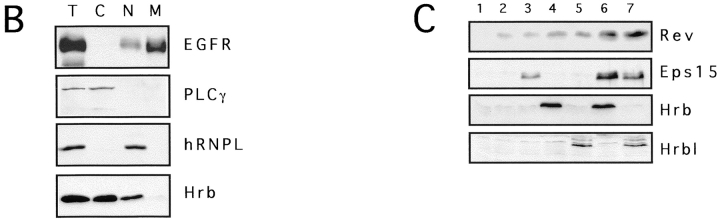Abstract
The Eps15 homology (EH) module is a protein–protein interaction domain that establishes a network of connections involved in various aspects of endocytosis and sorting. The finding that EH-containing proteins bind to Hrb (a cellular cofactor of the Rev protein) and to the related protein Hrbl raised the possibility that the EH network might also influence the so-called Rev export pathway, which mediates nucleocytoplasmic transfer of proteins and RNAs. In this study, we demonstrate that Eps15 and Eps15R, two EH-containing proteins, synergize with Hrb and Hrbl to enhance the function of Rev in the export pathway. In addition, the EH-mediated association between Eps15 and Hrb is required for the synergistic effect. The interaction between Eps15 and Hrb occurs in the cytoplasm, thus pointing to an unexpected site of action of Hrb, and to a possible role of the Eps15–Hrb complex in regulating the stability of Rev.
Keywords: EH, Eps15, Hrb, endocytosis, nucleocytoplasmic transport
The Eps15 homology (EH) domain is a protein–protein interaction module originally identified in the tyrosine kinase substrates Eps15 (Fazioli et al. 1993) and Eps15R (Wong et al. 1995; Coda et al. 1998). Several EH-binding proteins have been subsequently identified (Salcini et al. 1997). Biochemical and functional studies implicated proteins of the EH network in endocytosis and actin cytoskeleton organization (reviewed in Santolini et al. 1999). Other evidence points to additional functions. In particular, the EH network might be involved in the control of nucleocytoplasmic export, as suggested by the finding that three EH-containing proteins, Eps15, Eps15R, and intersectin, interact with Hrb (also called hRip or RAB, Human Gene Nomenclature Committee approved symbols are used in this paper), a cellular cofactor for the HIV-1 Rev protein (Bogerd et al. 1995; Fritz et al. 1995), and with the related protein Hrbl (Salcini et al. 1997; Yamabhai et al. 1998).
By shuttling between the nucleus and cytoplasm of an infected cell, Rev induces nucleocytoplasmic export of unspliced and partially spliced viral mRNAs that contain a unique Rev binding sequence (Rev response element, [RRE]; Cullen 1998). Rev functions through a cellular pathway, operationally defined as the Rev export pathway, which normally exports endogenous RNAs (Fischer et al. 1995), and proteins that, like Rev, contain nuclear export signals (NESs; Fridell et al. 1996; Fritz and Green 1996; Roth et al. 1998). Efforts to identify cellular cofactors mediating Rev export led to the isolation of the distantly related human Hrb (Bogerd et al. 1995; Fritz et al. 1995) and yeast Rip1p (Stutz et al. 1995) proteins, which were shown to enhance Rev function in cells. Both proteins also are related to nucleoporins, in that they display phenylalanine-glycine (FG) repeats, a common feature of this class of proteins. These observations led to the hypothesis that Rev recruits RRE-containing RNAs to the nuclear pore, through direct interaction with Hrb or other nucleoporins.
Recent observations suggest a more complex picture. The interaction between Hrb (or Rip1p) and Rev, observed in the yeast two-hybrid system, could not be demonstrated using purified proteins in vitro, suggesting that it might be indirect (Stutz et al. 1996; Henderson and Percipalle 1997; Neville et al. 1997). Indeed, Crm1, a shuttling protein that belongs to the transport receptor family, serves as a bridging factor between Hrb and Rev (reviewed in Ullman et al. 1997). Crm1 binds to NESs and mediates the export of NES-containing proteins (Fornerod et al. 1997; Stade et al. 1997). In addition, Crm1 interacts with the FG repeats of Hrb and of several FG-nucleoporins (Neville et al. 1997). Thus, Rev might be recruited to nuclear pores indirectly through sequential interactions with Crm1 and Hrb. There are, however, additional difficulties with this model. First, the FG-containing region of Rip1p was shown to make only limited contributions to Rev-mediated export (Stutz et al. 1997). Second, it is not yet certain whether Hrb is an authentic constituent of the nuclear pore complex (Bogerd et al. 1995; Fritz et al. 1995). Thus, while the finding that overexpression of Hrb enhances Rev activity, albeit modestly, suggests a role for Hrb in the Rev export pathway, the mechanisms remain to be determined. Based on the observation of a physical interaction between Eps15 (and Eps15R) with Hrb (and Hrbl; Salcini et al. 1997), this study was undertaken to test whether EH-mediated interactions are involved in the control of the Rev export pathway.
