Abstract
The promyelocytic leukemia (PML) nuclear body (also referred to as ND10, POD, and Kr body) is involved in oncogenesis and viral infection. This subnuclear domain has been reported to be rich in RNA and a site of nascent RNA synthesis, implicating its direct involvement in the regulation of gene expression. We used an analytical transmission electron microscopic method to determine the structure and composition of PML nuclear bodies and the surrounding nucleoplasm. Electron spectroscopic imaging (ESI) demonstrates that the core of the PML nuclear body is a dense, protein-based structure, 250 nm in diameter, which does not contain detectable nucleic acid. Although PML nuclear bodies contain neither chromatin nor nascent RNA, newly synthesized RNA is associated with the periphery of the PML nuclear body, and is found within the chromatin-depleted region of the nucleoplasm immediately surrounding the core of the PML nuclear body. We further show that the RNA does not accumulate in the protein core of the structure. Our results dismiss the hypothesis that the PML nuclear body is a site of transcription, but support the model in which the PML nuclear body may contribute to the formation of a favorable nuclear environment for the expression of specific genes.
Keywords: transcription, nuclear structure, acetylated chromatin, correlative microscopy, electron spectroscopic imaging
Introduction
The promyelocytic leukemia (PML) nuclear body is a nuclear matrix-associated structure 250–500 nm in diameter that is present in the nucleus of most cell lines ( Ascoli and Maul 1991; De Graaf et al. 1992; Stuurman et al. 1992; Chang et al. 1995; Grande et al. 1996). There are approximately ten of these structures per nucleus, though this can vary considerably depending on the cell type, cell cycle, and other factors ( Ascoli and Maul 1991). The first biochemical component to be identified was the Sp100 nuclear matrix associated protein, which is an autoantigen in some patients with primary biliary cirrhosis ( Szostecki et al. 1990). This protein has been found to trans-activate a variety of promoters ( Guldner et al. 1992; Xie et al. 1993) and has an MHC class I-like domain at its NH2 terminus ( Sternsdorf et al. 1997a). The promyelocytic leukemia gene product, PML, is also found in PML nuclear bodies. In some acute promyelocytic leukemias, a t(15;17) chromosomal translocation creates a fusion protein of PML and the retinoic acid receptor alpha ( Borrow et al. 1990; de The et al. 1990, de The et al. 1991; Kakizuka et al. 1991; Dyck et al. 1994; Koken et al. 1994). PML protein may have transcription regulation activity ( Xie et al. 1993; Vallian et al. 1997, Vallian et al. 1998a, Vallian et al. 1998b), may suppress growth and transformation ( Le et al. 1996; Fagioli et al. 1998), and is implicated in apoptosis ( Wang et al. 1998). Int-6 is another protein that may localize to the PML nuclear body, reported in the context of studies of the HTLV-1 virus ( Desbois et al. 1996). A ubiquitin homologue, PIC1/SUMO-1 can be covalently bound to PML and Sp100 protein, and partially colocalizes with the PML nuclear body ( Sternsdorf et al. 1997b, Sternsdorf et al. 1999). This posttranslational modification has been proposed to be responsible for the targeting of proteins to the PML nuclear domain ( Muller et al. 1998; Duprez et al. 1999).
PML nuclear bodies are also a target of a variety of viruses, and are thought to be involved in coordinating the expression and replication of viral genomes from phylogenetically distinct families (for a review, see Sternsdorf et al. 1997a). A number of DNA viruses, such as SV40, Ad5, and HSV-1, begin replication in the vicinity of a few, but not all, of the PML bodies in a nucleus. Increased amounts of infectious virus in a cell do not result in an increase in the number of nuclear sites of viral replication. This implies that PML nuclear bodies may not be identical ( De Bruyn-Kops and Knipe 1994; Maul 1998). Viral protein expression does not appear to be necessary for targeting viral genomes to these replication sites ( Ishov and Maul 1996). Transcription of the viral genome also takes place at the periphery of the PML nuclear bodies, perhaps because they create a suitable environment ( Ishov et al. 1997; Maul 1998). Finally, many viruses disassemble or degrade PML nuclear bodies before proceeding to the late phase in infection cycles ( Doucas et al. 1996; Maul et al. 1996; Ishov et al. 1997; Maul 1998). This may have a disruptive effect on host nuclear function.
The cellular role of the PML nuclear body remains uncertain, despite its obvious links to oncogenesis and viral infection (for reviews, see Sternsdorf et al. 1997a; Slack and Gallagher 1999). PML nuclear bodies may be sites of storage of transcription factors, sites of transcription, or sites of RNA accumulation. The presence of RNA, by fluorescein-UTP microinjection and EDTA-regressive staining, has been observed within the structure of the PML nuclear body ( LaMorte et al. 1998), indicating a potential role in the transcription of some genes. The high enrichment of the transcriptional coactivator/histone acetyltransferase, CBP, is further suggestive of transcriptional activity within this domain ( LaMorte et al. 1998; Doucas et al. 1999). Consequently, it is important to understand the composition of these structures and the composition and organization of the surrounding nucleoplasm. In this study, we used both indirect immunofluorescence microscopy and a novel analytical transmission electron microscope method, electron spectroscopic imaging (ESI), to address the composition of PML nuclear bodies. PML nuclear bodies did not stain with antibodies recognizing highly acetylated histones nor those recognizing fluorine-substituted uridine (FU). Furthermore, phosphorus mapping by ESI rules out the presence of chromatin or RNA within these structures. However, nascent RNA can be detected at the periphery, indicating that the surroundings of the PML nuclear bodies are sites of transcriptional activity.
