Abstract
The microtubule cytoskeleton plays a pivotal role in cytoplasmic organization, cell division, and the correct transmission of genetic information. In a screen designed to identify fission yeast genes required for chromosome segregation, we identified a strain that carries a point mutation in the SpRan GTPase. Ran is an evolutionarily conserved eukaryotic GTPase that directly participates in nucleocytoplasmic transport and whose loss affects many biological processes. Recently a transport-independent effect of Ran on spindle formation in vitro was demonstrated, but the in vivo relevance of these findings was unclear. Here, we report the characterization of a Schizosaccharomyces pombe Ran GTPase partial loss of function mutant in which nucleocytoplasmic protein transport is normal, but the microtubule cytoskeleton is defective, resulting in chromosome missegregation and abnormal cell shape. These abnormalities are exacerbated by microtubule destabilizing drugs, by loss of the spindle checkpoint protein Mph1p, and by mutations in the spindle pole body component Cut11p, indicating that SpRan influences microtubule integrity. As the SpRan mutant phenotype can be partially suppressed by the presence of extra Mal3p, we suggest that SpRan plays a role in microtubule stability.
Keywords: Ran GTPase, microtubules, chromosome segregation, mitosis, fission yeast
Introduction
Accurate duplication of chromosomes and the subsequent precise segregation of the sister chromatids into two daughter cells are essential processes in every eukaryotic cell cycle. The faithful inheritance of genetic information is a prerequisite for the survival of an organism and many diseases are associated with genetic instability (Cahill et al. 1999).
The microtubule cytoskeleton, as the major component of the mitotic spindle, is essential for the precise separation of the duplicated sister chromatids before cytokinesis.
Upon commitment to mitosis in most animal cells, the extensive interphase microtubule array, composed of long, relatively stable microtubules, is disassembled rapidly and reorganized into an elliptical bipolar spindle consisting of shorter and much more dynamic microtubules. The dynamic instability model predicts that microtubules have polymerization (growth) and depolymerization (shrinkage) phases and that switching between these two phases occurs with specific frequencies (Kirschner and Mitchison 1986). The factors regulating microtubule dynamics can be divided broadly into two groups: proteins with a stabilizing role, such as microtubule-associated proteins (MAPs), and proteins like XKCM1 and Op18 that have a microtubule destabilizing function (for reviews see Walczak 2000; Andersen 2000). Structural and regulatory proteins required for the rapid transition from the interphase microtubule cytoskeleton to the mitotic spindle structure have not been defined fully, but the importance of altered microtubule dynamics thought to result from the activation of the protein kinase complex M phase promoting factor has been extensively documented (for reviews see Walczak 2000; Andersen 2000).
In the fission yeast Schizosaccharomyces pombe a similar reorganization of the microtubule cytoskeleton occurs at the onset of mitosis (for review see Hagan 1998): the cytoplasmic microtubules disassemble and the mitotic microtubules are nucleated in the nucleus from the centrosome equivalents, the spindle pole bodies (SPBs), to generate a bipolar spindle.
We and others have identified several factors needed for chromosome segregation and spindle function by isolating mutants that display increased loss of a nonessential chromosome and are hypersensitive to microtubule destabilizing drugs (Takahashi et al. 1994; Fleig et al. 1996). The data presented here show that Spi1p is one of the proteins required for chromosome transmission fidelity in fission yeast as a mutant allele of spi1 + was identified in this screen. spi1 + codes for the fission yeast Ran GTPase, an evolutionarily conserved essential GTPase of the Ras superfamily (for review see Sazer 1996). Association with effector and regulatory proteins is dependent on the conformational state of Ran, which is a consequence of being bound to either GDP or GTP. Loss of Ran GTPase function affects many biological processes, but it is generally accepted that the primary function of the Ran GTPase cycle is in nucleocytoplasmic transport (Izaurralde and Adam 1998; Mattaj and Englmeier 1998). Several observations have pointed to a possible role of the Ran GTPase cycle in chromosome segregation and microtubule regulation, but it has remained unclear whether this was caused directly by perturbation of multiple downstream effector pathways or indirectly by transport defects (for review see Sazer and Dasso 2000). The finding of a human centrosomal Ran–GTP binding protein involved in microtubule nucleation (Nakamura et al. 1998) and the demonstration that Ran-GTP in vitro regulates microtubule spindle assembly in Xenopus M phase extracts in a transport-independent manner (Carazo-Salas et al. 1999; Kalab et al. 1999; Ohba et al. 1999; Wilde and Zheng 1999; Zhang et al. 1999) support a role for Ran in the regulation of bipolar spindle formation. However, a direct influence of Ran on spindle microtubule arrays in vivo has not been demonstrated previously and the mechanism by which Ran might regulate spindle assembly has remained unclear. Here, we report the characterization of a fission yeast Ran mutant, Spi1-25p, that causes microtubule defects in vivo. Strains carrying the defective spi1-25 allele have no transport defect but show defects in early spindle formation and function, have an activated spindle checkpoint, and are hypersensitive to microtubule destabilizing drugs. Interestingly, moderate overexpression of Mal3p, the fission yeast member of the evolutionarily conserved microtubule-associated EB1 family (Beinhauer et al. 1997), can partially rescue spi1-25 defects. The role of Ran with regard to spindle formation will be discussed in this context.
Materials and Methods
Media and Strains
The genotypes of strains used in this study are listed in Table . The S. pombe strains YP10.22, YP10.22a, UFY135, and UFY250 have been described (Fleig et al. 1996; Beinhauer et al. 1997). UFY250 was backcrossed three times with YP10.22a or YPKG246 resulting in strains UFY25OR and UFY25CX, respectively. The strains carrying cut11 alleles or cut11 +-GFP were a gift from the J.R. McIntosh lab (West et al. 1998). The mph1 + null, mad2 + null, and pim1-d1 strains have been described (Sazer and Nurse 1994; He et al. 1997, He et al. 1998b). The GFP-pap1 + expressing plasmid (Toone et al. 1998) was integrated into a wild-type strain, generating strain SS767. All double mutant strains were identified by tetrad analysis.
Table 1.
