Abstract
The single cytoplasmic dynein and five of the six kinesin-related proteins encoded by Saccharomyces cerevisiae participate in mitotic spindle function. Some of the motors operate within the nucleus to assemble and elongate the bipolar spindle. Others operate on the cytoplasmic microtubules to effect spindle and nuclear positioning within the cell. This study reveals that kinesin-related Kar3p and Kip3p are unique in that they perform roles both inside and outside the nucleus. Kar3p, like Kip3p, was found to be required for spindle positioning in the absence of dynein. The spindle positioning role of Kar3p is performed in concert with the Cik1p accessory factor, but not the homologous Vik1p. Kar3p and Kip3p were also found to overlap for a function essential for the structural integrity of the bipolar spindle. The cytoplasmic and nuclear roles of both these motors could be partially substituted for by the microtubule-destabilizing agent benomyl, suggesting that these motors perform an essential microtubule-destabilizing function. In addition, we found that yeast cell viability could be supported by as few as two microtubule-based motors: the BimC-type kinesin Cin8p, required for spindle structure, paired with either Kar3p or Kip3p, required for both spindle structure and positioning.
Keywords: kinesin, dynein, motor proteins, microtubules, mitotic spindle
Microtubules and their associated motor proteins accomplish many intracellular movements in eukaryotic cells. Two superfamilies of microtubule-based motor proteins have been identified, the kinesins and the dyneins. Members of each superfamily are defined by distinct force-producing domains that are conserved in amino acid sequence. Since all eukaryotic cells express multiple microtubule motors, a major research question concerns the cellular roles performed by each motor.
The budding yeast Saccharomyces cerevisiae has been particularly useful for the experimental dissection of microtubule-based motor roles (Hoyt and Geiser 1996). In addition to the experimental tractability of this organism, it possesses a relatively small and easy to manipulate set of microtubule motor genes. The S. cerevisiae genome encodes six kinesin-related gene products (Cin8p, Kip1–3p, Kar3p, and Smy1p) and a single dynein heavy chain (Dyn1p, a member of the cytoplasmic dynein subclass). Although these motor gene products are each distinct in amino acid sequence, none are individually essential for cell viability. All single deletion alleles are viable. This is due to extensive overlaps in functions between these motors. S. cerevisiae uses microtubules for only a single process essential for vegetative growth, mitotic spindle function. Previous studies have implicated six of these seven motors in spindle function (Hoyt et al. 1997; Stearns 1997). The single exception is kinesin-related Smy1p, which is involved in polarized cell growth (Lillie and Brown 1994, Lillie and Brown 1998). smy1 mutants display neither mitotic defects nor genetic interactions with any other microtubule motor gene deletion alleles (Cottingham and Hoyt 1997; Lillie and Brown 1998). In contrast, mutations in the remaining six motor genes cause mitotic phenotypes and display extensive genetic interactions. Examples of cooperative (or overlapping functions) and antagonistic interactions have been described and experimentally exploited to reveal essential spindle functions performed by these six motors (for reviews see Hoyt and Geiser 1996; Stearns 1997).
The mitotic division of S. cerevisiae cells requires microtubule-based activities operating on both sides of the nuclear envelope. The nuclear microtubules assemble into a bipolar spindle structure that separates replicated chromosomes in anaphase, primarily by elongation (anaphase B–type movement). The major and essential role for the cytoplasmic microtubules is positioning the spindle within the cell at the neck between mother and bud. Since this process results in the movement of the entire nuclear envelope and its contents, it is often referred to as nuclear migration. Proper spindle positioning ensures that anaphase spindle elongation segregates one progeny nucleus to the mother cell and the other to the bud.
Two of the S. cerevisiae kinesin-related proteins, Cin8p and Kip1p, are members of the BimC sequence subclass. BimC motors are required for bipolar spindle assembly in S. cerevisiae and other eukaryotic cell types (Hoyt and Geiser 1996; Kashina et al. 1998). Cin8p and Kip1p also make a major contribution to anaphase B spindle elongation (Saunders et al. 1995; Straight et al. 1998). These roles are most likely accomplished by the ability of BimC motors to cross-link and slide spindle midzone microtubules within the nucleus. Roles for Kar3p and Kip3p in the bipolar spindle have been implicated, but are not well understood. Kar3p clearly performs numerous but poorly defined roles during mitosis. kar3Δ mutant cultures display many aberrant spindles (Meluh and Rose 1990; Saunders et al. 1997a). Kar3p also antagonizes the pole-separating activity of the BimC motors during the process of spindle assembly (Saunders and Hoyt 1992; Hoyt et al. 1993; Saunders et al. 1997b). Kar3p accomplishes some of its mitotic functions in association with one of two homologous accessory subunits, Cik1p and Vik1p, which apparently target this motor to distinct cellular roles (Manning et al. 1999; Page et al. 1994). Kip3p has been localized to the nuclear microtubules (as well as cytoplasmic microtubules) and deletion mutants make slightly longer spindles (Cottingham and Hoyt 1997; DeZwaan et al. 1997; Miller et al. 1998; Straight et al. 1998).
Dynein is an important spindle positioning motor, but is not essential for cell viability (Eshel et al. 1993; Li et al. 1993). Functional overlap for spindle positioning between dynein and Kip3p was described previously (Cottingham and Hoyt 1997; DeZwaan et al. 1997). Interestingly, both dyn1 kip3 and dyn1 kar3 double mutants were severely compromised for growth, and in both cases the growth defect was suppressed by the deletion of KIP2, encoding another kinesin-related protein. This suggested that Kar3p may also overlap with dynein and Kip3p for spindle positioning roles. The studies presented here verify this hypothesis. In addition, functional overlap between Kar3p and Kip3p for a nuclear role essential for the structural integrity of bipolar spindles is described. Therefore, Kar3p and Kip3p overlap for essential roles both inside and outside the nucleus. Finally, our studies reveal that S. cerevisiae cells can perform mitosis with as many as five of the seven microtubule-based motor genes deleted.
Materials and Methods
Yeast Strains and Media
The S. cerevisiae strains used in these experiments are derivatives of S288C and are listed in Table . The dyn1::HIS3 (Geiser et al. 1997), dyn1::URA3 (Eshel et al. 1993), kar3::LEU2 (Meluh and Rose 1990), kip2::URA3 (Roof et al. 1992), kip3::kan and kip3-14 (Cottingham and Hoyt 1997), cik1::LEU2 (Page and Snyder 1992), and vik1::HIS3 (Manning et al. 1999) alleles were described previously. The unmarked dyn1-Δ10 allele was created by transforming a dyn1::URA3 strain with a DNA fragment of DYN1 that spanned the URA3 insertion but harbored an internal deletion. Ura− were selected on media containing 5-fluoro-orotic acid (5-FOA)1 (US Biological). The same strategy was used to create the unmarked kip2-Δ10 allele. The smy1-Δ10 unmarked deletion was created by PCR-targeted gene disruption (Wach et al. 1994). The PCR template DNA was URA3 flanked by a directly repeated sequence. Following transformation and selection for smy1::URA3, we were able to select, using 5-FOA, for segregants that had looped out URA3 by homologous recombination between the repeated sequence.
Table 1.
Yeast Strains and Plasmids
| Genotype | |
|---|---|
| Yeast strains | |
| MAY589 | MATa ade2 his3 leu2 ura3 |
| MAY591 | MATα lys2 his3 leu2 ura3 |
| MAY1089 | MATa ade2 lys2 his3 leu2 ura3 |
| MAY2268 | MATα lys2 his3 leu2 ura3 kar3::LEU2 |
| MAY3189 | MATα lys2 his3 leu2 ura3 dyn1::URA3 |
| MAY2269 | MATa ade2 his3 leu2 ura3 kar3::LEU2 |
| MAY4360 | MATa ade2 his3 leu2 ura3 (pRS313) |
| MAY4435 | MATα lys2 his3 leu2 ura3 cyh2 dyn1::HIS3 |
| MAY4518 | MATα lys2 his3 leu2 ura3 kip3::kan |
| MAY4560 | MATa ade2 lys2 his3 leu2 ura3 cyh2 kip3::kan kip2::URA3 dyn1::HIS3 |
| MAY4619 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan (pMA1223) |
| MAY4809 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan (pFC51) |
| MAY4908 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan (pFC51kip3-14) |
| MAY4921 | MATa lys2 his3 leu2 ura3 dyn1::HIS3 kip3::kan (pFC51kip3-14) |
| MAY4924 | MATa lys2 his3 leu2 ura3 dyn1::HIS3 kip3::kan (pFC51) |
| MAY5001 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan (pMA1428) |
| MAY5022 | MATa ade2 his3 leu2 ura3 kar3::LEU2 (pMA1428) |
| MAY5218 | MATa ade2 his3 leu2 ura3 kar3::LEU2 dyn1-Δ10 (pMA1428) |
| MAY5269 | MATa ade2 lys2 his3 leu2 ura3 cyh2 kar3::LEU2 kip3::kan kip2::URA3 (pMA1428) |
| MAY5360 | MATa ade2 his3 leu2 ura3 cik1::LEU2 |
| MAY5398 | MATa ade2 his3 leu2 ura3 kar3::LEU2 (pMA1428kar3-64) |
| MAY5399 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan (pMA1428kar3-64) |
| MAY5400 | MATa ade2 his3 leu2 ura3 kar3::LEU2 dyn1-Δ10 (pMA1428kar3-64) |
| MAY5464 | MATa ade2 his3 leu2 ura3 kar3::LEU2 dyn1-Δ10 kip2::URA3 (pMA1428) |
| MAY5465 | MATa ade2 his3 leu2 ura3 kar3::LEU2 dyn1-Δ10 kip2::URA3 (pMA1428kar3-64) |
| MAY5522 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan kip2::URA3 (pMA1428kar3-64) |
| MAY5742 | MATa ade2 his3 leu2 ura3 kar3::LEU2 (pRS313) |
| MAY5774 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan dyn1-Δ10 kip2-Δ10 (pMA1428) |
