Abstract
Recent studies have opened the possibility that quiescent, G0/G1 hematopoietic stem cells (HSC) can be gene transduced; lentiviruses (such as HIV type 1, HIV) encode proteins that permit transport of the viral genome into the nucleus of nondividing cells. We and others have recently demonstrated efficient transduction by using an HIV-1-based vector gene delivery system into various human cell types including human CD34+ cells or terminally differentiated neurons. Here we compare the transduction efficiency of two vectors, HIV-based and murine leukemia virus (MuLV)-based vectors, on untreated and highly purified human HSC subsets that are virtually all in G0/G1. The HIV vector, but not MuLV vector supernatants, transduced freshly isolated G0/G1 HSC from mobilized peripheral blood. Single-step transduction using replication-defective HIV resulted in HSC that expressed the green fluorescent protein (GFP) transgene while retaining their stem cell phenotype; clonal outgrowths of these GFP+ HSC on bone marrow stromal cells fully retained GFP expression for at least 5 weeks. MuLV-based vectors did not transduce resting HSC, as measured by transgene expression, but did so readily when the HSC were actively cycling after culture in vitro for 3 days in a cytokine cocktail. These results suggest that resting HSC may be transduced by lentiviral-based, but not MuLV, vectors and maintain their primitive phenotype, pluripotentiality, and at least in vitro, transgene expression.
Genetic modification of hematolymphoid cells relies on the successful transduction of hematopoietic stem cells (HSC), the only population of cells capable of life long self-renewal, and maturation to the various blood cell types (1, 2). The long-term subsets of HSC have a high capacity for self-renewal (1) and engraftment (3–5). Transduction of cells with replication-defective murine leukemia virus (MuLV) viral particles requires the orchestration of a number of factors: efficient binding and entry via specific cell-surface receptors, reverse transcription of the proviral RNA in the cytoplasm, integration of the viral cDNA into the host cell genome (6), and expression of viral proteins and the transgene product. MuLV gene transduction occurs in target cells (7) that are in the mitotic phase of the cell cycle, presumably because the viral cDNA can only gain access to the host at M phase, when the nuclear membrane disassembles. Unfortunately, in their native state, most long-term subsets of HSC are quiescent (1); and mobilized peripheral blood (MPB) HSC are almost exclusively in G0/G1 phases of the cell cycle (8–10). Attempts to stimulate long-term subsets of HSC into cycle by using various combinations of cytokines in vitro impairs their in vivo activity (11) and usually results in their differentiation out of the long-term subset of HSC pool.
Lentivirus (such as HIV)-based vectors have been shown to transduce a variety of nondividing cell types (12–17). HIV, the prototypical lentivirus, has at least two gene products that permits it to infect nondividing cells: The matrix at the N terminus of gene, has a canonical nuclear localization signal. In the absence of Vpr, it allows nuclear entry of the preintegration complex (18–20). Here, we used a replication-defective HIV vector pseudotyped with vesicular stomatitis virus G glycoprotein (VSV-G) envelope to transduce freshly isolated G0/G1 MPB HSC. It has been shown that purified CD34+ Thy-1+ Lin− MPB cells caused rapid and sustained engraftment when patients were transplanted with 3 × 105 to 3 × 106 cells/kg (21). Postsorted primitive CD38−/lo (and less primitive CD38lo/+) subsets of CD34+ Thy-1+ Lin− MPB cells were used to compare the transduction efficiency of the HIV and MuLV vectors that contained GFP transgenes. Efficient MuLV-mediated gene transfer to cycling MPB HSC occurs only if they were cultured in vitro for 3 days to activate cells into cycle. The same vector failed to transduce freshly isolated G0/G1 MPB HSC. In contrast, an HIV-based vector transduced both G0/G1 MPB CD34+ Thy-1+ Lin− subsets with high efficiency. Most of the CD38−/lo HSC subset transduced by HIV (VSV-G) pseudotyped particle retained their Thy-1+ phenotype. When single GFP+ HSC were resorted 3–4 days after HIV-mediated gene transduction and into long-term stroma-based bone marrow culture, virtually all progeny derived from GFP+ HSC retained GFP transgene expression in their CD45+ hematolymphoid progeny. These results demonstrate that HIV, and likely other Lentiviral vectors should allow the successful transduction of primitive, quiescent stem cells with a minimum of in vitro manipulation.
MATERIALS AND METHODS
Purification of Human HSC and Flow Cytometry Analysis.
MPB was obtained from leukophoresed volunteers that were pretreated with granulocyte colony-stimulating factor (G-CSF) for 5–6 days, and CD34+ cells were enriched on Baxter Isolex Magnetic Cell Separator (Baxter Healthcare, Deerfield, IL). This CD34 enriched fraction was further incubated with mAbs against Thy-1, CD38 and Lineage markers including CD2,CD14, CD15, CD16 and glycophorin A. CD34+ Lin− Thy-1+ CD38−/lo and CD34+ Lin− Thy-1+ CD38lo/+ cells were sorted by dual-laser flow cytometry (Becton Dickinson). After sorting, the purity of these Thy-1+ subsets was analyzed by flow cytometry. To determine the cell cycle status of HSC before gene transduction, Hoechst 33342 staining for DNA content analysis was performed as described (8).
In Vitro Long-Term Cobblestone Area-Forming Cells (CAFC) Assays.
