Abstract
Newly synthesized glycoproteins interact during folding and quality control in the ER with calnexin and calreticulin, two lectins specific for monoglucosylated oligosaccharides. Binding and release are regulated by two enzymes, glucosidase II and UDP-Glc:glycoprotein:glycosyltransferase (GT), which cyclically remove and reattach the essential glucose residues on the N-linked oligosaccharides. GT acts as a folding sensor in the cycle, selectively reglucosylating incompletely folded glycoproteins and promoting binding of its substrates to the lectins. To investigate how nonnative protein conformations are recognized and directed to this unique chaperone system, we analyzed the interaction of GT with a series of model substrates with well defined conformations derived from RNaseB. We found that conformations with slight perturbations were not reglucosylated by GT. In contrast, a partially structured nonnative form was efficiently recognized by the enzyme. When this form was converted back to a nativelike state, concomitant loss of recognition by GT occurred, reproducing the reglucosylation conditions observed in vivo with isolated components. Moreover, fully unfolded conformers were poorly recognized. The results indicated that GT is able to distinguish between different nonnative conformations with a distinct preference for partially structured conformers. The findings suggest that discrete populations of nonnative conformations are selectively reglucosylated to participate in the calnexin/calreticulin chaperone pathway.
Keywords: folding, glucosyltransferase, glucosidase II, calnexin, calreticulin
Introduction
The ER provides the folding and assembly environment for a multitude of soluble and membrane-bound proteins destined to diverse intra- and extracellular locations. Protein maturation in this organelle involves translocation, co- and posttranslational modifications, disulfide bond formation, and chaperone-assisted folding. In addition, newly synthesized proteins are subjected to quality control, which restricts forward transport to correctly folded and oligomerized conformers (Ellgard et al. 1999). Failing to reach the native conformations, proteins are targeted for selective degradation (Klausner and Sitia 1990).
Of the systems that assist and monitor protein maturation, the calnexin/calreticulin cycle is unique in that it directly depends on the structure of oligosaccharides on newly synthesized glycoproteins (Bergeron et al. 1994; Williams 1995; Helenius et al. 1997). Calnexin and calreticulin are homologous lectins that bind to the monoglucosylated forms of N-glycans Glc1Man6-9GlcNAc2. This association promotes correct folding and oligomerization, supports ER retention of folding intermediates and misfolded or incompletely assembled forms, and exposes them to the glycoprotein-specific thiol oxidoreductase ERp57 (Trombetta and Helenius 1998).
The monoglucosylated oligosaccharides central to this chaperone system are generated in two ways. Initially, they arise by the removal of the two outermost glucoses by the sequential action of glucosidases I and II, a rapid process which is independent of the folding status of the glycoproteins (Kornfeld and Kornfeld 1985). Monoglucosylated glycans are also generated by an ER lumenal enzyme, the UDP-glc:glycoprotein:glucosyltransferase (GT), which transfers a glucose residue from UDP-Glc to fully deglucosylated glycans, reestablishing the glucose-α(1,3)-mannose glycosidic bond recognized by calnexin and calreticulin. The opposing actions of glucosidase II and GT establish a deglucosylation/reglucosylation cycle that drives a cycle of calnexin and calreticulin binding and release (Hammond et al. 1993).
GT is a soluble glycoprotein of 170 kD with an ER localization signal ubiquitously expressed in the ER of most cell types, tissues, and species (Trombetta and Parodi 1992; Parker et al. 1995; Fernandez et al. 1996). Glycoprotein reglucosylation is a selective reaction that only occurs for incompletely folded proteins (Suh et al. 1989; Sousa et al. 1992; Trombetta and Parodi 1992; Fernandez et al. 1994; Hebert et al. 1995; Parker et al. 1995; Van Leeuwen and Kearse 1997; Wada et al. 1997; Cannon and Helenuis 1999; Labriola et al. 1999). While the in vitro studies do not rule out the possibility that it cooperates with other ER factors in vivo, they strongly suggest that the conformational sensor function is primarily, if not exclusively, performed by GT itself. Work by Parodi and co-workers has shown that to be reglucosylated, i.e., glycans have to be linked to the polypeptide backbone of a protein with a nonnative conformation, and the innermost GlcNAc bound to the Asn is a key element in the recognition process (Sousa et al. 1992; Trombetta and Parodi 1992; Sousa and Parodi 1995). It has been proposed that GT senses the exposure of hydrophobic regions normally buried in native conformations, in a way similar to HSP70 and other molecular chaperones (Sousa and Parodi 1995).
However, the mechanism whereby GT discriminates between native and nonnative protein conformations remains largely unexplored. This is in part due to the experimental challenge imposed by the nonnative nature of the GT substrates. Once proteins depart from their native conformation, they tend to aggregate and behave in ways that are difficult to control and characterize. To analyze the conformational requirements for glycoprotein reglucosylation we generated a panel of defined conformers of pancreatic RNaseB and studied their interaction with GT. Using monomeric variants of RNaseB as model substrates, we found that exposure of an otherwise buried GlcNAc-Asn bond, in a slightly perturbed conformer or in a completely unfolded polypeptide chain, was not a sufficient condition for reglucosylation. On the other hand, a partially structured conformation was better recognized by GT than a completely unfolded form, indicating that not all nonnative conformations can be equally reglucosylated by GT, which showed a preference for structured folding intermediates. These results show that distinct nonnative protein conformations are selectively reglucosylated to participate in the calnexin/calreticulin pathway.
