Abstract
The major target tissues for Epstein-Barr virus (EBV) infection are B lymphocytes and epithelial cells of the oropharyngeal zone. The product of the EBV BZLF1 early gene, EB1, a member of the basic leucine-zipper family of transcription factors, interacts with both viral and cellular promoters and transcription factors, modulating the reactivation of latent EBV infection. Here, we characterize a novel cellular protein interacting with the basic domains of EB1 and c-Jun, and competing of their binding to the AP1 consensus site. The transcript is present in a wide variety of human adult, fetal, and tumor tissues, and the protein is detected in the nuclei throughout the human epidermis and as either grainy or punctuate nuclear staining in the cultured keratinocytes. The overexpression of tagged cDNA constructs in keratinocytes revealed that the NH2 terminus is essential for the nuclear localization, while the central domain is responsible for the interaction with EB1 and for the phenotype of transfected keratinocytes similar to terminal differentiation. The gene was identified in tail-to-tail orientation with the periplakin gene (PPL) in human chromosome 16p13.3 and in a syntenic region in mouse chromosome 16. We designated this novel ubiquitously expressed nuclear protein as ubinuclein and the corresponding gene as UBN1.
Keywords: Epstein-Barr virus, epidermis, protein–protein interaction, keratinocyte differentiation, chromosome 16
Introduction
Epstein-Barr virus (EBV) is a human herpes virus that persists in latency for the lifetime of the infected host. Transmission by oral contact results in infection of the oral and/or nasopharyngeal epithelium, and subsequent infection of B lymphocytes in the adjacent lymphoid tissue ensues. EBV replication is highly dependent on the state of cell differentiation. In the oral epithelium, EBV replication is observed primarily in the upper spinous layer that consists of the terminally differentiated nondividing cells. EBV is also associated with human malignancies, such as Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and B and T cell lymphomas in immunocompromized individuals (for reference, see Rickinson and Kieff 1996). EBV-infected B lymphocytes from the peripheral blood or in vitro infected B lymphocytes can proliferate continuously in culture. While much investigation has sought to elucidate EBV's role in the immortalization and transformation of B lymphocytes, little is known about the molecular mechanisms underlying EBV's lytic infection of epithelial cells. Biological data are accumulating on a repertoire of EBV genes and molecular mechanisms by which the productive cycle is reactivated in latently infected epithelial cells, as well as the mechanisms by which reactivation can be suppressed.
During epithelial differentiation, major changes take place in the gene expression and morphology of the differentiating cells. The undifferentiated cells of the basal cell layer are responsible for the cell-basement membrane attachment through the adhesion complexes, called hemidesmosomes. When the cells lose their contact to the basement membrane, they start to travel towards the upper layers of the epithelium, changing their appearance as typical for the terminally differentiated cells. The cell–cell adhesion mediated through desmosomes and adhesion junctions plays an important role in the integrity of epithelium. The terminally differentiated keratinocytes flatten and develop the cornified envelope, the structure around the cell wall, which has a significant impact on the barrier function of epithelia. The pattern of genes expressed changes along the differentiation stage of the cells. Specific transcription factors and transcriptional regulators contribute to the differentiation of epithelial cells (Fuchs 1995; Eckert et al. 1997).
Many of the known transcriptional regulators have been identified through DNA–protein or protein–protein interactions. The EBV transcription factor EB1 (also called ZEBRA, Z, or Zta) is a member of the basic leucine-zipper (bZIP) family (Farrell et al. 1989; Chang et al. 1990; Lieberman and Berk 1990). The bZIP transcription factors, including Jun, Fos, ATF/CREB, and C/EBP family of proteins, form homo- and heterodimers through a coiled-coil domain, also called a leucine zipper, and bind DNA through a region rich in basic amino acids, located adjacent to the dimerization domain. The protein complexes known as AP1 and AP2 have an important function in the regulation of gene expression in the epithelial cells (Fuchs 1995; Eckert et al. 1997). EB1 is a sequence-specific DNA-binding protein that activates transcription from specific EBV promoters, as well as the promoters of some cellular genes, thereby initiating the viral lytic cascade. The first 167 NH2-terminal residues of EB1 are involved in the transcriptional activation, while the remaining 78 residues form the DNA binding and dimerization domains. The retinoic acid receptors, RARα and RXRα (Sista et al. 1995), as well as p53 and p65 (Gutsch et al. 1994; Zhang et al. 1994), have been shown to interact with EB1, suggesting that the viral protein EB1 may influence cellular regulatory pathways. The protein–protein interactions between the basic domain of EB1 and the key cell cycle control proteins are indeed involved in cell cycle arrest (Rodriguez et al. 1999). The molecular functions by which transcriptional regulators modulate polymerase II-mediated gene expression are initiated by the recruitment of components of the transcriptional preinitiation complex. The components are thought to be involved either individually or as holoenzyme complexes with coactivators and corepressors, and as complexes affecting chromatin organization (for references see Workman and Kingston 1998; Berger 1999; Berk 1999; Hampsey and Reinberg 1999). Activation of transcription by EB1 has been shown to occur in vitro by direct contact of TFIID (Lieberman et al. 1997). A direct interaction between the basic domain of EB1 and TBP has also been documented in vitro (Mikaelian et al. 1993b).
Two independent approaches reported in this study led to the biological characterization of a novel nuclear protein in human cells. Initially, partial cDNA was isolated through expression cloning while screening for cellular factors interacting with EBV EB1 protein. Independently, during characterization of the periplakin (PPL) locus (Aho et al. 1999), another previously unrecognized gene was identified. Further studies revealed that this gene is ubiquitously expressed and encodes a nuclear protein, capable for interacting with viral and cellular transcription factors. Consequently, we have designated this novel gene as ubinuclein (UBN1). Here, we detail cloning and chromosomal localization of the UBN1 gene, its tissue-wide expression, and structural and functional characteristics of the encoded protein, ubinuclein.
Materials and Methods
cDNA Cloning
A 1-kb partial cDNA clone was isolated through screening of the HeLa cell cDNA library in λgt11. About 4 × 105 phages were screened with biotinylated EB1, essentially as described by MacGregor et al. 1990. EB1 was produced in Escherichia coli BL21 (DE3) cells transformed by plasmid pET-3c (Novagen) carrying the EB1 cDNA Z2.25. The EB1 protein, which lacked the first 25 amino acid residues, was purified as insoluble granules by standard procedures. The purified granules were denatured for 10 min at 68°C in 50 mM Hepes, pH 7.9, 1% NP-40, 1% β-mercaptoethanol, 8 M urea and renatured by dialysis against buffer III (20 mM Hepes, pH 7.9, 20 mM KCl, 1 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 0.5 mM Na-Metabisulfate, and 20% glycerol). Purified EB1 was biotinylated in vitro by biotin-N-hydrosuccimide ester (EB1:ester 1/10) for 3.5 h at room temperature. Biotinylation was stopped by making the reaction 0.1 M in NH4Cl, followed by dialysis for 12 h at 4°C against buffer III.
Further screening of the λgt11 library with the 1-kb VT4 clone as a DNA probe resulted in isolation of the cDNA clone, ZAP5 (Z-associated protein). Further 5′-sequences were obtained through the 5′-RACE approach using a set of nested ZAP5 specific primers, a λgt11-specific primer, and the phage DNA from human placental λgt11 library (Stratagene) as a template for the PCR.
Genomic Cloning
The cloning and sequencing of the human periplakin gene (PPL) was described previously (Aho et al. 1999). During characterization of PPL, the P1 clone GS#14060 was used as a template for direct DNA sequencing, which revealed the 3′-end of UBN1. Genomic clones containing the UBN1 5′-exons were obtained by screening a human placental lambda-FIX II library (Stratagene) with a 284-bp PCR product from the 5′-end of cDNA, produced with primers 5′-AGA TGA CAC TTA TGA CAA GGA-3′ and 5′-CGT TGT CAG AAG TAG AGC CA-3′, and 32P-labeled by random priming. The exon–intron boundaries were confirmed by direct sequencing of the phage DNA using cDNA-specific primers. DNA sequences were obtained using the PRISM Ready Reaction DyeDeoxy Terminator Cycle sequencing Kit, and the Applied Biosystems Models 373A and 377 DNA sequencing systems. The DNA homology searches were performed with the BLAST algorithm (Altschul et al. 1997) and the sequences were edited and analyzed using MacVector 6.0 (Oxford Molecular).
Northern Analysis
Human cancer cell line multiple tissue Northern blot was obtained from Clontech. The 284-bp PCR product described above was 32P-labeled and used as a probe, and the RNA blot was hybridized according to the manufacturer's instructions.
