Abstract
The cAMP-dependent protein kinase (PKA) is localized to specific subcellular compartments by association with A-kinase anchoring proteins (AKAPs). AKAPs are a family of functionally related proteins that bind the regulatory (R) subunit of PKA with high affinity and target the kinase to specific subcellular organelles. Recently, AKAP18, a low molecular weight plasma membrane AKAP that facilitates PKA-mediated phosphorylation of the L-type Ca2+ channel, was cloned. We now report the cloning of two additional isoforms of AKAP18, which we have designated AKAP18β and AKAP18γ, that arise from alternative mRNA splicing. The AKAP18 isoforms share a common R subunit binding site, but have distinct targeting domains. The original AKAP18 (renamed AKAP18α) and AKAP18β target the plasma membrane when expressed in HEK-293 cells, while AKAP18γ is cytosolic. When expressed in epithelial cells, AKAP18α is targeted to lateral membranes, whereas AKAP18β is accumulated at the apical membrane. A 23-amino acid insert, following the plasma membrane targeting domain, facilitates the association of AKAP18β with the apical membrane. The data suggest that AKAP18 isoforms are differentially targeted to modulate distinct intracellular signaling events. Furthermore, the data suggest that plasma membrane AKAPs may be targeted to subdomains of the cell surface, adding additional specificity in intracellular signaling.
Keywords: protein kinase A, AKAP, epithelia, targeting, green fluorescent protein
Hormonally induced changes in intracellular cAMP influence many cellular processes, including growth and differentiation, vesicular trafficking, cellular metabolism, and ion channel activity (Taylor et al. 1990; Francis and Corbin 1994). These pleiotropic effects are predominantly due to activation of the cAMP-dependent protein kinase (PKA). The PKA holoenzyme is a tetramer containing two regulatory (R) and two catalytic (C) subunits; binding of cAMP causes dissociation of the R and C subunits, to reversibly activate the enzyme (Taylor et al. 1990). Although PKA has a broad substrate specificity in vitro, activation of cAMP-mediated processes by cell surface receptors results in phosphorylation of a specific and restricted set of protein substrates. Thus, compartmentalization of PKA in close proximity to specific targets may be crucial for controlling the specificity and efficiency of cAMP-mediated signaling in cells (Rubin 1994; Dell'Acqua and Scott 1997; Schillace and Scott 1999b). Furthermore, compartmentalization of PKA, together with other protein kinases or protein phosphatases, may facilitate appropriate cross-talk between signaling pathways (Pawson and Scott 1997).
The identification of a diverse family of A-kinase anchoring proteins (AKAPs) suggests that compartmentalization of PKA is a general mechanism for modulation of cAMP-mediated signaling. AKAPs bind with high affinity to the NH2 terminus of the type II R subunit (RII) dimer, via an amphipathic helix in each AKAP (Carr et al. 1991; Newlon et al. 1999). Recent data suggests that some AKAPs can also bind RI subunits (Huang et al. 1997b; Angelo and Rubin 1998), although the binding affinity is lower than for RII (Burton et al. 1997) and the physiological significance of the AKAP-RI interaction is yet to be established. In addition, each AKAP contains a unique targeting motif that directs the AKAP-mediated regulatory complex to the appropriate subcellular compartment (Glantz et al. 1993; Lester et al. 1996; Chen et al. 1997; Huang et al. 1999). Immunohistochemical and biochemical analyses have identified AKAPs that associate with cytoskeletal elements, mitochondria, ER, the Golgi complex, peroxisomes, and the centrosome (McCartney et al. 1995; Lester et al. 1996; Chen et al. 1997; Dransfield et al. 1997; Dong et al. 1998; Schmidt et al. 1999; Witczak et al. 1999).
AKAPs are implicated in the regulation of many plasma membrane-associated events, including modulation of ion channels and regulation of hormone secretion (Rosenmund et al. 1994; Gao et al. 1997; Lester et al. 1997; Ali et al. 1998; Fraser et al. 1998; Gray et al. 1998; Tibbs et al. 1998; Klussmann et al. 1999). Some AKAPs target the cell surface by association with submembranous cytoskeletal elements, and others by lipid modification or direct association with membrane phospholipids (Glantz et al. 1993; Dransfield et al. 1997; Dell'Acqua et al. 1998; Dong et al. 1998; Fraser et al. 1998; Gray et al. 1998).
AKAP18 (also named AKAP15) is a low molecular weight AKAP that associates with the cell membrane via NH2-terminal lipid modifications (Fraser et al. 1998; Gray et al. 1998). In rat skeletal muscle, AKAP18 is concentrated in transverse tubules and overlaps the distribution of the L-type Ca2+ channel (Gray et al. 1997, Gray et al. 1998). Coexpression of AKAP18 and cardiac L-type Ca2+ channels in fibroblasts resulted in a significant increase in cAMP-responsive Ca2+ currents (Fraser et al. 1998), and the two proteins can be copurified after expression in fibroblasts (Gray et al. 1998). Together, these data indicate that AKAP18 may serve to functionally couple PKA and ion channels at the cell surface. We cloned two novel AKAP18-related cDNAs (named AKAP18β and AKAP18γ) using expression library screening and PCR-based approaches. Therefore, we compared the expression of these AKAP18-related mRNAs and proteins in cells and examined their subcellular distributions in HEK-293 and epithelial cell lines. Our data suggest that AKAP18-related proteins arise secondary to alternative mRNA splicing of a single gene, to generate a family of proteins that are differentially targeted and contain a common RII binding site.
Materials and Methods
Cloning of AKAP18β and AKAP18γ
The pET11.RIIα plasmid was transformed into BL21(DE3) pLysS Escherichia coli and grown at 37°C; protein expression was induced by 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 3 h at 37°C, and the RIIα purified on cAMP agarose as described (Scott et al. 1990). The purified RIIα protein was dialyzed into 50 mM sodium bicarbonate, pH 8.5, and concentrated by centrifugation in a Biomax-10K centrifugal filter (Millipore Inc.). Purified RIIα (10 μM) was biotinylated by addition of 100 μM EZ-Link NHS-LC-Biotin (Pierce Chemical Co.). Excess biotin was removed by dialysis in 10 mM Tris-HCl, pH 7.4, + 0.15 M NaCl.
Biotin-RIIα was used as a probe to screen a λTriplEx human lung cDNA library (CLONTECH). Biotin-RIIα (10 nM) was prebound to 0.5 μg/ml streptavidin-alkaline phosphatase (SA-AP) in 50 ml TTBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20) for 4 h at 4°C, and the filters were incubated overnight at 4°C in TTBS containing the biotin-RIIα/SA-AP complex. After extensive washing in TTBS, bound RIIα/SA-AP complexes were visualized as described (Sparks et al. 1996). Plasmids were rescued from λTriplEx phage by in vivo excision, and inserts sequenced at the University of North Carolina Sequencing Facility.
An ∼2-kb cDNA clone encoding a novel AKAP was isolated from the library screen; after sequencing, this clone was designated AKAP18γ. To obtain upstream coding sequence, 5′ RACE was performed using Marathon-Ready human pancreas cDNA (CLONTECH); RACE products were cloned into pT-Adv (CLONTECH) and sequenced. AKAP18β was cloned using reverse transcriptase (RT)-PCR of human pancreas cDNA and KlenTaq DNA polymerase (CLONTECH), using oligonucleotides designed based on the previously reported human AKAP18 sequence. PCR products were excised from the gel and subcloned into pT-Adv (CLONTECH).
For RT-PCR analyses, total RNA from cultured cells was extracted using RNA STAT60 (Tel-test “B” Inc.) and treated with DNase (Promega). First strand cDNA was synthesized using Superscript II reverse transcriptase (GIBCO BRL) primed with random hexamers. PCR was performed using Taq DNA polymerase (GIBCO BRL) and AKAP18α specific primers. PCR products were purified, ligated into pT-Adv cloning vector, and sequenced.
Northern Blot, Southern Blot, and Screening of Genomic Libraries
A human multiple tissue Northern blot (CLONTECH) was probed with a 32P-labeled random-primed cDNA probe using the unique region of AKAP18γ (nucleotides [nt] 357–689). The blot was prehybridized at 68°C for 30 min and incubated with the probe at 68°C for 1 h in ExpressHyb (CLONTECH). After incubation, the blot was washed at room temperature for 30 min in 2× SSC + 0.05% SDS, followed by 0.1× SSC + 0.1% SDS for 40 min at 50°C. Blots were stripped and reprobed with 32P-labeled β-actin probe (CLONTECH). All blots were analyzed using a STORM840 PhosphorImager.
