Abstract
The multisubunit protein, dynactin, is a critical component of the cytoplasmic dynein motor machinery. Dynactin contains two distinct structural domains: a projecting sidearm that interacts with dynein and an actin-like minifilament backbone that is thought to bind cargo. Here, we use biochemical, ultrastructural, and molecular cloning techniques to obtain a comprehensive picture of dynactin composition and structure. Treatment of purified dynactin with recombinant dynamitin yields two assemblies: the actin-related protein, Arp1, minifilament and the p150Glued sidearm. Both contain dynamitin. Treatment of dynactin with the chaotropic salt, potassium iodide, completely depolymerizes the Arp1 minifilament to reveal multiple protein complexes that contain the remaining dynactin subunits. The shoulder/sidearm complex contains p150Glued, dynamitin, and p24 subunits and is ultrastructurally similar to dynactin's flexible projecting sidearm. The dynactin shoulder complex, which contains dynamitin and p24, is an elongated, flexible assembly that may link the shoulder/sidearm complex to the Arp1 minifilament. Pointed-end complex contains p62, p27, and p25 subunits, plus a novel actin-related protein, Arp11. p62, p27, and p25 contain predicted cargo-binding motifs, while the Arp11 sequence suggests a pointed-end capping activity. These isolated dynactin subdomains will be useful tools for further analysis of dynactin assembly and function.
Keywords: dynein, ultrastructure
The minus-end directed, microtubule-based motor, cytoplasmic dynein, drives a wide range of cytoplasmic motor activities. Dynein is required for the steady state localization of the Golgi complex and endosomal membranes near the center of the cell (Burkhardt et al. 1997; Harada et al. 1998) and for movement of ER-Golgi transport complexes (Presley et al. 1997), late endosomes, and lipid droplets (Valetti et al. 1999). The centripetal movement of virus capsids also appears to use the dynein motor (Sodeik et al. 1997; Suomalainen et al. 1999). Dynein contributes in a major way to microtubule organization during interphase (Quintyne et al. 1999) and mitosis (Compton 1998). Perturbation of dynein function during mitosis results in defective spindle pole separation (Vaisberg et al. 1993) and chromosome congression (Echeverri et al. 1996), causing pseudoprometaphase arrest. Assays of microtubule aster and spindle formation in vitro have shown dynein to be necessary for microtubule focusing and pole maintenance in these systems as well (Verde et al. 1991; Gaglio et al. 1996; Heald et al. 1996; Merdes et al. 1996). In some cells, such as budding yeast (Carminati and Stearns 1997), polarized epithelia (Busson et al. 1998) and Caenorhabditis elegans early embryos (Skop and White 1998; Gönczy et al. 1999), dynein also powers movements of the entire mitotic spindle and/or centrosomes relative to the cell cortex. In syncytial Drosophila embryos, dynein helps anchor and move centrosomes relative to nuclei (Robinson et al. 1999).
To drive such a variety of subcellular motile functions, cytoplasmic dynein works in concert with an accessory factor, dynactin. Like dynein (Holzbaur and Vallee 1994), dynactin is a massive protein complex that contains multiple polypeptide subunits (Schroer 1996). First discovered as an activator of cytoplasmic dynein-based vesicle motility in vitro (Gill et al. 1991; Schroer and Sheetz 1991), dynactin is now known to be required for all forms of dynein-based motility (Allan 1996; Vallee et al. 1996; Holleran et al. 1998). Dynein provides the “brawn” for movement, while dynactin appears to serve as the “brain” that directs and coordinates the activities of the dynein motor. Dynactin is thought to do this in two ways. First, it is proposed to function as an adapter that binds dynein to cargo structures. Second, dynactin may increase the efficiency of the cytoplasmic dynein motor by enhancing processivity (King, S.J., and T.A. Schroer, manuscript submitted for publication). Dynactin functions that are independent of cytoplasmic dynein, such as microtubule anchoring at centrosomes, have also been discovered (Quintyne et al. 1999).
In keeping with this diverse array of functions, dynactin has a unique and complicated structure. Previous biochemical and ultrastructural analyses (Schafer et al. 1994a) showed dynactin to contain at least nine different polypeptide species that are organized into two distinct structural domains, an actin-like minifilament and a pleomorphic projecting sidearm. The actin-like minifilament is a short, uniformly sized polymer of the actin-related protein (Arp),1 Arp1. This structure is capped on one end by capZ, a heterodimeric, barbed-end actin capping protein, and carries the p62 subunit at the other. The flexible dynactin sidearm contains the p150Glued subunit. In intact cells and subcellular fractions, exposure of dynactin to an excess of the dynactin subunit dynamitin (p50) causes both p150Glued (Echeverri et al. 1996; Wittman and Hyman 1999) and p24 (Karki et al. 1998) to be released from the Arp1 minifilament, suggesting that dynactin's two prominent structural domains are held together by dynamitin.
The dynactin structure has prompted much speculation and discussion as to how its different parts might contribute to function. A large body of evidence (Karki and Holzbaur 1995; Vaughan and Vallee 1995; Echeverri et al. 1996; Steffen et al. 1997; Quintyne et al. 1999) suggests that dynein's 74-kD intermediate chains bind directly to the projecting p150Glued sidearm. p150Glued can also bind microtubules (Waterman-Storer et al. 1995), an activity that appears to be important for dynactin's effects on dynein processivity (King, S.J., and T.A. Schroer, manuscript submitted for publication) and microtubule anchoring at centrosomes (Quintyne et al. 1999). Dynamitin can bind directly to the chromosomal protein zw10, which may provide a mechanism for dynactin binding to kinetochores (Starr et al. 1998). Dynactin's Arp1 minifilament is thought to be the domain that interacts with most cargoes. Golgi membrane binding is proposed to occur via a spectrin-like membrane skeleton (Schafer et al. 1994a; Holleran et al. 1996; Schroer et al. 1996; Vallee and Sheetz 1996; Holleran and Holzbaur 1998), while other cargoes use as yet undefined binding mechanisms.
As a first step toward analyzing binding interactions between specific dynactin subunits, dynein, and cargo organelles, we have isolated and characterized several different protein subcomplexes that comprise dynactin. Treatment of bovine brain dynactin with recombinant human dynamitin yields two stable domains, a p150Glued/p24 complex and an Arp1 minifilament, both of which contain dynamitin. Dissociation of the Arp1 minifilament with the chaotropic salt, potassium iodide (KI) (Bingham and Schroer 1999), yields four subcomplexes that can be separated by gel filtration chromatography. The largest of these, the dynactin shoulder/sidearm, contains the p150Glued, dynamitin, and p24 subunits. Dynamitin and p24, but not p150Glued, comprise the dynactin shoulder. The p62 and p27 subunits are associated with two previously undetected dynactin subunits, p25 and a novel actin-related protein, Arp11, in dynactin's pointed-end complex. The actin-capping protein, capZ, is recovered as the stable α/β2 heterodimer. Molecular cloning of the p62, Arp11, p27, and p25 subunits reveals a number of features that may contribute to interactions with membranous and other cargoes, including a RING-finger–like domain within p62 (a Neurospora Ropy-2 homologue) and the alkaline isoelectric points (pIs) of p62, Arp11, and p25. The Arp11 sequence predicts this protein will interact only with Arp1 or actin filament pointed ends, which may provide a novel mechanism for Arp1 filament capping and/or actin nucleation.
Materials and Methods
Dynactin Purification
Dynactin isolated from bovine brain and purified to 95% homogeneity as previously described (Bingham et al. 1998) was used in all experiments.
Dynamitin-mediated Disruption of Dynactin
20 μg of dynactin was combined with 144 μg of recombinant human dynamitin (Wittman and Hyman 1999) in 0.5 ml disruption buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA plus protease inhibitors) and incubated for 30 min on ice. Control dynactin samples that did not contain added dynamitin were incubated in parallel. Samples were diluted to 1.5 ml and the proteins were sedimented into 5–20% sucrose gradients at 35,000 rpm in an SW41 rotor for ∼17 h. 1-ml fractions were collected and the proteins were chloroform/methanol precipitated and subjected to SDS-PAGE followed by Coomassie blue staining (80% of the sample) or immunoblotting (20% of the sample).
For quantitation of the stoichiometry of subunits released by dynamitin, 200 μg of dynactin was disrupted with 1.4 mg dynamitin as described above. Sucrose gradient fractions 6 and 7, which contained the peak of p150Glued and p24, were combined and resedimented into a second 5–20% sucrose gradient. Fractions 6 and 7 from the second gradient were combined, precipitated, and subjected to SDS-PAGE followed by Coomassie blue staining. Shoulder/sidearm subunit stoichiometry was determined by densitometry (see below).
