Abstract
Background
Within mixtures, interactions between different xenobiotics may occur to give rise to additive, synergistic, inhibitory and/or stimulatory effects in target cells. The role that xenobiotics individually or in mixtures, and at environmental concentrations, play in the etiology of common human diseases often remains obscure.
Methods
In the presence or absence of lindane, chromosomal aberrations were detected in MCF-7 cells after 24-hr treatment with benzo[a]pyrene (B[a]P) or 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) using the cytokinesis-block micronucleus assay. Micronuclei were scored in 1,000 binucleate cells/treatment. We investigated intracellular responses using quantitative gene expression analyses of cyclin-dependent kinase inhibitor 1A [CDKN1A (P21WAF1/CIP1)], B-cell leukemia/lymphoma 2 (BCL-2), BCL-2–associated X (BAX), and isoforms of cytochrome P450 (CYP), CYP1A1, CYP1A2, and CYP1B1. Immunocytochemical analyses of p53, p21Waf1/Cip1, Bcl-2 and Bax protein expression in MCF-7 cells were also carried out.
Results
After exposure to binary mixtures of B[a]P plus lindane or PhIP plus lindane, a 10-fold increase in micronucleus formation resulted; these test agents individually induced 2- to 5-fold increases. Lindane increased the ratio of Bcl-2:Bax, as did 17β-estradiol (E2). Although treatment with B[a]P alone was found to elevate expression of P21WAF1/CIP1and CYP isoenzymes, it reduced the ratio of BCL-2:BAX mRNA transcripts. Treatment with a binary mixture of 10−8 M B[a]P plus 10−12 M lindane or 10−10 M E2 reversed B[a]P-induced reductions in the ratio of Bcl-2– to Bax-positive cells. In contrast, treatments with PhIP (known to possess hormonelike properties) plus lindane or E2 resulted in profound reductions in Bcl-2:Bax ratio.
Conclusions
Our results suggest that low-dose treatments (i.e., close to environmental levels) may increase DNA damage while influencing survival in exposed cells and that these effects may depend on the endocrine activity of test agents.
Keywords: 17β-estradiol, BAX, Bcl-2, benzo[a]pyrene, binary mixture, clonogenic assay, cytokinesis-block micronucleus assay, lindane, micronucleus, PhIP
Environmental exposures are likely to be a significant factor in the etiology of several diseases including neurodegenerative disease (Kamel and Hoppin 2004), coronary heart disease (Zhang et al. 1998), cancer (Grover and Martin 2002; Ragavan et al. 2006), and diabetes (Cranmer et al. 2000). Epidemiologic studies examining demographic differences in disease incidence and altered risk patterns among migrant populations (Peto 2001) lends weight to the notion that xenobiotics play an important role in disease causation (Safe 2004). Humans are variously and continuously exposed to mixtures of disease-implicated xenobiotics via environment and/or diet (Kalantzi et al. 2004c). Toxicity testing employing high-dose treatments with individual agents may be an inappropriate means of assessing low-dose exposures (Welshons et al. 2003). Different damage-induction mechanisms at low-dose versus high-dose concentrations may occur (Joosten et al. 2004; Kalantzi et al. 2004b). Following exposures to mixtures, additive, synergistic, inhibitory and/or stimulatory interactions between different xenobiotics may occur.
Candidate and ubiquitous xenobiotics include polycyclic aromatic hydrocarbons (PAHs) {e.g., benzo[a]pyrene (B[a]P)}, heterocyclic aromatic amines {e.g., 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)} and persistent organic pollutants (POPs) [e.g., lindane; γ-hexachlorocyclohexane (γ-HCH)] (Kalantzi et al. 2004b). B[a]P is a procarcinogen that requires metabolic activation to 7,8-diol 9,10-oxides (Sims et al., 1974); these ultimate carcinogenic and electrophilic species bind covalently with the N2 position of guanine (Cheng et al. 1989; Rubin 2001). PhIP is a protein-derived mutagen that appears to possess significant estrogenic properties (Felton et al. 2004; Gooderham et al. 2002; Lauber et al. 2004). It is metabolically activated via a two-step mechanism initially involving conversion to N-hydroxy-PhIP (Crofts et al. 1998; Zhao et al. 1994) followed by esterification to a metabolite that binds to the C8 position of guanine (Schut and Snyderwine 1999). Lindane is a lipophilic pesticide that moves through environmental compartments (Martin and Semple 2004) and may induce genomic alterations in the form of micronuclei (Kalantzi et al. 2004b).
Biotransformation is often facilitated by oxidative metabolism via the cytochrome P450 (CYP) mixed-function oxidase system. The CYP multigene family consists of constitutively-expressed, inducible isoenzymes that are found mainly in the liver but also to varying degrees in extrahepatic sites (Ragavan et al. 2004). The proteins of the B-cell leukemia/lymphoma-2 (Bcl-2) family regulate the permeability of the outer mitochondrial membrane (Vaux and Korsmeyer 1999) and apoptosis (Adams and Cory 1998), and also appear to be modulators of oncogenic progression (Theodorakis et al. 2002). Through a modulation of Bcl-2:Bcl-2-associated X (Bax), PhIP appears to delay the involution of the rat mammary gland (Venugopal et al. 1999); dysregulation effects that may not be dissimilar to those induced by 17β-estradiol (E2) (Leung and Wang 1999).
In previous studies, we demonstrated the genotoxicity of nanomolar concentrations of endogenous estrogens (E2, estrone, or estriol) (Yared et al. 2002) and low-dose lindane (Kalantzi et al. 2004b). Subsequently, it was shown that endogenous oestrogens or human milk-fat extracts, known to contain organo-chlorinated and brominated xenobiotics, markedly enhanced B[a]P-induced genotoxicity in clonogenic MCF-7 cells (Davis et al. 2002; Kalantzi et al. 2004a). Such exposures were associated with an elevation in the ratio of the anti-apoptotic Bcl-2 to the pro-apoptotic Bax proteins (Kalantzi et al. 2004b). If low-dose treatments (for the experiments described herein, less than or equal to nanomolar concentrations of test agent in culture medium) induce genotoxic and/or intracellular modulations, then quantifying and modeling co-exposure effects at such environmentally relevant levels are of utmost importance. In this study we examined the effects of various binary mixtures consisting of a DNA-reactive procarcinogen (B[a]P or PhIP) and hormone-like agent (E2 or lindane) to determine whether such in vitro exposures might markedly modulate genotoxicity and/or survival in damaged cells. Our goal was to determine whether low-dose exposure to such a binary mixture might give rise to a modulation both of genotoxicity and survival in target cells.
Materials and Methods
Chemicals and media
Unless otherwise stated, chemicals were obtained from Sigma Chemical Co. (Poole, UK), cell culture consumables from Invitrogen Life Technologies (Paisley, UK), and antibodies from Dako Cytomation (Ely, UK).
Cell culture
The estrogen-responsive breast carcinoma MCF-7 cell line was grown in Dulbecco’s modified essential medium supplemented with 10% heat-inactivated fetal calf serum, 100 U/mL penicillin and 100 μg/mL streptomycin. MCF-7 cells were cultured routinely in 75-cm2 flasks at 5% CO2 in air and 37°C in a humidified atmosphere and subcultured (1:10, vol/vol) twice weekly. Before subculture or incorporation into experiments, cultured cells were disaggregated, with 0.05% trypsin/0.02% EDTA.
The cytokinesis-block micronucleus (CBMN) assay
MCF-7 cells were treated with B[a]P, PhIP, E2, or lindane either individually or as a binary mixture, as indicated. Additions of test agent(s), in dimethylsulfoxide (DMSO), were made to a maximum concentration of 1% vol/vol.