For Southern blot analysis, human genomic DNA (CLONTECH) digested with 100 units of BamHI, EcoRV, Hind III, or XbaI, was electrophoresed on 1% agarose gels. Gels were soaked in 0.5 M NaOH/1.5 M NaCl for 20 min at room temperature and washed in neutralization solution. The DNA was transferred to GeneScreen (New England Nuclear Life Sciences) by capillary diffusion in 20× SSC overnight at room temperature. Hybridizations were carried out at 42°C in ExpressHyb (CLONTECH) using a 32P-labeled cDNA probe common to all known AKAP18 isoforms (nt 106–243 of AKAP18α), and membranes were washed as described above.
A genomic DNA library was created for the mouse strain ELM3 in the Lambda FIX II vector (Stratagene). The library was screened with an α[32P]dCTP random-primed probe representing full-length AKAP18α (nt 205–450) or the sequence common to the three identified AKAP18 isoforms (nt 256–450).
Generation of AKAP18 Plasmids
The coding sequences of AKAP18α, AKAP18β, and AKAP18γ were amplified by PCR using human pancreas cDNA as template. The sense primers incorporated an EcoRI site at the 5′ end and overlapped the initiator methionine of each AKAP18 isoform. The antisense primer overlapped the COOH terminus and stop codon, and incorporated a BamHI site. The PCR fragments were digested with EcoRI and BamHI, and subcloned into pcDNA3.1(−) (Invitrogen) digested with the same enzymes.
The cDNA encoding each AKAP18 isoform was also fused in-frame at the 3′ end with the cDNA encoding GFP. The coding regions of AKAP18α, AKAP18β, and AKAP18γ were amplified by PCR using the sense primers described above and a single antisense primer designed to remove the stop codon and incorporate a BamHI site at the 3′ end. Similarly, the 1-16αβ, 1-44β, and 17-44β constructs were generated by PCR using AKAP18β DNA as template. DNA sequencing confirmed the absence of mutations in all constructs generated by PCR.
Cell Culture and Transfection of Cells
CHO, human embryonic kidney (HEK-293), and MDCK type II were obtained from the American Type Culture Collection. The NIT-1 cell line, derived from mouse pancreatic β cells, was provided by Dr. Lloyd Fricker (Albert Einstein School of Medicine, Bronx, NY). Cultured cells were grown in the appropriate media as described previously (Chen et al. 1998; Short et al. 1998). CHO, HEK-293, or MDCK cells were grown to 30–50% confluency and transfected with the appropriate plasmid in Effectene reagent (Qiagen) for 15 h in complete media. Stable cell lines were selected for 3 wk in media containing 0.5 mg/ml G418, and cloned by serial dilution.
Western Blot and Immunoprecipitation
Rabbit antisera directed against AKAP18α was generated in rabbits using recombinant AKAP18α and affinity-purified as described (Fraser et al. 1998); these antisera also recognize AKAP18β and AKAP18γ. Transfected cells were washed once with PBS and lysed in ice-cold buffer (20 mM Hepes, pH 7.4, 20 mM NaCl, 5 mM EDTA, 2 μg/ml leupeptin, 1.6 μg/ml benzamidine, 0.3 μg/ml PMSF), with or without 1.0% Triton X-100. For some experiments, whole cell lysates were separated into soluble and particulate fractions by centrifugation at 40,000 g for 30 min at 4°C, and protein concentrations were determined using the BCA protein assay kit (Pierce Chemical Co.). For immunoblotting, proteins were resolved on 12.5 or 15% SDS-PAGE gels and transferred to Immobilon-P (Millipore Inc.). Western blots were performed as described previously (Fraser et al. 1998; Short et al. 1998) using rabbit anti-GFP (1:1,000; CLONTECH), affinity-purified rabbit anti-AKAP18 (VO57; 1 μg/ml), or mouse anti-C subunit (1 μg/ml; Transduction Laboratories). Immunoprecipitations were carried out overnight at 4°C using 500 μg of protein (whole cell lysate or soluble and particulate fractions) and 1 μg affinity-purified AKAP18 antiserum (Fraser et al. 1998). Immune complexes were collected on protein A agarose and electrophoresed on 12.5 or 15% SDS-PAGE gels. RII overlays were performed as described previously using 32P-labeled mouse RIIα (Fraser et al. 1998).
Confocal Microscopy
Rabbit antisera that specifically recognize AKAP18β (NC 257) were generated using residues 15–43 of AKAP18β coupled with keyhole limpet cyanin as immunogen. Complement proteins were removed from the whole serum by incubation with DEAE-Blue dextran (Pierce Chemical Co.). Transfected cells were grown on glass coverslips (HEK-293 and MDCK cells) or Transwell filters (MDCK cells). MDCK cells were grown until confluent monolayers were observed and transepithelial resistances were >1,000 ohm·cm2. Cells were washed once with PBS and then fixed for 20 min in fresh 4.0% paraformaldehyde prepared in PBS. For immunocytochemistry, cells were permeabilized for 10 min in acetone/methanol (1:1), washed three times with PBS, and blocked at room temperature in 4% nonfat dry milk, 2 mg/ml BSA, and 0.1% Triton X-100 in PBS. Cells were rinsed in PBS and incubated for 1 h at room temperature with affinity-purified antisera as noted in the figure legends. After extensive washing, Texas red-conjugated secondary antibodies were applied for 1 h at room temperature. Cells were washed and mounted with VectaShield mounting medium (Vector Laboratories) and analyzed by confocal microscopy as described (Chen et al. 1998).
Results
Cloning of Two Additional AKAP18 Isoforms
We screened a lung cDNA expression library using biotinylated RII as probe and identified a novel cDNA that shares sequence homology with AKAP18, a previously identified membrane-associated AKAP (Fraser et al. 1998; Gray et al. 1998). The cDNA was identical to AKAP18 at the 3′ end, but shared little homology with AKAP18 at the 5′ end (data not shown). Therefore, we designated the cDNA and protein product AKAP18γ, to indicate its relationship to other AKAP18 family members, including the previously reported AKAP18 (renamed AKAP18α) and AKAP18β (discussed below). The original AKAP18γ cDNA contained a single open reading frame, but no consensus ribosome binding site or initiator methionine. Therefore, we used rapid amplification of cDNA ends (RACE) with human pancreas cDNA as template to obtain the full-length AKAP18γ sequence. The longest AKAP18γ cDNA isolated was 2,917 nt in length with a single open reading frame from nt 107 to 1086 (these sequence data have been submitted to the GenBank/EMBL/DDBJ databases under the accession number AF152929). The upstream cDNA sequence contains three in-frame stop codons, and there are stop codons in the alternative reading frames (data not shown), suggesting that this is the correct open reading frame.
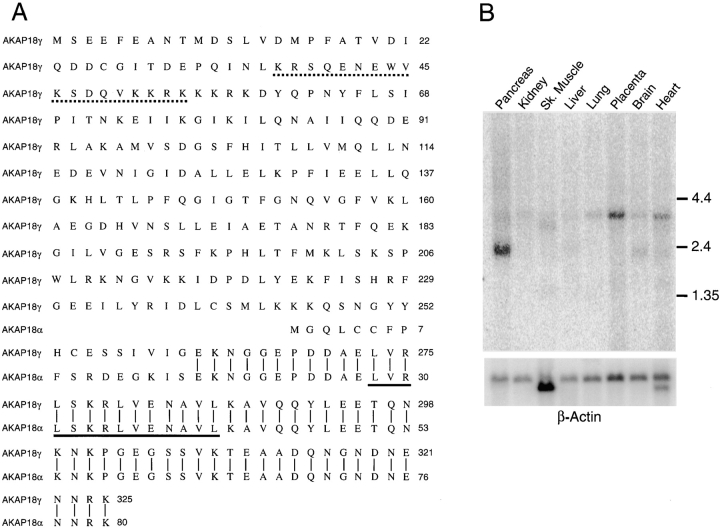
The AKAP18γ cDNA encodes a protein of 326 amino acids with a calculated molecular mass of 37 kD and a pI of 5.8. The first 262 amino acids are unique and do not share significant homology with any known proteins in GenBank/EMBL/DDBJ databases. However, the last 64 amino acids are identical to human AKAP18α and include a conserved RII binding site (Fig. 1 A). There are no consensus myristoylation or palmitoylation sites at the NH2 terminus of AKAP18γ, suggesting that the AKAP18γ protein is not modified by lipid side chains. We used PSORT (Nakai and Kanehisa 1992) to predict potential subcellular targeting sequences, and found that amino acids 37 to 54 of AKAP18γ fit the specifications for a consensus nuclear localization signal (Gerace 1992; Gorlich 1997; Nigg 1997).
