Abstract
The histone macroH2A1.2 has been implicated in X chromosome inactivation on the basis of its accumulation on the inactive X chromosome (Xi) of adult female mammals. We have established the timing of macroH2A1.2 association with the Xi relative to the onset of X-inactivation in differentiating murine embryonic stem (ES) cells using immuno-RNA fluorescence in situ hybridization (FISH). Before X-inactivation we observe a single macroH2A1.2-dense region in both undifferentiated XX and XY ES cells that does not colocalize with X inactive specific transcript (Xist) RNA, and thus appears not to associate with the X chromosome(s). This pattern persists through early stages of differentiation, up to day 7. Then the frequency of XY cells containing a macroH2A1.2-rich domain declines. In contrast, in XX cells there is a striking relocalization of macroH2A1.2 to the Xi. Relocalization occurs in a highly synchronized wave over a 2-d period, indicating a precisely regulated association. The timing of macroH2A1.2 accumulation on the Xi suggests it is not necessary for the initiation or propagation of random X-inactivation.
Keywords: histones, X inactive specific transcript (Xist) RNA, X chromosome inactivation, differentiation, nuclear structure
Mammalian genes on the X chromosome are expressed to the same level in males (XY) and females (XX) through genetic inactivation of one X chromosome in females (Lyon 1961; for review see Heard et al. 1997). This phenomenon, known as X-inactivation, occurs during embryogenesis. In early preimplantation XX mouse embryos, both X chromosomes are transcriptionally active. Immediately before gastrulation, either the maternally or the paternally derived X chromosome is inactivated in the embryo proper (Gardiner and Lyon 1971; McMahon et al. 1983). This random X-inactivation occurs in three stages: initiation of silencing in cis, propagation of inactivation along the length of the chromosome, and thereafter heritable maintenance of the silent state in the somatic lineages of the organism.
The X-linked X inactive specific transcript (Xist) gene is necessary for the initiation and propagation of silencing but not for maintenance (Brown and Willard 1994; Rack et al. 1994; Lee et al. 1996; Penny et al. 1996; Lee and Jaenisch 1997; Marahrens et al. 1997; Herzing et al. 1997; Csankovszki et al. 1999). Xist encodes large transcripts that lack a conserved open reading frame and are restricted to the nucleus (Brockdorff et al. 1992; Brown et al. 1992). Xist RNA expression is developmentally regulated, and multiple promoters have been described that can be classified as early or late (Brockdorff et al. 1992; Johnston et al. 1998; Lee et al. 1999). Before onset of X-inactivation, early promoters generate relatively unstable Xist RNA in XY and XX cells (Panning et al. 1997; Sheardown et al. 1997a; Johnston et al. 1998; Lee et al. 1999; Mise et al. 1999). At the onset of X-inactivation, late promoters are activated exclusively on the future inactive X chromosome (Xi) (Johnston et al. 1998). As X-inactivation advances, transcription from early promoters is shut off. Thus, in XY somatic cells no Xist RNA is expressed, whereas in XX cells Xist is continuously transcribed from the Xi. Late Xist transcripts are relatively stable and associate with the length of the Xi (Brown et al. 1992; Clemson et al. 1996; Lee et al. 1996; Panning and Jaenisch 1996; Sheardown et al. 1997a). This unique distribution is believed to be an essential step in the chromosome-wide silencing of transcription.
A fundamental aspect of X-inactivation is a change in the higher order chromatin structure of the Xi. This follows the initiation of late Xist RNA transcription and results in the Xi differing from its active homologue (Xa) in both composition and configuration (Eils et al. 1996). In interphase, the Xi is cytologically recognizable as the Barr body (Barr and Carr 1962). It replicates late in S phase, is underacetylated at histones H3 and H4, and many of its genes are methylated at CpG islands (Takagi 1974; Monk 1986; Jeppesen and Turner 1993; Belyaev et al. 1996; Boggs et al. 1996). These events occur sequentially and are thought to be involved in maintaining the silenced state (for reviews see Riggs and Pfeiffer 1992; Keohane et al. 1998).
Recently it has been shown that the Xi is enriched in the unusual core histone macroH2A1.2 (Costanzi and Pehrson 1998). The NH2-terminal third of macroH2A is very similar to a conventional histone 2A, whereas the larger part of the protein contains sequences unrelated to histones and is of unknown function (Pehrson and Fried 1992). At least two evolutionary conserved subtypes of macroH2A are known, differing in a single region that includes a putative leucine zipper protein–protein interaction domain (Pehrson and Fried 1992; Pehrson and Fuji 1998). Subtype 1.2 is expressed in somatic cells of both females and males and shows a diffuse nuclear distribution. However, in female cells, there is also a specific macroH2A1.2-dense region, the macrochromatin body (MCB), which colocalizes with the Xi but not its active counterpart (Costanzi and Pehrson 1998). A deletion of the Xist gene in embryonic fibroblasts in vitro disrupts the MCB on the Xi, indicating a link between Xist RNA and macroH2A1.2 (Csankovszki et al. 1999).
The precise function of macroH2A1.2 in the X-inactivation process is unknown. Studies to date have used cells in which X-inactivation has been completed. However, as generation of the Xi is a sequential process, we wished to determine the point at which the MCB participates in this pathway. Murine embryonic stem (ES) cells offer a powerful experimental model system that allows this analysis to be carried out directly (Rastan and Robertson 1985; Keohane et al. 1996). In undifferentiated XX ES cells, both X chromosomes are active. Random X-inactivation occurs during in vitro differentiation, recapitulating the events seen in the embryo proper. To analyze macroH2A1.2 localization during ES cell differentiation relative to the Xi and Xa, we developed a protocol for double-labeling of macroH2A1.2 and Xist RNA (immuno-RNA fluorescence in situ hybridization [FISH]). Our analysis reveals that a single macroH2A1.2-rich domain is present in both XX and XY undifferentiated ES cells, but that it does not colocalize with Xist RNA until initiation and establishment of X-inactivation has occurred in differentiating cells. These results indicate that significant nuclear reorganization takes place during differentiation, and that macroH2A1.2 accumulation on the Xi is not a key event for the initiation and propagation of random X-inactivation.
Materials and Methods
Cell Culture
Mouse somatic cell lines used in this study were C127, XX mammary gland tumor cells (provided by Dr. B.M. Turner, University of Birmingham Medical School, Birmingham, UK); Is1Ct and T37H, XX primary fibroblasts (Duthie et al. 1999); FSPE, XX female Mus spretus primary ear fibroblasts (provided by Dr. T.B. Nesterova, Medical Research Council, Hammersmith Hospital, London, UK); and MSPL, XY male M. spretus primary lung fibroblasts (provided by Dr. S.M. Duthie, Medical Research Council Hammersmith Hospital). Cells were maintained as described previously (Jeppesen and Turner 1993; Duthie et al. 1999).