After disaggregation and resuspension in complete medium, MCF-7 cells in 3-mL aliquots (≈ 1 × 104 cells) were seeded into 30-mm petri dishes containing 20-mm diameter coverslips (Sarstedt, UK). Cells were allowed to attach for 24 hr before the addition of test agent(s). After 24-hr treatment, the medium was replaced with fresh medium containing 2 μg/mL cytochalasin-B. Cultured for a further 24 hr, cells were fixed with 70% ethanol (EtOH) after washing the coverslips with phosphate-buffered saline (PBS). Cells were then stained with 5% Giemsa (in dH2O) before mounting the coverslips in DPX mountant (BDH Laboratory Supplies, Poole, UK) on microscope slides (Yared et al. 2002).
For each treatment condition, micronuclei (MNi) in 1,000 binucleate MCF-7 cells from a minimum of three experiments were objectively scored either as micronucleated binucleate cells (MN), as the total number of micronuclei (TMN) or as the distribution of MNi in binucleate cells. Mitotic rate was assessed as percentage of binucleate cells (mean ± SD, n = 3 counts of 500 cells). In 10 separate experiments, in the population of MCF-7 cells used in this study, ≤ 5% fluctuation in the micronucleus-forming activity of 10−6 M B[a]P was observed.
The clonogenic assay
MCF-7 cells were disaggregated and resuspended in complete medium. Aliquots (5 mL) containing ∼ 1 × 103 cells were seeded into 25-cm2 flasks with or without test agent(s) (B[a]P, PhIP, E2, or lindane) either individually or as a binary mixture, as indicated. Cells were incubated in 5% CO2 in air at 37°C in a humidified atmosphere for 24 hr. The medium was then replaced with fresh medium. Cells were cultured undisturbed for a further 7 days before removal of medium, washing with PBS and fixation with 70% EtOH. Colonies were then stained with 5% Giemsa and counted.
Immunohistochemical staining
Cells were disaggregated and resuspended in complete medium before seeding aliquots (5 mL; ∼ 1 × 105 cells) into 60-mm petri dishes containing 24-mm glass coverslips. After allowing 24 hr for attachment, cells were treated for 24 hr with test agents, as indicated. Medium was then removed and the cells washed with PBS before immediate fixation with CytoFixx fixative (CellPath plc, Skelmersdale, UK). The following antisera in bovine serum albumin (0.2%) diluted with Tris-buffered saline (pH 7.6) (BSAT) were used: cyclin-dependent kinase inhibitor 1A (CDKN1A (P21Waf1/Cip1) mouse monoclonal (SX118, Isotype: IgG1) in a 1:20 dilution; Bcl-2 mouse monoclonal (124, Isotype: IgG1) in a 1:100 dilution and Bax rabbit polyclonal in a 1:50 dilution. Fixative was removed by soaking coverslips in 95% industrial methylated spirits (IMS) for 30 min. After a 5-min wash with tap water, coverslips were incubated in 1:5 normal goat sera in Tris-buffered saline (TBS) (0.05 M, pH 7.6) for 15 min in a humidified environment. After removal of excess sera, the cover-slips were incubated with primary antibody for 1 hr at room temperature. Using the StreptABComplex duet kit (DakoCytomation), coverslips were then washed for 5 min with TBS, incubated for 30 min with secondary anti-sera (goat anti-mouse/rabbit) in BSAT and washed again for 5 min with TBS. Then coverslips were incubated with tertiary anti-sera (avidinbiotin complex) in BSAT for 30 min and washed with TBS for 5 min. 3,3′-Diaminobenzidine (DAB) chromogen in Tris/HCl buffer (0.05 M, pH 7.6) with hydrogen peroxide (0.1%) was applied to preparations for 15 min followed by another 5-min tap water wash. Finally, slides were transferred to a rack and stained (1 min) with hematoxylin (50%), rinsed with tap water, blued in Scott’s tap water for 15 sec and rinsed again. Preparations were stained for 1 min with eosin (0.1% in 0.1% calcium chloride), rinsed with tap water, and dehydrated with graded alcohol solutions through to xylene. Cell preparations were then mounted on microscope slides with Pertex mountant (CellPath plc, Newtown, UK). The percentage of positive cells was determined as the mean ± SD of five separate counts.
Quantitative real-time reverse transcriptase polymerase chain reaction (RT-PCR)
Routinely cultured cells were disaggregated and resuspended in complete medium before seeding aliquots (5 mL; ∼ 1 × 105 cells) into 60-mm petri dishes. After 24 hr, attached cells were treated for an additional 24 hr. Cells were then washed twice with PBS before lysis and total RNA extraction using the QIA-GEN RNeasy Kit in combination with the QIAGEN RNase free DNase kit (QIAGEN Ltd, Crawley, UK). DNase was incorporated into the extraction procedure to remove residual DNA, for example, pseudogene. RNA quality was routinely assessed in a 1.2% formaldehyde agarose gel; yield and purity were checked using a BioPhotometer (Eppendorf, Hamburg, Germany). RNA (0.4 μg) was reverse transcribed in a final volume of 20 μL containing Taqman reverse transcription reagents (Applied Biosystems, Warrington, UK): 1 × Taqman RT buffer; MgCl2 (5.5 mM); oligo d(T)16 (2.5 μM); dNTP mix (dGTP, dCTP, dATP, and dTTP; each at a concentration of 500 μM); RNase inhibitor (0.4 U/μL); reverse transcriptase (1.25 U/μL; Applied Biosystems, Warrington, UK); and RNase-free water. Reaction mixtures were then incubated at 25°C (10 min), 48°C (30 min), and 95°C (5 min).
cDNA samples were stored at –20°C before use. Primers (Table 1) for P21WAF1/CIP1, BCL-2, BAX, CYP1A1, CYP1A2, CYP1B1 and endogenous control or housekeeping gene cytoplasmic ACTB (β-ACTIN) were chosen using Primer Express software 2.0 (Applied Biosystems) and designed so that one primer spanned an exon boundary. Specificity was confirmed using the National Center for Biotechnology Information BLAST search tool (http://www.ncbi.nlm.nih.gov/blast/index.shtml). Quantitative real-time PCR was performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Reaction mixtures contained 1 × SYBR Green PCR master mix (Applied Biosystems); forward and reverse primers (Invitrogen) at a concentration of 300 nM (P21WAF1/CIP1, BCL-2, BAX, CYP1A1, CYP1A2, CYP1B1 or β-ACTIN); for P21WAF1/CIP1, BCL-2, BAX, CYP1A1, CYP1A2, or CYP1B1 amplification 20 ng cDNA template cDNA template or for β-ACTIN amplification 5 ng cDNA template; made to a total volume of 25 μL with sterile H2O. Thermal cycling parameters included activation at 95°C (1 min) followed by 40 cycles each of denaturation at 95°C (15 sec) and annealing/extending at 60°C (1 min). Each reaction was performed in triplicate and “no-template” controls were included in each experiment. Dissociation curves were run to eliminate nonspecific amplification, including primer dimers.
Table 1.
Primers used for quantitative real-time RT-PCR analyses.
| GenBank accession no. | Gene symbol | Primer | Sequence (5′→3′) |
|---|---|---|---|
| NM_078467 | P21WAF1/CIP1 |
P21WAF1/CIP1-F
P21WAF1/CIP1-R |
GAC CAG CAT GAC AGA TTT CTA CCA
TTC CTG TGG GCG GAT TAG G |
| NM_000633 | BCL-2 |
BCL-2-F
BCL-2-R |
GGC TGG GAT GCC TTT GTG
GCC AAA CTG AGC AGA GTC TTC AG |
| AF007826 | BAX |
BAX-F
BAX-R |
CCT GGG TTC AAG CGA TTC AC
GTG CAC AGG GCC TGT AAT CC |
| BC023019 | CYP1A1 |
CYP1A1-F
CYP1A1-R |
ACT TCA TCC CTA TTC TTC GCT ACC T
CGG ATG TGG CCC TTC TCA |
| NM_000761 | CYP1A2 |
CYP1A2-F
CYP1A2-R |
GAC ATC TTT GGA GCA GGA TTT GA
CTT CCT CTG TAT CTC AGG CTT GGT |
| NM_000104 | CYP1B1 |
CYP1B1-F
CYP1B1-R |
GTA CCG GCC ACT ATC ACT GAC A
CAC ATC AGG ATA CCT GGT GAA GAG |
| AK222925 | β-ACTIN | β-ACTIN-F
β-ACTIN-R |
CCT GGC ACC CAG CAC AAT
GCC GAT CCA CAC GGA GTA CT |
Abbreviations: F, forward primer; R, reverse primer. Nucleotide sequences were obtained from GenBank (2006).
