Abstract
With the Entamoeba genome essentially complete, the organism can be studied from a whole genome standpoint. The understanding of cellular mechanisms and interactions between cellular components is instrumental to the development of new effective drugs and vaccines. Metabolic pathway analysis is becoming increasingly important for assessing inherent network properties in reconstructed biochemical reaction networks. Metabolic pathways illustrate how proteins work in concert to produce cellular compounds or to transmit information at different levels. Identification of drug targets in E. histolytica through metabolic pathway analysis promises to be a novel approach in this direction. This article focuses on the identification of drug targets by subjecting the Entamoeba genome to BLAST with the e-value inclusion threshold set to 0.005 and choke point analysis. A total of 86.9 percent of proposed drug targets with biological evidence are chokepoint reactions in Entamoeba genome database.
Keywords: Entamoeba, metabolic pathway, chokepoint
Background
The amitochondrial protist Entamoeba histolytica, lacking both mitochondria and hydrogenosomes [1] is estimated to infect up to 10 percent of the world's population. Fifty million cases of amoebic dysentery and liver abscess are reported each year. [2] Between 40 and 100 thousand people die of amoebiasis each year. [3] Among parasitic infections, amoebiasis caused by E. histolytica ranks third worldwide, behind malaria and schistosomiasis, in lethal infections.
With the Entamoeba genome essentially complete [4] the organism can be studied from a whole genome standpoint. The knowledge of cellular mechanisms and interactions between cellular components is instrumental to the development of new effective drugs and vaccines. Metabolic pathways illustrate how proteins work in concert to produce cellular compounds or to transmit information at different levels. The view provided by the E. histolytica genome sequence is remarkable as it assists in the reconstruction of its metabolism and in the development of new antiamoebic drugs through the identification of its enzymes. The metabolism of E. histolytica has been shaped by an influx of bacterial genes through lateral gene transfer. [4]
Metabolic pathway analysis is very useful as it allows determining the overall capacity, i.e., theoretical maximum yield, of a cellular system and studying effects of any genetic modification. However, pathway analysis of large and highly entangled metabolic networks meets the problem of combinatorial explosion of possible routes across the networks. Here we propose a method to cope with this problem by two methods. In the present work, it has been tried to focus on the identification of drug targets by subjecting the Entamoeba genome to BLAST with the e-value inclusion threshold set to 0.005 and choke point analysis.
Methodology
Identification of potential drug targets through two methods
One feature that one can expect a good drug target to have is a lack of similarity to any human enzyme. The E. histolytica genome sequence is available from GenBank or from The Institute for Genomic Research website (http://www.tigr.org/tdb/e2k1/eha1/). KEGG [5] pathway database was used as a source of metabolic pathway information. Metabolic pathway identification numbers of the host H. sapiens and the pathogen E.histolytica were extracted from the KEGG database. Pathways which do not appear in the host but present in the pathogen according to KEGG database annotation have been identified as pathways unique to E.histolytica as compared to the host H. sapiens. The corresponding protein sequences were retrieved from the KEGG database. They were subjected to a BLASTp [6] search against the non-redundant database with the e-value inclusion threshold set to 0.005. The search was restricted to proteins from H. sapiens through an option available in the BLAST program, which allows the user to select the organism to which the search should be restricted. In the current context, the objective is to find only those targets, which do not have detectable human homologues. Enzymes, which do not have hits below the e-value inclusion threshold of 0.005, were picked out as potential drug targets.
Choke point analyses
Cells, whether free-living or as components of an organism, are regularly exposed to stimuli and perturbations to which they have to respond in a coordinated fashion. Such responses may vary widely in the numbers and types of identifiable steps that begin with sensing the stimulus and end with a change in biochemistry or physiology. Metabolic reconstruction of the organism can help in this direction. It is the purpose of this article to introduce alternative measures for this task and hence, the choke point analysis was one such measure. Here the choke point analysis method of Yeh et al. [7] has been adopted. It is being described below:
To identify potential drug targets, a chokepoint analysis of the metabolic network of E.histolytica is performed. A “chokepoint reaction” is a reaction that either uniquely consumes a specific substrate or uniquely produces a specific product in the Entamoeba metabolic network (Figure 1). As described by Yeh and colleagues [7] it is expected that the inhibition of an enzyme that consumes a unique substrate result in the accumulation of the unique substrate which is potentially toxic to the cell and the inhibition of an enzyme that produces a unique product to result in the starvation of the unique product which potentially cripple essential cell functions. Thus, it is believed that chokepoint enzymes may be essential to the parasite and are therefore potential drug targets.
Figure 1.
