Abstract
We characterized Apg8/Aut7p essential for autophagy in yeast. Apg8p was transcriptionally upregulated in response to starvation and mostly existed as a protein bound to membrane under both growing and starvation conditions. Immunofluorescence microscopy revealed that the intracellular localization of Apg8p changed drastically after shift to starvation. Apg8p resided on unidentified tiny dot structures dispersed in the cytoplasm at growing phase. During starvation, it was localized on large punctate structures, some of which were confirmed to be autophagosomes and autophagic bodies by immuno-EM. Besides these structures, we found that Apg8p was enriched on isolation membranes and in electron less-dense regions, which should contain Apg8p-localized membrane- or lipid-containing structures. These structures would represent intermediate structures during autophagosome formation. Here, we also showed that microtubule does not play an essential role in the autophagy in yeast. The result does not match with the previously proposed role of Apg8/Aut7p, delivery of autophagosome to the vacuole along microtubule. Moreover, it is revealed that autophagosome formation is severely impaired in the apg8 null mutant. Apg8p would play an important role in the autophagosome formation.
Keywords: autophagy, autophagosome, Apg8/Aut7p, Saccharomyces cerevisiae, vacuole
Autophagy is responsible for intracellular bulk protein degradation in the lytic organelle, lysosome/vacuole (Kopitz et al. 1990; Dunn 1994). It is induced when cells cannot ingest nutrients from the extracellular environment. By autophagy, cytoplasmic components are sequestered to the lysosome/vacuole to be degraded. In yeast, the defect in autophagy results in loss of viability during starvation, indicating that this protein degradation provides the minimal nutrients required for starved cells to survive (Tsukada and Ohsumi 1993). In addition, autophagy deficient mutants are defective in sporulation in yeast, suggesting that it is necessary for the cell differentiation (Tsukada and Ohsumi 1993).
A unique membrane dynamic is observed in the process of autophagy. The whole process seems to be well conserved through eukaryotes from yeast to mammal. In the case of yeast, autophagosome, a double-membrane structure surrounding a portion of the cytoplasm, is formed (Baba et al. 1994). Then, its outer membrane fuses to the vacuolar membrane depending upon the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) molecules (Baba et al. 1994; Darsow et al. 1997; Sato et al. 1998). Subsequently, autophagic body is released into the vacuolar lumen (Takeshige et al. 1992; Baba et al. 1994). The membrane of autophagic body is disrupted in the vacuole depending on proteinase A/proteinase B and the digestion of its contents, cytosolic materials, follows (Takeshige et al. 1992).
We isolated a group of autophagy defective mutants (apg) in the yeast Saccharomyces cerevisiae (Tsukada and Ohsumi 1993). Harding et al. 1995 isolated the cvt mutants defective in Cvt pathway, transport of precursor form of aminopeptidase I (proAPI)1 from the cytosol to the vacuole under nutrient-rich conditions (Harding et al. 1995). Its whole process is topologically the same as autophagy (Baba et al. 1997; Scott et al. 1997). Most cvt mutant is allelic to apg mutant (cvt5 is allelic to apg8/aut7), and every apg mutant is defective in Cvt pathway (Scott et al. 1996). Thus, Apg proteins participate in both autophagic and Cvt pathways.
We have characterized 13 Apg gene products for these years. APG1/AUT3 encodes a protein kinase, whose kinase activity is essential for autophagy (Matsuura et al. 1997; Straub et al. 1997). Its overexpression suppresses the defect in autophagy of the apg13 mutant (Funakoshi et al. 1997). Apg6/Vps30p and Apg14p form a protein complex on yet unidentified membrane structures (Kametaka et al. 1998). Apg12p is conjugated to Apg5p in a similar manner as ubiquitin (Mizushima et al. 1998). Apg7p and Apg10p are the enzymes catalyzing this conjugation reaction (Mizushima et al. 1998; Kim et al. 1999; Shintani et al. 1999; Tanida et al. 1999). Apg16p forms homooligomer and is bound to Apg5p–Apg12p conjugate, resulting in forming multimeric protein complex (Mizushima et al. 1999). It was reported that AUT1, AUT2, and AUT7, are essential for the autophagy (Schlumpberger et al. 1997; Lang et al. 1998). Aut2p is physically associated with Aut7p (Lang et al. 1998).
In the process of autophagy, formation of autophagosome has been poorly characterized so far. In mammalian cells, several groups proposed the potential source of the membrane of autophagosome such as ER, post-Golgi membrane, or yet uncharacterized organelle, phagophore (Dunn 1990; Seglen et al. 1990; Yamamoto et al. 1990a,Yamamoto et al. 1990b). Thus, the origin of autophagosome is still controversial. Moreover, it is unknown how the double membrane structure of autophagosome is constructed. This confusing situation may be due to lack of a specific marker for autophagosomal membrane. Even in yeast cells, a specific marker protein localized on autophagosome has not been identified, although many Apg proteins have been characterized so far. Since autophagosome is a transient structure in autophagic process, a marker molecule is prerequisite to trace the formation process.
Here, we report on the expression and intracellular localization of Apg8p, one of Apg proteins essential for autophagy, previously reported as Aut7p (Lang et al. 1998). Apg8p is localized on the autophagosomes and their precursor structures during starvation. Based upon the examination of the intermediate structures of autophagosome, we present a novel model for the formation process of autophagosome. We also report that apg8 mutant is defective in autophagosome formation. Apg8p may play an important role during autophagosome formation.
Materials and Methods
Yeast Strains and Media
Yeast strains used in this study are shown in Table . To construct the Δapg8 strains, TRP1, LEU2, and HIS3, which was obtained from pJJ288, pJJ252, and pJJ215, respectively, was substituted for the AccI–HpaI fragment containing 98% of the APG8 open reading frame (ORF). TK402, TK404, TK405, TK407, and KVY5 were obtained by selection on appropriate amino acid drop-out medium. Media used in this study were described previously, and SD medium supplemented with 0.5% casamino acid was referred as SD+CA medium (Shirahama et al. 1997).
Table 1.
Strains Used in this Study
| Strain | Genotype | Source |
|---|---|---|
| YW5-1B | MATa leu2 ura3 trp1 | Noda et al. 1995 |
| TK404 | MATa leu2 ura3 trp1 Δapg8::TRP1 | This study |
| TK201 | MATa leu2 ura3 trp1 Δapg8::TRP1 pTK201 | This study |
| TK405 | MATa leu2 ura3 trp1 Δapg8::LEU2 | This study |
| TK114 | MATa leu2 ura3 trp1 Δapg8::LEU2 pTK110 | This study |
| STY1 | MATa leu2 ura3 trp1 Δpep4::URA3 | Nakamura et al. 1997 |
| TK407 | MATa leu2 ura3 trp1 Δpep4::URA3 Δapg8::LEU2 | This study |
| TK116 | MATa leu2 ura3 trp1 Δpep4::URA3 Δapg8::LEU2 pTK110 | This study |
| TN124 | MATa leu2 ura3 trp1 Δpho8::PHO8 Δ60 Δpho13::LEU2 | Noda et al. 1995 |
| TK402 | MATa leu2 ura3 trp1 Δpho8::PHO8 Δ60 Δpho13::LEU2 Δapg8::TRP1 | This study |
| TK301 | MATa leu2 ura3 trp1 Δpho8::PHO8 Δ60 Δpho13::LEU2 Δapg8::TRP1 pTK101 | This study |
| TJ303 | MATa leu2 ura3 trp1 Δpho8::PHO8 Δ60 Δpho13::LEU2 Δapg8::TRP1 pRS316 | This study |
| TK307 | MATa leu2 ura3 trp1 Δpho8::PHO8 Δ60 Δpho13::LEU2 Δapg8::TRP1 pTK108 | This study |
| TK415 | MATa leu2 ura3 trp1 Δypt7::LEU2 | This study |
| TK411 | MATa leu2 ura3 trp1 Δypt7::LEU2 Δapg8::TRP1 | This study |
| KVY5 | MATα leu2 ura3 trp1 lys2 his3 suc2-Δ9 Δapg8::HIS3 | This study |
Cloning of APG8 and Plasmid Construction
APG8 was cloned according to the method previously reported (Kametaka et al. 1996). pAPG8317 was generated by cloning 2.6 kbp of XbaI–XbaI fragment, including APG8, into pBluescript II KS+. Using pAPG8317 as a template DNA, the SpeI–EcoRI fragment, including the 0.9 kbp of HincII–HincII sequence, was generated by PCR using following primers: APG8F1, 5′-GACTAGTAGGTCTCGCAAGAGAGC-3′; APG8R1, 5′-GGAATTCGAAATCTTGGCTCCGTTG-3′. pTK101 and pTK201 were generated by inserting this SpeI–EcoRI fragment into the yeast centromeric and multicopy vectors, pRS316 and pRS426 (Sikorski and Hieter 1989), respectively. For construction of 3 × HA (hemagglutinin)-tagged APG8 plasmids, a BamHI site was generated at the 5′ terminus of the APG8 ORF by PCR using APG8F1, APG8R1, and the following primers: 5′-GACATGGGATCCAAGTCTACATTTAAGTCTG-3′ and 5′-AGACTTGGATCCCATGTCTCTAGTAATTAT-3′. The resulting fragment was cloned into pBluescript II KS+, and a BamHI–BamHI fragment containing a 3 × HA sequence generated by PCR was inserted in the BamHI site at the 5′ terminus of the APG8 ORF. The 3 × HA-tagged APG8 plasmids, pTK110 and pTK108, were generated by cloning this SpeI–EcoRI fragment containing the 3 × HA-tagged APG8 gene into yeast centromeric plasmids, pRS314 and pRS316 (Sikorski and Hieter 1989), respectively.