| MAY5775 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan dyn1-Δ10 kip2-Δ10 (pMA1428kar3-64) |
| MAY5776 | MATa ade2 his3 leu2 ura3 nuf2::NUF2-sGFP (pRS313) |
| MAY5777 | MATa ade2 his3 leu2 ura3 nuf2::NUF2-sGFP kar3::LEU2 (pMA1428kar3-64) |
| MAY5778 | MATa ade2 lys2 his3 leu2 ura3 kar3::LEU2 kip3::kan nuf2::NUF2-sGFP (pMA1428kar3-64) |
| MAY5779 | MATa ade2 his3 leu2 ura3 kar3::LEU2 dyn1-Δ10 nuf2::NUF2-sGFP (pMA1428kar3-64) |
| MAY5802 | MATa lys2 his3 leu2 ura3 dyn1::HIS3 cik1::LEU2 kip2::URA3 |
| MAY5903 | MATa ade2 lys2 his3 leu2 ura3 dyn1-Δ10 kip2-Δ10 smy1-Δ10 kip1::URA3 kip3::kan kar3::LEU2 (pMA1428) |
| MAY5904 | MATa ade2 lys2 his3 leu2 ura3 dyn1-Δ10 kip2-Δ10 smy1-Δ10 kip1::URA3 kip3::kan kar3::LEU2 (pMA1428kar3-64) |
| MAY5905 | MATa ade2 lys2 his3 leu2 ura3 dyn1-Δ10 kip2-Δ10 smy1-Δ10 kip1::URA3 kip3::kan kar3::LEU2 (pFC82) |
| MAY5906 | MATa ade2 lys2 his3 leu2 ura3 dyn1-Δ10 kip2-Δ10 smy1-Δ10 kip1::URA3 kip3::kan kar3::LEU2 (pFC83) |
| MAY5940 | MATa ade2 lys2 his3 leu2 ura3 dyn1-Δ10 kip2-Δ10 smy1-Δ10 kip1::HIS3 kip3::kan cin8::LEU2 (pMA1260) |
| MAY5941 | MATa ade2 lys2 his3 leu2 ura3 dyn1-Δ10 kip2-Δ10 smy1-Δ10 kip1::HIS3 kip3::kan cin8::LEU2 (pTK199) |
| MAY6030 | MATa ade2 his3 leu2 ura3 kip3::kan nuf2::NUF2-sGFP (pFC83) |
| MAY6332 | MATa ade2 his3 ura3 dyn1::URA3 vik1::HIS3 |
| MAY6341 | MATa ade2 his3 leu2 ura3 trp1 kip3::kan vik1::HIS3 |
| MAY6417 | MATa ade2 his3 leu2 ura3 trp1 kip3::kan vik1::HIS3 cik1::LEU2 (pFC48) |
| Plasmids | |
| pFC48 | KIP3 URA3 (CEN) |
| pFC50 | KIP2 LYS2 (CEN) |
| pFC51 | KIP3 LYS2 (CEN) |
| pFC51kip3-14 | kip3-14 LYS2 (CEN) |
| pFC82 | KIP3 HIS3 (CEN) |
| pFC83 | kip3-14 HIS3 (CEN) |
| pMA1223 | KAR3 URA3 (CEN) |
| pMA1260 | CIN8 LYS2 (CEN) |
| pMA1428 | KAR3 HIS3 (CEN) |
| pMA1428kar3-64 | kar3-64 HIS3 (CEN) |
| pRS313 | HIS3 (CEN) |
| pTK199 | cin8-3 LYS2 (CEN) |
Multiple motor gene deletion mutants were constructed by standard genetic techniques. Strains with reduced spindle motor function often displayed evidence of aneuploidy (i.e., non-Mendelian segregation of genetic markers). Therefore, we routinely verified all seven microtubule motor gene alleles by PCR. For DYN1 and KIP2, we were able to use a primer set that flanked the genes and yielded distinct PCR products from both the wild-type and deletion alleles. For the remaining five microtubule motor genes, two PCR reactions were used to test each allelic form. Both reactions used an upstream primer with a sequence present in both wild-type and deletion mutant alleles. This was paired with downstream primers that were specific for either the wild-type allele DNA in one reaction or the deletion DNA in the other.
Rich (yeast extract, peptone, dextrose [YPD]) and minimal (synthetic dextrose [SD]) media were as described (Sherman et al. 1983). Benomyl (DuPont) was added to solid YPD medium from a 10 mg/ml stock in DMSO. Nocadazole (Sigma Chemical Co.) was added to liquid YPD medium from a 3.3 mg/ml stock in DMSO. For G1 synchronization, α-factor (Bachem Bioscience) was added to 4 μg/ml to log-phase cells in liquid YPD, pH 4.0, and incubated at 26°C until >80% of cells were unbudded. For arrest in S phase, hydroxyurea (Sigma Chemical Co.) was added to 0.1 M to log-phase cells in liquid YPD, pH 5.8, and incubated at 26°C until >70% of cells were large-budded.
DNA Manipulations
Temperature-sensitive alleles of KAR3 were generated by a mutagenic PCR procedure (Staples and Dieckmann 1993). The entire KAR3 gene, including 412 bp of upstream sequence and 560 bp of downstream sequence, was amplified under mutagenic PCR conditions. We used Taq polymerase according to the supplier's instructions (Stratagene), except that Mn2+ and Mg2+ were added to final concentrations of 0.25 and 4.5 mM, respectively. The PCR products were concentrated using QIAquick spin columns (Qiagen). pMA1428 was digested with NruI and StuI to create a gapped construct in which all but the first 210 bp of KAR3 coding sequence was removed. The gapped pMA1428 construct was gel-purified and cotransformed with the concentrated PCR products into strain MAY4619. This strain is unable to survive loss of a plasmid carrying KAR3 and URA3, and is therefore rendered sensitive to 5-FOA. Approximately 6,600 His+ transformants were selected at 26°C and then replica plated to 5-FOA–containing media at 26 and 35°C. Six colonies were selected that were viable on 5-FOA at 26 but not 35°C. Plasmids were isolated from the six temperature-sensitive transformants and retransformed into MAY4619. After growth on 5-FOA at 26°C, four of the six retransformed strains displayed reduced growth on YPD at 35°C. These four plasmids were designated pMA1428kar3-61 through pMA1428kar3-64. Of these, pMA1428kar3-64 caused the greatest reduction of growth for kar3Δ kip3Δ strains at 35°C, and was chosen for study in all subsequent experiments. pMA1428kar3-64 did not alter the temperature resistance of either wild-type or kip3Δ, dyn1Δ, and kar3Δ single mutant strains. Therefore, kar3-64 is a recessive and nonepistatic mutant.
Microscopy
To stain for DNA, cells were pelleted out of liquid media, resuspended in 70% ethanol, and stored on ice for 30 min. The ethanol-fixed cells were washed once with water and then resuspended in 0.3 μg/ml 4,6-diamidino-2-phenylindole (DAPI) containing 1 mg/ml p-phenylenediamine to prevent fading (Sigma Chemical Co.). To stain for chitin-containing bud scars (Pringle 1991), cells were fixed as above, washed once with water, resuspended in 1 mg/ml calcofluor solution (Sigma Chemical Co.), and stored at room temperature in the dark for 5 min. The cells were washed four times with water and then resuspended in 0.3 μg/ml DAPI plus 1 mg/ml p-phenylenediamine before viewing. Mother cell bodies were defined as those which possessed at least two calcofluor-stained chitin rings on their surface. For antitubulin immunofluorescence microscopy, cells were fixed by adding formaldehyde directly to the medium to a final concentration of 3.7%. Microtubules were visualized by a procedure described previously (Pringle et al. 1991) using the rat antitubulin antibody YOL1/34 (Harlan Bioproducts for Science) and rhodamine-conjugated goat anti–rat secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). Stained cells were examined with an inverted microscope (Axiovert 135; Carl Zeiss, Inc.) equipped with epifluorescent optics using a 100× objective. Digital images were captured with a cooled, slow-scan CCD camera. In cells expressing the spindle pole body (SPB)-associated fusion protein Nuf2p–green fluorescent protein (GFP) (Kahana et al. 1995), bipolar spindles were quantitated by counting the number of cells with two clearly separated GFP dots.
Electron microscopic examination of thin sections was performed as described by Byers and Goetsch 1991.
Quantitation of Nuclear Mislocalization
To quantitate nuclear mislocalization in cells, two assays were used. In the first, cells were arrested at 26°C with α-factor as described above. Cells were then washed once with water to remove the α-factor and resuspended in YPD media prewarmed to 35°C. The cultures were incubated at 35°C and at regular intervals samples were removed, fixed with ethanol, and stained with DAPI. The percentage of total cells that were large-budded (bud diameter greater than three quarters the diameter of the mother cell) with the nucleus away from the neck was determined for each time point. “Nucleus away from the neck” cells were defined as those in which the closest distance between the nucleus and the neck was greater than one half the diameter of the entire nuclear DNA mass, as judged by eye. In the second assay, cells were arrested at 26°C with hydroxyurea as described above. Next, cultures were shifted to 35°C, and at regular intervals samples were removed, fixed, and either stained with DAPI alone, or processed for both antitubulin immunofluorescence and DAPI staining. The percentage of large-budded cells with the nucleus away from the neck was determined for each time point.
Results
Characterization of a Temperature-Sensitive KAR3 Allele
In a previous study we obtained evidence of overlap for an essential function between Kar3p, Kip3p, and dynein. Although neither KAR3, KIP3, nor DYN1 (encoding the dynein heavy chain) are individually essential for cell viability, the double deletion mutant combinations are inviable, or in the case of dyn1Δ kip3Δ, grow very slowly (Cottingham and Hoyt 1997). To investigate the nature of this overlap and the mitotic role of Kar3p, we generated a recessive temperature-sensitive KAR3 mutant allele (see Materials and Methods). A plasmid carrying this allele, kar3-64, was identified in a strain deleted for KAR3 and KIP3. The kar3Δ kip3Δ (pkar3-64) strain was inviable at 33°C and higher (Fig. 1; compare row A10 to A9, 0 benomyl columns). kar3Δ dyn1Δ (pkar3-64) cells were also temperature-sensitive for growth, although the effect was not as extreme as for kip3Δ (Fig. 1; compare row A6 to A5). pkar3-64 also failed to complement the slight temperature sensitivity caused by kar3Δ (Fig. 1; compare row A4 to A3). In the absence of dynein, kinesin-related Kip2p acts antagonistically to Kar3p and Kip3p. Loss of Kip2p suppresses the growth defects of the dyn1Δ kip3Δ and dyn1Δ kar3Δ double mutants, but not that of kip3Δ kar3Δ (Cottingham and Hoyt 1997). Accordingly, we found that the kar3Δ dyn1Δ kip2Δ (pkar3-64) strain showed moderately improved growth at 35°C compared with the kar3Δ dyn1Δ (pkar3-64) strain (Fig. 1; compare row A8 to A6). However, the kar3Δ kip3Δ kip2Δ (pkar3-64) strain was just as temperature-sensitive as the corresponding strain possessing the wild-type KIP2 allele (Fig. 1; compare row A12 to A10).
Figure 1.