Isolation of Dynactin Subcomplexes
5 mg of bovine dynactin was disrupted by adding freshly prepared 6 M KI in H2O, to yield a final concentration of 0.7 M. The sample was incubated on ice for 25 min, and then chromatographed on a Superose12 (Pharmacia LKB Biotechnology Inc.) gel filtration column preequilibrated in column buffer (2 mM Tris-Cl, pH 8, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM DTT) containing 0.7 M KI. Individual column fractions (0.5 ml) were aliquoted and analyzed by SDS-PAGE to identify fractions of interest. The column (see Fig. 2 A) corresponds to a 500-μl sample run at 0.5 ml/min, but dynactin subcomplexes were no better resolved when the column was run at closer to ideal conditions (e.g., 50-μl sample load run at 0.2 ml/min).
Figure 2.

Purification of dynactin shoulder/sidearm and shoulder complexes. (A) Elution profile of KI-treated dynactin run on a Superose12 column. Individual column fractions were analyzed on a Coomassie blue–stained SDS gel. Protein peaks A–E are bracketed. Dynactin subunits are indicated on the left (DM, dynamitin; CPα and CPβ, α and β subunits of capZ). Asterisks in peak C indicate the novel 43- and 25-kD species. (B) Peak A (dynactin shoulder/sidearm) was dialyzed to remove KI, and then sedimented into a 5–20% sucrose gradient. Individual gradient fractions were analyzed on a Coomassie blue–stained SDS gel. The load and positions of sedimentation standards (run in parallel) are indicated. Molecular weight markers are on the left. (C) Peak B (dynactin shoulder) was dialyzed, sedimented, and analyzed as above.
To further purify shoulder/sidearm, shoulder, and pointed-end complexes, the relevant protein fractions were pooled, and then dialyzed briefly (30–60 min × 2,000 vol) in column buffer without KI. 1 ml of the different dialyzed pools was then loaded onto separate 11-ml sucrose gradients (5–20% sucrose in column buffer) and centrifuged at 34,000 rpm in an SW-41 rotor for 13.5 h. Sedimentation standards were run in parallel. 1-ml fractions were collected from the bottom of each tube and analyzed by SDS-PAGE. The pellet was resuspended in 1 ml of 1× sample buffer just before analysis. In some cases, fractions containing the peak of the protein of interest were reanalyzed by Superose12 chromatography in column buffer without KI or pooled and further fractionated by MonoS chromatography (pointed-end complex). The MonoS column was eluted with a linear gradient of 0–500 mM KCl over 20-column volumes. Efforts to improve the resolution of pointed-end complex subunits were unsuccessful; these proteins eluted as a very broad peak under all conditions.
Ultrastructural Analysis
Samples (2.5 μg protein in 0.5 ml column buffer) were adsorbed to freshly cleaved mica, rapidly pressure frozen, freeze fractured, and deep etched as previously described (Heuser 1983). Images of fields of molecules were made on conventional EM negatives; then molecules of interest were captured from projections of the negatives using a digital camera. Final galleries were assembled using Adobe Photoshop®, but the images were not digitally image processed except to add pseudocolor.
Antibodies
Monoclonal antibodies to p150Glued, Arp1, p62, and actin were previously described (anti–p150Glued: mAb 150B (Gaglio et al. 1996; Blocker et al. 1997); anti–Arp1: mAb 45A; anti–p62: mAb 62B (Schafer et al. 1994a); anti–actin mAb: C4 (Lessard 1988).
Gels, Blots, Densitometry, and Peptide Fingerprinting
PAGE was carried out as described (Laemmli 1970). Immunoblotting was performed as described (Towbin et al. 1979); appropriate alkaline phosphatase-conjugated secondary antibodies were detected by chemiluminescence (Tropix). For densitometry, gels were stained with Coomassie brilliant blue R-250, scanned on a flat-bed scanner (ScanMaker III; Microtek) and compared with a BSA standard dilution series using National Institutes of Health Image software. Molar ratios were determined based on cloned subunit Mrs. Peptide fingerprints were determined by in-gel proteolysis of gel-purified proteins, as described (Cleveland 1983).
Peptide Microsequencing
Dynactin subunits were separated by SDS-PAGE (12.5% acrylamide) and Coomassie stained at room temperature. For sequencing at EMBL (p62 and p27), stained gels were sent intact. For sequencing at Harvard University Microchemistry Facility (p25 and Arp11), gels were briefly destained in 10% acetic acid and individual bands were excised, washed twice in 50% acetonitrile, and frozen at −80°C. At both facilities, the proteins were subjected to trypsin digestion. p62 and p27 tryptic fragments were isolated by HPLC and their NH2-terminal sequences determined by Edman degradation. Arp11 and p25 sequences were obtained by in-line, tandem liquid chromatography and mass spectrometry, yielding multiple peptide sequences from a single digest.
p62 Cloning
A λ ZAP II rat brain library (Stratagene Inc.) was screened using mAb 62B (Schafer et al. 1994a). Immunopositive lambda ZAP clones were plaque purified and cDNAs were recovered in the pBluescript KS II+ vector by induction with the R408 helper phage (Stratagene Inc.).
Isolation of Full-length cDNAs
p62.
Rapid amplification of cDNA ends (RACE)–PCR was used to obtain the putative ATG start codon of the cDNA. A gene-specific primer, 62.05: 5′-ATGCACCAGCTCGTGGTCGCTGAAGCC-3′ was used to prime the synthesis of first-strand cDNA using poly A+ mRNA from adult rat brains. The reverse transcription was done at 42°C with Superscript reverse transcriptase (Life Technologies, Inc.). After dA tailing, the PCR reactions were performed essentially as described (Frohman et al. 1988). A primary PCR reaction was performed with the gene-specific primer, 62.03; 5′-TCCACTACATGAATAGTGTGTTGC-3′, and a primer matching the synthesized dA tail; 5′ AMP, 5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′.
The PCR products from the primary reaction were reamplified with a nested gene-specific primer, 62.06; 5′-CAAAGGCATGTAATTTCTTCGC-3′ and 5′ AMP. All PCR reactions were 30 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min using AmpliTaq (Perkin-Elmer Cetus Corp.). All PCR products were gel purified and ligated into the pCRII TA cloning vector (Invitrogen Corp.). The deduced amino acid sequence was verified by comparison with sequences obtained from tryptic peptides prepared from bovine brain p62 (available from GenBank/EMBL/DDBJ under accession number AF190798).
Arp11, p27, and p25.
cDNAs were obtained by searching the EST database (NCBI) using peptide sequences obtained from the native bovine proteins. Full-length mouse EST sequences containing the required in-frame stops upstream of the initiator methionines and polyadenlylation sites were obtained from the American Type Culture Collection. Mouse p25 and p27 were encoded by single ESTs (AA869597 and AA073653, respectively). pEGFP-p25 was constructed from AA869597 BamHI/HindIII and pEGFP-C2 BglII/HindIII. pEGFP-p27 was assembled from an EcoRI fragment containing the rat p27 open reading frame (clone 27.004) and pEGFP-C3 EcoRI. The Arp11 open reading frame was assembled from nonredundant fragments of three overlapping ESTs (the SalI/BstXI fragment from AA109989, the BstXI/EcoRI fragment from AA272131, and the EcoRI/HindIII fragment from AA475054) in three separate constructs (C, B, and A). Construct A contains the EcoRI/HindIII fragment of AA475054 in pEGFP-C2 HindIII/EcoRI. The EcoRI fragment of AA272131 was cloned into the EcoRI site of construct A to yield construct B (correct orientation was confirmed by restriction digest). Construct C (Arp11 ORF reverse) was finally assembled by ligating construct B digested with XhoI and BstXI with the SalI/BstXI fragment of AA109989. Bacterial strains were obtained from Genome Systems and large-scale plasmid preparations were CsCl-gradient purified before digests and subcloning. After cloning into the pEGFP vectors, plasmid inserts were completely sequenced from the EGFP-C primer (Clontech) and internal primers designed by the Johns Hopkins DNA Facility (Baltimore, MD). Sequences were visually confirmed from Editview 1.0 tracings of the sequencing gel output. The sequences of all three ESTs reported in the database were found to be correct (sequences are available from GenBank/EMBL/DDBJ under accession numbers: Dynactin p25, AF190795; DYNACT p27, AF190796; ARP1L, AF190797).
