Abstract
Impaired biosynthetic processing of the cystic fibrosis (CF) transmembrane conductance regulator (CFTR), a cAMP-regulated chloride channel, constitutes the most common cause of CF. Recently, we have identified a distinct category of mutation, caused by premature stop codons and frameshift mutations, which manifests in diminished expression of COOH-terminally truncated CFTR at the cell surface. Although the biosynthetic processing and plasma membrane targeting of truncated CFTRs are preserved, the turnover of the complex-glycosylated mutant is sixfold faster than its wild-type (wt) counterpart. Destabilization of the truncated CFTR coincides with its enhanced susceptibility to proteasome-dependent degradation from post-Golgi compartments globally, and the plasma membrane specifically, determined by pulse–chase analysis in conjunction with cell surface biotinylation. Proteolytic cleavage of the full-length complex-glycosylated wt and degradation intermediates derived from both T70 and wt CFTR requires endolysosomal proteases. The enhanced protease sensitivity in vitro and the decreased thermostability of the complex-glycosylated T70 CFTR in vivo suggest that structural destabilization may account for the increased proteasome susceptibility and the short residence time at the cell surface. These in turn are responsible, at least in part, for the phenotypic manifestation of CF. We propose that the proteasome-ubiquitin pathway may be involved in the peripheral quality control of other, partially unfolded membrane proteins as well.
Keywords: cystic fibrosis, lysosomal proteolysis, structural destabilization, functional complementation, ubiquitination
Introduction
Newly synthesized secretory and membrane proteins must attain their native conformation spontaneously or with the assistance of ER-resident and cytosolic chaperones before their export from the ER. The ER quality control mechanism assures that functionally incompetent, misfolded, or unassembled oligomeric proteins are retained and targeted for proteolysis via the ER-associated degradation (ERAD) pathway (Hurtley and Helnius 1989; Brodsky and McCracken 1997; Bonifacino and Weismann 1998; Ellgaard et al. 1999; Wickner et al. 1999). The ERAD involves the recognition, dislocation, and proteolysis of misfolded polypeptides by the 26S proteasome, a multicatalytic enzyme complex localized to the cytoplasm and nucleus (Baumeister et al. 1998; Bonifacino and Weismann 1998; Brodsky and McCracken 1999; Plemper and Wolf 1999). The ATP-dependent cleavage process is facilitated by multiple ubiquitin attachments to the ε-amino groups of lysine residues of the substrate and catalyzed by a cascade of enzymatic reactions involving ubiquitin-activating (E1) and -conjugating (E2) enzymes in addition to ubiquitin-protein ligases (E3) (Hershko and Ciechanover 1998).
Trafficking defects have been recognized as the underlying mechanism in a growing number of genetic diseases, including cystic fibrosis (CF), diabetes insipidus, and α1 antitrypsin deficiency (Thomas et al. 1995; Bonifacino and Weismann 1998; Hershko and Ciechanover 1998; Aridor and Balch 1999; Kopito 1999; Schwartz and Ciechanover 1999). CF, the most prevalent recessive genetic disorder in the caucasian population, is caused by the dysfunction of the CF transmembrane conductance regulator (CFTR), a member of the ATP-binding cassette transport protein family (Riordan et al. 1989; Rommens et al. 1989). CFTR is a cAMP-stimulated Cl− channel, residing predominantly at the cell surface and, less abundantly, in the endosomal compartment (Bradbury 1999). The channel consists of two homologous halves, each comprised of six transmembrane helices and a nucleotide binding domain (NBD1 and NBD2), which are connected by the regulatory domain (Riordan et al. 1989). This complex, multidomain structure conceivably renders the posttranslational folding of wild-type (wt) CFTR inefficient. Approximately 70% of the newly synthesized wt CFTR is trapped in the ER as core-glycosylated folding intermediate (apparent molecular weight ∼150, 000) and degraded with a t 1/2 of ∼30 min by the ERAD. Proteasome activity is responsible, at least in part, for the degradation of the co- and posttranslationally ubiquitinated and incompletely folded CFTR at the ER (Jensen et al. 1995; Ward et al. 1995; Xiong et al. 1999). However, inhibition of proteasomes has failed to promote the processing of core-glycosylated wt as well as the ΔF508 CFTR (Jensen et al. 1995; Ward et al. 1995).
Only 20–30% of the newly synthesized CFTR enters the secretory pathway after its ATP-dependent conformational maturation (Lukacs et al. 1994; Zhang et al. 1998) assisted by cytosolic and ER-resident chaperones (Yang et al. 1993; Pind et al. 1994; Loo et al. 1998; Meacham et al. 1999). Processing of the high mannose type N-linked glycan to complex type oligosaccharides in the cis/medial Golgi region is reflected by a decreased electrophoretic mobility (∼180 kD), providing a convenient method to monitor the trafficking of the CFTR (Cheng et al. 1990; Lukacs et al. 1994; Ward and Kopito 1994; Riordan 1999). Not only does the folded complex-glycosylated CFTR have a slow turnover in vivo (t 1/2 > 12 h), but it also displays a conformation which is more resistant to proteolysis in vitro (Zhang et al. 1998). The posttranslational folding or conformational maturation of CFTR is independent of ER to Golgi vesicular transport, implying that complex glycosylation is not a prerequisite to attain the native conformation (Lukacs et al. 1994; Ward and Kopito 1994; Zhang et al. 1998).
The majority of CF-associated point mutations, including the most common, ΔF508 CFTR, impair biosynthetic processing by disrupting posttranslational folding at the ER (Welsh and Smith 1993; Zielenski and Tsui 1995; Riordan 1999). Recently, we have identified a mechanistically distinct category of mutation, caused by premature terminations and frameshift mutations in the CF gene (Haardt et al. 1999). Although the ER processing of these COOH-terminally truncated CFTR variants missing their last 70–82 amino acid residues (T70 and T82 CFTR) is similar to that of the wt form, the biological half-life of the complex-glycosylated mutants is sixfold shorter than their wt counterpart (Haardt et al. 1999). Although the proteolytic elimination mechanism of the processing mutants entrapped in the ER has been extensively studied (Welsh and Smith 1993; Kopito 1999; Riordan 1999), the cellular processes responsible for the short residence time of the truncated CFTR in post-Golgi compartments remained obscure.
Here we show that deletion of the COOH terminus renders the complex-glycosylated truncated CFTR highly susceptible to proteasome-dependent proteolysis at post-Golgi compartments, including the plasma membrane. Structural destabilization, demonstrated by increased protease susceptibility in vitro and diminished thermostability of the complex-glycosylated mutants in vivo, may serve as signal for premature proteolysis. In contrast, lysosomal proteolysis appears to represent the rate-limiting step in the degradation of the complex-glycosylated wt CFTR. These results reveal a novel aspect of the proteasome-dependent degradation pathway in the elimination of nonnative CFTR after its escape from the ER quality control.
Materials and Methods
Plasmids
The influenza hemagglutinin (HA) epitope was attached to the NH2 terminus ubiquitin using PCR mutagenesis and the cDNA of the yeast ubiquitin as template (provided by Dr. R. Haguenauer-Tsapis, Jacques Monod, University of Paris VII, Paris, France). To generate the Tac-Lamp1 chimera, the extracellular and transmembrane segment of the interleukin 2 receptor (Tac) was fused to the cytoplasmic tail of Lamp-1. The constructs were sequenced to verify that no errors had been introduced. The cDNA, encoding for the Tac and Tac-TCRα, were provided by Dr. J. Bonifacino (National Institutes of Health, Bethesda, MD). Tac, Tac-TCRα, and Tac-Lamp1 were stably transfected in BHK-21 cells using the pNUT expression cassette (Haardt et al. 1999).
Cell Lines and Transfections
Mixtures of stably transfected BHK-21 cells expressing wt, truncated CFTR (missing the last 26, 70, 82, and 98 amino acid residues: T26, T70, T82, and T98 CFTR, respectively) or Tac, Tac-TCRα, and Tac-Lamp1, were generated and maintained as described (Haardt et al. 1999). Intestinal epithelia (T84 and CaCo-2) expressing endogenous CFTR were cultured as described (Lukacs et al. 1993, Lukacs et al. 1994). COS-1 cells were transiently transfected with Lipofectamine (Haardt et al. 1999).
Electrophoresis and Immunoblotting
Cells were washed with ice-cold PBS and lysed in RIPA buffer (150 mM NaCl, 20 mM Tris-HCl, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate, pH 8.0), containing 5 μg/ml of leupeptin and pepstatin A, 10 mM iodoacetamide, and 1 mM PMSF at 4°C. Nuclei and unbroken cells were removed by centrifugation (15,000 g, 15 min at 4°C). Proteins were denatured in Laemmli sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose membrane. Immunoblotting was performed as described previously, using mouse monoclonal L12B4 and M3A7 anti-CFTR antibodies (Abs; Kartner et al. 1992), mouse monoclonal anti-HA (Covance), and antiubiquitin Abs (Santa Cruz Biotechnology, Inc.; Lukacs et al. 1994). Primary Abs were visualized by horseradish peroxidase–conjugated sheep anti–mouse IgG and enhanced chemiluminescence (ECL Western blot kit; Amersham Pharmacia Biotech).
Metabolic Pulse–Chase Labeling and Immunoprecipitation
BHK or COS-1 cells were pulse labeled in the presence of [35S]methionine and [35S]cysteine (0.1–0.25 mCi/ml; Amersham Pharmacia Biotech) and chased in complete medium for the specified time at 37°C. CFTR was immunoprecipitated with a mixture of M3A7 and L12B4 anti-CFTR Abs, as described (Lukacs et al. 1994). The duration of pulse and chase periods and withdrawals of samples are indicated on schematic figures. Radioactivity associated with CFTR was visualized by fluorography and quantified by PhosphorImager (PDI) using the ImageQuant software (Haardt et al. 1999).
Cell Surface Biotinylation
To determine the expression levels of wt and truncated CFTR at the cell surface and early endosome, BHK cells were metabolically labeled to steady state overnight and rinsed with H buffer (154 mM NaCl, 10 mM Hepes, 3 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 10 mM glucose, pH 7.6). Cells were biotinylated in the presence of 1 mg/ml sulphosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (EZ-Link sulfo-NHS-SS-biotin; Pierce Chemical Co.) in H buffer at 37°C (Lukacs et al. 1997). Cells were washed with ice-cold PBS, supplemented with 0.1% BSA, 1 mM MgCl2, and 0.1 mM CaCl2, and solubilized in RIPA buffer. Biotinylated CFTR was isolated by immunoprecipitation with M3A7 and L12B4 anti-CFTR Abs and then on Streptavidin-Sepharose (Sigma-Aldrich). Biotinylated CFTR was visualized with fluorography and its radioactivity was measured by PhosphorImage analysis.