Materials and Methods
Proteins
RNaseB (Sigma R5870) and RNaseA (Sigma R5125) were purified by chromatography on a 2.5 × 60–cm column of BioRex70 (BioRad) by isocratic elution at 1 ml/min with 0.2 M sodium phosphate pH 6.5. Preparative digestions with EndoH (New England Biolabs 702S) or α-mannosidase (Sigma M7257) were performed in 0.1 M sodium acetate buffer, pH 5.0 at 20°C for 24 h. Digestions with subtilisin (Sigma P-4789) were performed at a protease/substrate ratio of 1:750 and a protein concentration of 10–50 mg/ml in 0.1 M Tris-HCl, pH 8.0. After 48 h on ice, pH was brought to 2.5 with ice-cold 1 N HCl. The acidified samples were incubated for 1 h on ice and the pH was brought back to 6.5 with ice-cold 5 N NH3. Samples were purified on BioRex70 as described above.
For preparation of RNaseBS-Prot, 1 ml of glacial acetic acid was added to the 1 ml of RNaseBS on ice. The acidified samples were run on Sephadex G-50 medium (Pharmacia; 1.5 × 45 cm), freshly equilibrated and run in 50% acetic acid at 4°C. RNaseBS-Prot eluted in the void was concentrated in Speed Vac to 0.5 ml and the buffer was exchanged by gel filtration into 50 mM NaCl and 20 mM Hepes, pH 7.5. To regenerate RNaseBS′ and RNaseBS′′, freshly prepared RNaseBS-Prot was incubated with a 5-M excess of S-peptides in 50 mM NaCl and 20 mM Hepes, pH 7.5, and purified in BioRex70 as described.
To prepare reduced and alkylated RNaseB, 10 mg of protein was reduced in 1 ml of 8 M urea, 10 mM DTT, and 0.1 M Tris-HCl, pH 8.0. After 30 min at 37°C, the alkylating agents were added at a final concentration of 80 mM, and samples were incubated for a further 30 min at 37°C, after which the samples were desalted into 50 mM NaCl and 20 mM Hepes, pH 7.5.
In all cases, the purity was analyzed by SDS-PAGE and by analytical chromatography on MonoS 5/5 (Pharmacia) in 20 mM Hepes, pH 7.5, eluted with a gradient from 0 to 150 mM NaCl during 45 min at 1 ml/min. Different ribonuclease samples were stored at −80°C in 0.2 M sodium phosphate, pH 6.5. To perform the assays, they were exchanged into 50 mM NaCl, 20 mM Hepes, pH 7.5, buffer and concentrated by ultrafiltration. In some experiments, the nonnative conformers were supplemented with 20% sucrose and 0.1% octylglucoside to prevent aggregation, which did not affect the glucosylation reactions.
To analyze the sensitivity of different conformers to EndoH, N-glycanase, or trypsin, 1 μg of protein was incubated with 250 U of EndoH (New England Biolabs 702), or 10 U of N-glycanase (New England Biolabs 704) or 0.01 μg of trypsin (Sigma T8642) in a final volume of 10 μl containing 10 mM CaCl2, 50 mM NaCl, 20 mM Hepes, pH 7.5. After 5–60 min at 30 or 37°C, samples were analyzed by SDS-PAGE and stained with Coomasie blue R-250.
Assays
GT was purified as previously described (Trombetta and Parodi 1992). Glucosylation reactions were performed in a final volume of 50 μl containing 100–500 μg of acceptor glycoproteins, 10 mM CaCl2, 50 mM NaCl, 20 mM Hepes, pH 7.5, 0.05–0.1 μM UDP[3H]Glc. Reactions were initiated by addition of 5 μl of GT (0.1–0.5 μg) and incubated at 30–37°C for 5–10 min. For inhibition studies, 50–100 μg of urea-denatured soybean agglutinin (SBA) was used as an acceptor, and ribonuclease forms were added at a one- to fivefold molar excess.
The ribonuclease activity was measured in a total volume of 140 μl containing 0.5% RNA (Sigma R6625), 20 mM NaCl and 50 mM Hepes buffer, pH 7.5. After equilibration at 30°C, reactions were started by the addition of 5 μl of ribonuclease (140 μg/ml). Aliquots of 20 μl were removed at 30-s intervals from the incubations and mixed with 10 μl of 25% perchloric acid and 0.8% uranylacetate. After 10 min at 20°C, samples were centrifuged for 5 min at 14,000 g, 10 μl of the supernatant was mixed with 300 μl of water, and the absorbance was measured at 260 nm.