Results
Dynamitin Disruption of Dynactin In Vitro
Overexpression of the dynactin subunit, dynamitin (p50), in cultured cells induces release of p150Glued, but not p62, from the dynactin Arp1 minifilament (Echeverri et al. 1996). This effect can be mimicked in vitro by adding an excess of recombinant dynamitin to a Xenopus egg cell extract (Wittman and Hyman 1999) or partially purified dynactin preparation (Karki et al. 1998). Because all these studies were performed in complex protein mixtures, it was not clear whether dynamitin alone was sufficient to disrupt dynactin or whether additional cellular factors were required. To explore this question further, and to characterize in more detail the dynactin components that were released by dynamitin, we mixed highly purified bovine brain dynactin with a 30-fold excess of purified, recombinant human dynamitin (Wittman and Hyman 1999). Sedimentation of the mixture into a sucrose density gradient revealed two major peaks of protein, one at ≈17S and one at ≈9S (Fig. 1). The subunit compositions of both pools were evaluated by SDS-PAGE staining (Fig. 1A and Fig. C) and immunoblot analysis with Abs to p150Glued and conventional actin (Fig. 1B and Fig. D). The 19S pool contained p62 and Arp1, as previously described (Echeverri et al. 1996), as well as conventional actin, the barbed-end actin-capping protein, capZ, and dynamitin, all in concentrations similar to the undisrupted control sample. That p62, Arp1, actin, and capZ subunits remained tightly associated in a complex that sedimented ≈19S suggested that excess dynamitin did not significantly perturb the structure of the Arp1 minifilament. Quantitative densitometry revealed the Arp1: dynamitin ratio to be similar in intact dynactin and the ≈19S complex produced by dynamitin treatment (1.97 ± 0.14:1 in intact dynactin vs. 1.80 ± 0.16:1 in the 19S complex). This finding suggests that dynamitin may contribute to the stability of the Arp1 minifilament.
Figure 1.
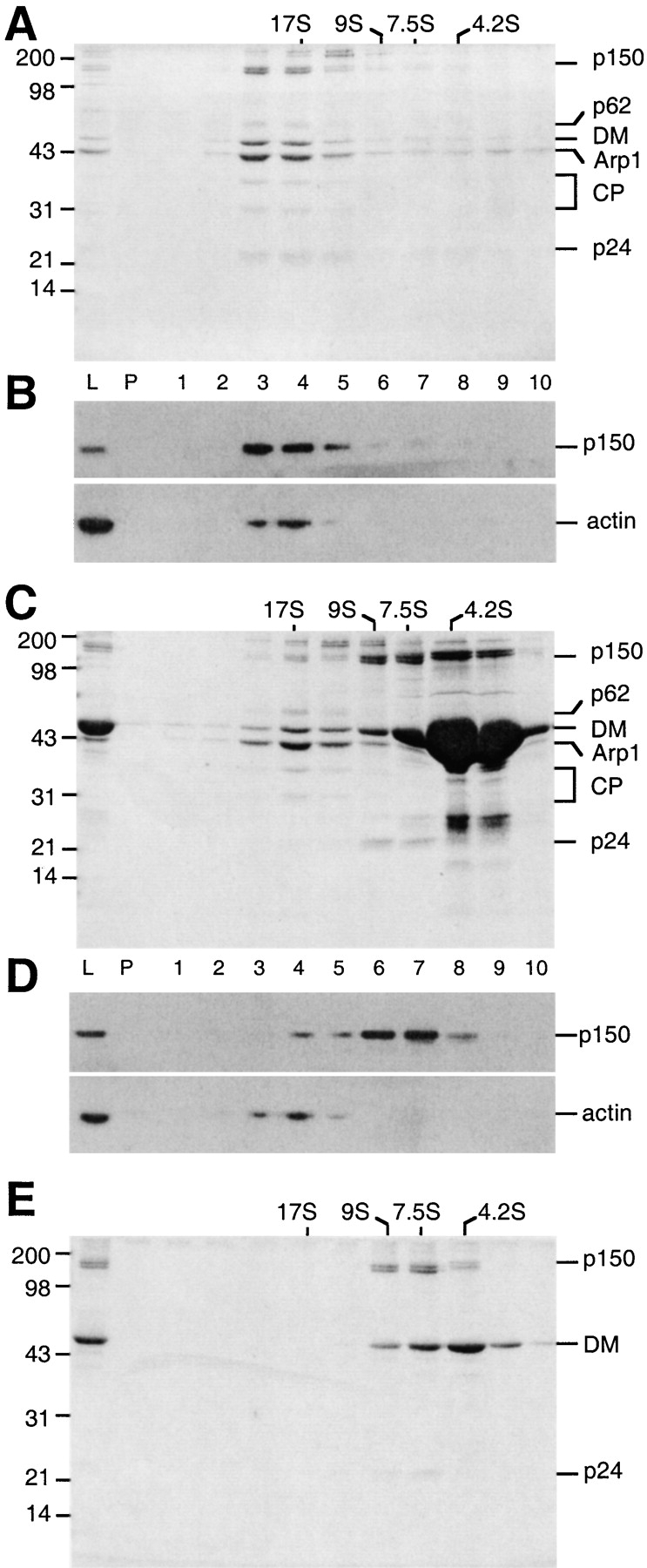
In vitro disruption of dynactin by recombinant dynamitin. A mock-treated dynactin control (A and B) and dynactin preincubated with recombinant dynamitin (C and D) were sedimented into 5–20% sucrose gradients. Gradient fractions (L, load; P, pellet; fractions 1–10) were analyzed on Coomassie blue–stained SDS gels (A and C) or immunoblots (B and D) with antibodies against p150Glued (mAb 150B and actin). Dynactin subunits are indicated on the right (DM, dynamitin; CP, capZ). Molecular weight markers are on the left. (E) Fractions 6 and 7 from a sample similar to C were pooled, diluted, resedimented into a second 5–20% sucrose gradient, and then analyzed by SDS-PAGE as above.
Dynamitin treatment of dynactin released a 9S protein complex containing p150Glued and p24 (Fig. 1 C, fractions 6 and 7), as expected from previous studies (Echeverri et al. 1996; Karki et al. 1998; Wittman and Hyman 1999). This material also appeared to contain dynamitin. However, the fractions most enriched in p150Glued and p24 (Fig. 1 C, lanes 6 and 7) were not well resolved from the bulk of free recombinant dynamitin (Fig. 1 C, lanes 8 and 9), making it impossible to estimate relative stoichiometries. To further characterize the sidearm structure released by dynamitin, 9S fractions (equivalent to fractions 6 and 7 in Fig. 1 C) were pooled, dialyzed, and sedimented into a second 5–20% sucrose gradient. p150Glued, p24, and some dynamitin cosedimented at ≈9S, while the residual free dynamitin sedimented at ≈7S (Fig. 1 E). Quantitative densitometry revealed the subunit stoichiometry of the 9S pool to be 2:4:2 p150Glued:dynamitin:p24, which is the same as in intact dynactin (see also Bingham et al. 1998). This result suggests that exogenous dynamitin effects disruption by introducing four additional dynamitin monomers at the Arp1/p150Glued junction. This generates an Arp1 minifilament and a complex of p150Glued and p24 subunits, both of which have four associated dynamitin subunits.
KI Disruption of Dynactin Reveals Additional Stable Subcomplexes
Dynactin's Arp1 minifilament can be depolymerized by treatment with the chaotropic salt, KI (Bingham and Schroer 1999). Gel filtration chromatography in the presence of 0.7 M KI yields primarily monomeric Arp1 and actin (Bingham and Schroer 1999), but other dynactin subunits elute earlier from the sizing column, suggesting they might be organized into higher order complexes (Fig. 2 A). Although the elution profile was complex, five overlapping peaks of protein (Fig. 2 A, A–E) were obvious. In highly concentrated samples, small amounts of Arp1 could be seen across the entire column profile, suggesting that some Arp1 might still be assembled into small multimers or associated with other dynactin subunits. Peak A contained p150Glued, dynamitin, and p24 and eluted with an approximate Stokes' radius of 10.7 nm. Peak B, which contained dynamitin and p24, but little or no p150Glued, eluted broadly from the column (mean Stokes' radius ≈6 nm). Peak C was enriched for the p62 and p27 subunits (Schafer et al. 1994a) and also contained a doublet of proteins at ≈43 kD, the larger of which comigrated with Arp1, and a protein of ≈25 kD that electrophoresed slightly slower than p24. Peak C eluted from the column with a Stokes' radius of 4.7 nm. Peak D was highly enriched for capZ. Arp1 was most concentrated in peak E, which eluted with the expected Stokes' radius of monomers.
The complex of p150Glued, dynamitin, and p24 that was liberated from dynactin by KI (Fig. 1 A, peak A) appeared similar in composition to the p150/dynamitin/p24 complex released by excess soluble dynamitin. However, KI treatment also appeared to yield a second, distinct subcomplex containing dynamitin and p24 subunits (peak B). To gain a better understanding of the relationship between these three protein complexes, peaks A and B were dialyzed to remove KI, and then subjected to velocity sedimentation into a sucrose gradient. The three polypeptides present in peak A (p150Glued, dynamitin, and p24) cosedimented at ≈9S (Fig. 2 B), much like the p150/dynamitin/p24 complex released by dynamitin. The small amount of Arp1 that was sometimes present in peak A did not sediment at 9S, but instead spread between 11S and >20S, demonstrating that Arp1 is not an integral component or necessary for the stability of this assembly. Densitometric analysis of Coomassie blue–stained gels suggested that each 9S complex contained p150Glued, dynamitin, and p24 in a ratio of 2:2:1 (Table ). This stoichiometry, together with the predicted molecular weight of each subunit from the cloned cDNAs, predicts a subcomplex of 401.5 kD. In good agreement with this estimate, a subcomplex of 396.6 kD was predicted from the Svedberg equation solved with sedimentation coefficient and Stokes' radius values (Table ).