Sorted CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+ cells subsets as well as CD34+ Lin− Thy-1− cells were resorted by automated cell-deposition unit into 96-well plates with a preestablished monolayer of stromal cell line SyS-1 (21). CAFC assay was performed as described (22), in the presence of LIF (10 ng/ml), thrombopoietin (50 ng/ml), and interleukin 6 (10 ng/ml). Cultures were screened every week for 6 weeks, and linear regression analysis of the proportion of negative wells at each cell concentration was used to determine the frequency of CAFC.
Plasmid Vector Constructions, Preparation of Vector Supernatant, and Gene Transduction.
pHIV-AP was obtained from N. Landau (23). It is derived from the HIV-1 NL4–3 isolate, has a frameshift in gp160, and human placental alkaline phophatase replaces Nef. pHIV-eGFP was derived from pHIV-AP by deleting the AflIII fragment (nucleotides 6,054–7,488) and the NdeI-EcoRI fragment (nucleotides 5,123–5,743) and replacing AP with the coding sequence of enhanced green fluorescent protein, (eGFP, CLONTECH). In this vector, Gag, Pol, Tat, and Rev are intact and Nef, Vif, Vpr, and Vpu are deleted. Typical end-point virus titer of HIV-eGFP (VSV-G) on adherent human cells was greater than 107 units/ml. pHIV-lacZ was made by substituting the NotI-XhoI fragment of pHIV-AP with a 3.8-kb nuclear localization signal-lacZ gene with compatible ends (a gift of S. Bartz, Fred Hutchinson Cancer Research Center, Seattle) and deleting the AflIII fragment (nucleotides 6,054–7,488). Typical end-point virus titer of HIV-lacZ (VSV-G) on adherent human cells was greater than 107 units/ml. The MLV-lacZ [amphotropic-Moloney MLV (A-Mo-MLV)] supernatants were made by using the Propak system (24) and titered on 293 cells (end-point virus titer 5 × 106 to 5 × 107 units/ml). pME VSV-G, encoding the VSV-G glycoprotein, was a gift of K. Maruyama (DNAX). Pseudotyped HIV viral supernatants were made essentially as described, without the addition of pcRev or butyrate (17). For the HIV-lacZ (VSV-G) transductions, previously frozen viral stocks were concentrated 10-fold by ultrafiltration by using Centriprep-10 units (Amicon) according to the manufacturer’s instructions and transductions were performed at unit gravity. As a negative control, an HIV vector without envelope was used (bald). MuLV vector was derived from pLN (25) and eGFP was inserted for transgene expression. Typical end-point virus titer on adherent human cells were 3 × 106 units/ml.
Sorted CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+ cells were transduced immediately by the spinoculation method for 4 hr (26, 27). After spinoculation, cells were resuspended in serum-free defined media Ex-Vivo 15 (BioWhittaker) in the presence of thrombopoietin (100 ng/ml), FLK2L (50 ng/ml), and SLF (50 ng/ml) for 4 days, and analyzed for GFP transgene and Thy-1 expression by flow cytometry. Alternatively, the sorted cells were cultured first in the same condition for 3 days then subsequently transduced by spinoculation methods. The cells were cultured for an additional 3–4 days before testing for GFP expression.
Evaluation of GFP Expression of Single Cell-Derived Progeny in Vitro.
Freshly isolated CD34+ Lin− Thy-1+ CD38−/lo, and CD38lo/+ cells were transduced by HIV (VSV-G) eGFP and cultured for 3–4 days as described above. The cultured cells were resorted based on GFP expression, using automated cell-deposition, into 96-well plates (1, 3, 6, or 10 cells/well) with a SyS-1 monolayer in the presence of thrombopoietin (50 ng/ml), SLF (50 ng/ml), and interleukin 6 (10 ng/ml). After 5–6 weeks, wells were scored for the appearance of human hematopoietic cells and CAFC. Wells originally plated with single cells were harvested to evaluate for the presence of Gag DNA sequences by DNA-PCR and for the expression of both human CD45 and GFP by flow cytometry. PCR was performed as described (28) by using the oligo primers for detection of Gag sequences; 5′-ATAATCCACCTATCCCAGTAGGAGAAAT-3′ and 5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′.
RESULTS
In Vitro Proliferative Potential of CD34+ Thy-1+ Lin− MPB Cells.
