for each cell.
To assay translation in vitro, nuclease-treated rabbit reticulocyte lysate and wheat germ extracts were purchased from Promega and reactions were performed according to the manufacturer's instructions. Translation was monitored by [S35]methionine (Amersham) incorporation. Translation products were separated by 12% SDS-PAGE. After electrophoresis, the gels were fixed, soaked in EN3HANCE (New England Nuclear), dried, and subjected to autoradiography to visualize labeled GFP polypeptide. The intensities of specific bands were integrated using ImageQuaNT.
Dequenching Assay for RNA Stability
Fluorescein-labeled GFP RNA and GFP/3′RTS RNA, prepared as previously described (Kwon and Carson 1998), were microinjected into the cytoplasm of neuroblastoma B104 cells. Images of injected cells were collected with a cooled CCD camera at 10-min intervals. Total intracellular fluorescence intensities were integrated using the ImageQuaNT program.
Antisense Oligonucleotide Treatment of B104 Cell Cultures
B104 cells or oligodendrocytes were incubated in defined culture medium containing 8 μM final concentration of phosphorothioate oligonucleotide obtained from the Molecular Core at UCHC) as described previously (Carson et al. 1997). The media containing the oligonucleotide was changed every 8 h for a period of 24 h. Antisense oligonucleotide (CTTTTCTCTCTCCATCGCGGA) was complementary to a region around the translation start site of human hnRNP A2 mRNA. The corresponding sense oligonucleotide was used as a control. Expression of hnRNP A2 in oligodendrocytes was evaluated by immunofluorescence (IF) using monoclonal antibody to hnRNP (1:100). Expression of hnRNP A2 in B104 cells was evaluated by Western blotting with antibody to hnRNP A2 (1:1,000) and rabbit polyclonal antibody to ribophorin I (1:1,000) as a control for loading.
RNA Transport Assay
The NaeI-HindIII fragment from pBluescript SK II(+) was inserted into NaeI-HindIII cut pGFP′A and pGFP′A/3′RTS to create pGT7 and pG3RT7, respectively. Digoxigenin-labeled GFP mRNA and GFP/3′RTS mRNA were generated by in vitro transcription using BsaWI cut pGT7 and pG3RT7 as a template. The RNA transport assay was performed as previously described (Ainger et al. 1993; Carson et al. 1997).
Results
The RTS Is a Translation Enhancer
A translation enhancer is a cis-acting RNA sequence that increases the translation efficiency of the mRNA containing it. Translation efficiency is a measure of the amount of protein translated from a certain amount of RNA. To determine translation efficiency for a particular RNA in vivo, it is necessary to measure both the concentration of that RNA and the amount of protein expressed from it in individual cells. Conventional transfection techniques are of limited use for this purpose because the amount of DNA or RNA introduced or expressed is variable and difficult to measure in individual cells. In some cells, the level of RNA may be saturating, in which case the rate of translation may be determined not by the concentration of RNA but by the activity of rate limiting components of the translation machinery (such as eIF4E) in the cell. Therefore, translation efficiency can only be accurately measured in cells where the exogenous RNA is present in the cell at subsaturating concentrations. Furthermore, cells with saturating levels of RNA generally have high levels of protein expression and, therefore, a disproportionate contribution to the level of protein expression measured for the population as a whole. This tends to obscure the contribution of cells with subsaturating levels of RNA and lower levels of protein expression which are the very cells that are informative for determining translation efficiency for a particular RNA. For these reasons it was necessary to develop a method to introduce subsaturating amounts of RNA into cells and to measure both RNA concentrations and protein translation levels in individual cells.
A ratiometric translation efficiency assay was developed using mRNA encoding green fluorescent protein (GFP) from Aequorea victoria as a reporter for translation and TRD (10 kD) as a reporter for the volume of microinjected RNA. Capped, polyadenylated mRNA encoding GFP was coinjected with TRD into the cytoplasm of B104 cells which were used instead of oligodendrocytes because they lack long processes so that translation of the injected RNA can be analyzed without the confounding variable of RNA transport. After incubation to allow time for GFP expression, injected cells were analyzed by dual channel confocal microscopy. TRD dispersed rapidly throughout the cytoplasm and nucleus, and total fluorescent intensity in each cell remained constant over 24 h (data not shown). GFP, which also dispersed throughout the cytoplasm and nucleus, was first detected by 2 h after injection, increased to a maximum by 9 h, and remained constant up to 24 h (data not shown). The relative amount of RNA injected into each cell was calculated based on the integrated TRD fluorescence intensity in the injected cell. The amount of GFP expression was quantified by integrating GFP fluorescence intensity. Since both TRD and GFP enter the nucleus by diffusion, fluorescence intensities were integrated in optical sections through the nucleus to avoid autofluorescent signal from the cytoplasm. The intensity of TRD fluorescence in each cell provides a measure of the volume of RNA injected. The intensity of GFP fluorescence provides a measure of the amount of protein translated from the injected RNA. The ratio of GFP to RNA provides a measure of the translation efficiency in each cell.
The RTS is a cis-acting sequence that mediates intracellular trafficking of MBP and other mRNAs in oligodendrocytes. To determine its effect on translation efficiency the RTS was inserted into the 3′UTR of GFP. Representative cells injected with GFP/RTS RNA or GFP RNA are shown in Fig. 1. The intensity in the red channel (which measures TRD concentration which is proportional to RNA concentration) was comparable in the cells injected with GFP/RTS RNA and GFP RNA (Fig. 1 A, i and ii) indicating that comparable volumes of RNA were injected into these cells. The intensity in the green channel (which measures GFP concentration) was greater in the cells injected with GFP/RTS RNA compared with cells injected with GFP RNA (Fig. 1 A, iii and iv), indicating that the RTS increases GFP expression. The values for intracellular RNA concentration (in nM) and GFP (expressed in arbitrary fluorescence intensity units) obtained from multiple individual cells are shown in Fig. 1 B. Overall, the level of GFP expression was greater in cells injected with GFP/RTS RNA compared with cells injected with GFP RNA. There was considerable variability in both the amount of RNA injected and the level of GFP expression per cell. Some of the variability in GFP expression may reflect injection of RNA into subcellular compartments with different translational activities. For example, RNA injected into the nucleus is not efficiently exported to the cytoplasm and, therefore, not translated efficiently.
Figure 1.
