Abstract
Introduction
Osteopontin (OPN) mediates cancer metastatis. Mechanisms regulating OPN expression in human colorectal cancer are unknown. Using SW480 colon adenocarcinoma cells, we hypothesized that transcription determines OPN expression.
Methods
SW480 constitutively express OPN. Transient transfection and deletion analysis of human OPN promoter (full-length 2.1 kb)-luciferase constructs identified cis-regulatory regions. Gelshift and chromatin immunoprecipitation (ChIP) assays identified the trans-regulatory nuclear protein. Using in vitro adhesion, migration and invasion studies, siRNA was utilized to determine the functional effect of decreased nuclear protein expression.
Results
A cis-regulatory promoter region, nt-80 to nt-108, upregulated OPN transcription. Gelshift assays demonstrated specific binding of nuclear proteins. Competition with unlabelled mutant oligonucleotides indicated that the region, nt-94 to nt-104 (TGGGCTGGGC), was essential for protein binding in gelshift assays. Confirmatory ChIP assays showed the corresponding nuclear protein to be Sp1. Sp1 expression was ablated with siRNA (si-Sp1) resulting in decreased OPN dependent adhesion, migration and invasion by 50%, 70% and 65%, respectively. Exogenous addition of OPN to si-Sp1 cells restored adhesion, migration and invasion indices.
Conclusions
In SW480 human colon cancer cells, we conclude that Sp1 mediated expression of the tumor metastasis protein, OPN, regulates in vitro functional correlates of tumor metastasis.
Introduction
Evidence indicates that OPN plays a major regulatory role in tumor metastasis. A correlation between high levels of OPN protein expression and malignant invasion was established when OPN was demonstrated within tumor cells and in the surrounding stroma of numerous human cancers.(1–4) In the setting of colon cancer, the role of OPN is less well characterized. In pooled sample expression profiling, OPN was identified as the lead marker of colon cancer progression in a screen of 12,000 human genes.(5) A significant association between the degree of OPN expression and advancing Astler Coller (AC) stage was confirmed by Northern blot analysis. Almost 10–20 fold OPN induction was observed in samples with liver metastases over normal mucosa.(5) Furthermore, Coppola and coworkers have subsequently shown that OPN protein expression correlates significantly with colon cancer stage.(6) In a murine liver metastasis model of CT26 cells, we have previously demonstrated that OPN mediates tumor metastasis by enhancing tumor cell invasion and migration through the extracellular matrix, independent of cellular proliferation and cell-matrix adhesion.(7) RNAi silencing of OPN significantly inhibited in vivo hepatic metastases, in vitro migration and invasion, and CT26 expression of matrix metalloproteinase-2.
Despite large amounts of correlative data, little is known of the regulatory mechanisms underlying OPN expression in colon cancer. Results from our lab and others indicate that OPN gene transcription is a critical regulatory component in the metastatic phenotype of colorectal cancer.(8) In this study, we map the differential cis- and trans- regulatory mechanisms of OPN gene transcription in the human SW480 colon cancer cell line and demonstrate functional in vitro correlates. Our results indicate that Sp1 is a critical determinant of OPN expression in this system.
Methods
Cell culture
The human colon carcinoma cell line SW480 was obtained from American Type Culture Collection (Manassas, Virginia, USA) and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Sigma-Aldrich, St Louis, Missouri, USA) and penicillin (100 media was collected after U/ml) at 37°C in 5% CO2. For secreted-protein analysis, serum-free and centrifuged at 600 × g for 5 min to remove cellular material; the supernatant was concentrated 100-fold through Ultrafree Centrifugal filters (Millipore, Bedford, MA).
Western blot analysis
Cells were lysed in buffer (0.8% NaCl, 0.02% KCl, 1% SDS, 10% Triton X-100, 0.5% sodium deoxycholic acid, 0.144% Na2HPO4, and 0.024% KH2PO4, 2 mM phenylmethylsulfonyl fluoride, pH 7.4) and centrifuged at 12,000 × g for 10 min at 4 °C. The protein concentration was determined by the Bio-Rad protein assay kit; the protein samples were separated by 4–20% SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes (Amersham Biosciences) by semi-dry transfer (Bio-Rad). The membranes were probed with goat OPN Ab (R&D Systems, Minneapolis, MN) for 1 h at room temperature and detected using the appropriate horseradish peroxidase-conjugated secondary antibody. The reactive proteins were visualized by means of the ECL kit (Amersham Bioscience). Relative protein expression was analyzed by laser densitometry and normalized to a β-actin standard.