Materials and Methods
Vectors
pDM128, pDM138, pDM121 (Huang et al. 1991), pCEVEps15, and pCEVEps15R (Wong et al. 1995; Coda et al. 1998) have been previously described. pMTHrb and pMTHrbl were generated by subcloning the open reading frame of the human cDNAs into the pMT2 vector. pCEVαEps15 harbors the Eps15 cDNA in the anti-sense orientation in the LTR-based pCEV vector. pMTHA-HrbNPV was generated by changing the sequences coding for the four NPF motifs to sequences coding for NPV in the pMTHA-Hrb vector (Salcini et al. 1997). pCEVEps15ΔEH, encoding a Eps15 protein devoid of its EH domains (and encompassing amino acids [aa] 342–897), was engineered by PCR and subcloned into pCEV. All constructs were sequenced in the regions that underwent genetic manipulations.
CAT Assays
CV-1 cells were transfected by the calcium phosphate method, with the CAT (chloramphenycol acetyltransferase) reporter plasmid pDM128 (100 ng) together with the Rev expression vector pDM121 (10 ng) and the pCMVβgal reporter plasmid (Clontech, 100 ng). Various combinations of pMT-Hrb (0.25 μg), pMT-Hrbl (0.25 μg), pCEVEps15 (1 μg), and pCEVEps15R (1 μg) were also transfected. 48 h after transfection, cellular lysates were tested for β-galactosidase reporter activity using a commercial kit (Promega). Cellular lysates, normalized for β-galactosidase activity, were tested for CAT activity using a commercial kit (Promega).
Protein Studies
Immunoprecipitation, subcellular fractionization, and immunoblotting were performed as previously described (Fazioli et al. 1993). Antibodies used were: anti-Eps15 (Fazioli et al. 1993) and anti-Eps15R (Coda et al. 1998) sera; polyclonal anti-Hrb IgGs directed against the last 19 amino acids of Hrb (Santa Cruz Biotechnology); a polyclonal anti-Hrbl serum recognizing amino acids 280–481 of the protein. Routinely, immunoblots were stripped and reprobed with an anti-tubulin antibody, to ensure equal protein loading in the various lanes (not shown).
Results
Eps15 and Eps15R Influence the Rev Export Pathway
To gain insight into the contribution of Eps15 and Eps15R to the nucleocytoplasmic export pathway used by Rev, we used a previously described assay based on cotransfection of Rev with the reporter plasmid pDM128 (Huang et al. 1991). Transcripts from this latter construct contain a CAT coding sequence within an intron from the env region of HIV-1, which includes an RRE. Upon cotransfection, Rev binds to the RRE, thus allowing cytoplasmic translocation and expression of the unspliced transcripts (Huang et al. 1991). In preliminary experiments (not shown), we transiently cotransfected CV-1 cells with pDM128 and increasing amounts of a Rev expression vector pDM121. Transactivation of CAT by Rev was linear in a range from 4–100-fold activation. For all subsequent experiments we used an amount of pDM121 yielding ∼20% of the maximal transactivation.
When expression vectors for Eps15 or Eps15R were cotransfected with Rev, an increase in CAT activity, ∼50% greater than the value obtained with Rev alone, was reproducibly detected (Fig. 1 A). Expression vectors for Hrb, a known cofactor of Rev, and for Hrbl yielded a comparable 50% increase in Rev activity (Fig. 1 A). All responses were Rev dependent, since they could be abolished by replacing pDM128 with pDM138, a variant construct lacking the RRE sequence (Fig. 1 A), or by omitting the pDM121 construct (not shown). The effects were also not due to influence of Eps15, Eps15R, Hrb, or Hrbl on transcription, since none of the corresponding vectors affected expression of an exonic CAT gene under the control of an SV-40 or RSV promoter in the absence of the RRE (not shown).
Figure 1.