Figure 1.
Cloning of human AKAP18γ. A, Comparison of human AKAP18γ and AKAP18α amino acid sequences. Common residues are indicated by a vertical line, a dashed line marks the putative nuclear localization signal, and the RII binding site is underlined. B, A human multiple tissue Northern blot was hybridized with a random primed 32P-labeled probe generated against the unique region of AKAP18γ (nt 357–689). The blot was stripped and rehybridized with a β-actin probe. mRNA size markers are shown in kb. Similar results were obtained in two separate blots.
In previous studies, mRNAs of ∼2.4-, 3.6-, and 4.3-kb were observed in rat and human tissues using the AKAP18α coding region as a probe (Fraser et al. 1998; Gray et al. 1998). We used Northern blot analysis to determine whether any of these mRNAs represented the AKAP18γ mRNA and to determine the tissue distribution of the message (Fig. 1 B). Using a radiolabeled probe directed against the unique region of AKAP18γ, we detected a dominant transcript of ∼4.3-kb in heart, brain, placenta, lung, and pancreas, and a smaller transcript of 2.4-kb, which is abundantly expressed in pancreas.
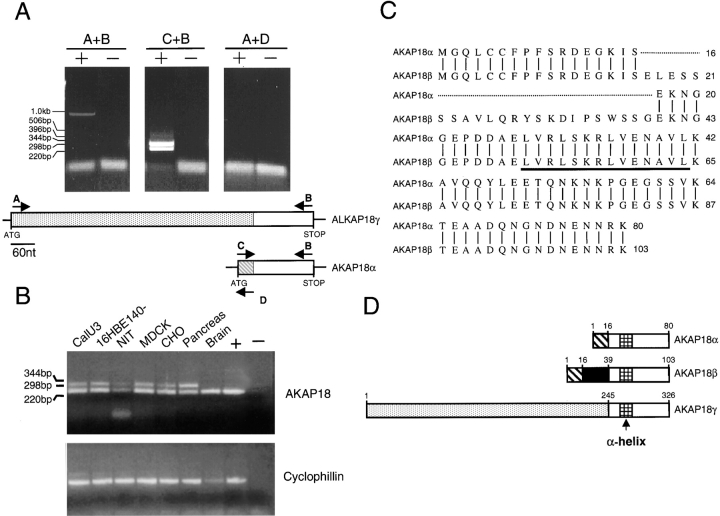
To further compare the expressions of AKAP18α and AKAP18γ, we used unique sense primers paired with a common antisense primer in RT-PCR reactions. Using a sense primer specific for the AKAP18γ cDNA, we obtained a 979-nt product whose sequence exactly matched that obtained from the cDNA library screen and 5′ RACE reactions (Fig. 2 A). Surprisingly, primers designed to specifically amplify AKAP18α consistently amplified two bands of 246 and 315 nt (Fig. 2 A). The 246-nt fragment was the expected size of the AKAP18α product, which was confirmed by DNA sequencing. The sequence of the 315-nt fragment matched AKAP18α at the 5′ and 3′ ends, but contained a 69-bp insert (these sequence data have been submitted to the GenBank/EMBL/ DDBJ databases under the accession number AF161075); we named this cDNA AKAP18β. We used RT-PCR to determine whether AKAP18α and -β are differentially expressed in cell lines and tissues. Both cDNAs were detected in fibroblast, endocrine, and epithelial cell lines, indicating that the two mRNAs are broadly expressed (Fig. 2 B). Although we were unable to reliably amplify the AKAP18β cDNA from human brain, a weak signal was observed in some reactions (data not shown).
Figure 2.
Cloning of AKAP18β. A, Primers used to amplify human pancreas cDNA are shown schematically; PCR was performed using human pancreas cDNA and the primer pairs indicated. + indicates addition of 1 ng cDNA and − indicates no cDNA. Samples were electrophoresed on 1% agarose gels and visualized with ethidium bromide. mRNA size markers are shown in kb. No PCR product was observed using primers A + D, indicating that cDNAs containing both AKAP18γ- and -α–specific sequence does not exist in this tissue. The data are representative of three PCR reactions using human pancreas or lung cDNA as template. B, RT-PCR with AKAP18α-specific primers (C + B) was performed using cDNA from cultured cell lines, human pancreas, and human brain. cDNA quality was verified by amplifying each sample with human cyclophillin primers. + indicates AKAP18α or cyclophilin plasmid control and – indicates no addition of template. Samples were electrophoresed on 1.0% agarose gels and visualized with ethidium bromide. DNA size markers are shown in kb. Data are representative of two separate experiments. C, Comparison of AKAP18α and AKAP18β amino acid sequences. Common residues are indicated by a vertical line and the RII binding site is underlined. D, Schematic of three AKAP18 isoforms. The proteins are drawn to scale and unique regions marked by different shadings.
The first 16 amino acids of human AKAP18β are identical to AKAP18α and are followed by an insert of 23 unique amino acids (Fig. 2 C). Distal to this 23-amino acid insert, AKAP18α and -β are identical to each other (Fig. 2C and Fig. D). Thus, we have identified three AKAP18 isoforms (named α, β, and γ) which share a common RII binding site, but have unique NH2-terminal sequences (Fig. 2 D).
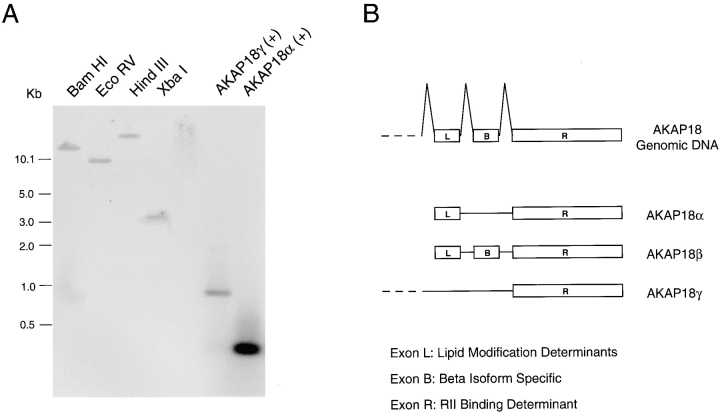
We used Southern blot analysis to determine whether these AKAP18 isoforms arise from a single gene. Genomic DNA was digested and hybridized with a probe common to all three AKAP18-related cDNAs. A single fragment was visualized on Southern blots, suggesting that AKAP18α, -β, and -γ mRNAs arise as alternate products of one gene (Fig. 3 A). This is consistent with the fact that the 3′ untranslated regions of AKAP18α, -β, and -γ are identical (data not shown).
Figure 3.
Alternative splicing gives rise to three distinct AKAP18 mRNAs. A, Human genomic DNA digested with the enzymes indicated, or positive control cDNAs were hybridized with a radiolabeled probe overlapping the common region of all three AKAP18 isoforms. DNA size standards are shown in kb. B, Schematic of the mouse AKAP18 gene and mRNAs encoding each AKAP18 isoform. Exons encoding the lipid modification determinant (Exon L), the AKAP18β-specific sequence (Exon B), and the RII binding region (Exon R) have been identified.
Preliminary analysis of mouse AKAP18 genomic sequence (Fig. 3 B) indicates that residues 1–16 of AKAP18α (also contained in AKAP18β) are encoded by a single exon; this exon contains the determinants for lipid modification. Another short exon encodes the 23-amino acid residues specific to AKAP18β. In addition, the COOH-terminal RII binding domain found in all AKAP18 isoforms is contained within a single exon. Taken together, the data indicate that alternative splicing of a single AKAP18 gene gives rise to (at least) three distinct AKAP18 mRNAs encoding different protein products.
