Abstract
The structural basis for the phosphoryla- tion-dependent regulation of smooth muscle myosin ATPase activity was investigated by forming two- dimensional (2-D) crystalline arrays of expressed unphosphorylated and thiophosphorylated smooth muscle heavy meromyosin (HMM) on positively charged lipid monolayers. A comparison of averaged 2-D projections of both forms at 2.3-nm resolution reveals distinct structural differences. In the active, thiophosphorylated form, the two heads of HMM interact intermolecularly with adjacent molecules. In the unphosphorylated or inhibited state, intramolecular interactions position the actin-binding interface of one head onto the converter domain of the second head, thus providing a mechanism whereby the activity of both heads could be inhibited.
Keywords: myosin light chains, phosphorylation, two-dimensional crystalline arrays, myosin regulation, myosin head interactions
The enzymatic activity and motor properties of smooth muscle myosin, or of its double-headed subfragment, heavy meromyosin (HMM), is controlled by phosphorylation of the regulatory light chain (RLC). The active, phosphorylated form displays high actin-activated ATPase activity and moves actin in an in vitro motility assay. The inhibited, unphosphorylated state shows several 100-fold lower actin-activated ATPase activity and does not act as a molecular motor (reviewed in Trybus 1991). In contrast, the single-headed subfragment 1 (S1; Ikebe and Hartshorne 1985), as well as the proteolytically prepared single-headed myosin (Cremo et al. 1995), are active regardless of the phosphorylation state. These studies indicate that interaction between the two myosin heads is required to obtain the inhibited state. It has not been established, however, if the key interaction to obtain the off state is between motor domains, between RLCs, between the head and the rod, or a combination of all three. A number of mutagenic studies show that regulation can be perturbed by mutations in each of these regions of the molecule (Trybus and Chatman 1993; Trybus et al. 1997, Trybus et al. 1998; Ikebe et al. 1998).
To fully understand phosphorylation-dependent regulation, a structural approach that directly shows the contacts between heads that give rise to the inhibited state is essential. Possible interactions between individual heads can be observed using electron microscopic techniques. However, images of actin decorated with HMM gave no insights regarding how the two heads might interact to inhibit product release (Craig et al. 1980). Metal-shadowed images of unphosphorylated smooth muscle myosin showed that the rod folds into thirds and the heads bend back toward the rod in the inhibited state and appear to closely abut each other, but these data were not of sufficient resolution to identify interaction sites that lead to lowered activity (Onishi and Wakabayashi 1982; Trybus et al. 1982; Trybus and Lowey 1984).
Formation of two-dimensional (2-D) crystals of soluble proteins on lipid monolayers provides a specimen suitable for the highest resolution imaging by EM (Kornberg and Ribi 1987; Chiu et al. 1997). Using this approach, we obtained projection maps from 2-D arrays of expressed smooth muscle HMM bound to lipid monolayers in both the unphosphorylated and thiophosphorylated states. Our data provide the first view of intramolecular interactions between the two heads of unphosphorylated HMM, which are not seen in thiophosphorylated HMM. These data suggest a possible mechanism for inhibition of product release from a regulated myosin. In addition, this approach allows the structures of other double-headed myosins to be determined without being dependent on the formation of large three-dimensional crystals.
Materials and Methods
Expression and Purification of Chicken Gizzard Smooth Muscle HMM
Recombinant baculovirus was isolated by conventional protocols (O'Reilly et al. 1992). For HMM expression, Sf9 cells in suspension culture were coinfected with two recombinant viral stocks, one coding for the heavy chain (amino acids 1–1175) and one coding for both the RLC and essential light chains (ELCs). The heavy chain was cloned with a FLAG-tag at the COOH terminus to facilitate purification (DYKDDDDK). The cells were harvested at 65–75 h, and the recombinant proteins isolated on an anti-FLAG affinity column (Sigma Chemical Co.). The expressed HMM yielded a homogeneous product with intact heavy and light chains. Thiophosphorylated HMM was prepared by incubation of unphosphorylated HMM with ATP-γ-S, CaCl2, myosin light chain kinase, and calmodulin.