Materials and Methods
Immunodetection of Nascent RNA and PML Nuclear Bodies
SK-N-SH cells were cultured directly on glass coverslips under conditions recommended by the American Type Culture Collection. Pulses of FU were performed by addition of FU to a final concentration of 2 mM in the culture medium. Cells were fixed with 1.0% paraformaldehyde in PBS, pH 7.5, at room temperature for 5 min. Subsequently, cells were permeabilized in PBS containing 0.5% Triton X-100 for 5 min. Antibodies against halogenated UTP (anti-BrdU; Sigma Chemical Co., catalog no. B-2531) were used to label nascent RNA. Acetylated chromatin was visualized using an antibody recognizing diacetylated histones H3 (Upstate Biotechnology, Inc., catalog no. 06-599). PML nuclear bodies were visualized using an anti-CBP NH2-terminal antibody (Santa Cruz, catalog no. sc-369; or Upstate Biotechnology, Inc., catalog no. 06-297) or an anti-PML antibody (5E10; Stuurman et al. 1992). Cells were then incubated with secondary antibody, goat anti–rabbit Cy3 (Chemicon International, Inc.) and goat anti–mouse (Alexa 488, Cedarlane Labs; or Cy2, Nycomed Amersham, Inc.). After rinsing, the samples were mounted in 1 mg/ml paraphenylenediamine in PBS/90% glycerol, containing DNA-specific staining DAPI (4′,6-diamidino-2-phenylindole) at 1 μg/ml. Digital deconvolution to reduce haze in optical sections was performed using a 14-bit cooled CCD camera (Princeton Instruments) mounted on a Leica DMRE immunofluorescence microscope. VayTek Microtome digital deconvolution software was used to remove out of focus contributions, and image stacks were projected into one image plane using Scion Image software. False coloring and superimposition was done with Adobe Photoshop 5.0.
Correlative Microscopy and Electron Microscopy
Detailed descriptions of the EM procedure are presented elsewhere ( Bazett-Jones and Hendzel 1999). In brief, cells were grown on polypropylene caps, fixed with 1.0% paraformaldehyde in PBS, and labeled for immunofluorescence as described above. Cells were then refixed in 2% glutaraldehyde in PBS for 5 min, dehydrated in ethanol, starting at 30%, before embedding in Quetol 651 resin. We have found that this fixation procedure preserves fine structural detail in chromatin. Moreover, chromatin-based and protein-based fibers are routinely observed ( Bazett-Jones and Hendzel 1999; Bazett-Jones et al. 1999; Hendzel et al. 1999). To reveal the detail seen with this protocol, a section from the same block as that used for data in this paper was stained with uranyl acetate and imaged in bright-field ( Fig. 1A and Fig. B). Elastic images recorded at 0-eV energy loss reveal well preserved heterochromatin masses along the nuclear envelope. Fine structural detail is particularly evident in the high magnification, uranium-enhanced image (recorded at 120 eV) of the region indicated in Fig. 1 C.
Figure 1.
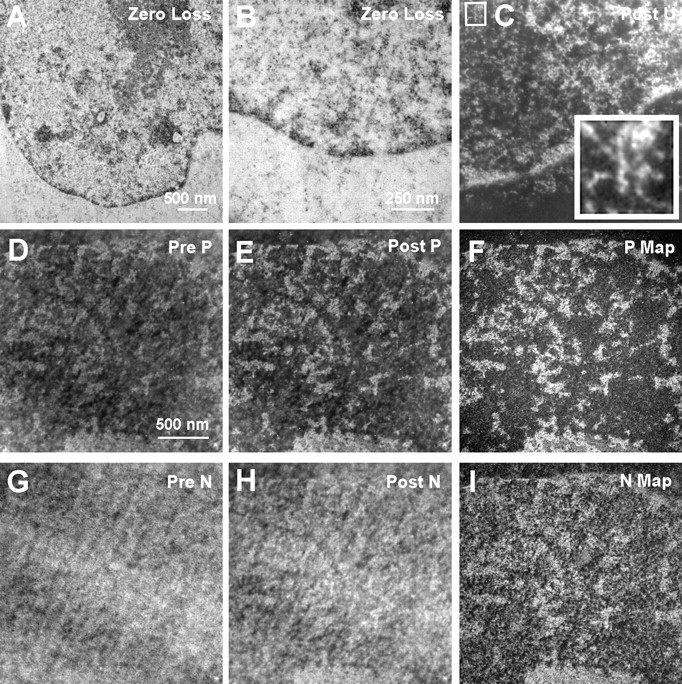
Energy-filtered electron micrographs of a 90-nm section stained with uranyl acetate (A–C). Elastic images, collected at 0-eV energy loss (A and B), show chromatin and other structures as black on a grey background. An energy-loss image recorded at a uranium edge (120 eV; C) shows chromatin and other structures as white on a black background. The inset (4×) shows a 10-nm-diam chromatin fiber with periodicity (vertical fiber at center of inset). Fibers of <50% of this thickness are also prevalent in this section. An unstained section from the same block used in A–C was used to map phosphorus and nitrogen. A phosphorus reference image (120 eV; D), a phosphorus-enhanced image (155eV; E), and a phosphorus map (F) are shown. Nitrogen reference (385 eV), enhanced (415 eV), and map are shown (G–I, respectively).
Sections of ∼30- and 90-nm thickness were obtained by ultramicrotomy with a diamond knife (Drukker), and were picked up onto finder grids. These lettered grids allow particular cells in the section, first imaged in the fluorescent microscope, to easily be found later in the electron microscope. Electron micrographs were obtained with a Gatan 14-bit slow scan cooled CCD detector on a Zeiss EM 902 transmission electron microscope equipped with an imaging spectrometer ( Bazett-Jones and Hendzel 1999). All images were corrected by dark current subtraction before being used for analysis. Energy filtered images recorded before an inner shell ionization edge do not contain element-specific information, but serve as a reference for the element-enhanced image recorded at an energy loss on, or just beyond, the inner shell ionization edge. In the case of phosphorus imaging, for example, an image recorded at an energy loss of 120 eV serves as a reference for the phosphorus-enhanced image recorded at 155 eV. A phosphorus map can be formed by subtracting the 120-eV image from the 155-eV image, after alignment to within a pixel, and normalization of the two images on a region that is known not to contain phosphorus; or, by dividing the 155-eV image by the 120-eV image after alignment of the two images. Similarly, a reference image for nitrogen is recorded at 385 eV and the nitrogen-enhanced image is recorded at an energy loss of 415 eV ( Bazett-Jones and Hendzel 1999). The sections are preirradiated before the images are recorded. Preirradiation stabilizes the section and produces a state of terminal mass loss. Therefore, no mass loss occurs during the recording of the complete set of reference and element-enhanced images. All images are recorded with nearly equal exposures measured over the background resin to insure equal signal counting statistics at the detector level for all images ( Bazett-Jones and Hendzel 1999; Bazett-Jones et al. 1999).
Examples of reference, element-enhanced, and element maps for phosphorus and nitrogen are shown in Fig. 1D-I. Blocks of chromatin on the nuclear envelope (top of image in Fig. 1, D–I), throughout the nucleoplasm, and at the surface of the nucleolus ( Fig. 2, Nu), are clearly seen in the phosphorus map. The granular component of the nucleolus has less contrast than the blocks of chromatin, but more contrast than the nucleoplasm or the cytoplasm. Blocks of chromatin can also be seen in nitrogen maps, but distinguishing chromatin from the granular component of the nucleolus, for example, is more difficult because nitrogen is present in both protein and nucleic acid. The granular component of the nucleolus has a higher protein/nucleic acid ratio than that of chromatin.