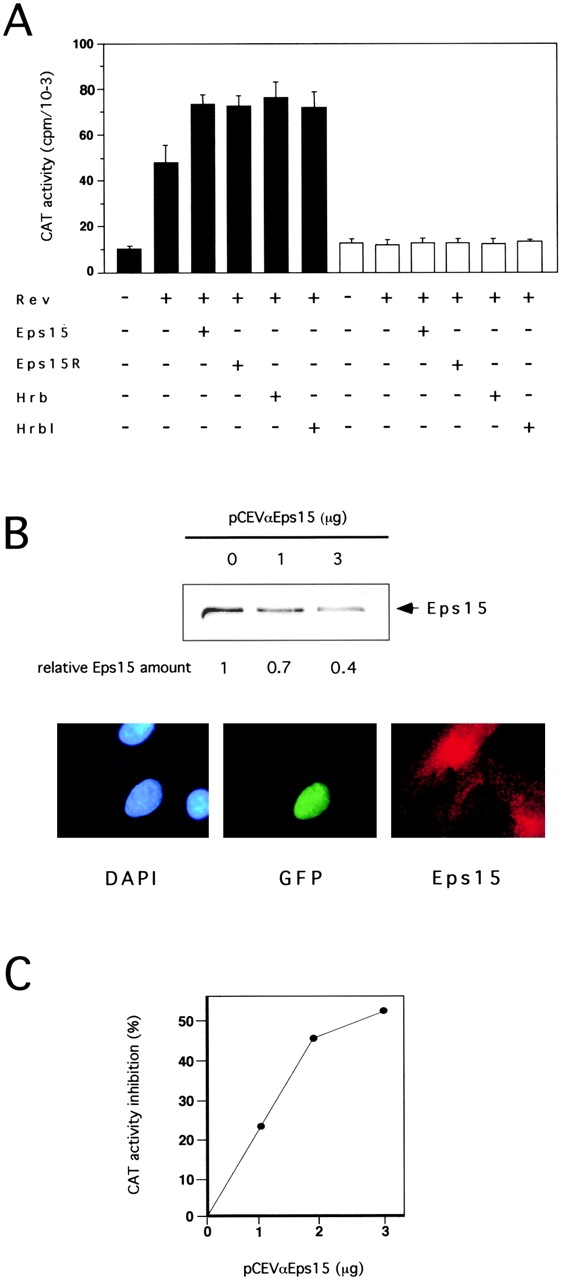
Eps15 and Eps15R influence Rev activity. (A) Rev-dependent CAT activity in cells transfected with the indicated vectors (underneath) and either the CAT reporter plasmid pDM128 (filled bars) or the control pDM138 (open bars). In this and all subsequent experiments, data are representative of at least three independent experiments, performed on individually transfected triplicates. (B, top) CV1 cells were transfected with the indicated amounts of pCEVαEps15. Cellular lysates (100 μg) were immunoblotted with an anti-Eps15 antibody and with an anti-tubulin antibody as a control (not shown). The levels of Eps15 protein, determined by densitometry and adjusted to account for variations in the level of tubulin, are indicated relative to the value in the mock-transfected lysate. (Bottom) CV1 cells were transfected with 0.5 μg of pNLS-GFP (as internal reference) and 5 μg of pCEVαEps15. Staining of cells was with rabbit anti-Eps15 antibody followed by CY3-conjugated anti-rabbit IgG and by nuclear counterstaining with DAPI. Photographs of the same field were taken with filters specific for DAPI-, GFP-, and Eps15(CY3)-specific fluorescences. The vast majority of cells expressing nuclear GFP, presumably expressing antisense Eps15 RNA, displayed a weaker signal for Eps15 as compared with untransfected cells. Cells transfected with pNLS-GFP alone or together with pCEV control vector showed normal levels of Eps15-specific staining (not shown). (C) CV1 cells were cotransfected with the pDM128 CAT reporter, Rev and with the indicated amounts of either pCEVαEps15 or pCEV empty vector. CAT activities are expressed as percent inhibition in pCEVαEps15 transfectants, compared with mock transfectants. In all above and subsequent experiments the total final amount of transfected DNA and the individual amounts of promoter units were kept constant, by adding appropriate amounts of control pDM, pMT, and pCEV empty vectors.