Subcellular Fractionation
Cells were washed with ice-cold PBS three times, resuspended in homogenization medium (0.25 M sucrose, 10 mM Hepes, 1 mM EDTA, 1 mM DTT, pH 7.2, supplemented with 5 μg/ml leupeptin, 5 μg/ml pepstatin A, and 2 mM PMSF). CHO-BQ1, CHO-K1, CaCo-2, and T84 cells were homogenized by nitrogen cavitation as described (Zhang et al. 1998), whereas BHK cells were homogenized in a Dounce homogenizer. After the sedimentation of unbroken cells and nuclei (2,500 g, 5 min at 4°C), mitochondria were pelleted by centrifugation (10,000 g, 10 min at 4°C) from the postnuclear supernatant. Microsomes were fractionated on self-forming Percoll density gradient (25% Percoll in 0.25 M sucrose, 10 mM Hepes and 1 mM EDTA, 5 μg/ml leupeptin, 5 μg/ml pepstatin A, pH 7.3) at 28,000 g for 100 min. The density profile of the gradient was verified with density-marker beads (Sigma-Aldrich) and fractions were downloaded as described (Lukacs et al. 1997). In some experiments lysosomes were labeled with the fluid-phase marker, fluorescein-dextran (0.5 mg/ml, 70 kD; Molecular Probes), overnight and chased in full medium for 3 h. Alkaline phosphatase, β-glucoronidase, and mannosidase II activity, specific markers of plasma membrane, lysosomes, and Golgi regions, respectively, were measured as described (Lukacs et al. 1994, Lukacs et al. 1997). The fluorescence associated with the fractions was determined with fluorescence spectrophotometry in the presence of 0.2% Triton X-100.
Limited Proteolysis
BHK cells expressing wt or T70 CFTR were incubated in the presence of cycloheximide (100 μg/ml) to ensure the degradation of the core-glycosylated wt and T70 CFTR (Lukacs et al. 1994). Isolation of ER-, Golgi complex–, and plasma membrane–enriched microsomes was performed as described (Zhang et al. 1998). The microsomes (0.8–1.5 mg/ml) were digested in HSE medium (10 mM Hepes, 0.25 M sucrose, pH 7.6) in the presence, at the indicated concentration, of trypsin or proteinase K for 15 min at 4°C (Zhang et al. 1998). Proteolysis was terminated by the addition of 1 mM PMSF. Samples were immediately denatured in 2× Laemmli sample buffer at 37°C for 20 min and probed by immunoblot analysis.
Measurement of the cAMP-stimulated Iodide Conductance of the Plasma Membrane
The plasma membrane cAMP-dependent halide conductance of BHK cells expressing T70 CFTR was determined with iodide efflux as described (Mohamed et al. 1997). Iodide efflux was initiated by replacing the loading buffer with efflux medium (composed of 136 mM nitrate in place of iodide). The extracellular medium was replaced every minute with efflux buffer (1 ml). After a steady state was reached, the intracellular cAMP level was raised by agonists (10 μM forskolin, 0.2 mM CTP-cAMP, and 0.2 mM isobutyl-methyl xanthane) to achieve maximal phosphorylation of the T70 CFTR. The collection of the efflux medium resumed for an additional 6–9 min. The amount of iodide in each sample was determined with an iodide-selective electrode (Orion).
Results
Cell Surface Delivery of the Truncated CFTR Is Preserved
We have demonstrated previously that the steady state expression level of the complex-glycosylated CFTR missing its last 70, 82, or 98 amino acids (designated as T70, T82, and T98 CFTR) was decreased by ∼90% compared with wt CFTR in heterologous expression systems (Haardt et al. 1999). In contrast, deletion of the last 26 amino acid residues (T26 CFTR) was without effect (Mickle et al. 1998; Haardt et al. 1999). It was also shown that neither the translational rate nor the biosynthetic maturation of the T70 and T82 CFTR was significantly impaired at the ER in transient COS-1 (Haardt et al. 1999) or in stable BHK expression systems (Benharouga, M., and G.L. Lukacs, unpublished data). We proposed that, in contrast to the most prevalent CF-associated processing mutations (e.g., ΔF508 CFTR), which compromise the biogenesis of CFTR, destabilization of the mature form at distal stages of the biosynthetic or endocytic pathway accounts for the phenotypic manifestation of truncated CFTR (Haardt et al. 1999).
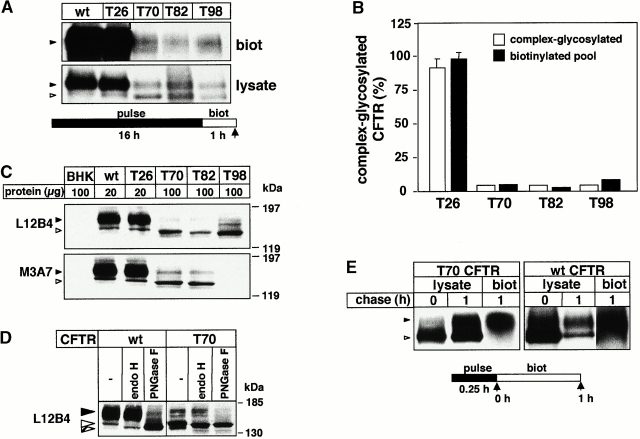
To examine whether impaired plasma membrane targeting of the complex-glycosylated truncated CFTR can attribute to accelerated degradation, the efficacy of cell surface delivery of the mutant was determined first. BHK cells constitutively expressing wt or truncated CFTR were metabolically labeled with [35S]methionine and [35S]cysteine overnight and cell surface–exposed CFTR was covalently tagged with sulfo-NHS-SS-biotin. After the solubilization and immunoprecipitation of CFTR, biotinylated polypeptides were isolated on Streptavidin-Sepharose and visualized by fluorography after separation by SDS-PAGE (Fig. 1 A). PhosphorImage analysis revealed that the abundance of biotinylated T70, T82, and T98 CFTR was only 5–8% relative to wt or T26 CFTR in the mixture of clones (Fig. 1 B). The steady state level of the complex-glycosylated T70, T82, and T98 CFTR pool was also diminished to 5–10% relative to that of wt or T26 CFTR, as determined by quantitative Western blotting using L12B4 or M3A7 anti-CFTR Abs (Fig. 1B and Fig. C). The complex and core-glycosylated forms of wt and truncated CFTR could be distinguished by their distinct electrophoretic mobility as well as their endoglycosidase H sensitivity as described previously and shown on Fig. 1C and Fig. D (Cheng et al. 1990). The parallel decrease in the size of the complex-glycosylated and biotinylated pools of the truncated CFTR suggests that the plasma membrane targeting is largely preserved. To confirm this notion, the cell surface delivery of newly synthesized T70 CFTR was measured.
Figure 1.
Steady state expression and cell surface targeting of wt and truncated CFTR. (A) Expression level of biotinylated CFTR. An identical number of BHK-21 cells expressing wt, T26, T70, T82, or T98 CFTR were metabolically labeled overnight with [35S]methionine and [35S]cysteine. Plasma membrane proteins biotinylated with 1 mg/ml sulfo-NHS-SS-biotin for 60 min at 37°C were isolated by immunoprecipitation with L12B4 and M3A7 anti-CFTR Abs and then on Streptavidin-Sepharose (top). 10% of the immunoprecipitate was directly loaded (bottom). To avoid clonal variations, this, and the experiments shown in C, were carried out on a mixture of clones. The sulfo-NHS-SS-biotin remains membrane impermeant during the labeling, indicated by the lack of biotinylated core-glycosylated forms. Complex- and core-glycosylated CFTR are indicated by black and white arrowheads, respectively. (B) The expression level of wt and truncated CFTR at the cell surface (biotinylated) and at post-ER compartments (complex glycosylated). The radioactivity of biotinylated CFTR was measured by PhosphorImage analysis in experiments as described in the legend to A. The level of the complex-glycosylated CFTR was determined by densitometry of immunoblots (C). Data represent means ± SEM (n = 3), and are expressed as the percentage of the wt, designated as 100%. (C) Expression of wt, T26, T70, T82, and T98 CFTR in BHK-21 cells. CFTR expression in cell lysate was assessed by immunoblotting using the L12B4 and M3A7 anti-CFTR Abs. BHK lane contains a lysate of the parental cells. (D) Glycosidase sensitivity of complex- and core-glycosylated wt and T70 CFTR. Cell lysates were incubated in the presence or absence of endoglycosidase H (endo H) or peptide-N-glycosidase F (PNGase F) for 3 h at 37°C. Polypeptides were separated by SDS-PAGE and probed by the L12B4 anti-CFTR Ab. Complex-, core-, and deglycosylated CFTR are indicated by black, white, and hatched arrowheads, respectively. (E) Targeting efficiency of newly synthesized CFTR to the cell surface. After the pulse labeling of wt or T70 CFTR for 20 min, plasma membrane insertion of the channels was determined by biotinylation during a 1-h chase at 37°C using freshly dissolved sulfo-NHS-SS-biotin (1 mg/ml) every 15 min. Biotinylated CFTR was precipitated with L12B4 and M3A7 Abs and then isolated on Streptavidin-Sepharose (biot). Labeled CFTR was visualized by fluorography. To monitor the pulse labeling of total CFTR pools, 10% of the precipitate was loaded directly (lysate).
Wt and T70 CFTR were pulse labeled for 15 min and those molecules that arrived at the cell surface were biotinylated during a 1-h chase. Biotinylated CFTR was affinity isolated and visualized by fluorography (Fig. 1 E, biot). Radioactivity of biotinylated forms was quantified with PhosphorImage analysis and expressed as a percentage of the respective pulse-labeled core-glycosylated CFTR (Fig. 1 E, lysate). The cell surface targeting efficiency of T70 CFTR was 74 ± 4% (mean ± SEM, n = 4) of wt CFTR. Since this measurement is compounded by the rapid degradation of the complex-glycosylated T70 CFTR (t 1/2 ∼ 1.5–2 h, see also Fig. 6 B; Haardt et al. 1999), the calculated targeting efficiency of the T70 CFTR represents an underestimate and suggests that the plasma membrane delivery of the complex-glycosylated T70 CFTR is largely preserved.