Strains Used in This Study
| Strain | Genotype | Source |
|---|---|---|
| YP10.22 | h− leu1-32 ade6-M210 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| YP10.22a | h+ ade6-M210 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| UFY135 | h+ mal3Δ::his3+ leu1-32 ade6-M210 ura4-D18 his3Δ | U. Fleig |
| UFY250 | h− spi1-25 leu1-32 ade6-M210 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| UFY25OR | h− spi1-25 leu1-32 ade6-M210 ura4-D6 | U. Fleig |
| UFY25CX | h+ spi1-25 leu1-32 ade6-M210 ura4-D6 his3Δ | U. Fleig |
| UFY156 | h− spi1+/kanR leu1-32 ade6-M210 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| UFY192 | h− spi1Δ::spi1-25cDNA/his3+ leu1-32 ade6-M210 ura4-D18 his3Δ | U. Fleig |
| UFY153 | h− spi1-25 mad2Δ::ura4+ ade6-M210 leu1-23 ura4-D18 | U. Fleig |
| UFY154 | h− spi1-25 mph1Δ::ura4+ ade6-M216 leu1-32 ura4-D18 | U. Fleig |
| UFY193 | h− spi1-25 mal3Δ ::his3+ ade6-M210 leu1-32 ura4-D18 his3Δ | U. Fleig |
| YPKG246 | h− leu1-32 ade6-M210 ura4-D18 his3Δ | K. Gould |
| 80 | h− cut11-2 leu1-32 ura4-D18 | J.R. McIntosh |
| 83 | h− cut11-3 leu1-32 ura4-D18 | J.R. McIntosh |
| 91 | h− cut11-7 leu1-32 ura4-D18 | J.R. McIntosh |
| 317 | h− cut11:GFP:ura4+ leu1-32 ura4-D18 | J.R. McIntosh |
| SS446 | h− leu1-32 ade6-M210 ura4-D18 | S. Sazer |
| SS767 | h− int:GFP-pap1/pRep41:leu+ leu1-32 ura4-D18 ade6-M210 | S. Sazer |
| SS893 | h− int:GFP-pap1/pRep41:leu+ spi1-25 leu1-32 ura4-D18 ade6-M216 | S. Sazer |
| SS131 | h− pim1-d1 leu1-32 ade6-704 | S. Sazer |
| SS560 | h− mph1Δ::ura4+ leu1-32 ade6-M216 ura4-D18 | S. Sazer |
| SS638 | h− mad2Δ::ura4+ leu1-32 ade6-M210 ura4-D18 | S. Sazer |
| SS482 | h− int:GFP-NLS-LacZ/pREP4X:ura4+ leu1-32 ade6-M216 ura4-D18 | S. Sazer |
| SS890 | h− spi1-25 int:GFP-NLS-LacZ/pREP4X:ura4+ leu1-32 ade6-M216 ura4-D18 | S. Sazer |
Strains were grown in rich (YE5S) or Edinburgh minimal medium (EMM) with appropriate supplements (Moreno et al. 1991). Sensitivity to thiabendazole (TBZ) was monitored at 24°C on YE5S plates or EMM plates containing 7 μg/ml of TBZ, while resistance to G418 (Calbiochem) was tested at 32°C on YE5S plates containing 100 μg/ml G418.
Microscopy
Photomicrographs of cells were taken with a ZEISS Axioskop or AxioplanII. Immunofluorescence images were processed as described previously (Beinhauer et al. 1997). For determination of morphological defects, a minimum of 250 cells were scored microscopically; for the Cut11-GFPp localization three different cultures with a total of 600 counted cells were analyzed. Processing of cells for immunofluorescence microscopy was carried out essentially as described (Hagan and Hyams 1988). For tubulin staining, we used the primary monoclonal antitubulin antibody TAT1 (Woods et al. 1989) followed by FITC-conjugated goat anti–mouse antibodies (EY labs). SPBs were stained using AP9.2 affinity-purified anti-Sad1p primary and Cy3-conjugated secondary sheep anti–rabbit antibodies (Sigma-Aldrich) (Hagan and Yanagida 1995).
Identification of mal25-1 Multicopy Suppressors and Linkage Analysis
Strain UFY25CX was transformed with an S. pombe genomic bank (Barbet et al. 1992) and Ura+ transformants were selected by incubation for 50 h at 24°C, followed by replica plating onto 7 μg/ml TBZ plates. From the 44,450 transformants, plasmids were isolated (Moreno et al. 1991) from the 21 surviving colonies and processed as described (Beinhauer et al. 1997). Two genomic DNA inserts were found several times: the spi1 + (Matsumoto and Beach 1991) and mal3 + (Beinhauer et al. 1997) genes were isolated five and four times, respectively. To determine allelism between mal25-1 and spi1 +, the kanR marker conferring resistance to the antibiotic G418 was inserted 6.28 kb away from the spi1 + open reading frame (ORF) via PCR-based gene targeting (Bahler et al. 1998). This strain, UFY156, was crossed with UFY25CX, and the resultant spores were analyzed via random spore analysis. After establishment of linkage, the spi1 + ORF in strain YP25CX was sequenced. Genomic DNA was PCR amplified using oligonucleotides 5′-CTCTCAGTTAGTTTAGGTGC-3′ and 5′-CTATCGTTACACAAGTC-3′ that flank the spi1 + ORF.
Recreation of mal25-1 Mutation In Vitro and Cloning into Expression Vectors
The 5′ end of the spi1-25 ORF from strain UFY25CX was amplified by PCR using the following primer pair: 5′-GGATGGACCTCAATACCCAAAGTTGCAATATC-3′ (italicized characters represent AvaII restriction site 132 bp downstream of the start codon; underlined characters represent mutated codon to generate spi1-25; underlined bold character represents base change to give mutated codon; bold and italicized characters represent sequence homologous to the spi1 + genomic template) and 5′-GAAAAGTTT CATATG GTCAACCACAAAACG-3′ (underlined characters represent NdeI restriction site; underlined and bold characters represent start codon; bold characters represent sequence homologous to the 5′ end of the spi1 + genomic template). This PCR product was cleaved AvaII-NdeI to generate the 5′ end of the spi1-25 ORF. The 3′ end was obtained by cleaving a spi1 clone (spi1-88/pAS1-CHY2; Sazer, S., unpublished data) with AvaII-BamHI. The 5′ and 3′ fragments were both cloned into NdeI-BamHI cut pAS1 (Harper et al. 1993) resulting in spi1-25/pAS1. Next, the sequenced spi1-25/pAS1 was cleaved with NdeI-BamHI and the spi1-25 ORF was subcloned into pETXHA (Elledge et al. 1992) to create spi1-25/pETXHA and into pTrcHis (Invitrogen) to create His-spi1-25/pTrcHis. The XhoI-HA-spi1-25-BamHI insert from the pETXHA-spi1-25 construct was subcloned into XhoI-BamHI cut pREP3X vector for expression of HA-spi1-25 in S. pombe.
Nuclear Protein Transport Assay
Nuclear protein transport was tested in strains with either an integrated copy of the gene encoding the GFP-tagged Pap1p protein (Toone et al. 1998) or a GFP-tagged LacZ construct fused to the SV-40 nuclear localization signal (NLS) (Demeter, J., and S. Sazer, unpublished data), constructed by subcloning the NLS-GFP-LacZ containing fragment from the Saccharomyces cerevisiae plasmid pPS817 (Lee et al. 1996) into pREP4X. spi1-25 and wild-type cells were grown to midlog phase at 25°C in EMM, then shifted to 36°C for 4 h. Live cells were observed microscopically either to determine the steady state localization of the GFP-SV40-NLS-LacZ protein, or to monitor the localization of the nucleocytoplasmic shuttling protein, GFP-Pap1p, before hydrogen peroxide addition (to test for nuclear protein export) or 15 min after addition of 0.8 mM hydrogen peroxide (to test for nuclear protein import).