Protein Analysis of AKAP18 Isoforms
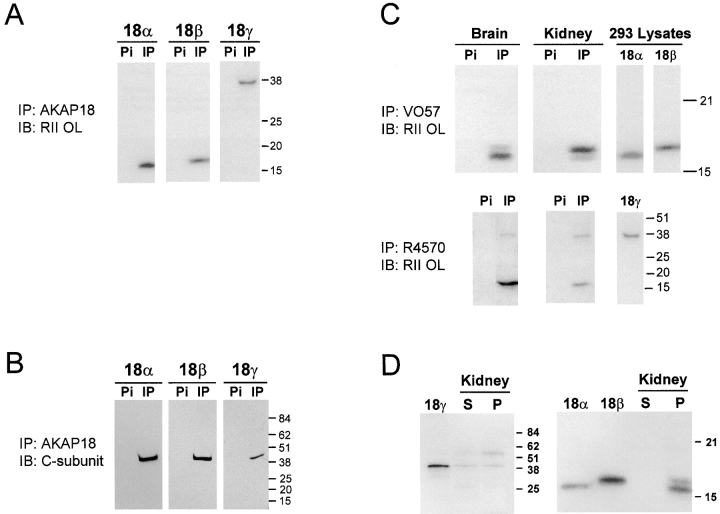
To determine whether each AKAP18 isoform is capable of binding PKA, we transiently transfected HEK-293 cells with cDNA encoding each isoform, and immunoprecipitated the expressed proteins with AKAP18-specific antisera. As expected, each of the immunoprecipitated AKAP18 isoforms was able to bind the RII subunit in overlay assays (Fig. 4 A). However, a new classification for AKAPs has been proposed, whereby the anchoring proteins must be able to interact with the PKA holoenzyme inside cells (Colledge and Scott 1999). Therefore, we also probed AKAP18-specific immunoprecipitates with antisera directed against the C subunit of PKA. The C subunit was detected in immunoprecipitates for each isoform, but was absent from control experiments with preimmune sera (Fig. 4 B). Thus, each of the AKAP18 isoforms functions as a bonafide AKAP in cells.
Figure 4.
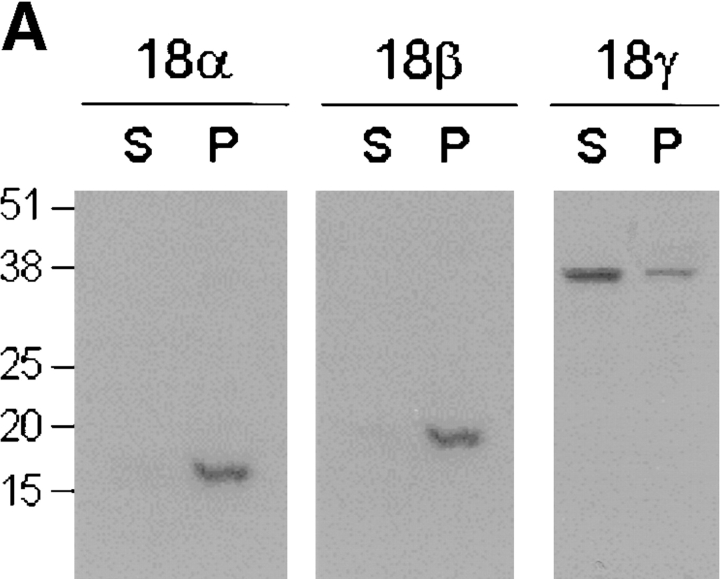
Analysis of AKAP18-related proteins. A, Lysates prepared from HEK-293 cells transiently expressing individual AKAP18 isoforms were immunoprecipitated with antisera directed against AKAP18. Samples were separated by SDS-PAGE and proteins visualized by RII overlay. B, Lysates prepared from HEK-293 cells transiently expressing individual AKAP18 isoforms were immunoprecipitated with rabbit antisera directed against AKAP18. Samples were separated by SDS-PAGE and blots were probed with mouse anti-PKA C subunit. C, Whole rat brain or kidney lysates were immunoprecipitated with rabbit antisera directed against AKAP18 as indicated. Samples were electrophoresed on SDS-PAGE and visualized by RII overlay. D, Whole rat kidney lysates were fractionated into soluble (S) and particulate (P) fractions in hypotonic buffers lacking detergent. Equal ratios of the soluble and particulate fractions were electrophoresed on SDS-PAGE and visualized by RII overlay. For C and D, lysates were also prepared from HEK-293 cells transiently transfected with AKAP18 isoforms, and samples were electrophoresed and analyzed in parallel with the tissue samples. For all panels, protein size standards are shown in kD and the data are representative of at least four similar experiments. IP, immunoprecipitation; IB, immunoblotting; Pi, preimmune sera.
We next determined whether each of the novel AKAP18 isoforms was expressed in rat tissues. We chose kidney as a tissue where mRNA was detected for all three AKAP18 isoforms, and brain as a tissue source where AKAP18β mRNA levels were low (Fig. 1 B and 2 B). Detergent soluble extracts were prepared from brain and kidney, immunoprecipitations were carried out with AKAP18 specific antisera (VO57 or R4570), and AKAPs were detected by RII overlay (Fig. 4 C). Although there was less AKAP18β protein in brain, two bands immunoprecipitated from both brain and kidney with VO57 antiserum corresponding to AKAP18α and -β. Both of these proteins were preferentially accumulated in the particulate fraction of rat kidney (Fig. 4 D). Bands corresponding to AKAP18α and -γ were immunoprecipitated from both tissues using R4570 antisera (Fig. 4 C). We also examined the distribution of AKAP18γ in rat kidney, and it was equally distributed in the soluble and particulate fractions (Fig. 4 D). Collectively, these results suggest that all three cloned AKAP18 isoforms are expressed as proteins in cells.
Localization of AKAP18 Isoforms in Cells
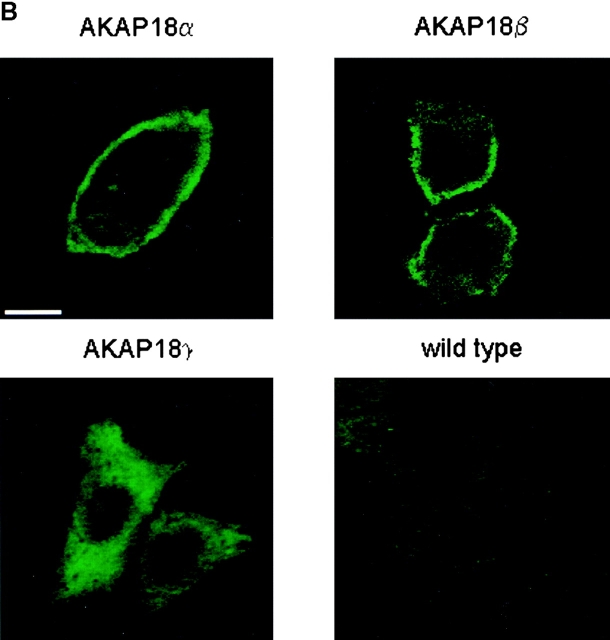
Accumulating evidence suggests that AKAPs compartmentalize PKA at discrete subcellular compartments to facilitate cAMP-responsive events and control the specificity of intracellular signaling (Colledge and Scott 1999). Therefore, each AKAP contains a targeting domain responsible for localizing PKA to specific organelles or subcellular compartments (Schillace and Scott 1999a). The targeting of AKAP18α is dependent upon lipid modification through myristylation of Gly and palmitoylation of Cys4 and Cys5 (Fraser et al. 1998). Accordingly, the first ten amino acids of AKAP18α encompass the minimal sequence necessary to target a reporter protein to the plasma membrane (Fraser et al. 1998). To determine whether AKAP18 isoforms are differentially targeted, we transiently transfected cDNAs encoding each construct into HEK-293 cells and compared the intracellular distribution of the proteins by differential fractionation and immunofluorescent microscopy. AKAP18α and -β fractionated exclusively with the cell membranes in buffers lacking detergent (Fig. 5 A) and both proteins were distributed at the cell surface (Fig. 5 B). This is consistent with the segregation of endogenous AKAP18α and -β with the particulate fraction of rat kidney (Fig. 4 D). In contrast, a significant fraction of the overexpressed AKAP18γ partitioned with the soluble fraction, although ∼20% was found in the particulate fraction (Fig. 5 A). The expressed AKAP18γ protein was visualized throughout the cytoplasm of cells, but did not significantly accumulate in the nucleus (Fig. 5 B). The even distribution of the endogenous AKAP18γ in the soluble and particulate fractions of rat kidney suggests that overexpression of AKAP18γ in HEK-293 cells saturates a protein–protein interaction required to maintain a particulate pool of this isoform.
Figure 5.
Targeting of AKAP18 isoforms in HEK-293 cells. A, HEK-293 cells were transiently transfected with cDNAs encoding each AKAP18 isoform. Cells were separated into soluble (S) and particulate (P) fractions and 25 μg of each sample was resolved by SDS-PAGE. The distribution of AKAP18 isoforms was determined by immunoblot analysis using rabbit antisera directed against AKAP18α. Protein size standards are shown in kD. The data are representative of three similar experiments. B, HEK-293 cells were transiently transfected with AKAP18 cDNAs and plated on glass coverslips. Eight hours after transfection, wild-type and transfected cells were stained with affinity-purified rabbit anti-AKAP18 IgG (VO57; 1 μg/ml) and FITC-conjugated secondary antibody. The cells imaged are representative of at least four independent transfection studies with each plasmid. Bar, 10 μm.