Alkaline Phosphatase Assay
Alkaline phosphatase (ALP) assay was performed as previously described (Noda et al. 1995; Noda and Ohsumi 1998).
Nocodazole Treatment
Cells growing in YEPD medium were treated with nocodazole (Sigma Chemical Co.) for 3 h at a final concentration of 10 μg/ml. Cells were transferred to 0.17% yeast nitrogen base without amino acids and ammonium sulfate supplemented with 1 mM PMSF and 10 μg/ml nocodazole, and further incubated for 4.5 h. The accumulation of autophagic bodies was observed under a microscope. For ALP assay, the nocodazole-treated cells in YEPD were starved in SD(−N) medium containing 10 μg/ml nocodazole for 4.5 h.
Production of Antiserum against Apg8p
A peptide including the NH2-terminal sequence of Apg8p, MKSTFKSEYPFEKC, was synthesized by a Model 433A peptide synthesizer (PE Applied Biosystems). The peptide was conjugated to Keyhole Limpet Hemocyanin (Sigma Chemical Co.) with sulfosuccinimidyl 4-(p-maleimidophenyl)butyrate (Pierce Chemical Co.) according to the method previously reported (Iwai et al. 1988). The resulting conjugates were immunized to a rabbit and anti-Apg8p antiserum was obtained.
Western Blotting of Apg8p
Cell lysates were prepared by breaking cells with the glass beads in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 mM PMSF, and the protease inhibitor cocktail™ (Boehringer Mannheim Corp.). Total protein (20 μg) was subjected to SDS-PAGE and transferred to PVDF (polyvinylidene fluoride) membrane (Millipore Corp.). The resulting membrane was incubated with a 1:10,000 dilution of the anti-Apg8p antibody for 1 h, followed by a 1:10,000 dilution of HRP-conjugated goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories) for 30 min. Signals were detected by the ECL kit (Nycomed Amersham, Inc.), which was used throughout this study. For the analysis of rapamycin treatment, growing cells were treated with 0.2 μg/ml rapamycin (Sigma Chemical Co.) for 2 h at 30°C in YEPD medium. Lysate preparation, followed by Western blotting, were performed as described above.
Northern Blotting
Cells were suspended in TES solution (10 mM Tris-HCl, pH 7.5, 10 mM EDTA, 0.5% SDS) and treated with acid phenol at 65°C for 1 h. Aqueous phase was collected and RNA was recovered by ethanol precipitation. mRNA was recovered by Oligotex-dT30 < super > (Takara) according to the manufacturer's protocol, separated on 1.2% agarose containing 2.2 M formaldehyde by electrophoresis, and transferred to NYTRAN membrane (Scleicher & Schuell, Inc.). 32P-labeled probes for APG8 and ACT1 mRNA were prepared from each ORF fragment using Megaprime DNA labeling system (Nycomed Amersham, Inc.) according to the appended protocol. The mRNA-transferred membrane was incubated with each probe at 42°C overnight, and signals were detected by autoradiography.
Subcellular Fractionation and Solubilization
Subcellular fractionation was performed as previously described by Horazdovsky and Emr 1993. YW5-1B cells were cultured in YEPD medium at 30°C to 1–2 × 107 cells/ml, shifted to SD(−N) medium for 3 h and converted to spheroplasts. Spheroplasting medium was composed of YEP, 1.2 M sorbitol, 20 mM Tris-HCl, pH 7.5, 1% glucose, and 25 μg/108 cells of Zymolyase 100T (Seikagaku Kogyo) for growing cells, or 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 1.2 M sorbitol, 20 mM Tris-HCl, pH 7.5, 1% glucose, and 37.5 μg/108 cells of Zymolyase 100T for starved cells. Spheroplasts were harvested and lysed with a lysis buffer containing 0.2 M sorbitol, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA and EGTA, 1 mM PMSF, and the protease inhibitor cocktail. Lysates (T) were generated by centrifugation at 500 g for 5 min and then spun at 13,000 g for 15 min to separate to pellet (LSP) and supernatant (LSS). LSS was then centrifuged at 100,000 g for 1 h, and pellet (HSP) and supernatant (HSS) were obtained. LSP and HSP were resuspended in equal volume of the lysis buffer to the original lysates. Equal volume of each sample was subjected to SDS-PAGE, transferred to PVDF membranes, and then incubated with a 1:5,000 dilution of the anti-Apg8p antibody.
For the solubilization experiment, lysates were prepared from YW5-1B cells growing or starved for 3 h as described above. Lysates were sonicated and spun at 100,000 g for 1 h to generate pellet. The pellet was suspended in the lysis buffer (without sorbitol) containing 2% Triton X-100, the suspension was chilled on ice for 30 min and centrifuged at 100,000 g for 1 h again to separate supernatant and pellet. Pellet was resuspended in an equal volume of the lysis buffer to the original sample. Proteins recovered in each fraction were precipitated with 10% TCA and resuspended in SDS sample buffer. Equal volume of the samples was subjected to Western blotting as described.
Immunofluorescence Microscopy
Immunofluorescent staining was performed according to the method previously described by Nishikawa et al. 1994. In this experiment, Δapg8 and Δapg8Δpep4 cells harboring 3 × HA-tagged APG8 plasmid were used. Spheroplasts prepared from the cells fixed with formaldehyde were laid on the multiwell slide glass (Cel-line Associates, Inc.) coated by polylysine (mol wt >300,000; Sigma Chemical Co.), and permeabilized by treatment with 0.5% Triton X-100 in PBS for 10 min. Permeabilized cells were incubated in PBS containing 1% BSA at room temperature for 10 min and treated with a 1:1,000 dilution of anti-HA mAb, 16B12 (Berkeley Antibody Co., Inc.), in the same buffer at room temperature for 1 h. The cells were then incubated with 10 μg/ml of anti-mouse Ig-fluorescein F(ab′)2 fragment (Boehringer Mannheim Corp.) at room temperature for 1 h and observed under a confocal microscope, LSM 510 (Zeiss).
Electron Microscopy
Cells were subjected to rapid freezing and freeze-substitution fixation, and observed as previously reported (Baba et al. 1997). For immunoelectron microscopy, ultrathin sections were collected onto formvar-coated nickel grids and blocked in PBS containing 2% BSA at room temperature for 15 min. Incubations were carried out by floating grids on a 20 μl drop of a 1:1,500 dilution of anti-HA mAb, 16B12, at room temperature for 1.5 h. After washing, the grids were incubated for 1 h with 5- or 10-nm gold-conjugated goat anti-mouse IgG (Bio Cell Lab.). The grids were washed several times in PBS followed by several drops of distilled water and fixed with 1% glutaraldehyde for 3 min. The sections were stained with 4% uranyl acetate for 7 min and examined.
Proteinase K Treatment
Cells were starved in SD(−N) medium for 4.5 h and converted to spheroplasts. Spheroplasts were lysed in 20 mM Pipes/KOH, pH 6.8, 0.2 M sorbitol, 0.5 mM PMSF, and the protease inhibitor cocktail. Cleared lysates were generated by centrifugation at 500 g for 5 min, and treated with 100 μg/ml of proteinase K with or without 2% Triton X-100 on ice for 30 min. Equal volume of 20% TCA was added to the lysates to stop the reaction. Precipitant was collected by centrifugation, suspended in SDS sample buffer, and subjected to immunoblotting. To detect the signal of API, 1:5,000 diluted anti-API antibody (gift from D. Klionsky, Section of Microbiology, UC Davis, CA) was used.
Results
APG8, an Essential Gene for Autophagy
We reported an apg8-1 mutant defective in the autophagy in yeast (Tsukada and Ohsumi 1993). To identify the APG8, cloning was done as previously described (Kametaka et al. 1996). Subcloning and sequencing revealed that APG8 is identical to the ORF named YBL078c, and encodes a hydrophilic protein of 117 amino acids. During this study, Lang et al. 1998 reported AUT7 as a novel autophagy-responsible gene, which was revealed to be identical to APG8.