GTP Binding Assays
Three E. coli BL21 cell–produced His(6)-tagged substrates His-Spi1p in pTrcHisB (Matynia et al. 1996), His-Spi1-25p in pTrcHisB, and S. cerevisiae His-UBC4 in pET (Novagen; construct a gift of J.W. Harper, Baylor College of Medicine, Houston, TX) were obtained using standard methods and purified on a nickel-nitrilotriacetic acid resin (QIAGEN) in sonication buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl) plus 10 mM imidazole, 0.1 mM PMSF, and 10 μg/ml leupeptin. Protein was determined by quantifying the intensity of Coomassie blue staining of the protein gel band using a DC-129 digital camera and Digital Science Electrophoresis Documentation and Analysis System 120 software (both from Eastman Kodak Co.). 10 μl of nickel resin with 3–5 μg His-tagged substrate was aliquoted into SpinX® (Costar) columns containing 180 μl GTP Binding Buffer (50 mM NaPO4, pH 7.0, 5 mM MgCl2) plus 10 mM imidazole, 0.1 mM PMSF, and 10 μg/ml leupeptin. The binding reactions were initiated by the addition of 10 μCi of [α-32P]GTP and stopped by spinning for 30 s and washing three times. Samples were added to Scintisafe Gel scintillant (Fisher Scientific) and bound radioactive nucleotide was quantified using a Beckman LS-3801 scintillation counter.
Results
Characterization of mal25-1 Mutant and Establishment of Allelism between mal25-1 and the Gene Encoding the SpRan GTPase Spi1p
From a previously described screen (Fleig et al. 1996), we isolated a mutation named mal25-1 that strongly decreased the transmission fidelity of a nonessential minichromosome that was identified as an increase in the number of red sectors in a white colony (Fig. 1 A) using an ade6-based colony color assay. Based on prior analysis of other mal mutants we estimate that presence of mal25-1 leads to an ∼400-fold increase in minichromosome loss (Fleig et al. 1996; Beinhauer et al. 1997). In addition, the mal25-1 strain was hypersensitive to the microtubule-destabilizing drug TBZ (Fig. 1 B), had reduced growth at ≥30°C (Fig. 1 C), and accumulated elongated, abnormally shaped cells (Fig. 1 D).
Figure 1.
Phenotypic characterization of the mal25-1 strain. (A) Sectoring phenotypes of wild-type (wt) and mal25-1 strains grown on indicator plates (Fleig et al. 1996) for 6 d at 24°C. (B) Serial dilution patch test for sensitivity to the microtubule-destabilizing drug TBZ. Dilutions shown were 10-fold. Although the isogenic wild-type strain (b and d) only shows slightly reduced growth at 24°C on rich medium containing 7 μg/ml TBZ, the mal25-1 strain (a and c) is unable to grow. (C) The mal25-1 strain shows growth defects at higher temperatures. Serial dilution patch tests of wild-type strain (a, c, and e, grown at 24°C, 30°C, and 36°C, respectively) and mal25-1 strain (b, d, and f grown at 24°C, 30°C, and 36°C, respectively) are shown. (D) Photomicrographs of wild-type (insert) and mal25-1 strain show an abnormal, elongated cell form of mutant strain. At 30°C, wild-type cells had an average length of 13.2 ± 1.2 μm at septa formation, whereas mal25-1 cells were 19.0 ± 3 μm in length. Arrows indicate septum. Bar, 10 μm.
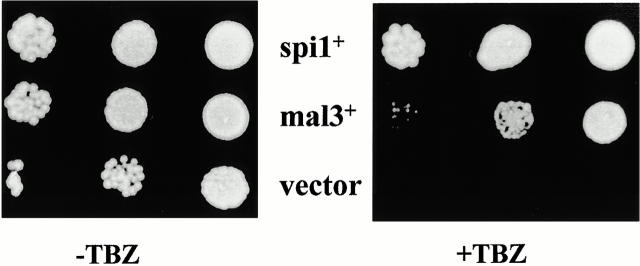
Multicopy plasmid-borne suppression of TBZ hypersensitivity of the mal25-1 strain by transformation with an S. pombe genomic library identified repeatedly the spi1 + and mal3 + ORFs (Fig. 2). spi1 + codes for the fission yeast Ran GTPase, whereas mal3 + encodes an MAP (Matsumoto and Beach 1991; Sazer and Nurse 1994; Beinhauer et al. 1997). The mal3 + gene only suppressed the TBZ hypersensitivity, whereas spi1 + could suppress all phenotypes associated with the mal25-1 mutation (data not shown). To determine via linkage analysis if mal25-1 and spi1 + were allelic, the mal25-1 strain was crossed to a strain carrying a wild-type spi1 + allele tagged with the kanR marker. Among 500 resulting mal25-1 spores tested by random spore analysis, none carried the kanR marker, indicating that mal25-1 and spi1 + were linked. Sequence analysis of the spi1 gene in the mal25-1 strain identified a single base pair change from G to A at position 315 of the ORF, resulting in a change from valine to isoleucine at position 44 of Spi1p that corresponds to V45 in mammalian Ran. This amino acid is in the Switch I region (Chook and Blobel 1999; Vetter et al. 1999) of Ran, which undergoes a substantial conformational change depending on whether the protein is bound to GDP or GTP and is important for the binding of factors of the importin β family and other effectors to Ran-GTP (Chook and Blobel 1999). To confirm that all phenotypes observed in the spi1-25 strain were caused by the single amino acid change in Spi1p, we remade the mutation in vitro in the spi1 + cDNA, tagged it with the S. pombe his3 + marker, and replaced the genomic copy of a wild-type S. pombe strain with this mutated version via homologous recombination. The resultant strain was indistinguishable in all phenotypes from the original mal25-1 strain (data not shown), confirming that the single mutation in the spi1 + ORF caused all phenotypes observed. mal25-1 was thus renamed spi1-25.
Figure 2.
The TBZ hypersensitivity of the mal25-1 strain is rescued by plasmid-borne copies of mal3 + and spi1 +. Left and right panels show serial dilution patch tests (104 to 102 cells) of mal25-1 transformants grown on selective minimal medium without TBZ or with 7 μg/ml TBZ, respectively. Vector control indicates plasmid without insert.
Interaction of Mutant Spi1p with Components of the SpRan GTPase System
Misregulation of the Ran system in fission yeast by mutation or overexpression of its regulators (Sazer and Nurse 1994; Matynia et al. 1996; He et al. 1998a) or by deletion of spi1 + (data not shown) results in a characteristic terminal phenotype. Cells arrest with normal size and shape, condensed postmitotic chromosomes, fragmented nuclear envelopes, and a wide medial septum.
The terminal phenotype of the spi1-25 strain is clearly different from the spi1 null strain, but the Spi1p protein levels are similar (data not shown), suggesting that Spi1-25p might be a separation of function mutant that retains normal interactions with some but not all binding partners. Alternatively, Spi1-25p could be a partial loss of function mutant in which the activity of the protein is lower than normal. To distinguish between these possibilities, we first asked whether the Spi1-25p mutant protein is able to interact with known regulatory and effector binding partners.