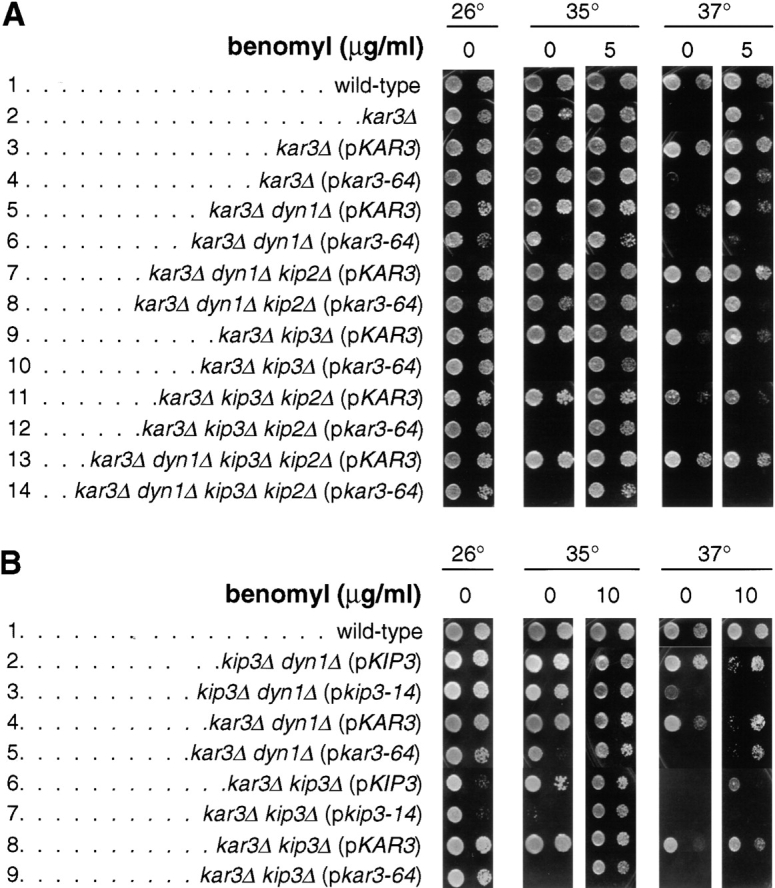
Growth properties of motor mutant strains. Yeast cells of the indicated genotype were spotted onto solid rich media containing the indicated concentration of benomyl, and incubated at the indicated temperature for 3 d. Two spots appear for each sample: the spot to the right is a 40-fold dilution of the sample on the left. Strains used (p indicates a centromere plasmid-carried gene): (A) wild-type (MAY4360), kar3Δ (MAY5742), kar3Δ (pKAR3) (MAY5022), kar3Δ (pkar3-64) (MAY5398), kar3Δ dyn1Δ (pKAR3) (MAY5218), kar3Δ dyn1Δ (pkar3-64) (MAY5400), kar3Δ dyn1Δ kip2Δ (pKAR3) (MAY5464), kar3Δ dyn1Δ kip2Δ (pkar3-64) (MAY5465), kar3Δ kip3Δ (pKAR3) (MAY5001), kar3Δ kip3Δ (pkar3-64) (MAY5399), kar3Δ kip3Δ kip2Δ (pKAR3) (MAY5269), kar3Δ kip3Δ kip2Δ (pkar3-64) (MAY5522), kar3Δ dyn1Δ kip3Δ kip2Δ (pKAR3) (MAY5774), and kar3Δ dyn1Δ kip3Δ kip2Δ (pkar3-64) (MAY5775). (B) wild-type (MAY589), kip3Δ dyn1Δ (pKIP3) (MAY4924), kip3Δ dyn1Δ (pkip3-14) (MAY4921), kar3Δ dyn1Δ (pKAR3) (MAY5218), kar3Δ dyn1Δ (pkar3-64) (MAY5400), kar3Δ kip3Δ (pKIP3) (MAY4809), kar3Δ kip3Δ (pkip3-14) (MAY4908), kar3Δ kip3Δ (pKAR3) (MAY5001), and kar3Δ kip3Δ (pkar3-64) (MAY5399).
Log-phase cultures of kar3Δ kip3Δ (pkar3-64), kar3Δ dyn1Δ (pkar3-64), and wild-type cells were shifted to 35°C for 2 h and examined by microscopy. The kar3Δ kip3Δ (pkar3-64) and kar3Δ dyn1Δ (pkar3-64) showed evidence of reduced proficiency to accomplish mitosis; 57 and 52%, respectively, of the mutant cells were large-budded and mononucleate, relative to 26% of the wild-type (n = 200 cells for each). The kar3Δ dyn1Δ (pkar3-64) cells often displayed mislocalized nuclei indicative of a spindle positioning defect (see next section).
Cells lacking KAR3 display a bilateral karyogamy defect; kar3Δ cells will form diploids with normal efficiency with KAR3 cells of the opposite mating type, but not with kar3Δ cells (Meluh and Rose 1990). In addition to its mitotic defects, kar3-64 caused a temperature-sensitive defect in diploid formation when mated to a kar3Δ strain, but not a KAR3 strain (data not shown).
A Role for Kar3p in Spindle Positioning Revealed in Cells Missing Dynein
In a previous study, the spindle positioning role of Kip3p was clearly revealed in cells missing dynein. Loss of Kip3 function in dynein-deficient cells caused spindle (and nuclear) mispositioning. The lethality of kar3 dyn1 and kar3 kip3 cells suggested that Kar3p may also participate in spindle positioning. To investigate this possibility, we assessed the proficiency of kar3-64 cells at positioning the nucleus at the mother–bud neck (a consequence of proper spindle positioning). Two related assays were used. In the first, cells were arrested in G1 with α-factor at 26°C and released to media at 35°C. The ability of the cells to translocate their nuclei to its proper position at the neck between mother and bud cell bodies was then assessed. In the second, spindle positioning was allowed to first occur at permissive temperature (26°C) in the presence of the DNA synthesis inhibitor hydroxyurea. Yeast cells treated with this inhibitor are still able to accomplish bipolar spindle assembly and spindle positioning (Pringle and Hartwell 1981). A temperature shift to 35°C was then imposed while the hydroxyurea block was maintained, and the ability of the cells to maintain the nucleus at the neck was assessed. In both assays, the inability to move the spindle or to maintain it at the neck was manifested by the appearance of large-budded cells with the nucleus located away from the neck (Fig. 2) (Cottingham and Hoyt 1997). Both assays yielded similar results (Fig. 3).
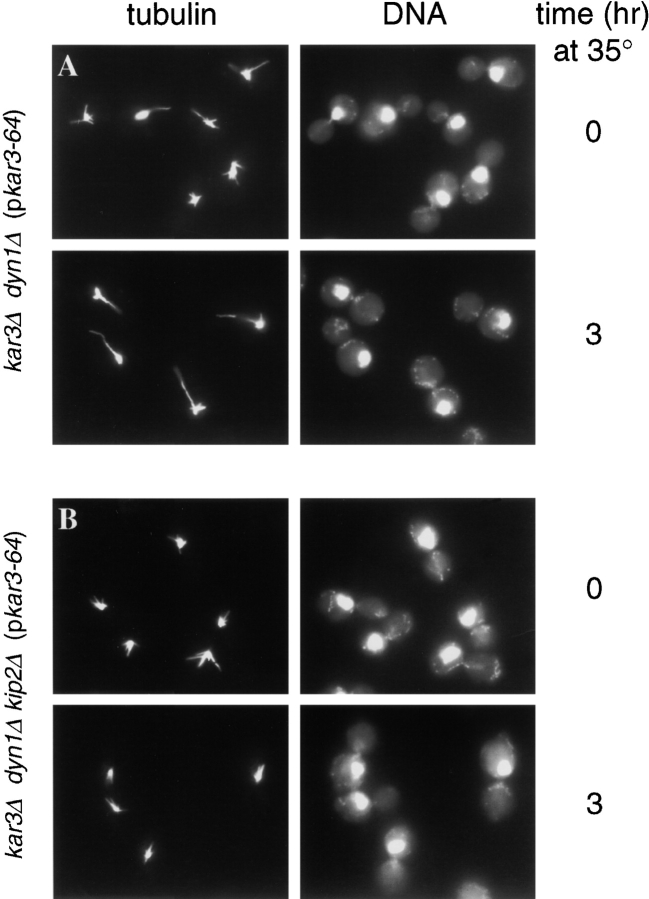
Figure 2.
Spindles and nuclei mislocalize in kar3-ts dyn1Δ cells. The nucleus and spindle were drawn away from the proper position at the mother–bud neck after loss of Kar3p function in the absence of dynein, but not in the absence of dynein and Kip2p. (A) kar3Δ dyn1Δ (pkar3-64) cells (MAY5400) were arrested with hydroxyurea at 26°C and then shifted to 35°C for 3 h in the continued presence of hydroxyurea. Samples taken before (top row) and after (bottom row) the temperature shift were processed for antitubulin immunofluorescence (left column) and DAPI staining of DNA (right column). (B) kar3Δ dyn1Δ kip2Δ (pkar3-64) cells (MAY5465) were treated as in A.
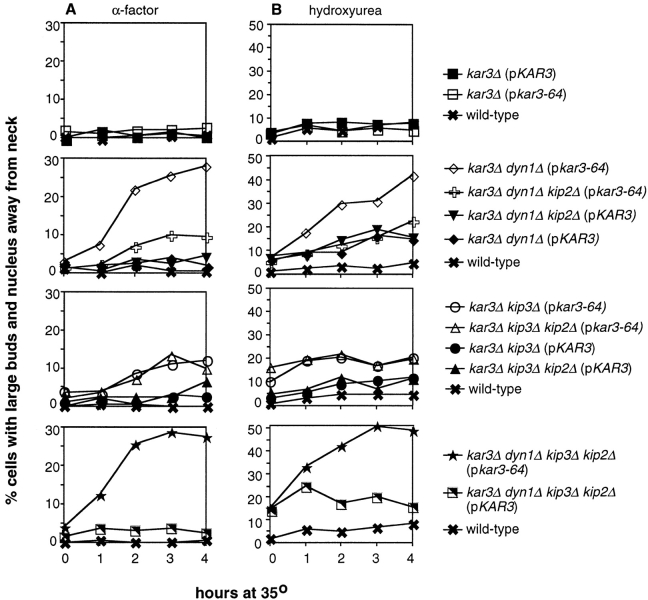
Figure 3.
Quantitation of nuclear positioning defects. (A) Nuclear positioning defects following α-factor synchronization at 26°C and release to 35°C. At the time points indicated, samples were removed, stained with DAPI, and the percentage of total cells that were large-budded with the nucleus positioned away from the bud neck was determined (defined as those in which the closest distance between the nucleus and the neck was greater than one half of the diameter of the entire nuclear DNA mass). (B) Nuclear positioning defects after hydroxyurea synchronization at 26°C and a shift to 35°C in the continued presence of hydroxyurea (also see Fig. 2). At the time points indicated, cells were analyzed for nuclear position as in A. Strains used are listed in legend of Fig. 1.