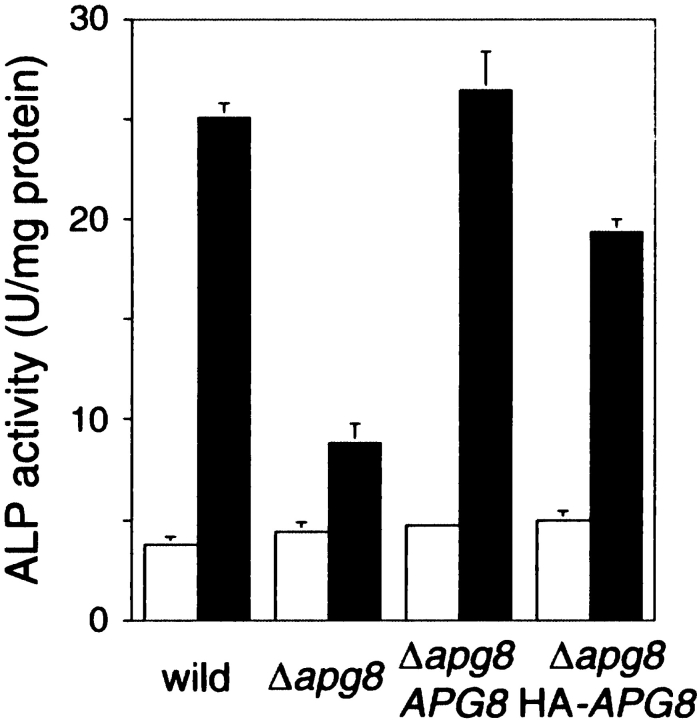
We constructed the apg8 null mutant and ascertained morphologically that Apg8p is essential for autophagy, judging from no accumulation of autophagic bodies in the vacuole (data not shown). Furthermore, using the ALP assay system (Noda et al. 1995), we confirmed biochemically that Apg8p is essential for autophagy. The cells used for this assay have a truncated form of Pho8p, Pho8Δ60p, in the cytosol as an inactive precursor form. During starvation, Pho8Δ60p is sequestered to the vacuole, depending upon autophagy, where it is processed by the vacuolar enzymes and acquires phosphatase activity. So, we can monitor the progress of the autophagy by the increase of the ALP activity. Wild-type (TN124), Δapg8 mutant, and the Δapg8 mutant harboring APG8 on a centromeric plasmid were grown in SD+CA medium until 1–2 × 107 cells/ml, and then shifted to SD(−N) medium for three hours. The ALP activity in the cell lysates prepared from each culture was measured (Fig. 1). In wild-type cells, the ALP activity increased in response to starvation, whereas in the Δapg8 cells its elevation was considerably reduced. The autophagic defect in the Δapg8 mutant was recovered by introducing APG8 on a centromeric plasmid. The result clearly shows that deletion of APG8 causes severe defect in autophagy.
Figure 1.
ALP assay. The cells expressing Pho8Δ60p were cultured in SD+CA medium (open bars) and shifted to SD(−N) medium for three hours (filled bars). The ALP activity of the lysates was measured to estimate autophagic activity. Wild-type cells, TN124; Δapg8 mutant harboring vector, TK303; Δapg8 mutant harboring APG8 on a centromeric vector, TK301; Δapg8 mutant harboring 3 × HA-tagged APG8 plasmid, TK307.
No Requirement of Microtubule for Autophagy
A homology search showed that Apg8p and its homologues make a large gene family through yeast to higher eukaryotes (Lang et al. 1998). One of the homologues has been reported as rat MAP1-LC3, which is directly bound to microtubule in vitro (Mann and Hammarback 1994). Furthermore, Lang et al. 1998 proposed that Apg8p is bound to microtubule via another protein, Aut2p, and functions on the delivery of autophagosomes to the vacuole. We have already cloned 13 APG genes, and realized that AUT2 is allelic to APG4, and that Apg8p physically interacts with Apg4p. However, Apg4p has an entirely different function from what they proposed (Kirisako, T., and Y. Ohsumi, manuscript in preparation).
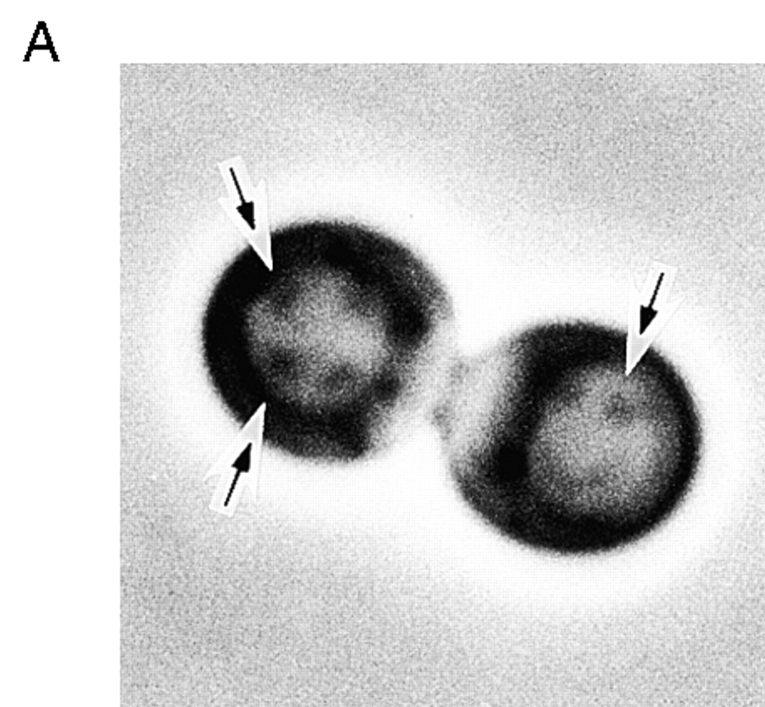
So far, there is no evidence showing that microtubules play a role in the autophagy in yeast. Nocodazole is a microtubule depolymerizing drug, which affects both spindle and cytoplasmic microtubules (Solomon 1991). We asked, morphologically and biochemically, whether autophagy proceeds normally in the presence of this drug. Wild-type cells were cultured until 107 cells/ml in YEPD medium at 30°C. Nocodazole was added to the culture and it was incubated at 30°C for three hours. After this treatment, >70% of the cells were arrested at G2 stage with a large bud. Then the cells were transferred to starvation medium containing 1 mM PMSF and 10 μg/ml nocodazole, and incubated further at 30°C for 4.5 h. As shown in Fig. 2 A, most large budded cells accumulated the autophagic bodies in their vacuole. TN124 cells were treated the same way, and the ALP activity was measured. Nocodazole treatment did not affect the increase of ALP activity (Fig. 2 B). Furthermore, in a tub2 mutant, autophagic bodies were normally accumulated in the vacuole (data not shown). These results indicate that depolymerization of microtubule does not affect the progress of autophagy. Hence, we concluded that microtubule is not necessary for the autophagy in yeast.
Figure 2.
Effect of nocodazole on autophagy. Vegetatively growing cells, YW5-1B and TN124, were treated with 10 μg/ml nocodazole in YEPD medium for three hours. A, YW5-1B cells were transferred to 0.17% yeast nitrogen base without amino acid and ammonium sulfate containing 1 mM PMSF and 10 μg/ml nocodazole, and further incubated for 4.5 h. The accumulation of autophagic bodies was examined under a light microscope. Arrows indicate autophagic bodies. B, TN124 cells were starved in SD(−N) containing 10 μg/ml nocodazole for 4.5 h. ALP activity was measured before (open bars) and after (filled bars) starvation.