Results
Generation of RNaseB Conformers
As a model substrate for GT, we selected bovine pancreatic ribonuclease B (RNaseB), a small (124 amino acids), soluble, and well characterized glycoprotein. Since it only differs from RNaseA by the presence of a single high mannose oligosaccharide on Asn34, we generated different conformers of RNaseB by adapting procedures previously described for RNaseA. We implemented a modification known as the ribonuclease S system (Richards 1958; Richards and Vithayathil 1959) in which the peptide bond between Ala20-Ser21 is cleaved by limited proteolysis, yielding an altered but fully active ribonuclease. While similar, intact RNaseA and the cleaved RNAseS differ in certain aspects: they can be separated chromatographically (Richards and Vithayathil 1959); RNaseS is sensitive to proteases that do not digest RNaseA (Allende and Richards 1962), and the region near the peptide bond cleaved in RNaseS is disordered and mobile (Kim et al. 1992).
Since the peptide bond cleaved in RNaseS is located near the glycosylated Asn34, we introduced a localized perturbation in the vicinity of the glycosylation site of RNaseB by reproducing the subtilisin treatment previously described for RNaseA. Cleavage was quantitative and the resulting form, hereafter designated RNaseBS, had full ribonuclease activity (Fig. 1 C). The subtilisin cleavage was also applied to RNaseB, which had been previously digested with endoglycosidase H to remove all but the innermost GlcNac residue of the N-linked glycan, and to RNaseB treated with α-mannosidase to remove the mannoses of the glycan (Fig. 1 B). RNaseA was also converted into RNaseS to serve as a nonglycosylated control.
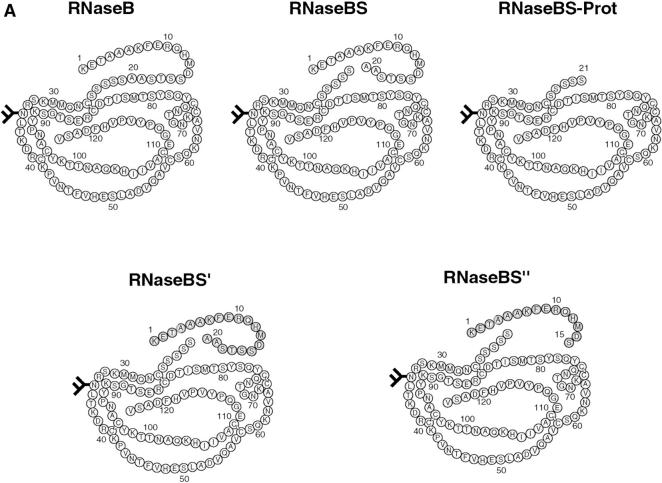
Figure 1.
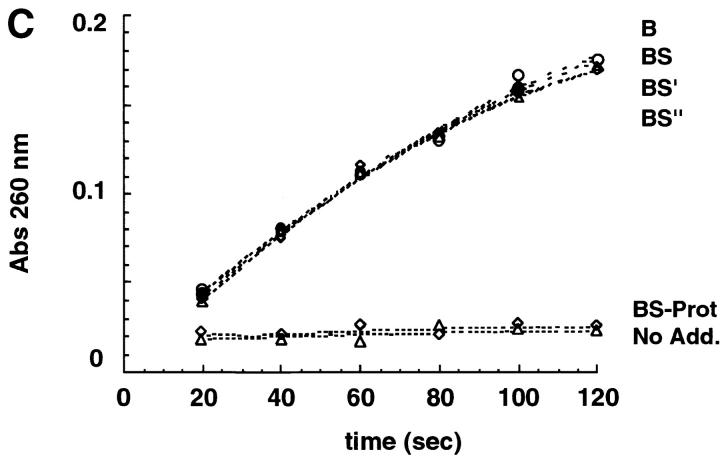
RNaseB conformers. (A) Schematic representation of some of the RNaseB conformers generated including RNaseB, RNaseBS, RNaseBS-Prot, RNase-B5′ and RNase-B5′′. Shaded parts depict peptides added to regenerate RNase-B5′, and RNase-B5′′. (B) RNase conformers analyzed by SDS-PAGE were as follows: (A) native intact RNaseB, (B) reduced and alkylated RNaseB, (C) RNaseBS, (D) RNaseBS-Prot, (E) RNaseBS′, (F) RNaseBS′′, (G) α-mannosidase–digested RNaseBS-Prot, (H) EndoH-digested RNaseBS-Prot, (I) reduced and alkylated RNaseA, (J) RNaseS, and (K) RNaseS-Prot. (C) Ribonuclease activity of the indicated RNaseB conformers. “No Add.” indicates reactions where no enzyme was added.
One key feature of the RNaseS system is the possibility of reversible removal and readdition of the NH2-terminal fragment (the S-peptide, residues 1–20), which remains bound to the protein after cleavage by subtilisin (Richards 1958). The protein, which is devoid of a peptide (called RNaseS-Prot) is soluble but enzymatically inactive (Richards 1958). Since it contains all four intrachain disulfide bonds, it retains much of the native structure (Richards and Wickoff 1971; Richards et al. 1972). We found that after removal of the NH2-terminal peptide from RNaseBS, the remaining core (RNaseBS-Prot) was well behaved, soluble, and monomeric (Fig. 2C and Fig. H). Like RNaseS-Prot, it avidly bound the NH2-terminal peptide (residues 1–20) to generate RNaseBS′. It also bound a shortened version of the peptide (residues 1–15) to generate RNaseBS′′. Both reconstituted forms possessed full ribonuclease activity (Fig. 1 C).
Figure 2.