Ran interacts with proteins that modulate its nucleotide bound state such as the guanine nucleotide exchange factor Pim1p, the GTPase activating protein Rna1p, and the Ran binding proteins including Sbp1p (Matsumoto and Beach 1991; Melchior et al. 1993; Sazer and Nurse 1994; He et al. 1998a). Both Spi1-25p and Spi1p interacted with Sbp1p in the two-hybrid assay (Table , data not shown), indicating that association of this Ran-GTP binding protein with Spi1-25p was possible. Furthermore, as has been shown for spi1 + wild-type strains (Matynia et al. 1996; He et al. 1998a), overexpression of sbp1 + or rna1 + from the regulatable nmt1 + promoter in a spi1-25 strain background was lethal (Table ), confirming the ability of the mutant protein to interact normally in vivo.
Table 2.
spi1-25 Interactions
| Two-hybrid interactions | |||
|---|---|---|---|
| Gene | Function | Interaction | |
| nda2+ | α-Tubulin | No | |
| sbp1+ | Ran-GTP BP | Yes | |
| Genetic interactions | |||
| Gene | Function | Phenotype | |
| pim1d | Ran GEF | Synthetic lethal | |
| cut11-2 | SPB component | Synthetic lethal | |
| cut11-3 | SPB component | Synthetic lethal | |
| cut11-7 | SPB component | Poor growth at 24°C, dead at 30°C | |
| mph1Δ | Spindle checkpoint component | Extremely poor growth at 24°C; increase in chromosome missegregation | |
| mad2Δ | Spindle checkpoint component | None | |
| mal3Δ | MAP | Very poor growth at 24°C; dead on sublethal doses of TBZ; increase in abnormal cell form; increase in chromosome missegregation | |
| Multicopy suppression | |||
| Overexpression | Function | Allele to be rescued | Phenotype |
| spb1+ | Ran-GTP BP | spi1-25 | Lethal as for spi1+ |
| rna1+ | Ran-GAP | spi1-25 | Lethal as for spi1+ |
| spi1+ | Ran | pim1d | Good rescue |
| spi1-25 | Ran | pim1d | Moderate rescue |
The nongrowth phenotype of strains carrying the pim1-d1 ts temperature-sensitive mutation can be rescued by spi1 + cDNA under the control of the full strength nmt1 + promoter at both low and high levels of expression (Sazer and Nurse 1994). We found that the spi1-25 cDNA could rescue the nongrowth phenotype of a pim1-d1 ts strain only with high level expression (data not shown), indicating either that the mutant protein has reduced activity or that only a portion of the protein is functional.
spi1-25 and pim1-d1 ts showed a strong genetic interaction as spores from double mutant strains grown at 25°C could germinate but then ceased growth (data not shown).
Spi1-25 Is Deficient in Nucleotide Binding
To further characterize the consequences of the V44I mutation, we asked whether the Spi1-25p protein is properly folded by monitoring its ability to bind nucleotide. Histidine-tagged wild-type Spi1p and Spi1-25p were incubated with radiolabeled GTP. By comparing the maximum amount of bound nucleotide per microgram of protein, we found that Spi1-25p is only 30% as efficient in GTP binding compared with Spi1p, but that the initial kinetics of binding appears similar to those of the wild-type protein (Fig. 3). These data demonstrate that although the kinetics of nucleotide binding is normal, only a portion of Spi1-25p is competent to bind nucleotide, perhaps due to misfolding.
Figure 3.
Comparison of the kinetics and efficiency of GTP binding between wild-type (wt) Spi1p and mutant Spi1-25p. His-tagged Spi1p, his-tagged Spi1-25p, or UBC4 as a negative control were incubated with [α-32P]GTP as described in Materials and Methods and the amount of bound nucleotide was quantitated. The initial binding kinetics of wild-type and mutant Spi1p are similar, but only 30% of Spi1-25p is competent to bind nucleotide. Data for each experiment were normalized for protein amount and calculated as percentage of maximal wild-type binding. Values from four independent experiments were averaged and graphed as percent of wild-type βmax using SigmaPlot (Jande Scientific) and curves were fit using the sigmoidal three parameter equation.
Mutant Spi1p Protein Does Not Cause Nucleocytoplasmic Transport Defects
To test if nucleocytoplasmic transport was affected in spi1-25 cells, we first monitored the localization of a GFP-β-galactosidase reporter protein constitutively targeted to the nucleus by the SV-40 NLS. In both spi1-25 and spi1 + cells, this reporter protein was predominantly localized to the nucleus (Fig. 4A and Fig. B). To more precisely monitor nucleocytoplasmic transport, we followed localization of a GFP-Pap1p fusion protein. Pap1p is an AP1-like transcription factor with a bipartite type1 nuclear localization sequence (Ding et al. 2000) and a nuclear export sequence (Kudo et al. 1999) that continually shuttles between the nucleus and the cytoplasm. At steady state it is actively exported from the nucleus and appears predominantly cytoplasmic; under oxidative stress conditions, such as the presence of hydrogen peroxide, it accumulates in the nucleus (Toone et al. 1998). In both untreated spi1 + or spi1-25 strains, the GFP-Pap1p fusion protein is actively exported to the cytoplasm and relocalizes to the nucleus upon oxidative stress (Fig. 4E and Fig. F), indicating that the mutant is competent for both nuclear protein export and import. The ratio of nuclear to cytoplasmic GFP-Pap1p fluorescence (see Materials and Methods) in spi1-25 cells was similar to that of wild-type cells: before hydrogen peroxide treatment, the ratios were 0.8 ± 0.07 for both strains; 15 min after hydrogen peroxide addition, the ratios were 3.8 ± 0.7 and 3.1 ± 0.8 for wild-type and spi1-25 cells, respectively.
Figure 4.

Nucleocytoplasmic transport is normal in spi1-25 cells. Wild-type or spi1-25 cells with either integrated GFP-SV40, NLS-LacZ, or GFP-pap1 were grown to midlog phase at 25°C then shifted to 36°C for 4 h. The GFP-SV40 NLS-β-galactosidase reporter protein is exclusively nuclear localized in wild-type (A) and spi1-25 cells (B). Without hydrogen peroxide treatment, GFP-Pap1p is exported from the nucleus in both wild-type (C) and spi1-25 cells (D). 15 min after the addition of 0.8 mM hydrogen peroxide, the fusion protein is imported into the nucleus in wild-type cells (E) and spi1-25 cells (F). Bar, 10 μm.
The spi1-25 Mutant Strain Shows Altered Interphase Microtubule Arrays
At 24°C, ∼10% of spi1-25 cells did not have wild-type cylindrical shape but were curved, bent, or branched (see Fig. 1 D and 8 B). At temperatures >30°C, this number increased to 21%. Because an abnormal microtubule cytoskeleton is known to cause changes in cell shape (Hiraoka et al. 1984; Verde et al. 1995; Beinhauer et al. 1997; Sawin and Nurse 1998), we analyzed microtubules in spi1-25 cells by indirect immunofluorescence. In wild-type cells, interphase microtubules are aligned along the long axis of the cell reaching the cell tips (Fig. 5 e). In contrast, the cytoplasmic microtubules in most branched and 10% of normal cylindrically shaped spi1-25 cells failed to reach the cell tips and in some cases were shorter than normal (Fig. 5, a–c). Cell elongation alone cannot explain the inability of the microtubules to reach the cell tips of spi1-25 cells because elongated cdc25 mutant cells have interphase microtubules that are much longer than those of wild-type cells and do reach the cell tips (Hagan and Hyams 1988).