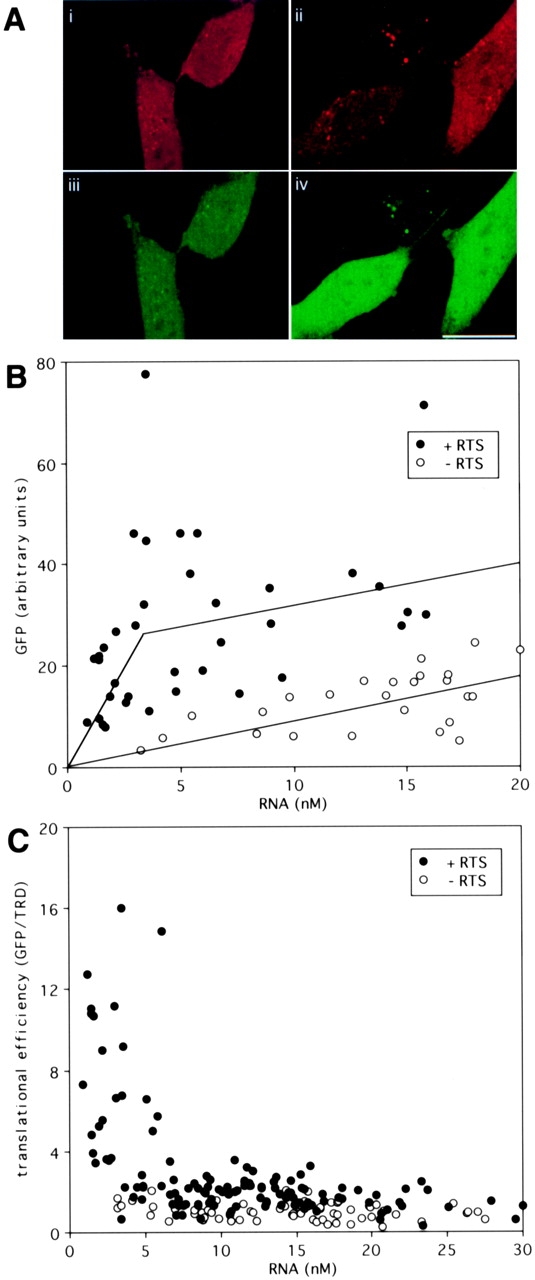
Ratiometric translation assay for GFP RNA and GFP/RTS RNA. (A) Dual channel confocal images of B104 neuroblastoma cells microinjected with a mixture of TRD and either GFP RNA (i and iii) or GFP/RTS RNA (ii and iv). After 24 h, the cells were imaged by dual channel confocal microscopy. i and ii show TRD images. iii and iv show GFP images. Bar, 10 μm. (B) GFP and TRD fluorescence in cells injected with GFP RNA (open circles) or GFP/RTS RNA (closed circles). For each cell the pixel values in the GFP and TRD channels were integrated over the nuclear area. TRD intensity provides a measure of the injected volume, from which the intracellular RNA concentration was calculated. GFP intensity provides a measure of GFP expression (in arbitrary units). (C) Translation efficiency in cells injected with GFP RNA (open circles) or GFP/RTS RNA (closed circles). The ratio of GFP to RNA is plotted versus the RNA concentration as a measure of the translation efficiency in each cell.
To quantify translation efficiency the ratio of GFP/RNA was calculated and plotted versus the amount of injected RNA for each cell. Fig. 1 C shows translation efficiencies for cells injected with different amounts of GFP/RTS RNA and GFP RNA. In cells injected with GFP RNA, translation efficiency was relatively constant (mean translation efficiency = 1.0 ± 0.3) over a wide range of injected RNA, indicating that the endogenous translation machinery in the cell is not saturated by the injected RNA. Cells injected with very low levels of GFP RNA (<5 nM) expressed very low levels of GFP. In some cases the level of GFP expression, while detectable, was too low to quantify accurately. These cells were not included in the graph. In cells injected with low amounts of GFP/RTS RNA (<5 nM), translation efficiency was enhanced (>10-fold in several cells and up to 16-fold in one cell) relative to GFP RNA. At higher amounts of injected RNA (>10 nM), translation efficiency for GFP/RTS RNA was comparable to GFP RNA. These results indicate that the RTS enhances expression of GFP mRNA at low RNA concentrations but the enhancer function is saturated at high RNA concentrations suggesting that RTS-mediated enhanced expression requires component(s) that are present in limiting amounts in the injected cells.
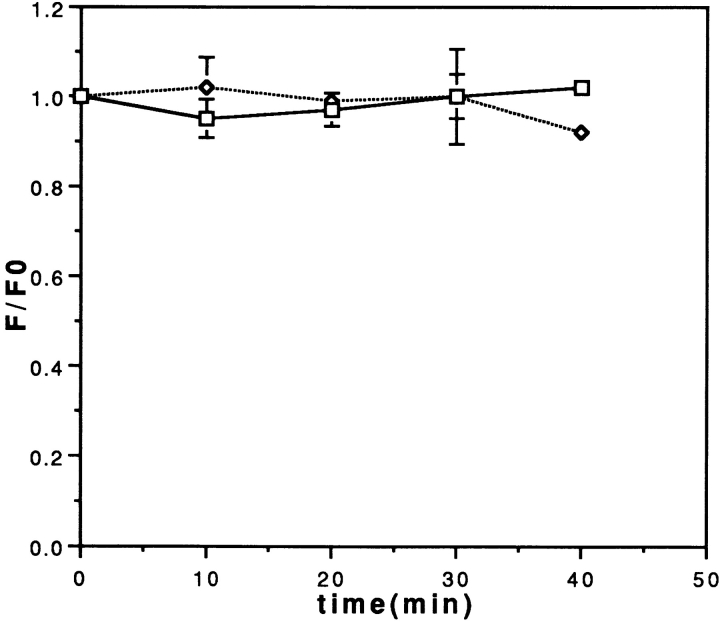
Increased GFP expression mediated by the RTS could be due to either enhanced translation efficiency or enhanced RNA stability in the injected cells. To measure the relative stabilities of GFP/RTS RNA and GFP mRNA in microinjected cells, a novel in vivo fluorescence dequenching assay (Kwon and Carson 1998) was used. Fluorescein-UTP was incorporated by in vitro transcription into the body of GFP/RTS and GFP RNA. In the intact RNA, fluorescence is reduced by intramolecular quenching between proximate fluorophores. The fluorescently labeled RNA was microinjected into cells and intracellular fluorescence was measured as a function of time after injection. In this assay, degradation of the fluorescent RNA causes an increase in total fluorescence due to relief of intramolecular quenching (dequenching) in the injected RNA. Complete degradation of the RNA results in an increase of ∼50% in the fluorescent quantum yield due to dequenching. The amount of dequenching provides a measure of the intracellular stability of the injected RNA. As shown in Fig. 2, there was no significant dequenching over a period of 40 min after injection with either GFP/RTS RNA or GFP RNA, indicating that the two RNAs are both relatively stable within this time frame. The assay was not extended beyond 40 min because translation of the injected RNA could produce GFP fluorescence that would interfere with measurement of the injected fluorescein-labeled RNA. The dequenching assay provides a measure of the rate of RNA degradation in the cell in the time period immediately after injection. Since there is a lag of up to 4 h between synthesis of GFP and appearance of GFP fluorescence in the cell, and since most accumulation of GFP fluorescence occurs within a few hours after injection, it follows that overall accumulation of GFP fluorescence reflects GFP translation within the first few hours after injection. Since there was no detectable difference in stability between GFP RNA and GFP/RTS RNA in the period immediately after injection, it is unlikely that the differential GFP expression observed with GFP/RTS RNA compared with GFP RNA is attributable to differences in initial RNA stability. Therefore, increased GFP expression must be due to enhanced translation efficiency mediated by the RTS.