Results
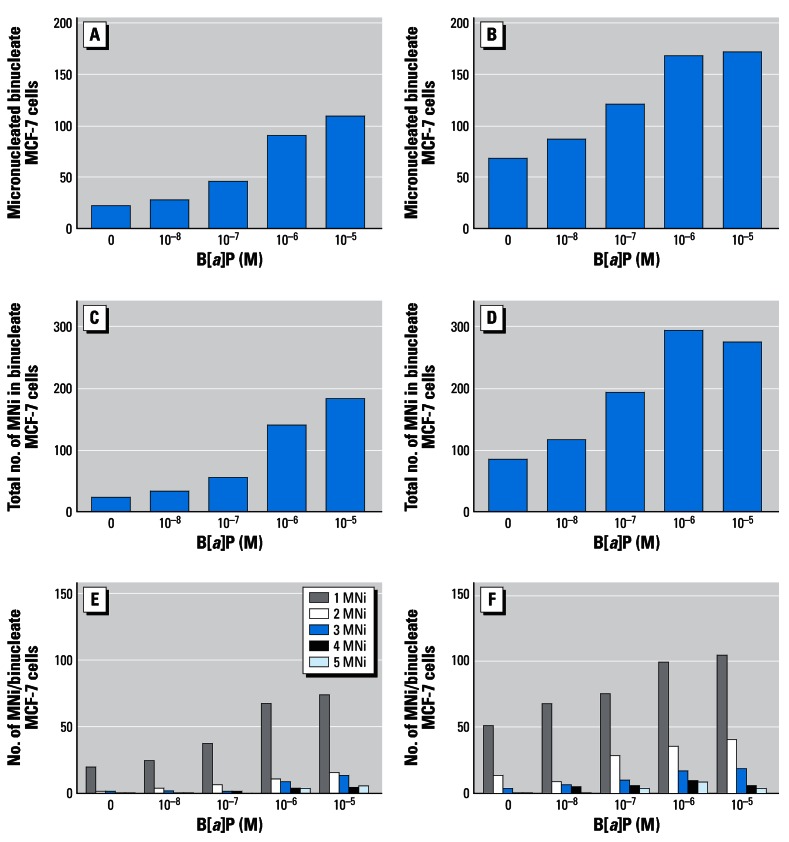
Table 2 shows the micronucleus-forming activity of B[a]P, lindane, or a binary mixture of both test agents in MCF-7 cells. Dose-related increases in the number of MN and the TMN were observed following treatment with either B[a]P or lindane alone. Induction of MNi following B[a]P treatment at concentrations of 10−7 M, 10−6 M, or 10−5 M resulted in approximately 2-, 3-, and 5-fold increases in micronucleus-forming activity, respectively. Lindane-induced increases were also observed following treatment with 10−12 M (~ 3-fold), 10−11 M (~ 3-fold), or 10−10 M (~ 4-fold). After treatment with a binary mixture, a marked elevation in micronucleus formation was induced compared with the effect of a single-agent exposure. In the presence of 10−12 M or 10−11 M lindane, clear dose-related increases in micronucleus formation were observed in the presence of B[a]P up to a concentration of 10−6 M; in combination with 10−6 M or 10−5 M B[a]P, these increases appeared to plateau (Table 2; Figure 1). Figure 1 shows that the micronucleus-forming effects of B[a]P treatment, in the absence or presence of 10−12 M lindane, were a consequence both of elevations in MN and TMN, namely, the summation of MNi in binucleate cells. In combination with 10−10 M lindane, dose-related increases in micronucleus formation in the presence of all B[a]P concentrations tested occurred; in the presence of 10−5 M B[a]P, approximately 10-fold increases in MNi compared with background control levels were observed (Table 2). Of note was the observation that micronucleus formation following treatment with 10−11 M lindane, in the absence or presence of B[a]P, was less than that observed with corresponding 10−12 M–induced effects; however, further increases in induced MNi were observed at the higher 10−10 M concentration (Table 2).
Table 2.
Micronucleus formation in MCF-7 cells treated with B[a]P and/or lindane.
| Lindane (γ-HCH) (M)
|
|||||
|---|---|---|---|---|---|
| B[a]P (M) | Micronuclei/1,000 binucleate cells | No lindane co-addition | 10−12 | 10−11 | 10−10 |
| Background | MN | 21 | 67 | 64 | 78 |
| TMN | 24 | 86 | 90 | 108 | |
| 10−8 | MN | 28 | 86 | 77 | 123 |
| TMN | 33 | 118 | 103 | 186 | |
| 10−7 | MN | 45 | 120 | 115 | 159 |
| TMN | 56 | 193 | 177 | 244 | |
| 10−6 | MN | 91 | 167 | 135 | 139 |
| TMN | 138 | 293 | 217 | 252 | |
| 10−5 | MN | 110 | 170 | 132 | 200 |
| TMN | 183 | 273 | 198 | 330 | |
Abbreviations: MN, the number of micronucleated binucleate cells scored per treatment; TMN, the total number of micronuclei scored per treatment. MCF-7 cells were treated for 24 hr in the presence or absence of test agents, as indicated, before cytokinesis block with cytochalasin-B. After further 24-hr incubation, cells were fixed and stained with 5% Giemsa as described in “Materials and Methods.”
Figure 1.
Micronucleus-forming activity of B[a]P with (A,C,E) or without lindane (B,D,F). Cell suspensions (3 mL, 1 × 104 cells) in 30-mm petri dishes were prepared and treated in the CBMN assay as described in “Materials and Methods.” Micronucleus formation was scored in 1,000 binucleate cells. (E,F) Number of MNi in binucleate MCF-7 cells were scored from 1 to 5, with 5 representing the maximum score for this study.
The effects of lindane on mitotic rate (percentage of binucleate cells) and clonogenic survival (percentage of plating efficiency) in B[a]P-treated cells are shown in Figure 2A,B. Treatment for 24 hr with B[a]P, in the absence of lindane, resulted in dose-related reductions in the percentage of binucleate cells from a background control level of 12.2% ± 0.7 to 6.6% ± 0.5 (10−5 M) (Figure 2A). In the presence of 10−12 M, 10−11 M, or 10−10 M lindane, concentration-dependent reductions in mitotic rate of approximately 20, 30, and 45%, respectively, were observed with or without B[a]P. Seven-day incubation, subsequent to a 24-hr treatment, resulted in B[a]P-induced reductions in the percentage of plating efficiency from a background control level of 35.4% ± 3.5 to 32.8% ± 4.7 (10−8 M), 28.1% ± 6.4 (10−7 M), 14.9% ± 4.2 (10−6 M), and 0 (10−5 M) (Figure 2B). Lindane, at the concentrations employed in this study, was not found to markedly alter clonogenic survival of MCF-7 cells in the absence or presence of B[a]P (Figure 2B).
Figure 2.