Table 1.
Hydrodynamic Properties, Composition, and Predicted Mass of Dynactin Subcomplexes
| Dynactin subcomplex | S value | Stokes' radius | Predicted dimensions | Mr 1 | Mr 2 | Subunit | Mr | Stoichiometry* |
|---|---|---|---|---|---|---|---|---|
| nm | nm | Da | Da | Da | ||||
| p150 | 137,757 | 2 | ||||||
| Shoulder/sidearm | 9 | 10.7 | 21.4 × 1.2 | 396,550 | 386,373 | Dynamitin | 44,941 | 2 |
| p24 | 20,977 | 1 | ||||||
| Shoulder | 4.4 | 6.1 | 12.2 × 1.0 | 110,780 | 110,859 | Dynamitin | 44,941 | 2 |
| p24 | 20,977 | 1 | ||||||
| p62 | 53,086 | 1 | ||||||
| Pointed-end complex | 7 | 4.7 | 9.4 × 1.9 | 135,570 | 140,108 | Arp11 | 46,230 | 1 |
| p27 | 20,653 | 1 | ||||||
| p25 | 20,139 | 1 |
Subcomplex S values and diffusion coefficients were determined from multiple runs and compared with sucrose-gradient or gel-filtration standards run in parallel. Elution peaks were identified by A280 and confirmed by SDS-PAGE. Stokes radii were calculated from the diffusion coefficients. Subcomplex dimensions were predicted from the Perrin factor as described (Bloom et al. 1988). Molecular weights were calculated using the Svedberg equation (Mr 1, left column) or from molecular weights predicted from cloned cDNAs (Mr 2, right column). Mouse EST clones were used to predict the size of Arp11, p27, p25 (this study), and p24 (Pfister et al. 1998). The p62 mass was predicted from the cloned rat sequence (this study), p50 (human; Echeverri et al. 1996), and p150 (human; Tokito and Holzbaur 1998) masses were predicted from the published sequences.
* Subunit molar ratios in each subcomplex, estimated by scanning densitometry of a dilution series of each purified subcomplex, compared with a BSA standard.
A parallel analysis of peak B (Fig. 2 C) revealed that dynamitin and p24 cosedimented at ≈5S. As was seen for peak A, the Arp1 in peak B sedimented cleanly away from the other polypeptides. The stoichiometric ratio of dynamitin to p24 in the 5S pool was estimated by densitometry to be 2:1 (Table ). Subunit molecular weights deduced from cloned cDNAs predict a complex mass of 110.9 kD, in good agreement with the 110.8 kD mass predicted from hydrodynamic measurements. Together, the complexes in peaks A and B sum to yield p150Glued, dynamitin, and p24 subunits in a ratio of 2:4:2, which is the same as in native dynactin.
The biochemical composition and hydrodynamic properties of the p150Glued/dynamitin/p24 assembly isolated by KI treatment strongly suggested that it corresponded to the flexible p150Glued sidearm that projects from the dynactin Arp1 filament (Fig. 3, top; Schafer et al. 1994a). To explore this possibility further, platinum replicas of quick-frozen, deep-etched (Heuser 1983) 9S complex were visualized by EM (Fig. 3, middle). The majority of molecules were elongated (>20 nm) and many contained a pair of globular heads similar in size and shape to those seen at the distal end of the dynactin sidearm. The structures were kinked in the middle to varying degrees, suggesting that, like the dynactin sidearm, this structure is flexible. Because the p150Glued/dynamitin/p24 assembly is biochemically and ultrastructurally similar to the projecting sidearm and shoulder domains of the intact dynactin molecule, we will refer to this protein complex as the dynactin shoulder/sidearm.
Figure 3.
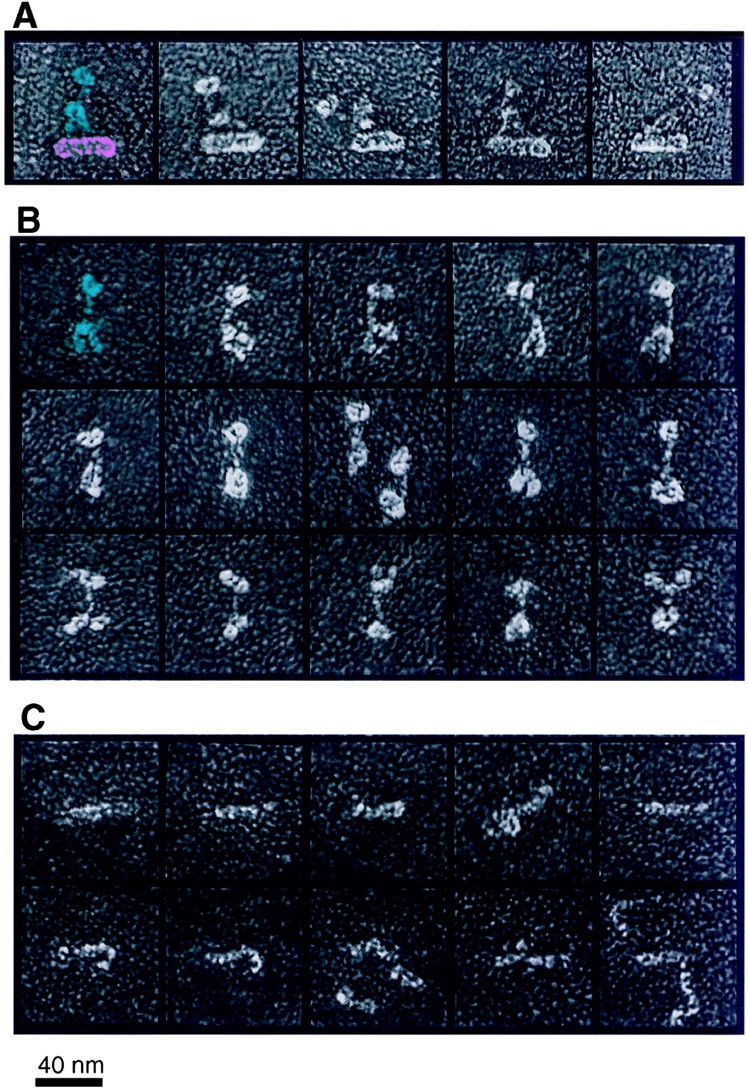
Ultrastructural comparison of intact dynactin (A), sucrose gradient-purified peak A molecules (shoulder/sidearm; B), and sucrose gradient-purified peak B molecules (shoulder; C). In the left-most dynactin image, the shoulder/sidearm structure and Arp1 minifilament domains are pseudocolored blue and pink for emphasis. One peak A molecule (isolated shoulder/sidearm) is colored blue accordingly. Bar, 40 nm.
The remarkably similar ultrastructures of the projecting sidearm seen for intact dynactin and the isolated shoulder/sidearm subcomplex raises the question of where the excess dynamitin/p24 present in peak B might be accommodated within the dynactin structure. Mass arguments, plus the fact that free dynamitin can dissociate the p150Glued sidearm from the Arp1 filament, suggest that dynamitin is contained within the highly flexible, elastic shoulder that tethers the filamentous p150Glued sidearm to the Arp1 minifilament. Previous attempts to label intact dynactin with antibodies to dynamitin were unsuccessful (Schafer, D.A., J.E. Heuser, and T.A. Schroer, unpublished data) and appropriate p24 antibodies are not available, so we could not address this question by direct labeling methods. Instead, we imaged isolated dynamitin/p24 heterotrimers as described above (Fig. 3, bottom). These molecules appeared as flexible, elongated structures that ranged in length from 11 to 30 nm and were occasionally seen to associate with each other end-to-end. The flexibility and apparent elasticity of these structures is consistent with a location within the dynactin shoulder. We will refer to the dynamitin/p24 heterotrimer as shoulder.
p62 Is Present in a Complex with Three Other Dynactin Subunits
KI disruption yielded two other protein peaks whose components behaved as protein complexes. Fig. 2 C, peak D, was most enriched in the capZ α/β heterodimer and was not pursued further. Peak C, in contrast, was enriched in p62, p27, and two previously unidentified dynactin subunits of 43 and 25 kD. This material eluted from the sizing column with an apparent Stokes' radius of 4.7 nm (Table ). These behaviors predict a globular protein of Mr 135,000 D. However, peak C also contained some capZ, so we were unsure as to whether the four predominant components (p62, p27, plus 43- and 25-kD species) comprised a single protein complex or they might be organized into multiple, independent protein assemblies. To explore this possibility further, the proteins in peak C were sedimented into a sucrose gradient (Fig. 4 A). CapZ sedimented at ≈5S, consistent with previous work (Cooper et al. 1984), while p62, p27, and the 43- and 25-kD proteins cosedimented at ≈7S. To further purify the 7S complex, it was subjected to ion exchange chromatography on a MonoS column (Fig. 4 B). p62 and the three associated polypeptides bound and coeluted in a broad peak, while other dynactin subunits (capZ, dynamitin, and p24) flowed through the column. The approximate stoichiometry of the p62, p27, and 43- and 25-kD polypeptides was 1:1:1:1 (Table ), as estimated by densitometry of Coomassie blue–stained gels.