Figure 2.
Fluorescence dequenching assay for stability of GFP RNA and GFP/RTS RNA in vivo. Fluorescein-labeled GFP RNA (squares, solid line) or GFP/RTS RNA (diamonds, dotted line) were microinjected into B104 cells. Cells were imaged by digital fluorescence microscopy at different times after injection and the intensity integrated over the whole cell area. The integrated fluorescence intensity at each time point (F) divided by the integrated intensity at time 0 (F0) is plotted versus time.
The Translation Enhancer Function of the RTS Is Position, Copy Number, and Cell Type Independent
To determine if the translation enhancer function of the RTS is dependent on its position in the mRNA, chimeric GFP mRNAs were synthesized containing the RTS inserted into either the 3′UTR, the 5′UTR, or both. Because there is an AUG within the RTS, insertion into the 5′UTR of GFP RNA may change the translation initiation site which could result in a 10–amino acid extension at the NH2 terminus of GFP. NH2-terminal extensions do not generally affect GFP fluorescence (Rizzuto et al. 1996; Miyawaki et al. 1997). Chimeric RNAs were microinjected into B104 cells and translation efficiencies were determined at subsaturating RNA concentrations as described in Fig. 1 (Table ). Translation was enhanced 4.19-fold with the RTS in the 3′UTR, 4.82-fold in the 5′UTR, and 3.27-fold with copies in both the 3′ and 5′UTRs. This indicates that the translation enhancer function of the RTS is position and copy number independent. The marginal decrease in translation enhancer activity with two copies of the RTS compared with a single copy may be due to titration of some limiting factor required for enhanced translation.
Table 1.
Translation Enhnacer Function of the RTS
| Cell | RNA | Translation efficiency | Average deviation |
|---|---|---|---|
| B104 | − RTS | 1 | 0.3 |
| B104 | 3′ RTS | 4.19 | 2.18 |
| B104 | 5′ RTS | 4.82 | 1.61 |
| B104 | 5′, 3′ RTS | 3.27 | 0.96 |
| CHO | − RTS | 1 | 0.46 |
| CHO | + RTS | 2.56 | 1.5 |
| Oligodendrocytes | − RTS | 1 | 0.36 |
| Oligodendrocytes | + RTS | 3.38 | 1.8 |
| B104 | Dicistronic | ||
| Cap-dependent | − RTS | 1 | 0.64 |
| Cap-dependent | + RTS | 3.61 | 1.61 |
| IRES-dependent | − RTS | 1 | 0.35 |
| IRES-dependent | + RTS | 1.40 | 0.77 |
To determine if the translation enhancer function of the RTS is cell type specific, translation efficiencies for GFP/RTS RNA and GFP RNA were compared in rat B104 cells, CHO cells, and mouse oligodendrocytes in primary culture (Table ). In CHO cells, the level of GFP fluorescence was more variable than in B104 cells. In some cells, the level of GFP fluorescence was so high that the image was saturated. These cells were not included in the calculation of translation efficiency because the intensity values were not within the dynamic range of the assay, resulting in an underestimate of the actual translation efficiency of GFP/RTS RNA in CHO cells. The variability in GFP expression in CHO cells suggests that factors required for the translation enhancer function of the RTS are expressed at variable levels in these cells. Wild-type mouse oligodendrocytes express high levels of endogenous MBP mRNA, which contains the RTS and which could compete with the microinjected exogenous GFP/RTS RNA. To minimize this potential competition, oligodendrocytes from shiverer mutant mice, which do not express endogenous MBP mRNA (Molineaux et al. 1986), were used for these experiments. The RTS enhanced translation 4.19-fold in B104 cells, 2.56-fold in CHO cells, and 3.38-fold in oligodendrocytes (Table ), indicating that the translation enhancer function is cell type independent, although the magnitude of the effect may vary among different cell types depending on the capacity of the translation machinery or the level of endogenous RTS-containing RNAs.