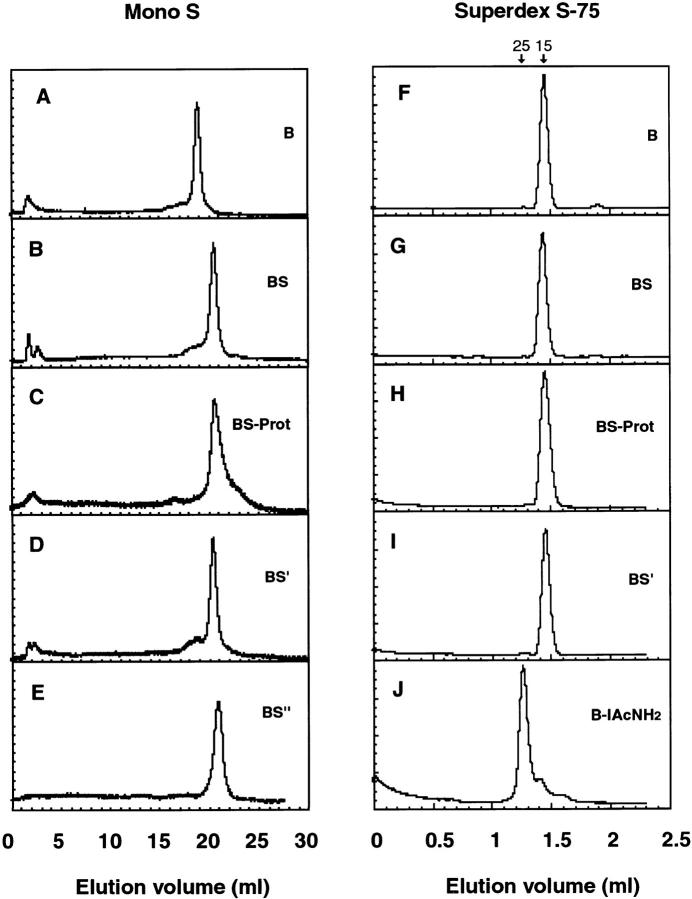
Characterization of RNaseB conformers. Elution profiles from ion exchange chromatography on MonoS (A–E) and Gel filtration chromatography on Superdex S-75 (F–J). (A and F) Native RNaseB, (B and G) RNaseBS, (C and H) RNaseBS-Prot, (D and I) RNaseBS′, (E) RNaseBS′′, and (J) RNaseB reduced and alkylated with iodoacetamide. Arrows indicate the elution peaks for 15- and 25-kD markers.
Since the disulfide bonds are critical for the folded structure, reduction of denatured RNaseA results in an unfolded polypeptide (Sela et al. 1957). To obtain such unstructured forms, RNaseB was denatured, reduced, and alkylated with iodoacetamide or iodoacetic acid, which produced soluble products (Fig. 1 and Fig. 2 J), or with N-ethylmaleimide or 4-vinylpyridine, resulting in insoluble forms.
Characterization of the Different Conformers of RNaseB
The procedures described above allowed us to generate a family of conformers and glycoforms, some of which are shown in Fig. 1. They were all soluble, with the exception of the NEM- and 4VP-alkylated forms. Most importantly, they were monomeric under the experimental conditions used (Fig. 2, F–J). Since reduced and alkylated RNase is monomeric (Harrington and Sela 1959), the increased Stokes radius (Fig. 2 J) is most likely due to a less compact structure consequence of unfolding and the loss of the disulfide bonds. Thus, they could be used to analyze the role of protein conformation in their interaction with GT, without the complications derived from substrate association and aggregation.
To evaluate the local environment around the glycosylated Asn34, we used endoglycosidases as macromolecular probes. In native glycoproteins, the innermost GlcNAc residue can be secluded by the surrounding polypeptide environment, preventing access of endoglycosidases. This is the case for intact RNaseB, in which the glycan can be readily cleaved by EndoH (Tarentino et al. 1974), but is much more refractive to cleavage by N-glycanase (Tarentino and Plummer 1982; Tarentino et al. 1985; Rudd and Dwek 1997), which cleaves between the Asn side chain and the first GlcNAc. We found that all the conformers generated were sensitive to EndoH (Fig. 3 A), indicating that the GlcNAc-GlcNAc bond remained accessible. However, with the exception of native RNaseB, all the other soluble conformers were readily degraded by N-glycanase (Fig. 3 B). This indicated that their Asn34 was more accessible than in native RNAseB. The environment around the glycosylation site had thus changed, even in RNaseBS, in which the S-peptide remained bound and the ribonuclease activity was unaffected.
Figure 3.
