Abstract
We discovered a nuclear import pathway mediated by the product of the previously identified Saccharomyces cerevisiae gene PDR6 (pleiotropic drug resistance). This gene product functions as a karyopherin (Kap) for nuclear import. Consistent with previously proposed nomenclature, we have renamed this gene KAP122. Kap122p was localized both to the cytoplasm and the nucleus. As a prominent import substrate of Kap122p, we identified the complex of the large and small subunit (Toa1p and Toa2p, respectively) of the general transcription factor IIA (TFIIA). Recombinant GST-Kap122p formed a complex with recombinant His6-Toa1p/Toa2p. In wild-type cells, Toa1p and Toa2p were localized to the nucleus. Consistent with Kap122p being the principal Kap for import of the Toa1p–Toa2p complex, we found that deletion of KAP122 results in increased cytoplasmic localization of both Toa1p and Toa2p. Deletion of KAP122 is not lethal, although deletion of TOA1 and TOA2 is. Together these data suggest that Kap122p is the major Kap for the import of Toa1p–Toa2p into the nucleus. Like other substrate–Kap complexes, the Toa1p/Toa2p/Kap122p complex isolated from yeast cytosol or reconstituted from recombinant proteins, was dissociated by RanGTP but not RanGDP. Kap122p bound to nucleoporins, specifically, to the peptide repeat–containing fragments of Nup1p and Nup2p.
Keywords: yeast, karyopherin β family, nuclear import, transcription factor IIA, RanGTP dissociation
The karyopherins (Kaps)1 (also termed importins, exportins, or transportins) are a structurally related family of proteins that function in transporting proteins, nucleic acids, and nucleoproteins into and out of the nucleus (for reviews see Pemberton et al. 1998; Wozniak et al. 1998). Comparative sequence analysis of the yeast genome indicated that this organism may have as many as 14 members of the karyopherin β family but only one representative of the karyopherin α family. Not all members of the Kap β family have been definitively shown to function as such (Pemberton et al. 1999; for reviews see Mattaj and Englmeier 1998; Pemberton et al. 1998; Wozniak et al. 1998). Each of the Kap β family members binds to a cognate signal in a transport substrate, and then docks the resulting complex to a subset of nucleoporins (Nups, collective term for nuclear pore complex [NPC] proteins). In contrast, Kap α functions as an adapter that binds to a classical nuclear localization sequence (NLS) (Conti et al. 1998) and to a member of the Kap β family, termed Kap95p or Kap β1. The small GTPase Ran and factors that regulate the GDP- or GTP-bound form of Ran are involved in transport across the NPC (Melchior et al. 1993; Moore and Blobel 1993; Rexach and Blobel 1995; Schwoebel et al. 1998; Englmeier et al. 1999; Ribbeck et al. 1999). Although several reactions that are likely to be relevant for transport have been reconstituted in vitro, the sequence of reactions leading to import or export of substrates remains to be elucidated.
The members of the Kap β family that have been identified so far function either in import or in export (for reviews see Pemberton et al. 1998; Wozniak et al. 1998), although it has not yet been excluded that a given Kap could function in both processes. The directionality of transport appears to depend on at least two nucleocytoplasmic asymmetries. First, the localization of RanGAP (GTPase-activating protein) in the cytoplasm and of RanGEF (GDP/GTP exchange factor) in the nucleus (for reviews see Corbett and Silver 1997; Moore 1998) is likely to yield a high ratio of RanGTP/RanGDP in the nucleus and a high ratio of RanGDP/RanGTP in the cytoplasm (Izaurralde et al. 1997; for review see Melchior and Gerace 1998). Second, certain nucleoporins that serve as docking sites for Kaps are asymmetrically located on either the cytoplasmic or the nucleoplasmic fibers of the NPC (Yang et al. 1998; for review see Ohno et al. 1998). Structural asymmetry of the NPC is a likely determinant for the directionality of transport, as certain Kaps appear to interact preferentially with certain Nups (Fornerod et al. 1997; Marelli et al. 1998).
All members of the Kap β family show a low level of sequence homology to each other (Görlich et al. 1997). Those that have been characterized so far can function without an adapter and bind directly to both a substrate and to a Nup (Aitchison et al. 1996; Pemberton et al. 1997; Rosenblum et al. 1997; Rout et al. 1997; Albertini et al. 1998; Senger et al. 1998). Even Kap β1 can bind to certain substrates directly, without the Kap α adapter (Jakel and Görlich 1998; Moore et al. 1999; Palmeri and Malim 1999; Truant and Cullen 1999). Several of the identified Kaps are not essential for viability, although often they transport substrates that are essential for viability (Rout et al. 1997; Schlenstedt et al. 1997; Pemberton et al. 1999). The explanation for this apparent paradox lies in overlapping substrate specificities, i.e., a given substrate can be transported by more than one Kap.
Here, we have characterized a putative Kap β homologue, Pdr6p. PDR6 was previously classified as a member of a gene family that is involved in pleiotropic drug resistance. Overexpression of PDR6's wild-type allele conferred sensitivity to cycloheximide, borrelidin, and hygromycin B in some drug resistant pdr1 mutants (Balzi et al. 1987; Chen et al. 1991). Pdr1p is a zinc finger–containing transcription factor that regulates the expression of several ATP binding cassette transporter–encoding genes, including PDR5, SNQ2, and YOR1 (Mahe et al. 1996). How Pdr6p overexpression suppresses mutations in the transcription factor Pdr1p is not known. Pdr6p is not essential for growth (Chen et al. 1991) and no function has yet been assigned to Pdr6p. Here, we show that Pdr6p functions as a Kap. In agreement with previously proposed nomenclature we, therefore, designated it Kap122p (because of Pdr6p's calculated M r of 123,529 D, it might have been designated either Kap123p or Kap124p; however, these terms had already been assigned to two other Kaps [for review see Pemberton et al. 1998]; therefore, we propose the name Kap122p for Pdr6p, knowing that the molecular masses of other putative Kap βs are not in this range). We have identified a complex of the large subunit (Toa1p) and the small subunit (Toa2p) of the general transcription factor IIA (TFIIA) as import substrate for Kap122p.
Materials and Methods
Yeast Strains and Methods
All strains used were derived from Saccharomyces cerevisiae wild-type DF5α (Finley et al. 1987) and its derivative kap122Δ (MATα, lys2-801, leu2-3, 112, ura3-52, his3-200, trp1-1 (am), pdr6::URA3). Yeast strains were grown at 30°C in yeast extract/peptone/glucose (YPD); all yeast manipulation was performed according to described protocols (Sherman et al. 1986).
Gene Replacement and Protein A Fusion Constructs
For deletion of PDR6 in wild-type strain DF5α, the HIS3 gene was used as a selective marker to replace the PDR6 open reading frame by integrative transformation in a haploid DF5α strain. HIS3 replacement cassette was generated by PCR amplification of markers from pRS 306 with primers that contained 60 nucleotides flanking the PDR6/KAP122 open reading frame from 5′ and 3′ ends (Aitchison et al. 1995). HIS3 marker was switched to URA3 marker by recombination (plasmids were gifts of Dr. F.R. Cross, Rockefeller University). Deletion of genes was confirmed by PCR on total yeast DNA with internal primers.
Carboxy-terminal genomic KAP122-PrA fusion constructs were created by integrative transformation of PCR-amplified constructs with four and a half IgG-binding domains of protein A immediately upstream of PDR6/KAP122 stop codon, followed by a not-in-frame HIS5 (Schizosaccharomyces pombe) selection marker (donated by Dr. R. Beckmann, Rockefeller University). TOA1 and TOA2 protein A tagging was similar to that of KAP122. Primers used for PCR amplification of protein A/HIS5 cassette contained 60 nucleotides directly upstream of the stop codon of the relevant gene for the 5′ primer and 60 nucleotides 150 bp downstream of the stop codon for the 3′ primer. Haploid yeast cells were transformed by electroporation. This resulted in expression of the chimeric protein A fusion construct under the control of the endogenous promoter.