The Translation Enhancer Function of the RTS Is Cap Dependent
The experiments in Fig. 1 and Table were performed with monocistronic RNA containing a 5′ cap. The results indicate that the RTS enhances cap-dependent translation. To determine if IRES-dependent translation is also enhanced, a dicistronic mRNA encoding blue fluorescent protein (BFP) in a cap-dependent cistron and GFP in an IRES-dependent cistron was constructed. The RTS was inserted into the 3′UTR of the dicistronic mRNA as shown in Fig. 3 A. Translation efficiencies for BFP and GFP were determined by microinjection into B104 cells. Triple channel images of representative injected cells are shown in Fig. 3 B. In the red channel, TRD intensities are comparable for cells injected with RTS and nonRTS RNA, indicating that the volume of injected RNA was comparable in the two cells shown. In the green channel, GFP intensities are comparable, indicating that IRES-dependent translation is not enhanced by the RTS. In the blue channel, BFP intensity is greater in the cell injected with RTS RNA compared with the cell injected with nonRTS RNA, indicating that cap-dependent translation is enhanced by the RTS. Translation efficiencies for cap-dependent and IRES-dependent translation in cells injected with various amounts of RNA were calculated from the ratio of blue to red and green to red intensities, respectively. In the case of cap-dependent translation, at subsaturating concentrations of RTS RNA, translation efficiency was enhanced 3.61-fold relative to nonRTS RNA (Fig. 3 C and Table ). This is consistent with the previous results for cap-dependent translation of monocistronic GFP RNA (Fig. 1 and Table ). In the case of IRES-dependent translation, translation efficiency was comparable between RTS and nonRTS RNA (Fig. 3 D and Table ) indicating that the translation enhancer function of the RTS is specific for cap-dependent translation and does not affect IRES-dependent translation. In addition, the observation that IRES-dependent translation efficiency was relatively constant over a wide range of RNA indicates that the machinery in the cell required for IRES-dependent translation is not saturated by high concentrations of exogenous RNA.
Figure 3.
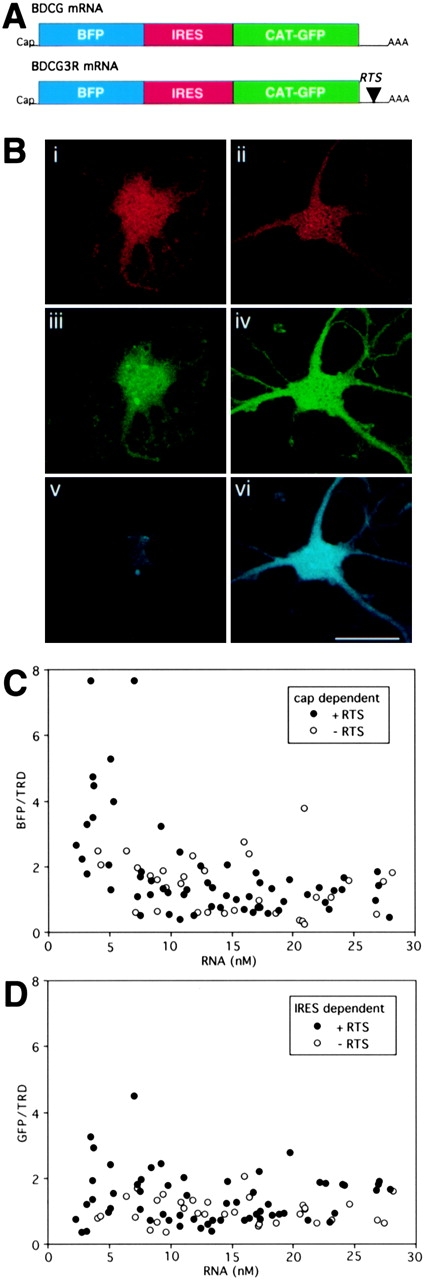
Ratiometric translation assay for discistronic RNA. (A) Structures of dicistronic RNAs. Dicistronic mRNAs encoding BFP in a cap-dependent cistron and GFP in an IRES-dependent cistron are shown. The RTS was inserted into the 3′UTR of the dicistronic RNA. (B) Triple channel confocal microscopy of oligodendrocytes injected with TRD and dicistronic RNA. Oligodendrocytes were microinjected with TRD and either nonRTS (left panels) or RTS-containing dicistronic RNA. After 24 h, the cells were analyzed by triple channel confocal microscopy. A representative cell is shown for each RNA. The TRD channel is shown in red (i and ii), the GFP channel in green (iii and iv), and the BFP channel in blue (v and vi). (C) Cap-dependent translation efficiency. B104 cells were microinjected with dicistronic RNA and imaged as described in B. For each triple channel image the pixel values in the three channels were integrated over the nucleus. The red channel provides a measure of injected TRD volume which is used to calculate intracellular RNA concentration. The blue channel provides a measure of expression of BFP. The ratio of BFP intensity to TRD intensity provides a measure of cap-dependent translation efficiency in each cell. Open circles indicate cells injected with nonRTS RNA. Closed circles indicate cells injected with RTS RNA. (D) IRES-dependent translation efficiency. B104 cells were microinjected with dicistronic RNA and imaged as described in B. For each triple channel image the pixel values in the three channels were integrated over the nucleus. The red channel provides a measure of injected TRD volume which is used to calculate intracellular RNA concentration. The green channel provides a measure of expression of GFP. The ratio of GFP intensity to TRD intensity provides a measure of IRES-dependent translation efficiency in each cell. Open circles indicate cells injected with nonRTS RNA. Closed circles indicate cells injected with RTS RNA.
Both the RNA Transport and Translation Enhancer Functions of the RTS Are hnRNP A2 Dependent
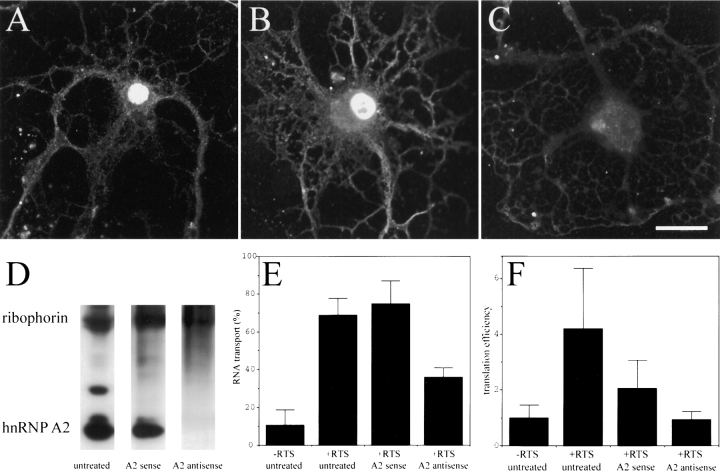
HnRNP A2 binds specifically to the RTS in vitro (Hoek et al. 1998). To determine if hnRNP A2 is required for either the RNA transport or translation enhancer functions of the RTS cells were treated with antisense oligonucleotide designed to hybridize with the translation start site of hnRNP A2 and suppress its expression. Control cells were treated with the corresponding sense oligonucleotide. The level of hnRNP A2 in oligodendrocytes was estimated by IF with monoclonal anti-hnRNP A2 antibody. In untreated oligodendrocytes (Fig. 4 A), hnRNP A2 is detected at high levels in the nucleus and also in granules in the perikaryon and processes indicating that hnRNP A2 is present in both the nucleus and the cytoplasm. To visualize the hnRNP A2 distribution in the cytoplasm, which is lower intensity than in the nucleus, images were collected under conditions where the nuclear signal was saturated. Both the nuclear and cytoplasmic staining are specific for hnRNP A2 and were not observed with control antibody (data not shown). In oligodendrocytes treated with hnRNP A2 sense oligonucleotide (Fig. 4 B), hnRNP A2 staining in both the cytoplasm and nucleus was comparable to the untreated cell indicating that hnRNP A2 expression is not affected. In oligodendrocytes treated with hnRNP A2 antisense oligonucleotide (Fig. 4 C), hnRNP A2 staining was reduced compared with either the untreated cell or the sense-treated cell, indicating that hnRNP A2 expression was suppressed. It appears that nuclear hnRNP A2 is decreased to a greater extent than cytoplasmic hnRNP A2, suggesting that in oligodendrocytes the nuclear pool of hnRNPA2 turns over more rapidly than the cytoplasmic pool. Since the image for untreated and sense-treated cells is saturated in the nuclear compartment, the extent of hnRNP A2 suppression in antisense-treated cells cannot be quantified from these images. However, in images collected under conditions where the nuclear compartment was not saturated (where cytoplasmic hnRNP A2 is not visualized; data not shown), suppression of nuclear hnRNP A2 in antisense-treated cells was >90%.