Figure 2.
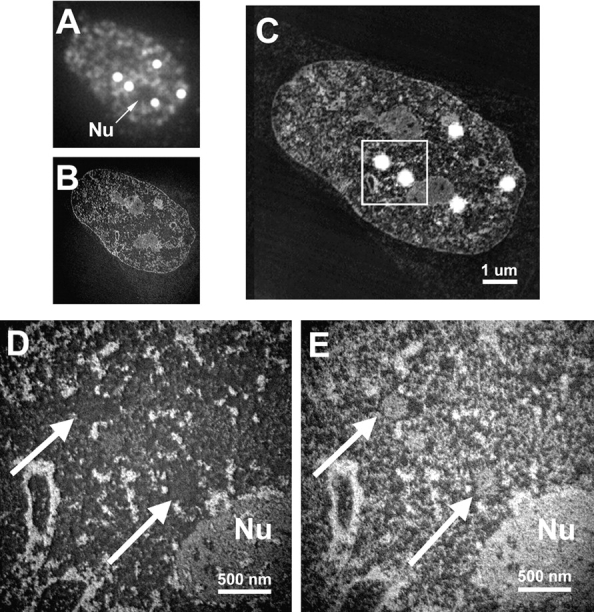
Correlative microscopy. Cells were labeled with anti–CBP-NT antibody, embedded, and sectioned. Sections are first imaged by immunofluorescence microscopy (A), followed by EM using ESI (B). The images are resized and rotated for proper alignment before being merged (C). Structures are represented as white objects on a black background. Immunofluorescence microscopy also serves to identify and locate structures of interest, so that these can be analyzed at the ultrastructural level by ESI, to map phosphorus (D) and nitrogen (E). The region indicated by the box in C is magnified in D and E. Nu, Nucleolus; arrows, the two PML nuclear bodies from C.
In this paper we used the two window method (one reference image and one post-edge, element-enhanced image) to obtain the elemental maps. We have shown that unstained, 30–40-nm thick sections do not contribute to detectable multiple scattering events or produce mass-density artifacts in the net images ( Bazett-Jones and Hendzel 1999; Bazett-Jones et al. 1999). It is always possible to find mass-dense structures, some of which contain no detectable element, and others with high levels of that element. Similarly, low mass-dense structures can be found that have either no detectable element or high amounts. Mass-density artifacts in the sections used in this study are only noticed where contaminating crystallites of salts are imaged.
A 600-μm condenser aperture and a 90-μm objective aperture were used. The energy-selecting slit aperture was set to correspond to 20 eV. To measure the relative amount of phosphorus or nitrogen in various morphologically defined structures in and around PML nuclear bodies, we used the technique called image EELS (electron energy-loss spectroscopy). Images of a selected field are collected at small steps (e.g., 10 eV) in the energy-loss spectrum, spanning, for example, the phosphorus L2,3 and nitrogen K edges. Masks are used to delineate objects of interest, and the mean signals under these masks are measured in every image. If a structure contains phosphorus, a steep increase in the energy-loss spectrum above 132 eV will emerge, along with a second steep increase beyond 150 eV, due to the delayed characteristic of this energy-loss event. The presence of nitrogen is more obvious than phosphorus, due to a sharper, less delayed event beginning at 385 eV (see Bazett-Jones et al. 1999). Measurements of signal intensities in the masked regions were made with ErgoVista 4.4 image analysis software.
Results
Structure of the PML Nuclear Body by Correlative Microscopy
It has been reported that PML nuclear bodies are sites of transcription and RNA accumulation ( LaMorte et al. 1998). We chose to test this model and to extend the model to higher resolution by direct visualization without using heavy atom contrast agents. We used ESI, a sensitive TEM method, to map protein-based and nucleic acid-based regions in and around PML nuclear bodies. One advantage of this technique is that comparisons of nitrogen and phosphorus maps provide the ability to directly distinguish protein from nucleic acids in situ. Second, it is possible to quantify mass contributions from both the protein and the nucleic acid components of a structure and to map the spatial relationships of protein- and nucleic acid-based structures within the complex nuclear environment ( Bazett-Jones and Hendzel 1999). Third, the heavy atom contrast agents, which limit resolution and stain different biochemical components in a nonuniform or predictable manner, can be avoided.
Positive identification of PML nuclear bodies amongst other nuclear structures with ESI alone is difficult. To identify these structures definitively, we first labeled cells with antibodies against the NH2 terminus of CBP, which we have shown to colocalize with PML in PML nuclear bodies in SK-N cells, by immunofluorescence microscopy. Indeed, in this cell line, CBP is as reliable a marker of PML nuclear bodies as for the PML protein itself. After embedding and thin sectioning, the same section can first be examined in the fluorescence microscope to identify the location of PML nuclear bodies in individual cells. The same section is then imaged at high resolution in the energy-filtering electron microscope ( Bazett-Jones et al. 1999). A thin section showing a nucleus imaged by immunofluorescence is shown in Fig. 2 A. Five PML nuclear bodies can easily be identified. Two are indicated ( Fig. 2A, Fig. D, and Fig. E, arrows) near the nucleolus ( Fig. 2 A, D, and E, Nu). The same section imaged at 155-eV energy loss is shown in Fig. 2 B at low magnification and superimposed with the immunofluorescence image in Fig. 2 C, to correlate the fluorescently labeled structures with the underlying ultrastructure. Two PML nuclear bodies are then detectable in the high magnification phosphorus ( Fig. 2 D) and nitrogen ( Fig. 2 E) maps. The cores of each of these nuclear bodies are well contrasted in the nitrogen map, whereas the phosphorus content is low. Notably, there is no evidence of small fibers or granules rich in phosphorus. Such signals are always observed when RNA or DNA is present, even as structures in the 2–10 nm range. These results indicate that the cores are mainly composed of protein.