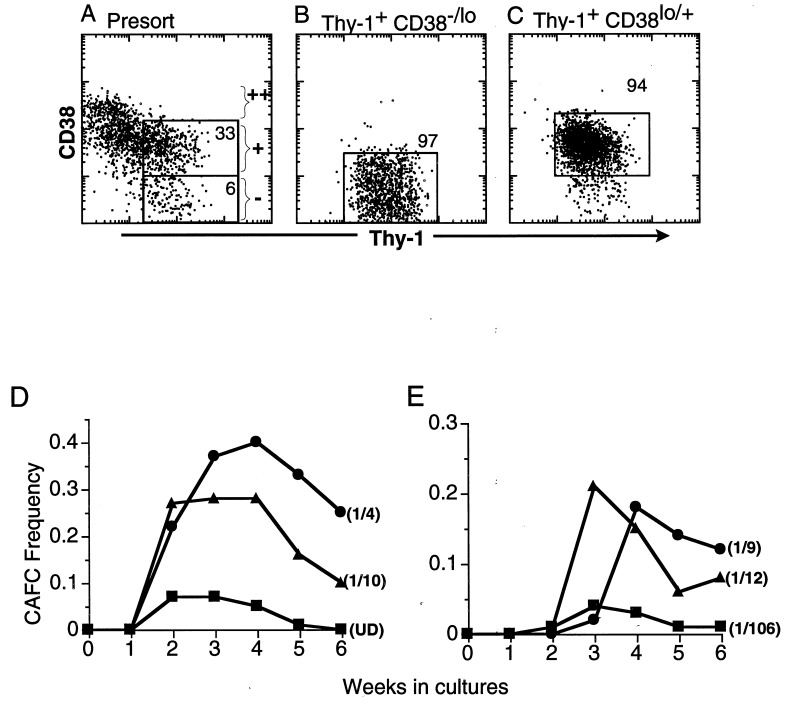
To compare human HSC subsets for biological activity and transducibility, highly purified CD34+Thy-1+Lin− MPB HSC were further separated into the CD38−/lo, and the CD38lo/+ subsets (Fig. 1 A–C). The CAFC assay was used as a measure of HSC biopotency. These Thy-1+ subsets were plated at limiting dilution (1, 2, 4, and 8 cells/well) on the cloned stromal line SyS-1 by automated cell-deposition. Although both subsets were highly enriched for long-term CAFC activity, as shown in Fig. 1 D and E, the CD38−/lo subset showed relatively low activity early on, but developed CAFC with higher frequency over time. After 6 weeks in culture, the level of CAFC activity in the CD38−/lo subset of the CD34+Thy-1+Lin− HSC was higher than the level from the CD38lo/+ subset, whereas the Thy-1− subset of CD34+ Lin− cells had little or no CAFC activity. The CAFC activity indicates that, among Thy-1+ cells, more primitive HSC were enriched in those that express negative to low levels of CD38.
Figure 1.
In vitro proliferative potential of CD34+ Thy-1+ Lin− MPB cells further subdivided based on CD38 expression. MPB CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+, and CD34+ Lin− Thy-1− cells were sorted as described. The Thy-1 vs. CD38 profile of CD34+ Lin−-gated cells revealed that the CD38 negative cells were highly enriched in Thy-1+ cells and CD38++ cells were virtually Thy-1− (A). Reanalysis of CD34+ Lin− Thy-1+ CD38−/lo (B) and CD34+ Lin− Thy-1+ CD38lo/+ (C) populations shows that we could not eliminate “CD38lo ” overlap, although sorting gates were defined to separate CD38− and CD38+ cells. With single-step sorts, the purity of CD38−/lo and CD38lo/+ subsets were 97 ± 1 (SE) % and 91 ± 2%, respectively, by the gates shown. These sorted cells were highly enriched for Thy-1+ expression (99 ± 0.08%, without CD38 gate). The mean fluorescent intensity of the Thy-1 profile on the CD38−/lo subset is consistently brighter (80 ± 8 fluorescence units) than the CD38lo/+ subset (53 ± 3) (P < 0.05). The CAFC frequency of CD34+ Lin− Thy-1+ CD38−/lo (•), CD34+ Lin− Thy-1+ CD38lo/+ (▴), and CD34+ Lin− Thy-1− (■) MPB cells from two independent experiments are shown in D and E. CAFC frequencies were calculated by linear regression analysis and the χ2 test was performed to validate linear regression analyses. The following CAFC frequencies at week 6 with lower and upper 95% confidence intervals were obtained; in experiment 1 (D) Thy-1+ CD38−/lo cells, 1/4 (1/2–1/5); Thy-1+ CD38lo/+ cells, 1/10 (1/4–1/24); and Thy-1− cells, undetectable, (UD). In experiment 2 (E), Thy-1+ CD38−/lo cells, 1/9 (1/6–1/13); Thy-1+ CD38lo/+ cells, 1/12 (1/8–1/21); and Thy-1− cells. 1/106 (1/49–1/248).
Transduction of Freshly Isolated CD38−/lo and CD38lo/+ CD34+ Thy-1+ Lin− Cells.
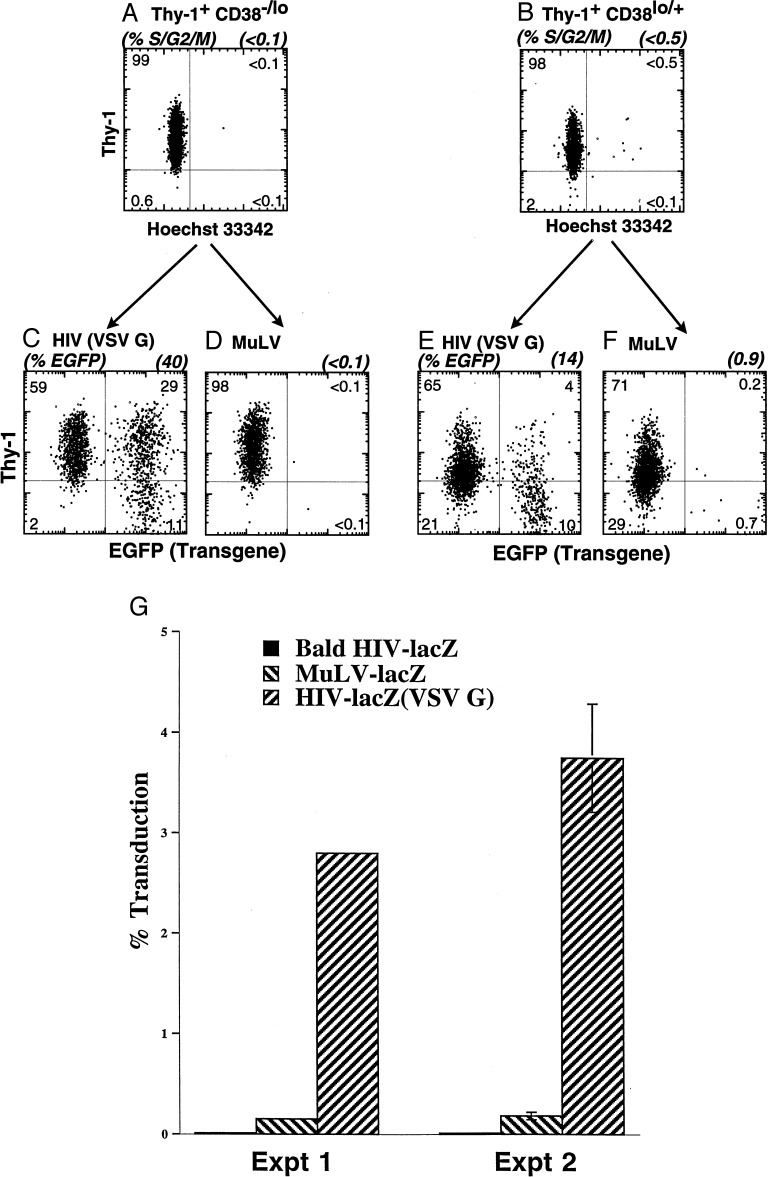