To prove the physiological relevance of Eps15 to the Rev export pathways, we used an antisense Eps15 construct (pCEVαEps15). Transfection of pCEVαEps15 into CV-1 cells significantly reduced the steady state levels of Eps15, as revealed by both immunoblotting and immunofluorescence analyses (Fig. 1 B). When pCEVαEps15 was cotransfected with pDM128 and the Rev expression vector, a dose-dependent reduction in the activity of Rev was observed (Fig. 1 C).
Eps15 and Eps15R Synergize with Hrb and Hrbl
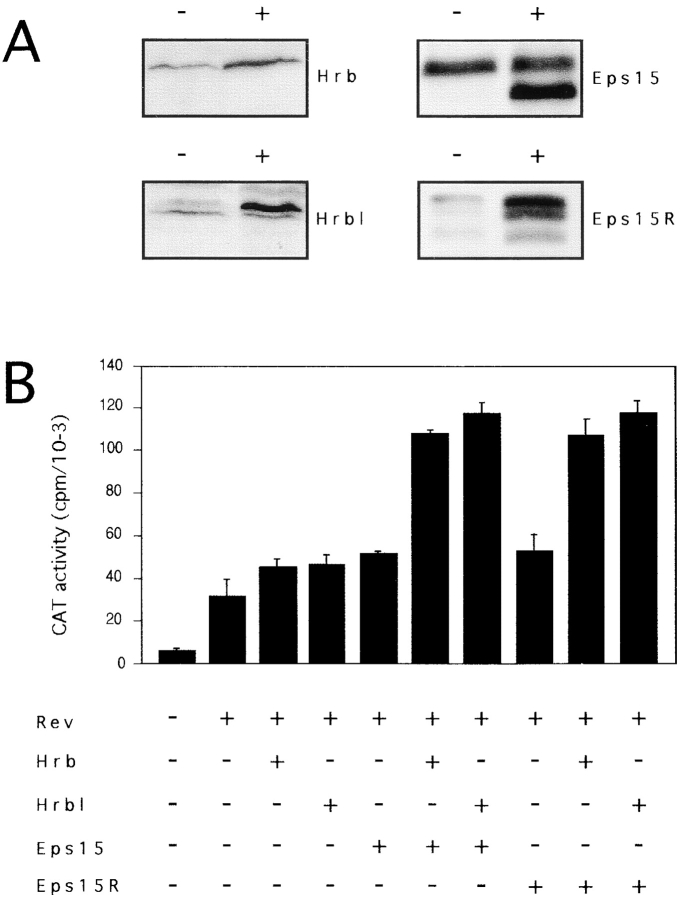
The finding that Eps15 and Eps15R can function as cofactors for the Rev export pathway prompted us to investigate whether they also synergize with Hrb and Hrbl. We cotransfected, together with Rev and the reporter pDM128, various combinations of the four molecules (Fig. 2). The transfected cells expressed three- to fivefold higher levels of these proteins than mock-transfected cells (Fig. 2 A). A synergistic effect of coexpressing Eps15 or Eps15R with either Hrb or Hrbl, was readily observable (Fig. 2 B). Whereas transfection of the individual plasmids (Eps15, Eps15R, Hrb, and Hrbl) brought about an ∼1.5-fold increase in CAT activity, compared with Rev alone, various combinations (Eps15/Hrb, Eps15/Hrbl, Eps15R/Hrb, and Eps15R/Hrbl) caused an ∼3.0–3.5-fold increase. Maximal activation was thus significantly superior to the sum of the individual effects, suggesting that Eps15 (or Eps15R) and Hrb (or Hrbl) can synergistically activate the Rev export pathway.
Figure 2.
Eps15 and Eps15R synergize with Hrb and Hrbl. (A) Immunoblotting analysis of Eps15, Eps15R, Hrb, and Hrbl overexpression. Cellular proteins (100 μg) of CV-1 cells, transfected with expression vectors for Eps15, Eps15R, Hrb, and Hrbl (+ lanes) or with the corresponding empty vector (− lanes) were analyzed by immunoblotting with antibodies specific for each protein, as indicated. The overexpressed human Eps15 migrates as a faster band (140 kD) compared with the endogenous monkey protein (150 kD). In repeated experiments, we observed 3–10-fold overexpression of Eps15, Eps15R, Hrb or Hrbl when the corresponding expression vectors were transfected either alone of in combination. (B) Rev-dependent CAT activity in CV-1 cells transfected with pDM128 and the indicated expression vectors.