Figure 5.
Antitubulin immunofluorescence images of interphase spi1-25 cells show aberrant cytoplasmic microtubule cytoskeleton. (a–d) spi1-25 cells cultured at 30°C; (e) a wild-type cell. Each cell is depicted as a phase–contrast image to visualize cell shape followed by two antitubulin immunofluorescence images representing two different focal planes. Although the cytoplasmic microtubules of wild-type cells (e) are aligned along the long axis of the cell reaching the cell tips, spi1-25 cells frequently have microtubules that fail to reach the cells tips (a and b) and in some cases are much shorter than wild-type interphase microtubules (c) or have a single microtubule bundle on one side of the cell (d) and cells bend away from this bundle. Bar, 10 μm.
Curved spi1-25 cells (Fig. 5 d) usually displayed a single microtubule bundle on the convex side of the cell as has been described for other curved S. pombe mutants (Verde et al. 1995). Therefore, a significant proportion of cells carrying the defective spi1-25 allele display defects of the cytoplasmic microtubule cytoskeleton.
The spi1-25 Mutation Affects Formation and Function of the Mitotic Spindle
At 25°C, the spi1-25 strain showed increased loss of a nonessential chromosome (see Fig. 1 A) and ∼2% of cells in a population showed abnormal mitosis as seen by staining of the chromatin with DAPI (data not shown). At temperatures >30°C spi1-25 cells showed reduced growth (see Fig. 1 C) and increased missegregation of chromosomes. We identified three phenotypic classes: (a) cytokinesis without prior completion of mitosis, resulting in a displaced nucleus and an anucleate daughter cell; (b) chromosomes segregated asymmetrically to the two ends of a cell; and (c) highly condensed chromatin in the middle fifth of the cell.
To determine the cause of the chromosome missegregation phenotype, the mitotic spindle in spi1-25 cells was analyzed. The spi1-25 allele is not a temperature-sensitive lethal mutation but shows significantly reduced growth and only 61% viability at 36°C. spi1-25 cells were incubated at 36°C to determine spindle structure, chromatin structure, and SPB localization. We found two main phenotypic aberrations in the formation of the mitotic spindle. One was a star- or fan-shaped tubulin staining pattern indicating multiple microtubule bundles that originated from a single focal point giving rise to a monopolar spindle (Fig. 6c and Fig. g). DAPI staining showed hypercondensed chromatin (Fig. 6b and Fig. f) in close proximity to the tubulin staining; occasionally, partially separated chromatin on either side of the monopolar spindle was observed. The second phenotype was a tiny pre-metaphase spindle (Fig. 6c and Fig. k) between barely separated SPBs (Fig. 6d and Fig. l). Cytoplasmic microtubules were absent but DAPI staining of these cells showed that the chromatin still had the typical hemispherical appearance of an interphase nucleus (Toda et al. 1981), indicating that this was a very early stage in spindle formation.
Figure 6.
spi1-25 cells show mitotic spindle defects. Early mitotic spindle defects in an asynchronous culture of the spi1-25 mutant held at 36°C for 3 h when most aberrant phenotypes were scored. Each panel shows four different images of the same cell: the first DIC with DAPI staining (a, e, and i) shows the cell outline and position of the chromatin; the second shows chromatin staining by DAPI (b, f, and j); the third and fourth show immunofluorescence of tubulin (c, g, and k) and the spindle component Sad1p (d, h, and l), respectively. The two predominant spindle defects are shown: star-shaped monopolar spindles and condensed chromatin (c and b, bottom cell; g and f) and a tiny premetaphase spindle (c, top cell; k) between duplicated and separated spindle pole bodies (d, top cell; l) and not visibly condensed chromatin (b, top cell; j). Tubulin staining of the nucleus in k shows nuclear import of tubulin. Bars, 10 μm.
The monopolar and tiny spindle phenotypes were not observed in wild-type cells (n ≈ 1,000) grown at 36°C. spi1-25 cells cultured asynchronously at 25°C showed no monopolar spindles, but 0.5% of cells showed the tiny spindle phenotype. Incubation of spi1-25 cells for 3 h at 36°C resulted in the appearance of 4.8% cells with tiny spindles and 4.5% with monopolar spindles. No further increase was observed at later time points. The number of spi1-25 cells with normal looking metaphase spindles was ∼2.5% of the population at all time points tested, as was found for wild-type cells (Hagan and Hyams 1988). However, at 3 h incubation at 36°C, >45% of cells with metaphase spindles showed hypercondensed chromatin, indicating a problem at this stage of mitosis (Toda et al. 1981; Hiraoka et al. 1984). Furthermore, although anaphase spindles looked normal, 42% of spi1-25 anaphase cells showed unequally divided chromatin or lagging chromosomes (data not shown).
The aberrant mitotic spindle phenotype seen in spi1-25 cells indicated that SpRan was required for an early stage in spindle formation. We used the SPB localization of the Cut11p protein in spi1-25 cells grown at 36°C to define this stage more precisely. In prophase, cells have a single bright Cut11-GFPp spot, whereas at later mitotic stages two spots are observed (West et al. 1998). 7.3 ± 2.1% spi1-25 cells grown for 1 h at 36°C showed a single bright spot compared with 0.8 ± 0.4% for wild-type cells, indicating that there is an approximately eightfold increase in prophase cells, whereas the percentages of mutant and wild-type cells with two close or separated spots were similar. Similar distributions were seen at longer incubation times.
spi1-25 Is Synthetically Lethal with cut11ts Alleles
Because the spi1-25 strain expressing cut11 +-GFP grew slower than the isogenic spi1-25 strain, possibly due to a subtle change in the properties of the Cut11p protein that was amplified in the spi1-25 strain, we tested whether cut11 + and spi1 + interact genetically by constructing spi1-25 cut11 ts double mutants. Three different temperature-sensitive cut11 ts alleles were used (see Table ) (West et al. 1998). The resultant double mutants showed allele specific synthetic lethal interactions (see Table ). At 25°C, both single mutants grow, but the cut11-2 spi1-25 and cut11-3 spi1-25 strains arrested after three to four cell divisions with highly elongated cells. The synthetic lethality between cut11-7 and spi1-25 was seen only at 30°C. These data indicate that spi1-25 and cut11 ts encoding an SPB component interact genetically.