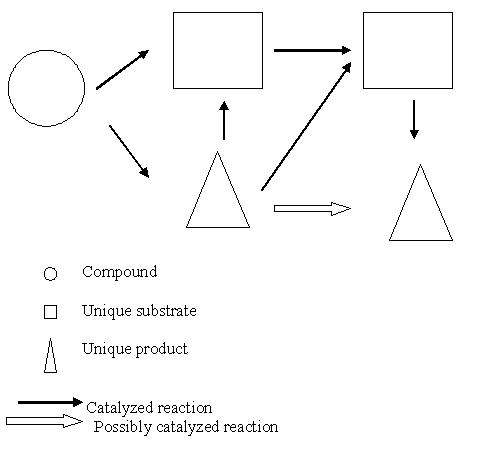
The thick arrows represent reactions that are catalyzed by enzymes, whereas the thin arrows represent reactions that are present (no evidence of the corresponding enzymes). While determining chokepoint reactions, only the catalyzed reaction are considered
Chokepoint analysis has several advantages. First, it allows us to test the consistency between experimental data and assumptions about the organization and regulation of the biochemical pathway and of its interdependencies with other processes. Second, it can be used to predict the consequences of various mutations or inhibitors.
Results and discussion
The targeting of metabolic pathways has several advantages on its own. Each step in the pathway is well validated as an essential function for pathogen growth. The target enzymes from the pathogen which are discarded and which share a similarity with the host proteins ensures that the targets have nothing in common with the host proteins, thereby, eliminating undesired host protein–drug interactions. Metabolic pathway analysis is becoming increasingly important for assessing inherent network properties in reconstructed biochemical reaction networks.
Pathways unique to E. histolytica when compared to the host H. sapiens
An important question to be addressed while choosing potential drug targets is whether the biochemical pathway to be targeted is unique to pathogen. The three pathways addressed in this study: peptidoglycan biosynthesis, streptomycin biosynthesis, and thiamine metabolism which are unique to the pathogen E.histolytica are discussed. Enzymes, from these pathways, which do not show similarity to any of the host proteins, are listed in Table 1 (supplementary material).
Peptidoglycan biosynthesis
Entamoeba histolytica was found to grow normally without producing glutathione and the main enzymes of glutathione metabolism, indicating that glutathione is not essential for many eukaryotic processes. This parasitic amoeba is an unusual eukaryote whose special features may help define the crucial functions of glutathione in those eukaryotes that do use it. Since Entamoeba histolytica lacks mitochondria and the usual aerobic respiratory pathways, the finding that it grows without glutathione and other evidence support the hypothesis that a primary function of glutathione in eukaryotes involves protection against oxygen toxicity associated with mitochondria and suggest that eukaryotes may have acquired glutathione metabolism at the time that they acquired mitochondria. [8] If this hypothesis is true, then the genes for GSH biosynthesis in eukaryotes should resemble those from alpha-proteobacteria, the modern relatives of the mitochondrial progenitor. [9,10]
Thiamine metabolism
The electrophoretic patterns of hexokinase and phosphoglucomutase strongly support the redefinition of pathogenic and nonpathogenic Entamoeba histolytica as two closely related species Entamoeba histolytica and Entamoeba dispar. [11] Thiamin pyrophosphokinase (TPK) catalyses the transfer of a pyrophosphate group from ATP to vitamin B1 (thiamin) to form the coenzyme thiamin pyrophosphate (TPP). Thus, TPK is important for the formation of a coenzyme required for central metabolic functions. The structure of thiamin pyrophosphokinase suggests that the enzyme may operate by a mechanism of pyrophosphoryl transfer similar to those described for pyrophosphokinases functioning in nucleotide biosynthesis. [12]
Streptomycin biosynthesis
Glycosylphosphatidylinositol (GPI)-anchored molecules such as cell surface Gal/GalNAc lectin and proteophosphoglycans of the protozoan parasite Entamoeba histolytica are thought to be involved in pathogenesis. [13] The cells of E.histolytica contained inorganic phosphate, pyrophosphate, nucleoside diphosphates, nucleoside triphosphates, NAD(P), phosphocholine, phosphoethanolamine, cytidine 5'-diphosphocholine and cytidine 5'- diphosphoethanolamine . The latter four compounds may act as intermediates in the salvage pathway for the synthesis of phosphatidylethanolamine and phosphatidylcholine. [14]
Carbohydrate metabolism
Studies have shown that E.histolytica grow or survive better in media containing glucose than in glucose deficient media, which is consistent with carbohydrates being the main energy substrates for anaerobic protists. Carbohydrates are the main source of energy for the parasite. The pathway involved in the fermentation of glucose includes some unusual enzymes that use pyrophosphate (PPi) rather than a nucleoside triphosphate as the phosphate donor. [15] Many of the genes encoding enzymes of the glycolytic pathway have been cloned and sequenced. Both D- glucose and D-galactose are transported and actively metabolized by E.histolytica. [15]
Lipid metabolism
Little is known about lipid metabolism in Entamoeba. Three phospholipase activities A1, A2 and L1 have been detected. [16] Entamoeba can also synthesise cholesterol and isoprenoids. [17] The only detectable polyamine present is putrescine at 9.5mM. [18] As it is present only in trace amounts in the growth medium, the amoeba must therefore synthesize it.