Expression of APG8
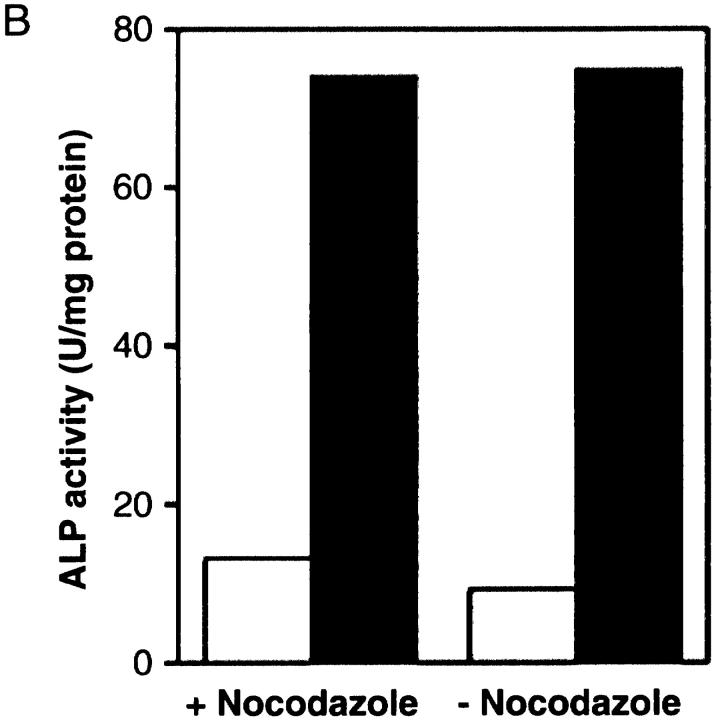
We raised an antiserum against Apg8p using a synthetic peptide of the NH2-terminal 13 amino acids of Apg8p. Using the anti-Apg8p antibody, we carried out Western blotting of the cell lysate prepared from logarithmically growing cells. In wild-type cells, the anti-Apg8p antibody brought out a single band at ∼15 kD (Fig. 3 A, lane 1), which is close to the predicted molecular mass of Apg8p (13.6 kD). Intensity of the band was enhanced in the cells harboring APG8 on a multicopy plasmid (Fig. 3 A, lane 2), whereas no signal was detected in the Δapg8 cells (Fig. 3 A, lane 3). The result revealed that Apg8p is expressed in the wild-type cells at growing phase. Since APG8 is required also for the transport of proAPI from the cytosol to the vacuole (Scott et al. 1996), Apg8p expressed during vegetative growth should play a role in this pathway. Next, we examined Apg8p levels before and after shift to nitrogen starvation. The wild-type cells growing in YEPD medium were shifted to SD(−N) medium for various periods of time, and the lysates prepared from these cell cultures were subjected to immunoblotting with the anti-Apg8p antibody. As shown in Fig. 3 B, the amount of Apg8p started to increase 30 min after shift to starvation and remained at a high level after two hours. Finally, the amount of Apg8p increased about eightfold in response to starvation (Fig. 3 B).
Figure 3.
Expression of APG8. A, The lysates prepared from the growing cells were subjected to immunoblotting with the anti-Apg8p antibody. Lane 1, wild-type cells (YW5-1B); lane 2, Δapg8 cells harboring APG8 on a multicopy vector (TK201); lane 3, Δapg8 cells (TK404). B, The wild-type (YW5-1B) cells were cultured in YEPD medium until 1–2 × 107 cells/ml (0 h) and shifted to SD(−N) medium for 0.5, 1, 2, 3, 4.5, and 6 h at 30°C. The amount of Apg8p was estimated by immunoblotting with anti-Apg8p antibody. Each lane has 10 μg of total protein. C, YW5-1B cells were cultured in YEPD medium until 1–2 × 107 cells/ml (0 h) and shifted to SD(−N) medium for 0.5, 1, 2, and 6 h at 30°C, and mRNA was prepared from each culture as described in Materials and Methods. APG8 mRNA and ACT1 mRNA were detected by Northern blotting with each specific probe. Each lane has 4 μg of total mRNA. D, YW5-1B cells were cultured in YEPD medium until 1 × 107 cells/ml at 30°C. Rapamycin was added to the culture at a final concentration of 0.2 μg/ml and the cells were further incubated in YEPD medium for two hours at 30°C. Western blotting was performed with the anti-Apg8p antibody. Each lane has 20 μg of total protein.
Next, we performed Northern blotting analysis. Total mRNA was prepared from wild-type cells growing in YEPD medium, and the cells shifted to SD(−N) medium for various periods of time. As shown in Fig. 3 C, the amount of APG8 mRNA increased drastically in response to starvation, and reached at a maximum level within 30 min after shift to starvation. After 30 min, the amount gradually decreased, but at six hours, it was still severalfold higher than that of the growing cells. This result indicates that the increase of Apg8p during starvation is regulated at transcription level. Among the 12 APG genes (APG1, 4–10, 12–14, and 16) characterized so far, APG8 is the first gene whose expression is enhanced by starvation.
Autophagy is known to be induced by inactivation of Tor, a phosphatidylinositol kinase homologue (Noda and Ohsumi 1998). We investigated the effect of Tor function on the expression of Apg8p. Wild-type cells were treated with rapamycin in YEPD medium for two hours, and the lysates prepared from the cells were subjected to immunoblotting with the anti-Apg8p antibody. As shown in Fig. 3 D, the expression of Apg8p was enhanced in response to the drug. Since rapamycin specifically inhibits the signal transduction mediated by Tor (for review see Thomas and Hall 1997), the transcription of APG8 should be under the control of Tor-mediated signal transduction.
In addition, we investigated whether other APG genes are necessary for the increase of Apg8p. Lysates were prepared from the other 14 apg mutant cells before and after shift to starvation and subjected to immunoblotting with the anti-Apg8p antibody. In every other apg mutant, Apg8p increased normally in response to starvation as the wild-type cells (data not shown), indicating that no other APG genes are required for the induction of Apg8p.
Cell Fractionation of Apg8p
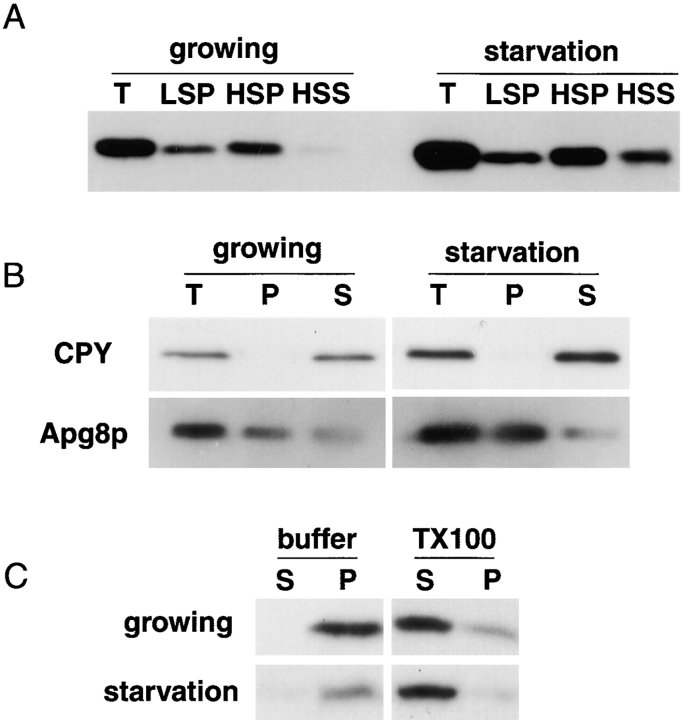
We carried out subcellular fractionation. Cell lysates were prepared from growing and starved wild-type cells. Cell lysates (T) were centrifuged at 13,000 g for 15 min and the pellet (LSP) fractions were obtained. The resulting supernatant fractions were spun at 100,000 g for one hour to generate supernatant (HSS) and pellet (HSP) fractions, and the distribution of Apg8p was examined by immunoblotting with the anti-Apg8p antibody. Apg8p was recovered mostly in LSP and HSP fractions under both growing and starvation conditions, although in starved cells it was more detectable in HSS fraction than in growing cells (Fig. 4 A). Next, the cell lysates were vigorously sonicated to exclude lumenal proteins out of organelles or other membrane structures, and was spun at 100,000 g for one hour to generate a pellet fraction. Under both growth conditions, Apg8p mostly remained in the pellet fraction, in contrast CPY, a vacuolar lumenal protein, was completely recovered in supernatant fraction (Fig. 4 B). It suggests that Apg8p is bound to membrane or associated with a pelletable large protein complex. The pellet fractions generated from the sonicated lysates were treated with 2% Triton X-100, incubated for 30 min on ice, and then separated to supernatant and pellet fractions by centrifugation at 100,000 g for another one hour. As shown in Fig. 4 C, 2% Triton X-100 efficiently solubilized Apg8p, indicating that most pelletable Apg8p is bound to some membrane structures under both growing and starvation conditions.
Figure 4.
Subcellular fractionation and solubilization of Apg8p. The wild-type cells (YW5-1B) growing logarithmically in YEPD medium or starved in SD(−N) medium for three hours were lysed. A, The lysates (T) were centrifuged at 13,000 g for 15 min to generate pellet (LSP) and supernatant (LSS). LSS fraction was further centrifuged at 100,000 g for one hour and separated to pellet (HSP) and supernatant (HSS). LSP and HSP were resuspended in an equal volume of the lysis buffer to the original lysates. Equal volume of each sample was applied to each well, and immunoblotting was performed with anti-Apg8p antibody. B, The lysates (T) were sonicated and centrifuged at 100,000 g for one hour to separate pellet (P) and supernatant (S). The distribution of CPY and Apg8p was examined by immunoblotting with each specific antibody, respectively. C, The sonicated lysates were centrifuged at 100,000 g for one hour to generate pellet fraction. The pellet fraction was treated with 2% Triton X-100 (TX-100) for 30 min on ice. The sample was centrifuged at 100,000 g for one hour and separated to supernatant (S) and pellet (P) fractions. The distribution of Apg8p was examined by immunoblotting with the anti-Apg8p antibody.