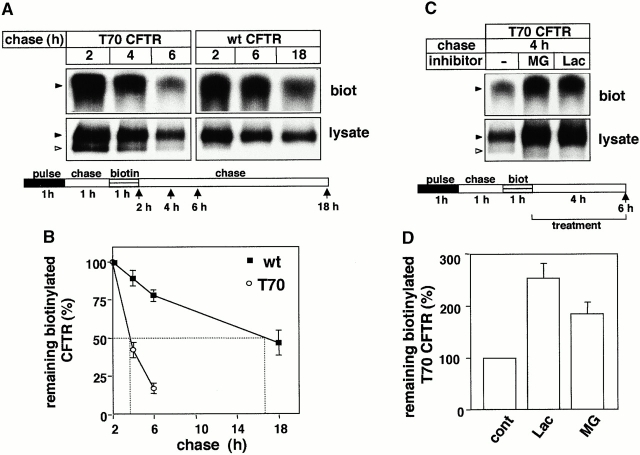
Figure 6.
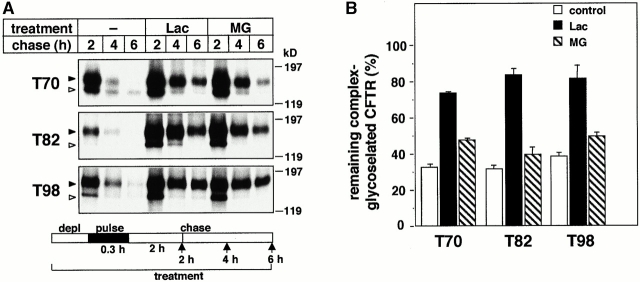
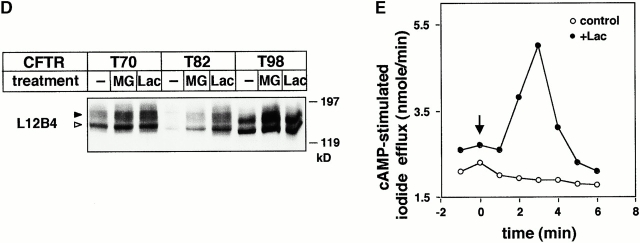
Residence time of cell surface biotinylated wt and T70 CFTR. (A) After metabolic labeling of the complex-glycosylated wt or T70 CFTR, cell surface resident proteins were biotinylated for 45 min and chased for the indicated time in complete medium. Biotinylated CFTR was immunoprecipitated by L12B4 and M3A7 anti-CFTR Abs and then isolated on Streptavidin-Sepharose (top). For comparison, 10% of the immunoprecipitate was loaded for fluorography (bottom). (B) Turnover rates of biotinylated wt and T70 CFTR. Radioactivity incorporated into the biotinylated CFTR was measured with PhosphorImage analysis and expressed as the percentage of the initial label. Data represent means ± SEM, n = 3. (C and D) Proteasome inhibitors delay the degradation of the biotinylated T70 CFTR. Radioactivity remaining in the biotinylated T70 CFTR was measured in pulse–chase experiments after 4 h of chase in the absence or presence of lactacystin (Lac; 10 μM) or MG132 (MG; 10 μM) by PhosphorImage analysis. Data are expressed as a percentage of the amount remaining for the control T70 CFTR and represent means ± SEM, n = 3.
Contribution of Endolysosomal Proteases to the Turnover of Complex-glycosylated Wt and Truncated CFTR
In light of efficient clathrin-dependent internalization of CFTR (Prince et al. 1994, Prince et al. 1999; Lukacs et al. 1997; Bradbury 1999) and proteolysis of other plasma membrane-resident ATP-binding cassette transporters in the vacuolar/lysosomal compartments (Loayza and Michaelis 1998; Hicke 1999; Katzmann et al. 1999), we hypothesized that COOH-terminal truncations may interfere with the recycling of CFTR and lead to premature endolysosomal proteolysis.
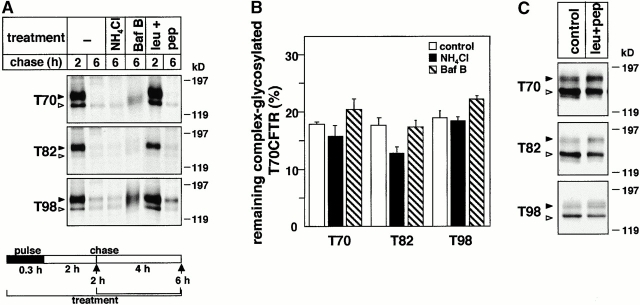
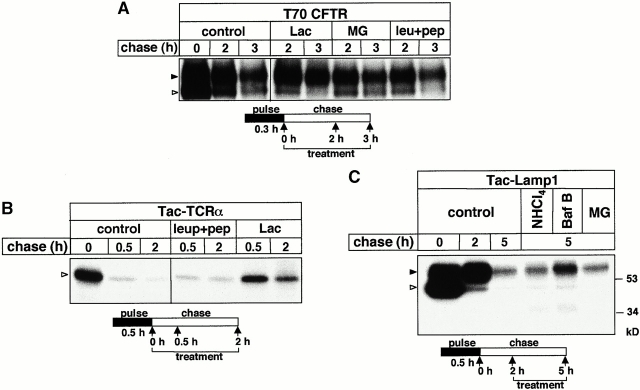
To test this hypothesis, the disposal rate of the metabolically labeled, complex-glycosylated T70 and T82 CFTR was measured in the presence of lysosomal protease inhibitors after allowing the conversion of the core- into complex-glycosylated form (Fig. 2 A). The degradation rate was unaltered upon exposing the cells to NH4Cl, chloroquin, or bafilomycin B, agents that inhibit endolysosomal proteolysis by dissipating the acidic luminal pH and interfering with cargo delivery (Fig. 2 B). Similarly, leupeptin and pepstatin A, inhibitors of cathepsins B, H, L, N, S, and T (Seglen 1983), have a negligible effect on the degradation rate (Fig. 2A and Fig. B) and the steady state expression of truncated CFTRs (Fig. 2 C). In contrast, these drugs stabilized the interleukin 2 receptor α chain (Tac), which is targeted for lysosomal degradation (Fig. 2 D; Hemar et al. 1995), validating their efficacy and suggesting that the initial proteolysis of the truncated CFTR is largely independent of endolysosomal proteases.
Figure 2.
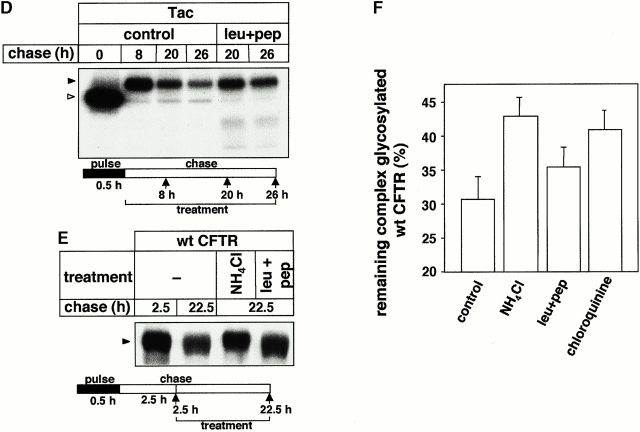
The role of lysosomal proteases in the metabolism of truncated and wt CFTR. (A) After the pulse labeling of T70, T82, and T98 CFTR expressors, BHK cells were chased for 2 h to ensure the conversion of core-glycosylated form to complex-glycosylated CFTR at 37°C. Subsequent chase was performed in the presence of NH4Cl (15 mM). Bafilomycin B (Baf B; 2 μM) and leupeptin plus pepstatin A (leu+pep; 50 μg/ml) were present during the entire pulse–chase. CFTR was immunoprecipitated and visualized by fluorography. (B) The radioactivity remaining in the complex-glycosylated T70, T82, and T98 CFTR after the chase was quantified by PhosphorImage analysis in experiments shown in A and expressed as the percentage of the initial amount present after 2 h of chase. Data represent means ± SEM, n = 3. (C) The effect of chathepsin inhibitors on the expression level of truncated CFTR. BHK transfectants were incubated with or without leupeptin and pepstatin A (50 μg/ml) for 4 h. Equal amounts of protein were probed by immunoblotting as described in the legend to Fig. 1 C. (D) The metabolic stability of Tac in BHK cells. The turnover of Tac was determined in the presence or absence of leupeptin and pepstatin A (50 μg/ml). Metabolically labeled cells were chased for the indicated time, and Tac was isolated by immunoprecipitation and visualized by fluorography. (E and F) Metabolic stability of the complex-glycosylated wt CFTR was determined in stably transfected CHO cells in the presence of NH4Cl (15 mM), chloroquin (200 μM), or leupeptin plus pepstatin A (leu+pep; 50 μg/ml) as described for A and B. Statistical analysis was performed on samples subjected to a 20-h chase (F). Data are means ± SEM, n = 3.
Pulse–chase analysis has revealed that the disposal of the complex-glycosylated wt CFTR was mitigated by NH4Cl and chloroquin (Fig. 2E and Fig. F). To further examine the discordant role of lysosomal proteases in the wt and T70 CFTR degradation, subcellular fractionation was performed, after the enrichment of short-lived proteolytic intermediates that may have been difficult to detect by treating the cells with the cathepsin inhibitors. Heavy and light microsomes, containing predominantly lysosomes and Golgi complex plasma membrane, and endosomes, respectively, were isolated on Percoll density gradient (Fig. 3 A).
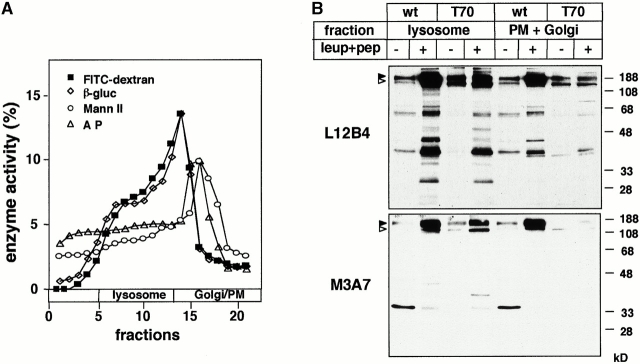
Figure 3.