PCR Analysis of Multiple Tissue cDNA Panels
Human multiple tissue cDNA panels (human MTC panels I and II, human fetal MTC panel, and human tumor MTC panel) were obtained from Clontech and used as templates for PCR analysis. The ubinuclein exon 1A-specific primer 5′-GGG ACC GGC ATG AGG AC-3′, exon 1B-specific primer 5′-GGT CAA GGA TCT AGG ATA CA-3′, and exon 2-specific primer 5′-AGA AGC CAT GCA GTG ACA C-3′, in combination with an exon 2-specific reverse primer 5′-AGC TCT GGG TAG AAG AAC T-3′, produced PCR fragments of 304, 483, and 243 bp, respectively. PCR conditions were: 2 min at 94°C, followed by 38 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 1 min. PCR was conducted using Taq DNA Polymerase and the Q-solution provided with the kit (Qiagen). G3PDH-primers, provided by Clontech with each MTC panel, were used as a control, and the PCR was performed for 30 s at 94°C, followed by 26 cycles of 94°C for 20 s and 68°C for 2 min. The PCR-products were separated on 1.5% agarose-TBE gels.
In Vitro Interaction Assays
Glutathione S-transferase (GST)–ZAP5 fusion protein was expressed in E. coli, and purified according to the instructions (Pharmacia Biotech). The GST–ZAP5 bound to glutathione agarose was incubated with in vitro-translated (Promega) 35S-labeled EB1, ZJ (Giot et al. 1991; Mikaelian et al. 1993a) and C/EBP (Landschulz et al. 1988), essentially as described by Manet et al. 1993. Bound proteins were loaded onto a 10% SDS-PAGE and visualized by autoradiography.
Electrophoretic Mobility Shift Assay (EMSA)
EMSAs were performed by incubating 4 × 104 cpm of 5′ 32P-labeled double-stranded oligonucleotide probe, 5′-TCG ACG CGA AGC ACT GAC TCA TGA AGG TGC-3′ (AP1 consensus site underlined) with 2 μl of in vitro-translated proteins for 30 min at room temperature in 20 mM Hepes, pH 7.9, 100 mM KCl, 1 mM MgCl2, 0.5 mM DTT, 10% glycerol, and 1 μg of poly(dI-dC) in a final volume of 20 μl. After incubation, the mixture was loaded onto a 4.5% (wt/vol) polyacrylamide gel (29 to 1 cross-linked), 0.2× TBE, and run at room temperature at 10 V/cm for 3 h. Protein–DNA complexes were visualized by autoradiography.
Expression Constructs
The CMV-based expression vectors for EB1 (pCMV-EB1) and EB1-GCN4 (pCMV-EB1/GCN4) have been described elsewhere (Segouffin et al. 1996). The Flag-tagged ubinuclein constructs were prepared using the expression vector pRc/CMV (Invitrogen). The EB1 constructs with point mutations in the basic domain or the basic domain replaced by the c-Jun basic domain (ZJ) have also been described elsewhere (Mikaelian et al. 1993a).
Cell Cultures and Transfection Studies
Primary foreskin keratinocytes and HaCaT keratinocytes were cultured in the KGM medium containing 0.15 mM Ca2+ (Clonetics). The transfections were executed using the FuGENE 6 transfection reagent according to the instructions provided by the manufacturer (Boehringer Mannheim Corp.). Cells were harvested 9 or 21 h after transfection, washed three times with PBS, fixed for 5 min in absolute methanol at −20°C, and processed for indirect immunofluorescence (IIF).
Immunological Analysis of Ubinuclein Protein
Rabbit polyclonal antibody was raised against the bacterially expressed ZAP5–GST fusion protein. Rabbit serum was passaged several times through a glutathione Sepharose 4B GST column to bind the anti-GST antibodies. The flow-through was loaded on the Glutathione Sepharose 4B GST–ZAP5 column to bind the antibodies, which were eluted at low pH. AZ125 mAb against EB1 has been described (Mikaelian et al. 1993a). Mouse anti–Flag-tag antibody M2 was purchased from Stratagene, and mouse monoclonal antiactin antibody from Boehringer Mannheim Corp.
For the Western blot, cells were harvested 24 h after transfection, washed three times with PBS, and dissolved directly into SDS-gel loading buffer. Proteins were separated by SDS-PAGE on 10% acrylamide, transferred onto nitrocellulose membrane, and detected by the affinity-purified ZAP5 antibody. The secondary antibody, conjugated with HRP, was detected with Renaissance Western blot chemiluminescence reagent (New England Nuclear Life Science Products).
A section of human newborn foreskin, embedded and frozen in the OCT compound, was cut into 7-μm cryosections, which were air-dried. Slides with tissue sections or with cultured keratinocytes were rinsed with PBS, fixed in methanol at −20°C for 5 min, cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature, washed three times with PBS, and blocked with 1% BSA in PBS for 1 h at room temperature. The primary antibodies were applied on the samples overnight at 4°C. After four washes with PBS, samples were incubated for 1 h at room temperature with the species-specific secondary antibody conjugated to Texas red or Fluorescein (Jackson Laboratories), washed four times with PBS, and mounted with Anti-Fade (Molecular Probes). Sections and slides were studied under fluorescent microscope (Axioskop; Carl Zeiss, Inc.), images were stored with ImagePro Plus 4.0 imaging software (Media Cybernetics) and processed with Photoshop 5.0 (Adobe Systems Inc.) and Canvas 5 (Deneba Software).
Results
Molecular Cloning of the Human Ubinuclein Gene and cDNA
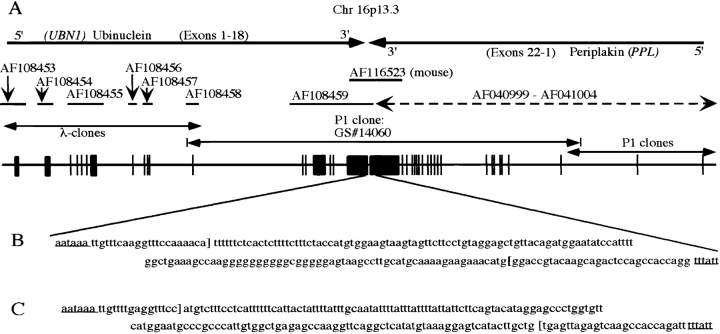
We recently reported characterization of periplakin cDNA and the corresponding gene (PPL) (Aho et al. 1998, Aho et al. 1999). DNA sequences downstream from the periplakin polyadenylation signal (AATAAA) obtained from the genomic P1 clone GS14060 (GenBank/EMBL/DDBJ AF041004 and AF108459) were subjected to the homology search by the BLAST algorithm (Altschul et al. 1997), which revealed an unexpected match with a large number of cDNA clones in the GenBank/EMBL/DDBJ EST database. Furthermore, sequencing of the opposite end of the P1 clone (GenBank/EMBL/DDBJ AF108458; Fig. 1 A) revealed a 146-bp sequence with complete match within the VT4 cDNA (GenBank/EMBL/DDBJ U19346), which has been isolated by screening a HeLa cell expression library for proteins that bind to a negative regulatory element (NRE1) in the long terminal repeat (LTR) of human immunodeficiency virus type 1 (HIV1; Tesmer et al. 1993; M. Bina, Purdue University, personal communication).
Figure 1.
Genomic organization of the periplakin/ubinuclein locus on human chromosome 16p13.3. A, Sequencing of the human genomic P1 clone, GS14060, containing PPL disclosed the presence of a novel gene, UBN1. The orientation of each gene is shown on top, and the accession numbers for the DNA sequences available from the GenBank/EMBL/DDBJ are denoted in the middle. The exons drawn and placed in scale are shown as black squares. B, Human UBN1 and PPL genes were found to reside in tail-to-tail orientation. The distance between the corresponding polyadenylation signals (AATAAA and TTTATT, underlined) is 194 bp. The ends of the cDNAs are indicated by brackets. C, The DNA sequencing of a mouse genomic λ-clone revealed that the arrangement of the mouse Ubn1/Ppl locus on the syntenic region of mouse chromosome 16 was similar to that of the human locus, and the distance between the polyadenylation signals is 197 bp.