Preparation of 2-D Crystals on Lipid Monolayers
Crystallization of both unphosphorylated and thiophosphorylated HMM was achieved using a positively charged lipid monolayer, essentially as described previously (Taylor and Taylor 1992). Crystals were found over a range of conditions in a phosphate buffer containing NaCl, MgCl2, EGTA, and ATP in combination with polyethylene glycol. The arrays typically appeared within 1–3 d, and degraded quickly thereafter. Specimens for EM were made by transferring the crystals to 200–300-mesh copper grids with reticulated carbon support film (Kubalek et al. 1991). Crystals were negatively stained with 2% uranyl acetate. After drying, the specimens were stabilized by vacuum deposition of a thin layer of carbon before examination.
Electron Microscopy and Image Processing
Micrographs where recorded on a Philips CM120 electron microscope at 120 kV acceleration voltage and a magnification of 28,000 and 35,000, respectively, under standard conditions. Electron micrographs initially were screened by optical diffraction and then digitized on a Perkin Elmer PDS1010M microdensitometer with a step size of 0.46 or 0.37 nm, with respect to the original object.
Correction for lattice distortions was performed using SPECTRA (Schmid et al. 1993) before calculating structure factors. The defocus was determined using ICE (Hardt et al. 1996) and corrected using CTFAPPPLY from the MRC package (Crowther et al. 1996). Structure factor data from the different images were merged using a modified version of an origin refinement program originally written by S.D. Fuller (EMBL, Heidelberg, Germany) and averaged in plane group P2. Maps were calculated using the CCP4 software (Collaborative Computational Project, 1994) and then imported into O (Jones et al. 1991; Kjeldgaard et al. 1993) for model building.
The ten micrographs of unphosphorylated HMM that were analyzed had average unit cell dimensions of a = 11.95 ± 0.25 nm, b = 28.20 ± 0.89 nm, and γ = 90.6° ± 0.8°. Structure factors were refined to a common phase origin and symmetrized in the plane group P2. This yielded a total of 76 averaged structure factors with a diffraction spot quality of IQ ≤ 4, as defined by Henderson et al. 1986. The averaged phase residual was 10°, extending to a resolution of 2.3 nm.
The 21 images of thiophosphorylated HMM that were analyzed had averaged unit cell dimensions of a = 15.53 ± 0.49 nm, b = 18.38 ± 0.24 nm, and γ = 80.2° ± 1.7°. A total of 65 unique averaged structure factors were obtained (IQ ≤ 4) with an average phase residual of 11°, extending to a resolution of 2.3 nm.
Model Building
We constructed an S1 model based on the high resolution X-ray structure of the smooth muscle motor domain plus ELC (MDE) with bound MgADP·AlF4 −, a transition state analogue (Protein Data Bank 1BR1; Dominguez et al. 1998). It should be noted that this structure was indistinguishable from one obtained with an ATP analogue, MgADP·BeFx. Thus, the MDE·MgADP·AlF4 − crystal structure should mimic the head conformation in our 2-D crystals produced with MgATP. The skeletal muscle RLC and the heavy chain associated with it (Rayment et al. 1993) were then modeled onto the smooth MDE crystal structure by aligning homologous regions of the light chain binding domain. Figures of the model were prepared using Bobscript 2.3 (Esnouf 1997).
Results
Unphosphorylated Heavy Meromyosin
2-D crystalline arrays of inhibited, unphosphorylated HMM on lipid monolayers were obtained in the presence of MgATP (see Materials and Methods). The arrangement of heads in unphosphorylated HMM is highly asymmetric. To further interpret the projection maps (Fig. 1 d), an atomic model for smooth muscle S1 was docked into the electron density. Even in projection, the docking is relatively unambiguous. All of the density seen in projection can be accounted for by the two heads. No obvious feature corresponding to the S2 fragment of the rod is visible. Without modifying the S1 model, a relatively good alignment can be achieved (Fig. 1 a). In this alignment, the long axis of one of the motor domains is oriented nearly perpendicular to the plane of the crystal, and the other is oriented nearly within the plane. The highest density in the projection occurs where the motor domain is oriented perpendicular to the crystal plane. However, the COOH-terminal ends of the heads are relatively far apart, and some density is unaccounted for by this model. To move the COOH termini closer together and achieve a better fit, it was necessary to modify the position of one of the light chain binding domains. An ∼30° rotation of the light chain binding domain about gly779 brings the light chain binding domain, in particular the region where the RLC is located, into density that had previously been unaccounted for, and at the same time brings the COOH-terminal residues closer together to form a vertex at the head–rod junction (Fig. 1b and Fig. c).