Mouse ES cell lines used were, PGK12.1, XX (Norris et al. 1994); LF2, XX and Efc-1, XY (both provided by Dr. A.G. Smith, Centre for Genome Research, Edinburgh, UK); and 129/1, XY (derived in-house by G.F. Kay). All but 129/1 ES lines were cultured in the absence of feeder cells on 0.1% gelatin-coated tissue culture plates. Cells were maintained in DME containing murine leukemia inhibitory factor (LIF) as described by Penny et al. 1996.
Differentiation of ES Cells
Differentiated cultures were generated by withdrawal of LIF from the ES media (Sheardown et al. 1997b). Details of this protocol and alternative methods of differentiation used are described in the legend of Fig. 6. Cells were harvested by trypsinization at selected timepoints during a 1–22-d differentiation period and analyzed by indirect immunofluorescence and Western blotting.
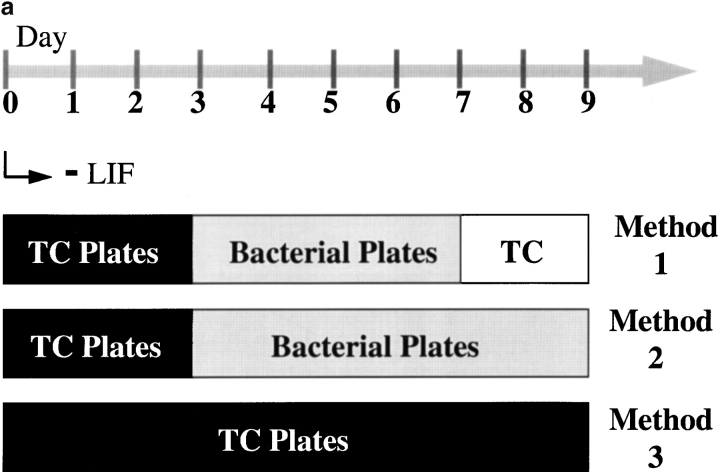
Figure 6.
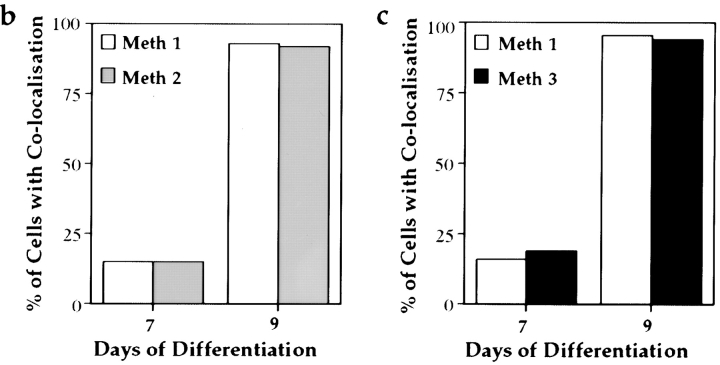
Timing and extent of MCB and Xist RNA colocalization is reproducible using three methods of differentiation. (a) ES cell differentiation was initiated by removal of the growth factor LIF from the medium (day 0). Subsequently, one of three plating patterns was applied. Method 1: cells were grown in tissue culture plates up to day 3 and replated onto bacterial grade petri dishes between day 3–7, during which they were unable to adhere and generated spherical aggregates of differentiating cell types (EBs). At day 7, EBs were transferred to standard tissue culture plates, where cells adhered and formed differentiating outgrowths. Method 2: cells were grown in bacterial dishes from day 3–9. Method 3: cells were grown in tissue culture plates from day 0–9. (b and c) Quantification of immuno-RNA FISH detection of macroH2A1.2 and Xist RNA. Over 100 cells were scored at day 7 and 9 of differentiation and results are given as percentage of colocalization. (b) A PGK12.1 culture was divided at day 3 of differentiation. One half was grown the conventional way (Method 1), whereas the other one was differentiated following Method 2. (c) One aliquot of a LF2 culture differentiated following Method 1 was compared with an aliquot differentiated by Method 3.
To confirm that progression of X-inactivation was occurring in XX cells, we assessed the H4 histone acetylation status of the X chromosome. Previously, an underacetylated X chromosome was detected from day 4 of differentiation onwards, frequencies increasing from 9% of metaphases to a maximum of 25% at day 7 (Keohane et al. 1996). We obtained similar results, with 28% (n = 105) of metaphases showing an underacetylated Xi at day 5 of differentiation.
Immunofluorescence
Indirect immunofluorescence labeling using an affinity-purified rabbit polyclonal antiserum against the nonhistone region of macroH2A1.2 was carried out as described by Costanzi and Pehrson, 1998. The incubation period with the primary antibody was reduced to 1 h at 37°C. Data using formaldehyde fixation (4%) are presented in all figures. Identical results were obtained using different fixation (0.5% glutaraldehyde or methanol/acetone) and permeabilization procedures (Almeida et al. 1998; Ferreira et al. 1997).
Immuno-RNA FISH
In protein and RNA double-labeling experiments, FISH preceded macroH2A1.2 immunostaining. Trypsinized cells were pipetted onto Superfrost Plus glass slides (BDH) and allowed to attach for 3–5 h. Slides were subsequently washed in PBS and fixed in 4% formaldehyde, 5% acetic acid, 0.9% NaCl for 30 min at room temperature, rinsed three times in PBS, dehydrated through a 70-90-100% ethanol series, air-dried, washed in 100% Xylene for 5 min, rehydrated to PBS, incubated with 0.01% pepsin in 0.01 M HCl for 30 s at 37°C, washed in PBS for 5 min, postfixed in 1% formaldehyde in PBS for 2.5 min at room temperature, washed in PBS, dehydrated through an ethanol series, and air-dried before applying 50–100 ng of nick-translated DNA probe.
Biotinylated GPT16, a 6-kb probe spanning most of murine Xist exon 1, was used. Hybridization and wash conditions were as described previously (Sheardown et al. 1997a). After probe detection with avidin–Texas red (TR) followed by biotinylated antiavidin and a final layer of avidin-TR, slides were washed twice for 3 min in 4× SSC, 0.1% Tween 20 at 37°C, followed by twice for 3 min in PBS, 0.1% Tween 20. After blocking in 5% nonfat dry milk at room temperature, slides were incubated for 1 h at 37°C with the primary antibody, anti-macroH2A1.2 rabbit polyclonal antiserum (Costanzi and Pehrson 1998). After three washes in PBS, 0.1% Tween 20 slides were incubated with FITC-labeled mouse anti–rabbit IgG for 30 min at 37°C, washed, and mounted in Vectashield antifade (Vector Labs, Inc.) containing 4,6-diamidino-2-phenylindole dihydrochloride (DAPI). Fluorescently labeled secondary antibodies, avidin-TR, and biotinylated antiavidin were from Vector Labs, Inc. Images were acquired on a Leica DMRB fluorescence microscope equipped with a Photometrics CCD camera using Smartcapture Software (Vysis Ltd.).
