Abstract
We identified the chromosomal addresses of a cohesin subunit, Mcd1p, in vivo by chromatin immunoprecipitation coupled with high resolution PCR-based chromosomal walking. The mapping of new Mcd1p-binding sites (cohesin-associated regions [CARs]) in single-copy sequences of several chromosomes establish their spacing (∼9 kb), their sequestration to intergenic regions, and their association with AT-rich sequences as general genomic properties of CARs. We show that cohesins are not excluded from telomere proximal regions, and the enrichment of cohesins at the centromere at mitosis reflects de novo loading. The average size of a CAR is 0.8–1.0 kb. They lie at the boundaries of transcriptionally silenced regions, suggesting they play a direct role in defining the silent chromatin domain. Finally, we identify CARs in tandem (rDNA) and interspersed repetitive DNA (Ty2 and subtelomeric repeats). Each 9-kb rDNA repeat has a single CAR proximal to the 5S gene. Thus, the periodicity of CARs in single-copy regions and the rDNA repeats is conserved. The presence and spacing of CARs in repetitive DNA has important implications for genomic stability and chromosome packaging/condensation.
Keywords: Mcd1p/Scc1p, chromatid cohesion, condensation, rDNA, transcription
Introduction
Higher order structural organization of mitotic chromosomes differs dramatically from interphase chromatin (Koshland and Strunnikov 1996). Accurate segregation of chromosomes during mitosis is aided by the acquisition of specific chromosomal structural attributes after DNA replication. One such attribute, sister chromatid cohesion, is established during S phase and results in the intimate association of newly replicated sister chromatids with each other at centromeres and along their entire lengths. Sister chromatids are more closely attached at centromeres than along arms such that centric regions are cytologically discernible as the site of the primary constriction in mammalian mitotic chromosomes. Differences in the extent of pairing and in the regulation of its dissolution, i.e., between centromere and arm regions of sister chromatids, suggest that cohesion is a complex and dynamic process (Bajer 1958; Miyazaki and Orr-Weaver 1994). Sister chromatid cohesion at the centromere orients sister kinetochores in an opposing configuration such that they can capture ends of microtubules emanating from opposite spindle poles. Regulated dissolution of sister chromatid cohesion at the metaphase to anaphase transition allows segregation of sister chromatids away from each other towards opposing spindle poles (Nicklas 1997).
Sister chromatid cohesion is mediated by cohesin, an evolutionarily conserved multisubunit complex (Losada et al. 1998; Koshland and Guacci 2000). This complex contains members of the evolutionarily conserved Smc (structural maintenance of chromosomes) family, which are abundant chromosomal proteins with long coiled-coil and ATP-binding domains, and also additional non-Smc subunits (for review see Hirano 1999) (Strunnikov and Jessberger 1999). The Saccharomyces cerevisiae cohesin complex contains Smc1p, Smc3p, Mcd1p/Scc1p, and Scc3p (Strunnikov et al. 1993; Guacci et al. 1997; Michaelis et al. 1997; Toth et al. 1999). Cohesin binds chromatin from S phase to mitosis, and the establishment of cohesion is coupled to DNA replication (Skibbens et al. 1999; Toth et al. 1999). Mutations in any of the cohesin subunits result in a precocious dissociation of sister chromatids. In addition, Mcd1p is required to maintain cohesion during mitosis, supporting a structural role for members of the cohesin complex as components of a molecular glue required for the maintenance of cohesion (Guacci et al. 1997).
Recent studies in budding yeast, using chromatin immunoprecipitation (ChIP), have established that cohesins bind to specific regions near centromeres and along arms, and this binding mediates cohesion (Blat and Kleckner 1999; Megee et al. 1999; Megee and Koshland 1999; Tanaka et al. 1999). However, questions remain. For example, what is the distribution of cohesin sites throughout the genome, including single-copy, tandemly repetitive, and dispersed repetitive DNA, and how does it influence their packaging and transmission? Also, differences have been observed in the behavior of cohesion at centromeres, telomeres, and arms. Do these regional differences reflect multiple distinct mechanisms of cohesion or differential regulation of a common cohesin-based mechanism? If the latter is true, then cohesin-binding sites should be present at centromeres, telomeres, and along arms.
The discovery that the origin recognition complex functions in silencing, as well as DNA replication, established precedent that chromosomal proteins may be usurped to function in several types of DNA metabolism (Bell et al. 1993; Foss et al. 1993). Indeed, this is likely to be the case for cohesins. Mutations in the Mcd1 and Smc1 cohesin subunits show defective condensation as well as precocious dissociation of sister chromatids (Guacci et al. 1997; Hogan, E., and D. Koshland, unpublished results). A mutation in the Smc1 cohesin subunit increases the size of the silent chromatin domain associated with HMR (Donze et al. 1999). To assess whether these phenotypes reflect a direct role of cohesins in condensation and gene silencing, it will be important to know whether the distribution of cohesin-binding sites is compatible with their function in these processes.
As a first step to examine cohesin site distribution in budding yeast, Blat and Kleckner 1999 performed an analysis of cohesin binding to the entire chromosome III. They hybridized probes made from DNA that coimmunoprecipitated with cohesins to a DNA array having ∼3-kb fragments of chromosome III (Blat and Kleckner 1999). This study helped to establish the existence of cohesin-binding sites, the periodicity between sites of ∼13 kb, the existence of an enrichment of cohesins near the centromere, and the absence of cohesins at telomeres. However, this approach was unable to address many of the questions raised above because of experimental limitations. The average size of the DNA fragments on the microarray was ∼3 kb, which is very large compared with many of the important genomic features like open reading frames (ORFs), intergenic regions, or interspersed repetitive DNA. In addition, it is unclear whether the microarray assay is sensitive enough to detect chromosomal regions that give low signal by ChIP, which complicates the interpretation of the absence of cohesin sites at telomeres. Finally, chromosome III lacks any tandem repetitive DNA. Thus, whereas the microarray approach has been very informative, many questions about cohesin site distribution remain unanswered.
To obtain a representative picture of mitotic chromosome organization at a high resolution and to examine the relationship of cohesin-binding sites with other chromosomal functional domains (e.g., transcription units, silenced domains, and replication units), we have used ChIP coupled with high resolution PCR-based chromosomal walking to construct a detailed and comprehensive map of Mcd1p-binding sites on ∼50 kb of DNA of selected regions of S. cerevisiae chromosomes, including single-copy DNA, tandem (RDN1) repeats, interspersed repeats (Ty long terminal repeats [LTRs]), centromeric and pericentric regions, subtelomeric repeats at chromosome ends, and single-copy (HMR) as well as repetitive (RDN1) silenced regions of chromosome III and XII arms.
Materials and Methods
Yeast Strains
The strains used were 1377A1-4B (MATa MCD1-6HA leu2 ura3) and 983-8A (MATa leu2 ura3). The 6X-HA–tagged allele of MCD1 is the same as reported previously (Megee et al. 1999).
ChIP and PCR Analysis
Exponentially dividing cell cultures were arrested in G1, S, or M phases by the addition of 10−6 M α-factor, 100 mM hydroxyurea (HU), or 15 μg/ml nocodazole, respectively. Cell cycle arrest was confirmed by scoring for distribution of various cell types.
ChIPs were done as described previously (Meluh and Koshland 1997), except that formaldehyde cross-links were reversed for 8 h at 65°C. Immunoprecipitations were done with 12CA5 anti-HA antibody from Babco or Boehringer.