The EH-mediated Interaction between Eps15 and Hrb Is Required in the Rev Export Pathway
Binding of Hrb to the EH domains of Eps15 depends on the presence of four NPF (asparagine-proline-phenylalanine)-containing motifs in its COOH terminus (Salcini et al. 1997). Mutagenesis of any of the residues in the NPF consensus abolishes the binding of EH domains to their targets (Salcini et al. 1997). We engineered an HA-tagged mutant of Hrb (HA-Hrb-NPV) in which the phenylalanines of its four NPF motifs were substituted with valines. The HA-Hrb-NPV mutant and a wild-type HA-tagged Hrb (HA-Hrb) could be expressed at comparable levels (Fig. 3 A, lanes 2 and 3). However, HA-Hrb-NPV could no longer be coimmunoprecipitated with Eps15 (Fig. 3 A, top, compare lane 7 with 9). HA-Hrb-NPV was also unable to increase CAT expression, when compared with Rev alone, and, most importantly, was unable to synergize with Eps15 (Fig. 3 B).
Figure 3.
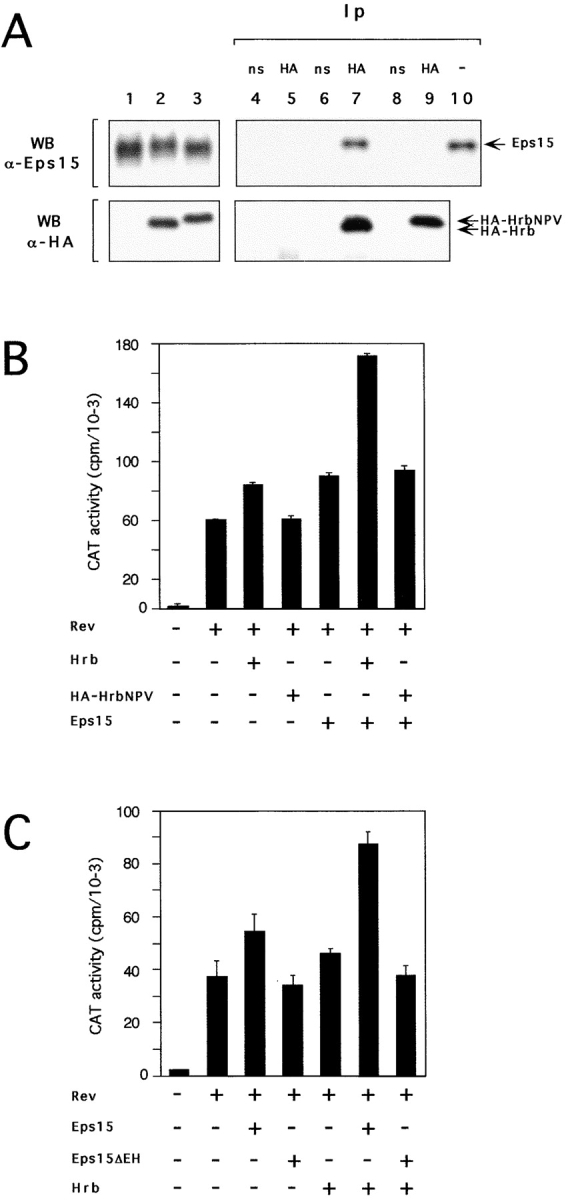
Interaction between Eps15 and Hrb is required for Rev activation. (A) C33A cells were transfected with 20 μg of pCEVEps15 together with 20 μg of either pMTHA-Hrb (lane 2) or pMTHA-HrbNPV (lane 3), or pMT control vector (lane 1). Cellular lysates were immunoblotted with an anti-Eps15 serum (top) or with an anti-HA antibody (bottom). We observed that, due to the presence of the four amino acid substitutions, HA-HrbNPV migrates as a faster band when compared with the corresponding HA-Hrb molecule. Cellular proteins from mock (lanes 4 and 5), HA-Hrb (lanes 6 and 7) or HA-HrbNPV (lanes 8 and 9) transfectants were immunoprecipitated with either an irrelevant monoclonal (ns-labeled lanes 4, 6, and 8) or with the anti-HA antibody (HA-labeled lanes 5, 7, and 9) and detected with an anti-Eps15 (top) or with the anti-HA (bottom) antibody. In the top panel, lane 10 corresponds to 50 μg of cellular lysate, to serve as a reference for positioning Eps15. (B and C) Rev-dependent CAT activity in CV-1 cells transfected with pDM128 and various mutants of Eps15 and Hrb (indicated underneath).