Localization of AKAP18α and AKAP18β in Epithelial Cells
Although both AKAP18α and -β are targeted to membranes in HEK-293 cells, recent data indicate that the formation of specialized plasma membrane microdomains is crucial for efficient intracellular signaling in many cell types (Shaul and Anderson 1998; Fanning and Anderson 1999; Ostrom and Insel 1999). Therefore, we considered whether the unique 23-amino acid insert present in AKAP18β directs this isoform to specific targets or microdomains of the plasma membrane. MDCK cells, a well-characterized kidney epithelial cell line, form a tight monolayer with distinct apical, basolateral, and junctional surfaces when grown on permeable filter supports. We stably expressed cDNAs encoding GFP-tagged AKAP18α and -β in MDCK cells to compare their distributions in polarized cells. Previous experiments established that the fusion of GFP to the COOH terminus of AKAP18α does not disrupt membrane targeting of the chimeric protein (Fraser et al. 1998).
Confocal microscopy performed on well-polarized MDCK cell cultures indicated that the distributions of GFP-tagged AKAP18α and -β differed. AKAP18α/GFP was accumulated predominantly along the lateral margins of the transfected cells. In contrast, AKAP18β/GFP was present at the apical membrane (Fig. 6 A), and overlapped the distribution of the apical membrane glycoprotein gp135 (Ojakian and Schwimmer 1988; Fig. 6 B). The AKAP18α/GFP and AKAP18β/GFP proteins were expressed at relatively equal levels, and both proteins were present in the particulate fraction when cells were lysed in hypotonic buffers lacking detergent (data not shown). The lateral membranes of polarized epithelial cells are comprised of two membrane specializations, the tight and adherens junctions. The distribution of AKAP18α significantly overlapped the distribution of β-catenin, a protein that accumulates at the adherens junctions (Nathke et al. 1994; Fig. 6 B). When the distribution of AKAP18α and -β was compared with the distribution of ZO-1, a marker for tight junctions (Willott et al. 1992; Itoh et al. 1993), neither protein was found to significantly overlap (Fig. 6 B). Lateral and apical distributions of AKAP18α/GFP and AKAP18β/GFP, respectively, were observed in several independent clonal cell lines and in transient transfection assays (data not shown). AKAP18β was observed at lateral surfaces of well-polarized MDCK cells in clones that expressed high levels of the transfected protein (data not shown), suggesting that a saturable protein–protein or protein–lipid interaction mediates the selective association of AKAP18β/GFP with apical membranes.
Figure 6.
AKAP18α and -β are differentially targeted in MDCK cells. A, MDCK cells stably expressing AKAP18α/GFP, AKAP18β/GFP, or GFP alone were grown on Transwell filters. Confluent monolayers were fixed in 4% paraformaldehyde and analyzed by confocal microscopy in XY and XZ planes. At least three individual cell lines expressing each construct were analyzed and similar results were obtained. Bar, 10 μm. B, The distributions of AKAP18α/GFP and AKAP18β/GFP were compared to the distribution of markers for tight junctions (ZO-1), adherens junctions (β-catenin), and apical membranes (gp135). Stably transfected cells were fixed and stained with rat anti ZO-1 (1:400), rabbit anti–β-catenin (1:400), or mouse anti-gp135 (1:50), followed by the appropriate Texas red-conjugated secondary antibody. Images are shown as XZ scans and are representative of images collected in two independent experiments.
The differential targeting of AKAP18α and -β was not due to overexpression of the proteins in the MDCK cells, since we found the endogenous proteins were also differentially distributed (Fig. 7). To compare the distributions of AKAP18α and -β in MDCK cells, we generated an antibody directed against residues 15–43 of AKAP18β (NC 257); this antibody specifically recognizes the overexpressed AKAP18β/GFP stably expressed in MDCK cells (Fig. 7 A). Furthermore, the antibody does not detect AKAP18α/GFP on the lateral borders of stably transfected MDCK cells, although we did observe apical membrane labeling of these cells, as well as some punctate staining towards the apical pole (Fig. 7 A). In contrast, antisera VO64 generated against recombinant AKAP18α, which detects multiple AKAP18 isoforms on Western blots (data not shown), recognized both AKAP18β/GFP and AKAP18α/GFP in stably transfected MDCK cells (Fig. 7 B). Having established that NC257 selectively recognized AKAP18β while VO64 recognized both AKAP18 isoforms, we stained wild-type MDCK cells with each antiserum and compared the distribution of endogenous AKAP18 in these cells. In cells stained with VO64, AKAP18 proteins were found distributed along the lateral cell membranes (Fig. 7 C) with a staining pattern resembling the distribution of the AKAP18α/GFP. Punctate staining was also observed at the apical cell surface in confocal sections (data not shown), which is consistent with the staining (presumably of endogenous AKAP18β) observed in AKAP18α/GFP cells (Fig. 7 B). In contrast, no staining of the lateral cell surface was observed when cells were stained with NC257 directed against the β-specific exon, although robust staining of subapical and apical vesicles was observed (Fig. 7 C). Therefore, we conclude that in polarized MDCK cells, endogenous AKAP18α and -β are differentially targeted to the lateral and apical cell surfaces, respectively. The formation of detergent-resistant membranes or lipid rafts is implicated in signal transduction and in the sorting of proteins to the apical cell surface (Brown and London 1998). Therefore, we tested whether the differential targeting of AKAP18α and -β correlated with the selective accumulation of AKAP18β in detergent-insoluble lipid rafts. To do this, we performed subcellular fractionation experiments in the presence of different concentrations of Triton X-100 and compared the solubilities of AKAP18α and -β. Since both proteins were easily extracted in buffers containing 0.2% Triton X-100 (data not shown), we conclude that AKAP18β is not associated with detergent insoluble complexes at the apical surface of MDCK cells.
Figure 7.
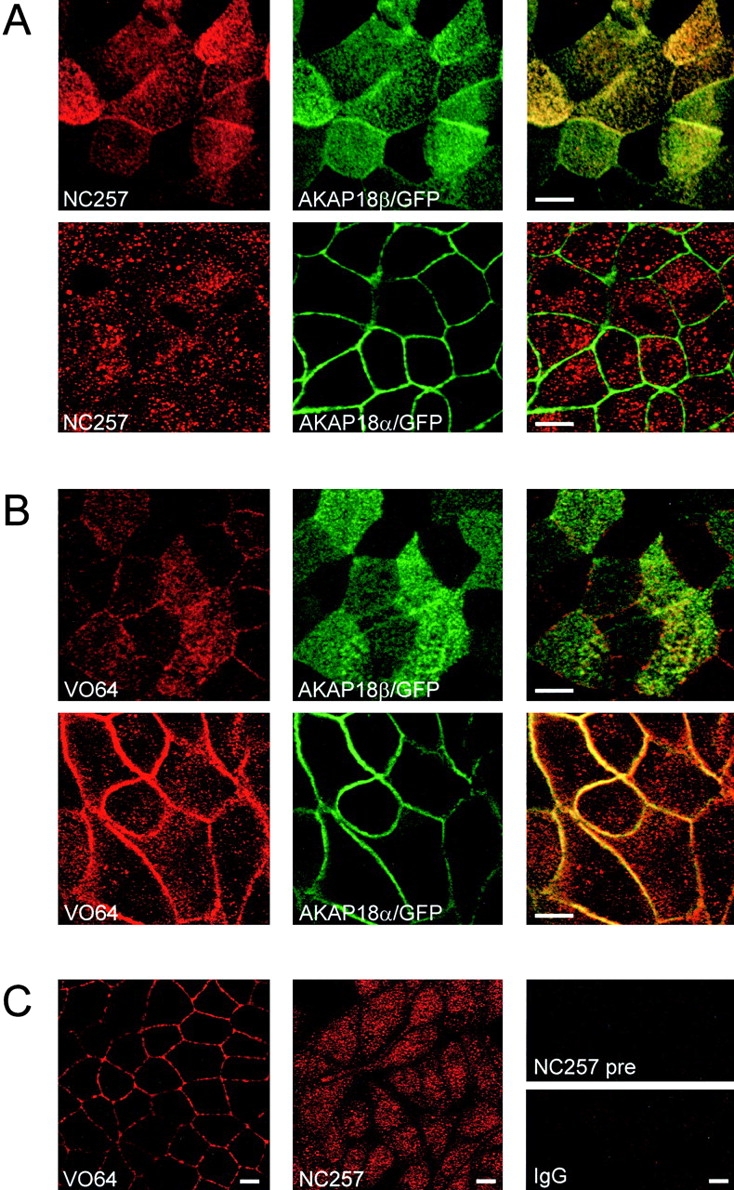
Localization of endogenous AKAP18 in MDCK cells. A, MDCK cells stably expressing AKAP18β/GFP or AKAP18α/GFP were grown on Transwell filters and confluent monolayers were fixed in 4% paraformaldehyde. Cells were permeabilized, blocked, and stained with NC257 (1:1,000 dilution) followed by Texas red-conjugated secondary antibody. Antibody staining was compared with the distribution of the GFP fusion proteins by confocal microscopy. B, MDCK cells stably expressing AKAP18β/GFP or AKAP18α/GFP were grown on Transwell filters and confluent monolayers were fixed and stained with VO64 (1 μg/ml) as described in A. For experiments in A and B, scanning in one channel was performed with the other laser off to assure that there was no bleed-through. C, Wild-type MDCK cells were grown on Transwell filters and confluent monolayers were fixed and stained with VO64 or NC257 as described above. Preimmune sera and normal rabbit IgG at the same concentrations failed to stain any structures in wild-type or transfected MDCK cells. Bar, 10 μm.