Protein metabolism
Protein metabolism in Entamoeba is not clearly elucidated. Amino acid utilization studies [19] showed that certain amino acids such as arginine, threonine, leucine, glutamine, phenylalanine and isoleucine could serve as an alternative energy source in the absence of glucose. The organism synthesizes cysteine, as cysteine synthase. [20]
Choke point analyses
If an enzyme catalyzes at least one chokepoint reaction, it is classified as a potential drug target. All the potential metabolic drug targets are listed in Table 2 (supplementary material). To assess the usefulness of identifying chokepoint enzymes for proposing drug targets, chokepoints and nonchokepoints against proposed drug targets from the literature is compared. A complete literature search for proposed amoebiasis drug targets is attempted that were metabolic enzymes and met the criteria discussed above. Of the 23 proposed targets with biological evidence, 20 are chokepoints in Table 2 (supplementary material). A total of 86.9% of proposed drug targets with biological evidence are chokepoint reactions in Entamoeba genome database. Chokepoints may not be essential. One reason could be that they create unique intermediates to an essential product which are not essential themselves and finally, there could be chokepoint reactions that are not essential due to other pathways that achieve the same metabolic goal within the organism. One example could be blocking the reaction that has no deleterious effects on the parasite. [7]
Due to the high percentage of enzymes identified as choke points, one additional criteria observed in addition to being a choke point enzyme for identifying potential metabolic drug targets is that an enzyme not having isozymes would make it more likely to be a good drug target. This is because one enzyme would be easier to inhibit than a family of enzymes. As for example, methionine γ – lyase in E. histolytica has two isozymes [40] and according to the criteria outlined above, it has not been included in the list of potential drug targets identified in Table 2 (supplementary material).
There are further aspects on which the list of potential drug targets can be narrowed down. The drug should adversely affect the parasite but not the human host which means that if the drug target has a homologous enzyme in human, it should not be essential or have differential inhibition in human. In other way, it can be said that potential drug targets should be expressed in the human stages of the parasite. The two most promising concepts for pathway analysis focused here are closely related. Assessing metabolic systems by the set of extreme pathways can, in general, give misleading results owing to the exclusion of possibly important routes. A full assessment of the proposed listed steps will require intense further effort. It is to be expected that some experiments may be significant which will stimulate the next phase of amendments and refinements. As it stands, it is hoped to serve the scientific community as a starting point for further data collection and experimentation in concert with, and based on, pathway analysis.
Conclusion
The genome sequence of E. histolytica has provided greater in-depth facts revealing that many pathways normally present in eukaryotes are absent from this organism. In addition, by identifying choke point reactions, identification of enzymes has been done that are essential to the parasite's survival. There is an enrichment of drug targets in chokepoints as compared with non-chokepoints. This leads to the conclusion that the classification of an enzyme as a chokepoint has some bearing on whether or not it would make a good drug target. The capabilities of the parasite to transport an accumulating metabolite out of the cell or a limiting metabolite into the cell have not been considered. It further limits the choke point analysis. The provisional targets which have been cited here needs to be examined further, both computationally and experimentally for these additional features. It can be said that biochemical reaction systems have become an important approach for understanding the functionality of metabolic networks reconstructed by genomic and biochemical data.