Cell Fractionation and Immunoisolation
Fractionation and immunoisolation of protein A fusion proteins were performed as described (Aitchison et al. 1996). For a typical isolation, 500 ml of postribosomal supernatant (cytosol) was prepared from a 6-liter YPD culture with a density of 1.7 at A600. Cytosol was incubated with 200 μl of rabbit IgG-Sepharose beads at 4°C overnight. After washing with transport buffer (TB: 20 mM Hepes-KOH, pH 7.5, 110 mM KOAc, 2 mM MgCl2, 1 mM DTT, 0.1% Tween 20), bound proteins were eluted with a step gradient of MgCl2 from 50 to 4,500 mM. Proteins were precipitated, resolved by SDS-PAGE on a 4–20% acrylamide gel (Novex), and stained with Coomassie blue. Proteins of interest were excised and prepared for MALDI-TOF spectrometry and/or sequencing.
Immunofluorescence Microscopy
Yeast cells were fixed in 3.7% formaldehyde for 15 min and cell walls were digested. Indirect immunofluorescence was carried out according to published protocols (Wente et al. 1992). Protein A moieties of fusion proteins were detected with rabbit IgG that had been preadsorbed to wild-type yeast spheroplasts; Nab2p was detected by rabbit polyclonal antiserum to Nab2p (Aitchison et al. 1996), Npl3p was detected by mouse monoclonal antiserum to Npl3p (Wilson et al. 1994), the appropriate Cy3-conjugated anti–rabbit or anti–mouse IgG was used for visualization. Nuclei were visualized with the DNA binding stain 4′,6-diamidino-2-phenylindole (DAPI).
Blot Overlay Assay
A Nup1p fragment containing the FXFG repeat region (amino acids 432–816) and a Nup2p fragment containing the FXFG repeat region (amino acid 186–561) were expressed as glutathione S-transferase (GST) fusion proteins as described (Enenkel et al. 1995; Rexach and Blobel 1995). Proteins of bacterial lysates were separated by SDS-PAGE and transferred to nitrocellulose. Overlay assays were performed as described (Aitchison et al. 1996). Yeast cytosol from the Kap122p-PrA–expressing strain was diluted 1:1 with TB-5% milk and incubated on the blot overnight at 4°C. Bound proteins were detected with rabbit antibodies to mouse IgG and HRP-conjugated donkey anti–rabbit antibodies and enhanced chemiluminescence.
Recombinant Protein Expression
TOA1, TOA2, and KAP122 open reading frames were amplified from yeast genomic DNA by PCR using synthetic oligonucleotide primers with incorporated restriction sites for subcloning into relevant Escherichia coli expression vectors.
TOA1 was subcloned into NdeI and BamHI sites of kanamycin resistance–conferring pET 28a (Novagen, Inc.) to allow expression of Toa1p with an amino-terminal His tag. TOA2 was subcloned into NcoI-BamHI sites of ampicillin resistance–conferring pET 19b (Novagen, Inc.) to allow coexpression of Toa2p with Toa1p.
E. coli strain BL21 gold (Stratagene) was cotransformed with both the His6-Toa1p– and the Toa2p-expressing plasmids, and E. coli cells coexpressing both subunits of TFIIA were grown at 37°C in LB medium containing 60 μg/ml kanamycin and 100 μg/ml ampicillin to a density of 0.7 at A600. Protein expression was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 10 h at 17°C. E. coli were lysed in TB with added protease inhibitor cocktails (Boehringer Mannheim) using the French pressure cell. The bacterial lysate was incubated with Talon™ resin (CLONTECH Laboratories) for 30 min at room temperature (RT). Talon™ beads were washed with lysis buffer (TB-0.1% Tween 20) and TB buffer containing 2 mM ATP, 10 mM Mg(OAc)2, and 10 mM imidazole. For protein binding assays, the His6-Toa1p/Toa2p complex was used either still bound to Talon™ beads or was eluted with TB containing 80 mM imidazole.
KAP122 was subcloned into BamHI-EcoRI sites of the derivative of pGeX4T3 (Pharmacia Biotech) with additional Tev protease cleavage site (Chook, Y.M., and G. Blobel, 1999), to allow expression of Kap122p with a potentially cleavable amino-terminal GST tag. Kap122p-expressing E. coli cells were grown in LB medium containing 100 μg/ml ampicillin to a density of 0.7 at A600 and GST-Kap122p expression was induced by 1 mM IPTG for 3 h at 30°C. GST-Kap122p fusion protein was purified from bacterial lysates according to the manufacturer's protocol (Pharmacia Biotech).
Solution Binding Assays
Interaction of Immobilized GST-Kap122p with Purified Recombinant Toa1p–Toa2p.
Binding assays were performed in TB with the addition of 10% glycerol and 0.1% casaminoacids (Invitrogen Corp.) to prevent nonspecific interactions. 10 μl of glutathione-Sepharose beads containing either 2 μg immobilized GST-Kap122p or 2 μg immobilized GST were incubated with 5 μg of purified His6-Toa1p/Toa2p complex in a total volume of 100 μl for 1 h at RT. Beads were collected by centrifugation at 1,000 g for 30 s, washed five times by mixing with 1 ml of TB followed by sedimentation, and were resuspended in 15 μl of sample buffer. Proteins in one half of each sample were resolved by SDS-PAGE on a 4–20% acrylamide gel (Novex, Inc.) and stained with Coomassie blue.
Interaction of Immobilized His6-Toa1p/Toa2p with Kap122p.
200 μl of GST-Kap122p E. coli lysate was incubated with 10 μl Talon™ resin at RT for 1 h to deplete the lysate of endogenous bacterial proteins that interact nonspecifically with Talon™ beads. Beads were sedimented at 1,000 g for 30 s, and the lysate was passed through a Micro Bio-Spin® chromatography column (Bio-Rad Laboratories) to exclude any remaining Talon™ beads. 50 μl of TB containing 50% glycerol and 0.5% casaminoacids was added to 200 μl of precleared lysate and the mixture was incubated for 1 h at RT with 2 μg of His6-Toa1p/Toa2p complex immobilized on 10 μl of Talon™ beads or with 10 μl of empty Talon™ beads as a control. At the end of incubation, beads were sedimented and washed five times with 1 ml of TB. Talon™ beads with bound proteins were resuspended in sample buffer and half of each sample was resolved by SDS-PAGE on a 4–20% acrylamide gel (Novex, Inc.) and stained with Coomassie blue.
In Vitro Dissociation of the Isolated Yeast Kap122p/Toa1p/Toa2p Complex by RanGTP
Recombinant Saccharomyces cerevisiae Ran was prepared and loaded with GDP or GTP as described (Floer and Blobel 1996). Protein A–tagged Toa2p with bound Kap122p was immunoisolated from a postribosomal supernatant as described above. After washing, IgG-Sepharose beads were resuspended in TB and divided into several equal fractions. These fractions were incubated either with TB alone, or with 4 μM RanGDP or RanGTP for 60 min at room temperature. 1 mM GTP was included, except when using RanGDP. Beads were transferred to a column, and the drained liquid, together with a subsequent 100-μl TB wash, was collected; this constituted the released material. After washing columns with TB, remaining bound proteins were eluted with 1,000 and 4,500 mM MgCl2 step gradient. Fractions eluted at 1,000 and 4,500 mM were combined and comprised the bound material. Proteins were precipitated, resolved by SDS-PAGE, and stained with Coomassie blue.
In Vitro Dissociation of the Recombinant Kap122p/Toa1p/Toa2p Complex by RanGTP
15 μl of Talon™ beads with 3 μg of immobilized His6-Toa1p/Toa2p complex were washed five times with 1 ml of TB, incubated with precleared GST-Kap122p lysate, as described above, to obtain a GST-Kap122p/His6-Toa1p/Toa2p complex. Three equal aliquots of beads were incubated with either 4 μM RanGTP, 4 μM RanGDP, or TB alone for 2 h at RT in a final volume of 40 μl. 1 mM GTP was included, except when using RanGDP. Beads were sedimented by centrifugation and 30 μl of supernatant were collected and passed through Micro Bio-Spin® chromatography column (Bio-Rad Laboratories) to exclude any contaminating Talon™ beads. The drained liquid represented the released material. Beads were washed five times with 1 ml of TB and resuspended in 15 μl of sample buffer. Proteins in one half of each bound and released sample were resolved by SDS-PAGE on 4–20% acrylamide gel (Novex, Inc.) and stained with Coomassie blue.
Results
In all cases described here, we found that the genomic replacement of a gene by its protein A–tagged version did not result in any apparent changes in growth rates under the conditions used in our experiments and when compared with those of an otherwise isogenic strain. This indicated that PrA tagging did not apparently interfere with the function of the essential proteins that were tested here; however, this cannot be confirmed for the Kap122p-PrA strain because of the absence of a discernible phenotype of the KAP122 deletion strain.