To compare the transduction efficiency of the HIV and MuLV vectors, we used the freshly isolated CD38−/lo and CD38lo/+ subsets characterized above. We have shown previously (8) that MPB HSC were virtually all in the G0/G1 phase of the cell cycle and displayed delayed cell cycle progression into S phase in the presence of multiple cytokines (>24 hr in vitro), compared with bone marrow (BM) CD34+ Thy-1+ Lin− HSC or the G0/G1 CD34+ Thy-1− Lin− progenitors from BM and MPB. Freshly isolated G0/G1 Thy-1+ CD38−/lo and CD38lo/+ cells (Fig. 2 A and B) were transduced with HIV or MuLV vectors encoding GFP. After transduction, cells were cultured in the presence of thrombopoietin, FLK-2 ligand, and Steel factor, in serum-free conditions.
Figure 2.
Transgene expression of CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+ cells transduced immediately after the sort. CD34+ Lin− Thy-1+ CD38−/lo (A) and CD38lo/+ (B) cells were sorted and analyzed for cell cycle status Hoechst 33342 (DNA content) and Thy-1 profile. These cells were transduced by with either HIV-eGFP (VSV-G) or MuLV-eGFP (A-Mo-MLV) by spinoculation for 4 hr without preculture as described in Materials and Methods; cells were cultured for 4 days. Five independent experiments revealed that an average of 36 ± 3% of CD34+ Lin− Thy-1+ CD38−/lo cells expressed GFP using HIV (VSV-G) (C), whereas virtually no cells expressed GFP (0.5 ± 0.2%) using the MuLV-eGFP (A-Mo-MLV) vector (D) (P < 0.0001). Similarly, an average of 16 ± 2% CD34+ Lin− Thy-1+ CD38lo/+ cells expressed GFP using HIV-eGFP (VSV-G) (E), 1 ± 0.5% expressed GFP using MuLV-eGFP(A-Mo-MLV) vector (F) (P < 0.0001). Thy-1+ CD38−/lo cells were transduced by HIV-eGFP(VSV-G) with higher efficiency than Thy-1+ CD38lo/+ cells (P < 0.0005). Sorted CD34+ Thy-1+ cells were transduced overnight by nit gravity in the presence into cytokine-containing medium with either MuLV-LacZ(VSV-G), MuLV-lacZ(A-Mo-MLV), ultrafiltrated bald HIV-lacZ, or ultrafiltrated HIV-lacZ(VSV-G) (G). Two days after coculture, fixed cells were stained for β-galactosidase and visually scored for blue color. In Exp. 1, MuLV-lacZ(A-Mo-MLV) was used, and in experiment 2, MuLV-lacZ(VSV-G). The mean and SD of triplicate results are shown for experiment 2. HIV-lacZ (VSV-G) was significantly better (P < 0.001) at transduction when, compared with MuLV-lacZ (VSV-G). The 20-fold difference between HIV and MuLV was reproduced in two other experiments, using different viral supernatant and stem cell preparations. The presence of Vpr, matrix, and integrase mostly facilitate nuclear pore entry of the lentiviral vectors in nondividing cells (51).
After transduction with HIV-eGFP(VSV-G), both the Thy-1+ CD38−/lo and CD38lo/+ HSC populations expressed GFP after 4 days in vitro (Fig. 2 C and E). As shown in Fig. 2C, 40% of the CD38−/lo HSC subset expressed GFP; 73% of the GFP+ cells retained Thy-1 expression (i.e., 29% of 40% GFP+). The parallel transduction of the CD38lo/+ subset resulted in only 14% of the cells expressing the transgene: the majority of GFP+ cells were Thy-1− (Fig. 2E). The CD38−/lo subset that expresses GFP retained the Thy-1+ phenotype of HSC significantly better that the CD38lo/+ subset. The former is likely to be more primitive than the latter population and is therefore more likely to retain its ability to self-renew (29).