The Core of the PML Nuclear Body Is a Protein-based Structure
Qualitative analysis of such images ( Fig. 2D and Fig. E) indicates that the core of the PML nuclear body is depleted in nucleic acids. Fig. 3 shows high magnification images of a PML nuclear body ( Fig. 3, left, phosphorous map, right, nitrogen map). The presence of RNA or DNA in this structure would likely resemble phosphorus-rich granules, as seen in the core of the interchromatin granule clusters and throughout the nucleoplasm ( Hendzel et al. 1998, Hendzel et al. 1999), or more extended fibrils, reflecting noncompacted nucleic acids ( Hendzel et al. 1999). Such structures are not apparent, even at high magnification ( Fig. 3). To extend the morphological analysis, we quantified the phosphorus and nitrogen content in different regions around the PML nuclear body ( Fig. 3B and Fig. D). The core of the PML nuclear body, a region corresponding to condensed chromatin, a background region in the nucleoplasm, and a background outside of the cell comprised of the embedding resin alone (not shown) were delineated with masks. The mean integrated signals from the energy-loss spectrum spanning the phosphorus L2,3 and nitrogen K edges were measured over the regions delineated by the masks ( Fig. 3B and Fig. D). No phosphorus or nitrogen can be detected in the embedding resin, indicated by the flat line in both spectra. The standard deviation in the phosphorus and nitrogen signal intensities was always <2% of the mean for the PML nuclear body, the chromatin, and the nucleoplasm. Error bars indicating these values would not show outside of the symbols on the graphs. The masks for the embedding resin, the chromatin, and the core of the PML nuclear body are approximately equal in area, whereas the mask for the nucleoplasm had to be smaller, ∼0.25 of the area of the others. However, the standard deviation of the means of numerous small regions of the nucleoplasm was also <0.5%, indicating that differences in sampling between nucleoplasm and nuclear structures do not account for the differences in element concentrations measured. The energy-loss spectra of regions in the nucleus are consistent with the presence of phosphorus, characterized by the strong delayed edge above 150 eV. The phosphorus content (155 eV) of the core of the PML nuclear body was ∼1.4% above the nucleoplasmic background, whereas the chromatin was 16% above the nucleoplasmic background, corresponding to a signal 11 times higher for the chromatin compared with the PML nuclear body. In contrast, the nitrogen signal (415 eV) for chromatin is 9% over the nucleoplasm and the PML body core is 10% over the nucleoplasmic background. The phosphorus signal in the nucleoplasmic background is likely derived predominantly from phosphorylated proteins. Because of a comparable phosphorus signal in the PML nuclear body core and the nucleoplasm, and the absence of morphologically recognizable phosphorus-rich complexes, we conclude that the core of this subnuclear structure does not contain nucleic acid (DNA and RNA) and is composed only of protein.
Figure 3.
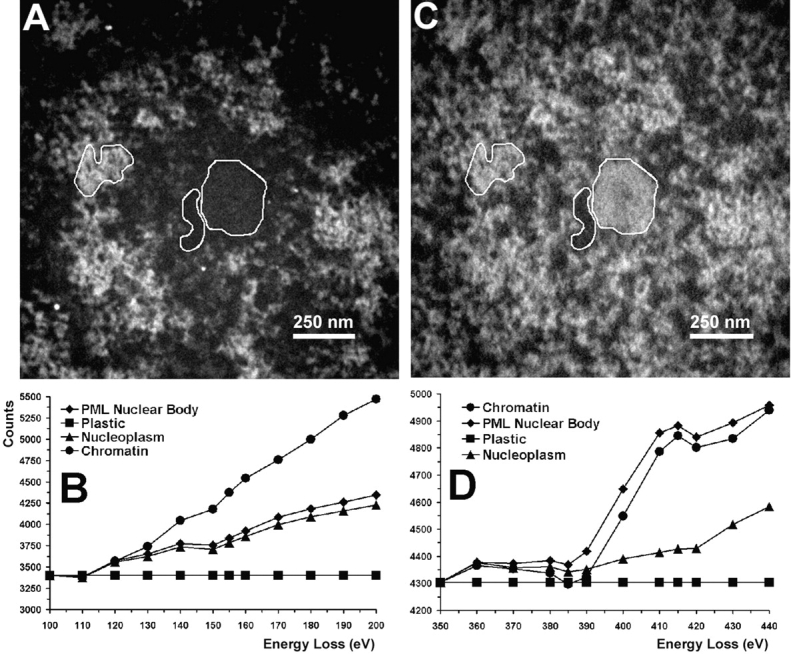
Quantification of phosphorus and nitrogen content of a PML nuclear body at high magnification. The three indicated areas were used for quantification of phosphorus and nitrogen. The left region corresponds to chromatin, the middle region to nucleoplasm, and the right region to the core of a PML nuclear body. A fourth region (not shown) corresponds to a region outside the cell, containing only the embedding resin. The signal over the resin was used to normalize all images and was adjusted to the same value for each image. Therefore, these represent background-stripped spectra on the basis of this normalization. The phosphorus map is shown in A and the nitrogen map in C. Energy-loss spectra of these regions spanning the phosphorus L2,3 edge (B) and the nitrogen K edge (D) are presented at 10-eV intervals. Structures are represented as white objects on a black background.
Phosphorus-rich Fibers Are Present at the Periphery of the Core of the PML Nuclear Body
Although the use of thin sections (30 nm) is necessary for quantitative elemental analysis by ESI (electron accelerating voltage of 80 kV), we have found that thicker sections stained with uranyl acetate still contain qualitatively useful elemental information (Boisvert, F.-M., preliminary observations). Thicker sections allow one to follow fibers that would otherwise rapidly leave the plane of the section if they are not parallel to the section. Fig. 4 shows consecutive 90-nm sections through a PML nuclear body. The center of the PML body core is characterized by a hole in the nitrogen map ( Fig. 4, Fig. 1, arrow). Blocks of condensed chromatin surrounding the core are evident in the phosphorus-enhanced maps ( Fig. 4, Fig. 1, Fig. 2, arrows; Fig. 3 A). Extended thin fibers measuring as little as 2 nm in diameter are consistently visualized at the immediate periphery of the core of the PML nuclear body when imaged with high resolution ( Fig. 5). Nascent RNA is the most abundant subnuclear structure that has been identified in an extended conformation ( Malatesta et al. 1994), and it is likely, therefore, that these phosphorus-rich fibers are RNA-based.
Figure 4.