The effect of leupeptin and pepstatin A on the metabolism of wt and T70 CFTR in lysosomes. (A) Separation of subcellular organelles on Percoll density gradient. Postnuclear supernatants of wt- and T70 CFTR–expressing BHK cells were fractionated on a 25% Percoll density gradient as described in Materials and Methods. Organellar distribution was established by the activity of specific marker enzymes (plasma membrane, alkaline phosphatase [A P]; Golgi, mannosidase II [Mann II]; and lysosomes, β-glucuronidase [β-gluc]) as described previously (Lukacs et al. 1997). Enrichment of lysosomes in the high density fractions was confirmed by the accumulation of fluorescein-dextran (70 kD, 1.5 mg/ml) as well after an overnight labeling and a 3-h chase. (B) BHK cells expressing wt or T70 CFTR were treated with leupeptin and pepstatin A overnight (50 μg/ml) and for 4 h (100 μg/ml), respectively. Microsomes were isolated on Percoll density gradient, and fractions 5–13 and 15–18, comprising the majority of lysosomes and plasma membrane, Golgi regions, and endosomes, respectively, were pooled. Equal amounts (50 μg) of protein were loaded and probed with NBD1 (L12B4) and NBD2 (M3A7) specific anti-CFTR Abs using ECL.
Although cathepsin inhibitors stabilized the complex-glycosylated wt CFTR by 7–10-fold in lysosomes as well as in the light-density fraction, only a modest accumulation of T70 CFTR could be documented by immunoblot analysis (Fig. 3 B). Similar results were obtained in lysosomes isolated by the calcium-precipitation method (data not shown; Kawashima et al. 1998). Surprisingly, leupeptin and pepstatin A provoked the accumulation of degradation intermediates of both wt and T70 CFTR predominantly in lysosomes (Fig. 3 B). These inhibitors also provoked the appearance of breakdown products in isolated lysosomes of intestinal epithelia (CaCo-2 and T84) expressing CFTR endogenously, implying that lysosomal proteolysis cannot be attributed to heterologous overexpession (data not shown). Considering that cathepsin inhibitors are unable to promote the ER processing of the core-glycosylated form (Lukacs et al. 1994; Ward and Kopito 1994), our results suggest that endolysosomal proteases are essential for the elimination of the complex-glycosylated full-length wt CFTR. In addition, endolysosomal proteases appear to be involved in the processing of the proteolytic intermediates derived from both T70 and wt CFTR.
Proteasome-dependent Degradation of the Complex-glycosylated Truncated CFTR
The 26S proteasome is responsible for the degradation of misfolded polypeptides recognized by the ER quality control, short-lived cytosolic, and some plasma membrane proteins after their ligand-induced internalization (Hochstrasser 1996; Bonifacino and Weismann 1998; Hershko and Ciechanover 1998; Brodsky and McCracken 1999; Plemper and Wolf 1999; Schwartz and Ciechanover 1999). To reveal the involvement of the 26S proteasome in the metabolism of the truncated CFTR, the impact of the peptide aldehyde, MG 132 (CBZ-leu-leu-leucinal), and lactacystin, a specific, irreversible inhibitor of the 20S subunit, was assessed (Fenteany et al. 1995).
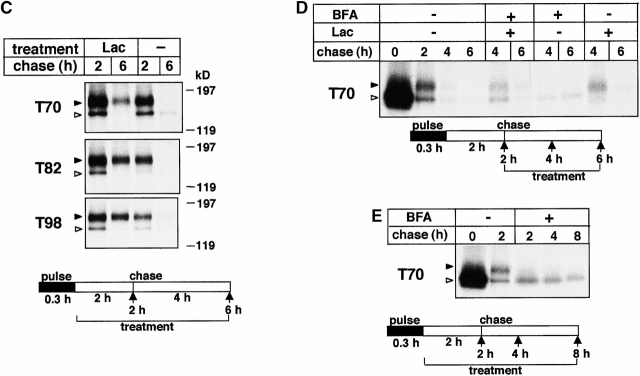
The disappearance of the complex-glycosylated, but not the core-glycosylated, T70, T82, and T98 CFTR was inhibited 2.3-, 2.6-, and 2.1-fold, respectively, by lactacystin when the drug was present during the pulse labeling and chase (Fig. 4A and Fig. B, and Fig. 5 A). MG132 exerted a less pronounced effect (Fig. 4 B). Comparable inhibition was observed when lactacystin was added after the pulse labeling (Fig. 4 C) or after a 2-h chase (Fig. 4 D). This latter protocol has ensured that augmented conversion of the core- to the complex-glycosylated form was not attributable to the lactacystin effect. Similarly, preventing ER to Golgi complex transport of the residual core-glycosylated T70 CFTR with brefeldin A (BFA) failed to influence the inhibitory effect of lactacystin (Fig. 4 D), substantiating the notion that disposal of the complex-glycosylated mutants is delayed, rather than that the escape of core-glycosylated form is facilitated, by lactacystin. In fact, neither lactacystin nor MG 132 promoted the processing of the core-glycosylated T70 CFTR (Fig. 5 A), similarly to other CFTR mutations identified in NBD1 (Jensen et al. 1995; Van Oene et al. 2000). The maturation efficiency of T70 CFTR (20 ± 2%, n = 9) was negligibly increased in the presence of lactacystin (23 ± 4%, n = 4).
Figure 4.
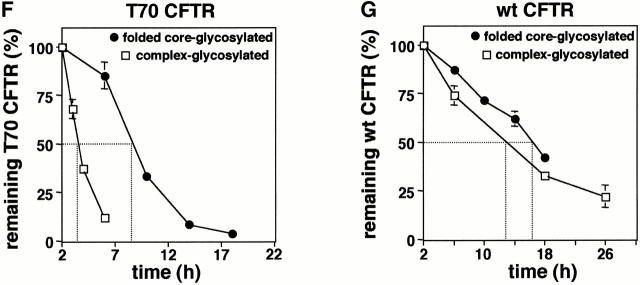
Biochemical rescue of the complex-glycosylated form of truncated CFTR. (A and B) The complex-glycosylated truncated CFTR is stabilized by proteasome inhibitors. BHK cells expressing T70, T82, and T98 CFTR were metabolically labeled and chased in the presence of lactacystin (Lac; 10 μM) or MG132 (MG; 10 μM). The stability of the complex-glycosylated form was monitored by immunoprecipitation and fluorography. The radioactivity remaining in the complex-glycosylated CFTR was measured by PhosphorImage analysis and expressed as the percentage of the amount after a 2-h chase (B). Data are means ± SEM, n = 3–4. (C and D) To rule out that lactacystin (10 μM) delays the degradation of the complex-glycosylated form by promoting the biosynthesis or the conversion of the core-glycosylated form, cells were treated with lactacystin after the pulse labeling (C) or after the conversion of the core- to complex-glycosylated T70 (D). This protocol provided similar inhibition of the complex-glycosylated turnover as reported for A. When indicated, BFA (5 μg/ml) was included to prevent the conversion of the residual core-glycosylated form (D and E). (E–G) Comparison of the turnover rate of folded wt and T70 CFTR in the ER and post-Golgi compartments. After the pulse labeling of BHK transfectants, the fully mature core-glycosylated T70 and wt CFTR were accumulated in the ER by BFA (5 μg/ml) during a 2-h chase. The disappearance of the folded core-glycosylated form was monitored during a subsequent 15-h chase in the presence of BFA and was expressed as the percentage of the amount detected after a 2-h chase (F and G). The stability of the complex-glycosylated T70 and wt CFTR, representing post-Golgi pools, was determined as described for A.
Figure 5.
Functional rescue of the T70 CFTR by lactacystin is exerted by selective inhibition of proteasomes. (A) Protease inhibitors have no effect on the disposal and maturation efficiency of the core-glycosylated T70 CFTR. Pulse-labeled (20 min) BHK transfectants were chased in the presence of lactacystin (10 μM), MG 132 (10 μM), or leupeptin and pepstatin A (50 μg/ml) as indicated. T70 CFTR was visualized by immunoprecipitation and fluorography. PhosphorImage analysis showed that none of the drug had significantly delayed the degradation of the core-glycosylated form (not shown). (B) Lactacystin (10 μM), but not leupeptin and pepstatin A (50 μg/ml), impedes the degradation of the core-glycosylated Tac-TCRα. Stable transfectants were pulse labeled for 15 min and chased in the presence of the indicated drugs. Fluorography was performed as described in the legend to Fig. 2 D. (C) The degradation of the Tac-Lamp1 chimera is sensitive to the inhibition of the vacuolar H+-ATPase with bafilomycin B (Baf B). BHK cells stably expressing Tac-Lamp1 were labeled (30 min) and chased for 2 h. After the addition of NH4Cl (10 mM), bafilomycin B (2 μM), or MG132 (10 μM) the cells were chased for an additional 3 h and Tac-Lamp1 was isolated by immunoprecipitation. (D) The effect of lactacystin and MG132 on the steady state expression of mutant CFTR. Equal amounts of whole cell lysate, obtained from cells exposed to lactacystin (Lac; 10 μM) and MG132 (10 μM) for 3 h, were separated by SDS-PAGE and immunoblotted with the L12B4 anti-CFTR Ab. Complex- and core-glycosylated CFTR are indicated by black and white arrowheads, respectively. (E) Functional rescue of T70 CFTR by lactacystin. The cAMP-activated halide conductance of T70 CFTR–expressing BHK cells was measured by the iodide efflux assay in the absence (control) or presence of lactacystin treatment (+Lac; 10 μM for 3 h). Data points are averages of triplicate determinations and show the difference of iodide release in the presence and absence of the protein kinase A agonist cocktail (10 μM forskolin, 0.5 mM dibutyryl-cAMP, and 0.2 mM IBMX), added as indicated.
If deletion of the COOH terminus destabilizes CFTR exclusively in post-Golgi compartments, retention of folded T70 CFTR in the ER would protect against degradation. Taking advantage of the fact that BFA can provoke the accumulation of folded CFTR in the ER without interfering with ERAD, the stability of folded T70 CFTR was measured after the elimination of the immature core-glycosylated form (Fig. 4 E). The turnover of the folded core-glycosylated T70 CFTR was fourfold slower (t 1/2 ∼ 7 h) than that of the complex-glycosylated T70 CFTR (t 1/2 ∼ 1.8 h; Fig. 4 F). In contrast, ER retention only marginally increased the half-life of the wt CFTR (t 1/2 ∼ 14 h vs. ∼11 h; Fig. 4 G), underlining our previous observations that destabilization of T70 CFTR primarily manifests at the distal stage of the secretory pathway. Similar stabilization was observed upon retaining the complex-glycosylated T70 CFTR in the distal Golgi region by aluminum fluoride (Benharouga, M., and G.L. Lukacs, unpublished observation).