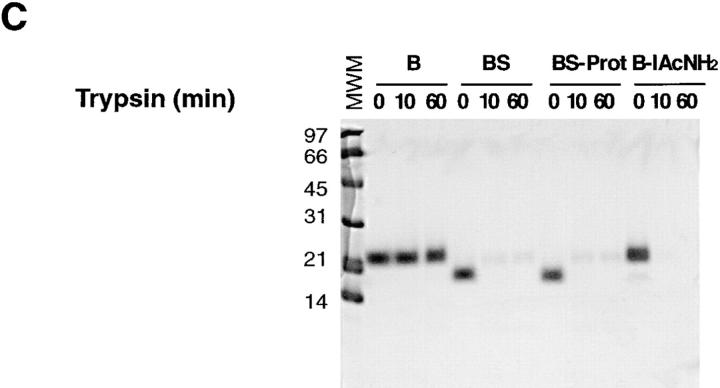
Characterization of RNaseB conformers. Sensitivity of indicated RNaseB conformers to EndoH (A), N-glycanase (B), and to trypsin (C). RNase samples were as follows: (B) intact RNaseB, (BS) RNaseBS, and (B-IAcNH2) RNaseB reduced and alkylated with iodoacetamide. Samples were digested as indicated in Materials and Methods and run in SDS-PAGE stained with Coomasie blue.
To further characterize the conformers generated, we tested their protease sensitivity. Native RNaseB was resistant to cleavage by trypsin, but all other conformers were readily digested, indicating that they had a more relaxed structure than native RNaseB (Fig. 3 C). The same pattern of protease sensitivity was obtained with chymotrypsin (not shown). Despite the marked susceptibility to protease digestion of the different conformers, the forms expected to have ribonuclease activity were fully active on RNA (Fig. 1 C).
Ion exchange chromatography was also used to monitor the different conformers. The technique was sensitive to changes in protein conformation and could differentiate between RNaseB, RNaseBS′, and RNaseBS′′ (Fig. 2, A–E). It also revealed that freshly isolated RNaseBS-Prot eluted as a rather homogeneous population. However, upon storage, it slowly populated different conformers and displayed more heterogeneous elution patterns. Although heterogeneous, they could still rebind S-peptide and recover full ribonuclease activity (not shown). Therefore, for the assays described below, RNaseBS-Prot was used immediately after removal of the S-peptide and neutralization.
Interaction of the Different Conformers with GT
To study the interaction of the various conformers with GT, we quantitated their capacity to be glucosylated in vitro by the isolated glucosyltransferase. The glucosylation assays used measured the rate of incorporation of [3H]Glc from UDP[3H]Glc into acceptor glycoproteins under conditions of initial velocity. Fig. 4 A shows that native RNaseB and mock-treated RNaseB were not recognized as substrates by GT as expected. Moreover, the enzyme was apparently unable to detect the altered conformation of RNaseBS. On the other hand, the form without the S-peptide, RNaseBS-Prot, was glucosylated by GT. As shown in Fig. 4 A and 5, the ability of RNaseBS-Prot to act as a substrate for GT was almost completely abolished upon addition of the NH2-terminal S-peptide (residues 1–20) or a truncated form of it (residues 1–15) to regenerate RNaseBS′ or RNaseBS′′, respectively. Neither peptide affected reglucosylation of other denatured glycoproteins (not shown). The loss of recognition by GT upon reconstitution of ribonuclease activity was, due to the restoration of a nativelike protein conformation upon rebinding of the S-peptides to the RNaseBS-Prot since it paralleled the stoichiometric regeneration of RNaseBS′ or RNaseBS′′ (Fig. 5).
Figure 4.

Recognition of RNaseB conformers by GT. (A) Different RNaseB conformers were tested as substrates of GT. In all samples, 350 μg of the indicated RNaseB conformers were used as substrates in glycoprotein reglucosylation reactions. (B) RNaseB conformers as inhibitors of GT. For inhibition experiments, the same incubations in A were conducted in the presence of 100 μg of denatured SBA.
Figure 5.
Reversible recognition of RNaseBSProt by GT. The indicated amounts of S-peptides 1–20 (solid diamonds) or 1–15 (solid circles) were added to samples containing 500 μg of RNaseBS-Prot in a total volume of 45 μl of 10 mM CaCl2, 50 mM NaCl, 20 mM Hepes, pH 7.5. After 5 min at 20°C, glucosylation reactions were started with 5 μl containing 0.3 μg of GT and 400,000 cpm of UDP[3H]Glc. Reactions were incubated for 5 min at 30°C, stopped with 1 ml of 10% TCA, and radioactivity incorporated in TCA insoluble material quantified. Parallel incubations lacking UDP[3H]Glc were used to measure ribonuclease activity (empty diamonds and circles) as described in Materials and Methods.
Interestingly, the unfolded conformations presented by the soluble derivatives obtained by reduction and alkylation with iodoacetamide or iodoacetic acid were poorly recognized by GT (Fig. 4 A). The aggregation of RNaseB by reduction and alkylation with NEM or 4VP, on the other hand, converted them into much better substrates for GT (Fig. 4 A).
RNaseB forms were less efficient substrates than denatured soybean agglutinin (SBA, not shown), a glycoprotein that contains a single Man9GlcNAc2 oligosaccharide. In contrast, RNaseB carries a mixture of oligosaccharide structures (Man4-9GlcNAc2), of which only a minor fraction has the terminal mannose acceptor for the Glc residue transferred by GT (Trombetta and Parodi 1992). Since the remaining glycoforms all contain a GlcNAc residue linked to Asn34, they have the potential to behave as competitive inhibitors in the glucosylation assay (Sousa et al. 1992). To test whether this was the case and to further evaluate the interaction of the different RNaseB forms with GT, we measured the ability of the different conformers and glycoforms to inhibit GT glucosylation of denatured SBA.