Northern blot analysis
Total RNA was isolated using a TRIzol kit according to the manufacturer’s instruction (Invitrogen). RNA (10 μg) was separated by electrophoresis through denaturing 1.2% agarose gel containing 1% formaldehyde and transferred onto Hybond N+ nylon membrane (Amersham Biosciences). The membrane was UV-cross-linked; hybridization was carried out with [α-32P]dCTP by random primers DNA labeling system (Invitrogen) to specific activities of 5 × 108 cpm/μg. A 32P-labeled 800-bp probe was constructed based upon the human OPN cDNA sequence (GenBank accession number ). After hybridization, the membranes were washed and exposed on film at −70 °C.
Transient transfection and luciferase assay
5′-Deletion fragments of the human OPN promoter were subcloned into pGL3 plasmid (Promega). The lengths of the osteopontin promoter fragments tested were: OPN −80 (nt −80 to nt +86), OPN −108 (nt −108 to nt +86), OPN −135 (nt −135 to nt +86), OPN −174 (nt −174 to nt +86), OPN −190 (nt −190 to nt +86), OPN −400 (nt −400 to nt +86), and OPN -Full (−2098 to +86). All constructs were confirmed by DNA sequencing. DNA transfections of SW480 colon cancer cells were carried out in 12-well plates using Lipofectin. Briefly, 1 × 105 cells were plated on a 12-well plate and allowed to grow for 24 h before the transfection. 2 μg of plasmid DNA and 24 μg of Lipofectin diluted in OPTI-DMEM were combined and incubated at room temperature for 20 min. The cells with transfection reagents were incubated for 4 h at 37 °C in a CO2 incubator. To control transfection efficiency between groups, 10 ng of pRL-SV40 was added to each well. Twenty-four hours after transfection, the cells were harvested in 0.4 ml of reporter lysis buffer (Promega), and dual luciferase reporter assays were performed. Transfection efficacy was normalized using the Renilla luciferase activity encoded by the co-transfected pRL-SV40 plasmid.
Nuclear extract preparation
Monolayers of SW480 cells were washed with phosphate-buffered saline and harvested by scraping into cold phosphate-buffered saline. The cell pellet obtained by centrifugation was resuspended in buffer containing 10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1.0 mM DTT and 0.5 mM phenylmethylsulfonyl fluoride; then 10% Nonidet P-40 was added and vortexed briefly; and the nuclei were pelleted by centrifugation. The nuclear proteins were extracted. Insoluble material was removed by centrifugation at 14000 rpm, and the supernatant containing the nuclear proteins was stored at −80 °C until use.
Gel shift assays
Gel shift assays were performed using nuclear cell extract fraction, as previously described. The target oligonucleotide (nt −108 to nt −80) used in gel shift was: 5′-AGGTGGGCTGGGCAGTGGCAGAAAACCT-3′. In specific competition binding assays, unlabeled target oligonucleotides or oligonucleotides containing a consensus Sp1 binding sequence (5′-ATTCGATCGGGGCGGGGCGAGC-3′) was added at 200 M excess. In nonspecific competition assays, unlabeled AP2 consensus oligonucleotides (Promega) were used. In Sp1-Mutant, 5′-AGGTGGGCTGGGCAGTGGCAGAAAACCT-3′ was mutated to 5′-AGGCAAATCAAACAGTGGCAGAAAACCT-3′. Supershift assays were performed by the addition of 0.5, 1, and 2 μl of SP1(H-225): sc-14027 a rabbit polyclonal antibody (Santa Cruz Biotechnology). The probe was prepared by end labeling the wild-type 28-bp double-stranded oligonucleotides with [32P]ATP (2500 Ci/mmol) using T4 polynucleotide kinase, followed by G-50 column purification. The reactions were resolved on a 6% nondenaturing acrylamide gel in 1x TBE buffer.