Immunolocalization of Kap122p-PrA
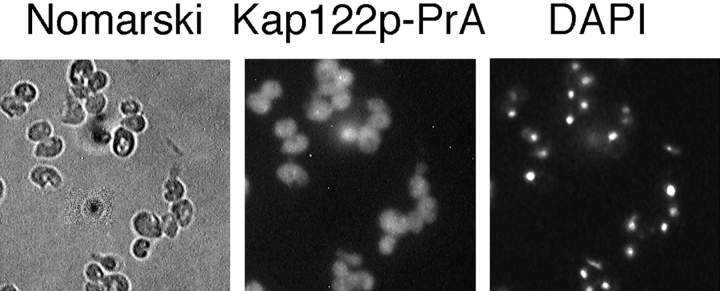
Immunofluorescence microscopy of a KAP122-PrA haploid strain showed that Kap122p-PrA is located both in the cytoplasm and the nucleus (Fig. 1), which is consistent with the localization of other protein A–tagged Kaps and their function in shuttling between the nucleus and the cytoplasm.
Figure 1.
Kap122p is diffusely located in both the cytoplasm and the nucleus. A haploid strain where endogenous Kap122p was replaced by protein A–tagged Kap122p (KAP122-PrA) was examined by Nomarski (left), by indirect immunofluorescence of the protein A tag (middle), and by staining of the DNA with DAPI (right).
Interaction of Kap122p with Repeat-containing Fragments of Nucleoporins Nup1p and Nup2p
Karyopherins have been previously shown to interact with peptide repeat–containing nucleoporins (Rexach and Blobel 1995; Aitchison et al. 1996; Fornerod et al. 1997; Rout et al. 1997; Pemberton et al. 1997; Rosenblum et al. 1997; Albertini et al. 1998; Marelli et al. 1998). To test whether Kap122p interacts with nucleoporins, we expressed FXFG repeat–containing fragments of the nucleoporins Nup1p and Nup2p in E. coli, separated proteins of bacterial lysates by SDS-PAGE, transferred them to nitrocellulose, and performed overlay assays with Kap122p-PrA cytosol. Kap122p-PrA interacted strongly and specifically both with Nup1p and Nup2p fragments (Fig. 2). This strongly suggests that Nup1p and Nup2p are among the nucleoporins that bind to Kap122p.
Figure 2.
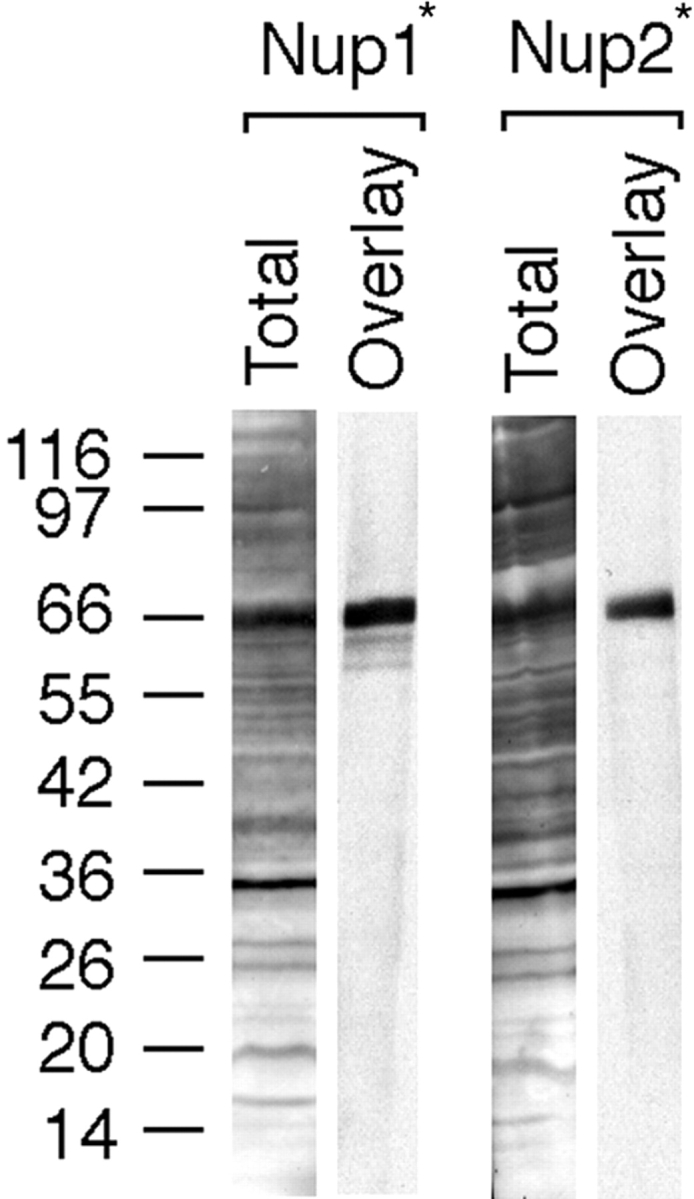
Kap122p interacts with FXFG repeat–containing nucleoporins Nup1p and Nup2p. Overlay assay demonstrates specific binding of Kap122-PrA to Nup1p and Nup2p repeat–containing fragments expressed in E. coli (Nup1* and Nup2*). Total bacterial lysates transferred to nitrocellulose were incubated with crude yeast cytosol from Kap122p-PrA strains. Bound proteins were detected via the PrA moiety (Overlay). Amido black-stained nitrocellulose strips with whole bacterial lysates from Nup1p- and Nup2p-expressing E. coli, respectively (Total).
Identification of Proteins Interacting with Kap122p-PrA
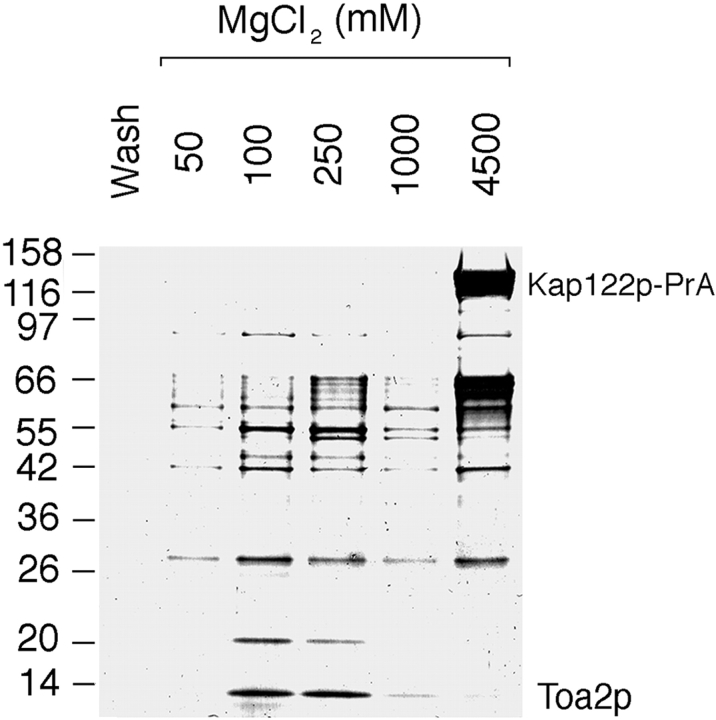
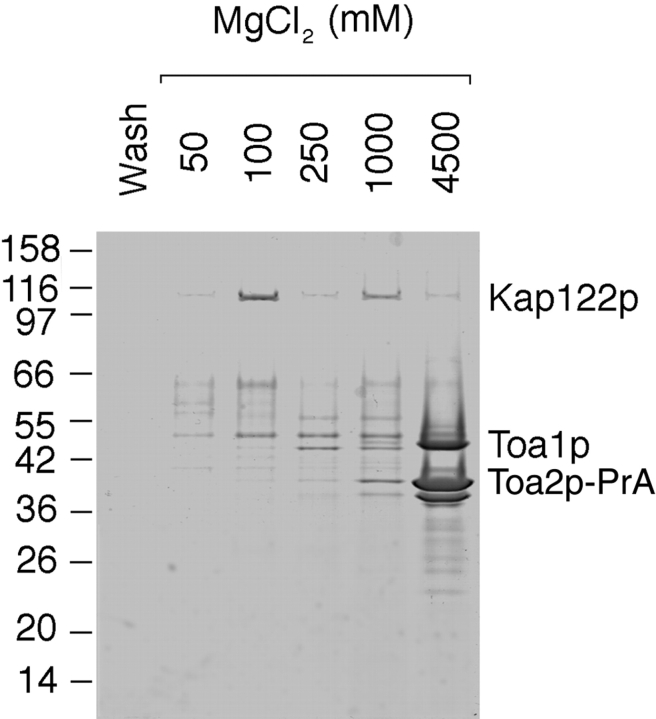
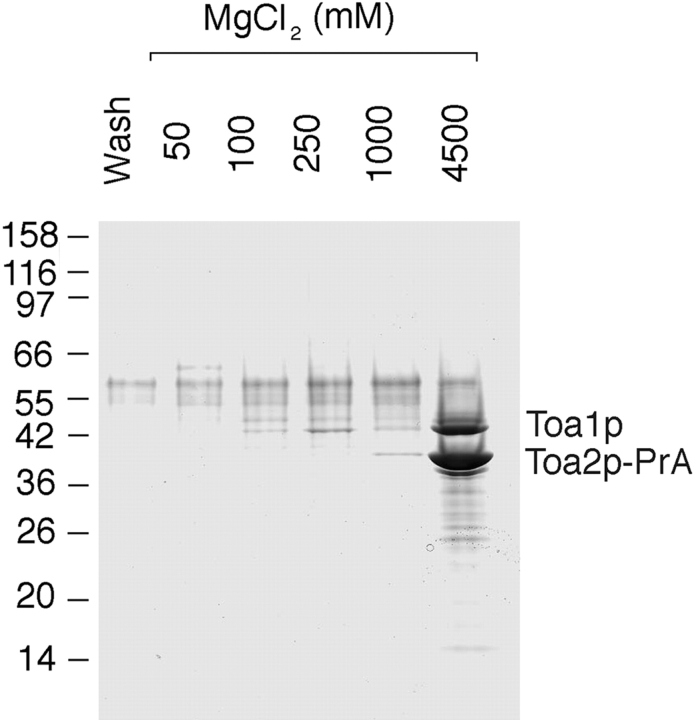
Kaps that function in protein import form stable complexes with their import substrates in the cytoplasm (Aitchison et al. 1996; Pemberton et al. 1997; Rosenblum et al. 1997; Rout et al. 1997; Albertini et al. 1998). To isolate such complexes, we prepared a postribosomal cytosol fraction from the KAP122-PrA haploid strain and incubated it with IgG-Sepharose. Bound proteins were eluted with a step gradient of MgCl2, separated by SDS-PAGE, and stained with Coomassie blue (Fig. 3). The Kap-bound substrates usually elute at MgCl2 concentration between 100–1,000 