In contrast to the HIV vector-mediated gene transfer, the MuLV-eGFP(A-Mo-MLV) vector did not transduce either subset of freshly isolated G0/G1 MPB HSC (Fig. 2 D and F). This difference may be related to the envelope used by these two viral supernatants. To exclude this possibility, freshly isolated MPB CD34+ Thy-1+ HSC were cultured for 2 days with MuLV-lacZ vectors pseudotyped with either amphotropic or VSV-G envelope. Less than 0.2% β-galactosidase expression was observed in either case, whereas a 20-fold greater rate was detected after transduction with HIV-lacZ(VSV-G) as measured by β-galactosidase expression (Fig. 2G).
Transduction of CD38−/lo and CD38lo/+ CD34+ Thy-1+ Lin− Cells after 3 Days in Vitro.
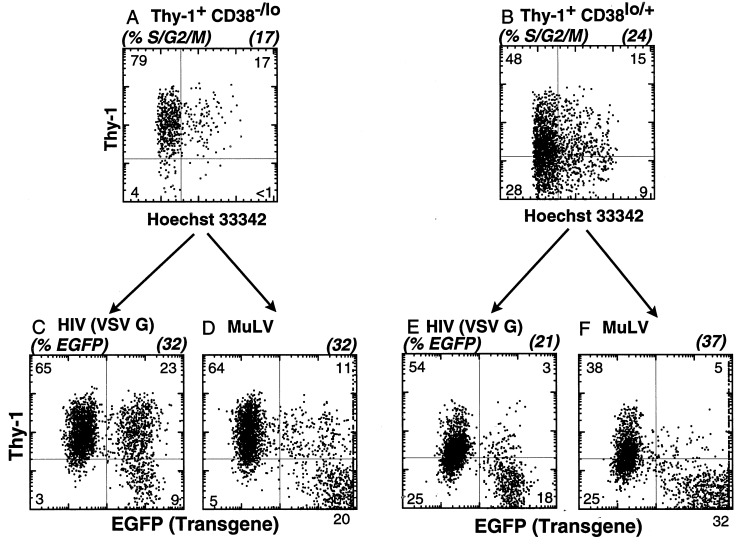
We tested whether cycling MPB HSC (previously cultured for 3 days in thrombopoietin, FLK-2 ligand, and Steel factor, in serum-free conditions) (Fig. 3 A and B) were equally transducable with either HIV or MuLV vectors. Cycling HSC subsets were transduced (as described in the Fig. 3 legend) for 4 hr, then cultured 3–4 days before analysis. As shown in Fig. 3C, the transduced progeny of the Thy-1+ CD38−/lo HSC subset expressed high levels of GFP with HIV-eGFP(VSV-G); 72% of the GFP+ cells retained Thy-1 expression (i.e., 23% of 32% GFP+). The parallel transduction of the Thy-1+CD38 lo/+ subset resulted in 21% of the cells expressing the transgene: the majority of GFP+ cells were Thy-1− (Fig. 3E). When the two subsets were transduced at day 3 with a MuLV-eGFP(A-Mo-MLV) vector, both populations were equally transduced as measured by expression of GFP (Fig. 3). Note that the GFP fluorescence of the MuLV-transduced cells was much higher compared with HIV-transduced cells, suggesting that the MuLV long-terminal repeat is more active in this subset.
Figure 3.
Transduction efficiency of CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+ cells after 3 days in vitro. CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+ cells were sorted and cultured for 3 days as described in Materials and Methods. Hoechst 33342 (DNA content) vs. Thy-1 profile of CD38−/lo (A) and CD38lo/+ (B) showed these cells were actively cycling. After 3-days in culture, cells were transduced either with HIV-eGFP (VSV-G) or MuLV-eGFP(A-Mo-MLV) by spinoculation for 4 hr. Cells were cultured for an additional 3–4 days before testing transgene expression. Five independent experiments revealed that a high but equal fraction of CD34+ Lin− Thy-1+ CD38−/lo cells resulted in an average of 34 ± 4% cells expressing GFP using HIV-eGFP (VSV-G) (C) compared with MuLV-eGFP(A-Mo-MLV) vector (32 ± 1%) (D) (P = 0.6; no statistically significant difference). Similarly, CD34+ Lin− Thy-1+ CD38lo/+ cells resulted in average of 27 ± 3% expressing GFP using HIV (VSV-G) (E) as well as by MuLV vector (33 ± 3%) (F) (P = 0.2, no significant difference).
Sustained GFP Expression in Progeny of Single Cell Derived HSC after 5–6 Weeks in Vitro.