Oligonucleotide primers for PCR amplification were synthesized by Genosys Biotechnology, Inc. Primers for amplification of centromere (CEN) and CEN-proximal sequences have been described previously (Meluh and Koshland 1997). PCRs were performed with 1:400 diluted total samples or with 1:4 diluted anti-HA immunoprecipitated samples using a PTC-200 Peltier thermal cycler from MJ Research. 26 cycles of PCR amplifications were done to analyze unique chromosomal sequences and 22 cycles to analyze RDN1 repeat sequences. Either 3 μl (for unique sequences) or 1 μl (for RDN1 sequences) of diluted template DNA was used to program a 25-μl PCR reaction. PCR products were resolved on 3% NuSieve® agarose gels in 1× TBE buffer with 0.08 μg/ml ethidium bromide. Gel images were acquired using the UVP Laboratory Products GDS 7500 digital imaging gel documentation system, and the band intensities were quantified by using ImageQuant software.
All experiments were done at least twice and one representative dataset is shown.
Sequence Analysis
All chromosomal sequences were downloaded from the Saccharomyces Genome Database website (http://genome-www.stanford.edu/Saccharomyces/). The coordinates for various genomic regions examined are as follows: chromosome XII (439,214–472,373), which includes two contiguous RDN1 repeats (RDN1-1 and RDN1-2, 451,474–468,931), chromosome III arm (88,966–102,771), chromosome III HMR (287,944–297,066), and the chromosome III right end (304,155–314,911).
Oligonucleotide primers for PCR-based chromosomal walks were designed using Macvector 6.5 software so as to generate a series of overlapping fragments ranging in size from ∼150–500 bp. Detailed information about the various primers is available upon request. The Saccharomyces Genome Database coordinates plotted in Mcd1p-binding distributions correspond to the midpoints of the PCR fragments.
The properties of DNA sequences corresponding to Mcd1p-binding peaks were further characterized by Macvector 6.5.
Results
A High Resolution Approach To Identify Chromosomal Addresses of Mcd1p
We searched for chromosomal sequences associated with Mcd1p in vivo using ChIP (Meluh and Koshland 1997) coupled with quantitative PCR-based chromosomal walking. Cells having an epitope-tagged functionally competent version of Mcd1p, Mcd1-6HAp (Megee et al. 1999), were arrested in mid-M phase with nocodazole, and then formaldehyde–cross-linked chromatin, which was sheared to a size of ∼0.2–1 kb, was prepared. Chromatin associated with Mcd1-6HAp was immunoprecipitated by mAbs recognizing the HA epitope. Specific DNA sequences coimmunoprecipitated with the Mcd1-6HAp–containing chromatin immunoprecipitates were detected by amplification using PCR. We designed sets of PCR primers so as to produce a series of adjacent, partially overlapping PCR products ranging in size from 150–500 bp spanning selected regions on chromosomes III and XII (Fig. 1 A). By using PCR products of this size, the resolution of cohesin mapping is as much as 10 times greater than the previous microarray analysis (Blat and Kleckner 1999). We quantified the abundance of various immunoprecipitated DNA sequences by comparing the band intensities of PCR fragments in the immunoprecipitates with PCR fragments generated using a 100-fold dilution of the original chromatin solution, such that signals from both samples were within the linear range of the assay.
Figure 1.
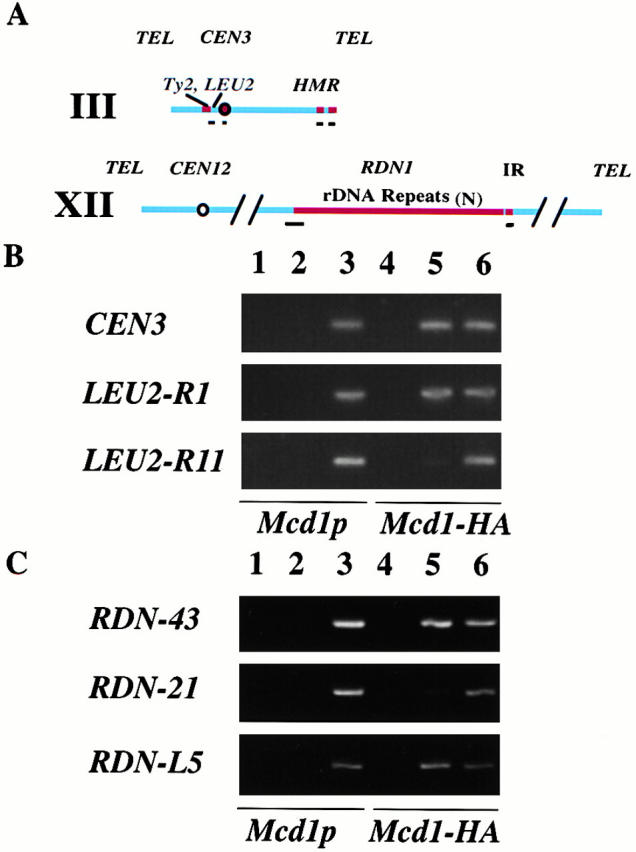
Mcd1p is associated specifically with chromosomal sequences. PCR analysis of total or coimmunoprecipitated DNA from ChIP of untagged (Mcd1p) and 6XHA-tagged MCD1 (Mcd1-HA) strains. (A) A map of chromosomes III and XII showing the regions examined. Black bars below the chromosome maps denote regions that were analyzed by PCR amplification of the chromatin immunoprecipitates. (B) The binding of Mcd1p to chromosome III centromere (CEN3) and arm (LEU2-R1 and R11) sequences. 26 cycles of PCR amplification were done using 4- or 400-fold diluted immunoprecipitated or total samples, respectively. (C) Binding of Mcd1p to selected sites (PCR fragments RDN-43 and RDN-21) on the highly repetitive rDNA and to a single-copy region (PCR fragment RDN-L5) to the left of RDN1 on chromosome XII. 22 cycles of PCR amplification were done using 4- or 400-fold diluted immunoprecipitated or total samples, respectively. Lanes: 1 and 4, mock immunoprecipitations; 2 and 5, α-HA immunoprecipitations; and 3 and 6, total chromatin solution (whole cell extract). No antibody was added in the mock immunoprecipitations.
Before performing an extensive analysis of Mcd1-6HAp binding to a large region, we first wanted to ensure that the binding we observed by ChIP was specific under our experimental conditions. Centromere DNA from chromosomes III (Fig. 1 B) and XVI (not shown) could be specifically coimmunoprecipitated with Mcd1-6HAp, as has been reported previously (Megee et al. 1999; Tanaka et al. 1999). We asked whether specific DNA fragments corresponding to a single-copy region on the chromosome III arm, near the LEU2 gene, were enriched in Mcd1-6HAp immunoprecipitates. A fragment corresponding to a region near the 3′ end of the LEU2 gene (R1) was specifically immunoprecipitated with anti-HA antibodies from the Mcd1-6HA strain, but was undetectable in mock immunoprecipitations and in mitotically arrested cells with untagged Mcd1p (Fig. 1 B). In contrast, DNA from an adjacent region (R11) could not be detected, establishing that the assay conditions were suitable for identifying Mcd1p-binding sites on a larger scale.
A similar study, which used fewer PCR amplification cycles with two PCR fragments corresponding to different regions in the RDN1 repeat, also established the possibility of identifying specific Mcd1-binding sites in this highly repetitive locus (Fig. 1 C). DNA corresponding to PCR fragment 43 was enriched, whereas PCR fragment 21 was relatively less abundant in Mcd1-6HAp immunoprecipitates. Similarly, a single-copy sequence on the chromosome XII arm also specifically coimmunoprecipitated with Mcd1-6HAp (Fig. 1 C, L5).