Chromatin immunoprecipitation (ChIP) assay
Chromatin from SW480 cells was fixed and immunoprecipitated using the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY). Sequences were identified for the human OPN promoter and primers constructed: Primer 1 (nt −261 to nt −240) AAGTGCTCTTCCTGGATGCTGA and Primer 2 (nt −8 to nt −29) AGCCCT CCCAGAATTTAAATGC. Purified chromatin was immunoprecipitated using 10 μg of anti-Sp1, 10 μg of anti-RNA Pol2 (Santa Cruz Biotechnology, Santa Cruz, CA) or 5 μl of rabbit nonimmune serum; eluted DNA fragments were purified to serve as templates. The input fraction corresponded to 0.1 and 0.05% of the chromatin solution before immunoprecipitation. The average size of the sonicated DNA fragments subjected to immunoprecipitation was 500 bp as determined by ethidium bromide gel electrophoresis. PCR primers utilized for Sp1 ChIP produced a 254 bp fragment.
Adhesion assay
Adhesion assays were performed on 96-well microtiter plates coated with 10 μg/ml matrigel. Cells were trypsinized and resuspended in DMEM with 1% bovine serum albumin, 1 mM MgCl2, and 0.5 mM CaCl2 at a concentration of 1 × 106 cells/ml. 1 × 105 cells (100 μl) were added into each well and placed for 30 min at 37 °C in 5% CO2 humidified air incubation. Non-adherent cells were removed by gently washing the wells three times with phosphate-buffered saline with 1 mM MgCl2 and 0.5 mM CaCl2. Adherent cells were fixed with 3.7% paraformaldehyde for 10 min at room temperature, followed by rinsing with phosphate-buffered saline, and stained with 0.4% crystal violet for 10 min. After extensive rinsing, the dye was released from the cells by addition of 30% acetic acid, and the microtiter plates were read in a microplate reader (Molecular Devices, Berkeley, CA) at 590 nm.
Migration and invasion assay
The migration and invasion assay were carried out in a Boyden Chamber system (Corning Glass-works). Cells were seeded at a density of 105 cells per well in triplicate in the upper chamber of 12-well transwells (8-μm pore). After being incubated at 37 °C with 5% CO2 for 24 h, the cells were fixed in 3.7% paraformaldehyde in phosphate-buffered saline for 10 min. The cells on the top surface of the filters were wiped off with cotton swabs. After three washes with phosphate-buffered saline, the filters were stained with 0.4% crystal violet for 10 min, and the dye was detected as the in vitro adhesion assay procedure. Statistical analysis: All data are presented as mean ± S.E. of three or four experiments. Analysis was performed using a Student’s t test. Values of p < 0.05 were considered significant.
Results
Regulation of OPN promoter activation
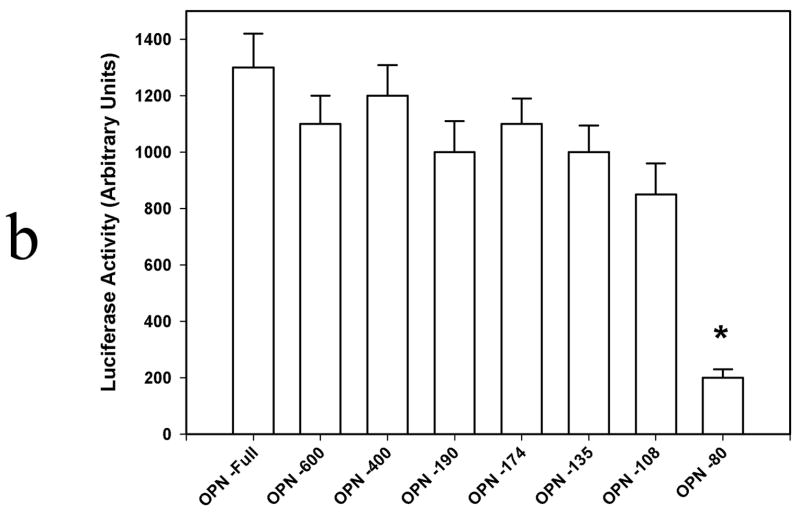
SW480 expression of OPN was first verified by Western blot analysis of cell lysate and media and Northern blot analysis of mRNA. (Figure 1a) OPN protein is readily detected in both cells and in the surrounding media. Transient transfection analysis of OPN promoter constructs was then performed. (Figure 1b) A significant step-off in activation was noted from OPN −80 and OPN −108, suggesting that a key regulatory element may reside in this location.
Figure 1. OPN expression in SW480 colon cancer cells.