In an independent approach, human HeLa cell λgt11 cDNA expression library was screened with the in vitro-translated and biotinylated EBV transcription factor EB1. Among the positive clones, one contained a 1-kb EcoRI insert, identical to the VT4 cDNA, with an open reading frame allowing the translation of the putative interacting fusion protein, but did not contain either translation initiation or termination codons. The 1-kb EcoRI fragment was used as a DNA probe in a second screening of the λgt11 library and a longer cDNA clone, ZAP5, was isolated. Rapid amplification of the cDNA library DNA with a pair of vector specific and cDNA-specific primers resulted in the extension of the 5′-end by 1.5 kb of cDNA sequences (Fig. 2). The 2.3-kb 3′-untranslated region (UTR), sequenced from a genomic P1 clone, was verified by DNA sequencing of the PCR-products generated using specific primers and a human keratinocyte cDNA library as a template. A single polyadenylation signal, AATAAA, was detected 2.3 kb downstream from the translation termination codon TGA. Human EST sequences cover almost 80% of the ubinuclein cDNA, including the 5′-end and the entire 3′-UTR (BLAST search of the dbEST 12/22/99).
Figure 2.
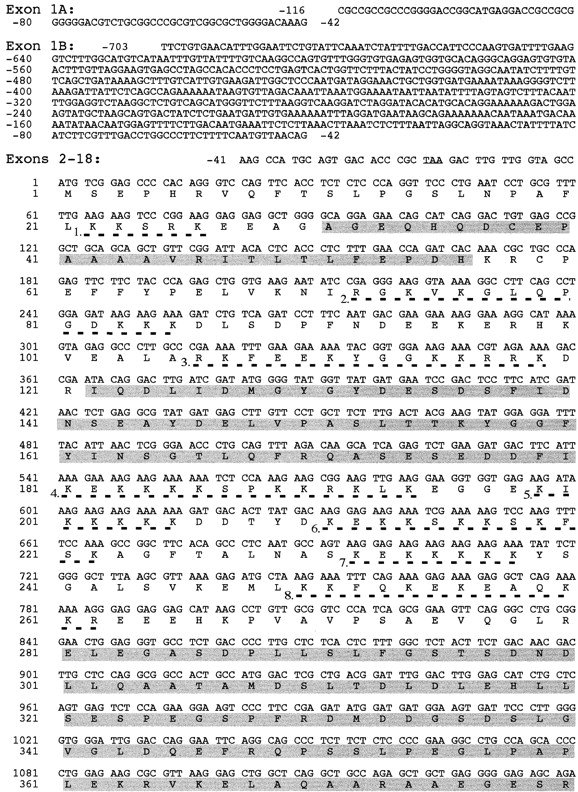


Nucleotide sequence of the cDNA encoding ubinuclein and the predicted amino acid sequence. Numbering of the cDNA sequence begins from the first nucleotide of the putative translation initiation codon, ATG, which is preceded by an in-frame stop codon, TAA, only six triplets upstream (underlined). Two alternative 5′-ends probably due to the multiple promoter usage, denoted Exon 1A and Exon 1B, were discovered. Five nucleolin-type acidic regions are highlighted by shading. Repeats of basic amino acids with homology to nuclear localization signals are underlined by dashed lines and numbered (1–11). The serine-rich region is denoted with dotted underlining and the ATP/GTP consensus binding sequence (amino acids 1119–1127) is underlined. The open reading frame encoding a polypeptide of 1134 amino acid residues terminates in a stop codon (indicated by an asterisk), which is followed by a consensus polyadenylation signal (AATAAA, underlined) at the end of the 2280-bp 3′-UTR. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers AF108460 and AF108461.
Genomic Organization of UBN1
Sequencing of the P1 clone, combined with the isolation and sequencing of overlapping genomic λ-clones, revealed that the human ubinuclein gene spans ∼70 kb of genomic DNA on the short arm of chromosome 16 (Fig. 1 A). The full-length ubinuclein cDNA sequence identified 18 distinct exons in the genomic DNA. Two alternative 5′-exons, preceded by putative promoter sequences, were identified from the genomic clone. The VT4 cDNA clone was found to match with ubinuclein exons 7–15 and the 146-bp matching sequence in the genomic DNA was subsequently identified as exon 12 of UBN1.
The two genes, PPL and UBN1, reside in a tail-to-tail orientation, the distance between the polyadenylation signals of the two human transcripts being 194 bp (GenBank/EMBL/DDBJ AF041004; Fig. 1 B). Although UBN1 and PPL genes together extend >150 kb of genomic sequences, the 23-kb central region of the P1 clone GS14060 contains 19 exons, which occupy 40% of the genomic DNA encoding 76% of the two cDNA sequences. Interestingly, a VT4 protein pseudogene was recently identified in clone 551E13 on human chromosome Xp11.2-11.3 (GenBank/EMBL/DDBJ AL022163).
DNA sequencing of a mouse genomic clone downstream from the 3′-end of the periplakin gene, Ppl (GenBank/EMBL/DDBJ AF116523), revealed high homology to the corresponding region in the human genomic clone. The BLAST search of the EST database with mouse genomic sequence identified a set of mouse cDNA clones, highly homologous to the human ubinuclein 3′-sequences, confirming a tail-to-tail arrangement for Ppl and Ubn1, similar to the human genes. Although human and mouse intergenic regions did not show sequence homology, the distance between the two polyadenylation signals in mouse was similar to that in human, 197 bp (Fig. 1 C).
Predicted Ubinuclein Amino Acid Sequence
The human ubinuclein open reading frame starts in exon 2 from an ATG codon which is preceded by an in-frame translation stop codon, TAA, only six triplets upstream (Fig. 2). The flanking sequence GtaGCCATGt surrounding the start codon is in agreement with the Kozak consensus sequence GCC(A/G)CCATGG (Kozak 1991, Kozak 1996).
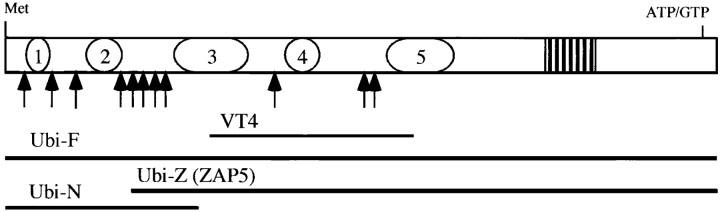
The open reading frame within the full-length UBN1 cDNA encodes a polypeptide, 1,134 amino acids long, with a calculated molecular mass of 121,529 D (Fig. 2). The entire protein is basic, pI 9.34, and rich in serine and lysine, 12.79% and 10.58%, respectively. The secondary structure prediction suggests that the NH2-terminal half of the protein, which is mainly composed of charged amino acids, forms several α helices, while the COOH-terminal half, with polar/nonpolar amino acids dominating, is flexible with high abundance of β-turns. Five acidic regions were identified within the NH2-terminal half of ubinuclein: amino acids 31–56 with pI 5.31; amino acids 122–180 with pI 3.52; amino acids 281–390 with pI 3.89; amino acids 450–503 with pI 5.01; and amino acids 621–720 with pI 4.71 (Fig. 2 and Fig. 6). The alternating acidic and basic domains have a potential to form secondary structures, which are known to interact with DNA. Indeed, the partial VT4 cDNA clone (GenBank/EMBL/DDBJ U19346), encoding ubinuclein amino acids 346–693, was cloned through DNA–protein interaction (see above; Tesmer et al. 1993).
Figure 6.
Organization of the deduced ubinuclein polypeptide. Acidic regions are shown as ovals 1–5, serine-rich domain is denoted as a striped box, and the basic nuclear localization signal-like regions are indicated by arrows. The expression constructs, a full-length ubinuclein (Ubi-F), NH2-terminal construct (Ubi-N), and NH2-terminally deleted cDNA (Ubi-Z/ZAP5), as well as the original VT4 clone (GenBank/EMBL/DDBJ U19346), are aligned with the amino acid sequence and denoted with horizontal bars.
Several clusters of basic amino acids, altogether 11, which fulfill the requirements of either SV40-type (Kalderon et al. 1984) or nucleolin-type (Dingwall et al. 1988) nuclear localization sequences, were identified. Especially, the amino acids 180–262 segment contains five repeats (Fig. 2 and Fig. 6). Computer prediction revealed abundance of possible phosphorylation sites, including consensus sequences for protein kinase C, casein kinase II, cAMP phosphokinase, and tyrosine phosphorylation. A domain rich in serines (DRS) was identified within the COOH-terminal region, amino acids 866–932, and a ATP/GTP binding site homologous to Walker A motif (Walker et al. 1982) was identified at the COOH terminus, amino acids 1119–1127.