Metaphase Analysis
Metaphase chromosome spreads were prepared as described by Keohane et al. 1996. Mitotic C127 cells and undifferentiated ES cells were harvested by mechanical shake-off, and differentiated ES cells were trypsinized after collection. C127 cells and ES cells were swollen in 0.075 and 0.1 M KCl, respectively. To assess the histone H4 acetylation status, immunolabeling was performed on unfixed metaphases with an antibody to acetylated lysine 8 of histone H4 (R232; gift of Dr. B.M. Turner, University of Birmingham Medical School) as described previously (Jeppesen and Turner 1993; Keohane et al. 1996). For the detection of macroH2A1.2, metaphases were fixed in 4% formaldehyde after swelling in KCl, followed by indirect immunofluorescence as described above, but without further permeabilization.
Extract Preparations and Immunoblotting
Total cell extracts were prepared by washing harvested cells twice with cold TB buffer (20 mM Hepes, pH 7.3, 110 mM KOAc, 5 mM NaOAc, 2 mM MgOAc, 1 mM EGTA, 2 mM DTT, protease inhibitors). They were then incubated for 40 min at 37°C in TB buffer with 0.1% NP-40, 10 mM MnCl2, and 20 μg/ml DNaseI followed by addition of SDS-PAGE loading buffer.
NP-40 permeabilization and salt extraction of cells was performed by washing harvested cells twice with cold TB buffer and incubating them on ice for 4 min in TB buffer containing 0.1% NP-40. The supernatant was recovered (S1) and pellets were resuspended in TB without (Ctrl) or with 0.5, 1.2, or 2 M NaCl and incubated for 20 min on a rotating wheel at 4°C. After centrifugation at 100,000 g for 40 min, supernatants were recovered (S2). Cell pellets were digested with 20 μg/ml DNaseI for 40 min at 37°C in TB buffer with 0.1% NP-40 and 10 mM MnCl2. SDS-PAGE loading buffer was added to both supernatants and cell pellets and samples were analyzed by SDS-PAGE and Western blotting.
For immunoblot analysis, protein was transferred to polyvinylidene difluoride membranes and incubated with antibodies as described by Pehrson et al. 1997. Antibodies were an affinity-purified rabbit polyclonal serum against the nonhistone region of macroH2A1.2 (Costanzi and Pehrson 1998), and a donkey anti–rabbit IgG conjugated to alkaline phosphatase (Pierce). For lactate dehydrogenase (LDH) detection, the goat primary antibody (Rockland) was followed by incubation with a rabbit anti–goat Ig conjugated with alkaline phosphatase (Sigma Chemical Co.). Alkaline phosphatase was detected using the 1-step NBT/BCIP kit (Pierce).
Results
Xist RNA and MCB Localization in Somatic Cells
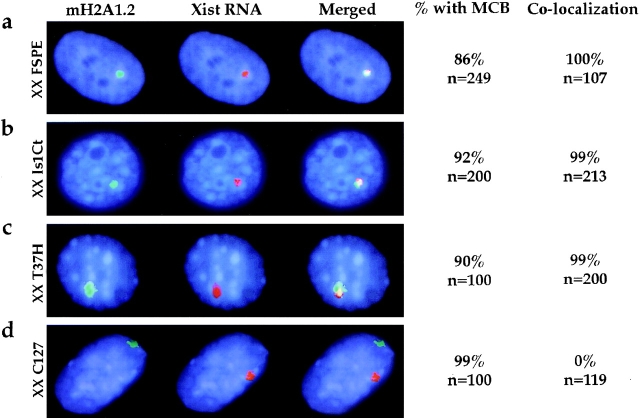
To analyze MCB localization during X-inactivation, it is essential to distinguish between the Xi and Xa. Previously, macroH2A1.2 association with the Xi was shown indirectly using combined immunofluorescence and chromosome painting (Costanzi and Pehrson 1998). Therefore, we developed a protocol for double-labeling of macroH2A1.2 and Xist RNA (immuno-RNA FISH) that allows us to directly correlate an MCB with the Xi. Initially four XX and one XY murine somatic cell lines were analyzed using this approach. In all XX cell lines, an antibody specific to the nonhistone region of macroH2A1.2 revealed diffuse staining throughout the nucleus in addition to an MCB. The MCB was detected in >86% of cells in every line using several different methods of fixation and permeabilization (Fig. 1, a–d, columns 1 and 4; see Materials and Methods). It localized to DAPI-bright regions in the nucleus, except in the case of C127 cells. Consistent with previous studies, the XY cell line exhibited diffuse macroH2A1.2 nuclear staining with nucleolar exclusion, and only infrequently did cells have an MCB (data not shown) (Costanzi and Pehrson 1998).
Figure 1.
MCB and Xist RNA localization in murine somatic cell lines. Immuno-RNA FISH detection of macroH2A1.2 protein (FITC, green) and Xist RNA (TR, red) in mouse somatic cells. DNA is stained with DAPI (blue). Representative examples of primary fibroblast cells (a) XX FSPE, (b) XX Is1Ct, (c) XX T37H, and of a mammary tumor line (d) XX C127 are shown. Column 1, macroH2A1.2, FITC signal. Control immunofluorescence experiments in which the primary antibody was omitted confirm that the staining of macroH2A1.2 is specific (data not shown). Column 2, Xist RNA, TR signal. Column 3, merged images of the first two panels. Column 4, the number of cells with an MCB was quantified and results are given as percentage of total cells (n) analyzed. Column 5, the percentage of cells with both RNA and protein signal detectable (n) exhibiting colocalization of Xist RNA with MCBs.
A simultaneous analysis of Xist RNA localization in the XX cell lines revealed a large Xist RNA signal that reflects accumulation of late Xist transcripts in cis (Fig. 1, a–d, column 2). In three primary fibroblast cell lines (FSPE, Is1Ct, and T37H) Xist RNA and the MCB colocalized in 99–100% of the cells as indicated by the overlap of the two signals (Fig. 1, a–c, columns 3 and 5). The degree of overlap was not always complete in all cells (Fig. 1 c and below). Colocalization was not observed in all cell types. In C127 cells, a mammary gland tumor cell line, the Xist RNA, and the MCB signals did not overlap; rather, they occupied separate territories (Fig. 1 d, columns 3 and 5). The presence of accumulated Xist RNA suggests that C127 cells have retained an Xi. In addition, C127 cells have an underacetylated chromosome corresponding to the Xi (data not shown) (Jeppesen and Turner 1993).
In summary, we show that immuno-RNA FISH can be used to analyze association of macroH2A1.2 with the Xi. We went on to use this methodology to examine the formation of the MCB and its localization with respect to Xist RNA before and during ES cell differentiation.