To rule out that cross talks between lysosomal and proteasomal degradation pathway account for the stabilization of the complex-glycosylated T70 CFTR, the specificity of the drugs was examined on BHK cells expressing Tac-TCRα or Tac-Lamp1 chimeras. Although the former polypeptide is a substrate of the ERAD (Bonifacino et al. 1990), the latter is targeted for lysosomal proteolysis (Marks et al. 1996). As we anticipated, proteasome inhibitors mitigated the disappearance of Tac-TCRα, but not Tac-Lamp1, whereas bafilomycin B, but not the proteasome inhibitors, stabilized Tac-Lamp1 (Fig. 5B and Fig. C). Consistent with the notion that proteasome activity accounts, at least in part, for the proteolysis of the complex-glycosylated truncated CFTR, proteasome inhibitors increased the steady state level of the complex-glycosylated T70, T82, and T98 CFTR by two- to fourfold according to densitometric analysis of immunoblots (Fig. 5 D). This was also reflected by the elevated cAMP-activated anion conductance of the plasma membrane (Fig. 5 E). Lactacystin treatment significantly increased the protein kinase A–stimulated iodide release from BHK cells constitutively expressing T70 CFTR as compared with nontreated cells (Fig. 5 E).
Proteasome-dependent Turnover of Cell Surface Resident T70 CFTR
To investigate the influence of proteasome inhibitors on the residence time of truncated CFTR at the cell surface, the turnover of biotinylated T70 CFTR was measured by the pulse–chase technique. The half-life of biotinylated T70 CFTR is sevenfold shorter (t 1/2 ∼ 1.9 h) than the wt CFTR (t 1/2 ∼ 14.5 h; Fig. 6A and Fig. B). Lactacystin and MG132 inhibited the turnover of biotinylated T70 CFTR by 2.6- and 1.9-fold, respectively. This observation provides direct support for our hypothesis that proteasomes, directly or indirectly, are involved in the proteolysis of complex-glycosylated T70 CFTR from the cell surface and endosomal compartments (Fig. 6C and Fig. D).
Ubiquitination of the Complex-glycosylated T70 CFTR
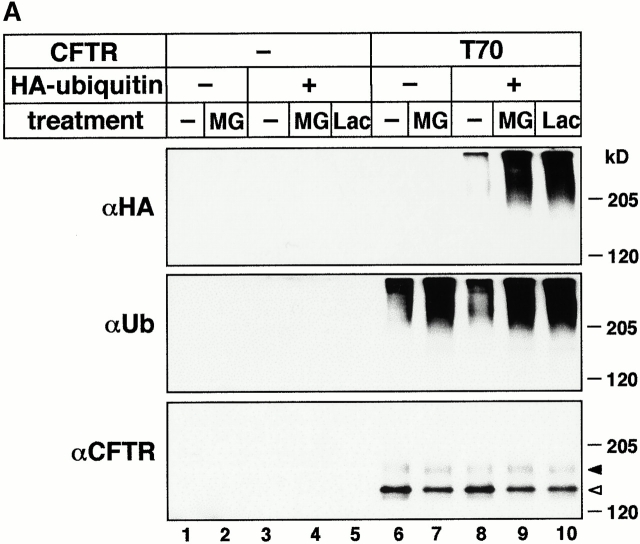
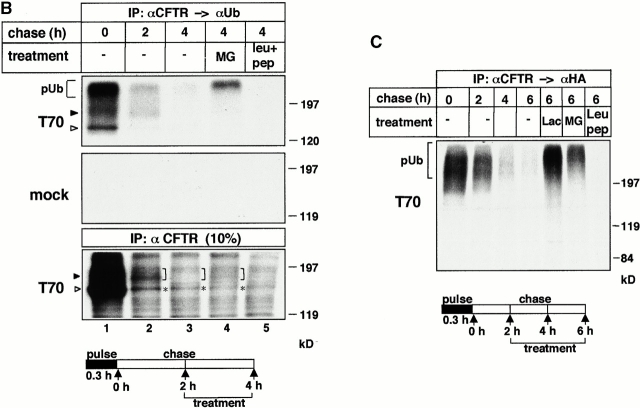
Since recognition of most of the substrates destined for destruction by the proteasome requires polyubiquitin modification (Hershko and Ciechanover 1998; Laney and Hochstrasser 1999; Plemper and Wolf 1999; Hirsh and Ploegh 2000), we examined whether T70 CFTR is subjected to ubiquitin attachments. COS-1 cells were transiently cotransfected with plasmids encoding for the wt or T70 CFTR and HA-tagged ubiquitin (HA-Ub). HA-Ub was detected with the monoclonal anti-HA Ab in immunoprecipitated T70 CFTR. The abundance of high molecular weight immunoreactive polypeptides was augmented by lactacystin and MG132, suggesting that proteasome activity is involved in the degradation of polyubiquitinated T70 CFTR (Fig. 7 A, top, lanes 8–10). Similar results were obtained by monitoring the conjugation of endogenous Ub with a monoclonal anti-Ub Ab (Fig. 7 A, middle, lanes 6–10). Ubiquitinated T70 CFTR could not be detected in cells expressing T70 CFTR or HA-Ub individually (Fig. 7 A, top, lanes 3–7) or after mock transfection (Fig. 7 A, lanes 1–3), validating the specificity of the assay. To discriminate whether polyubiquitinated T70 CFTR was derived from the core- and/or the complex-glycosylated T70 CFTR (Jensen et al. 1995; Ward et al. 1995; Xiong et al. 1999), double immunoprecipitation of metabolically labeled T70 CFTR was performed.
Figure 7.
Proteasome inhibitors induce the accumulation of the polyubiquitinated T70 CFTR. (A) COS-1 cells transiently expressing HA-Ub, T70 CFTR, or a combination of these, were treated with MG132 (MG; 10 μM) or lactacystin (Lac; 10 μM) for 3 h at 37°C. Equal amounts of cellular proteins were immunoprecipitated with L12B4 and M3A7 anti-CFTR Abs and immunoblotted with mouse monoclonal anti-HA (top) or antiubiquitin (middle) Ab. Expression of T70 CFTR was verified by probing 10% of the cell lysate with L12B4 anti-CFTR Ab (bottom). (B) Detection of polyubiquitinated complex-glycosylated T70 CFTR. After the pulse labeling (20 min) of T70 CFTR–expressing (T70) or nontransfected (mock) BHK cells, the degradation of the core-glycosylated T70 CFTR was ensured during a 2-h chase. An additional 2-h chase was performed in the presence of MG132 (MG; 10 μM) or leupeptin and pepstatin A (leu+pep; 50 μg/ml) (lanes 4 and 5). CFTR–ubiquitin conjugates were isolated with sequential immunoprecipitation (IP) using L12B4 and M3A7 Abs first and then antiubiquitin Ab (top and middle) and visualized by fluorography. The processing of the pulse-labeled T70 CFTR pool was monitored by visualizing 10% of the CFTR-immunoprecipitate (bottom). (C) Detection of polyubiquitinated complex-glycosylated T70 CFTR in COS-1 cells. The experimental protocols, described for B, were performed on transiently transfected COS-1 cells expressing T70 CFTR and HA-Ub. Incubation with protease inhibitors was extended to 4 h as indicated and lactacystin (Lac) was used at 20 μM. The second immunoprecipitation was done with anti-HA Abs.
Stably transfected BHK cells were pulse labeled for 20 min and chased for 2 h to ensure the elimination of core-glycosylated T70 CFTR, verified by fluorography (Fig. 7 B, bottom, lanes 1 and 2; see also Fig. 4 D). In a second set of samples, separation of ubiquitinated adducts was achieved by sequential immunoprecipitation with anti-CFTR and then with anti-Ub Ab. The results show that a negligible amount of polyubiquitinated and core-glycosylated T70 CFTR persisted after the initial 2-h chase (Fig. 7 B, top and bottom, lanes 1 and 2). In contrast, radioactivity appeared in the high molecular weight, polyubiquitinated T70 CFTR when MG132 was present during the last 2 h of chase (Fig. 7 B, top, lane 4). Accumulation of polyubiquitinated T70 CFTR was more pronounced in COS-1 cells transiently coexpressing T70 CFTR and HA-Ub (Fig. 7 C). This is most likely due to the higher immunoprecipitation efficiency of the anti-HA Ab. Ubiquitinated conjugates were not detectable upon inhibition of lysosomal proteolysis, or in mock transfected cells (Fig. 7 B, top and middle, and C). Considering that the turnover of the complex-glycosylated, but not the core-glycosylated T70 CFTR, is sensitive to proteasome inhibitors and the core-glycosylated form has been largely eliminated before proteasome inhibition, we conclude that at least a fraction of polyubiquitinated T70 CFTR originates from the complex-glycosylated T70 CFTR. Although we cannot rule out that a small amount of polyubiquitinated T70 CFTR is derived from the core-glycosylated form, the following observations substantiate the notion that the complex-glycosylated T70 CFTR can form ubiquitinated adducts. Ubiquitinated complex-glycosylated form was visualized by anti-Ub Abs after the separation of the core- and complex-glycosylated forms by immunoprecipitation with anti-CFTR Abs and wheat germ agglutinin affinity chromatography (data not shown). Furthermore, coexpression of the dominant negative K48RUb with T70 CFTR delayed the disposal of the complex-glycosylated mutant CFTR (data not shown).
Deletion of the COOH-terminal Tail Structurally Destabilizes CFTR
Nonnative or partially denatured soluble proteins are subjected to ubiquitination and/or proteasome-mediated degradation (Pacifici et al. 1993; Sadis et al. 1995; Michalek et al. 1996; Gilon et al. 1998; Laney and Hochstrasser 1999). A similar scenario may prevail for T70 CFTR if the COOH-terminal tail is engaged in the structural stabilization of CFTR in the post-ER compartment. A large body of evidence demonstrates the existence of intra- and intermolecular interactions in CFTR (Seibert et al. 1996; Ostedgaard et al. 1997, Ostedgaard et al. 2000; Hall et al. 1998; Neville et al. 1998; Zhang et al. 1998; Naren et al. 1999; Lu and Pedersen 2000).