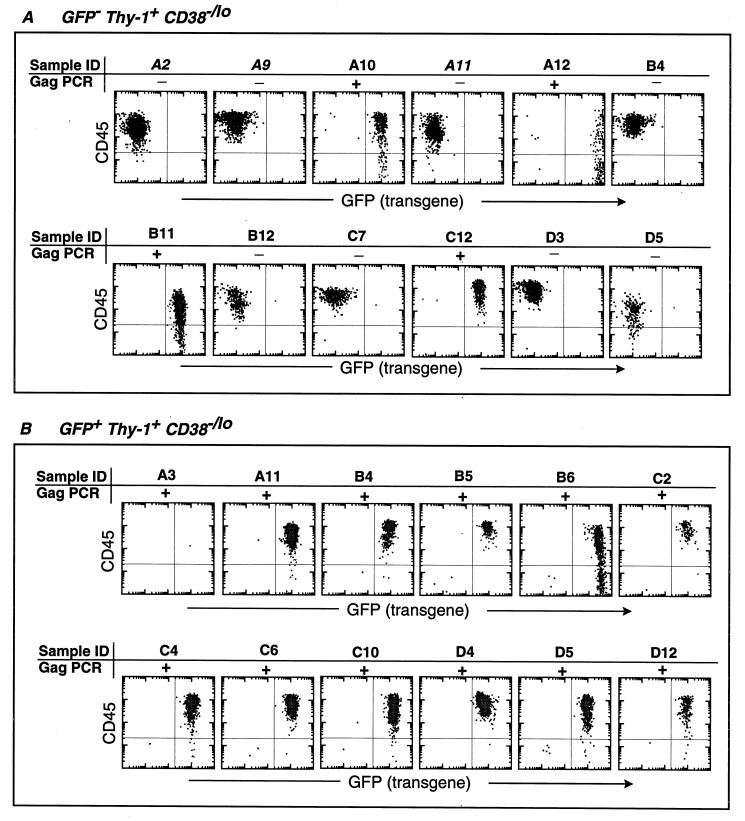
We wished to test whether the progeny of HIV eGFP(VSV-G) transduced-HSC continued to express GFP in vitro. Three to four days after HIV-mediated gene transduction, single GFP+ and GFP− HSC were sorted into wells containing SyS-1 mouse cloned stroma. Cells were harvested to evaluate the frequency of donor-derived CD45+ cells (Table 1). Both Thy-1+ CD38 −/lo and CD38 lo/+ clones derived from GFP+ cells retained GFP expression in their CD45+ progeny (Fig. 4 and Table 1). DNA-PCR analysis of clones, for HIV Gag sequences, revealed that all GFP+ clones had integrated viral Gag sequences. Surprisingly, some GFP− HSC gave rise to GFP+ clones after 5-6 wk of long-term BM culture (Fig. 4A); there was a perfect correlation between Gag+ sequence detection and GFP+ expression in these clones.
Table 1.
GFP transgene expression and Gag sequence detection from single cell-derived progenies isolated from long-term BM culture
| Population | No. wells screened | CD45+ | GFP+ | GFP+, % |
|---|---|---|---|---|
| Thy+ CD38lo/+ (bald control) | 42 | 23 | 0 | 0 |
| GFP− Thy+ CD38−/lo | 54 | 44 | 5 | 11 |
| GFP− Thy+ CD38lo/+ | 43 | 22 | 3 | 14 |
| GFP+ Thy+ CD38−/lo | 42 | 29 | 29 | 100 |
| GFP+ Thy+ CD38lo/+ | 26 | 7 | 7 | 100 |
| Total | 207 | 125 | 44 |
GFP and GFP+ Thy+ subsets after HIV-mediated gene transduction were resorted and single cells were plated on Sys-1 stromal cultures as described in Materials and Methods. After 5–6 wk of SyS-1 coculture, human hematopoietic cells were harvested to analyze for CD45 and GFP expression by flow cytometry. The same samples were also evaluated for the presence of Gag sequences by DNA-PCR. The results from three different tissues were combined. Virtually all CD45+ Gag+ progeny derived from GFP+ HSC expressed GFP. Among the CD45+ samples, no Gag sequences were detected in GFP− samples with single exception. Within the GFP+ populations, there were samples which no longer contained human CD45+ cells, yet displayed PCR Gag+.
Figure 4.
GFP expression of progeny from single cell derived HSC after 5–6 wk of in vitro culture. Freshly isolated CD34+ Lin− Thy-1+ CD38−/lo and CD38lo/+ cells were transduced by HIV-eGFP (VSV-G), cultured for 3–4 days and resorted based on GFP expression as described in Materials and Methods. After 5–6 weeks of SyS-1 coculture, proliferating human cells were scored and harvested to evaluate eGFP expressions. Human CD45 vs. GFP expression on progeny of single cells derived GFP− Thy-1+ CD38−/lo (A) and GFP+ Thy-1+ CD38−/lo (B) from the first 12 samples harvested were shown. These samples were also tested for the presence of Gag sequences by DNA-PCR analysis as indicated. In some samples, the down-regulation of CD45 expression was observed. It has been shown that thrombopoietin could induce differentiation of cells of the megakaryocyte lineage, and CD45 expression is down regulated as cells differentiate. In this experiment, the frequency of cells contributing to detectable levels of hematopoietic cell proliferation was 1 in 2.2 cells in GFP− Thy-1+ CD38−/lo and 1 in 5.9 cells from GFP+Thy-1+ CD38−/lo population. The frequency of CAFC was 1 in 3.9 cells in GFP− Thy-1+ CD38−/lo and 1 in 21.6 cells from GFP+ Thy-1+ CD38−/lo population.
DISCUSSION
The principal goal of gene therapy of HSC is to provide maximal integration and expression of transgenes without affecting the life span, self-renewal capacity, or differentiation of the transduced cells or their progeny. The barriers to achieving that goal include: (i) limitation of vector constructs to transduce human HSC with high efficiency; (ii) low frequency of transduced cells that have integrated the transgene into the genome of host cells; (iii) transgene expression “shut down” both in the short and long term; and (iv) the differentiation-inducing effects of hematopoietic cytokines on HSC in vitro. MuLV replicates poorly in noncycling cells because the breakdown of the nuclear membrane during M phase is required to allow the preintegration complex to gain access to host chromatin (30). Unfortunately, most HSC from cord blood, ABM, and MPB are in the G0/G1 phase of the cell cycle (8–10, 31). In experimental systems, two methods have been used to increase the efficiency of murine retrovirus transduction of primitive hematolymphoid cells by increasing the fraction of dividing cells. The HSC donor can be pretreated with 5-flurouracil, that depletes dividing cells and recruits HSC into cell cycle (32), or HSC can be cultured with cytokines capable of stimulating them into cell cycle before transduction (33, 34). Pretreatment of patients with chemotherapy such as 5-flurouracil is not without risk, especially when the patient suffers a hematolymphoid disorder for which gene therapy is sought. Similarly, the culture of HSC using purified cytokines has not yet resulted in expansion of HSC, and most of the cells that enter cell cycle, are committed to myeloerythroid differentiation pathways (35).