Both XX and XY ES Cells Express MacroH2A1.2 and Have a Single MCB Which Does Not Colocalize with Xist RNA
We anticipated that an MCB would not be evident in undifferentiated XX and XY ES cells since their X chromosomes are active. Surprisingly, in two independent XX ES cell lines we found that a single macroH2A1.2-rich domain was readily detectable in the majority of interphase cells (Fig. 2, a and b). The appearance of this macroH2A1.2-rich domain was similar to the MCB characterized in somatic cells, and thus we refer to it also as an MCB. In the two XY ES cell lines we analyzed, an MCB was also detected (Fig. 2c and Fig. d). Thus, the presence of a single MCB appears to be an intrinsic property of undifferentiated ES cells, regardless of X chromosome constitution. This contrasts with somatic cells, in which the MCB is found exclusively in XX cells despite macroH2A1.2 expression in both XX and XY cells (Costanzi and Pehrson 1998).
Figure 2.
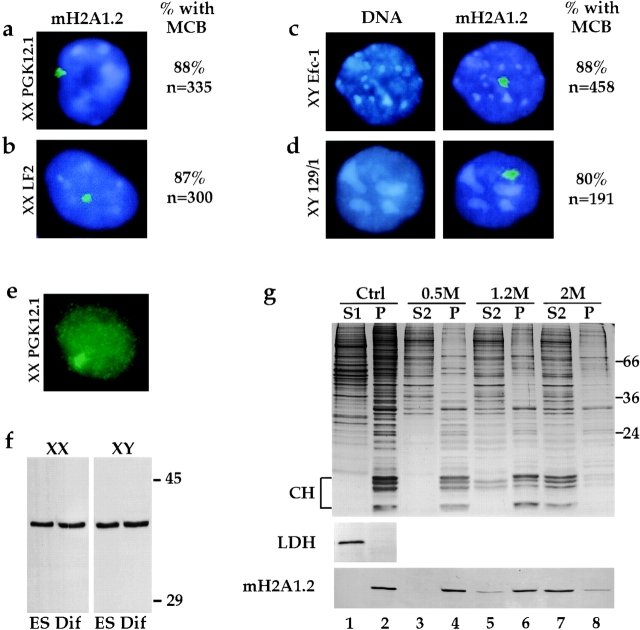
Both XX and XY ES cells express macroH2A1.2 protein that forms a single MCB. Indirect immunofluorescence was performed on ES cell lines detecting macroH2A1.2 (FITC, green) and staining DNA with DAPI (blue). Shown are representative examples of (a and e) XX PGK12.1 cells, (b) XX LF2 cells, (c) XY Efc-1, and (d) XY 129/1 cells. (c and d) Left panels show DAPI staining only. Right panels show combined DAPI and macroH2A1.2 staining. e shows FITC staining only and illustrates diffuse nuclear staining observed in addition to the MCB. Different fixation procedures, utilizing cross-linking or precipitating agents, gave the same result (data not shown). The number of cells with an MCB was quantified and results are given as a percentage of total cells (n) analyzed. (f) Immunoblot analysis of macroH2A1.2 in total extracts prepared from XX ES cells (PGK12.1) and XY ES cells (Efc-1), either undifferentiated (ES) or differentiated (Dif) for 10 or 9 d, respectively. The specificity of the antibody for macroH2A1.2 has been demonstrated by competition with the antigen (Costanzi, C., and J. Pehrson, manuscript submitted for publication). The position of the 45- and 29-kD molecular mass markers are indicated. (g) Undifferentiated XX ES cells (PGK12.1) were permeabilized with 0.1% NP-40, and supernatant (Ctrl S1, lane 1) and cell pellet (Ctrl P, lane 2) fractions were separated, followed by extraction of the pellet with 0.5 (lanes 3 and 4), 1.2 (lanes 5 and 6), or 2 M (lanes 7 and 8) NaCl. A 15% SDS-PAGE Coomassie-stained gel is shown, the position of the core histones (CH: H3, H2B, H2A, and H4 from top to bottom) and molecular mass markers are indicated. Supernatants and pellets were analyzed by Western blotting with antibodies against LDH and macroH2A1.2.
The single MCB was frequently found at the periphery of the nucleus (Fig. 2 a), close to the nuclear membrane as assessed by double-label immunofluorescence with an antibody against the nuclear pore complex (data not shown). Interestingly the MCB appeared to lie in a region that stained poorly with DAPI (Fig. 2c and Fig. d). In addition to the MCB we detected diffuse nuclear staining in XX and XY ES cells (Fig. 2 e). Thus, as is the case of XX and XY somatic cells (Costanzi and Pehrson 1998), the MCB is unlikely to represent the total macroH2A1.2 protein content.
We confirmed that the presence of an MCB in undifferentiated ES cells can be attributed to macroH2A1.2 protein using the same antibody as used for immunofluorescence detection in Western blots. A single protein with an expected electrophoretic mobility corresponding to that seen previously in somatic cells was detected in both XX and XY ES cells. The relative macroH2A1.2 protein content was similar not only between undifferentiated XX and XY ES cells but also between undifferentiated and differentiated ES cells (Fig. 2 f).
To investigate the characteristics of macroH2A1.2 in ES cells at the biochemical level, we permeabilized undifferentiated XX PGK12.1 cells with NP-40 to disrupt membranes. As reported previously (Falciola et al. 1997), this treatment brought about the complete release of LDH (Fig. 2 g, lanes 1 and 2). However, we found that macroH2A1.2 was quantitatively retained in the pellet (Fig. 2 g, lanes 1 and 2). Similarly, macroH2A1.2 protein remained associated with the insoluble fraction when either undifferentiated XY ES cells or XX ES cells differentiated for 10 d were treated with NP-40 (data not shown). Thus, macroH2A1.2 in undifferentiated ES cells is not present in a pool of soluble protein.
Next, the pellet fraction produced by NP-40 treatment was extracted with different concentrations of sodium chloride. Most nonhistone chromosomal proteins and histone H1 are extracted by 0.5 M sodium chloride, but not the core histones (CH; Fig. 2 g, lanes 3 and 4) (Hoffmann and Chalkley 1978). These conditions did not release macroH2A1.2 protein from the NP-40 pellet (Fig. 2 g, lanes 3 and 4). Similar results have been described for macroH2A1.2 in somatic cells (Pehrson and Fried 1992). Treatment of the NP-40 pellet fraction with 1.2 M sodium chloride released ∼50% of histones H2A and H2B (Fig. 2 g, compare lanes 5 and 6) but only a small fraction of macroH2A1.2. Complete extraction of all the core histones, including H3 and H4, is brought about with 2 M sodium chloride (Hoffmann and Chalkley 1978). This treatment also resulted in extraction of the vast majority of macroH2A1.2 (Fig. 2 g, compare lanes 7 and 8). The small fraction of the histones not extracted are probably trapped within the pellet (Fig. 2 g, lane 8).
Taken together, these results indicate that the association of macroH2A1.2 with the NP-40 pellet has the biochemical properties associated with a core histone and is consistent with the bulk of the protein being chromatin associated.