mM MgCl2, whereas the PrA-tagged Kap elutes at 4.5 M MgCl2 (Aitchison et al. 1996). Indeed, a major band with an apparent M r of 150 kD, which likely represented Kap122-PrA, eluted at 4.5 M MgCl2. The numerous minor bands in this fraction, as well as in fractions eluting at lower MgCl2 concentrations, are likely degradation products of Kap122-PrA retaining their PrA moiety, as confirmed by immunoblotting with rabbit IgG (data not shown). A number of proteins were present primarily in the 100- and 250-mM MgCl2 eluates and these are putative substrates for Kap122p. One of these bands migrating at ∼14 kD was excised, analyzed by mass spectrometry and peptide microsequencing after digestion with trypsin (Gharahdaghi et al. 1996; Fernandez et al. 1998), and thereby identified as the small subunit (Toa2p) of the general transcription factor IIA (TFIIA). These data suggested that Toa2p might be an import substrate for Kap122p.
Figure 3.
Immunoisolation of cytosolic proteins interacting with Kap122p-PrA. Cytosol from a haploid KAP122-PrA strain was incubated with IgG-Sepharose. The last wash fraction and fractions subsequently eluted with a step gradient of MgCl2 were analyzed by SDS-PAGE and Coomassie blue staining. Proteins eluting in the 100- and 250-mM MgCl2 fractions are potential binding partners for Kap122p. One of these bands, migrating like a protein of ∼14 kD, was identified by mass spectrometry as the small subunit (Toa2p) of the TFIIA. The major band eluting at 4.5 M MgCl2 is Kap122p-PrA. Relative molecular mass standards are indicated on the left.
Defective Nuclear Import of Toa2p-PrA in a kap122Δ Strain
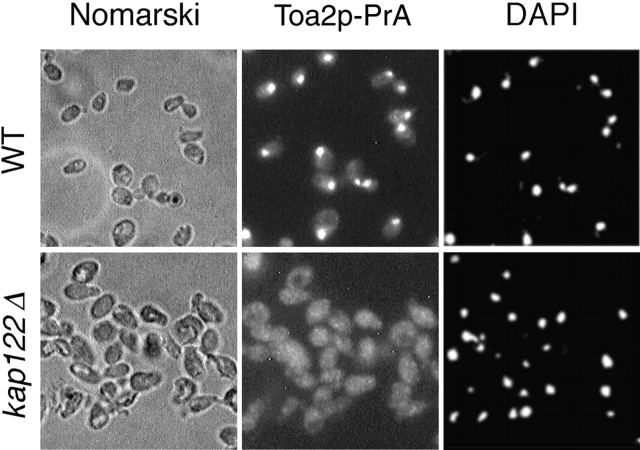
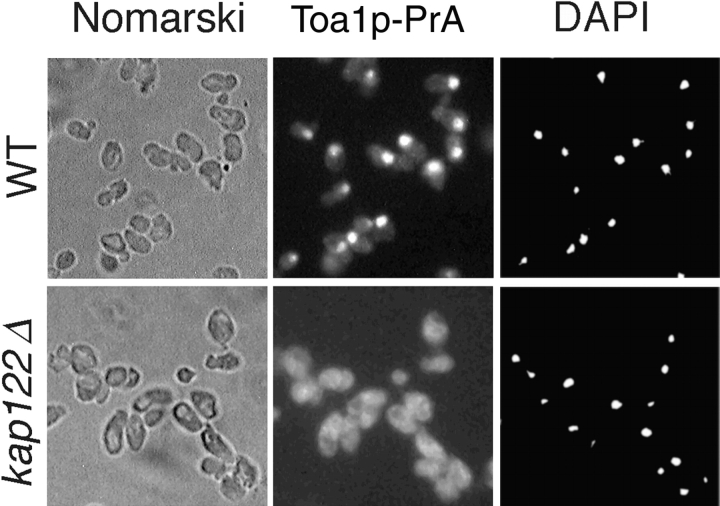
To determine whether Toa2p is indeed a transport substrate for Kap122p, we examined the localization of Toa2p in wild-type (WT) and KAP122 deletion (kap122Δ) strains. There were no apparent differences in growth rates between kap122Δ and an isogenic wt strain in YPD medium at 30°C. For these experiments, TOA2 was genomically tagged with PrA in isogenic wt and kap122Δ strains. Immunofluorescence microscopy showed that in wild-type cells Toa2p-PrA was located primarily in the nucleus, whereas in the kap122Δ cells Toa2p-PrA was mislocalized largely to the cytoplasm (Fig. 4). The diminished nuclear localization of Toa2p-PrA in the kap122Δ strain is consistent with Kap122p representing the principal Kap for import of Toa2p.
Figure 4.
Mislocalization of Toa2p-PrA in a kap122Δ strain. Haploid wild-type or kap122Δ cells, both with a genomic Toa2p-PrA fusion, were visualized by Nomarski (left), or stained by indirect immunofluorescence of the protein A tag (middle) and by DAPI (right). Note the primarily nuclear localization of Toa2p-PrA in wild-type cells and the shift to a diffuse cellular staining in kap122Δ.