As shown in Fig. 4 B, in native RNaseB, mock-treated RNaseB, RNaseBS and RNaseS-Prot, or the EndoH– or α-mannosidase–digested forms of RNaseBS did not inhibit GT. This indicated that they did not interfere with the substrate recognition site of the glucosyltransferase and did not affect its interaction with denatured SBA. In contrast, all forms that had a carbohydrate bound to Asn34 on the protein backbone of RNaseBS-Prot were potent inhibitors (Fig. 4 B). Although EndoH and α-mannosidase–digested RNaseBS-Prot were not glucose acceptors, they were efficient GT inhibitors, showing that sugar residues extending beyond the first Asn-bound GlcNAc had little effect on interaction with GT. The soluble reduced/alkylated RNaseB forms that were poorly recognized as substrates by GT were also less efficient inhibitors, whereas the insoluble derivatives that were better substrates were also better inhibitors of GT (Fig. 4 B).
Discussion
Reglucosylation of incompletely folded glycoproteins in the ER mediates their interaction with calnexin, calreticulin, and ERp57, which are part of a unique glycoprotein-specific chaperone system. Most glycoproteins are reglucosylated during their biogenesis (Gañan et al. 1991; Gotz et al. 1991), and several glycoproteins have been specifically shown to be reglucosylated in vivo during their maturation in the ER, including vesicular stomatitis G protein (Cannon and Helenuis 1999; Suh et al. 1989), influenza HA (Hebert et al. 1995), transferrin (Wada et al. 1997), T cell receptor subunits (Van Leeuwen and Kearse 1997), and cruzipain (Labriola et al. 1999). The reglucosylating enzyme uses only nonnative glycoproteins as substrates in vitro (Sousa et al. 1992; Trombetta and Parodi 1992; Fernandez et al. 1994; Parker et al. 1995), recognizing occupied glycosylation sites covalently attached to nonnative protein backbones (Sousa et al. 1992; Trombetta and Parodi 1992; Fernandez et al. 1994; Parker et al. 1995; Sousa and Parodi 1995). However, little is known about the mechanism by which GT distinguishes between native and nonnative protein structures. To investigate the principles underlying the conformational sensor in the lectin-based chaperone system, we analyzed the interaction of GT with a series of defined conformers of a naturally occurring glycoprotein, RNaseB.
Intact native RNaseB or a form submitted to mock treatments, were neither substrates nor inhibitors of GT as expected. The nicked form, RNaseBS, was also not recognized despite having a partially perturbed conformation, judging by its distinct chromatographic behavior and its susceptibility to proteases and N-glycanase digestion. The increased exposure of the glycosylated Asn in an altered protein conformation was not sufficient for reglucosylation.
Removal of the NH2-terminal S-peptide (residues 1–20) from RNaseBS resulted, however, in a conformation that was recognized both as a substrate and inhibitor by GT. This conformer is an interesting and well-defined substrate. The soluble and monomeric nature of the RNaseBS-Prot strongly indicates that some feature of its conformation is sensed as nonnative by GT. Like RNaseS-Prot (Richards and Wickoff 1971; Richards et al. 1972; Shindo et al. 1979; Rosa and Richards 1981), RNaseBS-Prot is likely to be highly structured. This is predicted by the constraints imposed by the four disulfide bonds and confirmed by its ability to rebind the S-peptide, which does not bind to denatured forms of RNaseBS-Prot (not shown). Although the affinity of the interaction is reduced, RNaseS-Prot is also known to bind RNase inhibitor (Neumann and Hofsteenge 1994), an intimate association requiring proper RNaseA folding (Kobe and Deisenhofer 1995). That GT specifically recognized the conformation of RNaseBS-Prot as nonnative, was shown by the stoichiometric correction of the conformation with the concomitant loss of the GT interaction by the rebinding of S-peptides. Therefore, the RNaseBS system reproduced the reglucosylation cycle observed in vivo using isolated components, underscoring the fact that it resembles the conformational requisites encountered in the lumen of the ER, where partially folded molecules become invisible to the enzyme upon proper folding.
Completely unfolded RNaseB molecules obtained by reduction and alkylation were poorly recognized by GT as substrates or inhibitors. That these were extensively denatured was shown by its enhanced susceptibility to proteases and N-glycanase, by their larger Stokes's radius observed by gel filtration, their ATP-dependent interaction with HSC70 (not shown), and their lack of ribonuclease activity. These results indicated that exposure of a glycosylated Asn, as in RNaseBS, even in combination with exposure of elements normally buried in native conformers, as in reduced and alkylated RNaseB, do not fulfill the requirements for recognition by GT.
Notably, we observed that GT interacted better with RNaseBS-Prot than with fully unfolded RNaseB. Some elements kept in close proximity in RNaseBS-Prot may become scattered in totally unfolded conformations, resulting in the observed loss of recognition by GT. Alternatively, GT may detect a structural scaffold in RNaseBS-Prot, resembling a partially structured or collapsed folding intermediate with limited flexibility, which would be largely absent in fully unfolded forms. If the loss of such structured conformation upon reduction of the disulfide bonds is what dampens recognition by GT, the enzyme could have a preference for structured folding intermediates as substrates. This possibility is indirectly supported by our observation that insoluble reduced/alkylated products of RNaseB interacted with GT better than the corresponding soluble forms. Also, in agreement with the observations presented here, using isolated components, reduced forms of glycosylated chimeras of bovine ribonuclease and bovine trypsin inhibitor are not reglucosylated when translated and translocated into microsomes in vitro (Rodan et al. 1996). Similarly, selective reglucosylation at advanced stages of folding after nearly complete oxidation-mediated binding of the trypanosomal protein cruzipain to calreticulin in vivo (Labriola et al. 1999).