Figure 4.
Purification and ultrastructural analysis of pointed-end complex. (A) Proteins in peak C were dialyzed, sedimented, and analyzed as described for shoulder/sidearm and shoulder complexes. Dynactin subunits are indicated on the right (CP denotes the α and β subunits of capZ). Molecular weight markers are on the left. (B) Purification of pointed-end complex by MonoS chromatography. The column load (L), flow through (FT), and individual column fractions are indicated. (C) Gallery of images of platinum replicas of sucrose gradient-purified peak C molecules (pointed-end complex). Bar, 40 nm.
The sample was then subjected to ultrastructural analysis (Fig. 4 C). Round particles were observed with an approximate diameter of 8 nm, in agreement with our gel filtration size estimates (Table ). The particles appeared to have a depression in the center and were reminiscent of the Arp2/3 complex (Mullins et al. 1997, Mullins et al. 1998), an important regulator of actin assembly that binds F-actin pointed ends (reviewed in Schafer and Schroer 1999). Our previous ultrastructural analysis suggested that p62 is associated with the pointed end of the Arp1 filament (Schafer et al. 1994a). This implies that the p62, p27, and the 43- and 25-kD assemblies are associated with the Arp1 filament pointed end. We will refer to this heterotetrameric protein complex as pointed-end complex.
Cloning of Pointed-End Complex Subunits
Though several dynactin subunits have been characterized at the amino acid sequence level (p150Glued, dynamitin, Arp1, actin, capZ, and p24; for review, see Holleran et al. 1998), the four components of the pointed-end complex have not. To complete the molecular characterization of dynactin, we cloned all four remaining subunits. A full-length p62 clone (Fig. 5 A) was isolated from a rat brain expression library using mAb 62B (Schafer et al. 1994a). The predicted protein has an Mr of 53,086 and pI of 7.4. Inspection of the sequence revealed a RING-finger–like motif (Saurin et al. 1996) and a series of predicted α helices with alternating acidic and basic character. Database searches indicate that p62 is homologous along its length with RO-2 (ropy-2), a Neurospora gene whose protein produce is also predicted to contain a metal binding motif (Vierula and Mais 1997). Analysis of ropy mutants reveals defects in dynein-dependent nuclear movement into growing hyphae. Since other RO genes encode subunits of dynein or dynactin (Plamann et al. 1994; Tinsley et al. 1996), it seems likely that p62 is the vertebrate RO-2 homologue.
Figure 5.
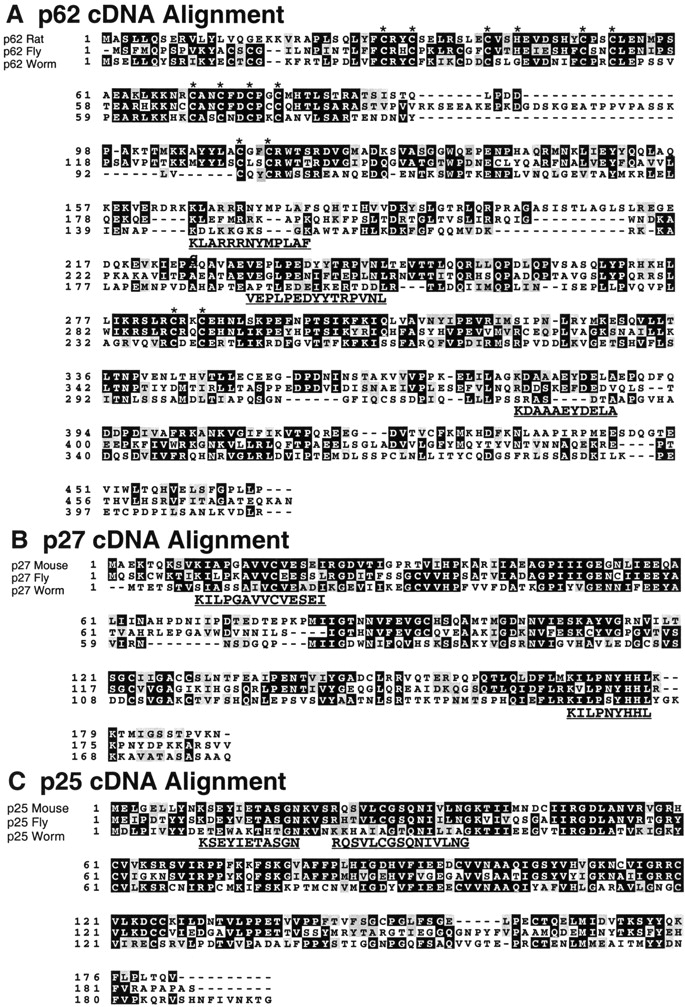
Predicted amino acid sequences of p62, p27, and p25. Tryptic peptide sequences obtained from each bovine protein are bold and underlined. (A) Rat p62 aligned with predicted sequences from fly (Drosophila melanogaster clone AC005447) and worm (C. elegans clone C26B2.4). The p62 sequence predicts RING finger–like metal-binding “knuckles” (CxxC or CxxH; indicated with asterisks). (B) Mouse p27 (AA073653) aligned with amino acid sequences deduced from fly (AC005705) and worm (Y54E10) genomic DNA sequences. (C) Mouse p25 (AA869597) aligned with amino acid sequences deduced from fly (AC002502) and worm (Y71F9AL) ESTs and genomic sequences.
In addition to p62, the pointed-end complex contains subunits of ≈25 and ≈43 kD that had not been observed previously. However, close inspection revealed a minor polypeptide migrating just above p24 in undisrupted bovine brain dynactin (Fig. 2 A, left). Immunoblotting showed the ≈25-kD species to be immunologically distinct from p24 (data not shown). In intact dynactin, the novel 43-kD species is obscured by Arp1, the major component of dynactin (one third by mass). The 43-kD polypeptide in purified pointed-end complex was determined to be distinct from both Arp1 and actin by peptide fingerprinting (Cleveland 1983) and immunoblotting (data not shown). The identification of the ≈25- and 43-kD species as novel dynactin components was verified by peptide microsequencing and cDNA cloning (see below).
To obtain clones for the 25- and 43-kD polypeptides, for which antibodies were not available, we screened genomic databases with tryptic peptide sequences from each protein. This yielded multiple EST clones that were assembled, as needed, to yield full-length sequences (Fig. 5B and Fig. C, and Fig. 6 A). Human p27 was previously identified by the Japanese genome sequencing project (Ichikawa et al. 1997) as a ubiquitously expressed gene. The mouse p27 gene encodes a protein of 20.6 kD with an isoelectric point of 6.0. p27 orthologs from fly and worm were identified using the predicted rat p27 amino acid sequence. Though the p27 gene appears to be evolutionarily conserved (Fig. 5 B), no novel features are revealed in an alignment of the sequences. The 25-kD subunit (p25; Fig. 5 C) is another novel protein with a predicted Mr of 20.1 kD and pI of 7.9. Orthologs were identified in fly and worm; both had basic isoelectric points. Although we did not detect chick embryo brain dynactin p25 on two-dimensional (2-D) gels (Schafer et al. 1994a), presumably due to its alkaline pI, a spot corresponding to bovine p25 can be seen at a pI of ≈8.0 by silver staining (data not shown). p25 is predicted to fold into a β structure.
Figure 6.