Binding Profile of Mcd1p on RDN1 and Adjoining Regions of Chromosome XII
The tandemly repeated rDNA locus on chromosome XII has been shown to be a target site for both cohesin and condensin action in vivo (Guacci et al. 1997; Freeman et al. 2000; Lavoie et al. 2000; Hogan, E., and D. Koshland, unpublished results). The rDNA locus (RDN1) on chromosome XII consists of a series of tandem iterations of 9.1-kb DNA segments repeated ∼100–150 times in different yeast strains. The RDN1 repeat contains the genes for 5S, 5.8S, 25S, and 18S rRNAs (Fig. 2 A). We designed a set of 96 primers to amplify 48 overlapping PCR fragments spanning the RDN1 repeat. These primers amplified PCR fragments from several RDN1 repeats, as demonstrated by the requirement for fewer PCR cycles to obtain a signal equivalent to single-copy regions (see Materials and Methods). The abundance of various fragments in Mcd1-6HA immunoprecipitates from mitotic cells was quantified relative to the total lysate, as in Fig. 1 C. The enrichment of fragments R39–R45 reveals the presence of a specific Mcd1-6HAp–binding site in the corresponding region (Fig. 2 B). A complete binding profile of Mcd1-6HAp on the RDN1 repeat (Fig. 2 C) demonstrates that there is only one prominent binding site per repeat. The site is located in the nontranscribed spacer (NTS)2, between the 5′ ends of the 35S and 5S RNA-encoding genes. The right slope overlaps with the 5S RNA gene. Because the signal corresponding to the RDN1 region represents an average of all the repeats, the observation that the height of this peak is of a similar magnitude (within 15–20%) to peaks on adjoining single-copy regions (Fig. 1 C and 2 C) suggests that this site must be occupied by Mcd1-6HAp on a majority of the RDN1 repeats.
Figure 2.
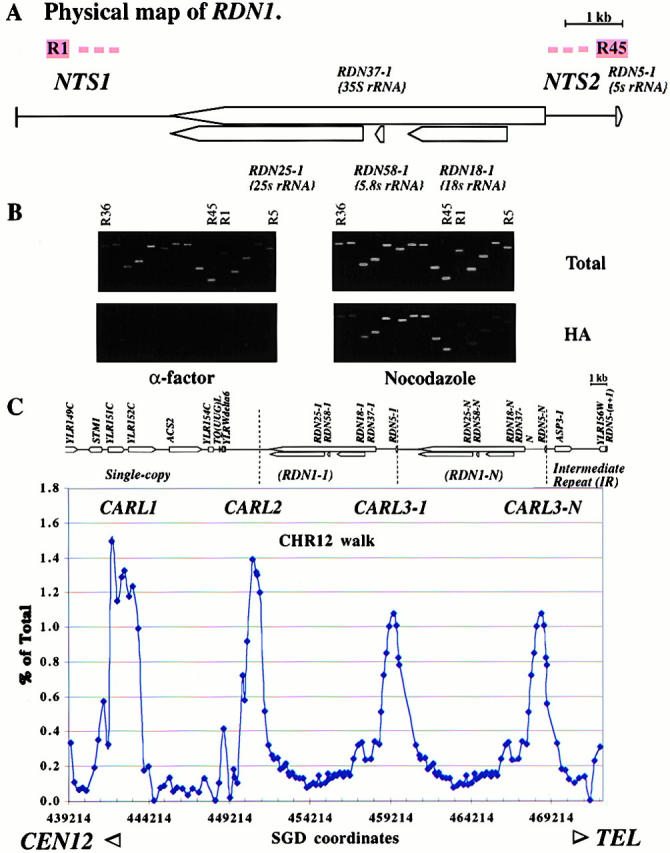
Distribution of Mcd1p-binding sites on RDN1 and neighboring unique and intermediate repetitive sequences on chromosome XII. (A) A physical map of the RDN1 repeat unit showing various sequence features. R1 and R45, PCR fragments; NTS1 and NTS2, nontranscribed spacers 1 and 2. (B) An Mcd1p-binding site on the repetitive rDNA locus, RDN1, on chromosome XII. PCR analysis of total or Mcd1p coimmunoprecipitated DNA (HA) from α-factor or nocodazole arrested cell lysates of a region of the rDNA repeat, including the Mcd1p-binding site (PCR fragments R39–R45). PCR fragments shown are R36, R37…R44, R45, and R1…R5. (C) Mcd1p binding profile on a region of chromosome XII including a single-copy region, RDN1, and an IR. The physical map of the selected region shows ORFs and other sequence features. Two copies of the RDN1 repeat are shown for clarity, assuming that a majority of repeats are occupied. The amount of DNA immunoprecipitated estimated as the percent of total DNA is plotted on the y axis versus the position of the midpoint of each fragment on chromosome XII in bp on the x axis.
We also extended our analysis of Mcd1-6HAp binding to an adjoining single-copy region on chromosome XII to the left of the RDN1 repeats and to a moderately repeated region (3.6-kb intermediate repeat [IR], including the ASP3 gene) to the right of the RDN1 repeats (Fig. 2 C). On the left side, two sites were identified. The first site lies in the nontranscribed region very near the junction of single-copy sequences with the beginning of RDN1 repeats (Fig. 2 C). The second site is located 8 kb away and overlaps with two ORFs. No significant binding site was seen in the IR. The binding profile of Mcd1-6HAp on the selected regions of chromosome XII (Fig. 2 C) shows a regular spacing of prominent peaks, separated by intervening sequences (putative loops) that range in size from 8–9 kb. The spacing is maintained from single-copy sequences, across the boundary of unique and repetitive DNA and into the tandemly repetitive RDN1 array.
We suggest a system of nomenclature for identifying various cohesin-binding sites on chromosomes. Cohesin-associated regions can be abbreviated as CARs (by analogy to previously defined scaffold-associated regions or SARs and matrix-associated regions or MARs, which are presumptive chromosomal structural determinants defined by virtue of their association with the chromosomal scaffold or matrix) followed by a letter designating the chromosome (e.g., 12th letter L for chromosome XII, 3rd letter C for chromosome III, etc.) and a number unique for a particular CAR. Accordingly, we have named the above Mcd1p-binding regions on chromosome XII as CARL1, CARL2, and CARL3 (Fig. 2 C).
Selective Association of Mcd1p with Specific Chromosome III Sequences
The binding profile of Mcd1-6HAp on chromosome III reveals several interesting features (Fig. 3 and Fig. 4). We identified two peaks on a 12-kb unique region of the left arm: one is located in the intergenic region between BUD3 and GBP2 (CARC1) and the other overlaps the 3′ ends and intergenic region between LEU2 and NFS1 (CARC2). In agreement with a previous study (Blat and Kleckner 1999), we noted a more pronounced binding to the BUD3/GBP2–associated peak, CARC1, which is located ∼10 kb to the left of CEN3, compared with the other peaks (Fig. 4 B). Our high resolution analysis reveals that more Mcd1-6HAp is also bound to the intervening region immediately to the left of CARC1, raising the possibility that the dramatic abundance of Mcd1-6HAp binding in pericentric regions (Blat and Kleckner 1999) may arise from a sum of signals from higher binding peaks as well as higher valleys in these regions (Fig. 3 B). We also analyzed Mcd1-6HAp binding to a part of the Ty2-1 retrotransposon, a dispersed repetitive element present in the yeast genome. One copy of this retrotransposon (YCLWTY2-1) is present on the left of the LEU2 gene on chromosome III. Mcd1-6HAp is associated with a region within the LTR of the Ty2-1 retrotransposon. Because the signal is an average from various dispersed copies of the retrotransposon, it is unclear whether the Mcd1p-coimmunoprecipitated signal comes from the copy of Ty2 on chromosome III or other copies elsewhere in the genome.
Figure 3.