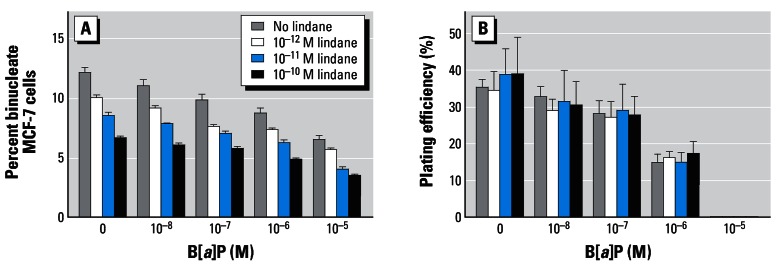
Effects of B[a]P, with or without lindane, on mitotic rate (A) and percentage of plating efficiency (B) in MCF-7 cells. After 24-hr treatment, cells were either blocked at cytokinesis before subsequent staining or cultured undisturbed for a further 7 days as described in “Materials and Methods.” Mitotic rate was estimated as percentage of binucleate cells from the mean ± SD of three separate counts (n = 500 cells). Clonogenic survival was calculated by estimating the percentage of colonies counted over the number of cells initially seeded.
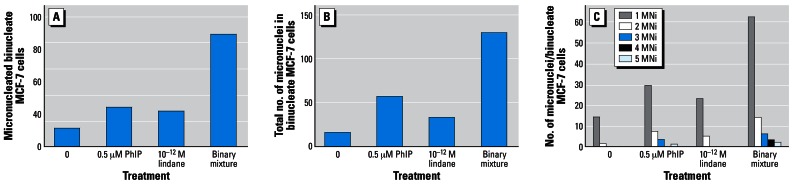
Figure 3 shows the micronucleus-forming activities, in MCF-7 cells of 24-hr treatments with PhIP (0.5 μM), lindane (10−12 M) or a binary mixture of both test agents. Both agents individually induced 2- to 4-fold increases in MNi. However, treatment with a binary mixture resulted in levels of MN and TMN of 87 and 130 (a 5- to 8-fold increase) compared with a background control level of 15 and 16, respectively (Figure 3).
Figure 3.
Micronucleus-forming activity of PhIP with or without lindane. Cell suspensions (3 mL, ~ 1 × 104 cells) in 30-mm petri dishes were prepared and treated in the CBMN assay as described in “Materials and Methods.” Micronucleus formation was scored in 1,000 binucleate cells. (C) Number of MNi in binucleate MCF-7 cells were scored from 1 to 5, with 5 representing the maximum score for this study.
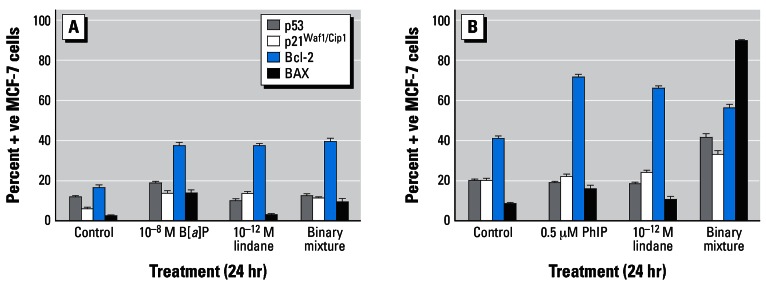
The effects of E2 (10−9 M), lindane (10−12 M) or of a binary mixture of both test agents on the immunocytochemical detection of p53, p21Waf1/Cip1, Bcl-2, and Bax proteins after 24-hr treatment are shown in Figure 4. E2 induced a modest increase in p53- and p21Waf1/Cip1-positive MCF-7 cells (17.8% ± 3.3 and 13.2% ± 1.5 compared with background control levels of 8.4% ± 1.5 and 8.2% ± 2.8, respectively). Lindane appeared to reduce levels of p21Waf1/Cip1-positive cells both in the absence (7.2% ± 1.5) or presence (5.6% ± 1.5) of E2. However, marked increases in Bcl-2–positive cells were induced by either E2 or lindane; the binary mixture elevated the percentage of both Bcl-2– and Bax-positive cells (Figure 4). From background control levels of 33.8% ± 3.1 (Bcl-2–positive) and 12.8% ± 1.9 (Bax-positive) cells, E2, or lindane treatment gave rise to percentages of cells staining positive for Bcl-2 of 58.8% ± 6.5 or 67.0% ± 4.3 and for Bax of 9.8% ± 2.8 or 6.4% ± 2.3, respectively. A binary mixture gave rise to levels of 74.6% ± 5.6 Bcl-2–positive cells and 55.0% ± 4.5 Bax-positive cells (Figure 4).
Figure 4.
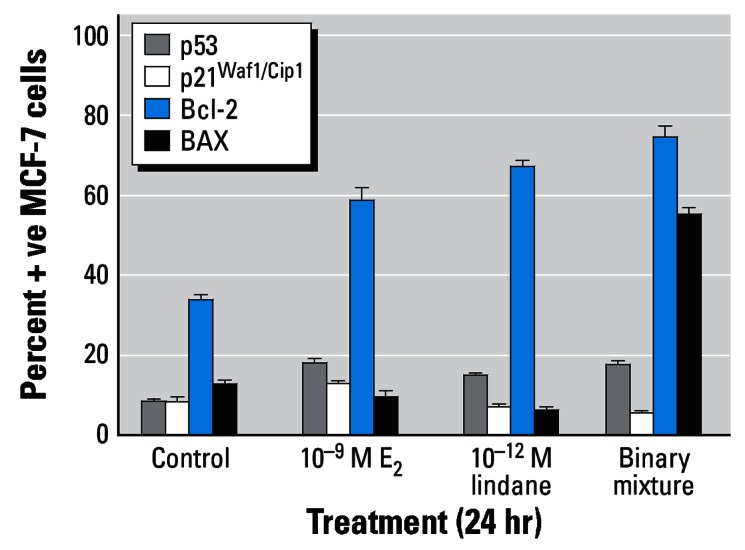
Immunocytochemical analysis of MCF-7 breast cells treated with E2 and/or lindane. Cells were treated as indicated for 24 hr on coverslips, after which they were analyzed for protein expression as described in “Materials and Methods.” The percentages of cells staining positive were determined after five separate counts of 100 cells and are presented as mean ± SD.
The effects of different treatments on the quantitative expression of six genes (P21WAF1/CIP1, BCL-2, BAX, CYP1A1, CYP1A2, and CYP1B1) were examined. To assess background fluctuation, we examined the variability in gene expression within control cell cultures (n = 6). Taking one control as the calibrator, marked intraexperimental variability was observed for relative levels of quantitative expression of CYP1A1 (0.7–1.5), CYP1A2 (0.5–1.7), and CYP1B1 (0.6–1.5). In contrast, calibrator-controlled quantitative expression levels for P21WAF1/CIP1 (1.1–1.4), BCL-2 (0.9–1.0), or BAX (0.9–1.3) appeared to be less variable.
In a panel of experiments (listed 1–5), the effects of a genotoxin (B[a]P or PhIP) in the absence or presence of 10−9 M E2 or 10−11 M lindane, on the quantitative gene expression of P21WAF1/CIP1, BCL-2, or BAX were examined (Table 3). Although 24-hr treatment with E2 or lindane (with or without 10−8 M B[a]P or 0.5 μM PhIP) was associated with a reduction in P21WAF1/CIP1 expression in experiments 1, 3, and 5, this was not apparent in experiments 2 and 4. In contrast, 24-hr treatment with 10−6 M B[a]P resulted in 36.1- and 56.3-fold elevations in P21WAF1/CIP1 expression in experiments 2 and 4, respectively; in the presence of E2 or lindane, small reductions in these increases in gene expression were observed (Table 3). No marked alterations in BCL-2 or BAX expression were observed following 24-hr treatment with 10−9 M E2 or 10−11 M lindane. However, 10−6 M B[a]P was observed to reduce the ratio of BCL-2:BAX expression to approximately 0.1 as observed in experiments 2 and 4; effects that were not markedly altered in the presence of a binary mixture (Table 3).
Table 3.