Drastic Change of Localization of Apg8p in Response to Starvation
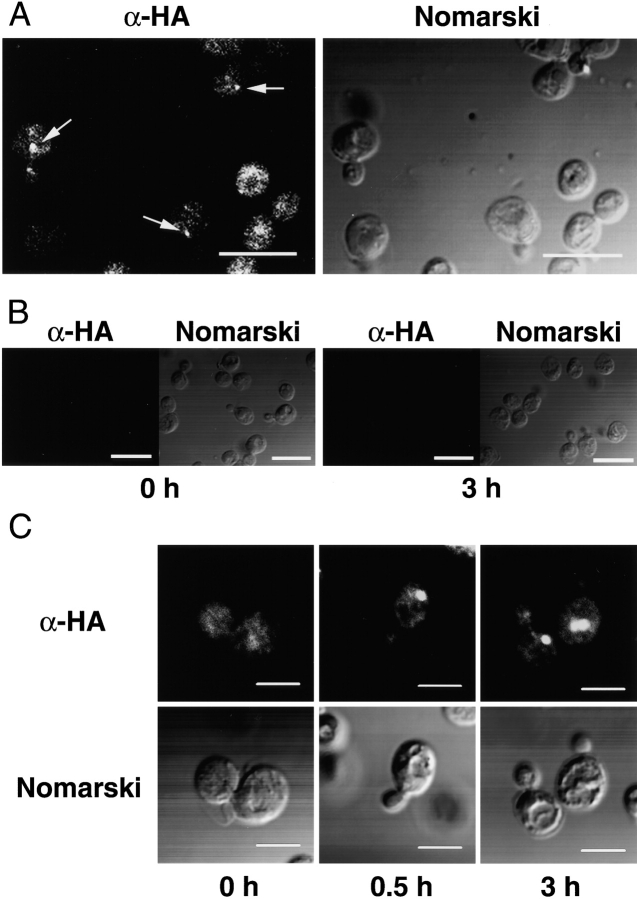
To examine intracellular localization of Apg8p, immunofluorescence microscopy was performed. First, we carried out this experiment using the anti-Apg8p antibody, but sufficient signal was not obtained. Then, we constructed a single copy 3 × HA-tagged APG8 plasmid and introduced it into the Δapg8 mutant. We ascertained that the expression level of the 3 × HA-tagged APG8 gene in the resulting transformant, TK114, was similar to that of authentic APG8 (data not shown), and that the transformant recovered the autophagic activity up to 70% of the wild-type cells (Fig. 1). The cells were grown in SD+CA medium until 2 × 107 cells/ml, and diluted fourfold in YEPD medium. The cells were grown for two generations, transferred to SD(−N) medium, and incubated for 0, 0.5, and 3 h. The localization of 3 × HA–Apg8p was examined with anti-HA mAb. In the cells growing in YEPD medium (t = 0), the fluorescence image consisted of many tiny dots dispersed throughout the cytoplasm, which was distinct from well known organelles such as ER, Golgi body, nucleus, vacuole, or plasma membrane (Fig. 5A and Fig. C, Fig. 010 h). In addition, we occasionally observed punctate signals distinct from the tiny dots (Fig. 5 A, arrows). 30 min after the shift to the starvation condition, the staining pattern drastically changed; one or two bright, large, punctate signals appeared in the cytoplasm (Fig. 5 C). These large punctate signals always resided in the cytoplasm, just beside the vacuole (Fig. 5 C). The number of these punctate signals was constantly 1–3/cell during starvation.
Figure 5.
Immunofluores-cent staining of Δapg8 cells expressing 3 × HA–Apg8p. A, The Δapg8 cells harboring 3 × HA-tagged APG8 plasmid, TK114 cells, were grown until logarithmic phase. The cells were fixed by formaldehyde and were treated with Zymolyase 100T to generate spheroplasts. The spheroplasts were permeabilized by 0.5% Triton X-100, and incubated with the anti-HA antibody, 16B12, followed with the FITC-conjugated anti–mouse IgG. Left, Fluorescence image of 3 × HA–Apg8p; right, Nomarski image of the cells. Arrows show punctate signals that are a little larger than tiny dot signals. Bars, 10 μm. B, Immunofluorescent staining of YW5-1B cells before (0 h) and after (3 h) shift to starvation (negative control). Left, Fluorescence image; right, Nomarski image. Bars, 10 μm. C, The cells were shifted to starvation for 0, 0.5, and 3 h. Upper panels, Fluorescence images of 3 × HA–Apg8p; lower panels, Nomarski images of the cell. Bars, 5 μm.
Figure 10.
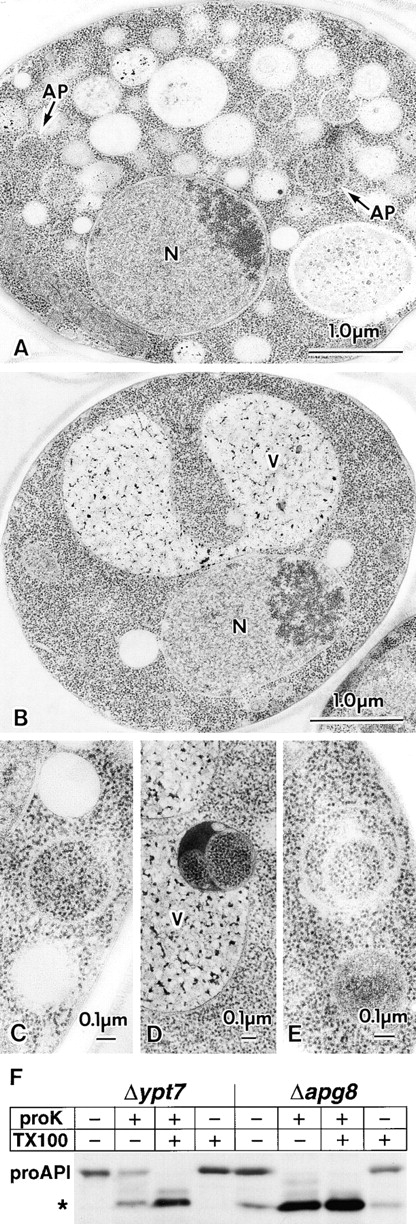
Fine morphology of Δypt7 and Δapg8 cells and proteinase-K accession to proAPI in these mutants. A, EM image of Δypt7 cell starved in SD(−N) medium for four hours. Autophagosomes (AP, arrows) were accumulated. B, EM image of the starved Δapg8 cell. C–E, The representatives of the membrane structures detected in the starved Δapg8 cells. C, Autophagosome-like structure indistinguishable from autophagosome. D and E, Aberrant multivesicular structures. N, nucleus; V, vacuole. F, Proteinase K-sensitivity of proAPI. Cell lysates were prepared from Δypt7 and Δapg8 cells starved in SD(−N) medium for 4.5 h. The lysates were treated with 100 μg/ml proteinase K in the presence or absence of Triton X-100. Asterisk shows mature API and digestion product of proAPI by proteinase K.
We further examined the localization of Apg8p in the background of Δpep4 mutant, in which the autophagic bodies accumulate in the vacuole during starvation (Takeshige et al. 1992). TK116 strain was generated by introducing the 3 × HA-tagged APG8 plasmid into the Δapg8Δpep4. As shown in Fig. 6 A, under growing condition, the fluorescence pattern was observed as tiny dots dispersed in the cytoplasm as TK114 cells. At 30 min after shift to the starvation condition, 1–3 large punctate signals per cell appeared next to the vacuole, also like wild-type cells (Fig. 6 B). At one hour, more punctate signals were observed than wild-type cells, and >50% were detected in the vacuole (Fig. 6 C). The punctate signals in the vacuole gradually increased and completely filled the vacuole at six hours later (Fig. 6, C–E). The signals coincided with autophagic bodies (Fig. 6C and Fig. H, arrows). As autophagic bodies are derived from autophagosomes, at least some population of Apg8p might be localized on or in the autophagosomes and delivered to the vacuole during starvation. Some of the large punctate signals in the cytoplasm (Fig. 5 C, 0.5 and 3 h, and Fig. 6) may represent autophagosomes.
Figure 6.