Interspecies Homology of the Ubinuclein Transcript
The homology between the human ubinuclein 5′-end with 557 bp of mouse EST sequences was 91%. In addition to human and mouse ESTs, a cDNA clone from Sus scrofa (emb/F23010/SSC18E02) showed 86% identity with the 5′-end of human sequence. Alignment of the human 3′-UTR sequences with the two contigs of mouse ESTs (696 and 907 bp) indicated 75% identity overall. An EST clone of Rattus norwegicus (GenBank/EMBL/DDBJ AI112799; Bonaldo et al. 1996) showed 96% identity to the 3′-UTR of the mouse ubinuclein. Collectively, the nucleotide sequence around the ATG codon and the 3′-UTR sequences, especially those immediately surrounding the AATAAA consensus signal, showed considerable homology between human, mouse, rat, and pig, significantly higher than would be expected from the 3′-UTR extending >2 kb beyond the translation termination codon.
Ubinuclein Transcript is Constitutively Expressed in Fetal and Adult Tissues
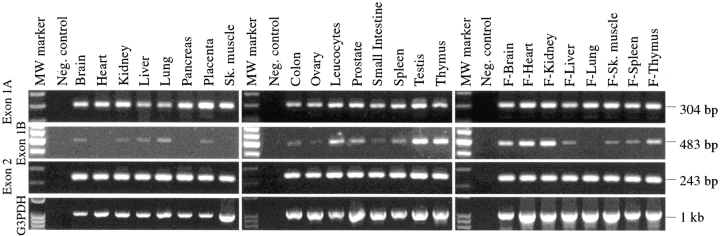
Human multiple tissue cDNA panels were used as PCR templates to study the ubinuclein expression in both adult and fetal tissues. With the exon 2-specific primers, the ubinuclein transcript was detected in all of the normal tissues, as well as in a set of human tumor tissues (Fig. 3 and Fig. 4 A). Ubinuclein cDNA cloning revealed two alternative 5′-ends, which were identified in the genomic clone as two alternative exons, 1A and 1B, encoding two alternative 5′-UTRs (Fig. 2). The primers specific for exon 1A–exon 2 produced a PCR product from all the templates used, while exon 1B–exon 2-specific primers produced a PCR product from tumor cells, fetal tissues, and a limited number of adult tissues (Fig. 3 and Fig. 4 A).
Figure 3.
Expression of ubinuclein transcripts in human adult and fetal tissues. The multiple tissue cDNA panels I and II (first two panels), and human fetal panel (last panel) were used as PCR templates. PCR primers specific for exon 1A–exon 2, exon 1B–exon 2, and exon 2 of ubinuclein, and for G3PDH as an internal standard, were used. cDNAs were derived from polyA+ RNA isolated from tissues indicated on the top and the quantity was normalized against the transcripts of several housekeeping genes. In most tissues, transcripts with exon 1A–exon 2-specific primers were detected at the equal level, whereas a transcript containing exon 1B was less abundant.
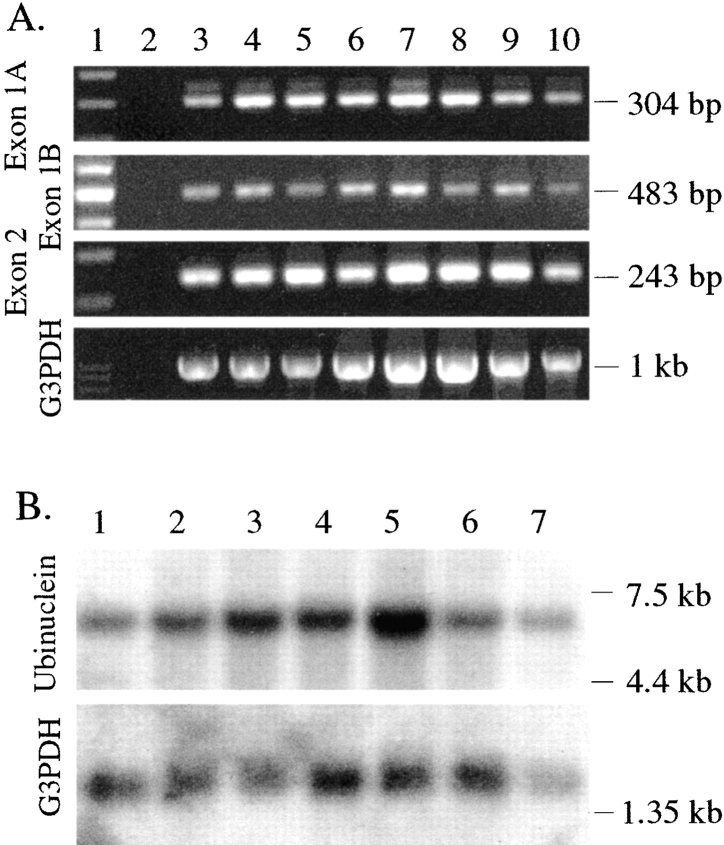
Figure 4.
Expression of ubinuclein transcript in human tumor tissues. A, Tumor tissue cDNA panel was used as a PCR template as described in Fig. 3. Lane 1, Molecular weight marker, 100-bp ladder; lane 2, negative control; lane 3, colon adenocarcinoma (GI-112); lane 4, colon adenocarcinoma (CX-1); lane 5, pancreatic adenocarcinoma (GI-103); lane 6, prostatic adenocarcinoma (PC-3); lane 7, lung carcinoma (GI-117); lane 8, lung carcinoma (LX-1); lane 9, breast carcinoma (GI-101); lane 10, ovarian carcinoma (GI-102). B, Northern blot analysis of human mRNA. Hybridization of the human cancer cell line multiple tissue Northern blot with the ubinuclein-specific probe revealed a transcript of 7 kb. G3PDH-specific probe was used as a control. Lane 1, Promyelocytic leukemia HL-60; lane 2, HeLa cells S3; lane 3, chronic myelogenous leukemia k-562; lane 4, lymphoblastic leukemia MOLT-4; lane 5, colorectal adenocarcinoma SW40; lane 6, lung carcinoma A549; lane 7, melanoma G361.
The hybridization of human cancer cell line multiple tissue Northern blot revealed that the transcript was present in various cancer cell lines (Fig. 4 B). The 7-kb transcript detected on the Northern blot is in agreement with the 6.3–7.0 kb of cloned cDNA sequences. On the Northern blot, only a single band was detected, suggesting that one of the alternatively used promoters was expressed on a relatively low level. The BLAST search identified human ESTs originating from a wide variety of tissues and cells, including heart, retina, gall bladder, liver, testis, and ovary; fetal heart, liver, and spleen; an ovarian tumor, an endometrial tumor, and T cell lymphoma; as well as Jurkat T cells and activated T cells. Furthermore, mouse EST clones derived from various adult tissues, as well as from unfertilized egg, 2-cell stage mouse embryo and 8.5-, 13.5-, and 19.5-d mouse embryos were identified. One of the pig 3′-UTR clones in the EST database has been identified by differential hybridization of a granulosa cell cDNA library (GenBank/EMBL/DDBJ no. X91689; Tosser-Klopp et al. 1997).
Demonstration of the Nuclear Localization of Ubinuclein Protein
Because PCR analysis revealed that ubinuclein is a relatively common transcript in human keratinocyte cDNA library (data not shown), we applied IIF analysis to study the expression and localization of ubinuclein protein in human skin and keratinocytes in culture. Frozen sections of human newborn foreskin revealed intense nuclear signal with ZAP5 antibody throughout the epidermis (Fig. 5 A). Human foreskin keratinocytes in culture showed staining both in fine granular nuclear pattern, as well as in distinct nuclear dots (Fig. 5 B).
Figure 5.
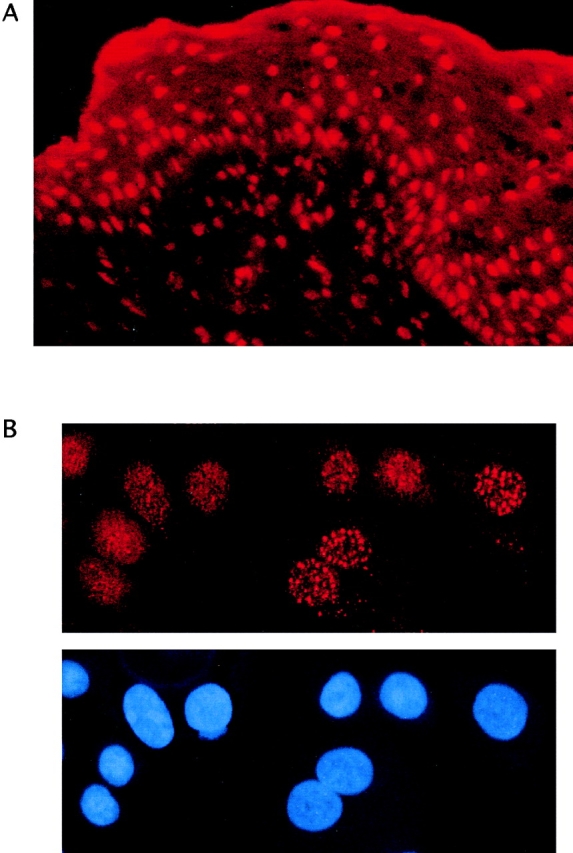
Nuclear localization of the ubinuclein protein. IIF of a newborn foreskin (A) and primary foreskin keratinocytes in culture (B). Top of B shows Ubi-Z/ZAP5 antibody staining visualized with Texas red, whereas nuclei are shown by DAPI in the bottom panel.