Derivatives of MuLV have been utilized as the principal gene transfer agents for both research purposes and clinical trials for gene therapy (36–41), especially for hematolymphoid cells. These oncoretroviral vectors have been useful for gene marking to trace the fate of transplanted cells (42, 43), correcting genetic defects (44, 45), modifying chemotherapeutic sensitively by insertion of drug resistance genes (e.g., mdr1 or dhfr) (46) and modulating the immune system (47, 48). Rates of MuLV-mediated gene transfer into HSC/progenitors in vitro have been improving in marking studies and animal models (mouse, dog, and nonhuman primates) (49, 50). However, clinical trials involving BM transplantation resulted in too low of a transduction efficiency in repopulating blood progeny in vivo in to be useful clinically.
We have demonstrated efficient gene transduction of freshly isolated human HSC by the use of lentiviral vectors. We and others have shown that initial integration and expression of transgenes in HSC does not guarantee expression of the transgenes in its progeny. In fact, MuLV transduction of mouse long-term HSC resulted in reproducible silencing of GFP integrants in differentiated progeny, whereas most, if not all, HSC express GFP within the same mice (C. Klug and I.L.W., unpublished results). In the experiments reported here HSC colonies grown on BM stroma retained full GFP expression in vitro. We observed that the CAFC activity of the GFP− sorted cells was consistently higher than that of the GFP+ sorted cells. Such results suggest that a gene product present in the vector HIV-eGFP, which includes Gag, Pol, Tat, Rev, and eGFP, may affect these HSC. Alternatively, more primitive (or dormant) cells may not be as easily transduced, even with lentiviral vectors. In these experiments, in vivo development (including T cells) and expression were not tested. It shall be important to study lentiviral vector mediated gene transduction and expression in vivo.
Acknowledgments
We would like to thank Drs. R. Rigg, S. Bartz, and V. Kewalramani for generous gifts of reagents, and Dr. A. Schlageter for helping in the preparation and review of the manuscript. We are grateful to Dr. S. Tamaki for stimulating discussions and reviewing the manuscript. R.E.S. was supported by National Institutes of Health Grant K08 CA71671.
ABBREVIATIONS
- MuLV
murine leukemia virus
- HSC
hematopoeitic stem cells
- A-Mo-MLV
amphotropic Moloney MLV
- MPB
mobilized peripheral blood
- BM
bone marrow
- CAFC
cobblestone area-forming cell
- GFP
green fluorescence protein
- VSV-G
vesicular stomatitis virus G glycoprotein
References
- 1. Morrison S J, Weissman I L. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 2.Spangrude G J, Heimfeld S, Weissman I L. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 3.Nibley W E, Spangrude G J. Bone Marrow Transplant. 1998;21:345–354. doi: 10.1038/sj.bmt.1701097. [DOI] [PubMed] [Google Scholar]
- 4.Zijlmans J M, Visser J W, Laterveer L, Kleiverda K, Heemskerk D P, Kluin P M, Willemze R, Fibbe W E. Proc Natl Acad Sci USA. 1998;95:725–729. doi: 10.1073/pnas.95.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uchida N, Tsukamoto A, He D, Friera A M, Scollay R, Weissman I L. J Clin Invest. 1998;101:961–966. doi: 10.1172/JCI1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller A D. Proc Natl Acad Sci USA. 1996;93:11407–11413. doi: 10.1073/pnas.93.21.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roe T, Reynolds T C, Yu G, Brown P O. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchida N, He D, Friera A, Reitsma M, Sasaki D, Chen B, Tsukamoto A. Blood. 1997;89:465–472. [PubMed] [Google Scholar]
- 9.Morrison S, Wright D, Weissman I. Proc Natl Acad Sci USA. 1997;94:1908–1913. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts A, Metcalf D. Blood. 1995;86:1600–1605. [PubMed] [Google Scholar]
- 11.Bodine D M, Crosier P S, Clark S C. Blood. 1991;78:914–920. [PubMed] [Google Scholar]
- 12.Akkina R K, Walton R M, Chen M L, Li Q X, Planelles V, Chen I S. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 14.Blomer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyoshi H, Takahashi M, Gage F H, Verma I M. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafri T, Blomer U, Peterson D A, Gage F H, Verma I M. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 17.Sutton R E, Wu H T M, Rigg R, Bohnlein E, Brown P O. J Virol. 1998;72:5781–5788. doi: 10.1128/jvi.72.7.5781-5788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallay P, Swingler S, Song J, Bushman F, Trono D. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 19.Gallay P, Swingler S, Aiken C, Trono D. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 20.von Schwedler U, Kornbluth R S, Trono D. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Archimbaud E I, Philip B, Coiffier M, Michallet G, Salles C, Sebban N, Sebban N, Roubi F, Lopez L, Bessueille P, Mazarrs P. Blood. 1996;88:595. (abstr.). [Google Scholar]