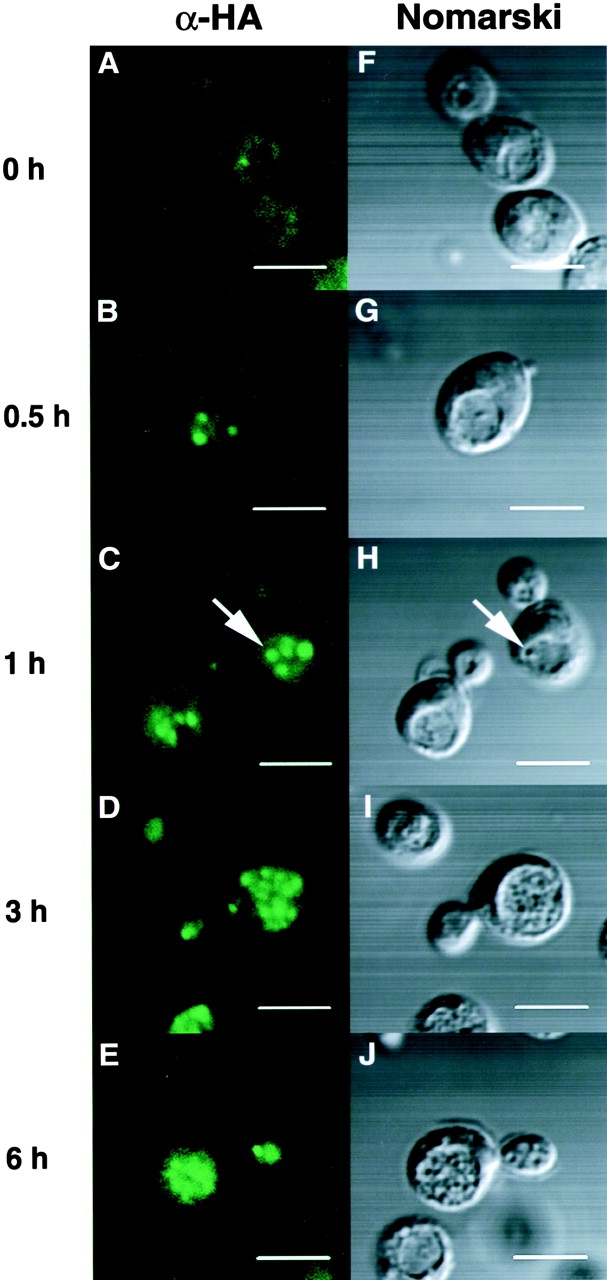
Change of Apg8p localization in Δpep4 cells after shift to starvation. The Δapg8Δpep4 cells harboring 3 × HA-tagged APG8 plasmid, TK116 cells, were shifted to starvation for 0, 0.5, 1, 3, and 6 h. Immunofluorescence microscopy was performed as described in Fig. 5. A–E, FITC staining of 3 × HA–Apg8p. F–J, Nomarski images of the cells. Arrows show an autophagic body. Bars, 5 μm.
Intermediate Structures Traced with Apg8p
To obtain further information about the intracellular localization of Apg8p during starvation, immunoelectron microscopic analysis was performed using the Δpep4 cells, TK116. The logarithmically growing cells were shifted to SD(−N) medium for one, two, or three hours. The localization of 3 × HA-Apg8p in the starved cells was examined with the anti-HA antibody. In the autophagic bodies, density of the gold particles was significantly higher than that in the cytoplasm (Fig. 7, arrows). This fact was in good agreement with the result obtained from immunofluorescence analysis. Moreover, it was revealed that most of Apg8p was in the lumen, but not on the membrane of autophagic bodies.
Figure 7.
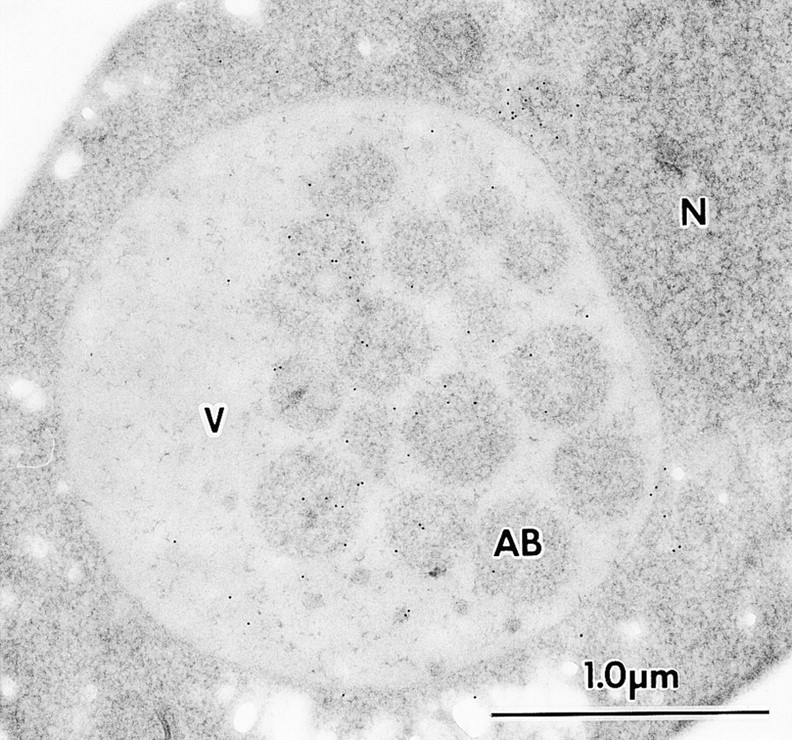
Immunostaining image of a cell containing autophagic bodies in the vacuole. The Δapg8Δpep4 cells expressing 3 × HA–Apg8p, TK116 cells, were shifted to SD(−N) medium for two hours. Cells were immunolabeled with anti-HA mAb, 16B12, followed by 10-nm gold-conjugated goat anti–mouse IgG. AB, autophagic body; N, nucleus; V, vacuole.
Next, we examined localization of Apg8p in the cytoplasm of the starved cell. As expected from the result of immunofluorescence microscopy, gold particles were associated with the autophagosomes (Fig. 8). Most mature autophagosomes with clear unit membranes and uniform intramembrane space were certainly stained, but not heavily (Fig. 8A and Fig. B), and, in some cases, the gold particles were mostly detected in their lumen (Fig. 8 B). As shown in Fig. 8C and Fig. D, some autophagosomes were heavily stained with the gold particles. In these autophagosomes, the gold particles were mostly detected along the double membrane. However, their intramembrane space was not homogenous, but partly swollen (Fig. 8C and Fig. D, arrows) not like the mature autophagosome. These structures may represent nascent autophagosomes or the latest structures in the autophagosome formation.
Figure 8.
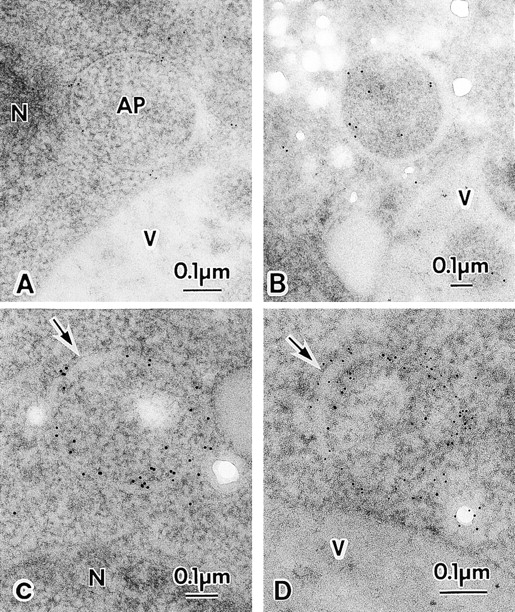
Immuno-EM of autophagosomes. A and B, Mature autophagosome. C and D, Premature autophagosome. Arrows show expanded regions of the intramembrane space of the premature autophagosomes. TK116 cells were incubated in SD(−N) medium for one hour (D), two hours (B and C), or three hours (A). The localization of 3 × HA–Apg8p was detected with anti-HA antibody, followed by the incubation with 5-nm gold- (A and D) or 10-nm gold- (B and C) conjugated goat anti–mouse IgG. AP, autophagosome; N, nucleus; V, vacuole.
Besides autophagosomes, we found heavily stained structures with immunogold in the cytoplasm. In Fig. 9 A, the gold particles are located along the surface on a curved membrane sac. Fig. 9 B shows that the gold particles were arranged in a rough circle, and some of them resided along membrane structures, which partly appear as clear double membrane. In Fig. 9 C, a semicircular isolation membrane (large arrow) and its open region (small arrow) were stained with gold particles. The gold particles were apparently more concentrated in the open region rather than on the isolation membrane. In addition, we found that gold particles were concentrated in a limited area close to the vacuole (Fig. 9 D). The electron density in this area was less than that of the cytosol. The image of this Apg8p-enriched area is similar to that of the open region of the semicircular isolation membrane shown in Fig. 9 C. We suppose that all these structures shown in Fig. 9 represent intermediate structures of autophagosome.
Figure 9.