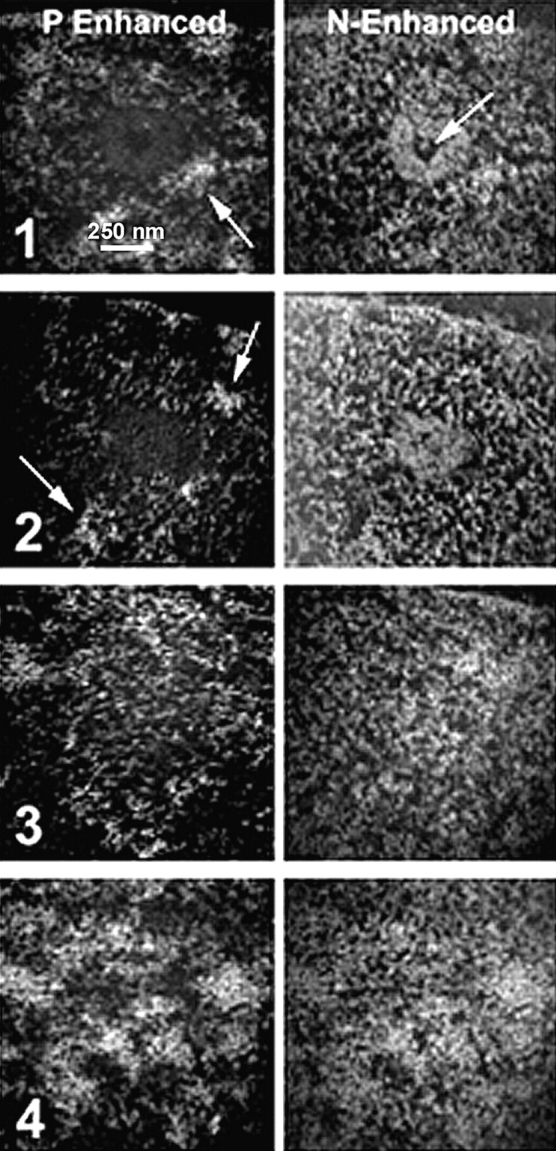
Serial sections of 90-nm thickness of a PML nuclear body stained with uranyl acetate. Images in the left column are phosphorus-enhanced, recorded at 155 eV, and those in the right column are nitrogen-enhanced, recorded at 415 eV. The middle of the PML nuclear body is shown in section 1. The protein core is visible in the nitrogen-enhanced image, and a hole on the core is indicated with the arrow in the nitrogen-enhanced image. Arrows in the phosphorus-enhanced images indicate blocks of condensed chromatin. Structures are represented as white objects on a black background. The next section (2) still shows the protein core, whereas section 3 is now out of the core of the nuclear body. The last section (4) shows the chromatin closing back over the structure.
Figure 5.
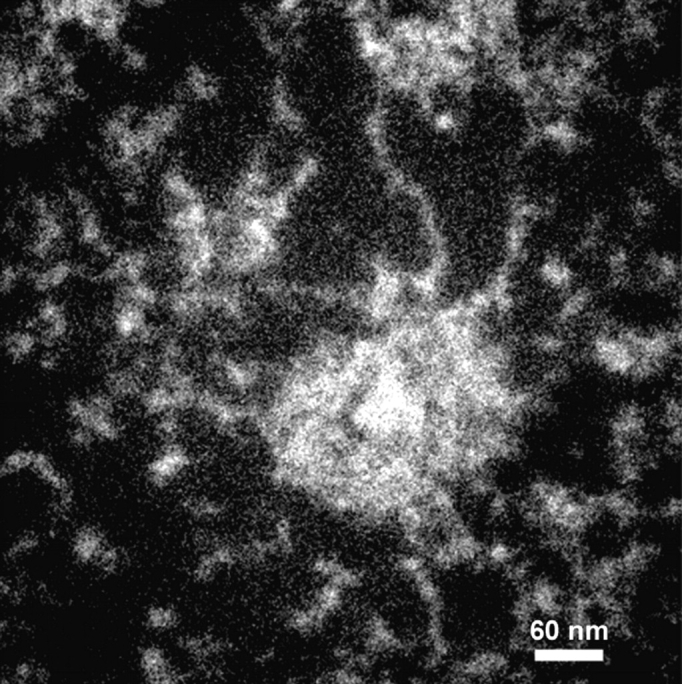
Thick section of a PML nuclear body, stained with uranyl acetate, recorded at 415 eV. Extended fibers are observed just outside the core of the PML nuclear body. Structures are represented as white objects on a black background. These fibers are ∼2–4 nm in diameter, and are also phosphorus-rich (not shown).
Localization of Nascent RNA at the Periphery of the PML Nuclear Bodies
Our ESI analysis indicates that the core of the PML nuclear body is composed mainly of protein, whereas the periphery contains DNA and/or RNA. In a previous report ( LaMorte et al. 1998), the entire PML body was reported to contain RNA, based on fluorescently labeled uridine in permeabilized cells. To determine whether newly transcribed RNA is present in the vicinity of PML bodies, we chose to label unperturbed cells with FU. Incorporation of FU in nascent RNA is rapid and specific and does not require a cell permeabilizing step. Signal visualized with antibodies against halogenated nucleotides can be detected throughout the nucleoplasm after two-minute pulses, with increasing signal over time ( Fig. 6C, Fig. H, and Fig. M). Strong, but less punctate, nucleolar labeling can also be seen after ten minutes, concordant with the high level of transcription therein. The diffuse nature of this signal causes an apparent attenuation of the signal relative to the punctuate foci found outside of the nucleolus in deconvolved optical sections. Before deconvolution, the nucleolar signal appears dominant. This nucleolar signal indicates that the penetration of the antibody and the FU is not inhibited by dense, compartmentalized structures, such as the nucleolus. To determine the relationship between PML nuclear bodies and RNA synthesis and accumulation, we colabeled cells with anti-CBP NH2-terminal antibodies ( Fig. 6B, Fig. G, and Fig. L, red) and anti-FU antibodies ( Fig. 6C, Fig. H, and Fig. M, green) after 2-, 10-, and 60-min pulses of FU. After only two minutes, most PML nuclear bodies have FU-incorporated RNA on their peripheries. This FU incorporation appears as a small number of foci (between 1 and 3) on the periphery of a PML nuclear body. When pulses were extended to longer times, RNA is still seen on the periphery in a small number of foci, and does not accumulate inside the structure as visualized by CBP labeling ( Fig. 6E, Fig. J, and Fig. O). The FU labeling is consistent with the ESI data, leading us to conclude that the core of the PML nuclear body is a protein-dense structure where RNA synthesis and RNA accumulation do not occur. Transcription does appear to occur near the periphery of PML nuclear bodies, but not within.
Figure 6.