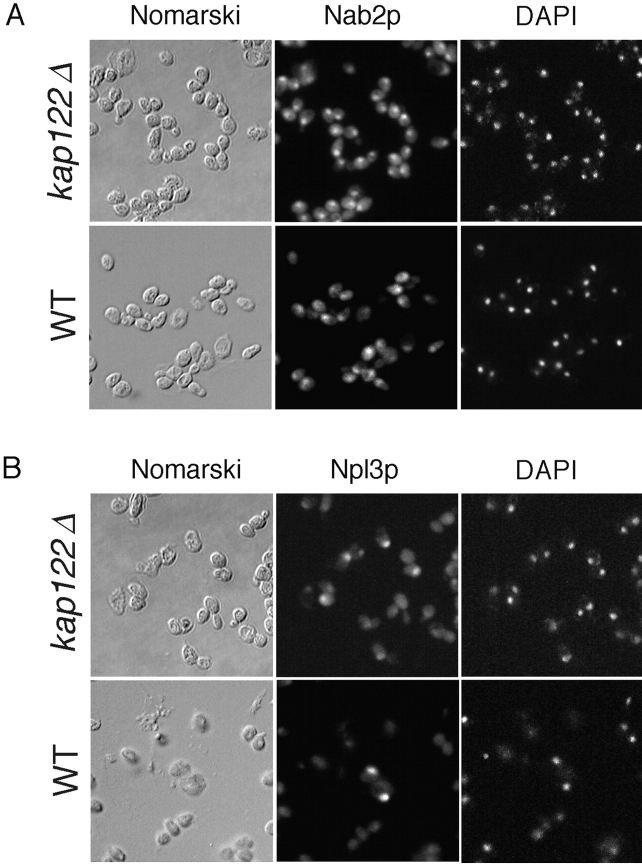
Localization of several other substrates, whose transport is mediated by distinct Kaps, was not affected by KAP122 deletion. The nuclear import of Npl3p, mediated by Kap111p (Pemberton et al. 1997; Senger et al. 1998) and Nab2p, mediated by Kap104p (Aitchison et al. 1996), appeared similar in both wild-type and kap122Δ strains (Fig. 5), showing that deletion of KAP122 does not generally affect nucleocytoplasmic transport.
Figure 5.
Nab2p and Npl3p nuclear import is not affected by KAP122 deletion. Haploid wild-type or kap122Δ cells were visualized by Nomarski (A and B, left), or stained with polyclonal antibodies to Nab2p (A, middle), or monoclonal Abs to Npl3p (B, middle). Nuclei were visualized with DAPI (A and B, right).
Toa2p and Toa1p Form a Cytoplasmic Complex that Binds to Kap122p
Yeast TFIIA has been shown to consist of two subunits: a small subunit (Toa2p, calculated M r 13.5 kD) and a large subunit (Toa1p, calculated M r 32 kD) that migrates as a 43-kD protein in SDS-PAGE (Hahn et al. 1989; Ranish and Hahn 1991; Ranish et al. 1992). Separately expressed Toa1p and Toa2p are insoluble and unable to complement transcription systems unless both subunits are renatured together (Ranish et al. 1992); this suggests that both subunits of TFIIA are unlikely to exist as separate entities in the cell (Ranish et al. 1992; Geiger et al. 1996). Crystallographic data of TFIIA bound to a DNA–TATA-binding protein (TBP) complex revealed that the two subunits are intertwined with each other to form a heterodimer, and that the heterodimer interacts with TBP and also with the phosphate backbone of DNA (Geiger et al. 1996; Tan et al. 1996). Hence, it is clear that in the nucleus Toa1p and Toa2p interact with the DNA–TBP complex as a heterodimer.
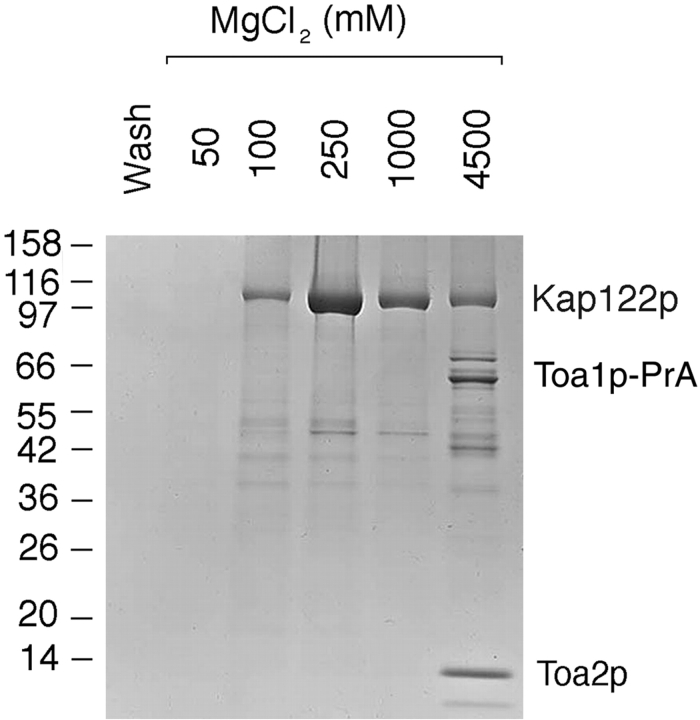
Our data above (Fig. 3) suggested that, in the cytoplasm, Toa2p existed in a complex with Kap122p-PrA and it was possible that Toa1p was part of this complex. As Toa2p and Toa1p bind as a dimer to the DNA–TBP in the nucleus, we investigated whether a Toa1p–Toa2p complex existed also in the cytoplasm. Therefore, we analyzed a postribosomal cytosol from a haploid TOA2-PrA strain by IgG-Sepharose affinity chromatography. A band migrating at ∼110 kD was eluted between 100 and 1,000 mM MgCl2 (Fig. 6), and this band was confirmed to be Kap122p by mass spectrometric analysis. As expected, Toa2p-PrA eluted at 4.5 M MgCl2. However, there was another major band, migrating at ∼45 kD, which coeluted with Toa2p-PrA (Fig. 6). By mass spectrometric analysis this band was identified as Toa1p. The elution of Toa1p at 4.5 M MgCl2 suggested that Toa1p and Toa2p form a tight cytoplasmic complex that interacts with Kap122p.
Figure 6.
Immunoisolation of cytosolic proteins interacting with Toa2p-PrA. Cytosol from a haploid TOA2-PrA strain was incubated with IgG-Sepharose and bound proteins were eluted and analyzed as in Fig. 3. The major band eluting at 100 mM and 1.0 M MgCl2 was identified by mass spectrometry as Kap122p. The band eluting above the Toa2p-PrA band in the 4.5-M MgCl2 eluate was identified by mass spectrometry as Toa1p. Relative molecular mass standards are indicated on the left.
To determine the cytoplasmic binding partners of Toa1p-PrA, we analyzed the postribosomal cytosol of a TOA1-PrA strain by IgG-Sepharose affinity chromatography. A band migrating at ∼110 kD and eluting between 100 and 1,000 mM MgCl2 was identified by mass spectrometric analysis as Kap122p. Toa2p, identified by mass spectrometric analysis, coeluted with the Toa1p-PrA at 4.5 M MgCl2 (Fig. 7), confirming the data in Fig. 6 that these two proteins form a tight cytoplasmic complex that interacts with Kap122p. Numerous degradation products of Toa1p-PrA, retaining their PrA moiety and identified by immunoblotting with rabbit IgG, were also visible in this fraction (data not shown). The apparent sensitivity of Toa1p to proteolysis could explain less than equimolar quantities of Toa1p that were isolated with either Kap122p-PrA (Fig. 3) or with Toa2p-PrA (Fig. 6, see also Fig. 10). Toa1p was similarly sensitive to proteolysis when coexpressed with Toa2p in E. coli (see Fig. 11 and Fig. 12). These smaller fragments of Toa1p that we observed when Toa1p was isolated from yeast or expressed in E. coli, may correspond to p55-related fragments observed when a human homologue of Toa1p was expressed in bacteria (DeJong and Roeder 1993).
Figure 7.
Immunoisolation of cytosolic proteins interacting with Toa1p-PrA. Cytosol from a haploid TOA1-PrA strain was incubated with IgG-Sepharose and bound proteins were eluted and analyzed as in Fig. 3. The major band eluting primarily at 250 mM and 1.0 M MgCl2 was identified by mass spectrometry as Kap122p. Eluting in the 4.5-M MgCl2 fraction is Toa1p-PrA and several lower M r bands that are likely to be degradation products of Toa1p-PrA. The major band migrating as a protein of ∼14 kD was identified by mass spectrometry as Toa2p. Relative molecular mass standards are indicated on the left.
Figure 10.