Flag epitope-tagged full-length ubinuclein cDNA, Ubi-F (amino acids 1–1134); an NH2-terminally deleted cDNA, Ubi-Z (amino acids 204–1134, ZAP5); and the NH2-terminal construct, Ubi-N (amino acids 1–347; Fig. 6); under the control of the CMV promoter, were transiently transfected into keratinocytes. Cells transfected with Ubi-F and Ubi-N showed strong nuclear signal with the ZAP5 antibody, but Ubi-Z/ZAP5 was distributed both in the nucleus and in the cytoplasm (Fig. 7 A). Double-labeling with Flag-tag antibody M2 showed colocalization with ZAP5 antibody staining in each case. However, transient transfections with Ubi-N and Ubi-Z resulted in the overexpression of the transfected construct, which, in microscopic images, masked the endogenous ubinuclein signal of the adjacent cells (Fig. 7 A). The strongest signal of overexpressed Ubi-F and Ubi-N within the nuclei colocalized with the signal obtained with an antibody against nucleoli (results not shown).
Figure 7.
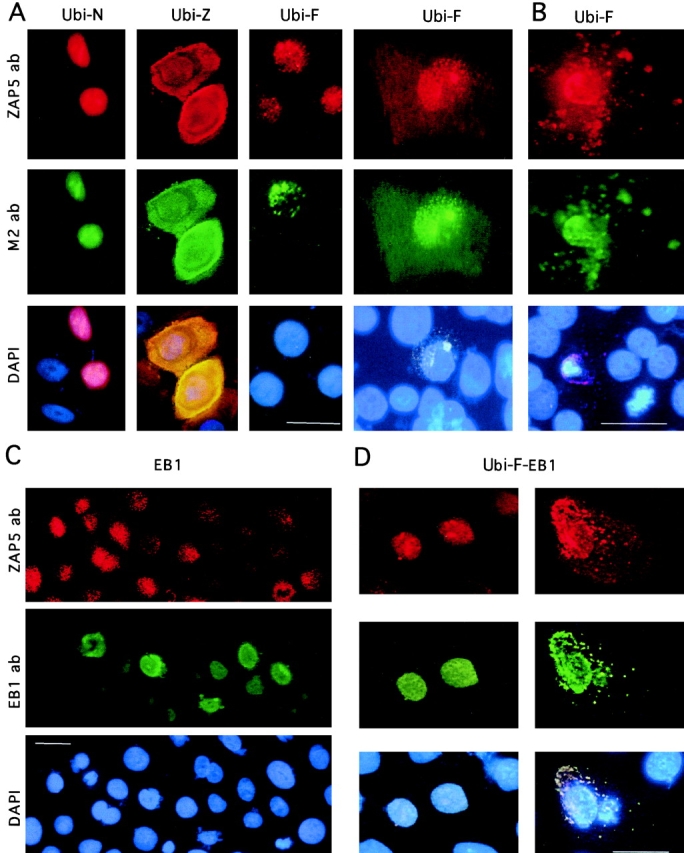
Expression of transiently transfected ubinuclein constructs Ubi-F, Ubi-N, and Ubi-Z/ZAP5, as well as EB1 in primary foreskin keratinocytes. The cultures were stained either with ZAP5 antibody detecting both endogenous and transfected ubinuclein, with M2 antibody recognizing the Flag-tag, or with DAPI for DNA to visualize the nuclei. A, NH2-terminal 345 amino acids contain sufficient information for the nuclear localization of ubinuclein. Cells were prepared for IIF 9 h after transfection of the expression constructs denoted on the top of each panel. Flag-tag signal (M2 ab) colocalized with the signal obtained with ZAP5 antibody. The endogenous signal was detectable with ZAP5 antibody in a panel transfected with Ubi-F, but the strong expression of the Ubi-N and Ubi-Z masked the endogenous signal in the adjacent cells. B, Cells were prepared for IIF 21 h after transfection with the full-length ubinuclein, Ubi-F. A and B, Ubi-F overexpression makes the transfected keratinocytes migrate from the basal cell layer, spread on the top of the basal cell layer, and finally disintegrate and shed off. C, EB1 overexpression downregulates the immunodetectable endogenous ubinuclein. Primary keratinocytes transiently transfected with the EB1 expression construct under the CMV promoter showed strictly nuclear localization in IIF for the EB1 protein. Only in a mitotic cell (see EB1 ab, green cell on the left) EB1 is temporarily released from the nucleus. D, The coexpression of EB1 and Ubi-F results in the cytoplasmic colocalization of EB1 and Ubi-F proteins. ZAP5 antibody was used to detect the ubinuclein protein (Texas red, A–D); M2 antibody was used to detect the Flag-epitope–tagged recombinant proteins (FITC, green, A and B); EB1 protein was detected with antibody AZ125 and visualized with anti-mouse–FITC (green, C and D). DNA in nuclei is demonstrated with DAPI staining and triple filter (blue staining in A–D). Bars, 10 μm.
Overexpression of Ubinuclein cDNA in Cultured Keratinocytes Results in Morphological Changes Resembling Terminal Differentiation
Overexpression of the full-length Ubi-F, but not Ubi-Z or Ubi-N, resulted, after the initial nuclear localization, in the spreading of the transfected cells and redistribution of the Flag-tagged Ubi-F into the cytoplasm. A circular pattern of dots radiating from the nucleus was often distinguished from the uniform cytoplasmic staining (Fig. 7 A, Ubi-F panel on right). Remains of cells, seen on the top of the basal cell layer and strongly staining with ZAP5 and M2 antibodies, suggested disintegration and shedding of the transfected cells, resembling the terminal differentiation of primary keratinocytes in culture (Fig. 7 B).
On the Western blot, the ZAP5 antibody detected a 150-kD band from the keratinocyte extract (Fig. 8). From the cell cultures expressing full-length construct Ubi-F, a single band of 150 kD was detected, while the polypeptide encoded by Ubi-Z/ZAP5 construct appeared as a slightly faster migrating band, and Ubi-N polypeptide was seen as a 50-kD band. In latter cases, the endogenous 150-kD band was also detected (Fig. 8).
Figure 8.
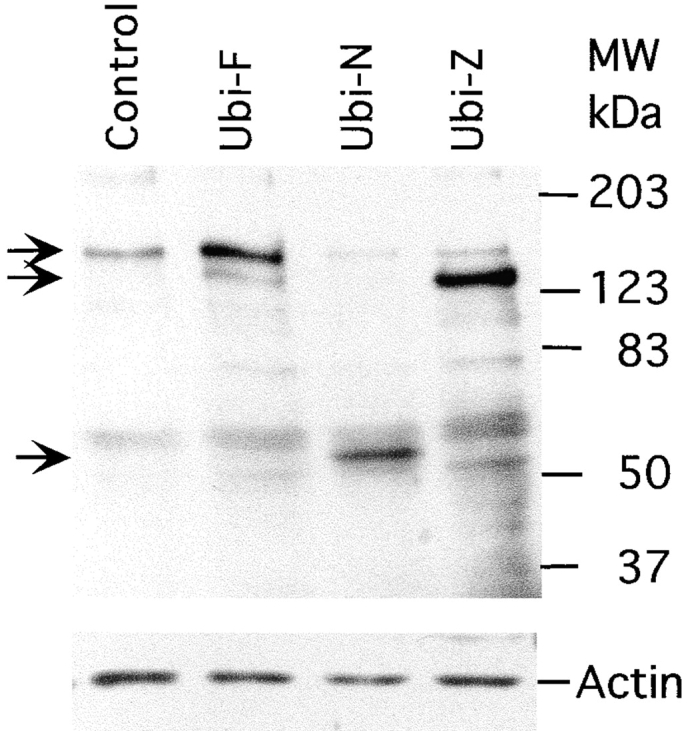
Western blot analysis of the endogenous ubinuclein and the polypeptides expressed from the transiently transfected expression constructs. Total extracts of HaCaT keratinocytes (control) and of those transfected with the Ubi-F, Ubi-N, and Ubi-Z/ZAP5 expression constructs were separated on a 10% SDS-PAGE and detected with ZAP5 antibody (top). The same blot was stained with antiactin antibody (bottom). The arrows point to the bands corresponding to the full-length ubinuclein (top), and polypeptides encoded by Ubi-Z/ZAP5 (middle) and Ubi-N (bottom) expression constructs.