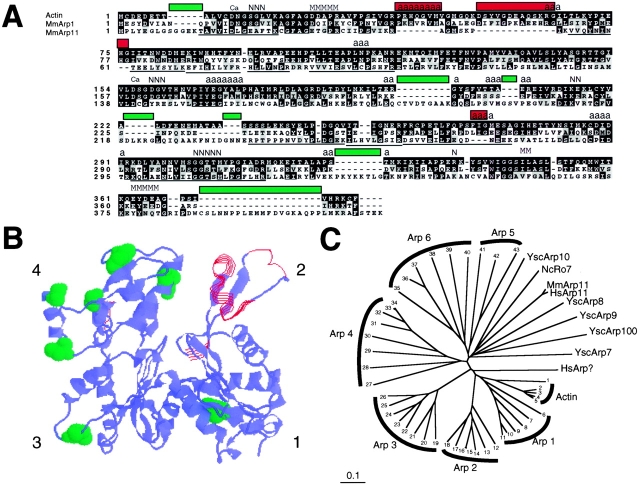
Comparison of Arp11 with actin and known Arps. (A) Comparison of mouse actin, Arp1, and Arp11 amino acid sequences. Mouse actin (AAA37164), Arp1(BAA24423), and Arp11 (this study) were aligned by CLUSTAL X. Actin residues implicated in calcium binding (Ca), nucleotide binding (N), myosin binding (M), or actin–actin contacts (a) are directly below labels. Residues identical to actin are shown in black, similar residues are shaded gray. Sequenced Arp11 tryptic peptides are underlined. Sites of insertions and deletions are indicated with red and green bars, respectively. (B) Arp11 changes presented in the context of actin crystal structure. Deletions are presented as red (open ribbons) and insertion points are labeled green (space-filling representation). Subdomains 1–4 are indicated. (C) Phylogenetic tree comparing actin and actin-related proteins from several species (Ac, Acanthamoeba castellanii; Ce, C. elegans; Dd, Dictyostelium discoideum; Dm, D. melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Nc, Neurospora crassa; Ysc, Saccharomyces cerevisiae; Ysp, Schizosaccharomyces pombe). Amino acid sequences were aligned by CLUSTAL X and plotted with Treeview PPC. Actin: 1: DmArp53D, P45891; 2: YscAct1p, BAA21512; 3: YspAct1p, P10989; 4: Hs alpha-actin, P02568; 5: Hs gamma-actin, P02571. Arp1: 6: YscArp1, P38696; 7: NcRopy-4, A54802; 8: CeArp1, Y53F4; 9: DmArp87C, P45889; 10: HsArp1, P42024; 11: MmArp1alpha, BAA24423. Arp2: 12: YscArp2, CAA98588; 13: CeArp2, P53489; 14: DmArp14D, P45888; 15: GgArp2, P53488; 16: HsArp2, NP_005713; 17: AcArp2, AAC46911; 18: DdArp2, AAC99776. Arp3: 19: YscArp3, P47117; 20: YspArp3, P32390; 21: NcArp3, P78712; 22: CeArp3, Y71F9A_294.A; 23: DmArp66B, P32392; 24: HsArp3, NP_005712; 25: DdArp3, P42528; 26: AcArp3, P53490. Arp4: 27: YscArp4, P80428; 28: YspC23D3.09, Q09849; 29: CeArp4, ZK616; 30: DmBAP55, AA949447; 31: HsActin-like gene, AB015906; 32: DrArp4, AI794230; 33: MmBaf53, AF041476; 34: HsBaf53a, ACTL6. Arp6: 35: Dm13E, P45890; 36: MmArp6, AA288832; 37: DrArp6, AI794429; 38: CeArp6, Q09443; 39: YscArp6, Q12509; 40: YspArp6, CAA19116. Arp5: 41: YscArp5, P53946; 42: MmArp5, AI550487; 43: DmArp5, AC006495. Other Arps: YscArp10, Q04549; NcRo-7 (M. Plamann, personal communication); MmArp11 (this study); HsArp11 (this study); YscArp8, Q12386; YscArp9, Q05123; YscArp100, CAA85175; YscArp7, Q12406.
Pointed-End Complex Contains Arp11, a Novel Actin-related Protein
The sequence of the 43-kD protein (Fig. 6 A) revealed it to be a novel Arp. A phylogenetic tree including representative actin and Arp sequences (Fig. 6 C) shows this new Arp to be less similar to actin than most other Arps. In keeping with existing nomenclature guidelines (Schroer et al. 1994; Poch and Winsor 1997), we suggest the name Arp11. Alignment of the Arp11 sequence with actin reveals overall conservation of the “actin fold,” a primordial core structure that contains nucleotide and metal binding elements (Kabsch and Holmes 1995). As in other actin-related proteins, sequence differences, when they are observed, are largely limited to surface residues. In addition to scattered amino acid substitutions, the alignment reveals several changes in surface loops (Fig. 6 B) that play key roles in actin–actin and actin–protein interactions (Page et al. 1998). The Arp11 barbed-end face is generally well conserved and is not predicted to contain large insertions that would prevent interactions with the pointed end of an Arp1 (or actin) filament. The NH2 and COOH termini (found at the barbed-end face of subdomain 1) are only slightly longer (5–20 amino acids) than in actin, which should interfere only slightly, if at all, with subunit–subunit interactions. In contrast, the pointed-end face of Arp11 is predicted to be dramatically different from that of actin (Fig. 6 B). The most conspicuous difference is a large deletion (actin residues 38–57) that eliminates an entire surface loop (Fig. 6 B, red) from subdomain 2. Subdomains 3 and 4 are predicted to contain insertions, some of which (Fig. 6 B, green) map within two important actin–actin interfaces inferred from the F-actin x-ray crystal structure (Holmes et al. 1990; Kabsch et al. 1990). The predicted structure of Arp11 suggests it will not form filaments by itself and will only interact with filaments of Arp1, or conventional actin, at their pointed ends.
Discussion
The work presented here provides significant new insight into dynactin composition and subunit organization, and suggests novel mechanisms for Arp1 filament length regulation and dynactin-membrane binding. The availability of purified dynactin shoulder/sidearm, shoulder, and pointed-end complexes will facilitate biochemical and biophysical analysis of dynein–dynactin and dynactin–cargo interactions. The four new dynactin subunit cDNAs we have isolated will be useful tools in studies of the assembly pathway of dynactin in vitro and should permit a comprehensive analysis of dynactin function in cells.
We are reasonably confident that the present effort represents the complete molecular characterization of dynactin subunits. 2-D gel analysis of isolated bovine brain dynactin reveals all of the proteins discovered thus far, including the novel subunits p25 and Arp11 (data not shown). Dynactin's predicted mass, based on the stoichiometries of its 11 different gene products and their predicted molecular weights, is 1.03 MD. This value agrees well with values (1.10 and 1.11 MD) predicted by hydrodynamic measurements (this study) and previous STEM analysis (Schafer et al. 1994a).
Dynactin's complicated, highly asymmetric structure suggests a multiplicity of functions. To serve its many roles in cells, dynactin must be able to interact with dynein, endomembranes, and other cytoplasmic particles, centrosomes, chromosomes, and the plasma membrane. While the present analysis does not provide any new information about these binding interactions specifically, it does allow us to better define the dynactin subdomains thought to underlie them.
The Shoulder/Sidearm and Shoulder Complexes
A large body of evidence indicates that dynein binds dynactin via the projecting p150Glued sidearm. p150Glued was first implicated as a dynein-binding subunit in blot overlay and affinity chromatography studies using recombinant proteins (Karki and Holzbaur 1995; Vaughan and Vallee 1995). We find that isolated dynactin shoulder/sidearm inhibits dynein-based microtubule aster formation in vitro, suggesting that this domain can act as a competitive inhibitor of the dynein–dynactin interaction. Moreover, overexpression of the dynein-binding domain of p150Glued profoundly interferes with dynein-based motility in cells (Quintyne et al. 1999), reinforcing the notion that this subunit interacts directly with dynein in vivo. Recombinant or overexpressed p150Glued binds cellular microtubules via an NH2-terminal microtubule binding site (Waterman-Storer et al. 1995) that maps to the globular heads at the distal end of the dynactin sidearm. Native dynactin and isolated shoulder/sidearm subcomplex also bind microtubules in vitro, although the binding is transient. The dynamic binding of dynactin to microtubules allows dynein to remain associated with the microtubule track throughout the dynein ATPase cycle, which is believed to enhance motor processivity (King, S.J., and T.A. Schroer, manuscript submitted for publication).
Together, dynactin's shoulder/sidearm and shoulder complexes form a flexible, elastic structure (Fig. 3, top) that can bind the Arp1 minifilament on one end, the microtubule substrate at the other, and dynein in between. The flexibility of the junction with the minifilament allows p150Glued to undergo a wide range of movement relative to the Arp1 backbone. This may facilitate passage of bulky cargoes through cytoplasm. The flexibility of this structure may also facilitate intercalation by exogenous dynamitin. Isolated shoulder/sidearm and shoulder complexes both exhibit a range of morphologies, much like the pleomorphic shoulder in intact dynactin. Two dynamitins in isolated shoulder complex are bound to one subunit of p24, suggesting that this may be a fundamental assembly unit of the dynactin molecule. However, dynamitin disruption leaves four dynamitin subunits on the Arp1 filament and four subunits on the p150Glued/p24 assembly, suggesting that dynamitin may be most stable in tetramer form. That KI dissociates the dynamitin tetramer into two dimers suggests a “fault line” at the dimer–dimer interface.
Pointed-End Complex
Our disruption studies have also provided new insights into subunit–subunit interactions in the Arp1 minifilament. Dynamitin treatment appears to leave this structure intact. CapZ remains associated, indicating that the barbed end is still capped. The pointed-end complex is also apparently retained. p62 is a component of the 19S complex (Echeverri et al. 1996; Fig. 1 B) and Arp11, p27, and p25 can be discerned by 2-D gel analysis (K.A. Melkonian, unpublished observations). The 19S structure also contains dynamitin, suggesting that this protein may contribute to Arp1 minifilament stability. KI disruption, in contrast, depolymerizes the Arp1 polymer, as previously seen for conventional F-actin (Szent-Gyorgi 1951), but leaves the capZ α/β heterodimer intact, as expected (Caldwell et al. 1989). Our KI disruption studies provide no new information about the location of the single actin monomer within the Arp1 minifilament, as actin elutes as monomer under these conditions (Bingham and Schroer 1999). However, association with the conventional actin-capping protein, capZ is a likely possibility.