Aside from the precise mechanism underlying the preference of GT for collapsed over fully unfolded conformations of RNaseB, we provide experimental evidence that GT not only distinguishes between native and nonnative conformations, but it can also discern between different nonnative conformations of the same glycoprotein. The finding that not all nonnative conformations of RNaseB are equally recognized by GT may be one of the reasons why protein unfolding, which was caused by the reduction of disulfide bonds, did not result in a generalized increase in protein reglucosylation in Schizosaccharomyces pombe (Fernandez et al. 1998).
Whereas chaperones of the Hsp70 family recognize short hydrophobic segments normally buried in native proteins (Zhu et al. 1996; Bukau and Horwich 1998), our results suggest that GT prefers, instead, partially structured conformations. This observation correlates well with cases where reglucosylation was shown to mediate the participation of glycoproteins in the calnexin/calreticulin pathway. Influenza HA does not bind to BiP during its maturation, but is reglucosylated and binds calnexin and calreticulin (Hebert et al. 1995). On the other hand, vesicular stomatitis virus glycoprotein binds BiP during early stages of folding, and afterwards reglucosylation mediates its binding to calnexin (Hammond and Helenius 1994). These examples show how the requirements for BiP binding and reglucosylation also differ in living cells. The development of defined substrates allows the design of novel experimental approaches to test these notions and to explore the mechanism underlying chaperone functions mediated by reglucosylation in the ER.
Acknowledgments
We thank the members of the Helenius-Mellman laboratory for helpful discussions. E.S. Trombetta was supported by the Jane Coffin Childs Fund and the Leukemia Society of America. These studies were supported by grants from the National Institutes of Health and the Swiss National Research Fund.
Footnotes
Abbreviations used in this paper: EndoH, endoglycosidase H; GT, UDP-Glc:glycoprotein:glycosyltransferase; SBA, soybean agglutinin.
References
- Allende J.E., Richards F.M. The action of trypsin of ribonuclease-S. Biochemistry. 1962;1:295–304. doi: 10.1021/bi00908a017. [DOI] [PubMed] [Google Scholar]
- Bergeron J.J., Brenner M.B., Thomas D.Y., Williams D.B. Calnexina membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Cannon K., Helenuis A. Trimming and readdition of glucose to N-linked oligosaccharides determines calnexin association of a substrate glycoprotein in living cells. J. Biol. Chem. 1999;274:7537–7544. doi: 10.1074/jbc.274.11.7537. [DOI] [PubMed] [Google Scholar]
- Ellgard L., Molinari M., Helenius A. Setting the standardsquality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Fernandez F., Jannatipour M., Hellman U., Rockeach L., Parodi A.J. A new stress proteinsynthesis of Schizosaccharomyces pombe UDP-Glc:glycoprotein glucosyltransferase mRNA is induced by stress conditions but the enzyme is not essential for cell viability. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:705–713. [PMC free article] [PubMed] [Google Scholar]
- Fernandez F., D'alessio C., Fanchiotti S., Parodi A.J. A misfolded protein conformation is not a sufficient condition for in vivo glucosylation by the UDP-Glc:glycoprotein glucosyltransferase. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:5877–5886. doi: 10.1093/emboj/17.20.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F.S., Trombetta S.E., Hellman U., Parodi A.J. Purification to homogeneity of UDP-glucoseglycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme from Saccharomyces cerevisiae . J. Biol. Chem. 1994;269:30701–30706. [PubMed] [Google Scholar]
- Gañan S., Cazzulo J.J., Parodi A.J. A major proportion of N-glycoproteins are transiently glucosylated in the endoplasmic reticulum. Biochemistry. 1991;30:3098–3104. doi: 10.1021/bi00226a017. [DOI] [PubMed] [Google Scholar]
- Gotz G., Ganan S., Parodi A.J. Glucosylation of glycoproteins in Crithidia fasciculata . Mol. Biochem. Parasitol. 1991;45:265–273. doi: 10.1016/0166-6851(91)90094-m. [DOI] [PubMed] [Google Scholar]
- Hammond C., Helenius A. Folding of VSV G proteinsequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA. 1993;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington W., Sela M. A comparison of the physical chemical properties of oxidized and reduced alkylated ribonuclease. Biochim. Biophys. Acta. 1959;31:427–434. doi: 10.1016/0006-3002(59)90017-4. [DOI] [PubMed] [Google Scholar]
- Hebert D.N., Foellmer B., Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Helenius A., Trombetta E.S., Hebert D., Simons J. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–199. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Kim E.E., Varadarajan R., Wyckoff H.W., Richards F.M. Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures. Biochemistry. 1992;31:12304–12314. doi: 10.1021/bi00164a004. [DOI] [PubMed] [Google Scholar]