The presence of an MCB before X-inactivation in undifferentiated cells raised the question of its localization relative to the X chromosome. Immuno-RNA FISH was performed on both XX and XY ES cells. In XX cells, early Xist transcripts were detected as two punctate dots, representing expression from both X chromosomes (Fig. 3, a and b, column 2) (Panning et al. 1997; Sheardown et al. 1997a). The MCBs observed lie in different focal planes with respect to Xist transcripts, clearly occupying different territories (Fig. 3 a, compare column 1 with 2). Identical results were obtained with another XX ES cell line, LF2 (Fig. 3 b, compare column 1 with 2). Thus the MCB in XX ES cells does not colocalize with Xist RNA expressed from either of the two active X chromosomes. Similarly, in XY ES cells, early Xist transcripts expressed from the single X chromosome were not associated with the MCB (Fig. 3c and Fig. d, columns 1 and 3). Although this does not rule out that the MCB is on the X chromosome but at a different locus, analysis of early differentiation stages indicate that this is not the case (see below).
Figure 3.
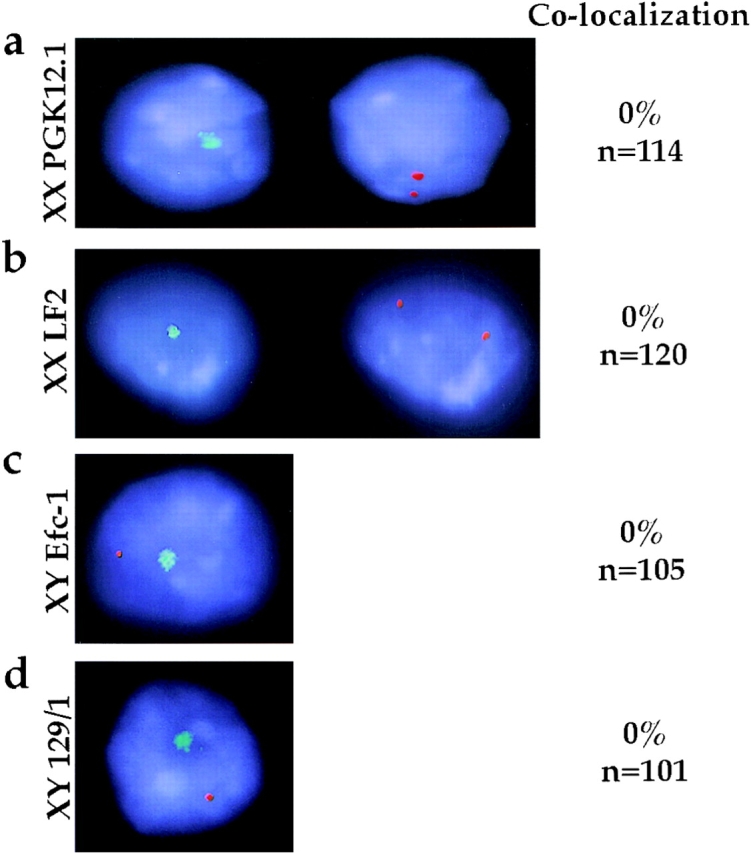
MCB and Xist RNA localization in undifferentiated ES cell lines. Immuno-RNA FISH detection of macroH2A1.2 protein (FITC, green) and Xist RNA (TR, red) in ES cells. DNA is stained with DAPI (blue). Representative examples of (a) XX PGK12.1, (b) XX LF2, (c) XY Efc-1, and (d) XY 129/1 cells are shown. Column 1, (a and b) macroH2A1.2, FITC signal, or (c and d) FITC signal combined with Xist RNA, TR signal. Column 2, (a and b) Xist RNA, TR signal in the same cells as column 1. Column 3, the percentage of cells with both RNA and protein signal detectable (n) exhibiting association of Xist RNA with the MCB.
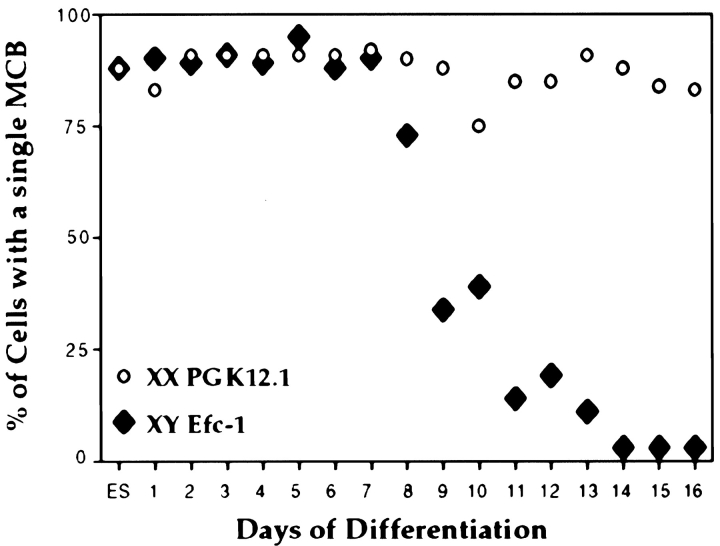
During Differentiation MCB Frequency Declines in XY but Not in XX Cells
Results presented above show that >80% of undifferentiated XY ES cells have a single MCB (Fig. 3). However, few or no XY somatic cells show an MCB (Costanzi and Pehrson 1998; data not shown). This suggests that the number of cells that contain an MCB declines during the process of differentiation. To determine whether this is the case, the number of XY Efc-1 cells displaying an MCB was scored over a 1–16-d differentiation time-course (Fig. 4). Early in differentiation (day 1–7), 88–95% of cells contained a single MCB. At progressively later timepoints (day 8–16), a clear decline in the number of cells containing an MCB was observed, reaching a minimum of 3% at day 14. This decline showed first order kinetics with a half-life of ∼1.7 d. For comparison, an identical analysis of XX ES cells was carried out. Fig. 4 shows that the percentages of cells containing an MCB (average 86%) remained unchanged during differentiation of PGK12.1 cells.
Figure 4.
MCBs in differentiating XX and XY ES cells. ES cells were stained with macroH2A1.2 before (day 0) and during differentiation (day 1–16). The number of cells with an MCB was quantified and results are given as percentage of total cells analyzed. XX ES cells (PGK12.1, circles) were compared with XY ES cells (Efc-1, diamonds). The MCB disappears in XY cells after day 7 of differentiation with a half-life of ∼1.7 d as determined by semi-log plot. More than 200 PGK12.1 cells and >300 Efc-1 cells were scored for each day of differentiation. The results represent the accumulated data from several replicate timecourse experiments. Similar results were obtained using other XX and XY ES cell lines (LF2 and 129/1; data not shown).
Diffuse nuclear macroH2A1.2 staining was observed throughout differentiation and is consistent with Western blot analysis showing that macroH2A1.2 protein is expressed not only in differentiating XX cells but also in differentiating XY cells (Fig. 2) (Pehrson et al. 1997).