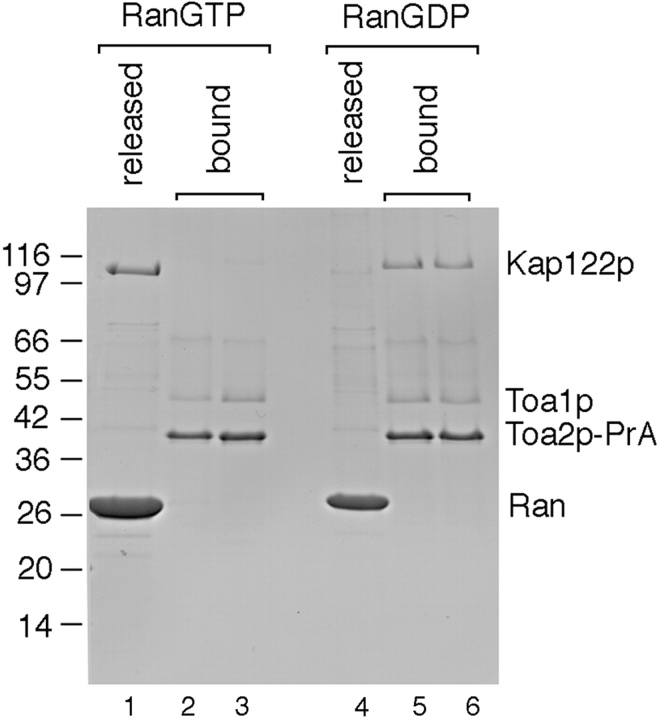
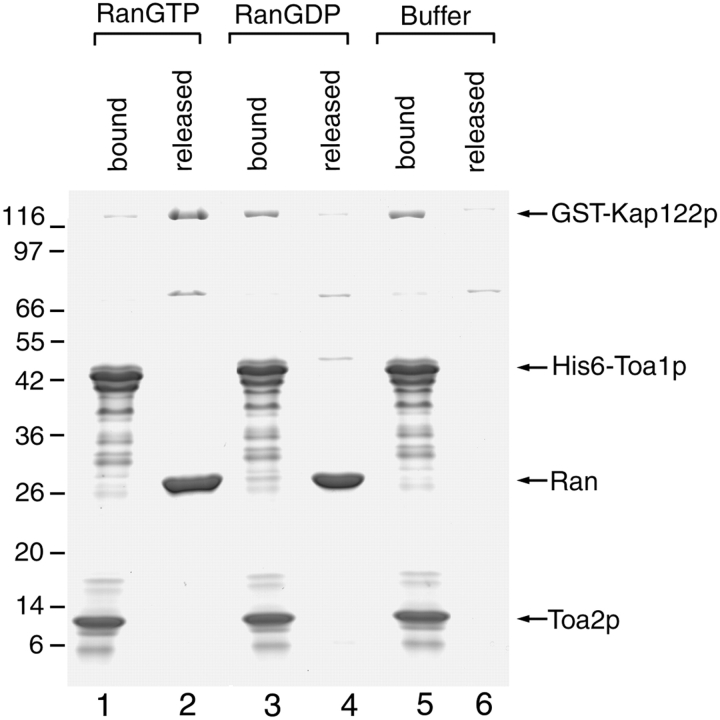
Dissociation of the Toa2p-PrA/Toa1p/Kap122p complex by RanGTP. Cytosol from the haploid TOA2-PrA strain was incubated with IgG-Sepharose. After washing, the IgG-Sepharose was incubated with either RanGTP or RanGDP. After incubation, released proteins were eluted with wash buffer and the remaining bound proteins were eluted with 1.0 and 4.5 M MgCl2. Proteins were separated by SDS-PAGE and stained with Coomassie blue. Relative molecular mass standards are indicated on the left. Note the complete release of Kap122p after incubation with RanGTP (lanes 1–3); RanGDP fails to dissociate Kap122p from Toa2p-PrA (lanes 4–6).
Figure 11.
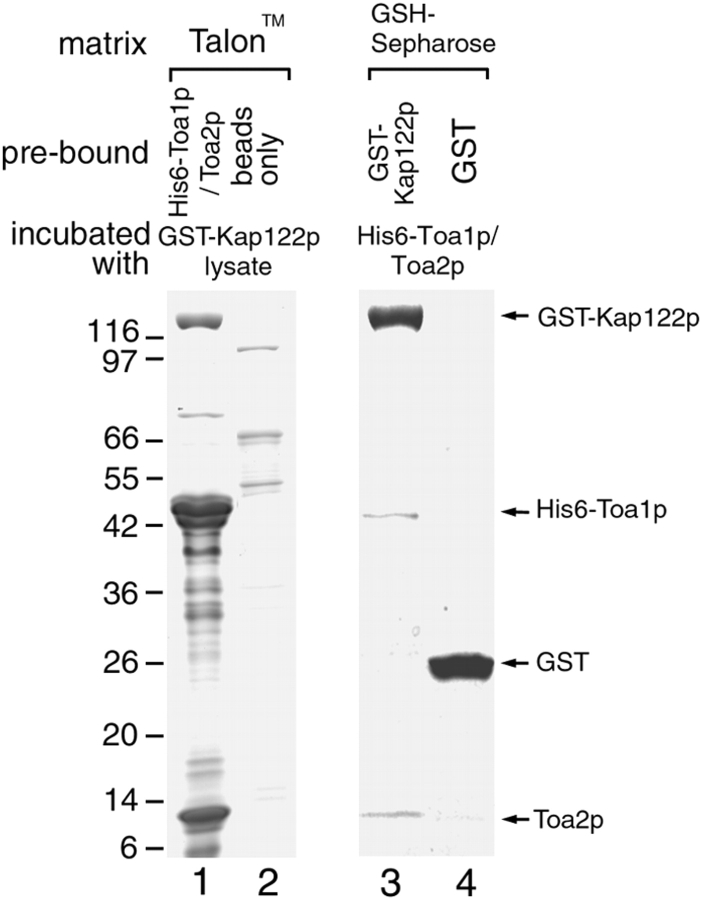
Recombinant Kap122p interacts directly with recombinant Toa1p–Toa2p complex. His6-Toa1p/Toa2p complex, immobilized on Talon™ beads, was incubated with GST-Kap122p present in a bacterial lysate. After extensive washing, bound proteins were eluted in sample buffer, separated by SDS-PAGE and stained with Coomassie blue (lane 1). Incubation of GST-Kap122p-containing E. coli lysate with empty Talon™ beads did not result in any GST-Kap122p binding (lane 2). GST-Kap122p, immobilized on GSH-Sepharose, bound purified His6-Toa1p/Toa2p complex (lane 3). A control with immobilized GST alone showed no binding of His6-Toa1p/Toa2p (lane 4).
Figure 12.
Dissociation of the recombinant GST-Kap122p/His6-Toa1p/Toa2p complex by RanGTP. Immobilized His6-Toa1p/Toa2p complex was incubated with GST-Kap122p E. coli lysate to obtain a GST-Kap122p/His6-Toa1p/Toa2p complex. Three equal aliquots of beads were incubated with either RanGTP, RanGDP, or buffer alone. After incubation, released and still bound proteins were separated by SDS-PAGE and stained with Coomassie blue. Note that RanGTP almost completely dissociated Kap122p from immobilized Toa1p–Toa2p complex (lanes 1 and 2), while Kap122p remained in the complex with Toa1p–Toa2p after incubation with RanGDP (lanes 3 and 4) or buffer alone (lanes 5 and 6).
As expected, immunofluorescence microscopy of Toa1p-PrA in a wt and a kap122Δ strain gave similar results to those obtained for Toa2p-PrA (Fig. 4): the primarily nuclear localization of Toa1p-PrA in the wt strain is diminished in the kap122Δ strain in favor of a diffuse cytoplasmic localization (Fig. 8).
Figure 8.
Mislocalization of Toa1-PrA in a kap122Δ strain. Haploid wild-type or kap122Δ cells, both with a genomic Toa1p-PrA fusion, were visualized by Nomarski (left), or stained by indirect immunofluorescence of the protein A tag (middle) and by DAPI (right). Note the primarily nuclear localization of Toa1-PrA in wt cells and the shift to a diffuse cellular staining in kap122Δ.