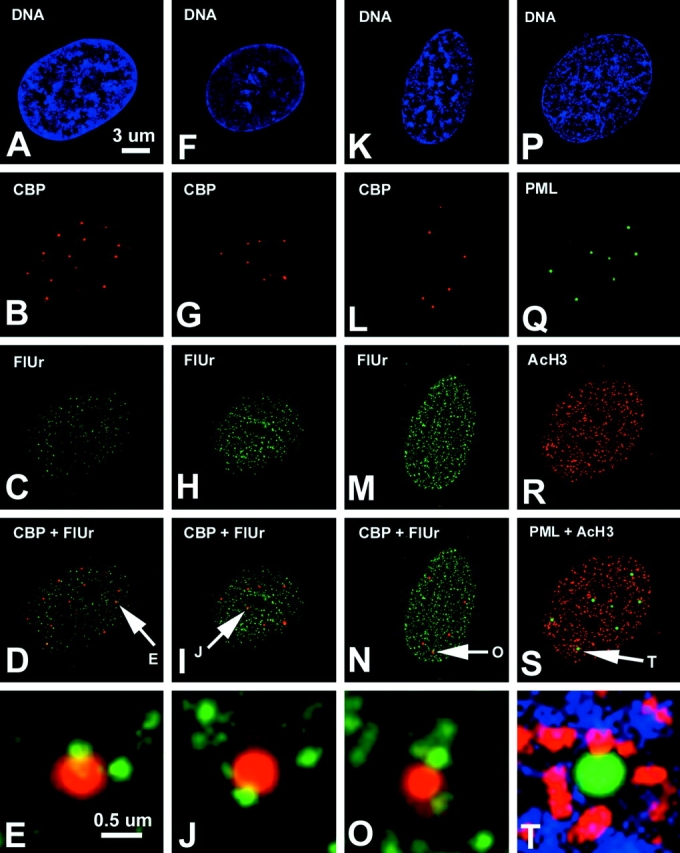
Digital deconvolution microscopy of SK-N cells pulsed with FU for 2 (A–E), 10 (F–J), or 60 (K–O) min. FU was labeled with an anti-BrdU antibody (green, C, H, and M) and anti–CBP-NT antibody (red, B, G, and L). Merged images are shown in D, I, and N, and a high magnification of one PML nuclear body for each time course is displayed in E, J, and O. DNA stained with DAPI shows the nucleus of each cell (A, F, K, and P). Some cells were labeled with an antibody against the highest acetylated form of histone H3 (green, R) to reveal transcriptionally active/competent chromatin. PML nuclear bodies were detected with an antibody against PML protein (5E10) shown in Q. Images Q and R are merged to form S, and the indicated PML nuclear body is magnified in T.
Highly Acetylated Chromatin Surrounds PML Nuclear Bodies, but Is Not Found Within
Phosphorus mapping by ESI and FU incorporation detected by fluorescence microscopy indicated that there is no nucleic acid in PML nuclear bodies. Nevertheless, we wished to strengthen our conclusion that there is no chromatin within the PML nuclear body, but also to determine whether transcription occurs on the periphery. For this experiment, we imaged PML nuclear bodies by deconvolution of optical sections, using an antibody recognizing the transcription-associated, highest acetylated species of histone H3 ( Fig. 6Q and Fig. R; Boggs et al. 1996). As expected from the FU incorporation experiments and ESI analysis, the cores of PML nuclear bodies do not show any evidence for the presence of highly acetylated euchromatin. All of the PML nuclear bodies, however, are within one diameter (250 nm) of a block of highly acetylated chromatin ( Fig. 6 S). Moreover, some PML nuclear bodies are associated with multiple blocks of acetylated chromatin ( Fig. 6 T; also see blocks of chromatin surrounding the PML nuclear body in the phosphorus map in Fig. 3 A). Further studies are required to determine whether transcriptionally active chromatin is indeed functionally associated with PML nuclear bodies, or whether the association is a random event. For example, it would be necessary to determine whether specific gene sequences associate with the periphery of PML nuclear bodies. From the ESI data and from the absence of highly acetylated chromatin or DAPI-stained chromatin within the core of the PML nuclear body, we conclude that the core of the structure is not a site of transcriptional activity.
Discussion
There has been an increasing interest in the PML nuclear bodies in the past few years because of their involvement in viral infection ( Sternsdorf et al. 1997b; Maul 1998) and their disruption and disappearance in acute promyelocytic leukemia ( Borrow et al. 1990; de The et al. 1990, de The et al. 1991; Kakizuka et al. 1991; Dyck et al. 1994; Koken et al. 1994). Whether this structure serves only as a storage site for regulatory factors, functions as a site for transcription by creating a suitable environment, or plays some other role will only be determined as its structure and composition are characterized. In this study, we have addressed several questions about the structure, organization, and function of the PML nuclear body.
First, we have shown that the core of the PML nuclear body is composed entirely, or almost entirely, of protein. There is no detectable nucleic acid in the 250-nm-diam core of the structure. This core is not a uniform sphere of protein, but rather has channels and pores throughout. Serial sections ( Fig. 4) indicate that the mass density of the very center is much lower than the outer regions of the core. Many proteins have been shown to localize to PML nuclear bodies, such as the transcription regulatory factor PML protein, the transcriptional coactivator CBP, and other proteins, such as Sp100. It is likely that such components of the PML nuclear body are localized in this core structure, and the core represents a specialized component of the karyoskeleton ( Hendzel et al. 1999).
The second major finding in this paper is that the protein-based core is surrounded by blocks of chromatin that are condensed to levels beyond the 30-nm fiber. In cross-sections, phosphorus maps illustrate a discontinuous ring of chromatin around the protein-based core, and by serial sectioning, phosphorus maps demonstrate that the blocks of chromatin completely surround the core. In future experiments, we will investigate whether some of this chromatin is participating in transcriptional activity.
The third major finding relates to the PML body as a site for RNA synthesis. Both ESI and long incubation times in the presence of fluorescently detected label reveal that the core of the structure always remains free of RNA. Nevertheless, phosphorus-rich fibers that are morphologically more consistent with RNA than chromatin are observed in the space between the protein core and the blocks of chromatin. It remains to be shown whether the apparent association of highly acetylated chromatin, FU incorporation, and the RNA fibers reflect a functional relationship to the PML nuclear body or reflect a random association. It might be expected that the high levels of CBP, located in PML nuclear bodies, would contribute to a favorable environment for chromatin remodeling and transcriptional activity. The presence of nascent RNA at the periphery of this structure is concordant with the observation by Ishov et al. 1997, who localized viral transcripts next to this nuclear body.