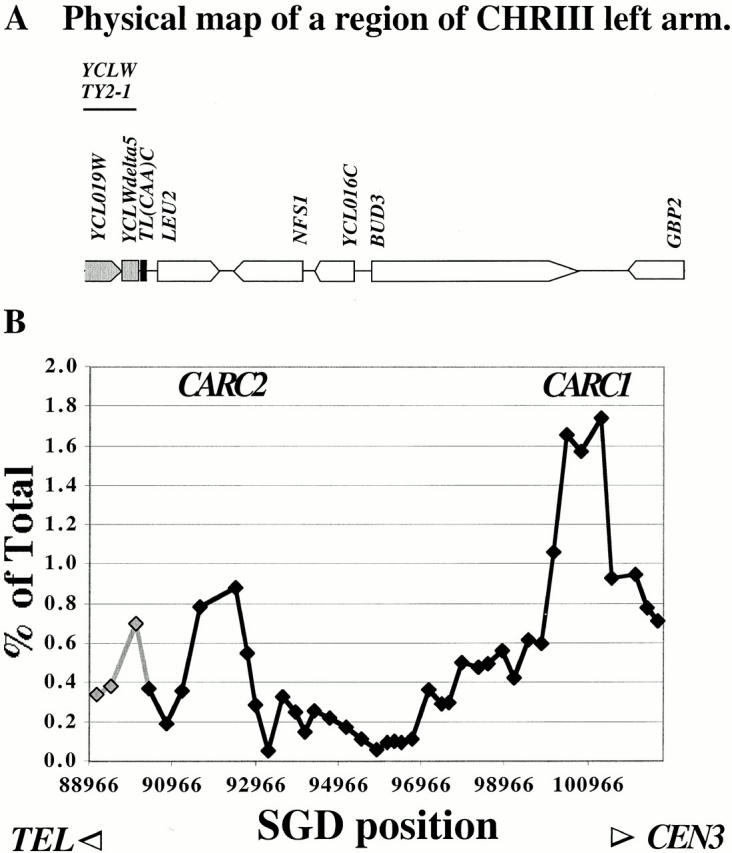
High resolution analysis of Mcd1p-binding sites on a region of chromosome III arm. (A) Physical Map of a region of the left arm of chromosome III, including LEU2 and other ORFs. Part of a full Ty2 retrotransposon inserted to the left of the LEU2 gene is shaded in gray. (B) Binding profile of Mcd1p on chromosome III arm. The amount of immunoprecipitated DNA (as percent of total) is plotted on the y axis versus DNA sequence coordinates on the x axis. The DNA sequence coordinates are aligned to the physical map in A. Gray shaded data points and line correspond to repetitive fragments within the Ty element.
Figure 4.
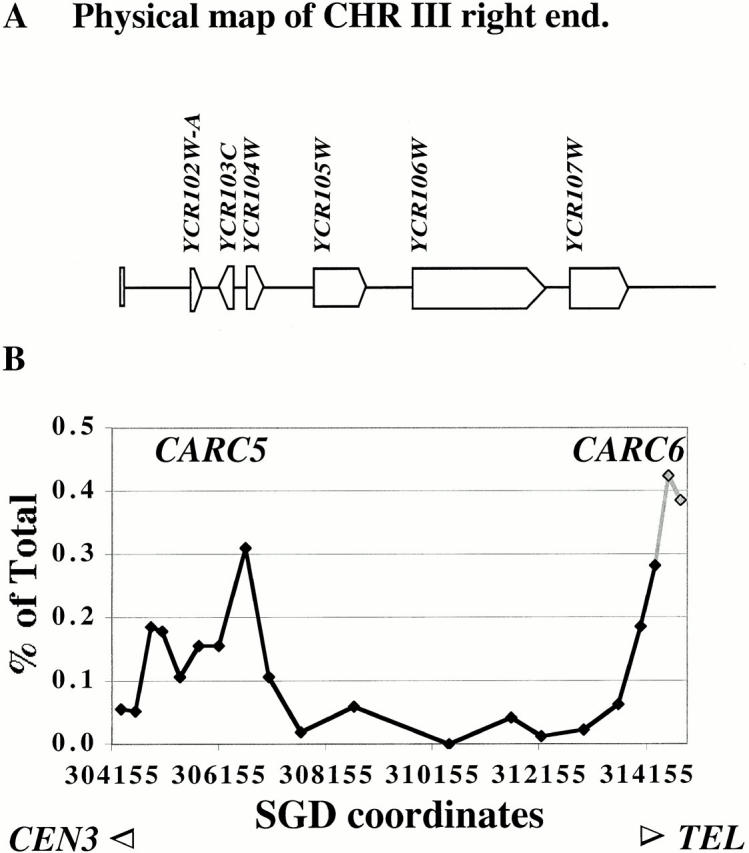
Binding of Mcd1p to subtelomeric sequences of the right arm of chromosome III. (A) Map of the right end of chromosome III showing various features. (B) The binding profile of Mcd1p on the right end of chromosome III. The amount of immunoprecipitated DNA (as percent of total) is plotted on the y axis versus DNA sequence coordinates on the x axis. The DNA sequence coordinates are aligned to the physical map in A. The gray shaded part of the line corresponds to the repeated segment.
We investigated Mcd1-6HAp binding to an ∼10-kb region near the right end of chromosome III at high resolution (Fig. 4). We detected very low binding in this region, but noted an enrichment of two subtelomeric repetitive fragments and adjacent single-copy fragments relative to other nearby fragments. This demonstrates that Mcd1-6HAp binds to the right end of chromosome III (CARC6) and also to a subtelomeric repetitive element present near the telomere of several chromosomes. A modest enrichment of fragments, which correspond to a region ∼8 kb away (CARC5) from the repeat element, was also observed, hinting at the presence of a less prominent binding site in this region. A previous study (Blat and Kleckner 1999) reported a low abundance of cohesin at the left end of chromosome III and suggested a conserved pattern of low cohesin binding at chromosome ends. Our results indicate that the overall organization (with respect to occurrence and distribution of cohesin-binding sites) towards the ends of chromosome arms is similar to other regions, but both binding peaks and intervening valleys show lower levels of cohesin binding.
Association of Mcd1p with the Silenced HMR Locus
Our identification of Mcd1-6HAp–binding sites at the border of the silenced RDN1 array (Fig. 2 C) and near the telomere (Fig. 4) was intriguing because both of these regions can exist in a silent chromatin state. This observation prompted us to further investigate the nature of Mcd1-6HAp association with silenced regions. The HMR locus is a well-characterized silenced locus in S. cerevisiae. HMR is flanked by two silencer elements, HMR-E and HMR-I, which recruit silent information regulator proteins to the silent loci. The binding of silent information regulator proteins to nucleosomes generates a heterochromatin domain that is transcriptionally repressed and spreads for a limited distance beyond the silencers. The spread of the silent heterochromatin domain is limited by the presence of boundary elements in the DNA flanking HMR (Donze et al. 1999). Boundary element function is compromised in an smc1-2 mutant, suggesting an involvement of another cohesin component Smc1p in limiting the spread of silenced heterochromatin (Donze et al. 1999).
We investigated the spatial relationship between Mcd1-6HAp binding and the previously mapped boundary elements by PCR amplification of fragments, which correspond to the left and right boundaries and within HMR, from DNA coimmunoprecipitated with Mcd1-6HAp (Fig. 5). Several PCR fragments corresponding to HMR-flanking regions overlapping the boundaries (corresponding to CARC3 and CARC4) were more abundant than a PCR fragment within HMR in the Mcd1-6HAp chromatin immunoprecipitates, which is consistent with the association of Mcd1-6HAp with HMR boundaries.
Figure 5.