Figure 4.
The effect of antisense suppression of hnRNPA2 on transport and translation of GFP RNA and GFP/RTS RNA. Oligodendrocytes or B104 cells were treated with antisense oligonucleotide to suppress hnRNP A2 or with sense oligonucleotide as a control. The amount of hnRNP A2 in the cells was analyzed by either IF or Western blotting with monoclonal antibody to hnRNP A2. (A) Untreated oligodendrocyte stained with anti-hnRNP A2; (B) oligodendrocyte treated with sense oligonucleotide stained with anti-hnRNP A2; (C) oligodendrocyte treated with antisense oligonucleotide stained with anti-hnRNP A2. Bar, 10 μm. (D) Western blot of B104 cells stained with anti-hnRNP A2 and anti-ribophorin I. The bands corresponding to hnRNP A2 and ribophorin are indicated. To measure RNA transport, oligodendrocytes were microinjected with digoxigenin-labeled GFP RNA or GFP/RTS RNA, and the intracellular distribution of the injected RNA was determined by IF and confocal microscopy. The percent of cells in which the injected RNA was transported is shown in E. Greater than 100 cells were analyzed for each treatment and for each RNA in three separate experiments. To measure translation efficiency B104 cells were microinjected with either GFP RNA or GFP/RTS RNA and the translation efficiency was determined for each cell as described in Fig. 1 and Table . The mean values for cells containing less than 5 nM RNA are given in F.
The effect of suppressing hnRNP A2 expression on RNA transport in oligodendrocytes was determined by microinjecting digoxigenin-labeled RNA into antisense-treated cells and measuring the percentage of cells in which the injected RNA was transported to the peripheral processes (Fig. 4 E). GFP/RTS RNA was transported in >70% of untreated or sense-treated oligodendrocytes but in <40% of antisense-treated oligodendrocytes, while GFP RNA was transported in <20% of treated or untreated cells. These results indicate that the RNA transport function of the RTS in oligodendrocytes is at least partially dependent on hnRNP A2 expression.
The level of hnRNP A2 in B104 cells was estimated by Western blotting with anti-hnRNP A2 antibody with anti-ribophorin antibody as a loading control (Fig. 4 D). In untreated cells, hnRNP A2 is detected as a major band with an apparent molecular mass of ∼36 kD, whereas ribophorin I is detected as a band with an apparent molecular mass of ∼65 kD. In some samples, the hnRNP A2 antibody detects two additional bands with molecular masses slightly larger than the major band at 36 kD, which may represent splicing variants. In cells treated with sense oligonucleotide, the relative intensities of the hnRNP A2 and ribophorin bands are comparable to untreated cells, whereas in cells treated with antisense oligonucleotide the intensity of the hnRNP A2 band is decreased relative to ribophorin. This indicates that treatment with antisense oligonucleotide suppresses hnRNP A2 expression in B104 cells.
The effect of suppressing hnRNP A2 expression on the translation enhancer function of the RTS was determined by measuring translation efficiencies for GFP/RTS and GFP RNA injected into antisense-treated B104 cells at subsaturating concentrations of RNA (Fig. 4 F). Compared with GFP RNA, the translation efficiency of GFP/RTS RNA was enhanced by 4.19-fold in untreated B104 cells, 2.0-fold in sense-treated cells, and was slightly inhibited (0.94-fold) in antisense-treated cells. These results indicate that the translation enhancer function of the RTS in B104 cells is dependent on hnRNPA2 expression. The partial reduction of translation efficiency in sense-treated cells may be due to nonspecific toxic effects of the oligonucleotides on the cells.
To determine if the RTS enhances translation in vitro, GFP/RTS and GFP mRNAs were translated in a rabbit reticulocyte lysate translation system (Fig. 5 A) and in wheat germ extract (data not shown). In both systems, the translation efficiencies were comparable for the two RNAs over a range of RNA concentrations (Fig. 5 B), indicating that the translation enhancer function of the RTS requires factors that are lacking in rabbit reticulocyte lysate and wheat germ extract. Addition of recombinant hnRNP A2 to rabbit reticulocyte lysate had little effect on translation of GFP RNA but enhanced translation of GFP/RTS RNA in a dose-dependent manner up to a maximum stimulation at 1 μg (Fig. 5 C). This experiment was repeated several times with different batches of reticulocyte lysate and different preparations of hnRNP A2. As shown in Fig. 5 D, the extent of stimulation by hnRNP A2 was variable, but in each experiment, translation of GFP/RTS RNA was enhanced relative to GFP RNA. These results indicate that the translation enhancer function of the RTS is hnRNP A2 dependent in rabbit reticulocyte lysate. The variability in the extent of stimulation may reflect variation in the activities of different preparations of recombinant hnRNP A2 or variation in the activities of different batches of reticulocyte lysate. Addition of recombinant human hnRNP A2 to wheat germ extract had no effect on translation of RTS RNA (data not shown), suggesting that the interaction between hnRNP A2 and component(s) of the translation machinery that enhances translation in rabbit reticulocyte lysate is specific to mammalian systems.