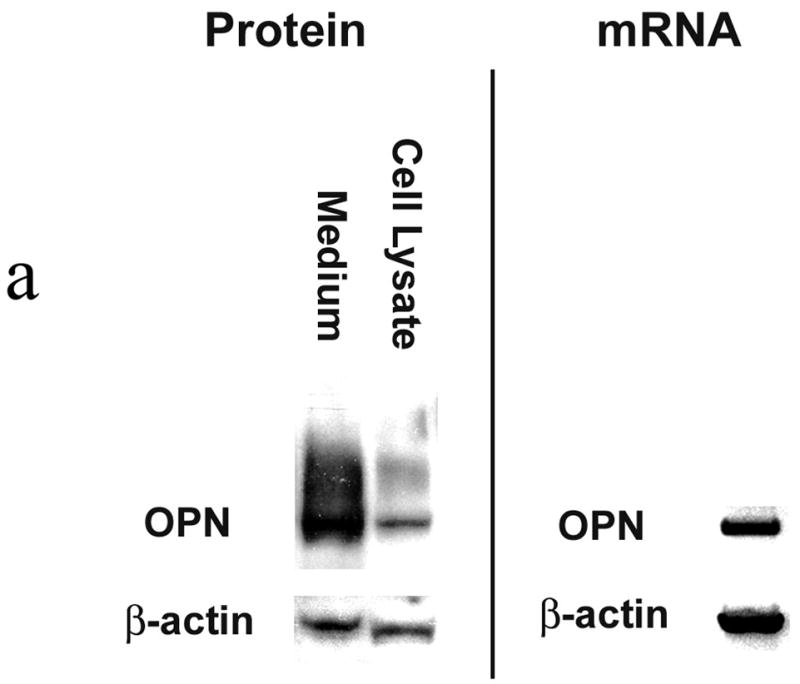
Figure 1a. OPN protein and mRNA expression.
Western blot analysis was performed using cell lysate and concentrated culture media from SW480 cells. Confirmation of OPN mRNA expression was determined using Northern blot analysis. Blots are representative of three experiments.
Figure 1b. Transient transfection analysis of OPN promoter constructs.
5′-Deletion fragments of the human OPN promoter tested were: OPN −80 (nt −80 to nt +86), OPN −108 (nt −108 to nt +86), OPN −135 (nt −135 to nt +86), OPN −174 (nt −174 to nt +86), OPN −190 (nt −190 to nt +86), OPN −400 (nt −400 to nt +86), and OPN -Full (−2098 to +86). Data are expressed as mean ± SEM of three experiments.
(* p<0.01 vs OPN −108, OPN −135, OPN −174, OPN −190, OPN −400, and OPN –Full)
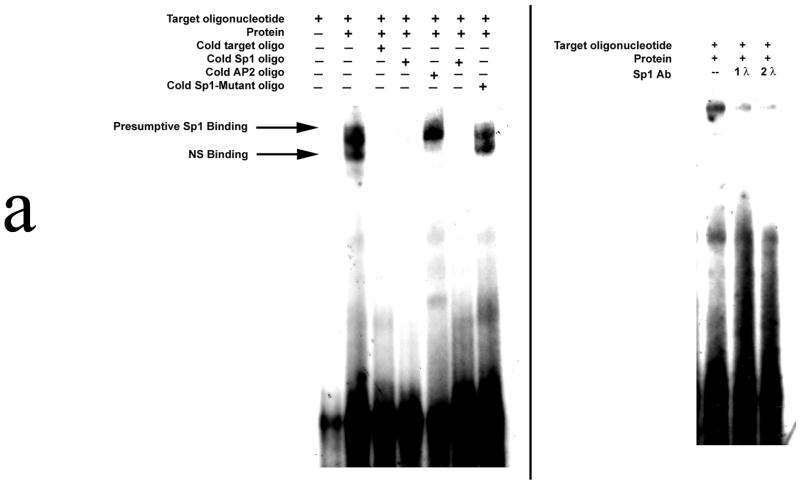
Electrophoretic mobility shift assays were then performed using the 28 bp nucleotide sequence as the target oligonucleotide. (Figure 2a) Our results demonstrate that SW480 nuclear protein binds to this oligonucleotide; binding is ablated in the presence of excess unlabelled target oligonucleotide. When an Sp1 consensus target oligonucleotide was initially utilized as the nonspecific competitor, binding was again abolished; however, when unlabelled AP2 target oligo was selected as an alternative nonspecific competitor, binding to the target oligonucleotide was maintained. The target sequence was then subjected to analysis, and an Sp1 binding was found: 5′-AGGTGGGCTGGGCAGTGGCAGAAAACCT-3′. When unlabelled Sp1-Mutant oligonucleotide was added as a competitor, no alteration in binding was noted. Confirmatory supershift studies using an Sp1 antibody were performed, and the extent of binding to the target oligonucleotide was significantly diminished. Finally, ChIP analysis also demonstrated in vivo Sp1 binding to this region of the SW480 promoter. (Figure 2b)
Figure 2. Sp1 binding to the OPN promoter.