Relative gene expression measured by quantitative real-time RT-PCR.
| Relative expression levels in MCF-7 cells
|
|||
|---|---|---|---|
| Treatment | P21WAF1/CIP1 | BCL-2 | BAX |
| Experiment 1 (mean ± SD, n = 5) | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−8 M B[a]P | 1.2 ± 0.4 | 0.9 ± 0.3 | 1.1 ± 0.2 |
| 10−9 M E2 | 0.6 ± 0.1 | 0.9 ± 0.3 | 1.2 ± 0.2 |
| Binary mixture | 0.9 ± 0.2 | 0.9 ± 0.3 | 0.6 ± 0.6 |
| Experiment 2 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−6 M B[a]P | 36.1 | 0.2 | 1.5 |
| 10−9 M E2 | 1.0 | 1.3 | 1.3 |
| Binary mixture | 35.5 | 0.1 | 1.4 |
| Experiment 3 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−8 M B[a]P | 1.1 | 1.3 | 1.0 |
| 10−11 M lindane | 0.7 | 1.2 | 1.0 |
| Binary mixture | 0.9 | 1.0 | 1.0 |
| Experiment 4 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−6 M B[a]P | 56.3 | 0.3 | 2.1 |
| 10−11 M lindane | 1.9 | 2.0 | 2.9 |
| Binary mixture | 47.9 | 0.3 | 2.2 |
| Experiment 5 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 0.5 μM PhIP | 1.4 | 1.2 | 1.4 |
| 10−9 M E2 | 0.8 | 1.0 | 1.6 |
| Binary mixture | 0.6 | 0.7 | 1.1 |
calibrator, which for the purposes of these experiments were untreated controls. Relative gene expression levels following 24-hr treatment in the presence or absence of test agents, as indicated. Reverse transcription of total RNA and subsequent amplification was carried out using primers as described in “Materials and Methods.” Within each experiment, reactions were performed in triplicate and “no-template” controls were included. Averaged threshold cycle (CT) values for each reaction were normalized to β-ACTIN values thus giving ΔCT values. Alterations in gene expression were determined by comparing treatment groups with the calibrator, giving ΔΔCT values. Finally, relative gene expression was calculated using the formula 2−ΔΔCT.
Figure 5A,B shows the effects of 24-hr treatment with B[a]P (10−8 M or 10−6 M), in the absence or presence of E2 (10−10 M or 10−9 M), on the immunocytochemical detection of p53, p21Waf1/Cip1, Bcl-2, and Bax proteins in MCF-7 cells. Although treatment with B[a]P was associated with dose-related increases in the percentage of p53- and p21Waf1/Cip1-positive cells, no such E2-induced alterations were observed. However, marked E2-induced alterations in the percentages of Bcl-2– and Bax-positive cells were observed. Treatment with 10−8 M B[a]P or 10−10 M E2 resulted in marked elevations in the percentage of Bcl-2–positive cells (52.4% ± 4.7 and 72.0% ± 4.0, respectively, compared with a background control level of 31.2% ± 3.7); E2 also reduced the percentage of Bax-positive cells from a background control level of 23.2% ± 4.1 to 6.2% ± 1.5 (Figure 5A). Similar modulations in the levels of Bcl-2–(47.8% ± 7.9) and Bax-positive (7.4% ± 3.0) cells were apparent after exposure to a binary mixture. The resulting ratios of Bcl-2–positive: Bax-positive cells were 1.3, 1.8, 11.6, and 6.5 in control, B[a]P-treated, E2-treated and binary mixture–treated cells, respectively (Figure 5A). In contrast, the ratio of Bcl-2–positive: Bax-positive cells following 24-hr treatment with 10−6 M B[a]P (58.0% ± 3.0 and 82.2% ± 1.9 Bcl-2– and Bax-positive cells, respectively) was observed to be 0.7 compared with a background control level of 2.4 (37.8% ± 5.1 and 16.0% ± 2.3 Bcl-2–and Bax-positive cells, respectively) (Figure 5B). The ratio of Bcl-2– positive: Bax-positive cells was again markedly elevated (8.4; from 78.6% ± 5.8 and 9.4% ± 2.1 Bcl-2– and Bax-positive cells, respectively) following 24-hr treatment with 10−9 M E2, whereas with a binary mixture (E2 plus B[a]P), a ratio of 0.9 (82.2% ± 2.8 and 87.0% ± 2.9 Bcl-2– and Bax-positive cells, respectively) was observed (Figure 5A).
Figure 5.
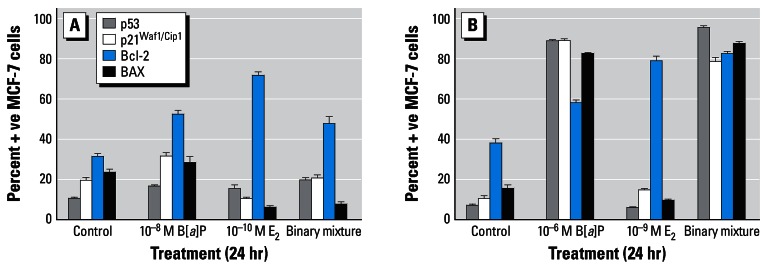
Immunocytochemical analysis of MCF-7 breast cells treated with B[a]P with or without E2. Cells were treated with 10−8 M B[a]P and/or 10−10 M E2 (A) or 10−6 M B[a]P and/or 10−9 M E2 (B) for 24 hr on cover-slips, after which they were analyzed for protein expression as described in “Materials and Methods.” The percentages of cells staining positive were determined after five separate counts of 100 cells and are presented as mean ± SD.
Treatment with 0.5 μM PhIP and/or 10−9 M E2 did not result in marked alterations in the levels of p53- or p21Waf1/Cip1-positive MCF-7 cells (Figure 6). However, both agents induced marked modulations in the levels of Bcl-2– and Bax-positive cells when tested either individually or in a binary mixture. Treatment with PhIP or E2 elevated the percentage of Bcl-2–positive cells (73.0% ± 22.0 and 78.6% ± 4.0, respectively, compared with a background control level of 14.2% ± 4.9); PhIP also increased the percentage of Bax-positive cells from a background control level of 3.4% ± 1.7 to 22.0% ± 4.8. Exposure of MCF-7 cells to a binary mixture (0.5 μM PhIP plus 10−9 M E2) resulted in increases in both Bcl-2– (55.0% ± 3.9) and Bax-positive (54.0% ± 4.6) MCF-7 cells. The resulting ratios of Bcl-2–positive:Bax-positive cells were 4.2, 3.3, 19.7, and 1.0 in control, PhIP-treated, E2-treated, and binary mixture–treated cells, respectively (Figure 6).
Figure 6.
Immunocytochemical analysis of MCF-7 breast cells treated with PhIP and/or E2. Cells were treated as indicated for 24 hr on coverslips, after which they were analyzed for protein expression as described in “Materials and Methods.” The percentages of cells staining positive were determined after five separate counts of 100 cells and are presented as mean ± SD.