Figure 1.
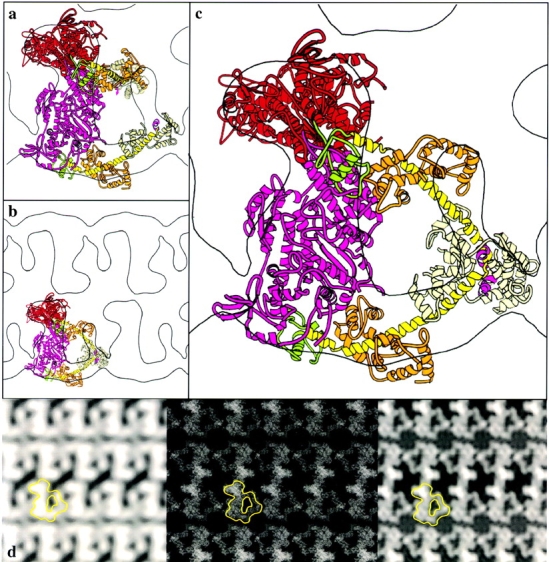
Docking of the S1 coordinates into the 2-D EM map of unphosphorylated HMM. The motor domain (red for one head, pink for the other), the converter domain (green), the long alpha helix (yellow, and magenta for the hook), the ELC (orange), and the RLC (cream) are indicated by a ribbon diagram in a–c. a, One contour from the 2-D map with two S1 molecules docked before modification of the light chain binding domain orientation. b, An overview of the same orientation shown in a, but after rebuilding the light chain binding domain of the upper S1 molecule. c, The arrangement of the unphosphorylated myosin heads is shown in the half unit cell of the map. d, The projection map (left) was obtained by averaging the structure factors obtained from ten electron micrographs after correction for the CTF. Middle, The atomic coordinates are projected into a 2-D image for comparison with the original electron density map. Right, This 2-D projection is shown filtered to 2-nm resolution. One HMM motif is outlined in each map in d.
The quality of the model was assessed by projecting the rebuilt X-ray coordinates into a 2-D image (Fig. 1 d, middle and right) for comparison with the initial electron density map (Fig. 1 d, left). The phase residual for this model when compared with the original data is 25°. This is reasonably good considering that solvent and negative stain effects have not been incorporated into the model.
The important molecular contacts within the crystal occur between the motor domains of the same unphosphorylated HMM. The actin-binding interface of one head (the blocked head) is located in close proximity to the converter domain of the companion head (the free head).
Thiophosphorylated Heavy Meromyosin
The unit cell is smaller in the crystals of thiophosphorylated HMM, but the projected density and twofold symmetry requires incorporation of two HMM molecules into the unit cell. Thus, the protein packing is much denser in these crystals than in the unphosphorylated HMM crystals. The docking of the smooth muscle S1 model into the thiophosphorylated HMM projection shown in Fig. 2, a–c, is the only way we have found to fit two HMM molecules into the unit cell and account for the mass in the electron density map, with the reasonable assumption that the two HMM molecules within the unit cell are the same.
Figure 2.