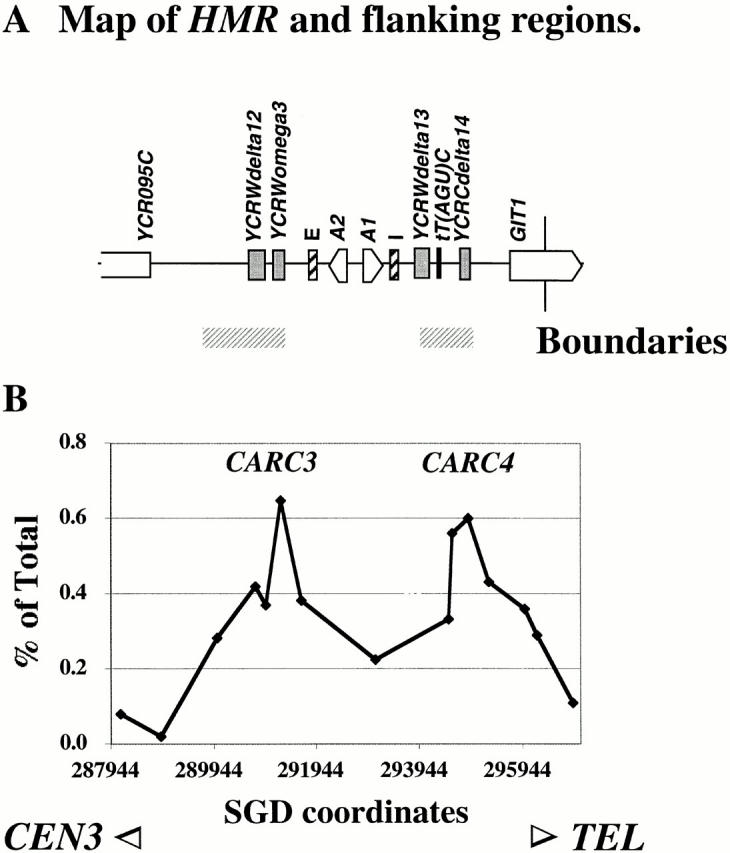
Association of HMR boundary elements with Mcd1p. (A) Map of HMR showing ORFs, silencers (I and E), and other sequence features. Previously mapped locations of the genetically defined boundaries are shown by horizontal gray bars below the map. (B) Quantification of Mcd1p binding to HMR and boundaries in mitotic cells. The amount of immunoprecipitated DNA (as percent of total) is plotted on the y axis versus DNA sequence coordinates on the x axis. The DNA sequence coordinates are aligned to the physical map in A.
Assessing the Size of the Mcd1p-binding Domain on Chromosomal Arm Sites
The Mcd1-6HAp–binding profiles defined by our high resolution PCR walk on chromosomal arms reveal that most prominent peaks span 0.8–1.0 kb, with an exceptional site of 1.5 kb. This broad range may be a real attribute of the association of Mcd1-6HAp with chromatin, resulting from the binding of cohesin to a broad region of DNA at the cohesin-binding site, or it may be apparent, due to a limitation of the method used for chromatin fragmentation before immunoprecipitation. The sheared chromatin prepared by sonication for the ChIP assay consists of a distribution of overlapping DNA fragments of various sizes (∼0.2–1 kb). It is possible that the broad distribution of Mcd1-6HAp at a binding site may arise from the immunoprecipitation of bigger chromatin fragments that are largely free of Mcd1-6HAp–binding sites, but have a small overlap at one end with a binding region, owing to their proximity to the binding site.
To resolve this issue, we compared the width of various CARs with the region associated with another chromosomal protein Mif2p, whose association with chromatin is known to be limited to a 120-bp centromere DNA (Meluh and Koshland 1997). Mif2p was immunoprecipitated from the same formaldehyde-fixed and sheared chromatin that was used for the high resolution localization of Mcd1-6HAp–binding sites. The association of Mif2p with CEN3 and flanking regions was measured by PCR amplification of a set of adjacent 300-bp fragments with small overlaps, corresponding to this region (Meluh and Koshland 1997). The width of the Mif2p-binding peak on the centromere was dramatically narrower than that of various Mcd1-6HAp–binding peaks on chromosome XII (Fig. 6). Mif2p binding to the CEN3 fragment was very specific and the level of Mif2p binding fell off precipitously on each side of the central CEN3 containing fragment. This demonstrates that the size distribution of chromatin fragments in our sheared chromatin preparations is appropriate for the detection of a highly specific protein–DNA interaction limited to a 300-bp DNA fragment. From these observations we conclude that the size of the cohesin sites is 0.8–1.0 kb ± 300 bp.
Figure 6.
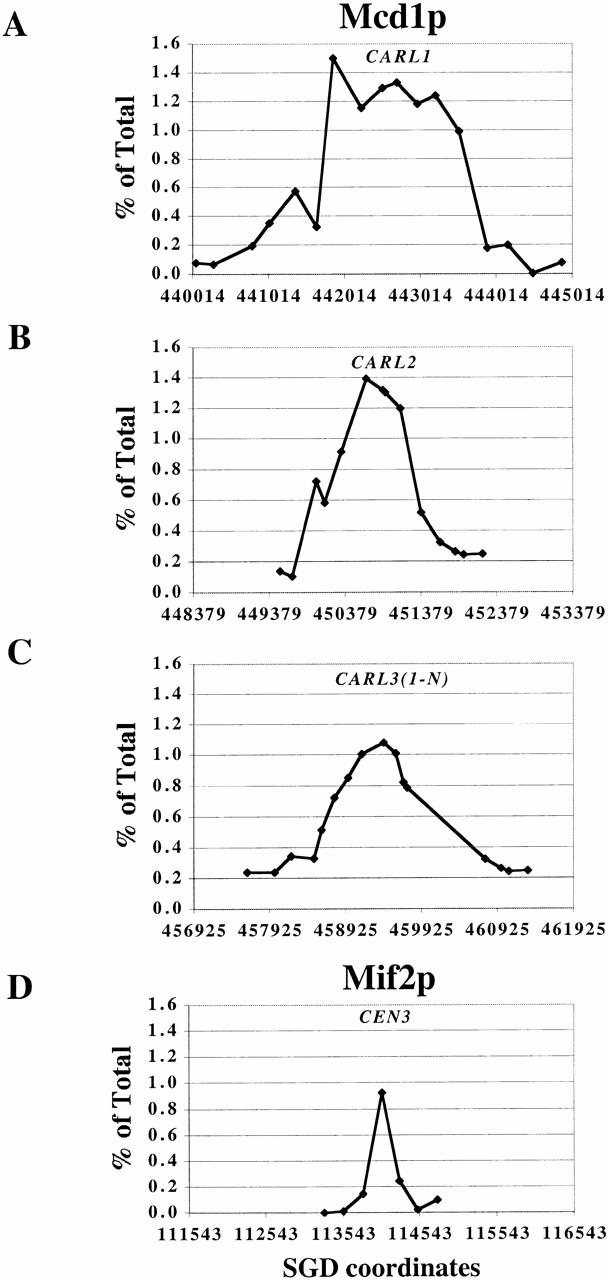
Spreading of Mcd1p on various chromosomal arm sites as revealed by comparing its binding profile (top three graphs) on three CARLs on chromosome XII to the distribution of Mif2p binding to CEN3 and flanking sequences (bottom graph). The amount of immunoprecipitated DNA (as percent of total) is plotted on the y axis versus DNA sequence coordinates on the x axis. The scale of the x axis is constant in all graphs (total: 5 kb; minor division ∼1 kb).