Search for Alternative Import Pathways
Toa1p and Toa2p are essential for viability, whereas Kap122p is not. One solution to this apparent paradox is that one or several Kap(s) other than Kap122p can import these proteins into the nucleus in a kap122Δ strain. However, the immunofluorescence data of Fig. 4 and Fig. 8 suggested that if Toa1p–Toa2p were imported by alternative Kaps, this import would be less efficient than that mediated by Kap122p. Nevertheless, to search for alternative Kaps, a cytosol from a kap122Δ/TOA2-PrA strain was analyzed by IgG-Sepharose affinity chromatography (Fig. 9). As expected, Toa2p-PrA was eluted together with Toa1p (and its numerous degradation products) in the 4.5-M MgCl2 fraction (Fig. 9). However, fractions eluted between 100 and 1,000 mM MgCl2 did not show any visible bands in the 100-kD region and above, where Kaps migrate. Hence, if Kaps other than Kap122p can import the Toa1p–Toa2p, they might do so with lower efficiency than Kap122p and, therefore, are likely to be below the limits of detection of this assay.
Figure 9.
Immunoisolation of cytosolic proteins interacting with Toa2p-PrA in a kap122Δ strain. Cytosol from kap122Δ strain containing the Toa2p-PrA fusion was incubated with IgG-Sepharose and bound proteins were eluted and analyzed as in Fig. 3. The major bands in the 4.5-M MgCl2 fraction were Toa2p-PrA and Toa1p, identified by mass spectrometry. The minor bands below the Toa2p-PrA band are likely degradation products of Toa1p. Note that no bands were visible in the 100-kD region of the 250-mM MgCl2 eluate, which would be expected to contain bound Kaps. Relative molecular mass standards are indicated on the left.
Dissociation of Kap122p from Toa2p-Toa1p by RanGTP
Import complexes of substrate–Kap are dissociated by RanGTP (Rexach and Blobel 1995; Schlenstedt et al. 1997; Albertini et al. 1998; Jakel and Gorlich, 1998; Kaffman et al. 1998; Senger et al. 1998). To investigate whether this is also the case for the Toa1p/Toa2p/Kap122p complex, we used postribosomal cytosol from a TOA2-PrA strain to prepare an IgG-Sepharose–bound complex of Toa2p-PrA/Toa1p/Kap122p. This complex was incubated with either transport buffer alone, RanGDP, or RanGTP, and the material that was released from the IgG-Sepharose after completion of incubation was collected. Thereafter, the remaining IgG-Sepharose–bound proteins were eluted at 1.0 and 4.5 M MgCl 2. Proteins were analyzed by SDS-PAGE and Coomassie blue staining (Fig. 10). Incubation with RanGTP clearly led to the release of most of the Kap122p (compare lanes 1–3). In contrast, incubation with RanGDP did not result in dissociation of Kap122p (lanes 4–6). These data indicated that the cytoplasmic Toa2p-PrA/Toa1p/Kap122p complex, like other import substrate–Kap complexes is sensitive to dissociation by RanGTP but not by RanGDP.
Recombinant Kap122p Interacts Directly with Recombinant Toa1p–Toa2p Complex
To determine whether Kap122p is able to interact directly with a Toa1p–Toa2p complex, we coexpressed both His6-Toa1p and Toa2p in E. coli. The TFIIA subunits formed a soluble complex and were purified from bacterial lysate via the His6 affinity tag at the amino terminus of Toa1p. Kap122p with an amino-terminal GST tag was also expressed in E. coli. We tested for binding between GST-Kap122p and the His6-Toa1p/Toa2p complex when either was immobilized on GSH-Sepharose or Talon™ beads, respectively.
We found that purified and immobilized His6-Toa1p/Toa2p complex bound GST-Kap122p present in a preincubated bacterial lysate depleted of its Talon™-binding proteins (Fig. 11, lane 1). Incubation of empty Talon™ resin with E. coli lysate containing GST-Kap122p did not result in any Kap122p binding (Fig. 11, lane 2). Likewise, GST-Kap122p, immobilized on GSH-Sepharose, bound the purified soluble His6-Toa1p/Toa2p complex (Fig. 11, lane 3). A control using immobilized GST alone showed no binding of His6-Toa1p/Toa2p (Fig. 11, lane 4).
These experiments using E. coli-expressed Toa1p–Toa2p and Kap122p show that Kap122p interacts directly with Toa1p–Toa2p complex.
Dissociation of Recombinant Kap122p and Toa1p–Toa2p Complex by RanGTP
We investigated whether the complex of recombinant GST-Kap122p/His6-Toa1p/Toa2p is sensitive to dissociation by RanGTP but not RanGDP. To this end, we prepared a Talon™-bound complex of GST-Kap122p/His6-Toa1p/Toa2p recombinant proteins (Fig. 11, lane 1). Beads were divided into three equal fractions and incubated with RanGTP, RanGDP, or TB alone. At the end of incubation, released material was collected, beads were extensively washed with TB, and bound and released proteins were analyzed by SDS-PAGE and Coomassie blue staining (Fig. 12). Incubation with RanGTP resulted in almost complete release of GST-Kap122p from Talon™-bound His6-Toa1p/Toa2p complex (Fig. 12, lanes 1 and 2). Incubation with RanGDP did not release GST-Kap122p from the Toa1p–Toa2p complex (Fig. 12, lanes 3 and 4), neither did control incubation with TB in the absence of Ran (Fig. 12, lanes 5 and 6). These results confirm the specificity of the interaction between recombinant Kap122p and the recombinant Toa1p–Toa2p complex as well as the sensitivity of this interaction to dissociation by RanGTP.
Discussion
Based on sequence similarity with karyopherin βs, the product of the PDR6 gene of S. cerevisiae was previously suggested to be a member of the Kap β family (for review see Pemberton et al. 1998). In this paper, we report that the hitherto uncharacterized product of the PDR6 gene does indeed function as a Kap and, therefore, named it Kap122p. We show that Kap122p functions in the nuclear import of the complex of large and small subunits, Toa1p, and Toa2p, of TFIIA. The relationship between the observed drug resistant phenotype of PDR6 and the function of Kap122p/Pdr6p in nuclear import of TFIIA (or of other proteins) remains to be elucidated.
We found that Kap122p is localized both in the cytoplasm and the nucleus, which is consistent with its function of shuttling between these two compartments. Cytosolic Kap122p exists as a complex with the small and large subunit of TFIIA. The precise stoichiometry of this complex remains to be determined. Based on biochemical and crystallographic data, it is unlikely that the two subunits exist as separate entities (Ranish et al. 1992; Geiger et al. 1996; Tan et al. 1996). Therefore, one possibility is that Kap122p also functions as a chaperone, and that immediately after synthesis in the cytoplasm, each subunit associates with Kap122p. In this scenario, each subunit would contain a Kap122p cognate NLS. Each of the subunit/Kap122p heterodimers would associate, via interaction between the two subunits, to form a tetramer, which is imported into the nucleus. Alternatively, only one of the subunits may contain a Kap122p-cognate NLS. After synthesis, this subunit could associate with Kap122p and with the other subunit to form a heterotrimer that would be imported into the nucleus. After import, in each of these two scenarios, RanGTP (Fig. 10 and Fig. 12) would dissociate the TFIIA heterodimer from Kap122p. Our data here argue against a third possibility, namely that a subunit/Kap122p heterodimer is imported separately because we found a stable interaction between the two subunits in the cytoplasm. In fact, while the Toa1p–Toa2p complex was dissociated from Kap122p by MgCl2 concentrations between 100 and 1,000 mM, the interaction between the two subunits resisted dissociation at these MgCl2 concentrations (Fig. 6, Fig. 7, Fig. 9, and Fig. 10).
PrA-tagged Kap122p was found to be associated in the cytoplasm with other proteins (Fig. 3). We do not yet know the identity of these proteins and whether they represent contaminants or alternative import substrates for Kap122p. However, it is clear that these other proteins are not part of the Toa1p/Toa2p/Kap122p complex as they were not copurified in a reverse pullout with PrA-tagged Toa1p or Toa2p (Fig. 6 and Fig. 7). As in the case of other PrA-tagged Kaps, several degradation products of Kap122p-PrA retaining their protein A moiety and eluting predominantly in the 4,500 mM MgCl2 fraction were observed and confirmed by immunoblotting (data not shown).
We were able to reconstitute the Kap122p/Toa1p/Toa2p complex from recombinant proteins (Fig. 11). Moreover, this complex was sensitive to dissociation by RanGTP but not RanGDP (Fig. 12). These findings support the conclusion that Kap122p interacts directly with the Toa1p–Toa2p complex and that this interaction, like Kap122p/Toa1p/Toa2p interaction in the yeast cytosol, is sensitive to dissociation by RanGTP but resists dissociation by RanGDP.