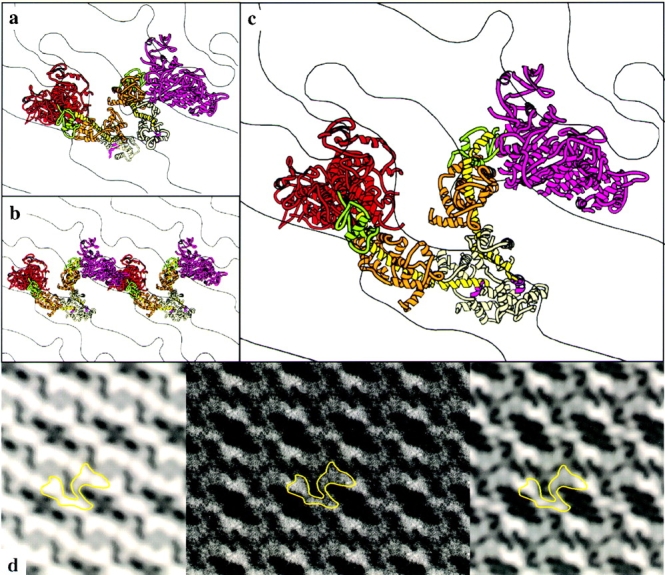
Docking of the S1 coordinates into the 2-D EM map of thiophosphorylated HMM. The myosin head structure in a–c is color coded as in Fig. 1. a, Two S1 molecules docked into one contour from the 2-D map. The same orientation is shown in b after rebuilding the light chain binding domain of the left S1 molecule. The lack of intramolecular interactions between two heads is shown in c. d, The projection map (left) was obtained from averaging the structure factors of 21 micrographs. The fitted coordinates were then projected into a 2-D image (middle) for comparison with the original electron density map and filtered to 2-nm resolution (right). One HMM molecule is outlined in each map in d.
As was found for the unphosphorylated HMM crystals, the long dimension of the motor domain of one of the heads extends out of the plane of the crystal, whereas the other lies more within the plane. As with the unphosphorylated HMM, it was necessary to rebuild the light chain binding domain of one of the heads to obtain a model where the two light chain binding domains connect to form the beginning of the S2 region (Fig. 2 c). The rod region is not identifiable in the projection. The agreement between the model and the reconstruction was evaluated by projecting the model coordinates (Fig. 2 d, middle and right) for comparison with the experimental electron density (Fig. 2 d, left). The phase residual for the model when cross-correlated with the original reconstruction is 31°.
In contrast to the model for the unphosphorylated HMM, the contacts between heads appear to be intermolecular, rather than intramolecular. Moreover, it is possible to build a head-to-head interaction in the thiophosphorylated HMM crystals (Fig. 2 b) roughly similar to that constructed in the unphosphorylated HMM crystals, but the interacting heads come from different HMM molecules.
Discussion
This is the first report of 2-D crystalline arrays of unphosphorylated and thiophosphorylated smooth muscle HMM, which provides the highest resolution structure of a double-headed myosin to date. Both 2-D arrays were formed in the presence of ATP, a state that cannot be analyzed using decorated F-actin because of its weak binding properties. The major new finding is the direct observation of interactions between the heads of one HMM molecule in the inhibited, unphosphorylated state. Such contacts have long been inferred from biochemical, structural, and mutagenic data. In contrast, only contacts between different HMM molecules are observed in the active thiophosphorylated HMM. The thiophosphorylated HMM structure is probably one of many conformations; the particular conformation obtained here is stabilized in the crystal by packing forces. Mobility of the two heads in the active state is a key feature of myosin's ability to search for actin monomers in adjacent thin filaments.
The docking of an atomic model for the myosin head into our 2-D projections allows a model for the disposition of the heads in the active and inhibited states to be proposed. The crystal structure used for docking into the electron density (MDE with a transition state analogue or an ATP analogue bound at the active site; Dominguez et al. 1998) is expected to be structurally similar to that formed in the 2-D crystals in the presence of MgATP. Consistent with this idea, one head of the HMM molecule in both the unphosphorylated and the thiophosphorylated HMM was fitted with this model without modification. The light chain binding domain of the second head required modification; this region of the molecule undergoes large rigid body movements during the power stroke. The arrangement of heads in both the unphosphorylated and the thiophosphorylated HMM is highly asymmetric. This differentiates our data from the only previous structural model for HMM (Offer and Knight 1996) that was obtained by modeling and energy-minimizing the head–tail junction of scallop myosin. In that model, the myosin heads are related by a molecular twofold rotation axis. The present results, though not excluding such a possibility under some conditions, favors asymmetric models and suggests that the heads are relatively independent of each other.