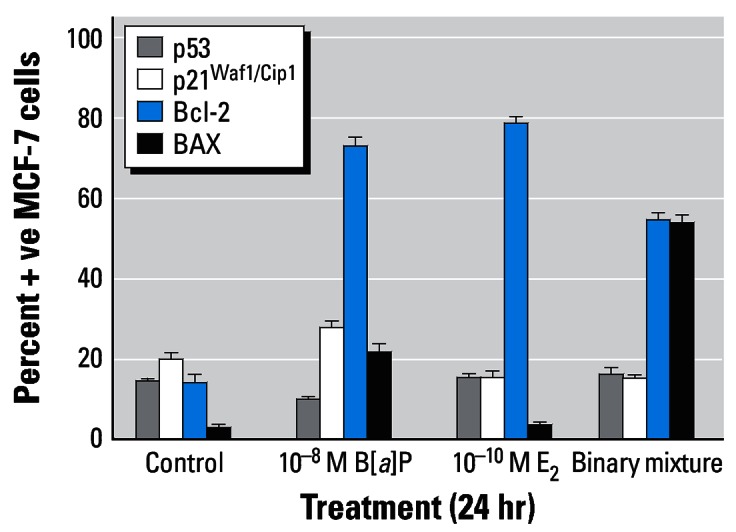
Treatment with 10−8 M B[a]P, 10−12 M lindane, or 0.5 μM PhIP was not observed to alter levels of p53- or p21Waf1/Cip1-positive cells; however, a binary mixture of lindane plus PhIP induced marked increases in p53- (41.4% ± 5.0) and p21Waf1/Cip1-positive (33.0% ± 4.4) cells compared with background control levels of 20.0% ± 1.9 and 20.2% ± 2.6, respectively (Figure 7A,B). Despite these observations, the low-dose treatments employed markedly altered levels of Bcl-2– and Bax-positive cells. B[a]P increased the levels of both Bcl-2– (37.6% ± 3.6) and Bax-positive (13.8% ± 3.5) cells giving rise to a ratio of 2.7 compared with a background control ratio of 7.5 (16.6% ± 2.7 and 2.2% ± 1.3 Bcl-2– and Bax-positive cells, respectively) (Figure 7A). In contrast, lindane-induced alterations in the ratio of Bcl-2–positive: Bax-positive cells were 13.4 (37.4% ± 2.9 and 2.8% ± 1.3 Bcl-2– and Bax-positive cells, respectively) (Figure 7A) and 6.1 (65.8% ± 2.8 and 10.8% ± 3.6 Bcl-2– and Bax-positive cells, respectively) (Figure 7B). The background control ratio of Bcl-2–positive: Bax-positive cells was 4.7 (40.8% ± 4.0 and 8.6% ± 1.8 Bcl-2– and Bax-positive cells, respectively) in Figure 7B. Exposure to a binary mixture resulted in ratios of Bcl-2–positive: Bax-positive cells of 4.2 (39.6% ± 3.8 and 9.4% ± 3.7 Bcl-2– and Bax-positive cells, respectively) (Figure 7A) and 0.6 (56.0% ± 4.9 and 89.6% ± 0.9 Bcl-2– and Bax-positive cells, respectively) (Figure 7B).
Figure 7.
Immunocytochemical analysis of MCF-7 breast cells treated with (A) B[a]P and/or lindane or (B) PhIP and/or lindane. Cells were treated as indicated for 24 hr on coverslips, after which they were analyzed for protein expression as described in “Materials and Methods.” The percentages of cells staining positive were determined after five separate counts of 100 cells and are presented as the mean ± SD.
Table 4 shows the results of a panel of experiments (listed 1–4) examining the effects of B[a]P (10−8 M or 10−6 M), E2 (10−10 M or 10−9 M), lindane (10−12 M, 10−11 M or 10−9 M), and various binary mixtures on the quantitative gene expression of CYP1A1, CYP1A2 or CYP1B1. Interexperimental variations in the ability of low-dose treatments to alter CYP expression were observed; in experiment 1, 10−8 M B[a]P and/or 10−9 M E2 did not induce the CYP isoenzymes examined, whereas in experiment 4, 3- to 4-fold increases in the CYP1A1 expression were observed. Similarly, lindane-associated inter-experimental variation was noted; in experiment 3, 10−11 M lindane treatment was associated with reductions in CYP expression, whereas in experiment 4, 24-hr treatment with either 10−12 M or 10−9 M lindane induced a 2- to 4-fold increase in CYP1A1. Treatment with 10−6 M B[a]P consistently induced marked elevations in CYP1A1, CYP1A2, and CYP1B1 expression in MCF-7 cells (Table 4).
Table 4.
Relative gene expression measured by quantitative real-time RT-PCR.
| Relative expression levels in MCF-7 cells
|
|||
|---|---|---|---|
| Treatment | CYP1A1 | CYP1A2 | CYP1B1 |
| Experiment 1 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−8 M B[a]P | 0.8 | 1.0 | 0.9 |
| 10−9 M E2 | 0.7 | 0.8 | 1.5 |
| Binary mixture | 0.9 | 0.5 | 1.8 |
| Experiment 2 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−10 M E2 | 1.4 | 0.3 | 0.9 |
| 10−8 M B[a]P | 1.6 | 1.2 | 1.4 |
| Binary mixture1 | 2.5 | 1.0 | 1.8 |
| 10−6 M B[a]P | 462.7 | 5.6 | 28.6 |
| Binary mixture 2 | 430.5 | 3.7 | 26.5 |
| Experiment 3 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−6 M B[a]P | 370.9 | 15.4 | 41.6 |
| 10−11 M lindane | 0.6 | 0.4 | 0.6 |
| Binary mixture | 183.9 | 12.0 | 23.3 |
| Experiment 4 | |||
| Control | 1 (c) | 1 (c) | 1 (c) |
| 10−9 M E2 | 3.6 | 0.9 | 1.6 |
| 10−8 M B[a]P | 3.6 | 1.4 | 1.3 |
| 10−6 M B[a]P | 457.7 | 9.4 | 23.9 |
| 10−12 M lindane | 2.0 | 0.8 | 1.0 |
| 10−9 M lindane | 3.6 | 1.3 | 1.4 |
calibrator, which for the purposes of these experiments were untreated controls. Where indicated binary mixture 1 = 10−10 M E2 + 10−8 M B[a]P; binary mixture 2 = 10−10 M E2 + 10−6 M B[a]P. Relative gene expression levels after 24-hr treatment in the presence or absence of test agents, as indicated. Reverse transcription of total RNA and subsequent amplification was carried out using primers as described in “Materials and Methods.” Within each experiment, reactions were performed in triplicate and “no-template” controls were included. Averaged threshold cycle (CT) values for each reaction were normalized to β-ACTIN values, thus giving ΔCT values. Alterations in gene expression were determined by comparing treatment groups with the calibrator, giving ΔΔCT values. Finally, relative gene expression was calculated using the formula 2−ΔΔCT.
Discussion
Humans may be exposed to different combinations of exogenous factors, including xenobiotics; evaluating only the toxicity of individual exposures might underestimate mixture effects (Culp et al. 2000; Maier et al. 2002; Marston et al. 2001; McLuckie et al. 2004). Apparently innocuous components of a mixture may significantly modulate underlying susceptibility of target cells to more toxic constituents (Slaga et al. 1979). Modulating effects may occur through ligand-activated, receptor-mediated mechanisms (Eltom et al. 1998) or interference with cell cycle control (Oikawa et al. 2001). Some xenobiotics may not only induce DNA damage but also may drive clonal expansion, often at low-dose levels not dissimilar to typical human exposures; such effects may explain the target-organ specificity of some procarcinogens (Lauber et al. 2004; Plíšková et al. 2004). Endocrine-active properties of xenobiotics may play a role in the etiology of much human pathology (Safe 2004) in hormone-responsive tissues (Ragavan et al. 2004).
B[a]P, PhIP, or lindane individually induced 2- to 5-fold increases in MNi; treatment with a binary mixture gave rise to approximately 10-fold increases (Table 2, Figures 1 and 3). Consistent with previous findings (Kalantzi et al. 2004b) that lindane (γ-HCH) induces a cell cycle arrest in MCF-7 cells, dose-related reductions in mitotic rate were observed following treatment with this agent (Figure 2A). However, in a binary mixture lindane was not observed to alter B[a]P-induced reductions in clonogenic survival (Figure 2B). At micromolar concentrations, B[a]P markedly altered gene expression by inducing up-regulation of P21WAF1/CIP1 and CYP isoenzymes and down-regulation of BCL-2 (Tables 4 and 5). However, with 24-hr treatment, no marked modulations in gene expression were apparent following low-dose treatments (less than or equal to nanomolar range) with either individual-agent or binary-mixture treatments (Tables 4 and 5). This was despite observations that 10−9 M E2 or lindane did induce a 3-fold elevation in CYP1A1 expression in one experiment (Table 4). Such observations suggest that modulations in expression of the candidate genes examined may be transient following low-dose treatment or that other factors are responsible for the effects observed, for example, oxidative damage (Jiao et al. 2007; Rajapakse et al. 2005).