Colocalization of Xist RNA and the MCB Occurs Late in XX ES Cell Differentiation
To address the key issue of when macroH2A1.2 accumulation on the Xi occurs, we carried out immuno-RNA FISH analysis on differentiating PGK12.1 XX ES cells. During day 1–3 Xist RNA undergoes a transition from a punctate (Fig. 5 a) to an accumulated (Fig. 5, b–i) signal. This reflects the switch from early transcripts to late transcripts that coat the inactivating X chromosome (Johnston et al. 1998; Lee et al. 1999). At early stages of differentiation, up to day 6, we observed no colocalization of the MCB with Xist RNA (Fig. 5, a and b). Because Xist RNA coats the entire Xi from day 3 onwards (Panning et al. 1997; Sheardown et al. 1997a), our results indicate that the MCB is not associated with any part of the Xi. Similarly, in XY ES cells differentiated for 3 d, no colocalization of punctate Xist RNA signal and the MCB was observed (n = 105, Efc-1).
Figure 5.
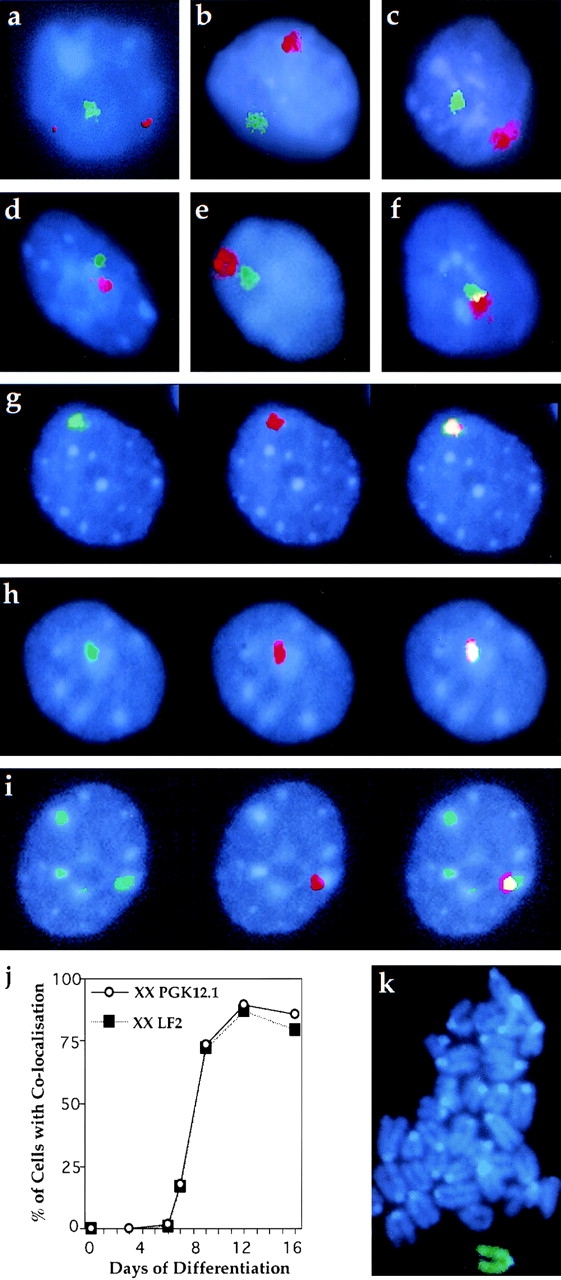
MCB and Xist RNA colocalize during XX ES cell differentiation. Immuno-RNA FISH detection of macroH2A1.2 protein (FITC, green) and Xist RNA (TR, red) in XX ES cells. Nuclear DNA is stained with DAPI (blue). PGK12.1 and LF2 cells were analyzed at day 3, 6, 7, 9, 12, and 16 of differentiation and representative images are shown. (a) At day 3, unstable Xist RNA is expressed from both alleles. It does not colocalize with the MCB. (b–i) Later in differentiation, cells display Xist RNA accumulation on the Xi. At day 6 (b) and 7 (c) Xist RNA and the MCB signals are clearly separate, but at day 7 they increasingly appear to be close together (d and e) or slightly overlapping (f). After day 7, Xist RNA and the MCB colocalize in most cells: (g) 9 d, (h) 12 d, (i) and 16 d. (j) Percentage of cells exhibiting MCB/Xist RNA colocalization during the differentiation time-course. Cell numbers scored for PGK12.1 (circles) and LF2 (squares), respectively, were: ES, n = 123/120; 3 d, n = 124; 6 d, n = 113/104; 7 d, n = 256/111; 9 d, n = 263/100; 12 d, n = 103/83; and 16 d, n = 172/125. (k) Metaphase spread of XX PGK12.1 cells differentiated for 10 d and stained for macroH2A1.2 (FITC, green).
Between day 7 and 9, we observed a striking change in localization patterns in XX ES cells. At day 7, Xist RNA and MCB signals were separate from one another in the majority of cells (Fig. 5 c). However, 48 h later colocalization was observed in most cell nuclei (Fig. 5 g). Within this transition period, cells frequently exhibited patterns of Xist RNA and MCB distribution with both signals close together (Fig. 5d and Fig. e) or showing a slight overlap (Fig. 5 f). We never observed nuclei with two MCBs during this period. At later timepoints (day 12, 16, and 22) the number of cells exhibiting colocalization did not change significantly (Fig. 5 h and data not shown). All the patterns described and their timing of appearance were highly reproducible. Importantly, a second XX cell line (LF2) gave identical results. A quantitative analysis of the data is presented in Fig. 5 j. This clearly demonstrates that colocalization occurs in most cells between day 7 and 9 of differentiation. To address whether this colocalization at interphase represents stable association of macroH2A1.2 with the chromatin of Xi, we analyzed metaphases of PGK12.1 cells differentiated for 10 d. We found that macroH2A1.2 is preferentially enriched on a single chromosome in each chromosome spread (Fig. 5 k).
From day 12 of differentiation onwards we observed an increasing number of nuclei with more than one MCB (Fig. 5 i). At day 16 up to eight MCBs could be detected in ∼50% of nuclei (PGK12.1 49%, n = 172; LF2 47%, n = 125). Xist RNA always colocalized to one of the MCBs. Less than 5% of nuclei had more than one Xist RNA signal, indicating that multiple MCBs are not attributable to polyploidy. Additional MCBs correlate with DAPI-dense staining regions in the nucleus.
Given the diversity of differentiating cells in an embryonic body (EB) (Keller 1995), the precise and reproducible formation of the MCB on the Xi between day 7 and 9 indicates that the process is independent of the developmental fate of individual cells. The occurrence of these changes coincides with the replating of EBs into tissue culture plates at day 7. To eliminate the possibility that these events are a consequence of the method of differentiation we tested two other differentiation procedures in which a replating step is avoided (Fig. 6 a). We found that the timing and extent of colocalization were identical, and thus independent of the method of differentiation used (Fig. 6b and Fig. c).