The Unique Sequence of AKAP18β Contains Apical Targeting Information
Our expression studies show that AKAP18α and -β are differentially targeted in polarized epithelial cells, yet they only differ by the presence of an alternative exon encoding 23 amino acids (Fig. 2 and Fig. 3). To further explore the function of this AKAP18β-specific sequence, we generated three GFP fusion proteins corresponding to exons in the AKAP18 gene: 1-16αβ/GFP, which encompasses the common membrane targeting domain; 17-44β/GFP, which encompasses the AKAP18β specific sequence; and 1-44β/GFP, which includes both exons (Fig. 8 A). We first transiently expressed each of the GFP chimeras in HEK-293 cells to compare the efficiency with which the expressed proteins were targeted to the cell surface. Both 1-16αβ/GFP and 1-44β/GFP were detected at the cell surface, whereas the 17-44β/GFP protein was uniformly distributed throughout the cell (Fig. 8 B). Similarly, when the 17-44β/GFP chimera was stably expressed in MDCK cells, the protein was distributed throughout the cytoplasm and nucleus (Fig. 8 C). These results indicate that the 23-amino acid insert unique to AKAP18β is not sufficient to mediate membrane targeting. However, the localization of the 1-16αβ/GFP and 1-44β/GFP proteins clearly differed when stably expressed in MDCK cells. The 1-16αβ/GFP protein targeted the plasma membrane, and most of the expressed protein was distributed along the lateral borders of the cells (Fig. 8 C). The 1-44β/GFP protein was also targeted to the plasma membrane in MDCK cells. However, a significant fraction of the 1-44β/GFP protein was present at the apical cell surface, although protein was detected along the lateral borders. Collectively, these data indicate that the 23-amino insert unique to AKAP18β facilitates targeting of AKAP18β to the apical membrane.
Figure 8.

The unique 23 amino acids in AKAP18β facilitate apical targeting. A, Schematic diagram of the three GFP fusion proteins expressed in HEK-293 and MDCK cells. GFP is not drawn to scale. The unique region of AKAP18β is filled in black. B, Each construct was expressed transiently in HEK-293 cells and the distributions of the expressed GFP-tagged proteins analyzed by confocal microscopy. Bar, 10 μm. The images are representative of three individual experiments. C, MDCK cells stably expressing the constructs shown in A were grown to confluence on glass coverslips and the distribution of the GFP chimeras was determined by confocal microscopy. Images were collected in the XY and XZ planes. Similar results were obtained in transient transfection assays and in at least three clonal cell lines for each construct. Bar, 10 μm.
Discussion
In this report, we describe the identification and characterization of two additional isoforms of AKAP18, a plasma membrane-associated AKAP suggested to play a role in modulation of L-type Ca2+ channels (Fraser et al. 1998; Gray et al. 1998). We propose to call the original AKAP18 cDNA AKAP18α, and the newly described cDNAs AKAP18β and -γ. Taken together, Southern blot analyses and partial sequencing of the mouse AKAP18 gene indicate that these cDNAs arise secondary to alternative splicing of exons in a single gene (Fig. 3). Although the sequencing of the mouse AKAP18 gene is not complete, we have already identified exons encoding the lipid modification domain found in AKAP18α and -β, the 23-amino acid insert found in AKAP18β, and the RII binding site in all three AKAP18 isoforms (Fig. 3 B).
Several other AKAPs are known to exist in multiple forms (Lin et al. 1995; Dong et al. 1998; Schmidt et al. 1999), and the generation of AKAP diversity by differential mRNA splicing may be a common occurrence. In theory, differential splicing of AKAP genes may result in the expression of proteins that are targeted to different subcellular compartments, or proteins that target the same compartment, but recruit additional binding partners. Some AKAPs bind other kinases or phosphatases (Klauck et al. 1996; Schillace and Scott 1999a), or contain additional putative interaction domains (Dong et al. 1998; Schmidt et al. 1999). Alternative splicing could, therefore, generate AKAP-mediated multiprotein complexes with different compositions. Although splicing of several AKAP genes is well documented, there are few examples where the function of the splicing is well established. For example, there are six known isoforms of AKAP-KL which share a common RII binding site, but the function of the unique sequence in each isoform is not known (Dong et al. 1998). Alternative splicing generates S-AKAP84, AKAP121, and D-AKAP1–related proteins (Lin et al. 1995; Chen et al. 1997; Huang et al. 1997a, Huang et al. 1999). Each of these proteins share a common RII binding site; however, while S-AKAP84 and AKAP121 are targeted to the outer mitochondrial membrane, D-AKAP1 splice variants may target mitochondria or the ER (Lin et al. 1995; Chen et al. 1997; Huang et al. 1997a, Huang et al. 1999). However, not all splicing generates AKAP proteins with the same RII binding domain. Splicing of a single gene on chromosome 7q21 generates three AKAP350-related proteins (with the same RII binding domain) and yotiao, which contains a different RII binding site (Lin et al. 1998; Schmidt et al. 1999; Witczak et al. 1999). Furthermore, AKAP350 targets PKA to the centrosome (Schmidt et al. 1999; Witczak et al. 1999), whereas yotiao is localized at neuronal synapses (Lin et al. 1998). Our data clearly demonstrate that alternative splicing of the AKAP18 gene generates proteins that may target distinct subcellular compartments, but contain a common RII binding determinant (Fig. 2 D). Furthermore, our data indicate that alternative splicing may dictate the targeting of AKAP18 isoforms to microdomains of the cell surface in some cell types (Fig. 6 Fig. 7 Fig. 8). However, it is likely that another consequence of this splicing is to regulate differential association with other cellular proteins.
The NH2-terminal targeting domain of AKAP18α and -β clearly directs the expressed proteins to the plasma membrane (Fig. 5, Fig. 6, and Fig. 8). Although computer-based resources for identifying organelle targeting signals predicted that AKAP18γ would be found in the nucleus (Fig. 1 A), we observed that AKAP18γ was distributed in cytoplasm of transiently transfected fibroblasts (Fig. 5 B) and stably transfected MDCK cells (data not shown). Approximately 50% of the native protein in rat kidney and 20% of the exogenously expressed AKAP18γ in HEK-293 cells was found in the particulate fraction (Fig. 4 D and 5 A), suggesting association with cellular membranes or cytoskeletal structures. Due to the increased proportion of soluble AKAP18γ in overexpression studies, we speculate that the targeting of the protein may rely strictly on association with an endogenous protein expressed at low levels in HEK-293 cells. However, we cannot rule out the possibility that AKAP18γ functions to bind PKA in the cytoplasm of cells. There is some precedence for cytosolic AKAPs, since treatment of ovarian granulosa cells with follicle stimulating hormone induced the expression of an ∼80-kD AKAP that was found predominantly in the cytosol of fractionated cells (Carr et al. 1993). The generation of AKAP18γ-specific antisera for immunohistochemical studies and the identification of proteins that associate with the unique region of AKAP18γ will hopefully resolve these questions.
