Abstract
Currently available codon usage analysis tools lack intuitive graphical user interface and are limited to inbuilt calculations. ACUA (Automated Codon Usage Tool) has been developed to perform high throughput sequence analysis aiding statistical profiling of codon usage. The results of ACUA are presented in a spreadsheet with all perquisite codon usage data required for statistical analysis, displayed in a graphical interface. The package is also capable of on-click sequence retrieval from the results interface, and this feature is unique to ACUA.
Availability
The package is available for non-commercial purposes and can be downloaded from: http://www.bioinsilico.com/acua.
Keywords: codon usage, ACUA, CAI, RSCU, statistical analysis
Background
The present work attempts to unify all prerequisite calculations required for codon usage analysis in a common graphical platform, enabling high throughput sequence analysis and statistical profiling. Development of ACUA has been made after comprehensive studies of tools such as CodonW [1], GCUA [2], EMBOSS [3], and Jemboss [4]. ACUA has been developed by integrating calculations like codon usage table, RSCU, CAI, Nc Value, selective positional AT, GC content and its skewness. [1] The package also provides on-click access of the sequence from the result interface, and is unique to ACUA.
Methodology
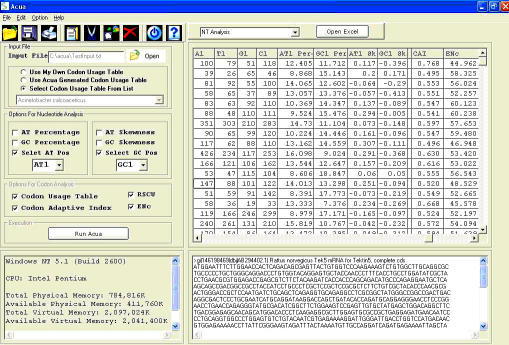
ACUA has been developed as a standalone package for codon usage analysis using Visual Basic, PERL and C++ programming languages. A snapshot of ACUA is shown in figure 1.
Figure 1.
ACUA interface showing the sample output
Program Input
ACUA requires FASTA formatted nucleotide sequence(s) in a plain text file as input. Additionally, for CAI calculation, user can select the reference set either from inbuilt codon usage tables or they can provide a customized codon usage table in Emboss .cut format. Furthermore, the user shall select their preferred calculations like CAI, RSCU, Nc value, AT and GC content with their positional preferences with an optional choice for excluding non synonymous and stop codons.
Program Output
ACUA consolidates all results into an MS-Excel File (.xls) with two worksheets. The first worksheet contains CAI value, Nc Value and all the Nucleotide Positional percentages with its respective skewness, whilst the second contains codon usage table and RSCU values facilitating statistical analysis. The user can sort the results within ACUA interface by right click of mouse. Furthermore, ACUA lists complete header information of each sequence in the results, which shall aid Gene ontology classifications. Moreover, user can access their preferred sequence as output by on-click access on result of their interest and these features are present only in ACUA.
Utility
ACUA has got the potential to serve as comprehensive platform, and also as a part of biologist's essential toolkit to extract all the prerequisite data required for statistical analysis of codon usage.
Caveats and future development
The present version of ACUA is only compatible for single processor machine. A MPI (Message Passing Interface) version of ACUA is being developed with R-language based inbuilt statistical functions, enabling cluster computing.
Acknowledgments
The authors are grateful to bioinsilico research team for their valuable suggestions and for hosting ACUA on web.
References
- 1. http://codonw.sourceforge.net//culong.html.
- 2.McInerney JO. Bioinformatics. 1998;14:372. doi: 10.1093/bioinformatics/14.4.372. [DOI] [PubMed] [Google Scholar]
- 3.Rice P, et al. Trends Genetics. 2000;16:276. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 4.Carver T, Bleasby A. Bioinformatics. 2003;19:1837. doi: 10.1093/bioinformatics/btg251. [DOI] [PubMed] [Google Scholar]