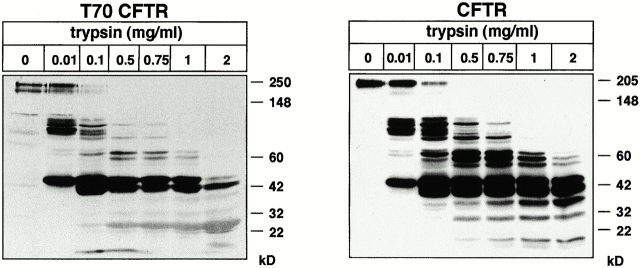
To compare the conformational stability of the complex-glycosylated wt and T70 CFTR, in situ protease susceptibility and in vivo thermostability assays were performed. The conformation of the cytosolic domains was probed with limited proteolytic digestion in conjunction with immunoblotting, a method we have implemented to reveal conformational difference between the wt and the ΔF508 CFTR (Zhang et al. 1998). Intact microsomes were isolated by differential centrifugation from BHK cells, enriched in the complex-glycosylated wt or T70 CFTR by cyclohexamide chase, and subjected to limited proteolysis in the presence of an increasing concentration of trypsin. Immunoblot analysis of the proteolytic fragments with the L12B4 anti-CFTR Ab recognizing the NBD1 demonstrates a distinct proteolytic digestion pattern and a moderate, but reproducibly increased protease sensitivity of the complex-glycosylated T70 CFTR compared with wt CFTR (Fig. 8). Similar results were obtained with proteinase K and using the NBD2-specific, M3A7 anti-CFTR Ab (data not shown), indicating that not only the NBD2, but also the NBD1 conformation was altered upon truncating the COOH terminus.
Figure 8.
Protease susceptibility of native wt and T70 CFTR. After treating the cells with cyclohexamide (100 μg/ml) for 2.5 h at 37°C to deplete the core-glycosylated forms, microsomes were isolated with differential centrifugation from BHK-21 cells stably expressing wt or T70 CFTR. Microsomes were subjected to limited proteolysis at the indicated concentrations of trypsin for 15 min at 4°C. The proteolytic digestion pattern was probed by L12B4 anti-CFTR Ab and ECL.
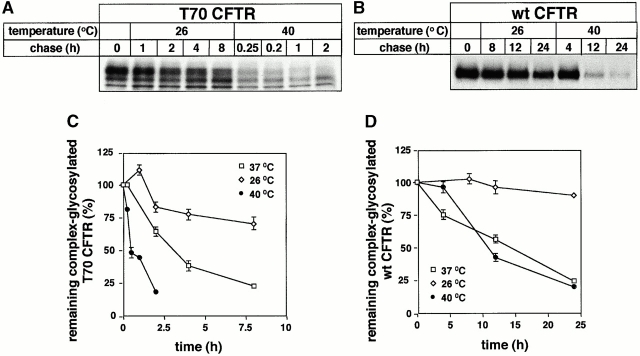
If the complex-glycosylated T70 CFTR was conformationally unstable, it was anticipated that the polypeptide in vivo thermostability would be compromised. The temperature dependence of the turnover of the complex-glycosylated wt and T70 CFTR was compared by metabolic pulse–chase studies. The turnover of the complex-glycosylated T70 CFTR was more than fivefold faster at 40°C (t 1/2 ∼ 0.5 h) than at 37°C (t 1/2 ∼ 2.7 h) (Fig. 9A and Fig. C). In sharp contrast, the turnover of the wt CFTR was only marginally affected at 40°C (t 1/2 ∼ 10.5 vs. 15 h at 37°C) (Fig. 9B and Fig. D). The simplest interpretation of these data is that the intrinsic structural instability of the T70 CFTR becomes more apparent at elevated temperature, leading to thermodenaturation and proteolysis, whereas the compact tertiary structures of wt CFTR are resistant to unfolding.
Figure 9.
Thermostability of the complex-glycosylated wt and T70 CFTR in vivo. (A and B) Temperature-dependent turnover of the complex-glycosylated T70 and wt CFTR. Metabolic stability of the pulse-labeled complex-glycosylated T70 and wt CFTR was monitored as described in the legend to Fig. 4. Chase was conducted at 26°C, 37°C, or 40°C. (C and D) The disappearance kinetics of complex-glycosylated T70 and wt CFTR were determined by PhosphorImage analysis in experiments shown in A and B. Data are means ± SEM, n = 3.
Discussion
A combination of transcriptional, cotranslational, and posttranslational mechanisms contributes to the ER quality control, preventing misfolded secretory and membrane proteins from traversing the secretory pathway (Ellgaard et al. 1999; Wickner et al. 1999; Mori 2000). Nevertheless, several examples demonstrate that the fidelity of the ER quality control is far from perfect. Nonfunctional polypeptides can escape ER retention, whereas mutants with partially or fully preserved biological function are being trapped, causing severe human diseases such as CF and alpha 1–protease inhibitor deficiency (Thomas et al. 1995; Zielenski and Tsui 1995; Aridor and Balch 1999; Schwartz and Ciechanover 1999). Nonnative polypeptides could also be generated in post-ER compartments as a result of unfolding upon environmental stresses or mutations that compromise the native form stability (Goldenberg 1992; Pacifici et al. 1993; Parsell and Lindquist 1993; Sitte et al. 1998). Little is known about the mechanism responsible for the degradation of abnormal membrane proteins from post-ER compartments.
In mammalian cells, aggregated furin, incompletely assembled Golgi coronavirus E1, and mutant connexin32 are destined for lysosomal degradation from the Golgi region without reaching the cell surface (Armstrong et al. 1990; Wolins et al. 1997; VanSlyke et al. 2000). A post-ER quality control mechanism appears to be responsible for targeting and vacuolar degradation of mutant plasma membrane ATPase, the late Golgi protease Kex2, and a model protein, comprising the secretory invertase and the NH2 terminus of the lambda repressor in Saccharomyces cerevisiae (Wilcox et al. 1992; Chang and Fink 1995; Hong et al. 1996). Aggregation of unoccupied class II major histocompatibility complex molecules and cross-linking of cell surface membrane proteins, such as the transferrin receptor, can evoke preferential lysosomal targeting and degradation (Weissman et al. 1986; Amigorena et al. 1995). Although most of these examples illustrate that nonnative membrane protein is targeted for lysosomal proteolysis either before their arrival to the cell surface or after their internalization, the complex-glycosylated truncated CFTR has a distinct intracellular fate.
We presented compelling evidence indicating that the dramatically decreased expression level and residence time of the complex-glycosylated truncated CFTR cannot be attributed to missorting for lysosomal degradation at the trans-Golgi or endosomal compartment. Instead, the truncated CFTR is targeted to the plasma membrane with comparable efficiency to its wt counterpart (Fig. 1). The turnover of the total and cell surface biotinylated T70 CFTR pools was insensitive to inhibitors of endolysosomal proteases. In contrast, weak bases and cathepsin inhibitors stabilized the full-length form and degradation intermediates of wt CFTR in transfected cells as well as in epithelia, highlighting the multiple role of endolysosomal proteolysis in the turnover of wt CFTR (Fig. 2 and Fig. 3).
Several criteria were used to establish that proteasomes are involved in the accelerated metabolism of the truncated CFTR from post-Golgi compartments, including the cell surface. First, the degradation rates of the complex-glycosylated T70, 82, and 98 CFTR were delayed in the presence of lactacystin, the most specific inhibitor of proteasome (Fenteany et al. 1995; Fig. 4). Second, a dramatic extension of the plasma membrane residence time of biotinylated T70 CFTR was evoked by lactacystin (Fig. 6), implying that proteasomes are indispensable in the proteolysis of the truncated CFTR from the cell surface and/or endosomal compartment. Since proteasome inhibitors had no impact on the degradation of Tac-Lamp1, an indirect effect of lactacystin on lysosomal proteolysis could be precluded (Fig. 5). Third, MG132 and lactacystin evoked the accumulation of polyubiqutinated adducts of the complex-glycosylated T70 CFTR, consistent with the notion that a fraction of the T70 CFTR is tagged by polyubiquitin chain(s) before degradation (Fig. 7). Finally, both MG132 and lactacystin partially restored the expression level of complex-glycosylated T70, T82, and T98 CFTR, as well as the plasma membrane cAMP-activated chloride conductance of T70 CFTR transfectants without facilitating the biogenesis of the mutants (Fig. 5D and Fig. E). Although these observations indicate that proteasome activity is one of the rate-limiting steps for the turnover of the full-length complex-glycosylated T70 CFTR, proteasome inhibitors failed to restore the mutant stability to that of the wt CFTR. This could be explained by incomplete inhibition of proteasome activity and/or the involvement of other, presently unidentified proteolytic mechanisms.
The COOH-terminal tail of CFTR might confer stability to the mature CFTR by several mechanisms or a combination thereof. Deletion of the COOH terminus, comprising a postsynaptic density-95, disc-large, and zonula occludens-1–binding motif (1476DTRL) that ensures the association of CFTR with EBP-50 (for ezrin-radixin-moesin–binding phosphoprotein 50) both in vitro and in vivo may facilitate lysosomal degradation by compromising endosomal sorting for recycling (Hall et al. 1998; Short et al. 1998; Moyer et al. 2000). A similar paradigm was proposed for the accelerated degradation of the β2-adrenergic receptor upon preventing its binding to EBP-50 (Cao et al. 1999). However, this possibility appears to be unlikely, since the steady state expression level and turnover rate of a CFTR variant lacking the COOH-terminal 26 amino acid residues are normal (Mickle et al. 1998; Haardt et al. 1999). Neither can aggregation of T70 CFTR at the trans-Golgi or at early endosomes target the mutant for lysosomal degradation. Finally, the absence of the COOH-terminal tail may structurally destabilize the folded CFTR, increasing the portion of nonnative molecules that are susceptible to proteolysis.
Evidence suggests that nonnative states of soluble polypeptides induced by mutations or thermal or chemical denaturation can lead to accelerated degradation (Parsell and Lindquist 1993; Hershko and Ciechanover 1998; Brodsky and McCracken 1999; Plemper and Wolf 1999), a paradigm that may apply to the degradation of nonnative plasma membrane proteins, including the truncated CFTR, as well. Based on the enhanced protease susceptibility and the decreased in vivo thermostability of T70 CFTR (Fig. 8 and Fig. 9), we speculate that structural destabilization of the native form is induced by disrupting interaction(s) of the COOH terminus. Using various yeast screening assays and site-directed mutagenesis of the Deg-1 proteolytic motif in the MATα2 transcription factor, it has been established that ubiquitin-conjugating enzymes (Ubc6 and Ubc7) can recognize hydrophobic stretches and exposed hydrophobic surfaces on amphipathic helices (Sadis et al. 1995; Gilon et al. 1998; Johnson et al. 1998). These or similar motifs, normally buried inside the globular domains of CFTR or inserted into the membrane plane, may serve as recognition signal(s). In support of this notion, the decreased plasma membrane residence time and impaired expression level of the β2- and the α2A-adrenergic receptors were also attributed to structural destabilization (Gether et al. 1997; Wilson and Limbird 2000). However, the mechanism responsible for the downregulation of these G protein–coupled receptors, which could be partially rescued with ligand stabilization, has remained obscure.