Significantly, in the unphosphorylated HMM, the actin-binding interface on one head is positioned in the middle of the other head near the converter domain. This arrangement leaves the actin-binding domain of one head free while the other is blocked. The interaction between heads seen with the unphosphorylated HMM can explain several existing pieces of experimental data. One is that the binding constant of unphosphorylated and phosphorylated HMM to actin is only three- to fourfold different, although phosphate release is regulated several 100-fold (Sellers et al. 1982). The model proposed here shows that only one of the two heads of unphosphorylated HMM would be capable of interacting with actin, and thus the difference in binding constant could reflect the difference between single-headed binding in the inhibited state versus double-headed binding in the active state. More importantly, the interaction of one head with the converter region of the adjacent head could prevent the domain motions that are required to open myosin's back door for phosphate release from that head (Yount et al. 1995). A large rotation (70°) of the converter region has been shown to occur as myosin goes from the closed to the open conformation (Rayment et al. 1993; Gulick and Rayment 1997; Dominguez et al. 1998). Mutagenesis studies also showed that conversion of the converter domain from a smooth to a skeletal muscle sequence produced a chimera that was not regulated by phosphorylation (Trybus et al. 1998). Although these experiments were interpreted as suggesting that sequences near the neck/motor domain interface are important for properly positioning the RLCs in an inhibitory position, the data could also accommodate the idea that there is a direct interaction between heads that involves the converter region.
The 2-D projection map is currently being extended into the third dimension, which will enable us to determine the interaction sites leading to the inhibited state more precisely. Nonetheless, this study has already provided a general mechanism by which product release could be inhibited from both heads (direct blocking of the actin-binding interface in one head, inhibition of converter rotation in the other), while maintaining one head's ability to bind to actin in the presence of MgATP.
Acknowledgments
We thank Gary Connors for assistance during the initial phase of the image processing and Ken Sale for providing us with a model of smooth muscle S1 carrying the RLC and the heavy chain associated with it.
This work was supported by National Institutes of Health grants to K. Trybus (HL-38113) and K. Taylor (AR-42872).
Footnotes
Abbreviations used in this paper: 2-D, two-dimensional; ELC, essential light chain; HMM, heavy meromyosin; MDE, motor domain-essential light chain complex; RLC, regulatory light chain; S1, myosin subfragment 1; S2, subfragment 2 region of the rod.
References
- Chiu W., Avila-Sakar A.J., Schmid M.F. Electron crystallography of macromolecular periodic arrays on phospholipid monolayers. Adv. Biophys. 1997;34:161–172. doi: 10.1016/s0065-227x(97)89638-4. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Cryst. D50:760–763. [DOI] [PubMed]
- Craig R., Szent-Györgyi A.G., Beese L., Flicker P., Vibert P., Cohen C. Electron microscopy of thin filaments decorated with a Ca2+-regulated myosin. J. Mol. Biol. 1980;140:35–55. doi: 10.1016/0022-2836(80)90355-1. [DOI] [PubMed] [Google Scholar]
- Cremo C.R., Sellers J.R., Facemyer K.C. Two heads are required for phosphorylation-dependent regulation of smooth muscle myosin. J. Biol. Chem. 1995;270:2171–2175. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- Crowther R.A., Henderson R., Smith J.M. MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Dominguez R., Freyzon Y., Trybus K.M., Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chainvisualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- Gulick A.M., Rayment I. Structural studies on myosin IIcommunication between distant protein domains. Bioassays. 1997;19:561–569. doi: 10.1002/bies.950190707. [DOI] [PubMed] [Google Scholar]
- Hardt S., Wang B., Schmid M. A brief description of I.C.E.the integrated crystallographic environment. J. Struct. Biol. 1996;116:68–70. doi: 10.1006/jsbi.1996.0012. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J.M., Downing K.H., Lepault J., Zemlin F. Structure of purple membrane from Halobacterium halobiumrecording, measurement and evaluation of electron micrographs at 3.5 Å resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- Ikebe M., Hartshorne D.J. Proteolysis of smooth muscle myosin by Staphylococcus aureus proteasepreparation of heavy meromyosin and subfragment 1 with intact 20000-dalton light chains. Biochemistry. 1985;24:2380–2387. doi: 10.1021/bi00330a038. [DOI] [PubMed] [Google Scholar]
- Ikebe M., Kambara T., Stafford W.F., Sata M., Katayama E., Ikebe R. A hinge at the central helix of the regulatory light chain of myosin is critical for phosphorylation-dependent regulation of smooth muscle myosin motor activity. J. Biol. Chem. 1998;273:17702–17707. doi: 10.1074/jbc.273.28.17702. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.-Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen P., Thirup S., Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- Kornberg, R.D., and H.O. Ribi. 1987. Formation of two-dimensional crystals of proteins on lipid layers. In Protein Structure, Folding and Design. 2. Alan R. Liss, Inc. NY. 175–186.