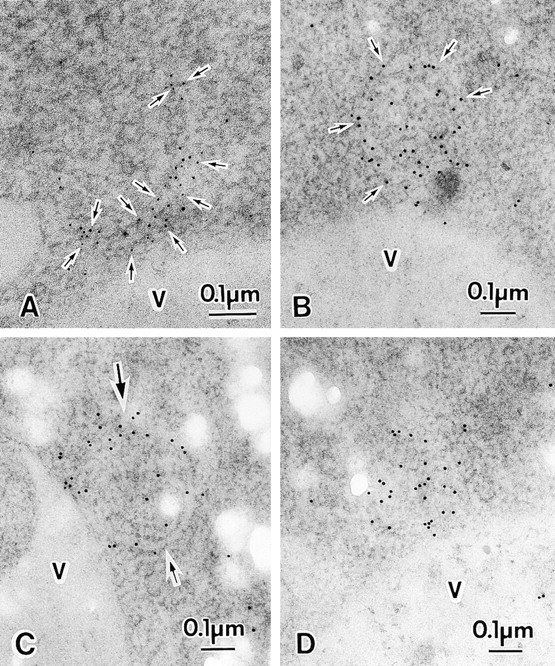
Possible intermediate structures of autophagosome. A, A membrane sac under construction. Arrows show gold particles. B and C, Isolation membranes. B, Isolation membrane is detected along the arrows. C, The small arrow shows a semicircular isolation membrane and a large arrow marks its open area. D, Apg8p-residing structures gathered in the area close to the vacuole. TK116 cells were incubated in SD(−N) medium for one hour (A and C), or two hours (B and D). The localization of 3 × HA–Apg8p was detected with anti-HA antibody, followed by the incubation with 5-nm gold- (A) or 10-nm gold- (B–D) conjugated goat anti–mouse IgG. V, vacuole.
Crucial Function of Apg8p in Autophagosome Formation
To elucidate the step at which Apg8p functions in the autophagic pathway, we used ypt7 mutant as a control. Ypt7p, a Rab family protein, is responsible for the fusion events to the vacuole (Schimmoller and Riezman 1993; Haas et al. 1995). It was reported that in Δypt7, Cvt vesicles become detectable (Kim et al. 1999). We studied by electron microscope, using rapid freezing and freeze-substitution-fixation method, whether Ypt7p is also required for the fusion of autophagosome to the vacuole. As shown in Fig. 10 A, Δypt7 cells under starvation had many fragmented vacuoles, and autophagosomes were much more detectable in the cytoplasm than in wild-type cells. Thus, the depletion of YPT7 causes the accumulation of autophagosomes in the cytoplasm.
The Δapg8 cells were examined also by EM. They were mostly normal with a few large vacuoles, but autophagosomes could be hardly detected in their cytoplasm (Fig. 10 B). However, at low frequency, membrane structures, having enclosed a portion of cytosol, were observed only under starvation conditions. Some were indistinguishable from the autophagosome (Fig. 10 C), but others showed more complicated structures, distinct from typical autophagosome, such as the structure having condensed contents or multivesicular structure (Fig. 10D and Fig. E). Next, Δapg8 cells having the background of Δypt7 were examined. The Δypt7Δapg8 cells did not accumulate autophagosomes in the cytosol, and at low frequency autophagosome-like structures were detected in their cytoplasm, like Δapg8 cells (data not shown). We counted the number of autophagosomes and autophagosome-like structures in Δypt7 cells and Δypt7Δapg8 cells. In the sections of 500 cells, we found 269 autophagosomes in Δypt7 cells, whereas in Δypt7Δapg8 cells, only 16 autophagosome-like structures were detected (5.9% of Δypt7 cells). The result clearly shows that autophagosomes are not accumulated in Δapg8 cells.
We reported that autophagy is not only responsible for nonselective degradation of cytosolic components, but also highly selective proAPI sequestration to the vacuole during starvation (Baba et al. 1997). ProAPI could be a marker as a cargo of the autophagosomes under starvation, just like Cvt vesicles under growing condition (Kim et al. 1999). Cell lysate was prepared from starved Δypt7 cells and analyzed by immunoblotting with anti-API antibody. As shown in Fig. 10 F, when the lysate was treated with proteinase K, ∼50% of proAPI was resistant to the proteinase, but it was completely digested in the presence of 2% Triton X-100, implying that the proteinase K-resistant proAPI resides in the lumen of autophagosomes. In contrast to Δypt7, proteinase K-resistant proAPI was not detected in Δapg8 cells (Fig. 10 F). This provides a biochemical evidence that Δapg8 cells do not accumulate autophagosome. Harding et al. 1995 reported that proteinase K is also completely accessible to proAPI at growing phase in cvt5/apg8 mutant, indicating that Cvt vesicles are not formed in apg8 mutants.
During this analysis, we found that proAPI was partially matured in the starved Δapg8 strain. This partial maturation was not detected in growing Δapg8 cells (data not shown). It is not a general feature of apg mutants. This suggests that there is some starvation-induced machinery delivering proAPI to the vacuole in apg8 null mutants. As shown in Fig. 10C–E, several kinds of autophagosome-like structures were observed in the starved Δapg8 cells. Although the frequency of these structures is very low, it is likely that these structures are responsible for the delivery of proAPI to the vacuole under starvation. ProAPI might be selectively sequestered to the vacuole via these membrane structures. However, because the Δapg8 mutant cannot carry out bulk protein degradation under starvation (Fig. 1), the structures must be insufficient for the bulk sequestration of cytoplasmic components to the vacuole. Maturation of proAPI should not reflect the nonselective bulk protein degradation exactly.
Lang et al. 1998 stated the accumulation of double membrane structure in aut7/apg8 mutant. Since the EM techniques that they used are quite different from ours, and the preservation of membrane structure is not good, it is hard to directly compare our morphological data with theirs. It is possible that their reported structure is one of autophagosome-like structures reported here. Our morphological and biochemical data demonstrate that Δapg8 strain never accumulates autophagosomes and strongly indicate that Apg8p plays a crucial role in autophagosome formation.
Discussion
Here, we report characterization of Apg8p, one of the APG gene products essential for autophagy. APG8 turned out to be identical to AUT7, recently reported (Lang et al. 1998). They proposed that Apg8p functions in the delivery of autophagosome to the vacuole along microtubule. However, we found that autophagy proceeds normally, even if the microtubule is depolymerized (Fig. 2). This result suggests that microtubule does not play an essential role in the autophagy in yeast. The proposal by Lang et al. 1998 was based upon the physical interactions of Apg4/Aut2p with tubulin and Apg8/Aut7p by in vitro and two-hybrid analyses. But, they did not demonstrate that Apg8p is bound to microtubule via Apg4/Aut2p. We have found that recombinant GST–Apg4p is bound to glutathione-Sepharose column nonspecifically (our unpublished result). It may be due to the acidic nature of Apg4p (predicted isoelectric point is 4.4). One possible interpretation of their result is that Apg4p is bound to tubulin nonspecifically. They also described qualitatively that the Δapg8 cells accumulate autophagosomes in the cytoplasm. Here, we demonstrated quantitatively that the apg8 null mutant does not accumulate autophagosomes in the cytoplasm. So, we concluded that Apg8p participates in the autophagosome formation. This conclusion is not affected whether Apg4/Aut2p is indeed bound to tubulin.
Intermediate structures of autophagosome formation have been poorly characterized in both yeast and mammalian cells because of their dynamic feature and the lack of specific marker of autophagosome. In yeast, only a cup-shape structure was detected as a possible intermediate structure of autophagosome (Baba et al. 1994). In this study, we found that most Apg8p is bound to membrane (Fig. 4) and localized in autophagosomes and autophagic bodies (Fig. 6 Fig. 7 Fig. 8). Apg8p must be a key molecule to identify intermediate structures of autophagosome formation. Actually, we found that Apg8p is localized on premature autophagosomes (Fig. 8C and Fig. D) and isolation membranes (Fig. 9, A–C). In addition, an Apg8p-enriched region was observed in the cytoplasm close to the vacuole (Fig. 9 D), and a similar image was obtained in the open region of the semicircular isolation membrane (Fig. 9 C, small arrow). These regions were always electron less-dense, indicating that they contain Apg8p-localized membrane- or lipid-containing structures, possible precursor structures of autophagosomal membrane. Based on our observations, we propose a model for the scheme of autophagosome formation as follows. The isolation membrane is formed by sequential assembly of the precursor structures. As the isolation membrane becomes spherical (Fig. 8C and Fig. D), its intramembrane space gradually becomes thin and homogenous, and finally a mature autophagosome is formed (Fig. 8A and Fig. B). Our assembly model is distinct from the one that autophagosome is formed by enclosing the cytosol with preexisting membrane cisterna, such as ER or Golgi body. Autophagosomal membrane shows a unique feature morphologically distinct from well-known organelles (Baba et al. 1994, Baba et al. 1995). It is observed as a thinner membrane and has a much lower density of intramembrane particles in freeze-fracture images than the membrane of other organelles. These features may be derived from the nature of the precursor structures. The dot structures observed under growing condition may be the same to them and to one of the sources of autophagosomal membrane.