The predicted structure of Arp11, based on alignment with conventional actin and other Arps, suggests it has the capacity to bind Arp1 directly. Earlier attempts to demonstrate binding between p62 and Arp1 were unsuccessful (S.R. Gill, unpublished observations), as would be expected if this interaction were mediated by Arp11. The positions of insertions and deletions (relative to actin) within the Arp11 sequence predict it will associate with the Arp1 filament exclusively at its pointed end. Dynactin Arp1 minifilaments are uniform in length (37 nm; Schafer et al. 1994a) and they do not readily depolymerize, self-associate, or nucleate assembly of conventional actin (J.B. Bingham, unpublished observations), suggesting that they are capped at both ends. In contrast, the filaments formed by purified Arp1 in vitro are heterogeneous in length (mean length, 52 nm) and can anneal end-to-end (Bingham and Schroer 1999). The conventional actin-capping protein α/β2 heterodimer associated with dynactin (Schafer et al. 1994a,Schafer et al. 1994b) is expected to prevent Arp1 growth and annealing at the Arp1 barbed end. Pointed-end complex may play a similar role at the opposite end of the filament and may also help template Arp1 assembly to ensure the filaments grow to a uniform size.
The association of p27 and p25 with p62 and Arp11 in the pointed-end complex appears to be somewhat labile. During KI disruption, p62 and Arp11 always copurify, but p27 and p25 sometimes copurify with monomeric Arp1 and actin (J.B. Bingham, Ph.D. thesis). When the resulting Arp1/actin/p27/p25 sample is further fractionated by MonoQ chromatography (Bingham and Schroer 1999), p25 and p27 again copurify (J.B. Bingham, Ph.D. thesis), suggesting they are tightly associated with one another. Thus, within the pointed-end complex, p62 and Arp11 appear to be binding partners, as do p27 and p25.
The primary sequences of the remaining three pointed-end complex subunits provide fertile ground for speculation about the assembly and function of the Arp1 minifilament, dynactin's proposed cargo-binding domain. The mechanism of dynactin-membrane binding remains obscure, though recent work suggests that Arp1 may interact with Golgi-associated spectrin (Holleran et al. 1996). However, the predicted biochemical properties of pointed-end complex subunits suggest that this structure may also participate in membrane binding. The conserved alkaline pIs of p62, Arp11, and p25 suggest that any or all may interact electrostatically with negatively charged membrane lipids or other acidic cargoes such as lipid droplets or viral nucleocapsids. p25 is predicted to fold into a β structure that may contain a lipid-binding, plekstrin homology domain. Thioacylation of p25 and/or p27 (both of which are cysteine rich) might further stabilize interactions between these subunits and membranes. Homologies of pointed-end complex subunits have been found in organisms as diverse as fly (all four subunits), worm (p62, p27, and p25), and Neurospora (p62 and Arp11), indicating that this structure is evolutionarily conserved. However, budding yeast, an organism that does not require cytoplasmic microtubules for endomembrane traffic, does not appear to contain pointed-end complex homologues. This suggests that yeast dynactin may not perform whatever function(s) this structure provides in other species and further supports a role in membrane interactions. But regardless of how dynactin binds membranes, it is important to keep in mind that dynein may have mechanisms for binding membranes that are independent of dynactin. Purified cytoplasmic dynein associates peripherally with purified synaptic vesicles and can bind purified phospholipid vesicles with similar affinity (Lacey and Haimo 1992, Lacey and Haimo 1994). The dynein light chain, TctexI, binds directly to the transmembrane protein rhodopsin (Tai et al. 1999). Thus the interaction of dynein and dynactin is likely to be complex and may involve multiple subunits of each protein.
Previous work indicates that all dynactin subunits except conventional actin and actin-capping protein are found in dynactin only (Paschal et al. 1993; Echeverri et al. 1996). We do not yet know if the same is true for Arp11, p27, or p25. It is intriguing that pointed-end complex ultrastructure so closely resembles the Arp 2/3 complex (Mullins et al. 1997, Mullins et al. 1998; J.E. Heuser, unpublished observations), a structure that plays an important role in events at conventional actin filament pointed ends (reviewed in Schafer and Schroer 1999). It will be interesting to determine whether pointed-end complex, or Arp11 alone, contributes to the dynamics of the conventional actin cytoskeleton. That Neurospora RO-2 mutants show defects in actin cytoskeletal organization (Vierula and Mais 1997) suggests that this may indeed be the case.
Further Directions
Dynactin is just one of a family of proteins proposed to mediate the interaction of membranes with microtubules. It shares a number of superficial similarities with CLIP-170, another protein in this class. CLIP-170 binds to and tracks with growing microtubule plus ends (Perez et al. 1999), where it may mediate static interactions between endosomes and microtubules (Pierre et al. 1992). As in the case of the shoulder/sidearm subunit p150Glued, microtubule binding occurs at the NH2 terminus via a conserved binding site whose activity appears to be subject to complex physiological regulation. The CLIP-170 COOH terminus contains predicted metal-binding motifs that are proposed to participate in cargo binding. p62's predicted metal-binding, RING-finger–like motif may serve a similar function. In both dynactin and CLIP-170, microtubule and cargo binding activities are positioned at opposite ends of the molecule. Like CLIP-170, dynactin accumulates at microtubule plus ends (Valetti et al. 1999; Vaughan et al. 1999; Habermann, A., T.A. Schroer, G. Griffiths, and J.K. Burkhardt, manuscript in preparation). This association appears to depend on CLIP-170 (Valetti et al. 1999), suggesting that CLIP-170 may recruit dynactin to microtubule plus ends to initiate dynein-based movement. How this process is coordinated and what other factors may contribute to cargo docking and motility will be an area of significant future work.
Acknowledgments
We are grateful to Dr. G. Griffiths (EMBL, Heidelberg, Germany) and Tony Houthaeve of the EMBL Peptide Sequencing facility for p62 and p27 microsequencing. We thank Robyn Roth and Kristen Harwick Poland for technical assistance.
H.V. Goodson was supported by a fellowship from the Helen Hay Whitney Foundation. J.E. Heuser was supported by a grant from the National Institutes of Health (NIH). T.A. Schroer was supported by a grant from the NIH (RO1 GM 44598), a Searle Scholars Award, and the Lucile and David Packard Fellowship for Science and Engineering.
Footnotes
1.used in this paper: 2-D, two dimensional; Arp, actin-related protein; pI, isoelectric point
Dr. Gill's current address is The Institute for Genomic Research, Rockville, MD 20850.