EB1 Expression Interferes with Endogenous Expression of Ubinuclein
As ubinuclein was initially identified through interaction with EB1 protein, we overexpressed EB1 in primary keratinocytes by transiently transfecting cells with an expression construct under the control of CMV promoter. IIF revealed that, in the cells where EB1 was overexpressed, the endogenous ubinuclein signal was either undetectable or weaker than in untransfected neighboring cells (Fig. 7 C). Cotransfection and subsequent overexpression of the full-length ubinuclein, Ubi-F, together with EB1, resulted in a similar appearance of cells as detected with the Ubi-F construct (Fig. 7 D). The colocalization of ubinuclein and EB1 was first limited to the nucleus. However, both proteins were also detected in the cytoplasm of the cells that appeared flat and were spread out on the top of the basal cell layer (Fig. 7 D), whereas the cells expressing only EB1 showed normal cellular morphology and exclusively nuclear staining with EB1 antibody (Fig. 7 C). The distinct cytoplasmic colocalization in a punctate pattern suggests that ubinuclein and EB1 are capable of interacting in vivo.
Ubinuclein Interacts In Vitro with the Basic Domain of both Cellular and Viral bZIP Transcription Factors
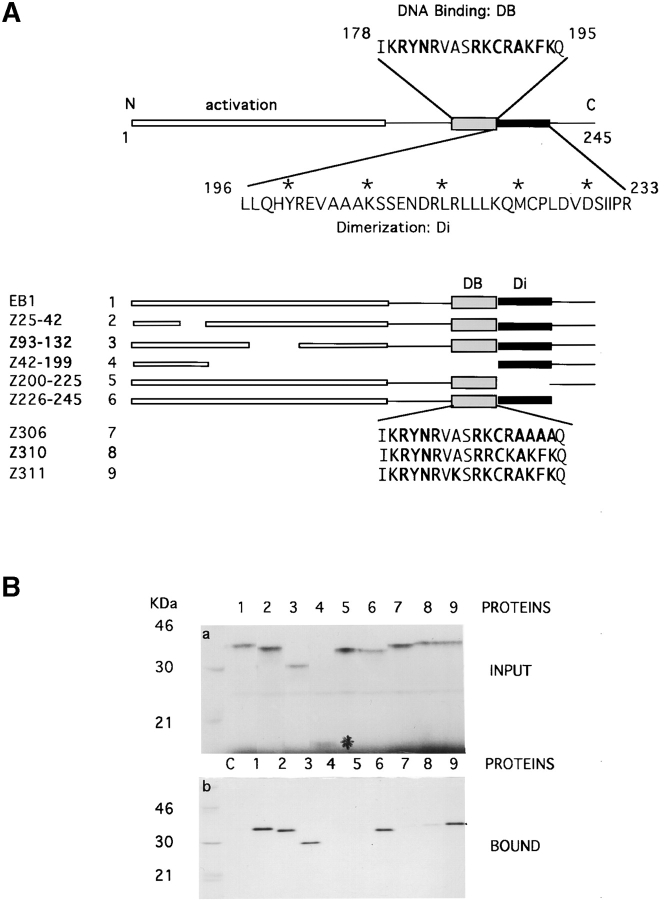
Because a partial ubinuclein cDNA clone was originally isolated through the interaction with EB1, we wanted to identify the domain in EB1 responsible for the interaction. A polypeptide, encoded by Ubi-Z/ZAP5 cDNA (Fig. 6), was expressed as a GST fusion protein in E. coli and immobilized to glutathione agarose particles. A series of in vitro-translated EB1 polypeptides with deletions within each domain and point mutations within the basic domain (Fig. 9 A) was tested for the affinity to the Ubi-Z/ZAP5 polypeptide. We observed that the interaction between Ubi-Z/ZAP5 and EB1 was impaired when the EB1 basic domain (Fig. 9 B, lane 4) or the EB1 dimerization domain (Fig. 9 B, lane 5) was deleted. To pinpoint the amino acids within the EB1 basic domain critical for the binding to Ubi-Z/ZAP5, we assessed the ability of three EB1 constructs carrying point mutations, Z306, Z310, and Z311, to interact with GST-Ubi-Z/ZAP5. As shown in Fig. 9 B, Z306 did not bind to Ubi-Z/ZAP5 at all (Fig. 9 B, lane 7) and Z310 bound only weakly (Fig. 9 B, lane 8), whereas Z311 bound as avidly as the wild-type EB1 (Fig. 9 B, lane 9). Thus, within the bipartite basic domain of EB1, the basic amino acid residues are essential for binding, but an alanine to lysine substitution between the two basic regions did not affect the binding.
Figure 9.
Ubinuclein binds to the basic domain of EB1. A, The EB1 protein is composed of the activation domain (amino acids 1–175), DNA-binding domain (DB; amino acids 178–195), and the dimerization domain (Di; amino acids 196–233). A set of deletion constructs and amino acid substitutions were prepared, as indicated in the bottom of A. B, The interaction between the in vitro-translated EB1 constructs (a) and Ubi-Z/ZAP5 (b) was impaired by the deletion of EB1 DNA-binding domain (lane 4), mutations in the EB1 DNA-binding domain (lanes 7 and 8), or the deletion of the dimerization domain (lane 5). Ubi-Z/ZAP5–GST fusion protein bound to the GT-agarose was used as the affinity matrix. Asterisk marks the low molecular weight protein, Z42-199 (a, lane 4).
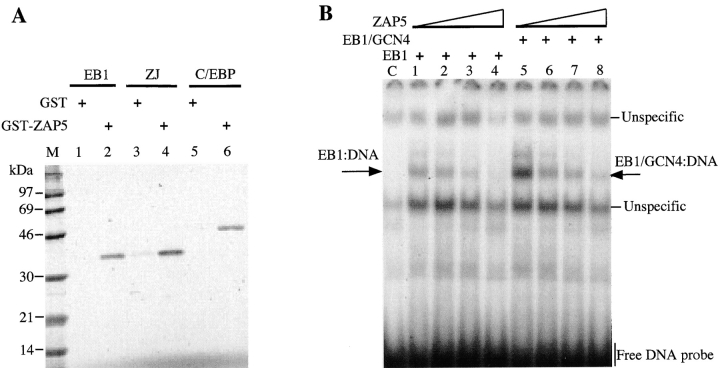
As shown above, EB1 interacts directly in vitro with Ubi-Z/ZAP5, and although the dimerization domain is required, this binding seems to involve specific residues in the region rich in basic amino acids. Therefore, we evaluated if this interaction was restricted to EB1 or if ubinuclein might also interact with other members of the bZIP family of proteins. As shown in the in vitro pull-down assay (Fig. 10 A), GST-Ubi-Z/ZAP5 showed significant affinity to the in vitro-translated EB1, to its derivative ZJ (the basic domain of EB1 replaced by the c-jun basic domain), and to C/EBP, whereas GST alone did not show interaction.
Figure 10.
Ubinuclein interacts with the basic domain of transcription factors. A, The Ubi-Z/ZAP5–GST fusion protein pull-down assay was performed with the in vitro-translated EB1, ZJ (the basic domain of EB1 replaced with the basic domain of c-Jun), and C/EBP. B, The EMSA assay showed that in vitro-translated ubinuclein (Ubi-Z/ZAP5) binds to in vitro-translated EB1 and EB1gcn4, competing with their binding to an oligonucleotide containing the AP1 consensus sequence.