Loss of Kar3p function alone did not cause nuclear mislocalization. In both assays, the kar3Δ (pkar3-64) strain behaved similar to wild-type (Fig. 3, top panels). This agrees with previous reports, in which the nuclear migration index (the shortest distance between the nucleus and the bud neck divided by the mother cell diameter) for a kar3Δ strain was found to be comparable to the nuclear migration index of a wild-type strain (DeZwaan et al. 1997). Although the kar3Δ (pkar3-64) strain did not exhibit nuclear mislocalization, this strain did accumulate a large percentage (∼50%) of large-budded mononucleate cells at 35°C, as reported previously for strains lacking Kar3p (Meluh and Rose 1990). In contrast, the elimination of Kar3p function in the absence of dynein resulted in a gradual but significant mislocalization of the nucleus (see kar3Δ dyn1Δ [pkar3-64] strain in Fig. 2 A and Fig. 3, second row of panels). The extent of this phenotype was always greater than loss of dynein (kar3Δ dyn1Δ [pKAR3] strain) or Kar3p (kar3Δ [pkar3-64] strain) alone. As was observed for loss of both Kip3p and dynein activities (Cottingham and Hoyt 1997), loss of both Kar3p and dynein caused the appearance of long cytoplasmic microtubules that extended into the bud. Bud scar staining with calcofluor was used to identify the mother bodies of cells in which Kar3p and dynein were inactivated (n = 200). This analysis revealed that all of the nuclear mislocalization events occurred exclusively in the mother cells, as was previously observed for kip3 dyn1 cells. Therefore, efficient spindle positioning in the absence of dynein requires Kar3p as well as Kip3p. Furthermore, loss of Kar3p function in dynein-deficient cells that had previously achieved nuclear positioning results in movement of the nucleus away from the neck toward the mother cell cortex. To ensure that the effects of loss of dynein and Kar3p function observed here were not an artifact of the cell cycle block used in the protocols, we also examined temperature-shifted log-phase cultures. After 2 h at 35°C, 28% of large-budded kar3Δ dyn1Δ (pkar3-64) cells exhibited mislocalized nuclei or two nuclei in one cell body, compared with 6, 8, and 0% of kar3Δ (pkar3-64), dyn1Δ, and wild-type, respectively (n = 200 cells for each).
The ability of the deletion of KIP2 to suppress the growth defect of kar3 dyn1 cells (see above) was also reflected in its effects on spindle positioning proficiency. In both assays, the kar3Δ dyn1Δ kip2Δ (pkar3-64) strain displayed a greatly attenuated nuclear positioning defect relative to the kar3Δ dyn1Δ (pkar3-64) strain (Fig. 2 B and Fig. 3, second row of panels). This indicates that the spindle mislocalization observed in the absence of Kar3p and dynein was caused by the action of Kip2p. kar3Δ dyn1Δ kip2Δ (pkar3-64) cells were found to have significantly shorter cytoplasmic microtubules compared with the kar3Δ dyn1Δ (pkar3-64) strain (Fig. 2). This is consistent with the previous observation that loss of Kip2p causes a reduction in cytoplasmic microtubule length (Cottingham and Hoyt 1997).
The spindle positioning defect caused by the absence of Kar3p and dynein, and its suppression by kip2Δ, was very similar to the defect observed previously for cells missing Kip3p and dynein (Cottingham and Hoyt 1997). Cells missing both Kar3p and Kip3p are inviable, possibly also due to a spindle positioning defect (i.e., eliminating any two of either dynein, Kar3p, or Kip3p would prevent spindle positioning). To test this possibility, we performed the two nuclear position assays on a kar3Δ kip3Δ (pkar3-64) strain. Both assays demonstrated that loss of Kar3p in the absence of Kip3p caused only relatively mild nuclear mislocalization (Fig. 3, third row of panels). However, this effect was greater than those observed in the absence of either Kar3p or Kip3p alone. Similar findings were obtained in experiments using a temperature-sensitive KIP3 allele in a kar3Δ strain (Cottingham, F.R., and M.A. Hoyt, unpublished observations). The minor nuclear position defect observed in the absence of Kar3p and Kip3p did not appear to be affected by the presence or absence of Kip2p, i.e., the kar3Δ kip3Δ kip2Δ (pkar3-64) strain behaved similarly to the kar3Δ kip3Δ (pkar3-64) strain in both nuclear position assays. These findings indicate that Kar3p and Kip3p together make only a minor contribution to spindle positioning when dynein is present. However, when dynein is absent, the contributions of both Kip3p (Cottingham and Hoyt 1997) and Kar3p (this study) become apparent. The fact that loss of both Kar3p and Kip3p caused only a mild spindle positioning defect, but that lethality was not suppressed by kip2Δ, suggests that they overlap for a different essential process. The nature of this process is addressed in the following section.
The findings presented here and elsewhere (Cottingham and Hoyt 1997) implicate four microtubule-based motors in the process of spindle and nuclear positioning: dynein, Kar3p, Kip3p, and Kip2p. To investigate the consequences of loss of all four of these motors, we performed the two nuclear position assays described above on a strain deleted for DYN1, KAR3, KIP3, and KIP2 and carrying the kar3-64 allele on a plasmid (Fig. 3, bottom row of panels). In both assays, the kar3Δ dyn1Δ kip3Δ kip2Δ (pkar3-64) strain exhibited a nuclear positioning defect at 35°C; it was not able to move the nucleus to the neck efficiently or maintain it there following hydroxyurea synchronization. The movement of nuclei away from the neck after hydroxyurea treatment was found to occur independent of microtubule function (Cottingham, F.R., and M.A. Hoyt, unpublished observations). Therefore, in this experiment, Kar3p was acting as a solitary and essential spindle positioning motor required to resist a microtubule-independent force that operates on the nucleus. In the last Results section, we demonstrate that either Kar3p or Kip3p alone is sufficient to perform spindle positioning.
Spindle Assembly and Integrity Requires Either Kar3p or Kip3p
As demonstrated above, loss of function of both Kar3p and Kip3p caused only a mild spindle positioning defect. Therefore, the inviability of kar3 kip3 double mutant cells may be due to a defect in another essential cellular process. In the course of our spindle positioning experiments, microscopy revealed frequent abnormal spindle morphologies in kar3 cells, as has also been observed in kar3 cells by others (e.g., Meluh and Rose 1990; Saunders et al. 1997a). The abnormal spindle phenotype appeared more severe when both Kar3p and Kip3p were absent. To assess bipolar spindle assembly proficiency, cells of various genotypes were synchronized in G1 with α-factor at 26°C and released into media at 35°C. Fig. 4 shows examples of spindles fixed and stained 1 h after release from the α-factor block. Most wild-type cells possessed spindles that were clearly bipolar; a bright bar of spindle microtubules was visible in the nuclei (in some cases the bar was long, indicating anaphase had initiated). Fewer bar structures were found in kar3Δ (pkar3-64) cells and even fewer in kar3Δ kip3Δ (pkar3-64) cells. Instead of bars of microtubules, kar3Δ kip3Δ (pkar3-64) cells often had a bright small mass of nuclear microtubules from which the cytoplasmic microtubules radiated. As described previously for cells deficient for Kar3p and Dyn1p or Kip3p and Dyn1p, the cytoplasmic microtubules of cells missing the functions of both Kar3p and Kip3p grew to much longer lengths than wild-type.
Figure 4.
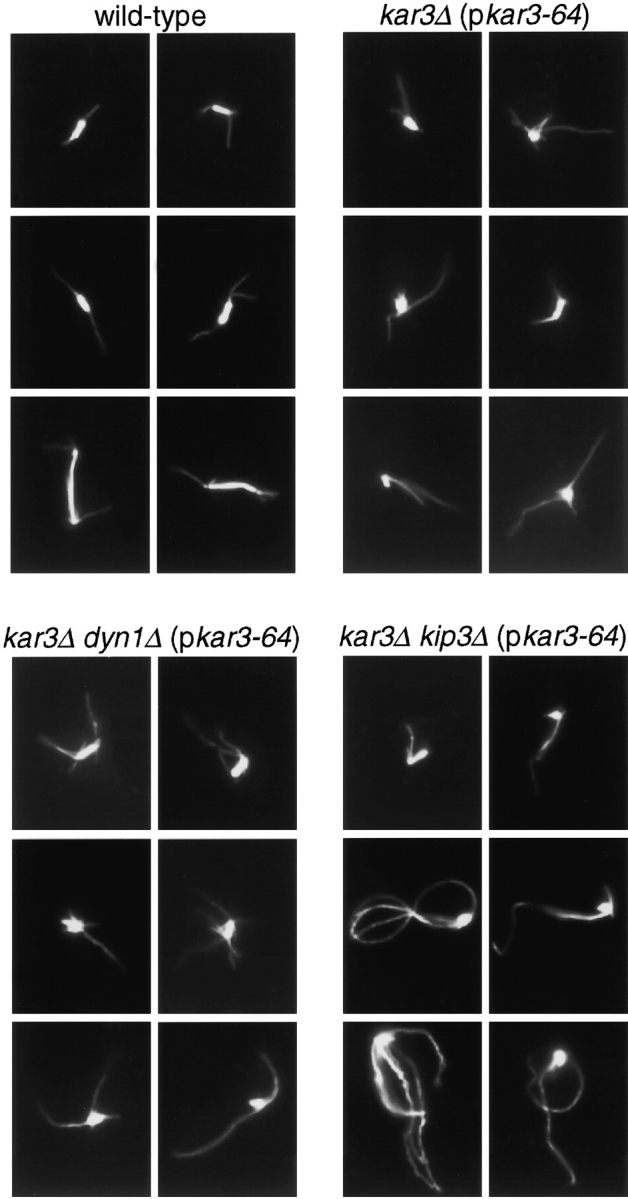
Spindle defects caused by loss of Kar3p function. Cell cultures of the genotypes indicated were synchronized with α-factor at 26°C and then released into medium at 35°C. After 60 min, samples were processed for antitubulin immunofluorescence microscopy. Microtubules of six representative cells are shown for each genotype. Note that the top left example of the kar3Δ kip3Δ (pkar3-64) strain represents a rare bipolar spindle observed for cells of this genotype. Strains used are listed in legend of Fig. 1.