- 22.Baum C M, Weissman I L, Tsukamoto A S, Buckle A M, Peault B. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Landau N R. J Virol. 1995;69:4587–4592. doi: 10.1128/jvi.69.7.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigg R J, Chen J, Dando J S, Forestell S P, Plavec I, Bohnlein E. Virology. 1996;218:290–295. doi: 10.1006/viro.1996.0194. [DOI] [PubMed] [Google Scholar]
- 25.Kiem H P, Darovsky B, von Kalle C, Goehle S, Stewart D, Graham T, Hackman R, Appelbaum F R, Deeg H J, Miller A D, et al. Blood. 1994;83:1467–1473. [PubMed] [Google Scholar]
- 26.Bahnson A B, Dunigan J T, Baysal B E, Mohney T, Atchison R W, Nimgaonkar M T, Ball E D, Barranger J A. J Virol Methods. 1995;54:131–143. doi: 10.1016/0166-0934(95)00035-s. [DOI] [PubMed] [Google Scholar]
- 27.Bonyhadi M L, Moss K, Voytovich A, Auten J, Kalfoglou C, Plavec I, Forestell S, Su L, Bohnlein E, Kaneshima H. J Virol. 1997;71:4707–4716. doi: 10.1128/jvi.71.6.4707-4716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akkina R K, Rosenblatt J D, Campbell A G, Chen I S, Zack J A. Blood. 1994;84:1393–1398. [PubMed] [Google Scholar]
- 29.Terstappen L W, Huang S, Safford M, Lansdorp P M, Loken M R. Blood. 1991;77:1218–1227. [PubMed] [Google Scholar]
- 30.Varmus H, Padgett T, Heasley S, Simon G, Bishop J. Cell. 1977;11:307–319. doi: 10.1016/0092-8674(77)90047-2. [DOI] [PubMed] [Google Scholar]
- 31.Traycoff C M, Abboud M R, Laver J, Clapp D W, Srour E F. Exp Hematol. 1994;22:1264–1272. [PubMed] [Google Scholar]
- 32.Randall T D, Weissman I L. Blood. 1997;89:3596–3606. [PubMed] [Google Scholar]
- 33.Bodine D M, McDonagh K T, Seidel N E, Nienhuis A W. Exp Hematol. 1991;19:206–212. [PubMed] [Google Scholar]
- 34.Tumas D B, Spangrude G J, Brooks D M, Williams C D, Chesebro B. Blood. 1996;87:509–517. [PubMed] [Google Scholar]
- 35.Szilvassy S, Weller K, Chen B, Juttner C, Tsukamoto A, Hoffman R. Blood. 1996;88:3642–3653. [PubMed] [Google Scholar]
- 36.Stamatoyannopoulos J A, Nienhuis A W. Annu Rev Med. 1992;43:497–521. doi: 10.1146/annurev.me.43.020192.002433. [DOI] [PubMed] [Google Scholar]
- 37.Miller A D. Nature (London) 1992;357:455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- 38.Dunbar C E, Young N S. Curr Opin Hematol. 1996;3:430–437. doi: 10.1097/00062752-199603060-00006. [DOI] [PubMed] [Google Scholar]
- 39.Brenner M K. Recent Results Cancer Res. 1998;144:60–69. [PubMed] [Google Scholar]
- 40.Kohn D B, Nolta J A, Weinthal J, Bahner I, Yu X J, Lilley J, Crooks G M. Hum Gene Ther. 1991;2:101–105. doi: 10.1089/hum.1991.2.2-101. [DOI] [PubMed] [Google Scholar]
- 41.Mulligan R C. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 42.Lemischka I R, Raulet D H, Mulligan R C. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- 43.Jordan C T, Lemischka I R. Genes Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 44.Blaese R M, Culver K W, Chang L, Anderson W F, Mullen C, Nienhuis A, Carter C, Dunbar C, Leitman S, Berger M, et al. Hum Gene Ther. 1993;4:521–527. doi: 10.1089/hum.1993.4.4-521. [DOI] [PubMed] [Google Scholar]
- 45.Anderson W F. Science. 1984;226:401–409. doi: 10.1126/science.6093246. [DOI] [PubMed] [Google Scholar]
- 46.Blakley R L, Sorrentino B P. Hum Mutat. 1998;11:259–263. doi: 10.1002/(SICI)1098-1004(1998)11:4<259::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 47.Bonini C, Ferrari G, Verzeletti S, Servida P, Zappone E, Ruggieri L, Ponzoni M, Rossini S, Mavilio F, Traversari C, et al. Science. 1997;276:1719–1724. doi: 10.1126/science.276.5319.1719. [DOI] [PubMed] [Google Scholar]
- 48.Nabel G J, Gordon D, Bishop D K, Nickoloff B J, Yang Z Y, Aruga A, Cameron M J, Nabel E G, Chang A E. Proc Natl Acad Sci USA. 1996;93:15388–15393. doi: 10.1073/pnas.93.26.15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiem H P, Darovsky B, von Kalle C, Goehle S, Graham T, Miller A D, Storb R, Schuening F G. Hum Gene Ther. 1996;7:89–96. doi: 10.1089/hum.1996.7.1-89. [DOI] [PubMed] [Google Scholar]
- 50.Dunbar C E, Seidel N E, Doren S, Sellers S, Cline A P, Metzger M E, Agricola B A, Donahue R E, Bodine D M. Proc Natl Acad Sci USA. 1996;93:11871–11876. doi: 10.1073/pnas.93.21.11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallay P, Hope T, Chi D, Trono D. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]