It is known that RanGTP dissociates import substrates from Kaps by binding to the Kap (Rexach and Blobel 1995). The X-ray crystal structure of RanGTP complexed to Kap β2 (transportin) and Kap β1 (importin) has been determined (Chook and Blobel 1999; Vetter et al. 1999). Ran binding to Kap122p/Pdr6p has been previously investigated and no Ran binding was detected (Görlich et al. 1997). Consistent with this report, we have so far not been able to demonstrate binding of RanGTP to Kap122p in overlay or solution binding assays (data not shown). This might indicate that RanGTP binds Kap122p with very low affinity. Stable binding of RanGTP to Kap122p may require additional proteins. However, the Kap122p/Toa1p/Toa2p complex isolated from yeast (Fig. 10) or reconstituted from recombinant proteins (Fig. 11 and Fig. 12) could be dissociated by RanGTP but not RanGDP. These data confirm that RanGTP is directly involved in dissociating Kap122p from the Toa1p–Toa2p complex and provide support for the existence of functionally relevant interaction between RanGTP and Kap122p.
It appears that Kap122p is the principal Kap dedicated to the import of TFIIA because in a strain where KAP122 had been deleted there was a significant mislocalization of the two TFIIA subunits from the nucleus to the cytoplasm (Fig. 4 and Fig. 8). Surprisingly, KAP122 deletion is not lethal, whereas deletion of either of the two TFIIA subunits is. Therefore, Kaps other than Kap122p are likely to be involved in nuclear import of the TFIIA subunits. However, so far we have failed to identify alternative Kaps by cytosolic pullout experiments with Toa2p-PrA in a kap122Δ strain (Fig. 9). It is likely that import of the TFIIA subunits by these putative alternative Kaps proceeds with much lower efficiency than import by Kap122p, based on the significant reduction in the nuclear localization of the TFIIA subunits observed in a kap122Δ strain. Nevertheless, import of these two essential proteins in the absence of Kap122p appears to be sufficient, as there is no apparent difference in the growth rate between the kap122Δ and an isogenic wt strain. Alternative Kap(s) may be difficult to detect in an immunoisolation assay as they may bind to the two TFIIA subunits with lower affinity. There are precedents for essential proteins being imported by several Kaps of which the principal one is not essential. For example, ribosomal proteins have been shown to be imported by the abundant Kap123p (Rout et al. 1997; Schlenstedt et al. 1997). However, Kap123p is not essential, but the essential Kap121p can back up ribosomal protein import in a kap123Δ strain (Rout et al. 1997; Seedorf and Silver 1997). Alternatively, the Toa1p–Toa2p heterodimer is small enough (<60 kD) that it might diffuse through the NPC.
Kap122p is the third Kap that so far has been shown to be dedicated to the import of a general transcription factor, the others being Kap119p, which is involved in the nuclear import of the nonessential transcription factor TFIIS (Albertini et al. 1998), and Kap114p, which is involved in the nuclear import of the TATA binding protein (Pemberton et al. 1999). The advantages of maintaining dedicated Kaps for the import of specific transcription factors are likely to be in the regulatory realm and remain to be elucidated.
Acknowledgments
A. Titov would like to dedicate this paper to Philip B. Royle, friend and mentor, in gratefulness for his encouragement, faith, and inspiration.
We thank Roland Beckmann, Lucy Pemberton, and Jonathan Rosenblum for advice and reading the manuscript, J. Fernandez (Rockefeller University Protein/DNA Technology Center, New York, NY) for protein sequence analysis, F. Gharahdaghi (PDTC) for mass spectral analysis, M. Floer for Ran and RanGAP, and E. Ellison and M. Misiak for technical assistance.
Footnotes
1.used in this paper: DAPI, 4′,6-diamidino-2-phenylindole; Kap, karyopherin; NLS, nuclear localization signal; NPC, nuclear pore complex; Nup, nucleoporin; PDR6, pleiotropic drug resistance; PrA, Staphylococcus aureus protein A; TBP, TATA-binding protein; TFIIA, transcription factor IIA
References
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup157pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J.D., Blobel G., Rout M.P. Kap104pa karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Albertini M., Pemberton L.F., Rosenblum J.S., Blobel G. A novel nuclear import pathway for the transcription factor TFIIS. J. Cell Biol. 1998;143:1447–1455. doi: 10.1083/jcb.143.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E., Chen W., Ulaszewski S., Capieaux E., Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae . J. Biol. Chem. 1987;262:16871–16879. [PubMed] [Google Scholar]
- Chen W.N., Balzi E., Capieaux E., Choder M., Goffeau A. The DNA sequencing of the 17 kb HindIII fragment spanning the LEU1 and ATE1 loci on chromosome VII from Saccharomyces cerevisiae reveals the PDR6 gene, a new member of the genetic network controlling pleiotropic drug resistance. Yeast. 1991;7:287–299. doi: 10.1002/yea.320070311. [DOI] [PubMed] [Google Scholar]
- Chook Y.M., Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran xGppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Corbett A.H., Silver P.A. Nucleocytoplasmic transport of macromolecules. Microbiol. Mol. Biol. Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong J., Roeder R.G. A single cDNA, hTFIIA/α, encodes both the p35 and p19 subunits of human TFIIA. Genes Dev. 1993;7:2220–2234. doi: 10.1101/gad.7.11.2220. [DOI] [PubMed] [Google Scholar]
- Enenkel C., Blobel G., Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo J.C., Mattaj I.W. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 1999;14:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Gharahdaghi F., Mische S.M. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS) Electrophoresis. 1998;19:1036–1045. doi: 10.1002/elps.1150190619. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- Fornerod M., van Deursen J., van Baal S., Reynolds A., Davis D., Murti K.G., Fransen J., Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger J.H., Hahn S., Lee S., Sigler P.B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F., Kirchner M., Fernandez J., Mische S.M. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal. Biochem. 1996;233:94–99. doi: 10.1006/abio.1996.0012. [DOI] [PubMed] [Google Scholar]
- Görlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P.A., Guarente L. Identification of a yeast protein homologous in function to the mammalian general transcription factor, TFIIA. EMBO (Eur. Mol. Biol. Organ.) J. 1989;8:3379–3382. doi: 10.1002/j.1460-2075.1989.tb08501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Kutay U., von Kobbe C., Mattaj I.W., Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Rank N.M., O'Shea E.K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe Y., Lemoine Y., Kuchler K. The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J. Biol. Chem. 1996;271:25167–25172. doi: 10.1074/jbc.271.41.25167. [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J.D., Wozniak R.W. Specific binding of the karyopherin kap121p to a subunit of the nuclear pore complex containing nup53p, nup59p, and nup170p. J. Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W., Englmeier L. Nucleocytoplasmic transportthe soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Melchior F., Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Melchior F., Paschal B., Evans J., Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.S. Ran and nuclear transport. J. Biol. Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Moore M.S., Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore J.D., Yang J., Truant R., Kornbluth S. Nuclear import of Cdk/cyclin complexesidentification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J. Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Fornerod M., Mattaj I.W. Nucleocytoplasmic transportthe last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Palmeri D., Malim M.H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S., Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel G., Rosenblum J.S. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S., Blobel G. Nuclear import of the TATA binding proteinmediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish J.A., Hahn S. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J. Biol. Chem. 1991;266:19320–19327. [PubMed] [Google Scholar]
- Ranish J.A., Lane W.S., Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nucleiassociation and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Kutay U., Paraskeva E., Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- Rosenblum J.S., Pemberton L.F., Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Schwoebel E.D., Talcott B., Cushman I., Moore M.S. Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J. Biol. Chem. 1998;273:35170–35175. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G., Smirnova E., Deane R., Solsbacher J., Kutay U., Görlich D., Ponstingl H., Bischoff F.R. Yrb4p, a yeast ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M., Silver P.A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B., Simos G., Bischoff F.R., Podtelejnikov A., Mann M., Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. Methods in Yeast Genetics 1986. Cold Spring Harbor Press; Cold Spring Harbor, NY: pp. 186 [Google Scholar]
- Tan S., Hunziker Y., Sargent D.F., Richmond T.J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–151. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- Truant R., Cullen B.R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I.R., Arndt A., Kutay U., Gorlich D., Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P., Blobel G. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Datar K.V., Paddy M.R., Swedlow J.R., Swanson M.S. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae . J. Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Rout M.P., Aitchison J.D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yang Q., Rout M.P., Akey C.W. Three-dimensional architecture of the isolated yeast nuclear pore complexfunctional and evolutionary implications. Mol. Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]