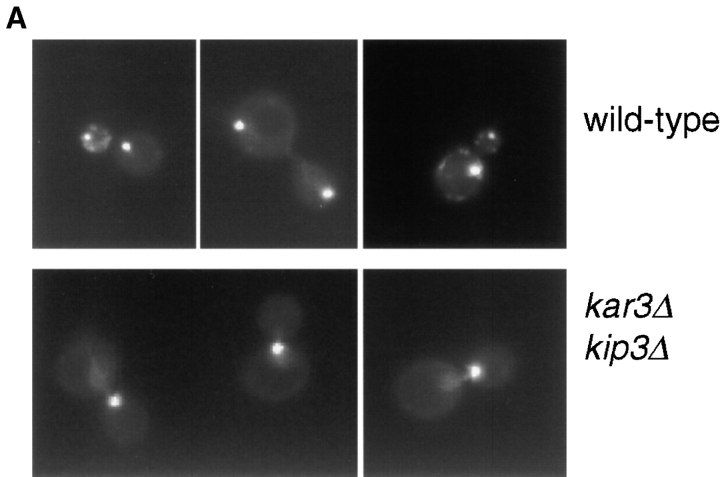
To quantitate the effects of loss of function of Kar3p and Kip3p on bipolar structure, we examined cells whose spindle poles were marked with the GFP. A construct encoding a GFP-tagged SPB protein, Nuf2p, was integrated into our motor mutant strains (Kahana et al. 1995). At various times after release from the α-factor block into 35°C, live cells were observed under the microscope and the percentage of cells with two clearly separated GFP dots was determined (Fig. 5a and Fig. b). Relative to the wild-type, the kar3Δ kip3Δ (pkar3-64) strain was severely compromised for its ability to generate cells with two distinct GFP dots. The majority of kar3Δ kip3Δ (pkar3-64) cells were large-budded, with a single GFP dot located at the bud neck (Fig. 5 A, bottom row). Therefore, Kip3p makes an important contribution to spindle assembly that is revealed when Kar3p activity is compromised. However, the activity provided by Kip3p cannot completely compensate for loss of Kar3p, since kar3-64 and kar3Δ (not shown) cells exhibited an intermediate reduction in spindle assembly proficiency, even when Kip3p was present. Dynein, on the other hand, does not appear to overlap with Kar3p for establishing spindle structure, since the kar3Δ dyn1Δ (pkar3-64) strain was no more severely affected than the kar3Δ (pkar3-64).
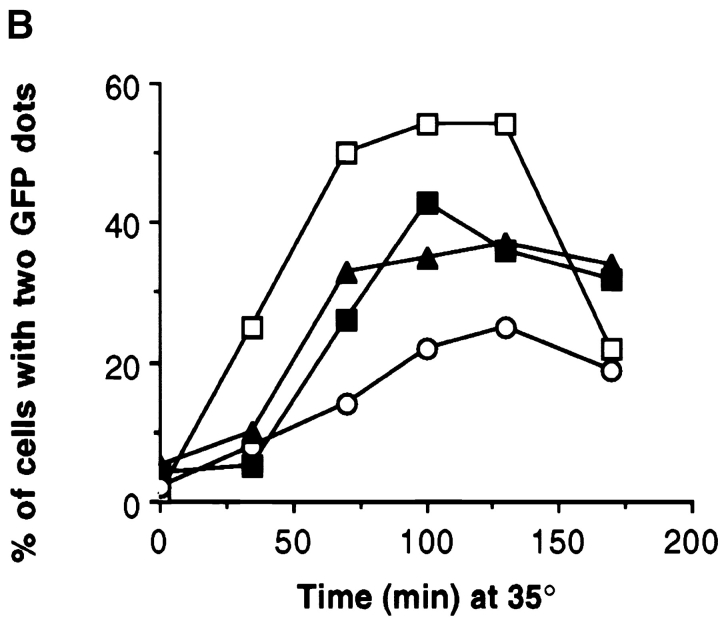
Figure 5.
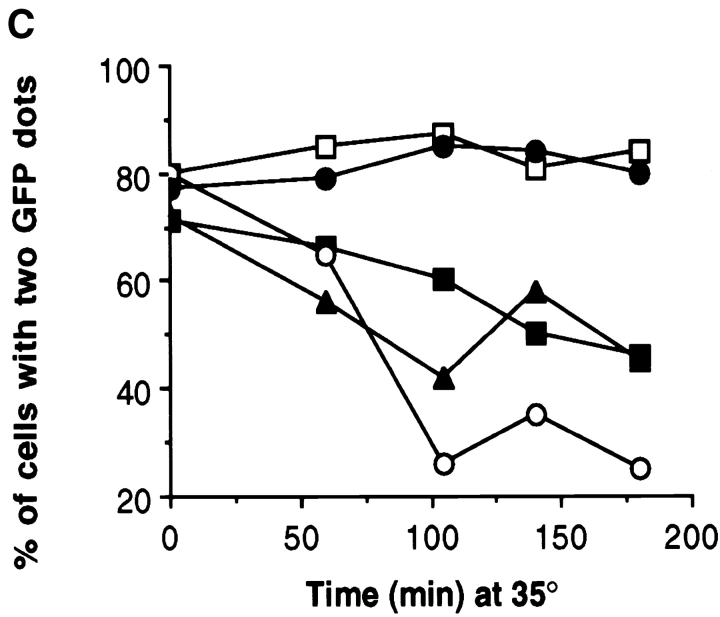
Bipolar spindle assembly and structural integrity requires either Kar3p or Kip3p. (A) Cells of the indicated genotypes also expressed the SPB component Nuf2p tagged with GFP. Cultures of the indicated genotypes were synchronized at 26°C with α-factor and released into media at 35°C. After 60 min, samples of live yeast cells were immediately observed under the microscope. Examples of wild-type (top row) and kar3Δ kip3Δ (pkar3-64) (bottom row) cells. (B) Quantitation of spindle assembly proficiency. Cells released from α-factor at 35°C, as in A, were observed and the percentage of cells exhibiting two clearly separated GFP dots was determined. The decreases observed in the later time point are due to cells that have entered the next cell cycle. (C) Quantitation of ability to maintain spindle structural integrity. Cells of the indicated genotype (and expressing Nuf2p-GFP) were synchronized at 26°C with hydroxyurea and shifted to 35°C for the indicated times, at which point the percentage of cells exhibiting two clearly separated GFP dots was determined. Strains used: wild-type (MAY5776), kar3Δ (pkar3-64) (MAY5777), kar3Δ kip3Δ (pkar3-64) (MAY5778), kar3Δ dyn1Δ (pkar3-64) (MAY5779), and kip3Δ (pkip3-14) (MAY6030).
To assess the ability of bipolar spindles to maintain their integrity after motor loss, we arrested the mutants with hydroxyurea at 26°C (a condition that allows bipolar spindle assembly) and then shifted to 35°C. Observation of the Nuf2p-GFP dots as a function of time revealed that wild-type and kip3-14 (a temperature-sensitive allele) cells maintained bipolar spindle structure (Fig. 5 C). In contrast, ∼80% of the kar3Δ (pkar3-64) cells displayed two Nuf2p-GFP dots before the temperature shift, but only ∼45% displayed two dots after 3 h at 35°C. The presence or absence of dynein in the kar3-64 cells made no apparent difference. Spindles also did not lose bipolarity when both Kip3p and dynein were inactivated (Cottingham and Hoyt 1997). However, the absence of Kip3p significantly enhanced the loss of bipolarity phenotype exhibited by kar3-64 cells; after 3 h, only ∼25% of kar3Δ kip3Δ (pkar3-64) cells had two clearly separated dots. Therefore, Kar3p and Kip3p overlap for a function required to maintain bipolar spindle integrity.
Thin-section electron microscopy was used to examine the spindle morphology of kar3Δ kip3Δ (pkar3-64) and wild-type cells treated with hydroxyurea and shifted to 35°C for 3 h (Fig. 6). The wild-type spindles were short and bipolar, with parallel SPBs separated by ∼1.5 μm and joined by a bundle of microtubules. No normal-appearing spindles were found in the kar3Δ kip3Δ (pkar3-64) culture. Of the spindles in which two poles could be visualized in one section, nine had SPBs located adjacent to one another. Six spindles had SPBs that were separated from between 0.2 and 1.2 μm (average = 0.53 ± 0.37 μm, SD), but were clearly defective. The poles of these spindles were not parallel. These findings agree with those from the Nuf2p-GFP analysis. Loss of Kar3p and Kip3p function caused preformed bipolar spindles to break with SPBs frequently moving close together.
Figure 6.
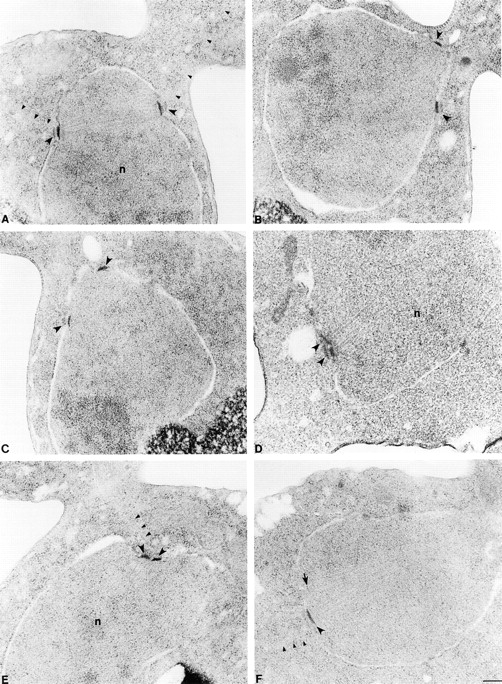
Electron microscopic analysis of kar3-64 kip3Δ cells. Wild-type (A) and kar3Δ kip3Δ (pkar3-64) cells (B–F) were arrested with hydroxyurea at 26°C, shifted to 35°C for 3 h, fixed, and sectioned for electron microscopy. A shows a normal preanaphase bipolar spindle. All the mutant spindles (B–F) appeared defective. In B and C, spindles have lost bipolarity. Although the poles are still slightly separated, they are not parallel. In D, E, and F spindles have collapsed with poles in a side by side orientation. All SPBs are marked with large arrowheads. Arrow in F marks a pole whose edge was just caught in this section. Small arrowheads mark cytoplasmic microtubules. n, nucleus. Bar, 0.2 μm. Strains used are listed in legend of Fig. 1.