References
- Allan V. Motor proteinsa dynamic duo. Curr. Biol. 1996;6:630–633. doi: 10.1016/s0960-9822(09)00434-5. [DOI] [PubMed] [Google Scholar]
- Bingham J.B., King S.J., Schroer T.A. Purification of dynein and dynactin from brain tissue. Methods Enzymol. 1998;298:171–184. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- Bingham J.B., Schroer T.A. Self-regulated polymerization of the actin-related protein, Arp1. Curr. Biol. 1999;9:223–226. doi: 10.1016/s0960-9822(99)80095-5. [DOI] [PubMed] [Google Scholar]
- Blocker A., Severin F.F., Burkhardt J.K., Bingham J.B., Yu H., Olivo J.-C., Schroer T.A., Hyman A.A., Griffiths G. Molecular requirements for bi-directional movement of phagosomes along microtubules. J. Cell Biol. 1997;137:113–129. doi: 10.1083/jcb.137.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G.S., Wagner M.C., Pfister K.K., Brady S.T. Native structure and physical properties of bovine brain kinesin and indentification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- Burkhardt J.K., Echeverri C.J., Nilsson T., Vallee R.B. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busson S., Dujardin D., Moreau A., Dompierre J., De Mey J.R. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- Caldwell J.E., Heiss S.G., Mermall V., Cooper J.A. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry. 1989;28:8506–8514. doi: 10.1021/bi00447a036. [DOI] [PubMed] [Google Scholar]
- Carminati J.L., Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W. Peptide mapping in one dimension by limited proteolysis of sodium dodecyl sulfate-solubilized proteins. Methods Enzymol. 1983;96:222–229. doi: 10.1016/s0076-6879(83)96020-2. [DOI] [PubMed] [Google Scholar]
- Compton D.A. Focusing on spindle poles. J. Cell Sci. 1998;111:1477–1481. doi: 10.1242/jcs.111.11.1477. [DOI] [PubMed] [Google Scholar]
- Cooper J.A., Blum J.D., Pollard T.D. Acanthamoeba castellanii capping proteinproperties, mechanism of action, immunologic cross-reactivity, and localization. J. Cell Biol. 1984;99:217–225. doi: 10.1083/jcb.99.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal B.M., Vaughan K.T., Vallee R.B. Molecular characterization of 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M.A., Dush M.K., Martin G.R. Rapid production of full-length cDNAs from rare transcriptsamplification using a single gene-specific oligonucleotide primer. Proc. Nat. Acad. Sci. USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J.B., Hasbani J., Gill S.R., Schroer T.A., Compton D.A. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.R., Schroer T.A., Szilak I., Steuer E.R., Sheetz M.P., Cleveland D.W. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Pilcher S., Kirkham M., Hyman A.A. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage C. elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A., Takei Y., Kanai Y., Tanaka Y., Nonaka S., Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J. Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heuser J.E. Procedure for freeze-drying molecules adsorbed to mica flakes. J. Mol. Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- Holleran E.A., Holzbaur E.L.F. Centractin (actin-related protein 1) binds directly to spectrin Mol. Biol. Cell. 9 1998. 274a(Abstr.) [Google Scholar]
- Holleran E.A., Karki S., Holzbaur E.L. The role of the dynactin complex in intracellular motility. Int. Rev. Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- Holleran E.A., Tokito M.K., Karki S., Holzbaur E.L.F. Centractin (Arp1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J. Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Holzbaur E.L., Vallee R.B. Dyneinsmolecular structure and cellular function. Annu. Rev. Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- Ichikawa K., Yamabe Y., Imamura O., Kuromitsu J., Sugawara K., Suzuki N., Shimamoto A., Matsumoto T., Tokutake Y., Kitao S. Cloning and characterization of a novel gene, WS-3, in human chromosome 8p11–p12. Gene (Amst.). 1997;189:277–287. doi: 10.1016/s0378-1119(96)00863-3. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Holmes K.C. The actin fold. FASEB J. 1995;9:167–174. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H.G., Suck D., Pai E.F., Holmes K.C. Atomic structure of the actinDNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Karki S., Holzbaur E.L.F. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- Karki S., LaMonte B., Holzbaur E.L.F. Characterization of the p22 subunit of dynactin reveals the localization of cytoplasmic dynein and dynactin to the midbody of dividing cells. J. Cell Biol. 1998;142:1023–1034. doi: 10.1083/jcb.142.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey M.L., Haimo L.T. Cytoplasmic dynein is a vesicle protein. J. Biol. Chem. 1992;267:4793–4798. [PubMed] [Google Scholar]
- Lacey M.L., Haimo L.T. Cytoplasmic dynein binds to phospholipid vesicles. Cell Motil. Cytoskelet. 1994;28:205–212. doi: 10.1002/cm.970280304. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lessard J.L. Two monoclonal antibodies to actinone muscle selective and one generally reactive. Cell Motil. Cytoskelet. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Heuser J.A., Pollard T.D. The interaction of Arp2/3 complex with actinnucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.D., Stafford W.F., Pollard T.D. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba . J. Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R., Lindberg U., Schutt C.E. Domain motions in actin. J. Mol. Biol. 1998;280:463–474. doi: 10.1006/jmbi.1998.1879. [DOI] [PubMed] [Google Scholar]
- Paschal B.M., Holzbaur E.L.F., Pfister K.K., Clark S., Meyer D.I., Vallee R.B. Characterization of a 50-kDa polypeptide in cytoplasmic dynein preparations reveals a complex with p150GLUED and a novel actin. J. Biol. Chem. 1993;268:15318–15323. [PubMed] [Google Scholar]
- Perez F., Diamantopoulos G.S., Stalder R., Kreis T.E. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Pfister K.K., Benashski S.E., Dillman J.F., III, Patel-King R.S., King S.M. Identification and molecular characterization of the p24 dynactin light chain. Cell Motil. Cytoskelet. 1998;41:154–167. doi: 10.1002/(SICI)1097-0169(1998)41:2<154::AID-CM6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Pierre P., Scheel J., Rickard J., Kreis T.E. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Plamann M., Minke P.F., Tinsley J.H., Bruno K.S. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Winsor B. Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997;13:1053–1058. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Presley J.F., Zaal K.J.M., Schroer T.A., Cole N.B., Lippincott-Schwartz J. ER to Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Quintyne N.J., Gill S.R., Eckley D.M., Crego C.L., Compton D.A., Schroer T.A. Dynactin is required for microtubule anchoring at fibroblast centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Wojcik E.J., Sander M.A., McGrail M., Hays T.S. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosome during mitosis in Drosophila . J. Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Borden K.L.B., Boddy M.N., Freemont P.S. Does this have a familiar RING? Trends Biochem. Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Schafer D.A., Gill S.R., Cooper J.A., Heuser J.E., Schroer T.A. Ultrastructural analysis of the dynactin complexAn actin-related protein is a component of a filament that resembles f-actin J. Cell Biol. 126 1994. 403 412a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Korshunova Y.O., Schroer T.A., Cooper J.A. Differential localization and sequence analysis of capping protein β-subunit isoforms of vertebrates J. Cell Biol. 127 1994. 453 465b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Schroer T.A. Actin-related proteins. Annu. Rev. Cell. Dev. Biol. 1999;In press doi: 10.1146/annurev.cellbio.15.1.341. [DOI] [PubMed] [Google Scholar]
- Schroer T.A. Structure and function of dynactin. Semin. Cell Biol. 1996;7:321–328. [Google Scholar]
- Schroer T.A., Bingham J.B., Gill S.R. Actin-related protein 1 and cytoplasmic dynein-based motility—what's the connection? Trends Cell Biol. 1996;6:212–215. doi: 10.1016/0962-8924(96)20014-5. [DOI] [PubMed] [Google Scholar]
- Schroer T.A., Fyrberg E., Cooper J.A., Waterston R.H., Helfman D., Pollard T.D., Meyer D.I. Actin-related protein nomenclature and classification. J. Cell Biol. 1994;127:1777–1778. doi: 10.1083/jcb.127.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T.A., Sheetz M.P. Two activators of microtubule-based vesicle transport. J. Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop A., White J. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B., Ebersold M.W., Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., Goldberg M.L. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen W., Karki S., Vaughan K.T., Vallee R.B., Holzbaur E.L., Weiss D.G., Kuznetsov S.A. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol. Biol. Cell. 1997;8:2077–2088. doi: 10.1091/mbc.8.10.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M., Nakano M.Y., Keller S., Boucke K., Stidwill R.P., Greber U.F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgi A. The reversible depolymerization of actin by potassium iodide. Arch. Biochem. Biophys. 1951;31:97–103. doi: 10.1016/0003-9861(51)90188-9. [DOI] [PubMed] [Google Scholar]
- Tai A.W., Chuang J.-Z., Bode C., Wolfrum U., Sung C.-H. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tinsley J.H., Minke P.F., Bruno K.S., Plamann M. p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol. Biol. Cell. 1996;7:731–742. doi: 10.1091/mbc.7.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokito M.K., Holzbaur E.L. The genomic structure of DCTN1, a candidate gene for limb-girdle muscular dystrophy (LGMD2B) Biochim. Biophys. Acta. 1998;1442:432–436. doi: 10.1016/s0167-4781(98)00195-x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Grodon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheetsprocedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg E.A., Koonce M.P., McIntosh J.R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C., Wetzel D.M., Schrader M., Hasbani M.J., Gill S.R., Kreis T.E., Schroer T.A. Role of dynactin in endocytic trafficeffects of dynamitin overexpression and colocalization with CLIP-170. Mol. Biol. Cell. 1999;In press doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Sheetz M.P. Targeting of motor proteins. Science. 1996;271:1539–1544. doi: 10.1126/science.271.5255.1539. [DOI] [PubMed] [Google Scholar]
- Vallee R.B., Vaughan K.T., Echeverri C.J. Targeting of cytoplasmic dynein to membranous organelles and kinetochores via dynactin. Cold Spring Harb. Symp. Quant. Biol. 1996;60:803–811. doi: 10.1101/sqb.1995.060.01.086. [DOI] [PubMed] [Google Scholar]
- Vaughan K.T., Tynan S.H., Faulkner N.E., Echeverri C.J., Vallee R.B. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J. Cell Sci. 1999;112:1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- Vaughan K.T., Vallee R.B. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued . J. Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Berrez J.-M., Antony C., Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggsrequirement for phosphorylated factors and cytoplasmic dynein. J. Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierula P.J., Mais J.M. A gene required for nuclear migration in Neurospora crassa codes for a protein with cysteine-rich, LIM/RING-like domains. Mol. Microbiol. 1997;24:331–340. doi: 10.1046/j.1365-2958.1997.3461704.x. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Karki S., Holzbaur E.L. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc. Natl. Acad. Sci. USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittman, T., and A. Hyman. 1999. Recombinant p50/dynamitin as a tool to examine the role of dynactin in intracellular processes. In Methods in Cell Biol. Academic Press, Inc., San Diego, CA. [DOI] [PubMed]