We also engineered an Eps15 mutant devoid of its EH domains (Eps15ΔEH). We have previously shown that the EH domains are necessary and sufficient for binding of Eps15 to Hrb (Salcini et al. 1997). In the Rev-dependent CAT assay, Eps15ΔEH was not able to potentiate Rev activity, nor was it able to synergize with wild-type Hrb (Fig. 3 C). Thus the EH-mediated interaction between Eps15 and Hrb is required for their synergistic effect in the Rev export pathway.
Eps15 and Hrb Might Control Rev Degradation in an Extranuclear Compartment
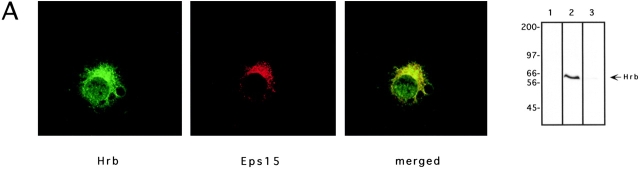
Hrb has been reported to have a nuclear localization, with either nucleolar accumulation (Bogerd et al. 1995) or a nucleoporin-like staining pattern (Fritz et al. 1995). The existence of a cytosolic fraction has also been reported (Bogerd et al. 1995). Such a fraction should in principle be the sole one available for interaction with Eps15, which is exclusively localized to the cytoplasm (Fazioli et al. 1993). We determined colocalization of Eps15 and Hrb by confocal microscopy. An affinity purified anti-Hrb peptide serum was used which recognized a single band in Western blot (Fig. 4 A). As previously described, Eps15 showed a punctate cytosolic distribution (Fig. 4 A). Hrb was present both in the nucleus and in extranuclear compartments (Fig. 4 A). The extranuclear fraction of Hrb displayed a punctate morphology. The majority of these punctate structures were also stained specifically by the anti-Eps15 antibody (Fig. 4 A). We do not know the nature of these structures, which do not contain AP2, AP1 and various markers for the ER and Golgi compartments (not show). The nuclear fraction of Hrb was homogeneously dispersed and did not show nucleolar accumulation or a clear nucleoporin-like pattern. A biochemical subcellular fractionization confirmed the nuclear and cytosolic localization of Hrb (Fig. 4 B). Of note, by both methods, the majority of Hrb appeared to be present in the cytosolic compartment.
Figure 4.
Eps15 and Hrb colocalize in the cytosol and increase Rev levels. (A) Subcellular localization of endogenous Eps15 and Hrb proteins in CV1 cells. Fluorescent cells were analyzed by optical scanning of horizontal sections using confocal microscopy. A representative horizontal section is shown (green, Hrb; red, Eps15; yellow, merged). The specificity of the anti-Hrb antibody was confirmed by immunoblot (right-most panel) on 100 μg of total proteins from CV-1 cells. Lane 1, staining with normal goat serum; lane 2, staining with anti-Hrb, lane 3, staining with anti-Hrb preincubated with the cognate peptide. Preincubation of the anti-Hrb antibody with the specific peptide also resulted in no signal in immunofluorescence analysis (not shown). (B) Subcellular fractionization. Subcellular fractions of CV-1 cells are identified as C (cytosolic), N (nuclear) and M (membrane), or T (total lysate). Aliquots of each fraction, representative of the same number of cells (1 × 106), were analyzed by immunoblot for the presence of EGFR (membrane marker), PLC-γ (cytosolic marker), hRNPL (nuclear marker) and Hrb. As evident C and M fraction are >95% pure, whereas the nuclear fraction was ∼10% contaminated by membranes. (C) Steady state levels of the Rev protein in various transfectants. Immunoblot analysis with the antibodies indicated on the right was performed on CV-1 cells (2.5 × 105) transfected with various combinations of plasmids. Transfectants were: 1, mock; 2, Rev; 3, Rev and Eps15; 4, Rev and Hrb; 5, Rev and Hrbl; 6, Rev, Eps15, and Hrb; 7, Rev, Eps15, and Hrbl.
The above results suggest that the action of a Eps15–Hrb complex must somehow be exerted at the cytosolic level. In search of a possible mechanism, we measured the steady state levels of Rev in the presence of Eps15 alone or in combination with Hrb/Hrbl. Transfection of Eps15 or Hrb or Hrbl, together with Rev, caused a modest increase in the steady state levels of Rev (Fig. 4 C). However, when Rev was cotransfected with Eps15 with either Hrb or Hrbl, a four- to sixfold increase in Rev levels was detected (Fig. 4 C). These differences were not due to variable transfection efficiency, since all cellular lysates displayed the same amount of β-galactosidase activity, which was expressed by the cotransfected plasmid pCMVβ-gal (see Materials and Methods), and used as an internal standard.