Sequence Characteristics of Mcd1p-binding Sites
A correlation between the distribution of cohesin-binding sites and AT-rich regions has been reported previously based on a global but low resolution study of cohesin binding to chromosome III (Blat and Kleckner 1999) and a higher resolution study of Mcd1-6HAp–binding profile on an artificial minichromosome (Megee et al. 1999). Our high resolution analysis of Mcd1-6HAp binding on the arms of chromosomes III and XII allowed us to take a magnified look at Mcd1-6HAp–binding sites and reexamine the robustness of this correlation in greater detail by using smaller (Fig. 7, 100 bp, bottom) sliding windows to estimate variations in AT content. Consistent with earlier studies, our study reveals that cohesin-binding sites are not found in regions having a consistently low AT content and most sites have peaks >60% AT. Although some sites are near peaks of high AT content (∼80% for CARL2, CARC1, and CARC6), this correlation is not very strong because such peaks of AT richness are also seen in several regions having very low or no cohesin binding. Some of the AT-rich peaks do not coincide with the middle of the Mcd1-6HAp–binding peaks, but are located towards one side (e.g., CARL2). Notably, in the case of the RDN1 repeat, an AT-rich peak (∼80% AT), which is located in nearly the middle of the repeat, shows very low cohesin binding, whereas the Mcd1-6HAp–binding site (CARL3) has slightly lower AT-rich peaks. Thus, the percent AT composition is not the only determinant of cohesin binding in the regions we examined, and other specific features of the sequence may contribute to cohesin binding.
Figure 7.
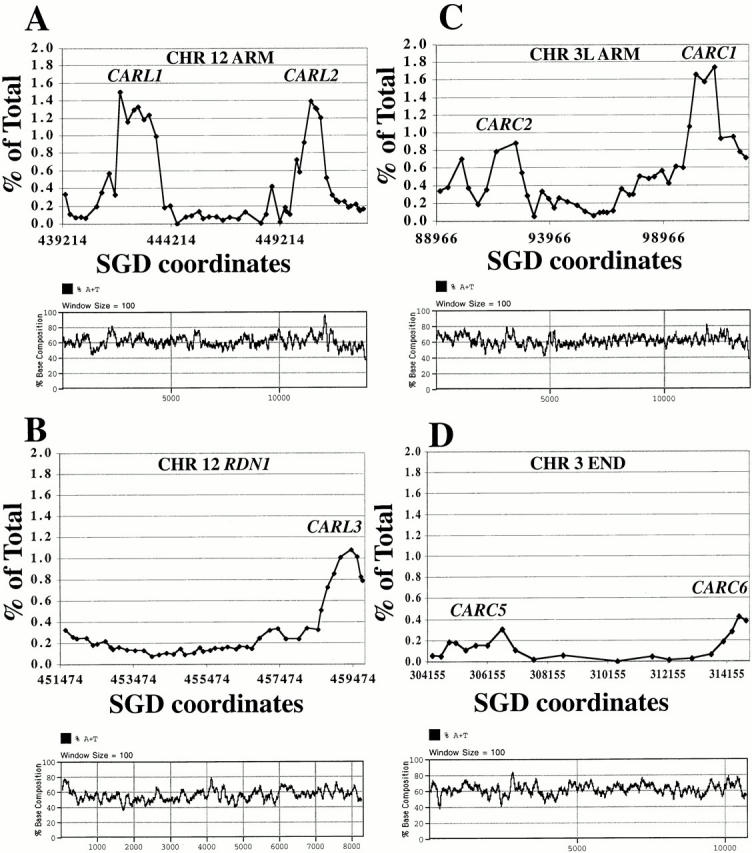
Comparison of characteristics of various Mcd1p-binding sites. AT content of various chromosomal regions aligned with respective Mcd1p-binding profiles. Top graphs are Mcd1p-binding profiles on selected regions of chromosomes XII (left) or 3 (right). Bottom graphs in each set show fluctuations in base composition (AT content on the y axis) revealed by using a sliding window of 100 bp (x axis).
Cell Cycle Dependence of Mcd1p Binding to Centromeres and Arm Sites
A previous global study of cohesin binding (Blat and Kleckner 1999) showed that cohesin binding at the centromere and pericentric regions of chromosome III is dramatically greater in M phase arrested cells compared with S phase arrested cells. It was suggested that this increase in centromere binding arises from the differential loss of cohesins from chromosome III arms. We reexamined this question by comparing high resolution Mcd1p-binding profiles in cell cycle arrested cells. In cells synchronized in G1, using α-factor, no specific chromosomal sequences were detectable in the Mcd1-6HAp chromatin immunoprecipitates (data not shown), presumably due to the undetectable levels of Mcd1p in α-factor arrested cells (Guacci et al. 1997). Mcd1-6HAp–specific chromatin immunoprecipitates from cells arrested in early S phase by HU or in mid M phase by nocodazole, a microtubule destabilizing agent, were analyzed by PCR to compare the levels of Mcd1-6HAp binding at CEN3 and pericentric regions (Fig. 8 A), chromosome III arm sites (Fig. 8 B), and within the RDN1 repeat (Fig. 8 C).
Figure 8.
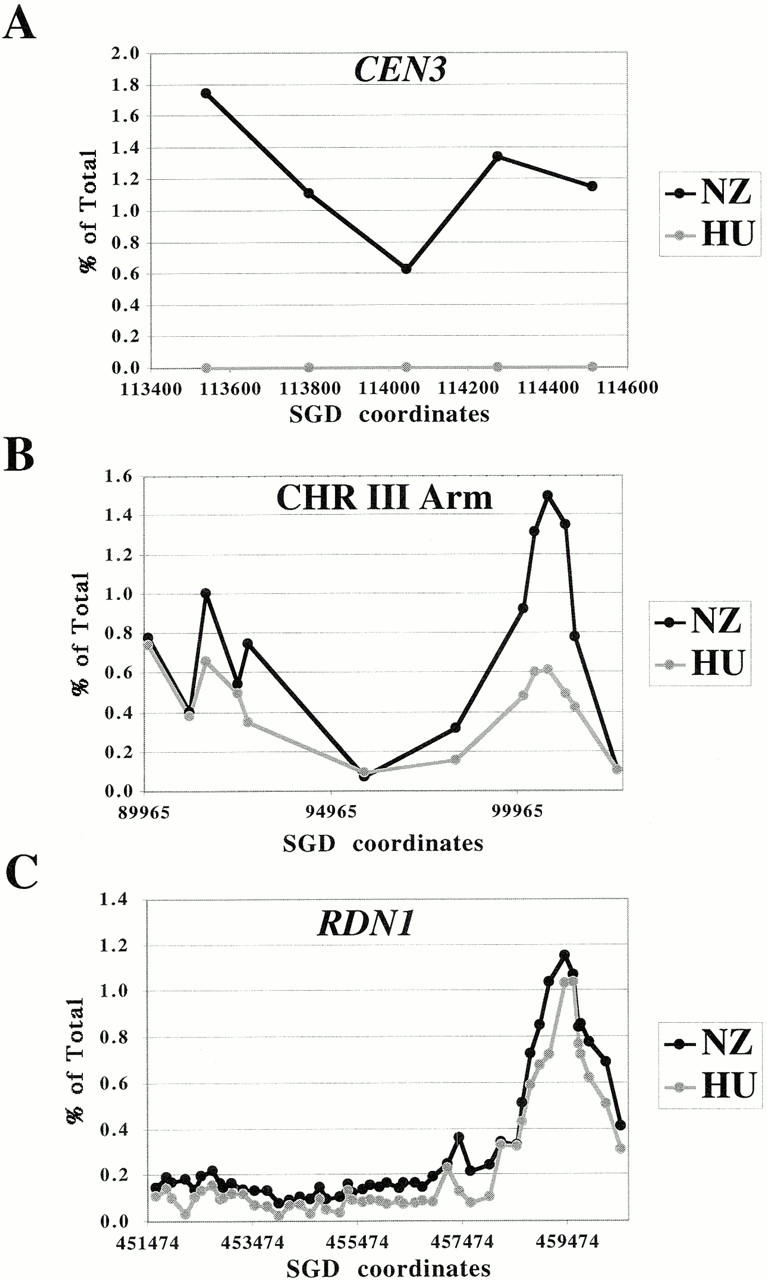
Association of Mcd1p with chromosomes in cell cycle arrested cultures. The immunoprecipitated DNA was quantified as a percent of the total lysate from HU (gray line, open circles) or nocodazole (black line, closed circles) arrested cultures. The amount of immunoprecipitated DNA (as percent of total) is plotted on the y axis versus DNA sequence coordinates on the x axis. (A) Cell cycle–dependent association of Mcd1p with CEN3 and flanking sequences. The middle fragment represents CEN3. The fold increase in binding in nocodazole arrested cells relative to HU arrested cells was tenfold or higher between different experiments. (B) Mcd1p binding to a part of the left arm of chromosome III in cell cycle arrested cells. (C) Comparison of Mcd1p binding to the entire RDN1 repeat in cell cycle arrested cells.