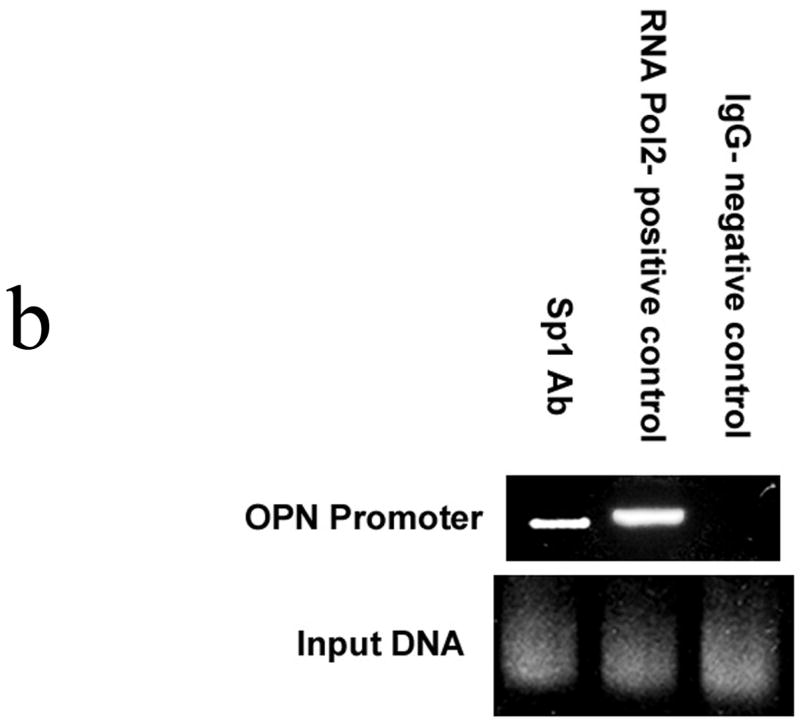
Figure 2a. Electrophoretic mobility shift assay.
The target oligonucleotide (nt −108 to nt −80) was: 5′-AGGTGGGCTGGGCAG TGGCAGAAAACCT-3′. In specific competition binding assays, unlabeled target oligonucleotides or oligonucleotides containing a consensus Sp1 binding sequence was added at 200 M excess. In nonspecific competition assays, unlabeled AP2 consensus oligonucleotide was used. In Sp1-Mutant, 5′-AGGTGGGCTGGGCAGTGGCAGAAAACCT-3′ was mutated to 5′-AGGCAAATCAAACAGTGGCAGAAAACCT-3′. Supershift assays were performed by the addition of 1 and 2 μl of SP1(H-225): sc-14027 a rabbit polyclonal antibody. Blots are representative of three experiments.
Figure 2b. Chromatin immunoprecipitation (ChIP) assay
Sequences were identified for the human OPN promoter and primers constructed: Primer 1 (nt −261 to nt −240) AAGTGCTCTTCCTGGATGCTGA and Primer 2 (nt −8 to nt −29) AGCCCT CCCAGAATTTAAATGC. Purified chromatin was immunoprecipitated using 10 μg of anti-Sp1, 10 μg of anti-RNA Pol2 (Santa Cruz Biotechnology, Santa Cruz, CA), or 5 μl of rabbit nonimmune serum; eluted DNA fragments were purified to serve as templates. The average size of the sonicated DNA fragments subjected to immunoprecipitation was 500 bp as determined by ethidium bromide gel electrophoresis. PCR primers utilized for Sp1 ChIP produced a 254 bp fragment. Gels are representative of three experiments.