Discussion
We have previously shown that Eps15 (and Eps15R) is associated in vivo with Hrb through its EH domain (Salcini et al. 1997). In this study, we showed, by an antisense approach, that Eps15 is physiologically required in the Rev export pathway. Likewise, overexpression of Eps15 (or of Eps15R) led to a small, yet reproducible, increase in Rev activity. The difference in the magnitude of the effects in the two types of experiments suggests that availability of endogenous Eps15 is not a limiting factor in the Rev export pathway. We further showed that Eps15 and Eps15R synergize with Hrb (and Hrbl) in the control of Rev activity and that they do so through their EH-mediated physical interaction. Since Hrb has been directly implicated in the control of the Rev export pathway, our results indicate that an Eps15–Hrb complex is involved in this pathway.
Hrb is thought to be a recruiter of Rev to the nuclear pore, either directly (Bogerd et al. 1995) or, more likely, through interaction with Crm1 (Neville et al. 1997). Hrb might, therefore, function as a nucleoporin, as also suggested by the presence of FG nucleoporin-like repeats. However, Bogerd et al. 1995 failed to detect a clear localization of Hrb in the nuclear membrane, suggesting that Rev might sequentially interact with Hrb and then with authentic nucleoporins. We also failed to observe a nucleoporin-like localization of Hrb. Our results instead point to an unexpected site of action of Hrb, i.e., in an extranuclear compartment where the majority of Hrb localizes to punctate structures where also Eps15 is present. Together with the findings of a physical interaction between Eps15 and Hrb (Salcini et al. 1997) and of its role in Rev activation (this paper), these results indicate that an important role of Hrb is exerted on the cytosolic side of nucleocytoplasmic transport.
The subcellular localization of Hrb is, therefore, critical to the understanding of its function. Discordant findings have been reported in literature, which, therefore, deserve further comment. Bogerd et al. 1995 reported significant levels of Hrb in the cytosolic fraction of HeLa cells, whereas Fritz et al. 1995 reported exclusive partitioning in the nuclear fraction in the same cells. Our results agree with those of Bogerd et al., except that the magnitude of the cytosolic pool appears relatively greater under our conditions of analysis. Our morphological analysis, however, diverges from previous literature in which Hrb was found predominantly in the nucleus by immunofluorescence (Bogerd et al. 1995; Fritz et al. 1995). We note that in previous studies polyclonal anti-Hrb antibodies were used that were generated against portions of Hrb containing multiple FG repeats and displaying homology to other nucleoporins. One might speculate that those sera were cross-reactive with authentic nucleoporins. In our case, we used an anti-peptide serum, which was generated against a portion of Hrb without significant homologies.
The mechanisms by which a cytosolic Eps15–Hrb complex participates to the regulation of the Rev export pathway remain to be elucidated. A direct effect on the rate of nucleocytoplasmic export of Rev might result in altered partitioning of Rev. However, attempts to verify whether overexpression of Eps15 and Hrb could alter the nuclear vs. cytosolic partitioning of Rev did not reveal any significant effect (not shown). However, we noticed, increased levels of immunoreactive Rev (not shown). This prompted us to perform a quantitative biochemical analysis of the steady state levels of Rev under conditions of overexpression of Eps15 and Hrb. Our finding of a significant increase in Rev levels upon concomitant overexpression of the two proteins provides a mechanistic basis for their synergistic effect on Rev activity.
Increased Rev levels might reflect stabilization of the protein, or of its mRNA, or an increased transcription/translation rate. The possibility that Eps15–Hrb affects Rev stability is an intriguing one. Rev is less stable in the cytosol than in the nucleus (Kubota et al. 1996), thus predicting the existence in the cytosol of a molecular machinery able to divert it from its metabolic destiny, to allow reimport into the nucleus. The Eps15–Hrb complex might be part of this machinery. The Rev export pathway has recently come into focus as a major mechanism involved in the regulation of levels of important proteins such as hdm2, p53, and NUMB, by targeting them to cellular compartments (nucleus or cytosol) appropriate for their degradation by the 26S proteasome (Juven-Gershon et al. 1998; Roth et al. 1998). It is thus possible that the Eps15–Hrb complex is involved in the regulation of the trafficking of cellular proteins whose localization and stability are controlled through the Rev export pathway.