- Kubalek E.W., Kornberg R.D., Darst S.A. Improved transfer of two-dimensional crystals from the air/water interface to specimen support grids for high-resolution analysis by electron microscopy. Ultramicroscopy. 1991;35:295–304. doi: 10.1016/0304-3991(91)90082-h. [DOI] [PubMed] [Google Scholar]
- O'Reilly D.R., Miller L.K., Luckow V.A. Baculovirus Expression Vectors, A Laboratory Manual. W.H. Freeman; New York: 1992. [Google Scholar]
- Offer G., Knight P. The structure of the head–tail junction of the myosin molecule. J. Mol. Biol. 1996;256:407–416. doi: 10.1006/jmbi.1996.0096. [DOI] [PubMed] [Google Scholar]
- Onishi H., Wakabayashi T. Electron microscopic studies of myosin molecules from chicken gizzard muscle Ithe formation of the intramolecular loop in the myosin tail. J. Biochem. 1982;92:871–879. doi: 10.1093/oxfordjournals.jbchem.a134001. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski W.R., Schmid-Bäse K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., Holden H.M. Three-dimensional structure of myosin subfragment-1a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Schmid M., Dargahi R., Tam M. SPECTRAa system for processing electron images of crystals. Ultramicroscopy. 1993;48:251–254. doi: 10.1016/0304-3991(93)90099-j. [DOI] [PubMed] [Google Scholar]
- Sellers J.R., Eisenberg E., Adelstein R.S. The binding of smooth muscle heavy meromyosin to actin in the presence of MgATP. J. Biol. Chem. 1982;257:1380–1383. [PubMed] [Google Scholar]
- Taylor K.A., Taylor D.W. Formation of 2-D paracrystals of F-actin on phospholipid layers mixed with quarternary ammonium surfactants. Struct. Biol. 1992;108:140–147. doi: 10.1016/1047-8477(92)90013-z. [DOI] [PubMed] [Google Scholar]
- Trybus K.M. Regulation of smooth muscle myosin. Cell Motil. Cytoskel. 1991;18:81–85. doi: 10.1002/cm.970180202. [DOI] [PubMed] [Google Scholar]
- Trybus K.M., Lowey S. Conformational states of smooth muscle myosin. J. Biol. Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- Trybus K.M., Chatman T.A. Chimeric regulatory light chains as probes of smooth muscle myosin function. J. Biol. Chem. 1993;268:4412–4419. [PubMed] [Google Scholar]
- Trybus K.M., Huiatt T.W., Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc. Natl. Acad. Sci. USA. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K.M., Freyzon Y., Faust L.Z., Sweeney H.L. Spare the rod, spoil the regulationnecessity for a myosin rod. Proc. Natl. Acad. Sci. USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K.M., Naroditskaya V., Sweeney H.L. The light chain-binding domain of the smooth muscle myosin heavy chain is not the only determinant of regulation. J. Biol. Chem. 1998;273:18423–18428. doi: 10.1074/jbc.273.29.18423. [DOI] [PubMed] [Google Scholar]
- Yount R.G., Lawson D., Rayment I. Is myosin a “back door” enzyme? Biophys. J. 1995;68:44s–49s. [PMC free article] [PubMed] [Google Scholar]