Genetic Interactions with Kar3p Accessory Factors
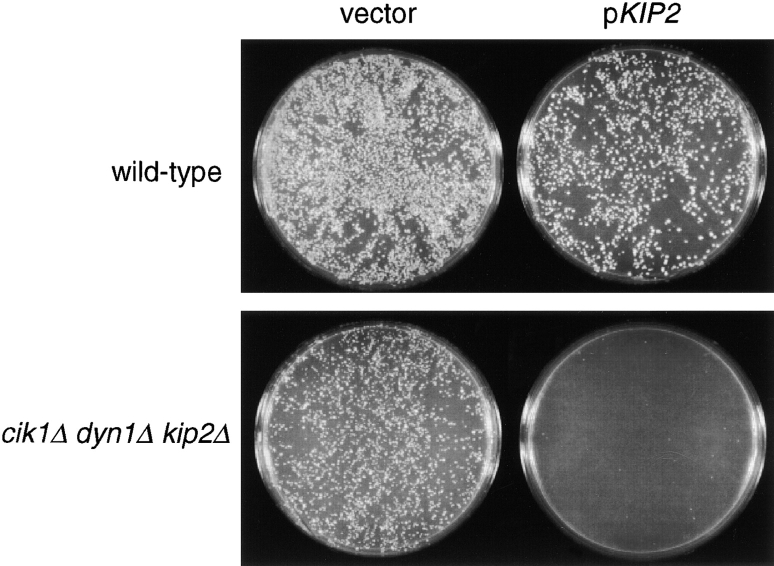
Kar3p forms distinct complexes with two homologous accessory factors, Cik1p and Vik1p (Page et al. 1994; Manning et al. 1999). cik1 and vik1 mutants display different phenotypes that each overlap in part with the phenotypes of kar3 cells. It is likely that these factors target Kar3p to different cellular roles, the precise nature of which have not been determined. We found that, similar to a kar3Δ mutant (Cottingham and Hoyt 1997), cik1Δ is lethal in combination with dyn1Δ, and this lethality is suppressed by kip2Δ. Viable cik1Δ dyn1Δ cells could only be created when KIP2 was deleted. Fig. 7 demonstrates that introduction of KIP2 by transformation into a cik1Δ dyn1Δ kip2Δ triple mutant prevented colony formation, indicating that lethality is due to the activity of KIP2. In this case, the similarity in genetic behavior of cik1Δ to kar3Δ suggests that it is the Cik1p–Kar3p complex that overlaps with dynein for an essential spindle positioning role. However, unlike kar3Δ, cik1Δ could be combined with kip3Δ yielding viable, healthy cells. Therefore, Cik1p–Kar3p complexes do not uniquely perform the function required for spindle integrity that overlaps with Kip3p. This supports the hypothesis that Cik1p is only required for a subset of Kar3p's roles (Manning et al. 1999).
Figure 7.
KIP2 is responsible for the growth defect of cik1Δ dyn1Δ cells. Cultures of wild-type (MAY591) and dyn1Δ cik1Δ kip2Δ (MAY5802) cells were split in two and transformed with either the KIP2-containing plasmid pFC50 or an empty LYS2 vector control (pRS317). Selective media (minus lysine) plates were incubated at 26°C for 3 d.
vik1Δ dyn1Δ and vik1Δ kip3Δ double mutant cells were also viable and healthy. Therefore, Vik1p–Kar3p complexes do not uniquely overlap for essential functions with either Dyn1p or Kip3p. However, cik1Δ vik1Δ kip3Δ triple mutants were inviable. Strain MAY6417 (cik1Δ vik1 kip3Δ [pKIP3 URA3]) could not survive loss of the KIP3 URA3 plasmid, as evidenced by its inability to segregate cells capable of growth on 5-FOA. This may indicate that the essential spindle integrity function that overlaps with Kip3p is performed by both Cik1p–Kar3p and Vik1p–Kar3p complexes, and that either alone is sufficient. Alternatively, the lethality of the cik1Δ vik1Δ kip3Δ triple mutant may reflect a nonspecific additive effect of combining three deleterious mutations.
Benomyl Can Partially Substitute for the Essential Function Performed by Either Kar3p or Kip3p
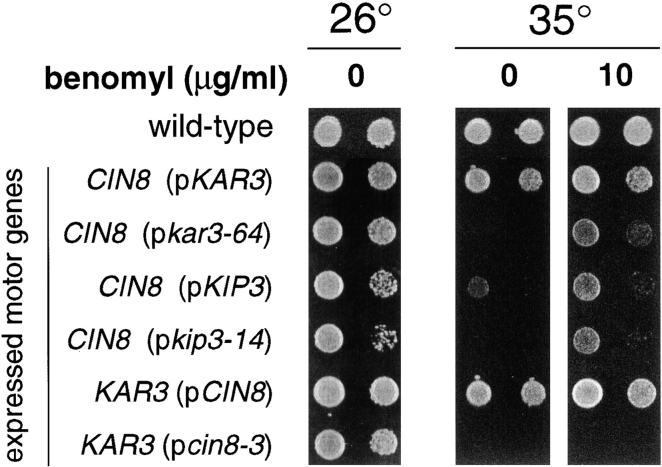
Kar3p can act as a microtubule destabilizer in vitro (Endow et al. 1994), and kar3Δ vegetative growth defects can be suppressed by the presence of the microtubule-destabilizing reagent benomyl (Roof et al. 1991; Saunders et al. 1997a) (Fig. 1, row A2; 37°C). We found that the temperature sensitivity of kar3Δ kip3Δ (pkar3-64) cells could be partially suppressed by the presence of 5–10 μg/ml benomyl in the media (Fig. 1, rows A10 and B9). The temperature sensitivity of kar3Δ kip3Δ (pkip3-14) cells could be suppressed by the same treatment as well (Fig. 1, row B7), similar to the suppression of kip3-30 (temperature-sensitive) kar3Δ reported by others (DeZwaan et al. 1997). This finding suggests that both Kar3p and Kip3p act to destabilize microtubules, and that this is an important aspect of the essential function for which they overlap (see Discussion). Indeed, all multiple motor mutant genotypes that are dependent upon either kar3-64 or kip3-14 for viability were suppressed for temperature-sensitive growth by benomyl (Fig. 1 and Fig. 8; also see below).
Figure 8.
Benomyl rescues the growth defects of cells expressing only CIN8 and either kar3-64 or kip3-14. Yeast cells were spotted onto solid rich media, with or without 10 μg/ml benomyl, and incubated at the indicated temperature for 3 d. The genotypes indicate the only two microtubule motor genes expressed in these cells (except for wild-type). Strains used (Note: p indicates a centromere plasmid-carried gene): wild-type (MAY1089), CIN8 pKAR3 (MAY5903), CIN8 pkar3-64 (MAY5904), CIN8 pKIP3 (MAY5905), CIN8 pkip3-14 (MAY5906), pCIN8 KAR3 (MAY5940), and pcin8-3 KAR3 (MAY5941).
Characterization of Minimal Microtubule-based Motor Strains
The S. cerevisiae genome encodes six kinesin-related motor genes and a single dynein heavy chain. In this and a previous study (Cottingham and Hoyt 1997) we described two viable triple mutant combinations. Cells deleted for DYN1 and either KAR3 or KIP3 were viable when the antagonistically acting KIP2 was deleted as well. Since a stripped-down or minimal system can help reveal how a process is accomplished, we attempted to reduce further the number of functional microtubule-based motor genes. In generating strains with reduced spindle motor function, evidence of aneuploidy was sometimes detected (i.e., non-Mendelian segregation of genetic markers). Therefore, we routinely checked all seven motor gene alleles by a PCR assay to verify that the genotypes constructed were correct (see Materials and Methods). We also note that for practical reasons, the minimal motor strains we created carried one of the active motor genes on a low-copy centromere plasmid. Centromere plasmids are usually present in one to two copies per haploid cell, but can increase slightly if selective pressure is applied (Futcher and Carbon 1986).
From the two viable triple mutants described above (dyn1Δ kip3Δ kip2Δ and dyn1Δ kar3Δ kip2Δ) we were able to delete an additional two genes encoding kinesin-related proteins, KIP1 and SMY1. KIP1 encodes the BimC family motor that is phenotypically less important (the more important BimC motor is encoded by CIN8) (Hoyt et al. 1992; Roof et al. 1992). SMY1 probably has no mitotic spindle role (see Introduction) but was deleted on account of its sequence relationship to kinesins. The strains created expressed only two of seven microtubule-based motor genes; CIN8 with either KAR3 or KIP3.
Cells expressing only CIN8 and KAR3 were quite healthy relative to wild-type, as judged by doubling time and other cell cycle criteria (Table ). Some difficulty in progression through mitosis was evident from the approximately twofold elevation of large-budded, mononucleate cells in log-phase cultures. Spindle positioning errors were also evident (by the appearance of bi and anucleate cell bodies), but at a level no higher than a dyn1 single mutant (Cottingham and Hoyt 1997). Cells expressing only CIN8 and KIP3 were considerably less well off. Their doubling time was significantly increased, and spindle positioning errors were approximately twice that of the CIN8 KAR3 cells. The plating efficiency of the CIN8 KIP3 cells was roughly half that of wild-type, indicating many dead cells in the culture. However, this figure is close to that observed for kar3 single mutants (Meluh and Rose 1990).
Table 2.