Discussion
We have shown that a single MCB is present before X-inactivation in both undifferentiated XX and XY ES cells; however, in neither case does it colocalize with Xist RNA. Only after the initiation and propagation of X-inactivation in differentiating XX ES cells do the MCB and Xist RNA colocalize. Concurrent with colocalization taking place in XX cells, the number of cells with an MCB declines significantly in differentiating XY cells. The timing of MCB and Xist RNA colocalization suggests that the accumulation of macroH2A1.2 on the Xi is unlikely to play a role in the primary events of X-inactivation.
MCB Appearance on the Xi and Progression of X-Inactivation
Previous analysis of the random X-inactivation process has shown that in differentiating XX ES cells the events leading to a fully inactivated X chromosome occur sequentially over a period of several days (Keohane et al. 1996, Keohane et al. 1998). The earliest detectable changes (expression of late Xist transcripts, late replication of the inactivating X, and a progressive increase in silencing X-linked genes) occur around day 2 of differentiation. These events are followed by overall histone H4 deacetylation on the Xi, which is essentially completed by day 7. We now have placed accumulation of macroH2A1.2 on the Xi in this pathway. We demonstrate that colocalization of the MCB with Xist RNA takes place after day 7 of differentiation in a highly synchronized wave. It occurs in all nuclei, is independent of the method of differentiation used or cell line studied, and is essentially completed within a 48-h period. Our analysis of H4 deacetylation (see Materials and Methods) and Xist expression during differentiation show that the timing of events in this study is identical to previous studies (Keohane et al. 1996, Keohane et al. 1998). Thus, MCB formation on Xi occurs subsequent to other steps in the X-inactivation process, with the exception of CpG island methylation. Therefore, we conclude that the MCB on the Xi cannot play a major role in initiation or propagation of random X-inactivation. Whether early events before day 7 are themselves necessary preconditions for MCB formation on the Xi is currently unknown.
Our data do not exclude a function for the Xi-MCB in maintenance of the inactive state, similar to CpG island methylation or global H4 underacetylation (Lock et al. 1987; Keohane et al. 1996). Consistent with such a possible role, an MCB is observed on the Xi in somatic cells and in a number of cell lines (Costanzi and Pehrson 1998; this study). However, the MCB is not absolutely required for maintenance. In C127 cells, no colocalization between Xist RNA and the MCB was observed, yet the X chromosome is underacetylated and coated with Xist RNA (Fig. 1 d). Furthermore, loss of the MCB did not affect the inactive state in embryonic fibroblasts (Csankovszki et al. 1999). Whether this simply reflects redundancy in maintenance mechanisms remains to be established. In this respect, it may resemble cases in which X-inactivation can be maintained in the absence of CpG island methylation in marsupials and in murine extra-embryonic lineages (Kratzer et al. 1983; Kaslow and Migeon 1987). These observations have previously led to the suggestion that there are a number of mechanisms that ensure maintenance of X-inactivation (Kaslow and Migeon 1987; Brown and Willard 1994). An alternative hypothesis is that localization of the MCB on the Xi is required only at a certain developmental time window, for example, to mark nucleosomes for subsequent event(s).
Possible Roles of the Non-Xi Associated MCB
The presence of a single MCB in undifferentiated XX ES cells before X-inactivation raises the possibility that it is involved in marking the X chromosome to be inactivated. Two results argue against this. Firstly, the MCB does not colocalize with Xist RNA in undifferentiated cells or in differentiating cells up to day 7. Xist RNA coats the entire Xi after day 3 of differentiation (Panning et al. 1997; Sheardown et al. 1997a), hence lack of colocalization in these cells indicates that the MCB is not on the Xi. Secondly, we show that undifferentiated XY cells also have an MCB, even though they never inactivate their single X chromosome.
In a previous study, macroH2A1.2 protein was readily detected in differentiated but not in undifferentiated ES cells (Pehrson et al. 1997). We have detected both macroH2A1.2 protein and an MCB in undifferentiated ES cells by using a significantly more sensitive antibody and a different method of extract preparation. The MCB was usually peripheral and did not correspond to DAPI-dense areas, suggesting that it is not associated with heterochromatin. Cell fractionation studies indicated that macroH2A1.2 in undifferentiated ES cells is not released by detergent (NP-40) lysis and is not in a pool of soluble cellular protein. However it is extracted by high concentrations of salt. In these respects, its behavior is similar to the core histones and is consistent with macroH2A1.2 protein being chromatin-associated in undifferentiated cells. However, the data do not rule out the possibility that the protein is associated with some other subcellular structure present in the NP-40 pellet. A more detailed analysis will now be required to understand the nature of the ES cell–MCB and whether its role at this stage of development is related to X-inactivation.
A single Xi-independent MCB was also observed in the mammary tumor cell line C127. It is not clear whether this structure, which does not correlate with obvious heterochromatic regions, is equivalent to that seen in undifferentiated and early differentiating ES cells. Presumably the MCB either never associated with the Xi or a previous association has been reversed. If the latter were true, it could reflect epigenetic remodelling processes associated with transformation. Investigation of other transformed and primary cell lines will be of interest in light of this discovery.
Possible Mechanisms for the Accumulation of MacroH2A1.2 on the Xi
A number of possible mechanisms may be postulated for the developmentally regulated formation of an MCB on the Xi. One mechanism involves the disassembly of the non-Xi MCB and import of newly synthesized macroH2A1.2 protein into the nucleus that is targeted to the Xi. Another possibility is that the non-Xi MCB disassembles, and macroH2A1.2 protein disperses and is reassembled at the Xi to form an MCB. A third possibility involves reorganization in the nucleus to bring the non-Xi MCB and the Xi in close proximity, allowing dynamic repositioning of macroH2A1.2 protein from the non-Xi MCB onto the Xi. Although we cannot yet distinguish between these and other possible mechanisms, we favor the latter model for the following reasons: The MCB is detectable throughout XX ES cell differentiation (Fig. 4). During the period when the MCB appears on the Xi, we never see more than one MCB per cell, arguing that the non-Xi MCB cannot be slowly disassembled concurrently with the Xi–MCB being formed. We do, however, observe an increase in the frequency of cells in which the non-Xi MCB is close to the Xi (Fig. 5). Furthermore, we detect cells in which Xist RNA and MCB signals are slightly overlapping. To investigate the repositioning hypothesis further, the precise dynamics of colocalization could be observed in living cells, for example, by using GFP-tagged macroH2A1.2.
Multiple MCBs in Gene Silencing
Evolutionary sequence conservation suggests that the function of macroH2A1.2 in X-inactivation may be an adaptation of a more general role of the protein in gene silencing (Pehrson and Fuji 1998). Thus, similar mechanisms and components might be used for silencing of autosomal genes and X-inactivation (Costanzi and Pehrson 1998). One example of an involvement of the MCB in silencing of autosomal genes could be the multiple MCBs observed in some cells from day 12 of differentiation onwards. They coincide with DAPI-dense regions of the nucleus, which may reflect incorporation of macroH2A1.2 into constitutive heterochromatin such as centromeres. Alternatively, these structures may represent clusters of loci with macroH2A1.2-enriched chromatin. Clustering of silenced genes with constitutive heterochromatin has been observed in recent studies on lymphocytes (Brown et al. 1997, Brown et al. 1999). Whether multiple MCBs are temporary structures or are related to the lineage or differentiated cell type is currently unknown. Interestingly, out of four murine somatic cell lines analyzed, only one displayed multiple MCBs (T37H; data not shown), whereas the other cell lines never had more than one MCB per diploid genome.