Our conclusions differ dramatically from those of LaMorte and coworkers ( LaMorte et al. 1998). These authors used EDTA-regressive staining technique, which preferentially labels RNA ( Bernhard 1969), and fluorescein-UTP incorporation in situ nuclear run-on experiments to conclude that very high concentrations of RNA are localized within the structure. Both experiments have limitations. The specificity of the EDTA-regressive staining procedure is not absolute, and, in the case of structures of unknown composition, cannot be interpreted to be RNA-based solely on the presence of staining after the EDTA chelation of uranium ions from chromatin ( Bernhard 1969). We report here that the core of the PML nuclear body contains only nucleoplasmic background levels of phosphorus and no discernible phosphorus-rich fibers. This holds for all PML nuclear bodies observed and does not characterize a small subset of these structures ( LaMorte et al. 1998). As a positive control for our observation that the core contains little or no RNA, subnuclear domains that are known to contain high amounts of RNA do have high phosphorus signals when imaged by ESI. These structures include 30-nm chromatin fibers, the nucleolus ( Fig. 2 D), interchromatin granule clusters ( Hendzel et al. 1998), or the Sam 68 nuclear bodies ( Chen et al. 1999). Because the sensitivity of these procedures has been shown to be very high ( Bazett-Jones and Hendzel 1999; Bazett-Jones et al. 1999; Hendzel et al. 1999), our results rule out the possibility that the intense staining of these structures using the EDTA-regressive staining method is the result of an abundance of RNA within the structure. Similarly, the nuclear run-on experiment, which shows labeling of the PML nuclear bodies, has a greater potential for experimental artifacts than does the direct incorporation into RNA through addition of FU into the tissue culture medium.
We present a number of independent methods that demonstrate an absence of: bulk chromatin; acetylated chromatin; newly synthesized RNA; or phosphorus-rich nucleic acids; in the core of the PML nuclear body. Therefore, we conclude that it is highly unlikely that the PML nuclear body represents a higher-order nuclear structure that nucleates or aggregates around specific clusters of transcriptionally active genes or even single, highly active genes. Instead, the presence of transcription regulatory factors within the PML nuclear body and the blocks of chromatin and nascent RNA in the immediate vicinity of the PML nuclear body, raise the importance of further experimentation to test whether these structures are responsible for creating or establishing a transcriptional domain. A nuclear-neighbor hypothesis for regulation of nuclear events has been presented to explain a number of similar relationships involving subnuclear domains ( Schul et al. 1998).
Subnuclear compartments that function to concentrate regulatory factors and complexes may be important in modulating gene expression ( Schul et al. 1998). The interchromatin granule cluster (IGC), for example, is able to concentrate transcriptional coactivators in the protein-rich core, and is surrounded by transcriptionally active chromatin ( Hendzel et al. 1998). Indeed, the peripheries of IGCs are sites of transcription of particular genes ( Smith et al. 1999). PML nuclear bodies, which, incidentally, are generally associated with IGCs ( Ishov et al. 1997), may also serve to concentrate regulatory factors that service transcriptional events on their surfaces. Genes that are implicated in requiring PML nuclear bodies for expression are early viral genes ( Guldner et al. 1992; Chelbi-Alix et al. 1995; Lavau et al. 1995; Grotzinger et al. 1996; Ishov et al. 1997) and, possibly, genes important for immune function, including MHC class I and II ( Kretsovali et al. 1998; Zheng et al. 1998; Fontes et al. 1999).
Acknowledgments
We thank Mr. Manfred Herfort for technical assistance with the EM. The 5E10 antibody was a generous gift from Dr. R. van Driel.
This work was funded by operating grants from the Cancer Research Society, Inc., and the Medical Research Council of Canada. The Microscopy and Imaging Facility, University of Calgary, is supported by a maintenance grant from the Medical Research Council of Canada.
Footnotes
Michael J. Hendzel's current address is Cross Cancer Institute, University of Alberta, Edmonton, Alberta, Canada T6G 1Z2.
Abbreviations used in this paper: CBP, transcriptional coactivator/histone acetyltransferase; DAPI, 4′,6-diamidino-2-phenylindole; ESI, electron spectroscopic imaging; FU, fluorine-substituted uridine; IGC, interchromatin granule cluster; PML, promyelocytic leukemia.
References
- Ascoli C.A., Maul G.G. Identification of a novel nuclear domain. J. Cell Biol. 1991;112:785–795 . doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Hendzel M.J. Electron spectroscopic imaging of chromatin. Companion Meth. Enzymol. 1999;17:188–200 . doi: 10.1006/meth.1998.0729. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Hendzel M.J., Kruhlak M.J. Stoichiometric analysis of protein- and nucleic acid-based structures in the cell nucleus. Micron. 1999;30:151–157 . doi: 10.1016/s0968-4328(99)00019-0. [DOI] [PubMed] [Google Scholar]
- Bernhard W. A new staining procedure for electron microscopical cytology. J. Ultrastruct. Res. 1969;27:250–265 . doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Connors B., Sobel R.E., Chinaud A.C., Allis C.D. Reduced levels of histone H3 acetylation on the inactive X chromosomes as shown by histone acetylation. Chromosoma. 1996;105:41–49 . doi: 10.1007/BF02524648. [DOI] [PubMed] [Google Scholar]
- Borrow J., Goddard A.D., Sheer D., Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580 . doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- Chang K.S., Fan Y.-H., Andreeff M., Liu J., Mu Z.-M. The PML gene encodes a phosphoprotein associated with the nuclear matrix. Blood. 1995;85:3646–3653 . [PubMed] [Google Scholar]
- Chelbi-Alix M.K., Pelicano L., Quignon F., Koken M.H.M., Venturini L., Stadler M., Pavlovic J., Degos L., de The H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033 . [PubMed] [Google Scholar]
- Chen T., Boisvert F.-M., Bazett-Jones D.P., Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell. 1999;10:3015–3033 . doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyn-Kops A., Knipe D.M. Preexisting nuclear architecture defines the intranuclear location of herpes virus DNA replication structures. J. Virol. 1994;68:3512–3526 . doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graaf A., Humbel B.M., Stuurman N., van Bergen en Henegouwen P.M.P., Verkleij A.J. Three-dimensional immunogold labeling of nuclear matrix proteins in permeabilized cells. Cell Biol. Int. Rep. 1992;16:827–836 . doi: 10.1016/s0309-1651(05)80025-6. [DOI] [PubMed] [Google Scholar]