spi1-25 Is Synthetically Lethal with the Spindle Checkpoint Mutant mph1Δ
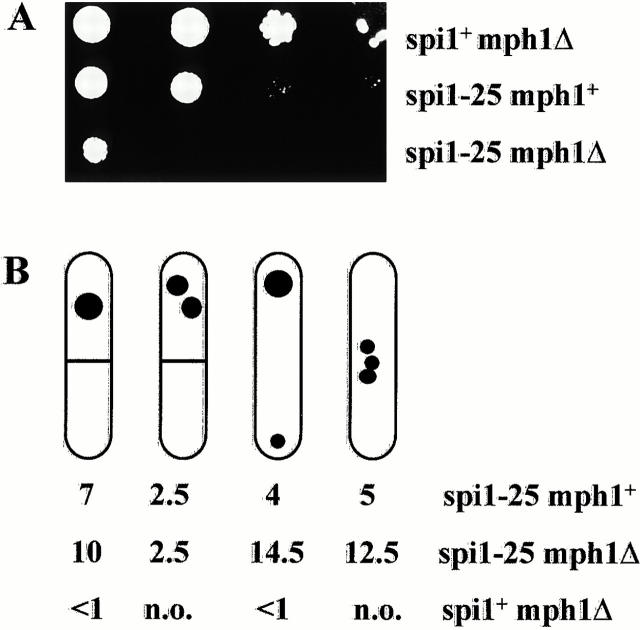
As the spi1-25 strain showed a variety of abnormal spindle phenotypes, we asked if the viability of the strain was dependent on the spindle checkpoint, which monitors the correct alignment of chromosomes on the spindle (for review see Gardner and Burke 2000), was activated by constructing double mutants of spi1-25 with mutant components of the spindle checkpoint pathway. We used deletion variants of mad2 + and mph1 + (He et al. 1997, He et al. 1998b), which are evolutionarily conserved components of this pathway. We found that the spi1-25 mad2Δ strains showed no apparent difference in growth or TBZ sensitivity compared with the single spi1-25 mutant (Table , data not shown). However, spi1-25 mph1Δ double mutant strains were severely affected. At 25°C, both single mutants grew, whereas the double mutant barely grew (Fig. 7 A). At temperatures >30°C or in the presence of sublethal doses of TBZ (6 μg/ml), the spi1-25 mph1Δ strain barely grew (data not shown). The severely reduced growth phenotype is most likely a consequence of increased aberrant mitosis in the double mutant strains. As shown diagrammatically in Fig. 7 B, absence of the mph1 + gene product gave rise to a significant increase of abnormal mitoses from 18% for the single spi1-25 mutant to 39% for the double mutant. These data indicate that the missegregation of chromosomes is reduced in spi1-25 cells due to the spindle checkpoint system.
Figure 7.
The mitotic spindle checkpoint is activated in spi1-25 cells. (A) Serial dilution patch tests (104 to 101 cells) show growth of mph1Δ and spi1-25 single mutants and spi1-25 mph1Δ double mutant at 25°C on rich medium for 4 d. The strains all come from the same tetrad. (B) Diagrammatic representation of chromatin missegregation in the strains described in A. Cells were grown at 30°C for 12 h and fixed, and the chromatin was analyzed by staining with DAPI. Given are the percentages for the various abnormal chromatin segregation phenotypes in an asynchronously growing cell population. n.o., not observed.
Mal3p and Spi1p Probably Act in Parallel Pathways
Mal3p is an evolutionary conserved microtubule-interacting protein that has been shown to associate with cytoplasmic and spindle microtubules (Beinhauer et al. 1997). Extra copies of mal3 + were able to rescue the TBZ hypersensitivity of the spi1-25 strain (see Fig. 2) but not the other phenotypes associated with spi1-25, indicating that extra Mal3p can partially complement spi1-25 malfunction. However, we were unable to coimmunoprecipitate Mal3p and Spi1p (data not shown), indicating that Mal3p and Spi1p probably do not interact physically.
We also found that nuclear transport of Mal3p was normal in the spi1-25 mutant by monitoring the in vivo localization of the Mal3p-GFP fusion protein (Beinhauer et al. 1997; data not shown).
Finally, genetic interactions between spi1 + and mal3 + were analyzed by constructing a spi1-25 mal3Δ double mutant. We found that the double mutant showed significantly reduced growth at 25°C, a temperature where mal3Δ single mutants grow normally, and spi1-25 mutants are affected only slightly (Fig. 8 A, left). Other phenotypes were also additive: the double mutant population had 53% cells with abnormal cell form, whereas the single mutants mal3Δ and spi1-25 showed 26.5 and 9.7% aberrant cells, respectively (Fig. 8 B). The mal3Δ spi1-25 strain was unable to grow on medium containing TBZ, whereas the single mutant strains showed reduced growth only (Fig. 8 A). The finding that spi1-25 and mal3Δ mutations are additive in terms of phenotypes (Table ) indicates that their gene products probably act in parallel pathways.
Figure 8.
Phenotypic analysis of the spi1-25 mal3Δ double mutant. (A) Serial dilution patch tests (104 to 101 cells) of wild-type (spi1 + mal3 +), single mutant (mal3Δ or spi1-25), and double mutant (spi1-25 mal3Δ) strains grown on rich medium with or without TBZ at 25°C. (B) The percentage of cells with abnormal cell morphology as shown diagrammatically was determined for the indicated strains. Strains were grown asynchronously in rich liquid medium at 25°C. Indicated is an abnormal cell type not seen in the single mutant strains (right).
Discussion
Spi1-25 Has a Mutation in the Switch I Region of Fission Yeast SpRan
The V44I mutation in Spi1-25p (V45 in mammalian Ran) is located in the Switch I effector binding region, which adopts a dramatically different structure depending on whether the protein is bound to GDP or GTP (Chook and Blobel 1999; Vetter et al. 1999). A mutation in the T42 residue within the Switch I region of the mammalian Ran protein is able to bind to some, but not all, of its known binding partners. Because the spi1-25 mutation is within the Switch I region, and because the phenotype of spi1-25 mutant cells is different from that of spi1 null cells, we first tested the possibility that, like T42A of human Ran, the V44I mutation in SpRan is a separation of function mutant. We found, however, that Spi1-25p is capable of interacting with several of its known binding partners in vivo and/or in vitro. These results, and the fact that spi1-25 cells are viable, indicated that spi1-25 might, in fact, be a partial loss of function mutant. Consistent with this possibility, we found that the level of Spi1p was similar in wild-type and mutant cells but that only 30% of the Spi1-25 protein was capable of binding GTP in vitro. We presume that the remaining 70% of protein is misfolded. The mutant protein is also less efficient than wild-type in its ability to rescue the temperature-sensitive lethality of the pim1-d1 ts SpRan-GEF mutant when overexpressed.
Even with a reduced level of active protein, spi1-25 cells are competent for nucleocytoplasmic transport of both the endogenous Pap1p protein and a fusion protein targeted to the nucleus by the classical SV-40 NLS. We cannot exclude the possibility that the mutant has subtle defects in transport not detected in our assays or that it is defective in the transport of only a small subset of proteins that affect microtubule function. However, we did rule out the possibility that the microtubule defect in spi1-25 was the result of an inability to import the MAP Mal3p, which rescues this defect at elevated levels of expression.
When the pool of functional SpRan protein is reduced by the spi1-25 mutation cells have a specific defect in microtubule integrity. This is in contrast to the observations that temperature-sensitive mutations in the Ran-GEF in both fission yeast and mammalian cells have no obvious effect on spindle formation. Overexpression of Ran and RanBP1 cause chromosome missegregation and TBZ sensitivity in budding yeast by an unknown mechanism (for review see Sazer and Dasso 2000).