Pleiotropic induction of unrelated phenotypic effects appears to be a feature of oestrogens, endocrine disruptors and PAHs (Jeffy et al. 2002; Moggs and Orphanides 2001; Mueller et al. 2004). Endogenous hormones facilitate cell growth/proliferation, differentiation, behavior, and activity in many tissues. In responsive cells, stimulatory effects are observable at less than or equal to nanomolar concentrations (Yared et al. 2002). Different xenobiotics may also induce effects through receptor-mediated processes (Kalantzi et al. 2004b; Lauber et al. 2004), for example, E2 appears to increase the percentage of Bcl-2–positive cells (Figures 4–6). A binary mixture of E2 and lindane induced a marked reduction in the ratio of Bcl-2–positive to Bax-positive cells (Figure 4); this was due to an apparently induced increase in Bax-positive cells while the percentage of Bcl-2–positive cells remained unchanged. 2,3,7,8-Tetra-chlorodibenzo-p-dioxin is cytostatic in MCF-7 cells through an ability to function as an antiestrogen and down-regulate a battery of E2-induced proliferative responses (Döhr et al. 1995; Wang et al. 1998); whether lindane may induce a similar effect through a reduction in Bcl-2:Bax ratio remains to be determined.
In a dose-related fashion, B[a]P elevated the percentage of p53-positive cells (Kaspin and Baird 1996) and the downstream cyclin-dependent kinase inhibitor, p21Waf1/Cip1 (Figure 5). PhIP (0.5 μM) apparently failed to increase the percentage of p53-positive or p21Waf1/Cip1-positive MCF-7 cells (Figures 6 and 7); at higher concentrations and after metabolic activation, PhIP has been shown to elevate these intracellular factors in MCF10A cells (Creton et al. 2005). However, PhIP markedly elevated the percentage of Bcl-2–positive cells while the levels of Bax-positive cells remained relatively low; although B[a]P also increased levels of Bcl-2–positive cells, marked increases in Bax-positive cells were also apparent (Figures 5–7). In a binary mixture with E2 or lindane, a reversal in B[a]P-induced reductions in the ratio of Bcl-2–positive to Bax-positive cells was observed (Figures 5 and 7). However, E2 or lindane in combination with PhIP appeared to reverse potential hormone-driven survival characteristics observed as marked reductions in the ratio of Bcl-2–positive to Bax-positive cells (Figures 6 and 7).
In this study we investigated whether binary mixtures of DNA-reactive procarcino-gens and hormonelike compounds at environmentally relevant low-dose concentrations give rise to markedly elevated DNA damage in target cells while also modulating survival. Our results suggest that this might indeed be the case.
Footnotes
This article is part of the monograph “Endocrine Disruptors—Exposure Assessment, Novel End Points, and Low-Dose and Mixture Effects.”
We gratefully acknowledge grant support from the North West Cancer Research Fund (R.H.) and Rosemere Cancer Foundation (F.L.M.). A.F. and P.J.L. were both recipients of a Fogarty Minority International Research Training Fellowship Program grant from the National Cancer Institute.
References
- Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- Cranmer M, Louie S, Kennedy RH, Kern PA, Fonseca VA. Exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is associated with hyperinsulinemia and insulin resistance. Toxicol Sci. 2000;56:431–436. doi: 10.1093/toxsci/56.2.431. [DOI] [PubMed] [Google Scholar]
- Creton S, Zhu H, Gooderham NJ. A mechanistic basis for the role of cycle arrest in the genetic toxicology of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Toxicol Sci. 2005;84:335–343. doi: 10.1093/toxsci/kfi075. [DOI] [PubMed] [Google Scholar]
- Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1, P4501A2 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- Culp SJ, Warbritton AR, Smith BA, Li EE, Beland FA. DNA adduct measurements, cell proliferation and tumor mutation induction in relation to tumor formation in B6C3F1 mice fed coal tar or benzo[a]pyrene. Carcinogenesis. 2000;21:1433–1440. [PubMed] [Google Scholar]
- Davis C, Bhana S, Shorrocks AJ, Martin FL. Oestrogens induce G1 arrest in benzo[a]pyrenetreated MCF-7 breast cells whilst enhancing genotoxicity and clonogenic survival. Mutagenesis. 2002;17:431–438. doi: 10.1093/mutage/17.5.431. [DOI] [PubMed] [Google Scholar]
- Döhr O, Vogel C, Abel J. Different response of 2,3,7,8-tetra-chlorodibenzo-p-dioxin (TCDD)-sensitive genes in human breast cancer MCF-7 and MDA-MB 231 cells. Arch Biochem Biophys. 1995;321:405–412. doi: 10.1006/abbi.1995.1411. [DOI] [PubMed] [Google Scholar]
- Eltom SE, Larsen MC, Jefcoate CR. Expression of CYP1B1 but not CYP1A1 by primary cultured human mammary stromal fibroblasts constitutively and in response to dioxin exposure: role of the Ah receptor. Carcinogenesis. 1998;19:1437–1444. doi: 10.1093/carcin/19.8.1437. [DOI] [PubMed] [Google Scholar]
- Felton JS, Knize MG, Bennett LM, Malfatti MA, Colvin ME, Kulp KS. Impact of environmental exposures on the mutagenicity/carcinogenicity of heterocyclic amines. Toxicology. 2004;198:135–145. doi: 10.1016/j.tox.2004.01.024. [DOI] [PubMed] [Google Scholar]
- GenBank. Bethesda, MD: National Center for Biotechnology Information; 2006. [accessed 24 July 2006]. Home Page. Available: http://www.ncbi.nlm.nih.gov/ [Google Scholar]
- Gooderham NJ, Zhu H, Lauber S, Boyce A, Creton S. Molecular and genetic toxicology of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Mutat Res. 2002;506–507:91–99. doi: 10.1016/s0027-5107(02)00155-0. [DOI] [PubMed] [Google Scholar]
- Grover PL, Martin FL. The initiation of breast and prostate cancer. Carcinogenesis. 2002;23:1095–1102. doi: 10.1093/carcin/23.7.1095. [DOI] [PubMed] [Google Scholar]
- Jeffy BD, Chirnomas RB, Romagnolo DF. Epigenetics of breast cancer: polycyclic aromatic hydrocarbons as risk factors. Environ Mol Mutagen. 2002;39:235–244. doi: 10.1002/em.10051. [DOI] [PubMed] [Google Scholar]
- Jiao H, Allinson SL, Walsh MJ, Hewitt R, Cole KJ, Phillips DH, Martin FL. Growth kinetics in MCF-7 cells modulate benzo[a]pyrene-induced CYP1A1 up-regulation. Mutagenesis. 2007;22:111–116. doi: 10.1093/mutage/gel060. [DOI] [PubMed] [Google Scholar]
- Joosten HF, van Acker FA, van den Dobbelsteen DJ, Horbach GJ, Krajnc EI. Genotoxicity of hormonal steroids. Toxicol Lett. 2004;151:113–134. doi: 10.1016/j.toxlet.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Kalantzi OI, Hewitt R, Ford KJ, Alcock RE, Thomas GO, Morris JA, et al. Inter-individual differences in the ability of human milk-fat extracts to enhance the genotoxic potential of the procarcinogen benzo[a]pyrene in MCF-7 breast cells. Environ Sci Technol. 2004a;38:3614–3622. doi: 10.1021/es035422y. [DOI] [PubMed] [Google Scholar]
- Kalantzi OI, Hewitt R, Ford KJ, Cooper L, Alcock RE, Thomas GO, et al. Low dose induction of micronuclei by lindane. Carcinogenesis. 2004b;25:613–622. doi: 10.1093/carcin/bgh048. [DOI] [PubMed] [Google Scholar]