- de The H., Chomienne C., Lanotte M., Degos L., Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–561 . doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- de The H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684 . doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- Desbois C., Rousset R., Bantignies F., Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I tax oncoprotein. Science. 1996;273:951–953 . doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- Doucas V., Ishov A.M., Romo A., Juguilon H., Weitzman M.D., Evans R.M., Maul G.G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207 . doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- Doucas V., Tini M., Egan D.A., Evans R.M. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA. 1999;96:2627–2632 . doi: 10.1073/pnas.96.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez E., Saurin A.J., Desterro J.M., Lallemand-Breitenbach V., Howe K., Boddy M.N., Solomon E., de The H., Hay R.T., Freemont P.S. SUMO-1 modification of the acute promyelocytic leukaemia protein PMLimplications for nuclear localisation. J. Cell Sci. 1999;112:381–393 . doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Dyck J.A., Maul G.G., Miller W.H., Jr., Chen J.D., Kakizuka A., Evans R.M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343 . doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Fagioli M., Alcalay M., Tomassoni L., Ferrucci P.F., Mencarelli A., Riganelli D., Grignani F., Pozzan T., Nicoletti I., Grignani F., Pelicci P.G. Cooperation between the RING + B1-B2 and coiled-coil domains of PML is necessary for its effects on cell survival. Oncogene. 1998;16:2905–2913 . doi: 10.1038/sj.onc.1201811. [DOI] [PubMed] [Google Scholar]
- Fontes J.D., Kanazawa S., Jean D., Peterlin B.M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol. Cell. Biol. 1999;19:941–947 . doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande M.A., van der Kraan I., van Steensel B., Schul W., de The H., van der Voort H.T.M., de Jong L., van Driel R. PML-containing nuclear bodiestheir spatial distribution in relation to other nuclear components. J. Cell. Biochem. 1996;63:280–291 . doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Grotzinger T., Sternsdorf T., Jensen K., Will H. Interferon-modulated expression of genes encoding the nuclear-dot–associated proteins Sp100 and promyelocytic leukemia protein (PML) Eur. J. Biochem. 1996;238:554–560 . doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- Guldner H.H., Szostecki C., Grotzinger T., Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol. 1992;149:4067–4073 . [PubMed] [Google Scholar]
- Hendzel M.J., Kruhlak M.J., Bazett-Jones D.P. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol. Biol. Cell. 1998;9:2491–2507 . doi: 10.1091/mbc.9.9.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Boisvert F.-M., Bazett-Jones D.P. Direct visualization of a protein nuclear architecture. Mol. Biol. Cell. 1999;10:2051–2062 . doi: 10.1091/mbc.10.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A.M., Maul G.G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 1996;134:815–826 . doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A.M., Stenberg R.M., Maul G.G. Human cytomegalovirus immediate early interaction with host nuclear structuresdefinition of an immediate transcript environment. J. Cell Biol. 1997;138:5–16 . doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka A., Miller W.H., Jr., Umesono K., Warrell R.P., Jr., Frankel S.R., Murty V.V., Dmitrovsky E., Evans R.M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674 . doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- Koken M.H.M., Puvion-Dutilleul F., Guillemin M.C., Viron A., Linares-Cruz G., Stuurman N., de Jong L., Szostecki C., Calvo F., Chomienne C. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:1073–1083 . doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsovali A., Agalioti T., Spilianakis C., Tzortzakaki E., Merika M., Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol. Cell. Biol. 1998;18:6777–6783 . doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMorte V.J., Dyck J.A., Ochs R.L., Evans R.M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA. 1998;95:4991–4996 . doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Marchio A., Fagioli M., Jansen J., Falini B., Lebon P., Grosveld F., Pandolfi P.P., Pelicci P.G., Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876 . [PubMed] [Google Scholar]
- Le X.F., Yang P., Chang K.S. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene, PML. J. Biol. Chem. 1996;271:130–135 . doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- Malatesta M., Zancanaro C., Martin T.E., Chan E.K., Amalric F., Luhrmann R., Vogel P., Fakan S. Is the coiled body involved in nucleolar functions? Exp. Cell Res. 1994;211:415–419 . doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- Maul G.G. Nuclear domain 10, the site of DNA virus transcription and replication. BioEssays. 1998;20:660–667 . doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Ishov A.M., Everett R.D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75 . doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- Muller S., Matunis M.J., Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:61–70 . doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W., de Jong L., van Driel R. Nuclear neighboursthe spatial and functional organization of genes and nuclear domains. J. Cell Biochem. 1998;70:159–171 . doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Slack J.L., Gallagher R.E. The molecular biology of acute promyelocytic leukemia. Cancer Treat. Res. 1999;99:75–124 . doi: 10.1007/978-0-585-38571-6_4. [DOI] [PubMed] [Google Scholar]
- Smith K.P., Moen P.T., Wydner K.L., Coleman J.R., Lawrence J.B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 1999;144:617–629 . doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T., Grotzinger T., Jensen K., Will H. Nuclear dotsactors on many stages Immunobiol. 198 1997. 307 331a [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1 J. Cell Biol 139 1997. 1621 1634b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Reich B., Will H. The nuclear dot protein Sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 1999;274:12555–12566 . doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- Stuurman N., de Graaf A., Floore A., Josso A., Humbel B., de Jong L., van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 1992;101:773–784 . doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Szostecki C., Guldner H.H., Netter H.J., Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol. 1990;145:4338–4347 . [PubMed] [Google Scholar]
- Vallian S., Gaken J.A., Trayner I.D., Gingold E.B., Kouzarides T., Chang K.S., Farzaneh F. Transcriptional repression by the promyelocytic leukemia protein, PML. Exp. Cell. Res. 1997;237:371–382 . doi: 10.1006/excr.1997.3801. [DOI] [PubMed] [Google Scholar]
- Vallian S., Chin K.V., Chang K.S. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter Mol. Cell. Biol 18 1998. 7147 7156a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallian S., Gaken J.A., Gingold E.B., Kouzarides T., Chang K.S., Farzaneh F. Modulation of Fos-mediated AP-1 transcription by the promyelocytic leukemia protein Oncogene. 16 1998. 2843 2853b [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., Pandolfi P.P. PML is essential for multiple apoptotic pathways. Nat. Genet. 1998;20:266–272 . doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- Xie K., Lambie E.J., Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol. Cell. Biol. 1993;13:6170–6179 . doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Guo Y., Niu Q., Levy D.E., Dyck J.A., Lu S., Sheiman L.A., Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]