To determine the functional relevance of Sp1 to OPN expression in SW480 cells, RNAi was used to knockdown Sp1 (si-Sp1) and in vitro adhesion, migration and invasion assays were performed. (Figure 3) Sp1 and OPN protein expression were both significantly decreased following siRNA treatment as determined by Western blot analysis. Thereafter, adhesion, migration and invasion in si-Sp1 were decreased by 50%, 70% and 65% in comparison to siRNA mismatch control (MM-Sp1) (p<0.01 for adhesion, migration and invasion) and wild-type SW480 (p<0.01 for adhesion, migration and invasion), respectively. Exogenous addition of OPN (50 μM) to si-Sp1 cells resulted in restoration of adhesion, migration and invasion indices to levels equivalent to those of wild type SW480 cells. These results indicate that: 1) Sp1 is a critical transcription factor for expression of OPN in SW480 colon cancer cells, and 2) Sp1-dependent OPN expression regulates in vitro adhesion, migration and invasion.
Figure 3. In vitro functional correlates of RNAi inhibition of Sp1 regulated OPN expression.
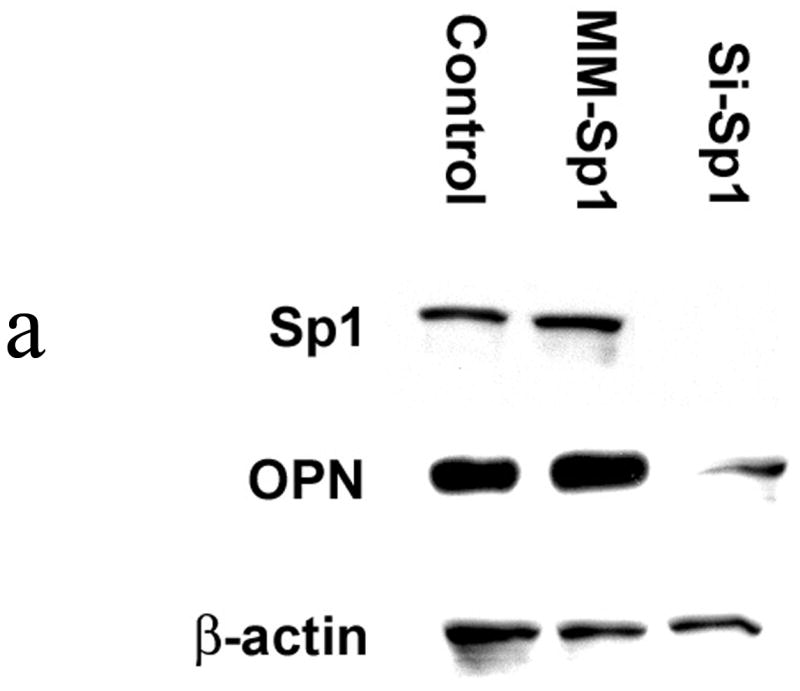
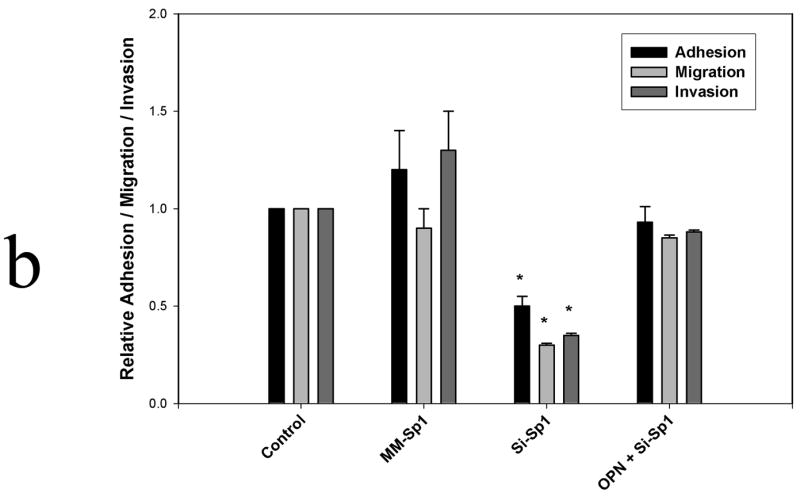
Figure 3a. Sp1 and OPN protein expression following RNAi
Western blot analysis for Sp1 and OPN protein was performed using cell lysate from SW480 cells at 24 hours following siRNA treatment. Mismatch siRNA (MM-Sp1) served as a negative control. Gel is representative of three experiments.