Figure 5.
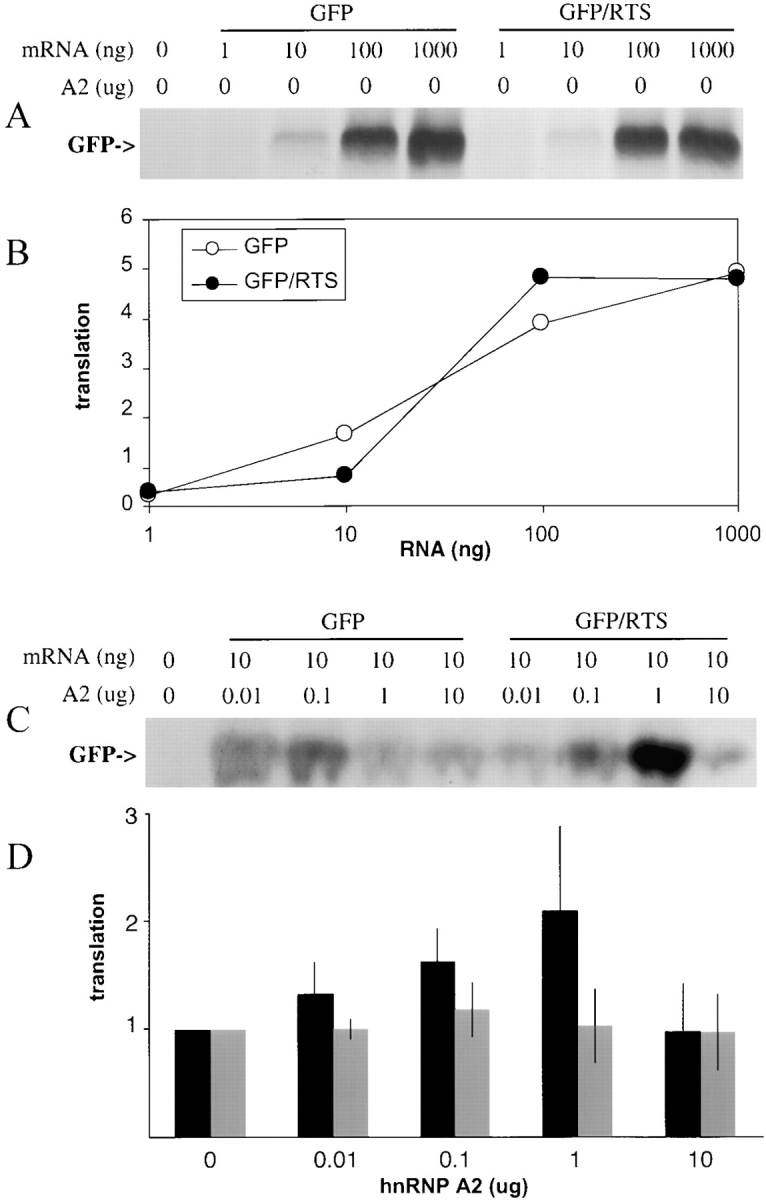
In vitro translation of GFP RNA and GFP/RTS RNA. (A) Increasing amounts of GFP RNA or GFP/RTS RNA were translated in rabbit reticulocyte lysate. Translation products were visualized by SDS PAGE and autoradiography. The band corresponding to GFP is indicated. (B) The intensities of the bands corresponding to GFP in the autoradiogram shown in A were quantitated and plotted in arbitrary translation units versus the amount of GFP RNA (open circles) or GFP/RTS RNA (closed circles) in the translation mixture. (C) Increasing amounts of recombinant hnRNPA2 were added to rabbit reticulocyte lysate along with either GFP RNA (10 ng) or GFP/RTS (10 ng). The translation products were analyzed by SDS-PAGE and autoradiography. The band corresponding to GFP is indicated. (D) The intensities of the bands corresponding to GFP in the autoradiograph shown in C and in three similar experiments were quantitated and are plotted versus the amount of hnRNP A2 added to the lysate. For each RNA the intensity value obtained without hnRNP A2 was set to 1 and the other values normalized accordingly and expressed as translation efficiency. GFP RNA (grey bars); GFP/RTS RNA (black bars).
Discussion
The RTS, originally identified as a cis-acting RNA trafficking sequence in MBP mRNA, is shown to function as an enhancer of cap-dependent translation in vivo and in vitro. This represents the first specific translation enhancer identified in a mammalian system. The translation enhancer function of the RTS is saturable with increasing amounts of RNA, position, copy number, and cell type independent, and cap and hnRNP dependent. A speculative model illustrating the proposed role of RTS/A2 cis/trans determinants in RNA transport and translation activation is shown in Fig. 6. According to this model, the RNA granule is transported along microtubule tracks using kinesin as a molecular motor (Carson et al. 1997). Association of the RNA granule cargo with the kinesin motor during transport requires RTS/A2 determinants. Once the RNA granule reaches its destination, translation is activated through interactions between RTS/A2 determinants and components of the translation machinery, possibly eIF4E at the 5′ cap of the mRNA. This provides an explanation for why the translation enhancer function of RTS/A2 is specific for cap-dependent translation and saturable at high concentrations of RNA, since eIF4E is required for cap-dependent initiation and is present in limiting amounts in most cells (Hiremath et al. 1985). The proposed interactions between RTS/A2 and kinesin during transport and between RTS/A2 and eIF4E during translation may be direct or indirect, mediated through unknown adapter molecules. The model does not specify a detailed molecular mechanism for either the transport or translation enhancer functions of RTS/A2 determinants. However, several possible mechanisms are suggested by structural features of the RTS and properties of hnRNP A2.
Figure 6.