A key question that remains to be resolved is whether ubiquitination is a prerequisite for proteasome-dependent degradation of the mutant CFTR from the cell surface or endosomal compartment. Several tyrosine kinase and G protein–coupled receptors have been observed to undergo ligand-induced ubiquitination and downregulation via lysosomal and/or proteasomal degradation (Bonifacino and Weismann 1998; Hershko and Ciechanover 1998; Hicke 1999; Strous and Govers 1999). Ubiquitination also appears to be necessary and sufficient to induce the internalization and downregulation of plasma membrane receptors and transporters (e.g., FUR4, GAP1, STE2, STE3, and STE6) via vacuolar proteolysis in yeast (Hicke 1999). The constitutive downregulation of the epithelial sodium channel (ENaC), in association with the Nedd4 ubiquitin-ligase, was proposed to follow a similar degradation route, in contrast to the unassembled subunits of the channel, which were degraded by a proteasome-dependent mechanism at the ER (Staub et al. 1997).
Since both covalent and noncovalent attachment of ubiquitins can serve as a degradation signal of plasma membrane proteins via internalization (Govers et al. 1999; Shih et al. 2000), one possible scenario is that augmented ubiquitination of CFTR, or an associated protein, facilitates the internalization of the mutant. On the other hand, in light of multiple tyrosine- and dileucine-based endocytic signals in CFTR, ubiquitin attachment may not be a prerequisite for endocytosis (Lukacs et al. 1997; Prince et al. 1999; Hu et al. 2001). Consistent with the dispensable nature of ubiquitination, downregulation of the β2-adrenergic and mutant m2-muscarinic receptors can proceed without endocytosis, presumably by direct association with proteasome and/or other, presently unidentified proteases (Jockers et al. 1999; Wilson and Limbird 2000). The recognition that both 26S proteasome and P700 subunit are able to degrade nonnative polypeptides lacking polyubiquitin conjugates supports the notion that a ubiquitin-independent degradation mechanism cannot be ruled out (Murakami 1992; Braun et al. 1999; Sheaff et al. 2000; Strickland et al. 2000). Importantly, these degradation pathways are not mutually exclusive and may function in parallel.
Since the retrograde translocation of the T70 CFTR via the translocon is unlikely to occur in post-Golgi compartments (Xiong et al. 1999), the coordinated action of multiple proteolytic systems is invoked to resolve the topological problem of disposing transmembrane segments and exofacial loops of the T70 CFTR. Degradation appears to be initiated by the unfolding of cytosolic domains in a proteasome-dependent manner. The biochemical as well as functional rescue of cell surface resident T70, but not wt CFTR (data not shown), by proteasome inhibitors substantiate this hypothesis. Whereas the initial cleavage of the mature wt CFTR, as opposed to mutant, relies on the activity of lysosmal proteases, subsequent degradation of wt and T70 CFTR converges in endolysosomes, demonstrated by their overlapping proteolytic fragmentation pattern in the presence of cathepsin inhibitors (Fig. 3 B).
Although the truncated CFTR is one of the first mutant plasma membrane proteins subjected to sequential proteasomal and lysosomal breakdown, the coordinated action of these proteolytic systems is not without precedent. Although both lysosomes and proteasomes are involved in the constitutive and ligand-induced disposal of connexin43, the renal Na/Pi cotransporter, and the PDGF β receptor, respectively (Mori et al. 1995; Laing et al. 1997; Pfister et al. 1998), the recognition mechanism of these substrates by the proteasome remains unknown. Considering that deletion of the COOH-terminal tail appears to destabilize CFTR structurally, we propose that the ubiquitin-proteasome pathway may play a role in the recognition and elimination of not only the T70 CFTR, but also other nonnative or partially unfolded plasma membrane proteins.
Acknowledgments
We thank R. Haguenauer-Tsapis (Jacques Monod, University of Paris VII, Paris, France), J. Bonifacino (National Institutes of Health, Bethesda, MD), and M. Fukuda (The Burnham Institute, La Jolla, CA) for their generous gifts of cDNA. The authors are indebted to J. Brodsky and A. Weissman for helpful suggestions and J. Szapor for critical reading of the manuscript.
This work was supported by the Medical Research Council of Canada, the Canadian Cystic Fibrosis Foundation, and the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases. Instrumentation was covered in part by a block term grant from the Ontario Thoracic Society. M. Benharouga was supported by a Canadian Cystic Fibrosis Foundation Postdoctoral Fellowship. G.L. Lukacs was a scholar of the Medical Research Council of Canada.
Footnotes
Abbreviations used in this paper: Ab, antibody; BFA, brefeldin A; CF, cystic fibrosis; CFTR, CF transmembrane conductance regulator; ERAD, ER-associated degradation; HA, hemagglutinin; HA-Ub, HA-tagged ubiquitin; NBD, nuclear binding domain; wild-type, wt.
References
- Amigorena S., Webster P., Drake J., Newcomb J., Cresswell P., Mellman I. Invariant chain cleavage and peptide loading in major histocompatibility complex class II vesicles. J. Exp. Med. 1995;181:1729–1741. doi: 10.1084/jem.181.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M., Balch W.E. Integration of endoplasmic reticulum signaling in health and disease. Nat. Med. 1999;5:745–751. doi: 10.1038/10466. [DOI] [PubMed] [Google Scholar]
- Armstrong J., Patel S., Riddle P. Lysosomal sorting mutants of coronavirus E1 protein, a Golgi membane protein. J. Cell Sci. 1990;95:191–197. doi: 10.1242/jcs.95.2.191. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Walz J., Zuhi F., Seemuller E. The proteasomeparadigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Weismann A.M. Ubiquitin and the control of protein fate in the secretory and endocytic pathway. Ann. Rev. Cell Dev. Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Suzuki C.K., Klausner R.D. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science. 1990;247:79–82. doi: 10.1126/science.2294595. [DOI] [PubMed] [Google Scholar]
- Bradbury N.A. Intracellular CFTRlocalization and function. Physiol. Rev. 1999;79:S175–S192. doi: 10.1152/physrev.1999.79.1.S175. [DOI] [PubMed] [Google Scholar]
- Braun B.C., Glickman M., Kraft R., Dahlmann B., Kloetzel P.-M., Finley D., Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., McCracken A.A. ER-associated and proteasome-mediated protein degradationhow two topologically restricted events came together. Trends Cell Biol. 1997;7:151–156. doi: 10.1016/S0962-8924(97)01020-9. [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., McCracken A.A. ER protein quality control and proteasome-mediated protein degradation. Cell Dev. Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Cao T.T., Deacon H.W., Reczek D., Bretscher A., von Zastrov M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- Chang A., Fink G.R. targeting of the yeast plasma membrane H+-ATPasea novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.H., Gregory R.J., Marshall J., Paul S., Souza D.W., White G.A., O'Riordan C.R., Smith A.E. Defective intracellular transport and processing of CFTR is the molecule basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Molinari M., Helenius A. Setting the standardsquality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Fenteany G., Standaert R.F., Lane W.S., Chois S., Corey E.J., Schreiber S.L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Gether U., Ballesteros J.A., Seifert R., Sanders-Bush E., Weinstein H., Kobilka B.K. Structural instability of a constitutively active G protein-coupled receptor. J. Biol. Chem. 1997;272:2587–2590. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- Gilon T., Chomsky O., Kulka R.G. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae . EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg D.P. Mutational analysis of protein folding and stability. In: Creighton T.E., editor. Protein Folding. W.H. Freeman and Co; New York: 1992. pp. 353–403. [Google Scholar]
- Govers R., ten Broeke T., van Kerkhof P., Schwartz A.L., Strous G.J. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 1999;18:28–36. doi: 10.1093/emboj/18.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haardt M., Benharouga M., Lechardeur D., Kartner N., Lukacs G. C-terminal truncations destabilize the CFTR without impairing its biogenesis. J. Biol. Chem. 1999;274:21873–21877. doi: 10.1074/jbc.274.31.21873. [DOI] [PubMed] [Google Scholar]
- Hall R.A., Ostedgaard L.S., Premont R.T., Blitzer J.T., Rahman N., Welsh M.J., Lefkowitz R.J. A C-terminal motif found in the beta2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc. Natl. Acad. Sci. USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemar A., Subtil A., Lieb M., Morelon E., Hellio R., Dautry-Varsat A. Endocytosis of interleukin 2 receptors in human T lymphocytesdistinct intracellular localization and fate of the receptor α, β, and γ chains. J. Cell Biol. 1995;129:55–64. doi: 10.1083/jcb.129.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L. Gettin' down with ubiquitinturning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- Hirsh C., Ploegh H.L. Intracellular targeting of the proteasome. Trends Cell Biol. 2000;10:268–272. doi: 10.1016/s0962-8924(00)01768-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hong E., Davidson A.R., Kaiser C.A. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Howard M., Lukacs G. Multiple endocytic signals in the C-terminal tail of the CFTR. Biochem. J. 2001;354:561–572. doi: 10.1042/0264-6021:3540561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley S.M., Helnius A. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- Jensen T.J., Loo M.A., Pind S., Williams D.B., Goldberg A.L., Riordan J.R. Multiple proteolytic systems including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Jockers R., Angers S., Silva A.D., Benaroch P., Strosberg A.D., Bouvier M., Marullo S. β2-adrenergic receptor down-regulation. Evidence for a pathway that does not require endocytosis. J. Biol. Chem. 1999;274:28900–28908. doi: 10.1074/jbc.274.41.28900. [DOI] [PubMed] [Google Scholar]
- Johnson P.R., Swanson R., Rakhilina L., Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- Kartner N., Augustinas O., Jensen T.J., Naismith A.L., Riordan J.R. Mislocalization of DF508 CFTR in cystic fibrosis sweat gland. Nat. Genet. 1992;1:321–327. doi: 10.1038/ng0892-321. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Epping E.A., Moye-Rowley W.S. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 1999;19:2998–3009. doi: 10.1128/mcb.19.4.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima A., Sato A., Kawashima M., Nitta K., Yumura W., Sugino N., Nihei H., Natori Y. A simple procedure for the isolation of rat kidney lysosomes. Kidney Int. 1998;54:275–278. doi: 10.1046/j.1523-1755.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. Biosynthesis and degradation of CFTR. Rev. Physiol. 1999;79:S167–S173. doi: 10.1152/physrev.1999.79.1.S167. [DOI] [PubMed] [Google Scholar]