AKAP18α and -β are well situated to modulate cAMP-mediated signaling at the plasma membrane, since both proteins accumulate at the cell surface when expressed in cells (Fig. 5 B and 6). This is not surprising since residues 1–16, present in both isoforms, contain membrane targeting information (Fraser et al. 1998; Gray et al. 1998). Acting alone, the 23-amino acid insert unique to AKAP18β does not function to redistribute a GFP reporter protein to the plasma membrane (Fig. 8). Therefore, these amino acids do not contain plasma membrane targeting information. However, AKAP18α is restricted to the lateral surfaces of polarized MDCK cells, whereas AKAP18β is preferentially localized apically (Fig. 6). Indeed, our data demonstrate that acting in tandem with residues 1–16, the unique sequence (residues 17–39) facilitates apical targeting (Fig. 8 C). Although we considered the possibility that association with apical membrane lipid rafts explained the preferential apical distribution of AKAP18β, the protein showed no difference in its solubility compared with AKAP18α. Therefore, we speculate that the selective targeting of AKAP18β to the apical membrane is due to specific protein–protein interactions involving residues 17–39.
In epithelial cells, AKAP18α is restricted to the lateral cell membranes overlapping the distribution of β-catenin (Fig. 6 B). Endogenous AKAP18 was also observed along the lateral membranes of cells, but only when cells were stained with VO64 antiserum, which recognizes multiple AKAP18 isoforms, including AKAP18α (Fig. 7). Although these data strongly support our AKAP18α/GFP studies (Fig. 6 and Fig. 8), additional immunohistochemical analyses of intact human tissues will help further characterize the subcellular localization of AKAP18α. Nonetheless, our data suggest that AKAP18α may localize PKA to sites of cell–cell contact, where PKA is known to play a role (together with other protein kinases) in regulation of junctional stability (Citi 1992; Nilsson et al. 1996; Collares-Buzato et al. 1998; Kovbasnjuk et al. 1998). In contrast, AKAP18β is predicted to serve a different function in polarized cells. Many ion channels and transporters at the apical cell surface are regulated by PKA-mediated phosphorylation, and recent data implicate AKAPs in several events restricted to the apical cell surface. For example, AKAPs facilitate vasopressin-induced translocation of aquaporin-2 water channels in kidney (Klussmann et al. 1999) and modulation of the cystic fibrosis transmembrane conductance regulator Cl− channels in airway epithelia (Huang et al. 1999). In MDCK cells, endogenous AKAP18β is distributed at the apical cell surface, although some staining was also observed on intracellular vesicles (Fig. 7). Thus, AKAP18β may be required for PKA-mediated regulation of apical ion or water transport, and may also be involved in vesicular trafficking to this membrane. It will be important to compare the distributions of AKAP18α and -β in different epithelial tissues and in other tissues containing specialized plasma membrane domains, including neurons and skeletal or cardiac muscle. In addition, it will be important to determine whether a specific AKAP18 isoform targets L-type Ca2+ channels. The identification of proteins that associate specifically with AKAP18β, and the generation of reagents to selectively disrupt a single isoform, will hopefully elucidate the function of each AKAP18 isoform.
Acknowledgments
We thank Dr. George Ojakian (State University of New York, Brooklyn, NY) for providing antibodies to gp135 and Dr. Lloyd Fricker (Albert Einstein School of Medicine, Bronx, NY) for providing the NIT-1 cell line. The Milgram lab thanks Dr. Marvin Adams and Lihong Chen for their advice, Peter Mohler for help with confocal microscopy and transfections of MDCK cells, and Mark Larson for help during the initial screening of cDNA expression libraries. The Scott lab is grateful to John Scarborough for help with preliminary genomic DNA library screens, Lorene Langeberg for confocal microscopy support, and Ann Westphal for assistance with cell culture.
Supported by grants from the National Institutes of Health (HL60280 to M.J. Stutts and S.L. Milgram; and GM48231 to J.D. Scott), the Cystic Fibrosis Foundation (MILGR9710), and the Wellcome Trust (049076/Z/96 to I.D.C. Fraser).
Footnotes
Kevin W. Trotter and Iain D.C. Fraser contributed equally to this work.
Abbreviations used in this paper: AKAP, A-kinase anchoring protein; C, catalytic; nt, nucleotides; PKA, cAMP-dependent protein kinase; R, regulatory; RII, type II R subunit; RT, reverse transcriptase.
References
- Ali S., Chen X., Lu M., Xu J.Z., Lerea K.M., Hebert S.C., Wang W.H. The A kinase anchoring protein is required for mediating the effect of protein kinase A on ROMK1 channels. Proc. Natl. Acad. Sci. USA. 1998;95:10274–10278. doi: 10.1073/pnas.95.17.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo R., Rubin C.S. Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans . J. Biol. Chem. 1998;273:14633–14643. doi: 10.1074/jbc.273.23.14633. [DOI] [PubMed] [Google Scholar]
- Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Burton K.A., Johnson B.D., Hausken Z.E., Westenbroek R.E., Idzerda R.L., Scheuer T., Scott J.D., Catterall W.A., McKnight G.S. Type II regulatory subunits are not required for the anchoring-dependent modulation of Ca2+ channel activity by cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1997;94:11067–11072. doi: 10.1073/pnas.94.20.11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn R.E., Fraser I.D., Bishop S.M., Acott T.S., Brennan R.G., Scott J.D. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- Carr D.W., DeManno D.A., Atwood A., Hunzicker-Dunn M., Scott J.D. Follicle-stimulating hormone regulation of A-kinase anchoring proteins in granulosa cells. J. Biol. Chem. 1993;268:20729–20732. [PubMed] [Google Scholar]
- Chen L., Johnson R.C., Milgram S.L. P-CIP1, a novel protein that interacts with the cytosolic domain of peptidylglycine alpha-amidating monooxygenase, is associated with endosomes. J. Biol. Chem. 1998;273:33524–33532. doi: 10.1074/jbc.273.50.33524. [DOI] [PubMed] [Google Scholar]
- Chen Q., Lin R.Y., Rubin C.S. Organelle-specific targeting of protein kinase AII (PKAII). Molecular and in situ characterization of murine A kinase anchor proteins that recruit regulatory subunits of PKAII to the cytoplasmic surface of mitochondria. J. Biol. Chem. 1997;272:15247–15257. doi: 10.1074/jbc.272.24.15247. [DOI] [PubMed] [Google Scholar]
- Citi S. Protein kinase inhibitors prevent junction dissociation induced by low extracellular calcium in MDCK epithelial cells. J. Cell Biol. 1992;117:169–178. doi: 10.1083/jcb.117.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collares-Buzato C.B., Jepson M.A., Simmons N.L., Hirst B.H. Increased tyrosine phosphorylation causes redistribution of adherens junction and tight junction proteins and perturbs paracellular barrier function in MDCK epithelia. Eur. J. Cell Biol. 1998;76:85–92. doi: 10.1016/S0171-9335(98)80020-4. [DOI] [PubMed] [Google Scholar]
- Colledge M., Scott J.D. AKAPsfrom structure to function. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- Dell'Acqua M.L., Scott J.D. Protein kinase A anchoring. J. Biol. Chem. 1997;272:12881–12884. doi: 10.1074/jbc.272.20.12881. [DOI] [PubMed] [Google Scholar]
- Dell'Acqua M.L., Faux M.C., Thorburn J., Thorburn A., Scott J.D. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4,5-bisphosphate. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Feldmesser M., Casadevall A., Rubin C.S. Molecular characterization of a cDNA that encodes six isoforms of a novel murine A kinase anchor protein. J. Biol. Chem. 1998;273:6533–6541. doi: 10.1074/jbc.273.11.6533. [DOI] [PubMed] [Google Scholar]
- Dransfield D.T., Bradford A.J., Smith J., Martin M., Roy C., Mangeat P.H., Goldenring J.R. Ezrin is a cyclic AMP-dependent protein kinase anchoring protein. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:35–43. doi: 10.1093/emboj/16.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning A.S., Anderson J.M. PDZ domainsfundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis S.H., Corbin J.D. Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 1994;56:237–272. doi: 10.1146/annurev.ph.56.030194.001321. [DOI] [PubMed] [Google Scholar]