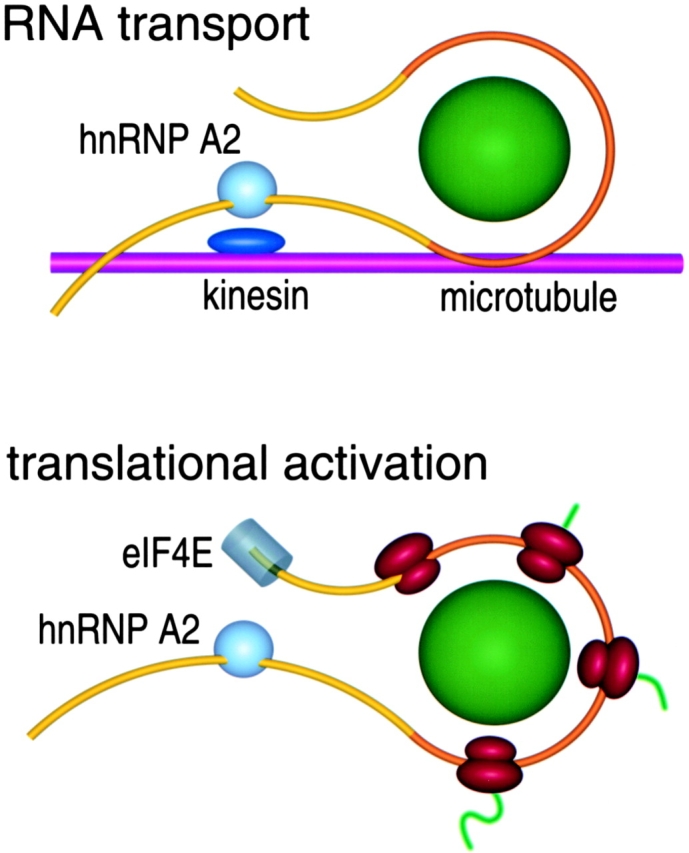
Model for RTS/hnRNP A2 functions in RNA transport and translation. RTS-containing mRNA is shown in yellow with the coding region in orange. hnRNP A2 is shown as a light blue sphere. Core granule components (aminoacyl tRNA synthetases, elongation factors) are shown as a green sphere. A microtubule is shown in magenta. Kinesin is shown in dark blue. eIF4E is shown in grey. Ribosomes are shown in red. Nascent polypeptide chains are shown in green. Specific molecular components are not drawn to scale and juxtaposition of specific components does not necessarily imply direct molecular interactions.
One possible mechanism is based on sequence homology between the RTS and the consensus Kozak sequence. In the scanning model of eukaryotic translation initiation, the 40S ribosomal subunit binds initially at the 5′ end of mRNA and scans in the 3′ direction, initiating translation at the first AUG codon in a favorable context (Kozak 1989). In vertebrates, efficiently used AUG start codons are embedded in a consensus sequence motif, termed the Kozak sequence. As shown in Table the consensus Kozak sequence is partially homologous to the RTS suggesting that the Kozak sequence and the RTS use similar mechanisms for enhancing translation. There are several caveats to this hypothesis. The Kozak sequence is position dependent, since it always encompasses the initiation codon, whereas the RTS is position independent, although it does contain an AUG. In addition, the Kozak sequence is known to interact with two proteins (∼50 and 100 kD) from HeLa cells (McBratney and Sarnow 1996), while the RTS interacts with hnRNP A2 (36 kD), suggesting that the Kozak sequence and the RTS use different trans-acting factors to enhance translation.
Table 2.
Homology between the RTS and the Kozak Consensus Sequence
| RTS | 5′ | GCCAAGGAGCCAGAGAGCAUG | 3′ |
|---|---|---|---|
| Kozak | 5′ | GCCGCCACCAUGG | 3′ |
A second possible mechanism involves the RNA helix destabilizing and annealing properties of hnRNP A2 (Monroe and Dong, 1992). The presence of secondary structure in the 5′UTR of mRNA tends to inhibit translation. After association of the eIF4F complex (consisting of initiation factors eIF4A, eIF4E, and eIF4G) with the cap of the mRNA, the interaction of eIF4B and the RNA helicase eIF4A causes ATP-dependent melting of secondary structure in the 5′UTR. This activity is believed to facilitate binding of the 40S preinitiation complex to the mRNA. Recently, eIF4B was shown to stimulate eIF4A-mediated melting of RNA secondary structure in vitro (Altman et al., 1995). In a similar fashion, binding of hnRNP A2 to the RTS might promote unwinding of secondary structure in the mRNA to reduce a kinetic barrier to formation of the translation initiation complex. Melting of RNA secondary structure by hnRNP A2 could facilitate binding of the 40S preinitiation complex to the mRNA and scanning the 5′UTR in translation initiation. Besides melting secondary structures, the RNA annealing activity of hnRNP A2 could also play a role in recognition of the start codon. Base pairing between the AUG initiation codon and the anticodon of the initiator tRNA, which is a major determinant for AUG codon recognition, could be facilitated by the RNA annealing activity of hnRNP A2, thus, enhancing translation efficiency.
A third possible mechanism involves potential eIF4E-binding motifs in hnRNP A2. A recently determined x-ray structure of murine eIF4E, bound to a cap analogue (7-methyl-GDP), provides a basis for analyzing contacts between eIF4E and proteins that interact with eIF4E during translation initiation (Marcotrigiano et al. 1997). The eIF4E-binding sites in eIF4E-binding proteins, 4E-BP1, 4E-BP2, and eIF4G, share a common motif, Y-X-X-X-X-L-L, suggesting that these proteins compete for the same site in eIF4E. The RBD1 domain of hnRNP A2 contains a peptide sequence, Y-E-Q-W-G-K-L, which is similar to the putative eIF4E binding motif and might be involved in eIF4E binding. This could affect formation of the initiation complex. It will be important to determine whether there is direct biochemical interaction between hnRNP A2 and eIF4E.
Whatever the molecular mechanism(s) for translation activation by the RTS and hnRNP A2, the fact that the same cis/trans determinants that mediate multiple steps in the RNA trafficking pathway also enhance translation underscores the integral role of translation regulation in RNA trafficking. The RTS and hnRNP A2 comprise the first translation enhancer identified in a mammalian system. This may prove useful in applications where maximal expression is critical.
Acknowledgments
We thank Dr. R.Y. Tsien, Dr. G.G. Carmichael, and Dr. D.D. Moser for plasmids and Dr. R. Smith for recombinant hnRNP A2 protein.
This work was supported by National Institutes of Health grant number NS15190 to J.H. Carson.