Phenotypes of Strains Expressing Only Two Microtubule Motor Genes
| Genotype | |||||
|---|---|---|---|---|---|
| Active genomic motor gene | Motor gene on plasmid | Doubling time | Plating efficiency | % Large-budded, mononucleate cells | % Bi or anucleate cells |
| h | % | ||||
| wt | – | 2.0 | 100 | 15 | 0 |
| CIN8 | KAR3 | 2.5 | 89 | 37 | 6 |
| KAR3 | CIN8 | 2.2 | 114 | 34.5 | 3.5 |
| CIN8 | KIP3 | 3.4 | 41 | 32 | 12 |
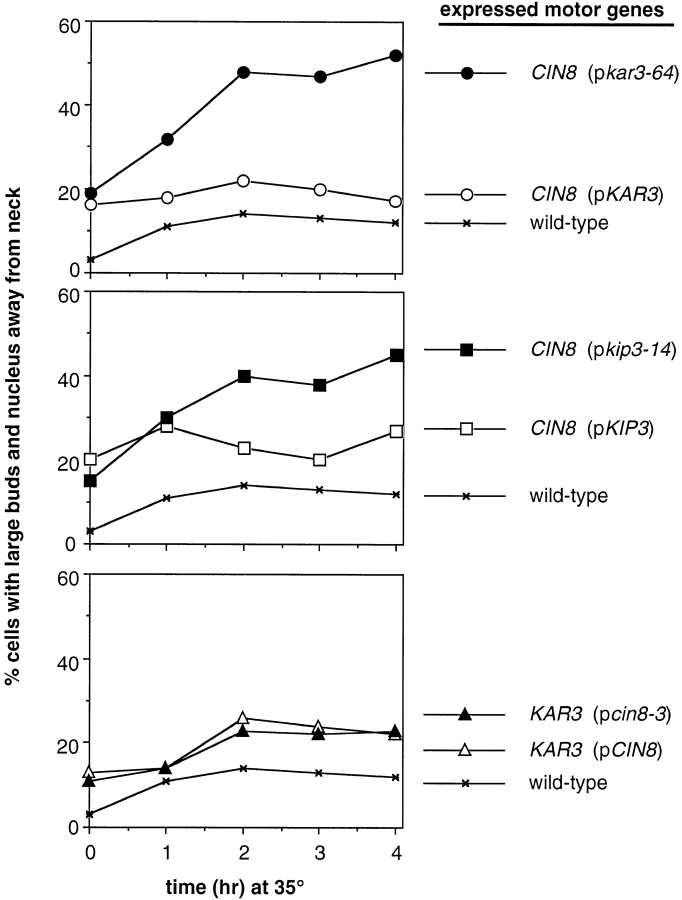
To determine if Cin8p Kar3p or Cin8p Kip3p represent the lowest possible motor complements, we created strains in which one of the two active motor genes was replaced with a temperature-sensitive allele. While cells surviving with only CIN8 KAR3 or CIN8 KIP3 were able to grow at 35°C, neither CIN8 kar3-64, CIN8 kip3-14, nor cin8-3 KAR3 cells grew at this elevated temperature (Fig. 8). Therefore, under normal growth conditions, Cin8p–Kar3p or Cin8p–Kip3p were the minimal motor sets that supported cell viability (but see below). We examined the effects on spindle integrity and positioning caused by elimination of the function of one of the two active motors in the two-motor strains. Wild-type, CIN8 kar3-64, CIN8 kip3-14, and cin8-3 KAR3 cells were arrested with hydroxyurea at 26°C and then shifted to 35°C for 3 h, maintaining the hydroxyurea block. The motor mutant strains exhibited greatly reduced numbers of bipolar spindles as judged by antitubulin immunofluorescence microscopy (78% for wild-type; 17% for CIN8 kar3-64; 9% for CIN8 kip3-14; and 7% for cin8-3 KAR3). This was an expected finding, because a BimC motor (either Cin8p or Kip1p) plus either Kar3p or Kip3p is required for bipolar spindle integrity. However, examination of the nuclear positioning proficiency of these strains did reveal differences. After hydroxyurea synchronization, the shift to 35°C caused nuclei in CIN8 kar3-64 and CIN8 kip3-14 cells, but not cin8-3 KAR3 cells, to mislocalize (Fig. 9). Therefore, it is reasonable to conclude that Kar3p or Kip3p are the only motors performing spindle positioning in their respective two-motor strains, and that Cin8p makes no detectable contribution to this process.
Figure 9.
Nuclear positioning proficiency of cells expressing only two microtubule motor genes. Cultures were arrested with hydroxyurea, shifted to 35°C and analyzed as in Fig. 2 and Fig. 3 B. The genotypes indicate the only two microtubule motor genes expressed in these cells (except for wild-type). Strains used are the same as in Fig. 8.
Finally, we found further support for a microtubule-destabilizing role for Kar3p and Kip3p in vivo. Addition of benomyl to the media could suppress the temperature-sensitive growth of cells surviving with only CIN8 kar3-64 or CIN8 kip3-14 (Fig. 8). The slow growth of the CIN8 KIP3 strain was exacerbated at elevated temperature, a phenotype that was also suppressed by benomyl. In contrast, the temperature sensitivity of the cin8-3 KAR3 strain was not relieved by benomyl. Therefore, although microtubule destabilization may be an important function of Kar3p and Kip3p, this does not appear to be a role for the BimC motor Cin8p.
Discussion
Kar3p and Kip3p Overlap for Spindle Structural and Positioning Functions
Our findings demonstrate that S. cerevisiae Kar3p and Kip3p motors perform overlapping roles contributing to both mitotic spindle positioning and spindle structural integrity. These two activities are primarily accomplished by distinct sets of microtubules, cytoplasmic and nuclear, respectively. Therefore, we conclude that Kar3p and Kip3p act on both sides of the nuclear envelope, a property that may be unique among the seven S. cerevisiae microtubule-based motors. We suggest that a major aspect of the nuclear and cytoplasmic functions of both of these motors is to destabilize microtubules.
In the absence of dynein, both Kar3p and Kip3p are required for spindle positioning and vigorous cell growth. Similar to cells in which dynein and Kip3p were eliminated (Cottingham and Hoyt 1997), we demonstrated here that elimination of dynein and Kar3p caused spindle mispositioning and the appearance of extremely long cytoplasmic microtubules. In both cases, the long microtubule phenotype and the spindle positioning defect were suppressed by the elimination of Kip2p. In cells missing both dynein and Kip2p, either Kar3p or Kip3p would now suffice for viability, probably becoming the sole spindle positioning motor in these cells. We demonstrated that in cells surviving with only Cin8p and Kar3p or Kip3p, all spindle positioning function appeared to be accomplished by Kar3p or Kip3p. It is not clear whether Kar3p and Kip3p normally act in distinct spindle positioning pathways or if they redundantly perform an activity required for the same pathway.
In previous studies, Kar3p was visualized only upon microtubules in the nucleus of mitotic cells (Page et al. 1994; Saunders et al. 1997a). However, our finding that Kar3p participates in spindle positioning indicates that it must be acting upon cytoplasmic microtubules as well. Two other findings indicate that Kar3p can act upon cytoplasmic microtubules: Kar3p influences the dynamics of cytoplasmic microtubules in mitotic cells (this study; Saunders et al. 1997a; Tirnauer et al. 1999), and acts upon cytoplasmic microtubules in karyogamy to draw together the nuclei of mated cells (Meluh and Rose 1990; Page et al. 1994).
Our studies also demonstrate a role for Kar3p and Kip3p in the nucleus, overlapping for a function essential for bipolar spindle assembly and structural integrity. Previous observations of spindle abnormalities in kar3Δ cells suggested a spindle integrity role for Kar3p (Meluh and Rose 1990; Saunders et al. 1997a). A nuclear role for Kip3p was suggested by its localization to nuclear (as well as cytoplasmic) microtubules (DeZwaan et al. 1997; Miller et al. 1998) and the extra long pre and postanaphase spindles observed in kip3Δ cells (Cottingham and Hoyt 1997; Straight et al. 1998). We demonstrated here that the structural integrity of the nuclear bipolar spindle requires the actions of at least one of these motors. Since loss of both motors is lethal, but causes only modest effects on spindle positioning, it is likely that spindle integrity is the essential activity for which they overlap. When both motors were inactivated, bipolar spindle assembly and the structural integrity of preformed spindles were compromised. The collapse of preformed spindles was slow, requiring over an hour to reach the maximal effect (Fig. 5 C). Spindle poles moved together, but many poles were still slightly separated. This contrasts with the rapid spindle collapse observed when the two BimC motors, Cin8p and Kip1p, were inactivated; all spindle poles were pulled to an adjacent position after as little as 5 min at the nonpermissive temperature (Saunders and Hoyt 1992). This indicates that Kar3p and Kip3p probably function differently from the BimC motors in maintaining bipolar spindle structure.
Kar3p forms functionally distinct complexes with two homologous accessory factors, Cik1p and Vik1p (Page and Snyder 1992; Page et al. 1994; Manning et al. 1999). Cik1p, but not Vik1p, is required for karyogamy, and Kar3p and Cik1p are interdependent for localization to cytoplasmic microtubules in mating cells. We found that, similar to a kar3Δ mutant, cik1Δ is lethal in combination with dyn1Δ, and this lethality is suppressed by kip2Δ. This overlap in function between Cik1p and dynein suggests that Kar3p complexed to Cik1p performs the cytoplasmic spindle positioning function. Our genetic studies indicated that neither Cik1p–Kar3p nor Vik1p–Kar3p complexes uniquely perform the function essential for spindle integrity that overlaps with Kip3p. However, since cik1Δ vik1Δ kip3Δ triple mutants were inviable, it remains possible that these two complexes redundantly provide this activity. Our findings lend further support to the hypothesis that Cik1p and Vik1p target the Kar3p motor to distinct cellular functions (Manning et al. 1999).
Spindle Motors Regulate Microtubule Dynamics
The molecular nature of the spindle positioning and structural defects caused by loss of Kar3p and Kip3p is likely to involve a defect in microtubule polymer length regulation. Recent studies have implicated microtubule motors as important regulators of microtubule dynamics. Notably, members of the XKCM1/MCAK family of vertebrate kinesins have been demonstrated to cause microtubule instability by promoting catastrophe events (Desai et al. 1999; McNally 1999). A microtubule-destabilizing activity for Kar3p has been detected in vitro (Endow et al. 1994). Findings reported here, combined with those of others, clearly demonstrate that Kar3p, Kip3p, and dynein either directly or indirectly act to shorten microtubules in vivo (Roof et al. 1991; Carminati and Stearns 1997; Cottingham and Hoyt 1997; DeZwaan et al. 1997; Saunders et al. 1997a; Huyett et al. 1998; Miller et al. 1998; Straight et al. 1998; Tirnauer et al. 1999). These observations can be generally summarized as follows: mutants deficient for any one of these motors display elongated microtubules and/or phenotypes that are suppressible by the antimicrotubule compound benomyl. Double mutants are much more severely affected; they are either inviable or very sick and display extremely long microtubule structures. In the studies reported here, numerous multiple motor mutant genotypes were created that were missing Kar3p or Kip3p and relied upon a temperature-sensitive form of the other for viability. All of these strains were suppressed for temperature-sensitive growth by the addition of the antimicrotubule compound benomyl to the media. Although the mechanism of suppression could be indirect, it is probable that benomyl is substituting for an activity normally accomplished by Kar3p and Kip3p. Depending upon the genotype tested, benomyl suppressed either the lethal spindle structural defect, the lethal spindle positioning defect, or both (i.e., for cells surviving only with CIN8 and kar3-64). Therefore, both the spindle structural and positioning defects caused by kar3 and kip3 mutations involve effects on microtubule stability.
It is not clear how increased microtubule stability or length caused the observed kar3 kip3 mutant spindle structural and positioning defects. The dynamic behavior of the ends of microtubules is probably important for functions such as interactions with kinetochores, antiparallel microtubules, and the cell cortex. Any of these processes may be disrupted by the inability to control the growth of microtubules. A less direct possibility for the kar3 kip3 spindle structural defect is that the longer or less dynamic spindle microtubules do not form proper interactions with the BimC motors, leading ultimately to spindle collapse. For the spindle positioning defect, it is possible that the extremely long cytoplasmic microtubules act to prohibit migration of the nucleus up to the cell neck or actually push the nucleus away.
The functional overlap between Kar3p and Kip3p could not have been predicted from the amino acid sequence of these gene products, since each is distinct. Kar3p defines a unique subclass of kinesin-related proteins characterized by their COOH terminus–located motor domain and their minus end–directed force production on microtubules (Meluh and Rose 1990; Endow et al. 1994; Middleton and Carbon 1994). The motor domain of Kip3p is located at its NH2 terminus. The directionality of Kip3p has not been determined, but a related motor from Drosophila has been found to exhibit plus end–directed activity (Pereira et al. 1997). The directionality of these motors may relate to activities for which they need not necessarily overlap. We predict that the only essential overlap between these motors is their ability to shorten both nuclear and cytoplasmic microtubules.