Supplementary material
References
- 1.Muller M. Evolutionary Relationships among Protozoa. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. p. 109. [Google Scholar]
- 2.Ravdin JI. Clinical Infectious Diseases. 1995;20:1453. doi: 10.1093/clinids/20.6.1453. [DOI] [PubMed] [Google Scholar]
- 3.Jackson TFHG. Amebiasis. London: Imperial College Press; 2000. p. 47. [Google Scholar]
- 4.Loftus B, et al. Nature. 2005;433:865. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 5.Kanehisa M, et al. Nucleic Acids Res. 2002;30:42. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul SF, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh I, et al. Genome Res. 2004;14:917. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahey RC, et al. Science. 1984;224:70. doi: 10.1126/science.6322306. [DOI] [PubMed] [Google Scholar]
- 9.Yang D, et al. Proc Natl Acad Sci. 1985;82:4443. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray MY. Curr Opin Genet Dev. 1993;3:884. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- 11.Ortner S, et al. Mol Biochem Parasitol. 1997;86:85. doi: 10.1016/s0166-6851(97)90008-6. [DOI] [PubMed] [Google Scholar]
- 12.Baker LJ, et al. Structure. 2001;9:539. doi: 10.1016/s0969-2126(01)00615-3. [DOI] [PubMed] [Google Scholar]
- 13.Vats D, et al. Inf and Immun. 2005;73:8381. doi: 10.1128/IAI.73.12.8381-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin JB, et al. Eur J Biochem. 1993;214:711. doi: 10.1111/j.1432-1033.1993.tb17972.x. [DOI] [PubMed]
- 15.Tanaka T, et al. Biochem Biophys Res Comm. 1997;236:611. doi: 10.1006/bbrc.1997.7019. [DOI] [PubMed] [Google Scholar]
- 16.Villarreal JV, et al. Parasitology Research. 1995;81:320. doi: 10.1007/BF00931538. [DOI] [PubMed] [Google Scholar]
- 17.Lujan HD, Diamond LS. Arch Med Res. 1997;28:96. [PubMed] [Google Scholar]
- 18.Bakker-Grunwald T, et al. J Eukar Microbiol. 1995;42:346. doi: 10.1111/j.1550-7408.1995.tb01592.x. [DOI] [PubMed] [Google Scholar]
- 19.Zuo X, Coombs GH. FEMS Microbiol Lett. 1995;130:253. doi: 10.1111/j.1574-6968.1995.tb07728.x. [DOI] [PubMed] [Google Scholar]
- 20.Nozaki T, et al. Mol. Biochem Parasitol. 1998;97:33. doi: 10.1016/s0166-6851(98)00129-7. [DOI] [PubMed] [Google Scholar]
- 21.Garrett CE. Mol Biochem Parasitol. 1984;11:257. doi: 10.1016/0166-6851(84)90070-7. [DOI] [PubMed] [Google Scholar]
- 22.Becker K, et al. Eur J Biochem. 2000;267:6118. doi: 10.1046/j.1432-1327.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 23.Nixon JEJ, et al. Biol Bull. 2003;204:1. doi: 10.2307/1543490. [DOI] [PubMed] [Google Scholar]
- 24.MacFarlane RC, Singh U. Infect Immun. 2006;74:340. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller S, et al. Trends in Parasitol. 2001;17:242. doi: 10.1016/s1471-4922(01)01908-0. [DOI] [PubMed] [Google Scholar]
- 26.Shuman S. Nat Rev Mol Cell Biol. 2002;3:619. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya AK, et al. Current Sci. 1999;77:564. [Google Scholar]
- 28.Hassan HF, Coombs GH. Mol Biochem Parasitol. 1986;19:19. doi: 10.1016/0166-6851(86)90061-7. [DOI] [PubMed] [Google Scholar]
- 29.Rosenthal B, et al. J Bacteriol. 1997;179:3736. doi: 10.1128/jb.179.11.3736-3745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller S, et al. Trends in Parasitol. 2003;19:320. doi: 10.1016/s1471-4922(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan G, et al. Mol Biochem Parasitol. 2004;138:205. doi: 10.1016/j.molbiopara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Copley SD, Dhillon JK. Genome Biol. 2002;3:1. doi: 10.1186/gb-2002-3-5-research0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhtar H. Exp Parasitology. 1969;26:135. doi: 10.1016/0014-4894(69)90104-0. [DOI] [PubMed] [Google Scholar]
- 34.Kumagai M, et al. J Biol Chem. 2004;279:2316. doi: 10.1074/jbc.M311478200. [DOI] [PubMed] [Google Scholar]
- 35.Bruchhaus I, Tannich E. Biochem J. 1994;303:743. doi: 10.1042/bj3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tachibana H, et al. J Clin Microbiol. 1991;29:2234. doi: 10.1128/jcm.29.10.2234-2239.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullu E, et al. Cell Microbiol. 2004;6:509. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 38.Kaur G, Lohia A. Biochem Biophys Res Commun. 2004;320:1118. doi: 10.1016/j.bbrc.2004.06.064. [DOI] [PubMed] [Google Scholar]
- 39.Stechmann A, Cavalier-Smith T. Science. 2002;297:89. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- 40.Tokoro M, et al. J Biol Chem. 2003;278:42727. doi: 10.1074/jbc.M212414200. [DOI] [PubMed] [Google Scholar]
- 41. ftp://ftp.ncbi.nlm.nih.gov/blast/executables/BLASTexecutables.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