The pattern of Mcd1-6HAp binding (i.e., locations of binding sites) on chromosomal arms in HU arrested cells is similar to mitotic cells. A dramatic increase (ranging from tenfold or higher) in the magnitude of binding of Mcd1-6HAp to the centromere and pericentric regions was observed in mitotic cells relative to cells arrested in early S phase (Fig. 8 A). A smaller (2.5-fold) but consistent increase in Mcd1-6HAp binding was also detected at the cen-proximal chromosome III arm site, CARC1, which is located ∼10 kb to the left of CEN3 (Fig. 8 B). Binding sites further away from the centromere, such as the LEU2 proximal peak and Ty2-1 LTR, show lower or no significant increase in Mcd1-6HAp binding in mitotic cells. We also analyzed the association of Mcd1-6HAp with the entire RDN1 repeat in S phase arrested and mitotic cells. No significant difference in binding of Mcd1-6HAp to the RDN1-binding peak or valleys could be detected in mitotic cells relative to early S phase cells. These results suggest that the enhancement in cohesin loading observed in mitotic cells relative to S phase arrested cells may be limited to centromeres and pericentric regions and occurs upon or subsequent to progression through S phase. Enhanced binding to CEN and pericentric regions may arise from de novo loading of cohesin on CEN and proximal regions.
Discussion
Here, we define new cohesin-binding sites in single-copy sequences (CARL1 and CARL2) and repetitive sequences, such as Ty2 retrotransposon and rDNA repeats (CARL3). We also define sites adjacent to telomeres (CARC5 and CARC6) and HMR, the silent mating type locus (CARC3 and CARC4). These sites, along with three previously defined sites (CARC1, CARC2, and CEN3), were analyzed to identify parameters that control cohesin binding and to establish potential links between cohesins and genomic determinants that control gene expression, chromosome structure, and genome stability.
Several characteristics of cohesin-binding sites in single-copy sequence have been suggested based upon limited observations, including the spacing of sites, their positioning relative to genes, and their AT-biased base composition (Kleckner et al. 1977; Megee et al. 1999; Tanaka et al. 1999). Here, the identification and high resolution analysis of cohesin sites in single-copy sequence have established these characteristics as general genomic properties of cohesin sites. First, we observe a spacing between sites of ∼9 kb for chromosomes III and XII, which is slightly less than the 13–15-kb spacing for chromosome III reported earlier (Blat and Kleckner 1999). Since chromosome III was known to have unusual recombination and replication features (McCarroll and Fangman 1988; Wu and Haber 1996), it was important to establish that the close proximity of cohesin sites (9 kb) was not unique to that chromosome. Second, we observe that seven of nine new CARs are centered within intergenic regions. This greatly strengthens the idea that cohesin binding and transcription might be incompatible, which was suggested previously based upon the analysis of just two arm cohesin sites (Tanaka et al. 1999). Finally, we observe a preference for cohesin-binding sites in AT-rich sequences, as reported earlier (Blat and Kleckner 1999; Megee et al. 1999). However, our results suggest two serious caveats regarding the relevance of AT-rich sequences. The AT bias of cohesin sites may be an indirect consequence of their sequestration to intergenic regions that are inherently more AT rich. Also, we observe that the AT content is constant at the boundaries of the cohesin domain associated with CARC1. This is inconsistent with the model that spikes of GC-rich DNA are necessary to limit cohesin-binding domains (Megee et al. 1999)
Mcd1p binding to CEN and pericentric regions is greater in mitotic cells than S phase cells (Blat and Kleckner 1999; this study). It was thought that this enrichment resulted from the recruitment of Mcd1p that was previously associated with chromosomal arms in S phase (Blat and Kleckner 1999). However, we do not observe a decrease in binding to any chromosomal arm sites or intervening sequences during mitosis. Based on this observation, we suggest that the increase in Mcd1p on centromeres and pericentric regions in mitotic cells results from de novo targeting of Mcd1p to these regions, rather than the recruitment from arm regions. This de novo loading demonstrates that cohesin binding at CARs and centromeres must be differentially regulated.
In a previous study, the failure to detect cohesin binding near the left telomere of chromosome III led to the suggestion that telomeres might be devoid of cohesin-binding sites (Blat and Kleckner 1999). However, we detect cohesin binding at the right end of chromosome III. The periodicity of these telomere proximal sites is similar to other parts of the chromosomes. The lower peaks and valleys of cohesin binding at the right telomere may reflect less frequent occupancy, fewer proteins bound per site, or masking of the bound Mcd1p epitope by the specialized structure of the end. Whatever the cause of the decreased signal for cohesins at telomeres, it will be interesting to determine whether it reflects a distinct function in telomere biology.
In addition to these contributions, we define new characteristics of cohesin-binding sites and their genomic distribution. First, we find that Mcd1p associates with 0.8–1 kb of chromosomal DNA on many prominent arm sites. In previous studies, the size of arm sites could not be determined either because the resolution of the micoarray approach (3 kb) was too crude (Blat and Kleckner 1999), or the resolution of the ChIP was not calibrated by comparing cohesin binding to a known localized protein, like Mif2 (Tanaka et al. 1999). The more precise value of 0.8–1 kb is intriguing because it is clearly greater than the domain size for conventional protein–DNA complexes, but not as large as the centromere-associated cohesin domain. It is possible that arm domains arise from limited spreading of Mcd1p-containing complexes on either side of a nucleation site, as reported earlier for Mcd1p binding to centromere flanking DNA on minichromosomes (Megee et al. 1999). Alternatively, the Smc subunits of cohesin are long rod-shaped molecules that can span ∼50–100 nm (Melby et al. 1998). This length is more than sufficient to spread across a 1.0-kb arm site, assuming it is packaged in chromatin. Hence an arm site, unlike centromeres, may recruit only a single cohesin complex. The existence of rare (1.5 kb) arm sites, like CARL1, does not invalidate this second model as the broad nature of CARL1 may arise from the fortuitous cluster of two such binding sites.
A second new discovery reported here, is the presence of cohesin-binding sites at the junction of the left end of the rDNA array and adjoining single-copy sequences, the right end of each rDNA repeat, a subtelomeric repeat element near the end of the right arm of chromosome III, and the boundaries of the HMR locus. These sites are proximal to the physical boundaries between silenced and nonrepressed chromosomal domains (Gottschling et al. 1990; Bryk et al. 1997; Pryde and Louis 1997, Pryde and Louis 1999; Smith and Boeke 1997; Fourel et al. 1999). Interestingly, we observed the specific association of Mcd1p with the LTR of the Ty2 retrotransposon. Retrotransposon LTRs have been shown to contribute to boundary element function in the HMR boundaries (Donze et al. 1999). Furthermore, genetic data suggest that the spread of the silenced chromatin at the HMR locus is limited by the Smc1-cohesin component (Donze et al. 1999). Together with these genetic results, our mapping of a cohesin component to the HMR boundary strongly supports a model in which cohesins have a direct role in defining the boundaries of silent chromatin.