In contrast to Kar3p, Kip3p, and dynein, the Kip2p motor acts to increase the length of cytoplasmic microtubules. Loss of function causes extremely short cytoplasmic microtubules (Cottingham and Hoyt 1997; Huyett et al. 1998; Miller et al. 1998), whereas overexpression causes an extra long microtubule phenotype (Huyett et al. 1998; Cottingham, F.R., and M.A. Hoyt, unpublished observations). In addition to shortening the microtubules of kar3 dyn1 and kip3 dyn1 cells, kip2Δ also suppresses their lethal spindle positioning defect (this study; Cottingham and Hoyt 1997). The shortened microtubules may be the direct cause of the suppression, allowing spindles now to migrate unimpeded to the neck region. However, the possibility that other activities of Kip2p may antagonize spindle positioning in the absence of dynein cannot be ruled out. In contrast, the elimination of Kip2p did not suppress the lethal spindle structural defect of kar3 kip3 cells. This is consistent with its localization to cytoplasmic microtubules only (Huyett et al. 1998; Miller et al. 1998), as well as our failure as yet to detect any nuclear role for this motor.
The Minimal Motor Requirement for S. cerevisiae
Cytological descriptions of mitotic spindle function have revealed a complex series of motility events that affect the segregation of replicated chromatids. These events include bipolar spindle assembly, kinetochore capture and congression, and two chromosome-separating movements, anaphase A (movement of kinetochores towards spindle poles) and anaphase B (separation of spindle poles). In considering the minimal microtubule-based motor set required for S. cerevisiae viability, it first must be recognized that unlike higher eukaryotic cells, this yeast requires microtubules for only one essential role, mitotic spindle function. Nonetheless, our finding that S. cerevisiae can survive with only two microtubule-based motors is unexpected in light of the observed complexities of spindle motile behavior.
We were able to create viable strains expressing only two of seven microtubule-based motors; the BimC-type motor Cin8p combined with either Kar3p or Kip3p. Attempts to reduce motor function further caused expected deleterious phenotypic consequences and revealed that these represented the minimal sets required for viability. Removing the function of Kar3p (from Cin8p Kar3p-ts cells) or Kip3p (from Cin8p Kip3-ts cells) caused spindles to both collapse and misposition. Removing the function of Cin8p caused spindle collapse, an expected consequence since these cells were missing the other BimC motor, Kip1p (Saunders and Hoyt 1992). However, elimination of Cin8p function caused no detectable spindle positioning defect. Therefore, it seems likely that in these two-motor strains, all nuclear microtubule motor functions were accomplished by the combination of Cin8p and either Kar3p or Kip3p, and cytoplasmic microtubule motor functions accomplished by either Kar3p or Kip3p.
A few caveats must be considered regarding the conclusion that only two motors are required for viability. First, we cannot exclude the possibility that the observed vigorous growth of the two-motor cells could have been aided by suppressing mutations. The genotypes of created strains were verified with respect to the seven motor loci, so suppressors would necessarily affect genes that do not encode dyneins or kinesins. Second, we must strongly consider a contribution from microtubule-based motility mechanisms that do not include dyneins and kinesins. We are not aware of any force-producing microtubule-based ATPases outside of the dynein and kinesin families. However, an undiscovered class of motors cannot be excluded. All kinesins, dyneins, and myosins (as well as many other proteins) possess a domain with a nucleotide-binding sequence (G-X-X-X-X-G-K-T/S; (Saraste et al. 1990) and a distinct region of coiled α-helical coil sequence. An examination of the S. cerevisiae genome reveals gene products of unknown function that contain both of these sequence motifs (i.e., the gene products specified by YLR106C and YPL217C). Another possibility is a motility mechanism that uses a different form of force generation. For example, a protein that is able to couple the dynamic behavior at the ends of microtubules to force production (Koshland et al. 1988; Coue et al. 1991). Such a coupler need not necessarily be a nucleotidease. Therefore, the proper conclusion for our findings is that of the six kinesins plus one dynein encoded in the genome, only two kinesins are required for S. cerevisiae viability.
The presence of benomyl in the media partially suppressed the temperature-sensitive growth defects of the Cin8p Kar3p-ts and Cin8p Kip3p ts two-motor strains. However, we have been unable to find a condition allowing vigorous growth in the complete absence of Kar3p and Kip3p (cells surviving on Cin8p alone). Therefore, the temperature-sensitive forms of these motors may retain some activity at the elevated temperature that is required in addition to the microtubule-destabilizing activity of benomyl. Nonetheless, these findings further indicate that microtubule-destabilization is an important activity of Kar3p and Kip3p.
In summary, S. cerevisiae cells only require two microtubule-based motors to accomplish mitosis. The BimC motor Cin8p is required to assemble and elongate the bipolar spindle, probably by virtue of its ability to cross-link and slide microtubules. The second motor, either Kar3p or Kip3p, acts to promote microtubule shortening both within the nucleus and in the cytoplasm. Until the possible discovery of other spindle motility-producing mechanisms, we must entertain models in which all essential spindle motile activities can be accomplished by this limited set of motor proteins.
Acknowledgments
The authors wish to thank Laura Totis for technical assistance; Michael McCaffery for performing electron microscopy; Brendan Manning and Michael Snyder for strains; and Cindy Dougherty, Emily Hildebrandt, and Kevin McCabe for comments on the manuscript.
These experiments were supported by National Institutes of Health grant GM40714, awarded to M.A. Hoyt.
Footnotes
1.used in this paper: DAPI, 4,6-diamidino-2-phenylindole; GFP, green fluorescent protein; SPB, spindle pole body; 5-FOA, 5-fluoro-orotic acid
The first two authors contributed equally to this study.
References
- Byers B., Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Carminati J.L., Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F.R., Hoyt M.A. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically-acting microtubule motor proteins. J. Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M., Lombillo V.A., McIntosh J.R. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- DeZwaan T.M., Ellingson E., Pellman D., Roof D.M. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A., Kang S.J., Satterwhite L.L., Rose M.D., Skeen V.P., Salmon E.D. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D., Urrestarazu L.A., Vissers S., Jauniaux J.C., van Vliet-Reedijk J.C., Planta R.J., Gibbons I.R. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B., Carbon J. Toxic effects of excess cloned centromeres. Mol. Cell. Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J.R., Schott E.J., Kingsbury T.J., Cole N.B., Totis L.J., Bhattacharyya G., He L., Hoyt M.A. S. cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Geiser J.R. Genetic analysis of the mitotic spindle. Annu. Rev. Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., He L., Loo K.K., Saunders W.S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., He L., Totis L., Saunders W.S. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutants. Genetics. 1993;135:35–44. doi: 10.1093/genetics/135.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Hyman A.A., Bähler M. Motor proteins of the eukaryotic cytoskeleton. Proc. Natl. Acad. Sci. USA. 1997;94:12747–12748. doi: 10.1073/pnas.94.24.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyett A., Kahana J., Silver P., Zeng X., Saunders W.S. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 1998;111:295–301. doi: 10.1242/jcs.111.3.295. [DOI] [PubMed] [Google Scholar]
- Kahana J.A., Schnapp B.J., Silver P.A. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina A.S., Rogers G.C., Scholey J.M. The bimC family of kinesinsessential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta. 1998;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Koshland D.E., Mitchison T.J., Kirschner M.W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Yeh E., Hays T., Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S.H., Brown S.S. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae . J. Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S.H., Brown S.S. Smy1p, a kinesin-related protein that does not require microtubules. J. Cell Biol. 1998;140:873–883. doi: 10.1083/jcb.140.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B.D., Barrett J.G., Wallace J.A., Granok H., Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J. Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J. Microtubule dynamicscontrolling split ends. Curr. Biol. 1999;9:R274–R276. doi: 10.1016/s0960-9822(99)80177-8. [DOI] [PubMed] [Google Scholar]
- Meluh P.B., Rose M.D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Middleton K., Carbon J. KAR3-encoded kinesin is a minus-end-directed motor that functions with centromere binding proteins (CBF3) on an in vitro yeast kinetochore. Proc. Natl. Acad. Sci. USA. 1994;91:7212–7216. doi: 10.1073/pnas.91.15.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Heller K.K., Frisen L., Wallack D.L., Loayza D., Gammie A.E., Rose M.D. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell. 1998;9:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page B.D., Snyder M. CIK1a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae . Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Page B.D., Satterwhite L.L., Rose M.D., Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J. Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A.J., Dalby B., Stewart R.J., Doxsey S.J., Goldstein L.S.B. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila . J. Cell Biol. 1997;136:1081–1090. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J.R. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Pringle J.R., Hartwell L.H. The Saccharomyces cerevisiae cell cycle. In: Strathern J.N., Jones E.W., Broach J.R., editors. The Molecular Biology of the Yeast SaccharomycesLife Cycle and Inheritance. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1981. pp. 97–142. [Google Scholar]
- Pringle J.R., Adams A.E.M., Drubin D.G., Haarer B.K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Roof D.M., Meluh P.B., Rose M.D. Multiple kinesin-related proteins in yeast mitosis. Cold Spring Harbor Symp. Quant. Biol. 1991;56:693–703. doi: 10.1101/sqb.1991.056.01.078. [DOI] [PubMed] [Google Scholar]
- Roof D.M., Meluh P.B., Rose M.D. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P.R., Wittinghofer A. The P-loop, a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Saunders W.S., Hoyt M.A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders W.S., Koshland D., Eshel D., Gibbons I.R., Hoyt M.A. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S., Hornack D., Lengyel V., Deng D. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules J. Cell Biol. 137 1997. 417 431a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S., Lengyel V., Hoyt M.A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors Mol. Biol. Cell. 8 1997. 1025 1033b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. Methods in Yeast Genetics 1983. Cold Spring Harbor Press; Cold Spring Harbor, NY: pp. 120 [Google Scholar]
- Staples R.R., Dieckmann C.L. Generation of temperature-sensitive cbp1 strains of Saccharomyces cerevisiae by PCR mutagenesis and in vivo recombinationcharacteristics of the mutant strains imply that CBP1 is involved in stabilization and processing of cytochrome b pre-mRNA. Genetics. 1993;135:981–991. doi: 10.1093/genetics/135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Motoring to the finishkinesin and dynein work together to orient the mitotic spindle. J. Cell Biol. 1997;138:957–960. doi: 10.1083/jcb.138.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., O'Toole E., Berrueta L., Bierer B.E., Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachet A., Pöhlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]