- Laing J.G., Tadros P.N., Westphale E.M., Beyer E.C. Degradation of connexin43 gap junctions involves both the proteasome and the lysosome. Exp. Cell Res. 1997;236:482–492. doi: 10.1006/excr.1997.3747. [DOI] [PubMed] [Google Scholar]
- Laney J.D., Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Loayza D., Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae . Mol. Cell. Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo M.A., Jensen T.J., Cui L., Hou Y.-X., Chang X.-B., Riordan J.R. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the protesome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N.T., Pedersen P.L. CFTRthe purified NBF1+R protein interacts with the purified NBF2 domain to form a stable NBF1+R/NBF2 complex while inducing a conformational change transmitted to the C-terminal region. Arch. Biochem. Biophys. 2000;375:7–20. doi: 10.1006/abbi.1999.1656. [DOI] [PubMed] [Google Scholar]
- Lukacs G.L., Chang X.B., Bear C., Kartner N., Mohamed A., Riordan J.R., Grinstein S. The DF508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. J. Biol. Chem. 1993;268:21592–21598. [PubMed] [Google Scholar]
- Lukacs G.L., Mohamed A., Kartner N., Chang X.B., Riordan J.R., Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (DF508) occurs in the ER and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs G.L., Segal G., Kartner N., Grinstein S., Zhang F. Constitutive internalization of cystic fibrosis transmembrane conductance regulator occurs via clathrin-dependent endocytosis and is regulated by protein phosphorylation. Biochem. J. 1997;328:353–361. doi: 10.1042/bj3280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M.S., Woodruff L., Ohno H., Bonifacino J.S. Protein targeting by tyrosine- and di-leucine–based signalsevidence for distinct saturable components. J. Cell Biol. 1996;135:341–354. doi: 10.1083/jcb.135.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G.C., Lu Z., King S., Sorscher E., Tousson A., Cyr D.M. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek M.T., Grant E.P., Rock K.L. Chemical denaturation and modification of ovalbumin alters its dependence on ubiquitin conjugation for class 1 antigen presentation. J. Immunol. 1996;157:617–624. [PubMed] [Google Scholar]
- Mickle J.E., Macek M., Jr., Fulmer-Smentek S.B., Egan M.M., Schwiebert E., Guggino W., Moss R., Cutting G.R. A mutation in the cystic fibrosis transmembrane conductance regulator gene associated with elevated sweat chloride concentration in the absence of cystic fibrosis. Hum. Mol. Genet. 1998;7:729–735. doi: 10.1093/hmg/7.4.729. [DOI] [PubMed] [Google Scholar]
- Mohamed A., Ferguson D., Seibert F.S., Cai H.M., Kartner N., Grinstein S., Riordan J.R., Lukacs G.L. Functional expression and apical localization of the cystic fibrosis transmembrane conductance regulator in MDCK I cells. Biochem. J. 1997;322:259–265. doi: 10.1042/bj3220259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A., Tanaka K., Omura S., Saito Y. Degradation process of ligand-stimulated platelet-derived growth factor β-receptor involves ubiquitin-proteasome proteolytic pathway. J. Biol. Chem. 1995;270:29447–29452. doi: 10.1074/jbc.270.49.29447. [DOI] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Moyer B.D., Duhaime M., Shaw C., Denton J., Reynolds D., Karlson K.H., Pfeiffer J., Wang S., Mickle J.E., Milewski M. The PDZ-interacting domain of CFTR is required for functional expression in the apical plasma membrane. J. Biol. Chem. 2000;275:27069–27074. doi: 10.1074/jbc.M004951200. [DOI] [PubMed] [Google Scholar]
- Murakami Y. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:348–351. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Naren A.P., Cormwt-Boyaka E., Fu J., Villain M., Blalock J.E., Quick M.W., Kirk K.L. CFTR chloride channel regulation by an interdomain interaction. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- Neville D.C., Rozans C.R., Tulk B.M., Townsend R.R., Verkman A.S. Expression and characterization of the NBD1-R domain region of CFTRevidence for subunit-subunit interactions. Biochemistry. 1998;37:2401–2409. doi: 10.1021/bi972021k. [DOI] [PubMed] [Google Scholar]
- Ostedgaard L.S., Rich D.P., DeBerg L.G., Welsh M.J. Association of domains within the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1997;36:1287–1294. doi: 10.1021/bi962174s. [DOI] [PubMed] [Google Scholar]
- Ostedgaard L.S., Puldarsoon O., Vomeer D.V., Welsh M.J., Gittes G.K. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc. Natl. Acad. Sci. USA. 2000;97:5657–5662. doi: 10.1073/pnas.100588797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici R.E., Kono Y., Davies K.J.A. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1993;268:15405–15411. [PubMed] [Google Scholar]
- Parsell D.A., Lindquist S. The function of heat-shock proteins in stress tolerancedegradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Pfister M.F., Ruf I., Stang G., Ziegler U., Lederer E., Biber J., Murer H. Parathyroid hormone leads to the lysosomal degradation of the renal type II Na+/Pi cotransporter. Proc. Natl. Acad. Sci. USA. 1998;95:1909–1914. doi: 10.1073/pnas.95.4.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pind S., Riordan J.R., Williams D.B. Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]
- Plemper R.K., Wolf D.H. Retrograde protein translocationERADication of secretory proteins in health and disease. Trends Biochem. Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- Prince L.S., Workman R.B., Jr., Marchase R.B. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc. Natl. Acad. Sci. USA. 1994;91:5192–5196. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince L.S., Peter K., Hatton S.R., Zaliauskiene L., Cotlin L.F., Clancy J.P., Marchase R.B., Collawn J.F. Efficient endocytosis of the CFTR requires tyrosine-based signal. J. Biol. Chem. 1999;274:3602–3609. doi: 10.1074/jbc.274.6.3602. [DOI] [PubMed] [Google Scholar]
- Riordan J.R. Cystic fibrosis as a disease of misprocessing of the cystic fibrosis transmembrane conductance regulator glycoprotein. Am. J. Hum. Genet. 1999;64:1499–1504. doi: 10.1086/302429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J.R., Rommens J.M., Kerem B.S., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L. Identification of the cystic fibrosis genecloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rommens J.M., Iannuzzi M.C., Kerem B.S., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N. Identification of the cystic fibrosis genechromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Sadis S., Atienza C.J., Finley D. Synthetic signals for ubiquitin-dependent proteolysis. Mol. Cell. Biol. 1995;15:4086–4094. doi: 10.1128/mcb.15.8.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A.L., Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Biochem. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- Seglen P.O. Inhibitors of lysosomal functions. Methods Enzymol. 1983;96:737–764. doi: 10.1016/s0076-6879(83)96063-9. [DOI] [PubMed] [Google Scholar]
- Seibert F., Linsdell P., Loo T.W., Hanrahan J.W., Riordan J.R., Clarke D.M. Cytoplasmic loop three of cystic fibrosis transmembrane conductance regulator contributes to the regulation of chloride channel activity. J. Biol. Chem. 1996;271:15139–15145. doi: 10.1074/jbc.271.44.27493. [DOI] [PubMed] [Google Scholar]
- Sheaff R.J., Singer J.D., Swanger J., Smitherman M., Roberts J.M., Clurman B.E. Proteasomal turnover of p21Clip1 does not require p21Clip1 ubiquitination. Mol. Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- Shih S.C., Sloper-Mould K.E., Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short D.B., Trotter K.W., Reczek D., Kreda S.M., Bretscher A., Boucher R.C., Stutts M.J., Milgram S.L. An apical PDZ protein anchors the CFTR to the cytoskeleton. J. Biol. Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- Sitte N., Merker K., Grune T. Proteasome-dependent degradation of oxidized proteins in MRC-5 fibroblasts. FEBS Lett. 1998;440:399–402. doi: 10.1016/s0014-5793(98)01495-1. [DOI] [PubMed] [Google Scholar]
- Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. Regulation of stability and function of the epithelial Na-channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland E., Hakala K., Thomas P.J., DeMartin G.N. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26 S proteasome. J. Biol. Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- Strous G.J., Govers R. The ubiquitin-proteasome system and endocytosis. J. Cell Sci. 1999;112:1417–1423. doi: 10.1242/jcs.112.10.1417. [DOI] [PubMed] [Google Scholar]
- Thomas P.J., Qu B.-H., Pedersen P.L. Defective protein folding as a basis of human disease. Trends Biochem. Sci. 1995;20:456–459. doi: 10.1016/s0968-0004(00)89100-8. [DOI] [PubMed] [Google Scholar]
- Van Oene M., Lukacs G.L., Rommens J.M. Cystic fibrosis mutations lead to carboxyl-terminal fragments that highlight an early biogenesis step of the CFTR. J. Biol. Chem. 2000;275:19577–19584. doi: 10.1074/jbc.M002186200. [DOI] [PubMed] [Google Scholar]
- VanSlyke J.K., Deschenes S.M., Musil L.S. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.L., Kopito R.R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- Ward C.L., Omura S., Kopito R.R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Weissman A.M., Klausner R.D., Rao K., Harford J.B. Exposure of K562 cells to anti-receptor monoclonal antibody OKT9 results in rapid redistribution and enhanced degradation of the transferrin receptor. J. Cell Biol. 1986;102:951–958. doi: 10.1083/jcb.102.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M.J., Smith A.E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- Wickner S., Maurizi M.R., Gottesman S. Posttranslational quality controlfolding, refolding and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Wilcox C.A., Redding K., Wright R., Fuller R.S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol. Biol. Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.H., Limbird L.E. Mechanisms regulating the cell surface residence time of the α2A-adrenergic receptor. Biochemistry. 2000;39:693–700. doi: 10.1021/bi9920275. [DOI] [PubMed] [Google Scholar]
- Wolins N., Bosshart H., Kuster H., Bonifacino J.S. Aggregation as a determinant of protein fate in post-Golgi compartmentsrole of the luminal domain of furin in lysosomal targeting. J. Cell Biol. 1997;139:1735–1745. doi: 10.1083/jcb.139.7.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Chong E., Skach W.R. Evidence that endoplasmic reticulum (ER)-associated degradation of CFTR is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Yang Y., Janich S., Cohn J.A., Wilson J.M. The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA. 1993;90:9480–9484. doi: 10.1073/pnas.90.20.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Kartner F., Lukacs G.L. Limited proteolysis as a probe for arrested conformational maturation of the F508 CFTR. Nat. Struct. Biol. 1998;5:180–183. doi: 10.1038/nsb0398-180. [DOI] [PubMed] [Google Scholar]
- Zielenski J., Tsui L.C. Cystic fibrosisgenotypic and phenotypic variations. Annu. Rev. Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]