- Klausner R.D., Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990;62:611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- Kobe B., Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Labriola C., Cazzulo J.J., Parodi A.J. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Mol. Biol. Cell. 1999;5:1381–1394. doi: 10.1091/mbc.10.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann U., Hofsteenge J. Interaction of semisynthetic variants of RNase A with ribonuclease inhibitor. Prot. Sci. 1994;3:248–256. doi: 10.1002/pro.5560030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K., Fessler L., Nelson R., Fessler J. Drosophila UDP-glucose:glycoprotein glucosyltransferasesequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:1294–1303. doi: 10.1002/j.1460-2075.1995.tb07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F.M. On the enzymatic activity of subtilisin modified ribonuclease. Proc. Natl. Acad. Sci. USA. 1958;44:162–166. doi: 10.1073/pnas.44.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F.M., Vithayathil P.J. The preparation of subtilisin-modified ribonuclease and the separation of the peptide and protein components. J. Biol. Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- Richards F.M., Wickoff W.W. Bovine pancreatic ribonucleases. In: Boyer P.D., editor. The Enzymes. Vol. IV. Academic Press; New York: 1971. pp. 647–806. [Google Scholar]
- Richards F.M., Wyckoff H.W., Carlson W.D., Allewell N.M., Lee B., Mitsui Y. Protein structure, ribonuclease-S and nucleotide interactions. Cold Spring Harb. Symp. Quant. Biol. 1972;36:35–43. doi: 10.1101/sqb.1972.036.01.008. [DOI] [PubMed] [Google Scholar]
- Rodan A.R., Simons J.F., Trombetta E.S., Helenius A. N-linked oligosaccharides are necessary and sufficient for association of glycosylated forms of bovine RNase with calnexin and calreticulin. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:6921–6930. [PMC free article] [PubMed] [Google Scholar]
- Rosa J.H., Richards F.M. Hydrogen exchange from identified regions of the S-protein component of ribonuclease as a function of temperature, pH, and the binding of S-peptide. J. Mol. Biol. 1981;145:835–851. doi: 10.1016/0022-2836(81)90318-1. [DOI] [PubMed] [Google Scholar]
- Rudd P.M., Dwek R.A. Glycosylationheterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 1997;32:1–100. doi: 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- Sela M., White F.H., Anfinsen C.B. Reductive cleavage of disulfide bridges in ribonuclease. Science. 1957;125:691–692. doi: 10.1126/science.125.3250.691. [DOI] [PubMed] [Google Scholar]
- Shindo H., Matsuura S., Cohen J.S. Conformation of ribonuclease S-protein. Experientia. 1979;35:1284–1285. doi: 10.1007/BF01963958. [DOI] [PubMed] [Google Scholar]
- Sousa M., Parodi A.J. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:4196–4203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M.C., Ferrero G.M., Parodi A.J. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- Suh K., Bergmann J.E., Gabel C.A. Selective retention of monoglucosylated high mannose oligosaccharides by a class of mutant vesicular stomatitis virus G proteins. J. Cell Biol. 1989;108:811–819. doi: 10.1083/jcb.108.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino A.L., Plummer T.H. Oligosaccharide accessibility to peptideN-glycosidase as promoted by protein-unfolding reagents. J. Biol. Chem. 1982;257:10776–10780. [PubMed] [Google Scholar]
- Tarentino A.L., Plummer T.H., Maley F. The release of intact oligosaccharides from specific glycoproteins by Endo-β-N-acetylglucosaminidase H. J. Biol. Chem. 1974;249:818–824. [PubMed] [Google Scholar]
- Tarentino A.L., Gomez C.M., Plummer T.H. Deglycosylation of asparagine-linked glycans by peptideN-glycosidase F. Biochemistry. 1985;24:4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- Trombetta E.S., Helenius A. Lectins as chaperones in glycoprotein folding. Curr. Opin. Struct. Biol. 1998;8:587–592. doi: 10.1016/s0959-440x(98)80148-6. [DOI] [PubMed] [Google Scholar]
- Trombetta S.E., Parodi A.J. Purification to apparent homogeneity and partial characterization of rat liver UDP-glucose:glycoprotein glucosyltransferase. J. Biol. Chem. 1992;267:9236–9240. [PubMed] [Google Scholar]
- Van Leeuwen J.E., Kearse K.P. Reglucosylation of N-linked glycans is critical for calnexin assembly with T cell receptor (TCR) alpha proteins but not TCRbeta proteins. J. Biol. Chem. 1997;272:4179–4186. doi: 10.1074/jbc.272.7.4179. [DOI] [PubMed] [Google Scholar]
- Wada I., Kai M., Imai S., Sakane F., Kanoh H. Promotion of transferrin folding by cyclic interactions with calnexin and calreticulin. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5420–5432. doi: 10.1093/emboj/16.17.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.B. Calnexina molecular chaperone with a taste for carbohydrate. Biochem. Cell Biol. 1995;73:123–132. doi: 10.1139/o95-015. [DOI] [PubMed] [Google Scholar]
- Zhu X., Zhao X., Burkholder W.F., Gragerov A., Ogata C., Gotesman M., Hendrickson W. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]