The EH network is implicated in the control of endocytosis and actin cytoskeleton organization (reviewed in Santolini et al. 1999). Our results indicate additional levels of involvement, i.e., modulation of nucleocytoplasmic shuttling and possibly of protein degradation. We note that both Eps15 and Eps15R possess FG repeats in their COOH termini. In addition, another component of the clathrin coat, AP-180 also displays FG repeats. It will be of interest, therefore, to analyze whether endocytic proteins interact with members of the importin family. If true, this would unveil an unexpected convergence of molecular machineries governing vesicle sorting and nucleocytoplasmic shuttling.
Acknowledgments
We are indebted to I. Nicoletti for help with confocal microscopy.
This work was supported by grants from Istituto Superiore della Sanita' (AIDS 1998), from the National Institutes of Health (AI40317), from Associazione Italiana Ricerca sul Cancro, from Fondazione Telethon, and from the Armenise-Harvard Foundation. M. Doria is the recipient of a fellowship from the Fondazione Vollaro.
Footnotes
Dr. Doria's present address is Ospedale Pediatrico Bambino Gesù, D0133 Rome, Italy.
Abbreviations used in this paper: aa, amino acids; EH, Eps15 homology; FG, phenylalanine-glycine; NESs, nuclear export signals; RRE, Rev response element.
References
- Bogerd H.P., Fridell R.A., Madore S., Cullen B.R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Coda L., Salcini A.E., Confalonieri S., Pelicci G., Sorkina T., Sorkin A., Pelicci P.G., Di Fiore P.P. Eps15R is a tyrosine kinase substrate with characteristics of a docking protein possibly involved in coated pits-mediated internalization. J. Biol. Chem. 1998;273:3003–3012. doi: 10.1074/jbc.273.5.3003. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. HIV-1 auxiliary proteinsmaking connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- Fazioli F., Minichiello L., Matoskova B., Wong W.T., Di Fiore P.P. eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol. Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Huber J., Boelens W.C., Mattaj I.W., Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fridell R.A., Bogerd H.P., Cullen B.R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc. Natl. Acad. Sci. USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz C.C., Green M.R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr. Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- Fritz C.C., Zapp M.L., Green M.R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- Henderson B.R., Percipalle P. Interactions between HIV Rev and nuclear import and export factorsthe Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- Huang X., Hope T.J., Bond B.L., McDonald D., Grahl K., Parslow T.G. Minimal Rev-response element for type 1 human immunodeficiency virus. J. Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T., Shifman O., Unger T., Elkeles A., Haupt Y., Oren M. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell Biol. 1998;18:3974–3982. doi: 10.1128/mcb.18.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S., Duan L., Furuta R.A., Hatanaka M., Pomerantz R.J. Nuclear preservation and cytoplasmic degradation of human immunodeficiency virus type 1 Rev protein. J. Virol. 1996;70:1282–1287. doi: 10.1128/jvi.70.2.1282-1287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M., Stutz F., Lee L., Davis L.I., Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Roth J., Dobbelstein M., Freedman D.A., Shenk T., Levine A.J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcini A.E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P.G., Di Fiore P.P. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E., Salcini A.E., Kay B.K., Yamabhai M., Di Fiore P.P. The EH network. Exp. Cell Res. 1999;253:186–209. doi: 10.1006/excr.1999.4694. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford C.S., Guthrie C., Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stutz F., Izaurralde E., Mattaj I.W., Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol. Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Kantor J., Zhang D., McCarthy T., Neville M., Rosbash M. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 1997;11:2857–2868. doi: 10.1101/gad.11.21.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Neville M., Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Ullman K.S., Powers M.A., Forbes D.J. Nuclear export receptorsfrom importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- Wong W.T., Schumacher C., Salcini A.E., Romano A., Castagnino P., Pelicci P.G., Di Fiore P. A protein-binding domain, EH, identified in the receptor tyrosine kinase substrate Eps15 and conserved in evolution. Proc. Natl. Acad. Sci. USA. 1995;92:9530–9534. doi: 10.1073/pnas.92.21.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M., Hoffman N.G., Hardison N.L., McPherson P.S., Castagnoli L., Cesareni G., Kay B.K. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J. Biol. Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]