- Kalantzi OI, Martin FL, Thomas GO, Alcock RE, Tang HR, Drury SC, et al. Different levels of polybrominated diphenyl ethers (PBDEs) and chlorinated compounds in breast milk from two U.K. regions. Environ Health Perspect. 2004c;112:1085–1091. doi: 10.1289/ehp.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel F, Hoppin JA. Association of pesticide exposure with neurologic dysfunction and disease. Environ Health Perspect. 2004;112:950–958. doi: 10.1289/ehp.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspin LC, Baird WM. Anti-benzo[a]pyrene-7,8-diol-9,10-epoxide treatment increases levels of proteins p53 and p21WAF1 in the human carcinoma cell line MCF-7. Polycyclic Aromatic Compounds. 1996;10:299–306. [Google Scholar]
- Lauber SN, Ali S, Gooderham NJ. The cooked food derived carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine is a potent oestrogen: a mechanistic basis for its tissue-specific carcinogenicity. Carcinogenesis. 2004;25:2509–2517. doi: 10.1093/carcin/bgh268. [DOI] [PubMed] [Google Scholar]
- Leung LK, Wang TT. Paradoxical regulation of Bcl-2 family proteins by 17β-oestradiol in human breast cancer cells MCF-7. Br J Cancer. 1999;81:387–392. doi: 10.1038/sj.bjc.6690706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Schumann BL, Chang X, Talaska G, Puga A. Arsenic co-exposure potentiates benzo[a]pyrene genotoxicity. Mutat Res. 2002;517:101–111. doi: 10.1016/s1383-5718(02)00057-8. [DOI] [PubMed] [Google Scholar]
- Marston CP, Pereira C, Ferguson J, Fischer K, Hedstrom O, Dashwood W, et al. Effect of a complex environmental mixture from coal tar containing polycyclic aromatic hydrocarbons (PAH) on the tumor initiation, PAH-DNA binding and metabolic activation of carcinogenic PAH in mouse epidermis. Carcinogenesis. 2001;22:1077–1086. doi: 10.1093/carcin/22.7.1077. [DOI] [PubMed] [Google Scholar]
- Martin FL, Semple KT. Environmental health impacts: occurrence, exposure and significance, Lancaster University, UK, 9–10 September 2003. Mutagenesis. 2004;19:423–429. doi: 10.1093/mutage/geh046. [DOI] [PubMed] [Google Scholar]
- McLuckie KIE, Gaskell M, Farmer PB, Martin EA, Jones GDD, Routledge MN. Effects of the order of exposure to a binary mixture of mutagens on the induced mutation spectra in the supF gene. Mutagenesis. 2004;19:137–141. doi: 10.1093/mutage/geh011. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Orphanides G. Estrogen receptors: orchestrators of pleiotropic cellular responses. EMBO Reports. 2001;2:775–781. doi: 10.1093/embo-reports/kve185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SO, Simon S, Chae K, Metzler M, Korach KS. Phytoestrogens and their human metabolites show distinct agonistic and antagonistic properties on estrogen receptor alpha (ER) and ERβ in human cells. Toxicol Sci. 2004;80:14–25. doi: 10.1093/toxsci/kfh147. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Ohbayashi T, Mimura J, Iwata R, Kameta A, Evine K, et al. Dioxin suppresses the checkpoint protein, MAD2, by aryl hydrocarbon receptor-independent pathway. Cancer Res. 2001;61:5707–5709. [PubMed] [Google Scholar]
- Peto J. Cancer epidemiology in the last century and the next decade. Nature. 2001;411:390–395. doi: 10.1038/35077256. [DOI] [PubMed] [Google Scholar]
- Plíšková M, Vondráček J, Vojtěšek B, Kozubík A, Machala M. Deregulation of cell proliferation by polycyclic aromatic hydrocarbons in human breast carcinoma MCF-7 cells reflects both genotoxic and nongenotoxic events. Toxicol Sci. 2004;83:246–256. doi: 10.1093/toxsci/kfi040. [DOI] [PubMed] [Google Scholar]
- Ragavan N, Grover PL, Balasubramanian SP, Hindley AC, Matanhelia SS, Martin FL. An observational study of cancers among female partners of UK-resident prostate cancer patients. Cancer Lett. 2006;242:88–94. doi: 10.1016/j.canlet.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Ragavan N, Hewitt R, Cooper LJ, Ashton KM, Hindley AC, Nicholson CM, et al. CYP1B1 expression in prostate is higher in the peripheral than in the transition zone. Cancer Lett. 2004;215:69–78. doi: 10.1016/j.canlet.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Rajapakse N, Butterworth M, Kortenkamp A. Detection of DNA strand breaks and oxidized DNA bases at the single-cell level resulting from exposure to estradiol and hydroxylated metabolites. Environ Mol Mutagen. 2005;45:397–404. doi: 10.1002/em.20104. [DOI] [PubMed] [Google Scholar]
- Rubin H. Synergistic mechanisms in carcinogenesis by polycyclic aromatic hydrocarbons and by tobacco smoke: a bio-historical perspective with updates. Carcinogenesis. 2001;22:1903–1930. doi: 10.1093/carcin/22.12.1903. [DOI] [PubMed] [Google Scholar]
- Safe S. Endocrine disruptors and human health: is there a problem. Toxicology. 2004;205:3–10. doi: 10.1016/j.tox.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Schut HA, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- Sims P, Grover PL, Swaisland A, Pal K, Hewer A. Metabolic activation of benzo(a)pyrene proceeds by a diolepoxide. Nature. 1974;252:326–328. doi: 10.1038/252326a0. [DOI] [PubMed] [Google Scholar]
- Slaga TJ, Jecker L, Braken WM, Weeks CE. The effects of weak or non-carcinogenic polycyclic hydrocarbons on 7,12-dimethylbenz[a]anthracene and benzo[a]pyrene skin tumor-initiation. Cancer Lett. 1979;7:51–59. doi: 10.1016/s0304-3835(79)80076-2. [DOI] [PubMed] [Google Scholar]
- Theodorakis P, Lomonosova E, Chinnadurai G. Critical requirement of BAX for manifestation of apoptosis induced by multiple stimuli in human epithelial cancer cells. Cancer Res. 2002;62:3373–3376. [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Venugopal M, Callaway A, Snyderwine EG. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) retards mammary gland involution in lactating Sprague-Dawley rats. Carcinogenesis. 1999;20:1309–1314. doi: 10.1093/carcin/20.7.1309. [DOI] [PubMed] [Google Scholar]
- Wang W, Smith R, Safe S. Aryl hydrocarbon receptor-mediated antiestrogenicity in MCF-7 cells: modulation of hormone-induced cell cycle enzymes. Arch Biochem Biophys. 1998;356:239–248. doi: 10.1006/abbi.1998.0782. [DOI] [PubMed] [Google Scholar]
- Welshons WV, Thayer KA, Judy BM, Taylor JA, Curran EM, vom Saal FS. Large effects from small exposures. I. Mechanisms for endocrine-disrupting chemicals with estrogenic activity. Environ Health Perspect. 2003;111:994–1006. doi: 10.1289/ehp.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yared E, McMillan TJ, Martin FL. Genotoxic effects of oestrogens in breast cells detected by the micronucleus and the Comet assay. Mutagenesis. 2002;17:345–352. doi: 10.1093/mutage/17.4.345. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Weksler BB, Wang L, Schwartz J, Santella RM. Immunohistochemical detection of polycyclic aromatic hydrocarbon-DNA damage in human blood vessels of smokers and non-smokers. Atherosclerosis. 1998;140:325–331. doi: 10.1016/s0021-9150(98)00136-1. [DOI] [PubMed] [Google Scholar]
- Zhao K, Murray S, Davies DS, Boobis AR, Gooderham NJ. Metabolism of the food derived mutagen and carcinogen 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine (PhIP) by human liver microsomes. Carcinogenesis. 1994;15:1285–1288. doi: 10.1093/carcin/15.6.1285. [DOI] [PubMed] [Google Scholar]