Our results suggest the possibility that fission yeast SpRan has multiple independent functions which are differentially sensitive to loss of function of the GTPase system. The following data are consistent with this possibility: (a) our finding that imp2, a gene that encodes a protein that destabilizes the actin ring during septation is a high copy suppressor of the lethality of the RanGEF mutant pim1-d1 ts at its semipermissive temperature of 34°C but not its restrictive temperature of 36°C (Demeter and Sazer 1998); and (b) the observation that diploid fission yeast cells with a single copy of the wild-type spi1+ gene lose chromosomes and haploidize (Matsumoto and Beach 1991). We are currently testing this model by monitoring the phenotypes of cells with different levels of functional SpRan.
SpRan Is Involved in the Integrity of Interphase Microtubules
spi1-25 cells showed a variety of abnormal cell morphologies instead of the normal linear rod shape (see Fig. 1 D) that were apparent at all temperatures but became more prominent with increasing temperature. Cytoplasmic microtubules play an important role in fission yeast cell morphogenesis (Verde et al. 1995; Mata and Nurse 1997; Sawin and Nurse 1998), and mutations causing altered interphase microtubule arrays lead to misshapen cells. Tubulin mutants (Toda et al. 1984), microtubule destabilizing drugs (Walker 1982; Sawin and Nurse 1998), or mutations in genes such as tea2 + and mal3 + (Verde et al. 1995; Beinhauer et al. 1997) that give rise to abnormally short interphase microtubules all lead to aberrantly shaped cells. In wild-type cells, interphase microtubules extend along the long axis of the cell, reaching the cell tips (Hagan and Hyams 1988). In contrast, the cytoplasmic microtubules in spi1-25 cells were often abnormally short or were positioned aberrantly (see Fig. 5). These data imply that SpRan is involved in the integrity of the cytoplasmic microtubule cytoskeleton.
SpRan Affects the Formation and Function of the Mitotic Spindle
We found two main aberrations in the formation of the mitotic spindle in spi1-25 cells. One phenotype was a star- or fan-shaped tubulin staining pattern, indicating that multiple microtubule bundles originate from a single focal point. Such staining patterns are typical of mutants with a defective SPB component (Hagan and Yanagida 1995; Bridge et al. 1998; West et al. 1998) or mitotic motor protein (Hagan and Yanagida 1992). Condensed chromatin was found in close proximity to the tubulin staining. The second phenotype was a tiny bipolar premetaphase spindle between barely separated SPBs. The chromatin in these cells still had the hemispherical appearance of an interphase nucleus, indicating a delay and/or arrest in very early spindle formation. We attempted to clarify these spindle phenotypes by various methods of cell synchronization but were unsuccessful due to the heterogeneous cell shapes of spi1-25 cells and their tendency to clump. Using the mitotic stage–specific localization pattern of Cut11-GFPp (West et al. 1998), we found that spi1-25 cells have an eightfold increase in prophase cells. In addition, spi1-25 was synthetically lethal, with mutant alleles of cut11 + encoding an SPB component required for bipolar spindle formation (West et al. 1998). Cells expressing this particular mutant SpRan protein are thus able to nucleate spindle microtubules but have problems with the establishment of a bipolar spindle or show a delay and/or arrest in the transition from a premetaphase to metaphase spindle.
Our phenotypic in vivo data are in accordance with recently obtained in vitro results demonstrating that Ran regulates spindle assembly in M phase Xenopus egg extracts by a yet unknown mechanism independent of nucleocytoplasmic transport (Carazo-Salas et al. 1999; Kalab et al. 1999; Ohba et al. 1999; Wilde and Zheng 1999; Zhang et al. 1999). When Ran-GTP levels were lowered in these extracts, spindle assembly was blocked, whereas high Ran-GTP levels promoted formation of spindle structures. We do not know whether the balance between Ran-GDP and Ran-GTP is altered in spi1-25 cells, but there is a strong genetic interaction between spi1-25 and the pim1-d1 ts Ran-GEF mutant, which is predicted to have low Ran-GTP levels.
spi1-25 Interacts Genetically with the Spindle Checkpoint Pathway
The spindle checkpoint pathway arrests cells at the metaphase to anaphase transition when chromosomes are not attached properly to the mitotic spindle (for review see Gardner and Burke 2000). Although several mitotic spindles in spi1-25 cells appear morphologically normal, the increase in chromosome loss and missegregation indicates that these spindles are functionally defective. We confirmed this by showing that loss of the spindle checkpoint gene mph1 (He et al. 1998b) exacerbates the growth and chromosome missegregation defects of spi1-25. Because Mph1p is required for checkpoint activation, but unlike its S. cerevisiae homologue Mps1p is not essential for viability or spindle pole body duplication (He et al. 1998b), our results suggest that the spindle defects in spi1-25 are monitored by the spindle checkpoint pathway which transiently arrests cells in metaphase until the defects are corrected.
Although we cannot rule out the possibility that Mad2p is not required for this cell cycle delay, the lack of interaction between mad2Δ (He et al. 1997) and spi1-25 most likely reflects these facts: (a) mad2Δ is less sensitive to microtubule destabilizing drugs than mph1Δ (Kadura, S., and S. Sazer, unpublished results); (b) Mph1p acts upstream of two branches of the checkpoint pathway, only one of which includes Mad2p (for review see Taylor 1999); and (c) the interactions among components of the checkpoint pathway that have been placed in a linear genetic pathway are complex and dynamic (Brady and Hardwick 2000).
Mal3p Can Suppress the TBZ Hypersensitivity of spi1-25 Cells
Although the exact role of the SpRan GTPase in spindle formation is not yet known, the finding that extra copies of mal3 +, encoding a MAP, were able to partially complement spi1-25 malfunction points to a role of Spi1p in microtubule integrity. Mal3p may act by stabilizing microtubules since the lethal overexpression phenotype of mal3 + can be rescued by decreasing microtubule stability by various means (Beinhauer et al. 1997), and the absence of Mal3p in vivo affects microtubule dynamics by leading to a reduction in the microtubule growth rate (Ding, D.-Q., and Y. Hiraoka, personal communication).
spi1-25 and mal3Δ mutations cause different spindle defects and are additive in all phenotypes analyzed, suggesting that these two gene products act in parallel pathways. The exact role of Mal3p on mitotic spindle formation and/or function is at present unclear. Absence of mal3Δ leads to greatly reduced spindle staining and an increased chromosome condensation index (Beinhauer et al. 1997). The mal3 + homologue BIM1/YEB1 in S. cerevisiae also influences formation and function of the mitotic spindle (Schwartz et al. 1997; Muhua et al. 1998; Tirnauer et al. 1999). Bim1 was isolated in a budding yeast SPB preparation (Wigge et al. 1998), indicating an affinity of this protein family for the SPB. In this context, it is interesting to note that we have identified a novel, evolutionary conserved SPB component in a suppressor screen of mal3-1 mutant phenotypes (Decker, S., and U. Fleig, unpublished data).