Since ubinuclein appeared to interact with the bZIP protein basic domain, which makes direct contact with DNA, we postulated that Ubi-Z/ZAP5 might impair the binding of EB1 to DNA. Indeed, in an EMSA, increasing amounts of the in vitro-translated Ubi-Z/ZAP5 impaired the binding of in vitro-translated EB1 to an oligonucleotide containing an AP1 consensus binding site (Fig. 10 B). Since the dimerization domain of EB1 is essential for the formation of Ubi-Z/ZAP5-EB1 complex in vitro, we examined whether specific amino acids within the dimerization domain were required for the formation of this complex. The dimerization domain of EB1 was replaced by the homodimerization domain of the yeast transcriptional activator, GCN4. The in vitro-translated hybrid protein EB1gcn4 bound to the AP-1 consensus oligonucleotide, and the complex formation was competed by increasing amounts of in vitro-translated Ubi-Z/ZAP5 (Fig. 10 B). Collectively, our results strongly suggest that the dimerization of EB1 is necessary for the binding, whereas critical amino acid residues within the basic domain of EB1 are specifically involved in binding to ubinuclein.
Discussion
In this study, we report the molecular cloning and biological characterization of a novel gene actively expressed in a variety of tissues and cell lines. The protein is able to bind directly to both viral and cellular transcription factors, and both the endogenous protein and Flag-tagged recombinant protein encoded by an expression construct show nuclear localization. Hence, the protein was termed ubinuclein (ubiquitously expressed nuclear protein), and the corresponding human gene was termed UBN1. A most interesting morphological observation in keratinocyte cultures was that transfection with the full-length ubinuclein construct (Ubi-F) was associated with migration and spreading of the cells in vitro. The spread-out, flattened cells showed ubinuclein staining, in addition to nucleus, also in the cytoplasm. Eventually, these cells appeared to disintegrate and shed off from the top layer of the cultured cells. These observations, which were only noted with the full-length construct, but not with shortened constructs (Ubi-N and Ubi-Z), suggest a selective and specific biological role for ubinuclein in the differentiation of epidermal keratinocytes.
Ubinuclein Is Ubiquitously Expressed in Various Tissues
Ubinuclein is actively transcribed in essentially all tissues and cells, a notion supported by abundance of cDNA clones in the EST database. The high degree of homology between EST sequences representing different species and especially including the sequences within the 3′-UTR, suggests potential importance for this novel transcript and the corresponding protein. The usage of two alternative promoters, characterized from the genomic DNA sequence and detected within the mRNA population through RT-PCR analysis, suggests the possibility for tissue-specific and constitutive or inducible expression of the transcripts.
Ubinuclein Shares Structural Features with Nuclear Proteins
The isolation of a cDNA clone encoding a partial ubinuclein polypeptide through binding to an upstream element, NRE1, in the LTR of the HIV1 promoter, and through the interaction with an EBV transcription factor, EB1, suggested the presence of functional binding domains within the central region of ubinuclein. The detailed analysis of full-length sequence revealed that ubinuclein indeed shares features with nuclear proteins.
Nucleolin is an acidic protein with specific domains that allow its binding with RNA, DNA, and proteins (see references in Ginisty et al. 1999). Although ubinuclein is an unusually basic protein, the presence of highly acidic regions separated from each other by basic sequences at the NH2-terminal region makes it structurally related to nucleolin. The alternating acidic and basic domains potentially form loops that facilitate binding to both DNA and other proteins. The difference between the predicted and the apparent (SDS-PAGE–derived) molecular mass of nucleolin has been attributed to a high content of negatively charged amino acids. Also, the calculated molecular mass for ubinuclein, 121,529 D, is lower than 150 kD, derived from SDS-PAGE. The discrepancy between the predicted and observed molecular mass may also be due to posttranslational modifications, especially to the numerous potential phosphorylation sites.
The NH2-terminal domain of ubinuclein showed sequence homology with the Drosophila yemanuclein-α both on DNA and amino acid levels. In fact, the majority of the ubinuclein 5′-end EST clones had been identified as similar to the Drosophila yemanuclein-α (GenBank/EMBL/DDBJ P25992). Comparison of the human ubinuclein and Drosophila yemanuclein-α amino acid sequences revealed within the 247 NH2-terminal amino acid residues 32.4% identity and 46.6% similarity. Another region of ubinuclein, amino acids 351–624, revealed 22.6% identity and 44.9% similarity to yemanuclein-α. It was reported recently that the 5′-coding sequence, but not the 5′- or 3′-UTR, is necessary for the early accumulation of yemanuclein-α mRNA in the oocyte and for its localization pattern during oogenesis (Capri et al. 1997). The first 500 bp of the ubinuclein open reading frame showed 52.6% identity with the corresponding region of yemanuclein-α cDNA. The function of this region of ubinuclein transcript remains to be studied. The Drosophila yemanuclein-α is a basic, serine-rich nuclear protein, which was originally isolated through search for differentially expressed maternal genes in Drosophila (Aït-Ahmed et al. 1987). The yemanuclein-α transcript is specifically expressed in the female germ cells at early oogenic stages and displays a graded distribution along the antero–posterior axis of the oocyte, thus potentially playing a role in egg organization (Aït-Ahmed et al. 1992). However, in contrast to the yemanuclein-α transcript, the ubinuclein transcript was detected in essentially all adult and fetal tissues.
Functional analysis by cell transfections showed that the NH2 terminus of 346 amino acids, which includes the majority of the putative nuclear localization signals, is sufficient for the exclusive nuclear localization of ubinuclein in keratinocytes. However, the NH2-terminally deleted Ubi-Z/ZAP5, which also contains several putative nuclear localization signals, was found both in the cytoplasm and in the nucleus of the keratinocytes.
Within the COOH-terminal domain of ubinuclein, a region rich in hydroxy amino acid residues, notably serines, was detected. This type of sequence domain is characteristic for nuclear proteins. For example, another speckled-type nuclear protein, also known as pinin, belongs to the class of proteins termed domain rich in serines (DRS) proteins (Brandner et al. 1998).
Viral versus Cellular Transcription Factors
The EBV EB1 protein is a transcription factor belonging to the bZIP family of nuclear proteins, which serve as components for dimeric transcription factors collectively called AP1 (Speck et al. 1997). Our results showed that in vitro-translated ubinuclein (Ubi-Z/ZAP5) prevented EB1 from binding to DNA. The dimerization of EB1, either through the authentic dimerization domain or through the GCN4 dimerization domain, is a prerequisite for binding. Specific amino acid residues in the EB1 dimerization domain are not essential for the in vitro interaction between ubinuclein and EB1. Rather, it seems that a specific conformation of the basic domain imposed by the dimerization event is the prerequisite for binding. Recently, a nuclear chaperone regulating the dimerization of bZIP proteins has been characterized (Virbasius et al. 1999). Also, a possible function for ubinuclein in vivo might be that it acts as a chaperone for bZIP factors, in which case ubinuclein mediates impairment of EB1 and EB1gcn4 binding to DNA. It has been shown that phosphorylation of a serine residue at position 186 in the basic domain of EB1 by protein kinase C, although it impairs EB1 binding to DNA in vitro, increases EB1-mediated activation of transcription (Baumann et al. 1998). Other cellular bZIP proteins are also regulated by phosphorylation/dephosphorylation at or near the basic domain (see references in Karin 1994; Su and Karin 1996). Whether ubinuclein is an active coregulator in these processes remains to be evaluated.
The immediate early transcription factor EB1 is responsible for the initiation of the lytic cycle of EBV in human epithelial keratinocytes. Overexpression of the EB1 in primary keratinocytes in culture seemed to downregulate the level of the endogenous ubinuclein. The consequence of the overexpression of ubinuclein, after the initial nuclear and nucleolar localization, was seen as the cytoplasmic distribution of the protein, followed by spreading and shedding of the transfected cells, which resembled the premature terminal differentiation event. The overexpression of transfected ubinuclein in the same cell with EB1 resulted in the cytoplasmic distribution of EB1 as well. These results suggest that ubinuclein and EB1 are able to interact in vivo, which was detected as colocalization, and is responsible for redistribution of otherwise exclusively nuclear EB1 protein. Cellular concentration, as well as distribution, of EB1 and ubinuclein proteins may be important factors in the reactivation of the productive cycle in latently infected epithelial cells.
Acknowledgments
We thank Kyle Rothenberger for excellent technical assistance, Dr. Hans-Jürg Alder (Kimmel Cancer Center, Thomas Jefferson University) for DNA sequencing, and Sue Gotta for immunofluorescence imaging. Dr. Minou Bina (Purdue University), Dr. Ulrich Rodeck (Thomas Jefferson University), and Dr. Gerd Maul (Wistar Institute), are acknowledged for helpful discussions.
This study was supported by the United States Public Health Service, National Institutes of Health grant RO1-AR41438 (to J. Uitto), by the Dermatology Foundation (to S. Aho), and by grants from Institut National de la Santé et de la Recherche Médicale and Association pour la Recherche sur le Cancer, France (contract No. 9439 to A. Sargeant).