Immuno-EM analysis showed that Apg8p was enriched on the cytoplasmic faces of premature autophagosomes and intermediate structures (Fig. 8C and Fig. D, and Fig. 9). These data indicate that Apg8p bound to membrane plays its role in the formation of autophagosome. In mature autophagosomes, Apg8p was observed less than in those intermediate structures (Fig. 8A and Fig. B). In some typical autophagosomes, it was detected more readily in the lumen than on the membrane (Fig. 8 B). In the autophagic bodies, most Apg8p was detected in the lumen (Fig. 7). Furthermore, it was scarcely detected on the vacuolar membrane to which the outer membrane of the autophagosome had fused (Fig. 7). These results suggest that Apg8p starts to dissociate from the membrane of autophagosome after finishing its role. This may be a reason why more Apg8p was detected in HSS fraction during starvation (Fig. 4 A). Apg8p dissociated from the inner membrane would be entrapped in the lumen of autophagosome and transported to the vacuole, and Apg8p detached from the outer membrane may be recycled for the next autophagosome formation. Since Apg8p is not an integral membrane protein, it would be able to attach and detach from membrane. We found that some portion of Apg8p is tightly bound to membrane (data not shown). However, it is still unclear how Apg8p interacts with membrane structures, particularly autophagosomal membrane. The interaction of Apg8p with membrane is now under investigation.
The apg8 null mutation severely impairs autophagosome formation, leading to the defect in bulk nonselective protein transport and degradation (Fig. 1 and Fig. 10). However, a small amount of mature API was detected during starvation. ProAPI may be transported to the vacuole via structures rarely observed in Δapg8 cells (Fig. 10, C–E). Among them, there are the structures indistinguishable from autophagosome (Fig. 10 C), suggesting that a small number of autophagosomes are built up in the absence of Apg8p. Thus, Apg8p may modulate the efficiency of autophagosome formation. In addition, we found the starvation-induced structures showing abnormal morphology in the Δapg8 cells (Fig. 10D and Fig. E). Alternatively, Apg8p might regulate the morphogenesis of autophagosome. In any case, Apg8p would play an important role in the assembly of the precursor structures into autophagosomal membrane.
The Cvt pathway proceeds with topologically the same membrane dynamics to the autophagy (Baba et al. 1997; Scott et al. 1997). Proteinase K is accessible to proAPI without detergent in the growing cvt5/apg8 mutant cells (Harding et al. 1995), indicating that Apg8p also is required for the formation of Cvt vesicle. Since Apg8p is localized on autophagosomes, it would be reasonable to speculate that it is localized on the Cvt vesicles also. In the growing cells, small punctate signals of 3 × HA–Apg8p may represent the Cvt vesicles (Fig. 5 A; arrows). Between the two pathways, the most apparent difference is the size of vesicles. The autophagosomes are 300–900-nm in diam, while the Cvt vesicles are 140–160-nm (Baba et al. 1994, Baba et al. 1997). Surface area of autophagosomal membrane is calculated at ∼16-fold of Cvt vesicle, on average. Therefore, the autophagosome formation would require more Apg8p than the Cvt vesicle formation. Moreover, a significant amount of Apg8p is transported to the vacuole upon autophagy during starvation. These may be the reasons why Apg8p increases during starvation. However, it is still an open question whether the increase of Apg8p is actually necessary for the autophagy. At least, it is clear that the increase of Apg8p is not sufficient for the induction of autophagy, because overexpression of Apg8p did not induce autophagy under growing condition (data not shown). In addition, we showed that Apg8p is transcriptionally upregulated by inactivation of Tor signaling cascade. We found STRE-like sequences in the promoter region of APG8. STREs are found in the genes induced in response to various stresses, such as CTT1, encoding cytosolic catalase T, and play roles as positive control element (Belazzi et al. 1991; Schuller et al. 1994; Moskvina et al. 1998). Marchler et al. 1993 reported the mutation in the STREs of CTT1 (CTT1-23), which causes loss of induction activity. The corresponding mutation was introduced in the STRE-like sequences of APG8. However, it led to the increase of Apg8p at growing phase instead of the loss of the induction during starvation (data not shown), suggesting that a negative regulator is bound to the cis-elements to repress the expression under nutrient rich condition.
Apg8p is the first identified molecule that is localized on the autophagy-related membrane structures. Now it becomes a useful marker for further analysis on the whole process of membrane dynamics in autophagy. Primary structures of Apg8p are highly conserved among homologues in other organisms (Lang et al. 1998). We anticipate that some Apg8p homologues may function in the process of formation of autophagosome in higher eukaryotes. Further study of Apg8p will provide a breakthrough for elucidating the molecular mechanism of autophagy.
Acknowledgments
We are grateful to Dr. Masako Osumi for the use of EM facilities, Dr. Klionsky for the gift of anti-API antibody, and Dr Mizushima for the gift of the ypt7 null mutant.
This work was supported in part by Grants-in-Aid for the Ministry of Education, Science, and Culture of Japan.
Footnotes
1.used in this paper: ALP, alkaline phosphatase; API, aminopeptidase I; HA, hemagglutinin; ORF, open reading frame; proAPI, precursor form of aminopeptidase I
References
- Baba M., Takeshige K., Baba N., Ohsumi Y. Ultrastructural analysis of the autophagic process in yeastdetection of autophagosomes and their characterization. J. Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi M., Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S.V., Klionsky D.J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belazzi T., Wagner A., Wieser R., Schanz M., Adam G., Hartig A., Ruis H. Negative regulation of transcription of the Saccharomyces cerevisiae catalase T (CTT1) gene by cAMP is mediated by a positive control element. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:585–592. doi: 10.1002/j.1460-2075.1991.tb07985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Rieder S.E., Emr S.D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr. Studies on the mechanisms of autophagyformation of the autophagic vacuole. J. Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura A., Noda T., Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae . Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T.M., Morano K.A., Scott S.V., Klionsky D.J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky B.F., Emr S.D. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 1993;268:4953–4962. [PubMed] [Google Scholar]
- Iwai K., Fukuoka S., Fushiki T., Kido K., Sengoku Y., Semba T. Preparation of a verifiable peptide–protein immunogendirection-controlled conjugation of a synthetic fragment of the monitor peptide with myoglobin and application for sequence analysis. Anal. Biochem. 1988;171:277–282. doi: 10.1016/0003-2697(88)90486-1. [DOI] [PubMed] [Google Scholar]
- Kametaka S., Matsuura A., Wada Y., Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–143. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae . J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kim J., Dalton V.M., Eggerton K.P., Scott S.V., Klionsky D.J. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopitz J., Kisen G.O., Gordon P.B., Bohley P., Seglen P.O. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J. Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Schaeffeler E., Bernreuther D., Bredschneider M., Wolf D.H., Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S.S., Hammarback J.A. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- Marchler G., Schuller C., Adam G., Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae . Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Ohsumi Y. Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina E., Schuller C., Maurer C.T., Mager W.H., Ruis H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast. 1998;14:1041–1050. doi: 10.1002/(SICI)1097-0061(199808)14:11<1041::AID-YEA296>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Hirata A., Ohsumi Y., Wada Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae . J. Biol. Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Hirata A., Nakano A. Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol. Biol. Cell. 1994;5:1129–1143. doi: 10.1091/mbc.5.10.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae . Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Sato T.K., Darsow T., Emr S.D. Vam7p, a SNAP-25–like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Biol. Cell. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmoller F., Riezman H. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J. Cell Sci. 1993;106:823–830. doi: 10.1242/jcs.106.3.823. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M., Schaeffeler E., Straub M., Bredschneider M., Wolf D.H., Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae . J. Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller C., Brewster J.L., Alexander M.R., Gustin M.C., Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Hefner-Gravink A., Morano K.A., Noda T., Ohsumi Y., Klionsky D.J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Baba M., Ohsumi Y., Klionsky D.J. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O., Gordon P.B., Holen I. Non-selective autophagy. Semin. Cell Biol. 1990;1:441–448. [PubMed] [Google Scholar]
- Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1999;In press doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama K., Noda T., Ohsumi Y. Mutational analysis of Csc1/Vps4pinvolvement of endosome in regulation of autophagy in yeast. Cell Struct. Funct. 1997;22:501–509. doi: 10.1247/csf.22.501. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon F. Analyses of the cytoskeleton in Saccharomyces cerevisiae . Annu. Rev. Cell Biol. 1991;7:633–662. doi: 10.1146/annurev.cb.07.110191.003221. [DOI] [PubMed] [Google Scholar]
- Straub M., Bredschneider M., Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae . J. Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. Apg7p/Cvt2pa novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Hall M.N. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae . FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Masaki R., Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry J. Histochem. Cytochem. 38 1990. 573 580a [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Masaki R., Fukui Y., Tashiro Y. Absence of cytochrome P-450 and presence of autolysosomal membrane antigens on the isolation membranes and autophagosomal membranes in rat hepatocytes J. Histochem. Cytochem. 38 1990. 1571 1581b [DOI] [PubMed] [Google Scholar]