Finally, we have observed Mcd1 binding with interspersed repetitive DNA (Ty2) and the subtelomeric repeats. We also observed a single Mcd1p-binding site overlapping with the NTS of the 9-kb RDN1 repeat unit. Since the average height of the peak within the RDN1 repeat is of a similar magnitude as peaks in single-copy regions, Mcd1p is likely to be bound to this site in a majority of the RDN1 repeats. This suggests that the tandemly repeated RDN1 locus must have a highly ordered organization, with uniformly spaced cohesin complexes bound to the rDNA NTS and 5S RNA gene, generating loops (or intervening regions) of uniform sizes.
The presence of CARs in repetitive DNA has important implications for the stability of the mosaic eukaryotic genome. Cohesion at an interspersed repeat element may enhance the likelihood that the repeat on the sister is used as a template for recombination/repair chromatid, rather than on another chromosome. In this way, sister chromatid cohesion may prevent chromosomal rearrangements after DNA strand breaks within dispersed repetitive sequences (e.g., retrotransposons). Similarly, cohesion at each rDNA repeat may promote stability of the rDNA region by increasing the chances that the same repeat on the sister chromatid would be used as the preferred template for recombination/repair of a DNA break within one repeat. This equal sister chromatid exchange would maintain the copy number and genomic stability. In the absence of a cohesin-binding site in every repeat, there would be more structural flexibility. A different repeat may be used as the template, resulting in an unequal sister chromatid exchange and the expansion or contraction in the length of the RDN1 array.
The periodicity of cohesin-binding sites in the RDN1 array is likely to be directly relevant to chromosome condensation, since mcd1 mutants are also defective in mitotic chromosome condensation, including condensation of the RDN1 (Guacci et al. 1997). This link between cohesion and condensation has led to a model in which the spacing of cohesin sites constrains the size of the putative loops of condensed chromosomes (Guacci et al. 1997; Hartman et al. 2000). Indeed, the high density (about every 9 kb) of cohesin-binding sites throughout the yeast genome would account for the low level of condensation in this organism, i.e., ∼5 times less compact than mammalian chromosomes (Guacci et al. 1994). In addition, the strikingly similar periodicity of cohesin-binding sites on single-copy sequence and the tandemly repeated rDNA suggests that budding yeast may have a preference to maintain a constant loop size to facilitate packaging of its chromosomes.
Is this preference general to other eukaryotes? If it were, one would predict that the rDNA repeat in other eukaryotic cells would contain a conserved cohesin-binding site, and the length of the rDNA repeat might increase, correlating with the size of the euchromatic metaphase loops. Whereas cohesin-binding sites have yet to be mapped in other eukaryotes, several relevant observations about mammalian rDNA have been made. A fragment of the nontranscribed spacer, which also includes the major site of replication initiation in the human rDNA repeat and a matrix-association site, was localized close to the axial region of extracted metaphase chromosomes (Bickmore and Oghene 1996). This organization suggests that this region may be functionally analogous to the yeast NTS2 of RDN1, which includes an Mcd1p-binding region (this study) and an autonomously replicating sequence element (Miller and Kowalski 1993). The axial position of this spacer fragment is the expected position for a cohesin-binding site, according to the previously proposed models of mitotic chromosome structure (Saitoh and Laemmli 1994; Saitoh et al. 1995; Guacci et al. 1997). Therefore, these observations are collectively consistent with the notion of the rDNA spacer having a conserved cohesin-binding site. In addition, the metaphase loops in human cells are thought to be much larger than those in yeast, and the rDNA repeat in human cells is 41 kb rather than 9 kb, as in budding yeast. This extra length results from the addition of noncoding sequences, as the genes within the rDNA of mammals and yeast are highly conserved. Thus, it is possible that abundant noncoding DNA found throughout the genome of most eukaryotic organisms is not junk, but rather performs an important structural function as a spacer to facilitate folding of mitotic chromosomes.
Acknowledgments
We thank Pamela Meluh for the ChIP protocol and Cathy Mistrot for technical assistance in preparation of replicate ChIP samples in 8. We thank Amy Rubinstein and members of the Koshland lab for comments on the manuscript. We thank Alan Weiner for discussion and Shiv Grewal for pointing out valuable references.
This work was funded by grants from the Howard Hughes Medical Institutes and in part by a grant from the National Institutes of Health (GM1718) to D. Koshland.
Footnotes
Abbreviations used in this paper: CAR, cohesion-associated region; CEN, centromere; ChIP, chromatin immunoprecipitation; HU, hydroxyurea; IR, intermediate repeat; LTR, long terminal repeat; NTS, nontranscribed spacer; ORF, open reading frame; Smc, structural maintenance of chromosomes.
References
- Bajer A. Ciné-micrographic studies on chromosome movements in beta-irradiated cells. Chromosoma. 1958;9:319–331. [PubMed] [Google Scholar]
- Bell S.P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Bickmore W.A., Oghene K. Visualizing the spatial relationships between defined DNA sequences and the axial region of extracted metaphase chromosomes. Cell. 1996;84:95–104. doi: 10.1016/s0092-8674(00)80996-4. [DOI] [PubMed] [Google Scholar]
- Blat Y., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Donze D., Adams C.R., Rine J., Kamakaka R.T. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae . Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss M., McNally F.J., Laurenson P., Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae . Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Fourel G., Revardel E., Koering C.E., Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide L., Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S. cerevisiae telomeresreversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae . Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T., Stead K., Koshland D., Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae . J. Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. SMC-mediated chromosome mechanicsa conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. J. Mol. Biol. 1977;116:125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Koshland D., Strunnikov A. Mitotic chromosome condensation. Annu. Rev. Cell. Dev. Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Koshland D.E., Guacci V. Sister chromatid cohesionthe beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 2000;12:297–301. doi: 10.1016/s0955-0674(00)00092-2. [DOI] [PubMed] [Google Scholar]
- Lavoie B.D., Tuffo K.M., Oh S., Koshland D., Holm C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of barren . Mol. Biol. Cell. 2000;11:1293–1304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll R.M., Fangman W.L. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Koshland D. A functional assay for centromere-associated sister chromatid cohesion. Science. 1999;285:254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Mistrot C., Guacci V., Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Melby T.E., Ciampaglio C.N., Briscoe G., Erickson H.P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteinslong, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesinschromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miller C.A., Kowalski D. Cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:5360–5369. doi: 10.1128/mcb.13.9.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki W., Orr-Weaver T. Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Pryde F.E., Louis E.J. Saccharomyces cerevisiae telomeres. A review. Biochemistry (Mosc.) 1997;62:1232–1241. [PubMed] [Google Scholar]
- Pryde F.E., Louis E.J. Limitations of silencing at native yeast telomeres. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y., Laemmli U.K. Metaphase chromosome structurebands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Goldberg I., Earnshaw W.C. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. Bioessays. 1995;17:759–766. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- Skibbens R.V., Corson L.B., Koshland D., Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Strunnikov A.V., Jessberger R. Structural maintenance of chromosomes (SMC) proteinsconserved molecular properties for multiple biological functions. Eur. J. Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- Strunnikov A.V., Larionov V.L., Koshland D. SMC1an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Cosma M.P., Wirth K., Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Toth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Haber J.E. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]