- Fraser I.D., Tavalin S.J., Lester L.B., Langeberg L.K., Westphal A.M., Dean R.A., Marrion N.V., Scott J.D. A novel lipid-anchored A-kinase anchoring protein facilitates cAMP-responsive membrane events. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2261–2272. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Yatani A., Dell'Acqua M.L., Sako H., Green S.A., Dascal N., Scott J.D., Hosey M.M. cAMP-dependent regulation of cardiac L-type Ca2+ channels requires membrane targeting of PKA and phosphorylation of channel subunits. Neuron. 1997;19:185–196. doi: 10.1016/s0896-6273(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Gerace L. Molecular trafficking across the nuclear pore complex. Curr. Opin. Cell Biol. 1992;4:637–645. doi: 10.1016/0955-0674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- Glantz S.B., Li Y., Rubin C.S. Characterization of distinct tethering and intracellular targeting domains in AKAP75, a protein that links cAMP-dependent protein kinase II beta to the cytoskeleton. J. Biol. Chem. 1993;268:12796–12804. [PubMed] [Google Scholar]
- Gorlich D. Nuclear protein import. Curr. Opin. Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Gray P.C., Tibbs V.C., Catterall W.A., Murphy B.J. Identification of a 15-kDa cAMP-dependent protein kinase-anchoring protein associated with skeletal muscle L-type calcium channels. J. Biol. Chem. 1997;272:6297–6302. doi: 10.1074/jbc.272.10.6297. [DOI] [PubMed] [Google Scholar]
- Gray P.C., Johnson B.D., Westenbroek R.E., Hays L.G., Yates J.R., 3rd, Scheuer T., Catterall W.A., Murphy B.J. Primary structure and function of an A kinase anchoring protein associated with calcium channels. Neuron. 1998;20:1017–1026. doi: 10.1016/s0896-6273(00)80482-1. [DOI] [PubMed] [Google Scholar]
- Huang L.J., Durick K., Weiner J.A., Chun J., Taylor S.S. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain Proc. Natl. Acad. Sci. USA 94 1997. 11184 11189a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.J., Durick K., Weiner J.A., Chun J., Taylor S.S. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits J. Biol. Chem 272 1997. 8057 8064b [DOI] [PubMed] [Google Scholar]
- Huang L.J., Wang L., Ma Y., Durick K., Perkins G., Deerinck T.J., Ellisman M.H., Taylor S.S. NH2-terminal targeting motifs direct dual specificity A-kinase-anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J. Cell Biol. 1999;145:951–959. doi: 10.1083/jcb.145.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Trotter K.W., Boucher R.C., Milgram S.L., Stutts M.J. PKA holoenzyme is functionally coupled to CFTR by AKAPs. Am. J. Physiol. (Cell). 1999;In press doi: 10.1152/ajpcell.2000.278.2.C417. [DOI] [PubMed] [Google Scholar]
- Itoh M., Nagafuchi A., Yonemura S., Kitani-Yasuda T., Tsukita S. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight juntion-associated protein in epithelial cellscDNA cloning and immunoelectron microscopy. J. Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck T.M., Faux M.C., Labudda K., Langeberg L.K., Jaken S., Scott J.D. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- Klussmann E., Maric K., Wiesner B., Beyermann M., Rosenthal W. Protein kinase A anchoring proteins are required for vasopressin-mediated translocation of aquaporin-2 into cell membranes of renal principal cells. J. Biol. Chem. 1999;274:4934–4938. doi: 10.1074/jbc.274.8.4934. [DOI] [PubMed] [Google Scholar]
- Kovbasnjuk O.N., Szmulowicz U., Spring K.R. Regulation of the MDCK cell tight junction. J. Membr. Biol. 1998;161:93–104. doi: 10.1007/s002329900317. [DOI] [PubMed] [Google Scholar]
- Lester L.B., Coghlan V.M., Nauert B., Scott J.D. Cloning and characterization of a novel A-kinase anchoring protein. AKAP 220, association with testicular peroxisomes. J. Biol. Chem. 1996;271:9460–9465. doi: 10.1074/jbc.271.16.9460. [DOI] [PubMed] [Google Scholar]
- Lester L.B., Langeberg L.K., Scott J.D. Anchoring of protein kinase A facilitates hormone-mediated insulin secretion. Proc. Natl. Acad. Sci. USA. 1997;94:14942–14947. doi: 10.1073/pnas.94.26.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.W., Wyszynski M., Madhavan R., Sealock R., Kim J.U., Sheng M. Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J. Neurosci. 1998;18:2017–2027. doi: 10.1523/JNEUROSCI.18-06-02017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R.Y., Moss S.B., Rubin C.S. Characterization of S-AKAP84, a novel developmentally regulated A kinase anchor protein of male germ cells. J. Biol. Chem. 1995;270:27804–27811. doi: 10.1074/jbc.270.46.27804. [DOI] [PubMed] [Google Scholar]
- McCartney S., Little B.M., Langeberg L.K., Scott J.D. Cloning and characterization of A-kinase anchor protein 100 (AKAP100). A protein that targets A-kinase to the sarcoplasmic reticulum. J. Biol. Chem. 1995;270:9327–9333. doi: 10.1074/jbc.270.16.9327. [DOI] [PubMed] [Google Scholar]
- Nakai K., Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke I.S., Hinck L., Swedlow J.R., Papkoff J., Nelson W.J. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J. Cell Biol. 1994;125:1341–1352. doi: 10.1083/jcb.125.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon M.G., Roy M., Morikis D., Hausken Z.E., Coghlan V., Scott J.D., Jennings P.A. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. Nucleocytoplasmic transportsignals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Fagman H., Ericson L.E. Ca2+-dependent and Ca2+-independent regulation of the thyroid epithelial junction complex by protein kinases. Exp. Cell Res. 1996;225:1–11. doi: 10.1006/excr.1996.0151. [DOI] [PubMed] [Google Scholar]
- Ojakian G.K., Schwimmer R. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 1988;107:2377–2387. doi: 10.1083/jcb.107.6.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom R.S., Insel P.A. Caveolar microdomains of the sarcolemmacompartmentation of signaling molecules comes of age. Circ. Res. 1999;84:1110–1112. doi: 10.1161/01.res.84.9.1110. [DOI] [PubMed] [Google Scholar]
- Pawson T., Scott J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Rosenmund C., Carr D.W., Bergeson S.E., Nilaver G., Scott J.D., Westbrook G.L. Anchoring of protein kinase A is required for modulation of AMPA/kainate receptors on hippocampal neurons. Nature. 1994;368:853–856. doi: 10.1038/368853a0. [DOI] [PubMed] [Google Scholar]
- Rubin C.S. A kinase anchor proteins and the intracellular targeting of signals carried by cyclic AMP. Biochim. Biophys. Acta. 1994;1224:467–479. [PubMed] [Google Scholar]
- Schillace R.V., Scott J.D. Association of the type 1 protein phosphatase PP1 with the A-kinase anchoring protein AKAP220 Curr. Biol 9 1999. 321 324a [DOI] [PubMed] [Google Scholar]
- Schillace R.V., Scott J.D. Organization of kinases, phosphatases, and receptor signaling complexes J. Clin. Invest 103 1999. 761 765b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P.H., Dransfield D.T., Claudio J.O., Hawley R.G., Trotter K.W., Milgram S.L., Goldenring J.R. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem. 1999;274:3055–3066. doi: 10.1074/jbc.274.5.3055. [DOI] [PubMed] [Google Scholar]
- Scott J.D., Stofko R.E., McDonald J.R., Comer J.D., Vitalis E.A., Mangili J.A. Type II regulatory subunit dimerization determines the subcellular localization of the cAMP-dependent protein kinase. J. Biol. Chem. 1990;265:21561–21566. [PubMed] [Google Scholar]
- Shaul P.W., Anderson R.G. Role of plasmalemmal caveolae in signal transduction. Am. J. Physiol. 1998;275:L843–L851. doi: 10.1152/ajplung.1998.275.5.L843. [DOI] [PubMed] [Google Scholar]
- Short D.B., Trotter K.W., Reczek D., Kreda S.M., Bretscher A., Boucher R.C., Stutts M.J., Milgram S.L. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J. Biol. Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- Sparks A.B., Hoffman N.G., McConnell S.J., Fowlkes D.M., Kay B.K. Cloning of ligand targetssystematic isolation of SH3 domain-containing proteins. Nat. Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Buechler J.A., Yonemoto W. cAMP-dependent protein kinaseframework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- Tibbs V.C., Gray P.C., Catterall W.A., Murphy B.J. AKAP15 anchors cAMP-dependent protein kinase to brain sodium channels. J. Biol. Chem. 1998;273:25783–25788. doi: 10.1074/jbc.273.40.25783. [DOI] [PubMed] [Google Scholar]
- Willott E., Balda M.S., Heintzelman M., Jameson B., Anderson J.M. Localization and differential expression of two isoforms of the tight junction protein ZO-1. Am. J. Physiol. 1992;262:1119–1124. doi: 10.1152/ajpcell.1992.262.5.C1119. [DOI] [PubMed] [Google Scholar]
- Witczak O., Skalhegg B.S., Keryer G., Bornens M., Tasken K., Jahnsen T., Orstavik S. Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:1858–1868. doi: 10.1093/emboj/18.7.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]