Footnotes
The sequence data from this article has been deposited with the GenBank/EMBL/DDBJ Data Library under accession nos: AF108453, AF108454, AF108455, AF108456, AF108457, AF108458, and AF108459 (human UBN1 gene); AF108460 and AF108461 (human UBN1 cDNA); AF116523 (mouse Ubn1 gene, 3′-end). Symbols for ubinuclein, UBN1 (human) and Ubn1 (mouse) have been approved by HUGO Nomenclature Committee (www.gene.ucl.ac.uk/nomenclature/).
Abbreviations used in this paper: bZIP, basic leucine-zipper; EBV, Epstein-Barr virus; EB1, product of the EBV BZLF1 early gene; EMSA, electrophoretic mobility shift assay; GST, glutathione S-transferase; IIF, indirect immunofluorescence; PPL, periplakin gene; Ubi-F, full-length ubinuclein construct; Ubi-N, NH2-terminal ubinuclein construct; Ubi-Z, NH2-terminally deleted cDNA; UBN1, ubinuclein gene; UTR, untranslated region; ZAP5, Z-associated protein.
References
- Aho S., McLean W.H.I., Li K., Uitto J. cDNA cloning, mRNA expression, and chromosomal mapping of human and mouse periplakin gene. Genomics. 1998;48:242–247. doi: 10.1006/geno.1997.5188. [DOI] [PubMed] [Google Scholar]
- Aho S., Rothenberger K., Tan E.M.L., Ryoo Y.W., Cho B.H., McLean W.H.I., Uitto J. Human periplakingenomic organization in a clonally unstable region of chromosome 16p with an abundance of repetitive sequence elements. Genomics. 1999;56:160–168. doi: 10.1006/geno.1998.5704. [DOI] [PubMed] [Google Scholar]
- Aït-Ahmed O., Thomas-Cavallin M., Rosset R. Isolation and characterization of the Drosophila genome which contains a cluster of differentially expressed maternal genes (yema gene region) Dev. Biol. 1987;122:153–162. doi: 10.1016/0012-1606(87)90341-1. [DOI] [PubMed] [Google Scholar]
- Aït-Ahmed O., Bellon B., Capri M., Joblet C., Thomas-Delaage M. The yemanuclein-αa new Drosophila DNA binding protein specific for the oocyte nucleus. Mech. Develop. 1992;37:69–80. doi: 10.1016/0925-4773(92)90016-d. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Stephen F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLASTa new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M., Mischak H., Dammeier S., Kolch W., Gires O., Pich D., Zeidler R., Delecluse H.J., Hammerschmidt W. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate–induced phosphorylation. J. Virol. 1998;72:8105–8114. doi: 10.1128/jvi.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. Gene activation by histone and factor acetyltransferases. Curr. Opin. Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- Berk A.J. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 1999;11:330–335. doi: 10.1016/S0955-0674(99)80045-3. [DOI] [PubMed] [Google Scholar]
- Bonaldo M.F., Lennon G., Soares M.B. Normalization and subtractiontwo approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Brandner J.M., Reidenbach S., Kuhn C., Franke W.W. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the “speckle” type. Eur. J. Cell Biol. 1998;75:295–308. doi: 10.1016/S0171-9335(98)80063-0. [DOI] [PubMed] [Google Scholar]
- Capri M., Santoni M.-J., Thomas-Delaage M., Aït-Ahmed O. Implication of a 5′ coding sequence in targeting maternal mRNA to the Drosophila oocyte. Mech. Develop. 1997;68:91–100. doi: 10.1016/s0925-4773(97)00130-5. [DOI] [PubMed] [Google Scholar]
- Chang Y.N., Dong D., Hayward G., Hayward S.D. The Epstein-Barr virus Zta transactivatora member of the bZip family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Robbins J., Dilworth S.M., Roberts B., Richardson W.D. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 1988;107:841–849. doi: 10.1083/jcb.107.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R.L., Crish J.F., Banks E.B., Welter J.F. The epidermisgenes on - genes off. J. Invest. Dermatol. 1997;109:501–509. doi: 10.1111/1523-1747.ep12336477. [DOI] [PubMed] [Google Scholar]
- Farrell P.J., Rowe D.T., Rooney C.M., Kouzarides T. Epstein-Barr virus BZLF1 transactivator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO (Eur. Mol. Biol. Organ.) J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Keratins and the skin. Annu. Rev. Cell. Dev. Biol. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Giot J.-F., Mikaelian I., Buisson M., Manet E., Joab I., Nicolas J.-C., Sergeant A. Transcriptional interference between the EBV transcription factors EB1 and Rboth DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 1991;19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsch D.E., Holley-Guthrie E.A., Zhang Q., Stein B., Blanar M.A., Baldwin A.S., Kenney S.C. The bZIP transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-kappa B. Mol. Cell. Biol. 1994;14:1939–1948. doi: 10.1128/mcb.14.3.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M., Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Karin M. Signal transduction from the cell surface to the nucleus through the phosphorylation of transcription factors. Curr. Opin. Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic messenger RNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kozak M. Interpreting cDNA sequencessome insights from studies on translation. Mamm. Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- Landschulz W.H., Johnson P.F., Adashi E.Y., Graves B.J., McKnight S.L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Lieberman P., Berk A. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P.M., Ozer J., Gursel D.B. Requirement for transcription factor IIA (TFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol. Cell. Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor P.F., Abate C., Curran T. Direct cloning of leucine zipper proteinsJun binds cooperatively to the CRE with CRE-BP1. Oncogene. 1990;5:451–458. [PubMed] [Google Scholar]
- Manet E., Allera C., Gruffat H., Mikaelian I., Rigolet A., Sergeant A. The acidic activation domain of the Epstein-Barr virus transcription factor R interacts in vitro with both TBP and TFIIB and is cell-specifically potentiated by a proline-rich region. Gene Expr. 1993;3:49–59. [PMC free article] [PubMed] [Google Scholar]
- Mikaelian I., Drouet E., Marechal V., Denoyel G., Nicolas J.-C., Sergeant A. The DNA-binding domain of two bZIP transcription factors, the Epstein-Barr virus switch gene product EB1 and Jun, is a bipartite nuclear targeting sequence J. Virol. 67 1993. 734 742a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelian I., Manet E., Sergeant A. The bZip motif of the Epstein-Barr virus (EBV) transcription factor EB1 mediates a direct interaction with TBP C.R. Acad. Sci. III 316 1993. 1424 1432b [PubMed] [Google Scholar]
- Rickinson A.B., Kieff E. Epstein-Barr virus. In: Fields B.N., Knipe D.M., Howley P.M., editors. Fields Virology. Third Edition. Lippincott-Raven Publishers; Philadelphia, PA: 1996. pp. 2397–2445. [Google Scholar]
- Rodriguez A., Armstrong M., Dwyer D., Flemington E. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J. Virol. 1999;73:9029–9038. doi: 10.1128/jvi.73.11.9029-9038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segouffin C., Gruffat H., Sergeant A. Repression by RAZ of Epstein-Barr virus bZIP transcription factor EB1 is dimerization independent. J. Gen. Virol. 1996;77:1529–1536. doi: 10.1099/0022-1317-77-7-1529. [DOI] [PubMed] [Google Scholar]
- Sista N.D., Barry C., Sampson K., Pagano J. Physical and functional interaction of the Epstein-Barr virus BZLF1 transactivator with the retinoic acid receptors RARα and RXRα. Nucleic Acids Res. 1995;23:1729–1736. doi: 10.1093/nar/23.10.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S.H., Chatila T., Flemington E. Reactivation of Epstein-Barr virusregulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- Su B., Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr. Opin. Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- Tesmer V.M., Rajadhyaksha A., Babin J., Bina M. NF-IL-6–mediated transcriptional activation of the long terminal repeat of the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA. 1993;90:7298–7302. doi: 10.1073/pnas.90.15.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosser-Klopp G., Benne F., Bonnet A., Mulsant P., Gasser F., Hatey F. A first catalog of genes involved in pig ovarian follicular differentiation. Mamm. Genome. 1997;8:250–254. doi: 10.1007/s003359900403. [DOI] [PubMed] [Google Scholar]
- Virbasius C.M., Wagner S., Green M.R. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell. 1999;4:219–228. doi: 10.1016/s1097-2765(00)80369-x. [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO (Eur. Mol. Biol. Organ.) J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Gutsch D., Kenney S.C. Functional and physical interactions between p53 and BZLF1implications for Epstein-Barr virus latency. Mol. Cell. Biol. 1994;14:1929–1939. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]