Footnotes
1.used in this paper: BFP, blue fluorescent protein; GFP, green fluorescent protein; hnRNP, heterogeneous nuclear RNA binding protein; IF, immunofluorescence; IRES, internal ribosome entry site; MBP, myelin basic protein; RTS, RNA trafficking signal; TRD, Texas red–conjugated dextran
S. Kwon's current address is Department of Cell Biology, Harvard Medical School, Boston, MA 02115.
References
- Ainger K., Avossa D., Morgan F., Hill S.J., Barry C., Barbarese E., Carson J.H. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K., Avossa D., Diana A.S., Barry C., Barbarese E., Carson J.H. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Wittmer B., Methot N., Sonenberg N., Trachsel H. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homolog, eIF-4B, have RNA annealing activity. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:3820–3827. doi: 10.1002/j.1460-2075.1995.tb00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge M.B., Bunge R.P., Pappas G.D. Electron microscopic demonstrations of connections between glia and myelin sheets in the developing mammalian central nervous system. J. Cell Biol. 1962;12:448–457. doi: 10.1083/jcb.12.2.448. [DOI] [PubMed] [Google Scholar]
- Carson J.H., Worboys K., Ainger K., Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskel. 1997;38:318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Carson J.H., Kwon S., Barbarese E. RNA trafficking in myelinating cells. Curr. Opin. Neurobiol. 1998;8:607–612. doi: 10.1016/s0959-4388(98)80088-3. [DOI] [PubMed] [Google Scholar]
- Dahanukar A., Wharton R.P. The Nanos gradient in Drosophila embryos is generated by translation regulation. Genes Dev. 1996;10:2610–2620. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- Deshler J.O., Highett M.I., Schnapp B.J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Hentze M., Lamond A.I. From transcript to protein. Cell. 1996;85:963–972. doi: 10.1016/s0092-8674(00)81298-2. [DOI] [PubMed] [Google Scholar]
- Ephrussi A.L., Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Gavis E.R., Lunsford L., Bergsten S.E., Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA Development. 122 1996. 2791 2800a [DOI] [PubMed] [Google Scholar]
- Gavis E.R., Curtis D., Lehmann R. Identification of cis-acting sequences that control nanos RNA localization Dev. Biol. 176 1996. 36 50b [DOI] [PubMed] [Google Scholar]
- Gavis E.R. Expeditions to the poleRNA localization in Xenopus and Drosophila . Trends Cell Biol. 1997;7:485–492. doi: 10.1016/S0962-8924(97)01162-8. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. Translational regulationversatile mechanisms for metabolic and developmental control. Curr. Opin. Cell Biol. 1995;7:393–398. doi: 10.1016/0955-0674(95)80095-6. [DOI] [PubMed] [Google Scholar]
- Hiremath L.S., Webb N.R., Rhoads R.E. Immunological detection of the messenger RNA cap-binding protein. J. Biol. Chem. 1985;260:7843–7849. [PubMed] [Google Scholar]
- Hoek K.S., Kidd G.J., Carson J.H., Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr. Opin. Genet. Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Webster P.J., Smith J.L., Macdonald P.M. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences upstream from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translationan update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich G. E.E., Marcantonio, Sabatini D.D. Ribophorin I and II-membrane proteins characteristic of rough endoplasmic reticulum. Methods Enzymol. 1983;96:520–530. doi: 10.1016/s0076-6879(83)96045-7. [DOI] [PubMed] [Google Scholar]
- Kwon S., Carson J.H. Fluorescence quenching and dequenching analysis of RNA interactions in vitro and in vivo. Anal. Biochem. 1998;264:133–140. doi: 10.1006/abio.1998.2846. [DOI] [PubMed] [Google Scholar]
- McCarthy J.E.G., Kollmus H. Cytoplasmic mRNA-protein interactions in eukaryotic gene expression. Trends Biochem. Sci. 1995;29:191–197. doi: 10.1016/s0968-0004(00)89006-4. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M., Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M., Kerr K., Smith J.L., Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- McBratney S., Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol. Cell. Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikoshiba K., Okano H., Tamura T.-A., Ikenaka K. Structure and function of myelin protein genes. Annu. Rev. Neurosci. 1991;14:201–217. doi: 10.1146/annurev.ne.14.030191.001221. [DOI] [PubMed] [Google Scholar]
- Miyawaki A., Llopis J., Heim H., McCaffery J.M., Adams J.A., Ikura M., Tsien R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Molineaux S.M., Engh H., De Ferra F., Hudson L., Lazzarini R.A. Recombination within the myelin basic protein gene created the dysmyelinating shiverer mouse mutation. Proc. Natl. Acad. Sci. USA. 1986;83:7542–7546. doi: 10.1073/pnas.83.19.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K.L., Melton D.A. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Rizzuto R., Brini M., De Giorgi F., Rossi R., Heim R., Tsien R.Y., Pozzan T. Double labeling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr. Biol. 1996;6:183–188. doi: 10.1016/s0960-9822(02)00451-7. [DOI] [PubMed] [Google Scholar]
- Ross A.F., Oleynikov Y., Kislauskis E.H., Taneja K.L., Singer R.H. Characterization of a β-actin mRNA zip code-binding protein. Mol. Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow P., Hentze M.W. Starting at the beginning, middle, and endtranslation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Seydoux G. Mechanism of translational control in early development. Curr. Opin. Genes Dev. 1996;6:555–561. doi: 10.1016/s0959-437x(96)80083-9. [DOI] [PubMed] [Google Scholar]
- Simbert C.A., Wilson J.E., Kerr K., Macdonald P.M. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Meerovitch K., Lee H.S., Dholakia J.N., Kenan D.J., Agol V.I., Sonenberg N. Internal translation initiation on poliovirus RNAfurther characterization of La function in poliovirus translation in vitro. J. Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Ovchinnikov L.P., Dreyfuss G., Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E., Connell J.E., Macdonald P.M. Aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Tsuji H., Obokata J. 5′-Leader of a photosystem I gene in Nicotiana sylvestris, psaDb, contains a translational enhancer. J. Biol. Chem. 1995;270:12466–12470. doi: 10.1074/jbc.270.21.12466. [DOI] [PubMed] [Google Scholar]
- Zhou B., Boudreau N., Coulber C., Hammarback J., Rabinovitch M. Microtubule-associated protein 1 light chain 3 is a fibronectin mRNA-binding protein linked to mRNA translation in lamb vascular smooth muscle cells. J. Clin. Invest. 1997;100:3070–3082. doi: 10.1172/JCI119862. [DOI] [PMC free article] [PubMed] [Google Scholar]