Acknowledgments
The authors are particularly grateful to Drs. S. Duthie and T. Nesterova for providing primary cell lines. We also thank Drs. C. Dingwall, A. Fisher, A. Merdes, P. Varga-Weisz, and members of the X-Inactivation Group for comments on the manuscript.
This work was supported by the Medical Research Council.
Footnotes
Abbreviations used in this paper: DAPI, 4,6-diamidino-2-phenylindole dihydrochloride; EB, embryonic body; ES, embryonic stem; FISH, fluorescence in situ hybridization; LDH, lactate dehydrogenase; LIF, leukemia inhibitory factor; MCB, macrochromatin body; TR, Texas red; Xa, active X chromosome; Xi, inactive X chromosome; Xist, X inactive specific transcript.
References
- Almeida F., Saffrich R., Ansorge W., Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M.L., Carr D.H. Correlation between sex chromatin and sex chromosomes. Acta Cytol. 1962;6:34–45. [PubMed] [Google Scholar]
- Belyaev N.D., Keohane A.M., Turner B.M. Histone H4 acetylation and replication timing in Chinese hamster chromosomes. Exp. Cell Res. 1996;225:277–285. doi: 10.1006/excr.1996.0177. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Connors B., Sobel R.E., Chinault A.C., Allis C.D. Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma. 1996;105:303–309. doi: 10.1007/BF02524648. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Willard H.F. The human X inactivation center is not required for maintenance of X chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F. The human XIST geneanalysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Baxter J., Graf D., Merkenschlager M., Fisher A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphaseevidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C., Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Panning B., Bates B., Pehrson J.R., Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Duthie S.M., Nesterova T.B., Formstone E.J., Keohane A.M., Turner B.M., Zakian S.M., Brockdorff N. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum. Mol. Gen. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- Eils R., Dietzel S., Bertin E., Schrock E., Speicher M.R., Ried T., Robert-Nicaud M., Cremer C., Cremer T. Three-dimensional reconstruction of painted human interphase chromosomesactive and inactive X chromosome territories have similar volumes but differ in shape and structure. J. Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciola L., Spada F., Calogero S., Längst G., Voit R., Grummt I., Bianchi M. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol. 1997;137:19–26. doi: 10.1083/jcb.137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J., Paolella G., Ramos C., Lamond A.I. Spatial organization of large scale chromatin domains in the nucleusa magnified view of single chromosome territories. J. Cell Biol. 1997;139:1579–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner R.L., Lyon M.F. X-chromosome inactivation studied by injection of a single cell into the mouse blastocyst. Nature. 1971;231:385–386. doi: 10.1038/231385a0. [DOI] [PubMed] [Google Scholar]
- Heard E., Clerc P., Avner P. X-chromosome inactivation in mammals. Annu. Rev. Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Herzing L.B., Romer J.T., Horn J.M., Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann P., Chalkley R. Procedures for minimizing protease activity during isolation of nuclei, chromatin, and the histones. Meth. Cell Biol. 1978;17:1–12. doi: 10.1016/s0091-679x(08)61130-9. [DOI] [PubMed] [Google Scholar]
- Jeppesen P., Turner B.M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene-expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Johnston C.M., Nesterova T.B., Formstone E.J., Newall A.E.T., Duthie S.M., Sheardown S.A., Brockdorff N. Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell. 1998;94:809–817. doi: 10.1016/s0092-8674(00)81739-0. [DOI] [PubMed] [Google Scholar]
- Kaslow D.C., Migeon B.R. DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupialsevidence for multistep maintenance of mammalian X dosage compensation. Proc. Natl. Acad. Sci. USA. 1987;84:6210–6214. doi: 10.1073/pnas.84.17.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G.M. In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Keohane A.M., O'Neill L.P., Belyaev N.D., Lavender J.S., Turner B.M. X-inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Keohane A.M., Lavender J.S., O'Neill L.P., Turner B.M. Histone acetylation and X inactivation. Dev. Genet. 1998;22:65–73. doi: 10.1002/(SICI)1520-6408(1998)22:1<65::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kratzer P.G., Chapman V.M., Lambert H., Evans R.E., Liskay R.M. Differences in the DNA of the inactive X chromosome of fetal and extraembryonic tissues of mice. Cell. 1983;33:37–42. doi: 10.1016/0092-8674(83)90332-x. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Strauss W.M., Dausman J.A., Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Davidow L.S., Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lock L.F., Takagi N., Martin G.R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- McMahon A., Fosten M., Monk M. X chromosome inactivation mosaicism in the three germ layers and the germ line of the mouse embryo. J. Exp. Morphol. 1983;46:53–64. [PubMed] [Google Scholar]
- Mise N., Goto Y., Nakajima N., Takagi N. Molecular cloning of antisense transcripts of the mouse Xist gene. Biochem. Biophys. Res. Commun. 1999;258:537–541. doi: 10.1006/bbrc.1999.0681. [DOI] [PubMed] [Google Scholar]
- Monk M. Methylation and the X chromosome. Bioessays. 1986;4:204–208. doi: 10.1002/bies.950040505. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Patel D., Kay G.F., Penny G.D., Brockdorff N., Sheardown S.A., Rastan S. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Panning B., Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Panning B., Dausman J., Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Pehrson J.R., Fuji R.N. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J.R., Costanzi C., Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Rack K.A., Chelly J., Gibbons R.J., Rider S., Benjamin D., Lafreniere R.G., Oscier D., Hendriks R.W., Craig I.W., Willard H.F. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet. 1994;3:1053–1059. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- Rastan S., Robertson E.J. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J. Embryol. Exp. Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- Riggs A.D., Pfeiffer G.P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Sheardown S.A., Duthie S.M., Johnston C.M., Newall A.E.T., Formstone E.J., Arkell R.M., Nesterova T.B., Alghisi G.-C., Rastan S., Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation Cell. 91 1997. 99 107a [DOI] [PubMed] [Google Scholar]
- Sheardown S.A., Newall A.E.T., Norris D.P., Rastan S., Brockdorff N. Regulatory elements in the minimal promoter region of the mouse Xist gene Gene. 203 1997. 159 168b [DOI] [PubMed] [Google Scholar]
- Takagi N. Differentiation of the X chromosome in early female mouse embryos. Exp. Cell Res. 1974;86:127–135. doi: 10.1016/0014-4827(74)90657-0. [DOI] [PubMed] [Google Scholar]