Given the observation that Mal3p can partially suppress Spi1-25p malfunction together with the mitotic spindle defects seen in spi1-25 cells, we propose that the SpRan GTPase is required for the very early stages of spindle formation by possibly exerting a microtubule stabilizing function. We do not propose that SpRan is involved directly in microtubule integrity, as purified Ran has no effect on microtubule polymerization (Wilde and Zheng 1999), and Spi1p does not interact with tubulin (see Table ), but suggest that it regulates a component(s) required for microtubule dynamics. Analysis of our remaining spi1-25 multicopy suppressors might help in the identification of such a component.
Acknowledgments
We are very grateful to Iain Hagan for his generous help with 6 and the anti-Sad1p antibody; to Ted Wensel for advice on the GTP-binding assays and analysis; and to Ngoctoyen Ong for the two-hybrid assays and excellent technical assistance. We thank Dick McIntosh and Robert West (University of Colorado, Boulder, CO) for the cut11 strains; Kathy Gould for the his3Δ strain; Keith Gull (University of Manchester, Manchester UK) for the Tat1 antibody; Tony Carr for the genomic DNA library; Jens Beinhauer for Western blot analysis; Wade Harper for the UBC4 clone; and Johannes Hegemann for support.
This work was supported by the Deutsche Forschungsgemeinschaft (HE1383/7-1 to U. Fleig) and by the National Institutes of Health (GM49119 to S. Sazer).
Footnotes
Abbreviations used in this paper: EMM, Edinburgh minimal medium; ORF, open reading frame; MAP, microtubule-associated protein; NLS, nuclear localization signal; SPB, spindle pole body; TBZ, thiabendazole.
References
- Andersen S.S. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–267. doi: 10.1016/s0962-8924(00)01786-4. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Barbet N., Muriel W.J., Carr A.M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe . Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Beinhauer J.D., Hagan I.M., Hegemann J.H., Fleig U. Mal3, the fission yeast homologue of the human APC–interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady D.M., Hardwick K.G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Morphew M., Bartlett R., Hagan I.M. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D.P., Kinzler K.W., Vogelstein B., Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:57–60. [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chook Y.M., Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Demeter J., Sazer S. imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe . J. Cell Biol. 1998;143:415–427. doi: 10.1083/jcb.143.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.Q., Tomita Y., Yamamoto A., Chikashige Y., Haraguchi T., Hiraoka Y. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells. 2000;5:169–190. doi: 10.1046/j.1365-2443.2000.00317.x. [DOI] [PubMed] [Google Scholar]
- Elledge S.J., Richman R., Hall F.L., Williams R.T., Lodgson N., Harper J.W. CDK2 encodes a 33-kDa cyclin A-associated protein kinase and is expressed before CDC2 in the cell cycle. Proc. Natl. Acad. Sci. USA. 1992;89:2907–2911. doi: 10.1073/pnas.89.7.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U., Sen-Gupta M., Hegemann J.H. Fission yeast mal2+ is required for chromosome segregation. Mol. Cell. Biol. 1996;16:6169–6177. doi: 10.1128/mcb.16.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.D., Burke D.J. The spindle checkpointtwo transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M. The fission yeast microtubule cytoskeleton. J. Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hagan I.M., Hyams J.S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe . J. Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- He X., Patterson T.E., Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Hayashi N., Walcott N.G., Azuma Y., Patterson T.E., Bischoff F.R., Nishimoto T., Sazer S. The identification of cDNAs that affect the mitosis-to-interphase transition in Schizosaccharomyces pombe, including sbp1, which encodes a spi1p-GTP-binding protein Genetics 148 1998. 645 656a [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Jones M.H., Winey M., Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe J. Cell Sci 111 1998. 1635 1647b [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Toda T., Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulina cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R.T., Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. Beyond self-assemblyfrom microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Kudo N., Taoka H., Toda T., Yoshida M., Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Henry M., Silver P.A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Mata J., Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Beach D. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell. 1991;66:347–360. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W., Englmeier L. Nucleocytoplasmic transportthe soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Matynia A., Dimitrov K., Mueller U., He X., Sazer S. Perturbations in the spi1p GTPase cycle of Schizosaccharomyces pombe through its GTPase-activating protein and guanine nucleotide exchange factor components result in similar phenotypic consequences. Mol. Cell. Biol. 1996;16:6352–6362. doi: 10.1128/mcb.16.11.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F., Weber K., Gerke V. A functional homologue of the RNA1 gene product in Schizosaccharomyces pombepurification, biochemical characterization, and identification of a leucine-rich repeat motif. Mol. Biol. Cell. 1993;4:569–581. doi: 10.1091/mbc.4.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Muhua L., Adames N.R., Murphy M.D., Shields C.R., Cooper J.A. A cytokinesis checkpoint requiring the yeast homologue of an APC-binding protein. Nature. 1998;393:487–491. doi: 10.1038/31014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Masuda H., Horii J., Kuma K., Yokoyama N., Ohba T., Nishitani H., Miyata T., Tanaka M., Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Sawin K.E., Nurse P. Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S. The search for the primary function of the Ran GTPase continues. Trends Cell Biol. 1996;6:81–85. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- Sazer S., Dasso M. The ran decathlonmultiple roles of Ran. J. Cell Sci. 2000;113:1111–1118. doi: 10.1242/jcs.113.7.1111. [DOI] [PubMed] [Google Scholar]
- Sazer S., Nurse P. A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:606–615. doi: 10.1002/j.1460-2075.1994.tb06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K., Richards K., Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamada H., Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S. Chromosome segregationdual control ensures fidelity. Curr. Biol. 1999;9:R562–R564. doi: 10.1016/s0960-9822(99)80355-8. [DOI] [PubMed] [Google Scholar]
- Tirnauer J.S., O'Toole E., Berrueta L., Bierer B.E., Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Yamamoto M., Yanagida M. Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombevideo fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J. Cell Sci. 1981;52:271–287. doi: 10.1242/jcs.52.1.271. [DOI] [PubMed] [Google Scholar]
- Toda T., Adachi Y., Hiraoka Y., Yanagida M. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different alpha-tubulin genes in Schizosaccharomyces pombe . Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Toone W.M., Kuge S., Samuels M., Morgan B.A., Toda T., Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stressrequirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Mata J., Nurse P. Fission yeast cell morphogenesisidentification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I.R., Arndt A., Kutay U., Gorlich D., Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 Å resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Walczak C.E. Microtubule dynamics and tubulin interacting proteins. Curr. Opin. Cell Biol. 2000;12:52–56. doi: 10.1016/s0955-0674(99)00056-3. [DOI] [PubMed] [Google Scholar]
- Walker G.M. Cell cycle specificity of certain antimicrotubular drugs in Schizosaccharomyces pombe . J. Gen. Microbiol. 1982;128:61–71. doi: 10.1099/00221287-128-1-61. [DOI] [PubMed] [Google Scholar]
- West R.R., Vaisberg E.V., Ding R., Nurse P., McIntosh J.R. cut11(+)a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe . Mol. Biol. Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen O.N., Holmes S., Soues S., Mann M., Kilmartin J.V. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Zhang C., Hughes M., Clarke P.R. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J. Cell Sci. 1999;112:2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]