Figure 3b. Adhesion, migration and invasion
Adhesion, migration and invasion were determined as described in Methods. The absorbance obtained for control and experimental groups were each divided by the absorbance obtained for controls and expressed as an adhesion, migration or invasion index. In selected instances, exogenous OPN (50 μM) was added to si-Sp1 cells. By definition, untreated SW480 controls were assigned an index of 1. Data are expressed as mean ± SEM of three experiments.
(* p<0.01 vs Control, MM-Sp1 and OPN + si-Sp1)
Discussion
Colorectal cancer remains a significant public health concern. Eighty percent of patients are without distant metastases at the time of presentation and undergo potentially curative surgical procedures; however, two-thirds experience either a locoregional or distance recurrence. The liver is the most common site of the distant colorectal metastases and is involved in two-thirds of those patients with metastatic disease. The majority of cancer deaths in patients with colorectal cancer are directly related to the hepatic component of their disease.(9–11) Clearly, locoregional therapies for cancer are effective only in the absence of metastasis.
Metastasis is defined as the formation of progressively growing secondary tumor foci at sites discontinuous from the primary lesion. Tumor metastasis is the result of complex, cellular cascades which control tumor cell invasion into local tissue, entry into the vascular or lymphatic system, transport to and arrest within the microcirculation of distant organs, invasion through basement membrane into stroma and proliferation at this secondary site. Critical cellular mechanisms that facilitate several of these steps during metastatic transformation include (a) tumor cell attachment to basement membrane through cell-surface adhesion molecules (b) proteolytic degradation of the extracellular matrix (ECM) by tumor-derived proteinases and (c) tumor cell migration through the ECM. The formation of clinically important metastases depends upon the completion of every step of this cascade, the last of which is metastatic colonization.(12–14) The molecular mechanisms which regulate the metastatic cascade in various cancers remain largely undefined.
In this regard, a substantial body of data indicates that OPN plays a major regulatory role in tumor metastasis. A correlation between high levels of OPN protein expression and malignant invasion was established when OPN was demonstrated within tumor cells and in the surrounding stroma of numerous human cancers.(1;15;16) Transcriptional regulation of OPN expression in these settings has been the focus of much effort. The human, porcine and murine OPN promoters show a diverse number of consensus regulatory sequences.(17–22) Potential regulatory sequences in the human OPN promoter include TATA-like (−27 to −22 nt) and CCAAT-like (−73 to −68 nt) sequences, vitamin-D-responsive (VDR)-like motifs (−1892 to −1878 and −698 to −684 nt), GATA-1 (−851 to −847 nt), AP-1 binding sequence (TGACACA, −78 to −72 nt), PEA3 (−1695 to −1690 and −1418 to −1413 nt), Ets-1 (−47 to −39 nt) and multiple TCF-1 recognition sequences.
In the present study, we demonstrate an essential role for Sp1 in OPN gene transcription in human SW480 colon cancer cells. Specificity protein 1 (Sp1) was the first transcription factor identified and cloned, and shown to be a sequence-specific DNA-binding protein that activated a broad and diverse spectrum of mammalian and viral genes.(23) Sp1 protein recognizes GC/GT boxes and interacts with DNA through three C2H2-type zinc fingers located at the C-terminal domain. Based on results of crystal structure and NMR studies, each of the three zinc fingers in Sp1 recognizes three bases in one strand, and a single base in the complementary strand of the GC-rich elements where the consensus Sp1 binding site is 5′-(G/T)GGGCGG(G/A)(G/A)(C/T)-3′; Sp family proteins regulate expression of genes involved in multiple functions in both normal and cancerous tissues. Genes that regulate growth and cell cycle progression frequently contain proximal GC-rich promoter sequences, and their interactions with Sp proteins and other transcription factors are critical for their expression. Cancer-related Sp targets include VEGF, TGF-β, cyclin D1, E2F1, c-fos, and transforming growth factor α. Based upon our findings, OPN may be yet another Sp1 regulated protein relevant to the metastatic biology of cancer. Clearly, these findings will need to be verified in other human colon cancer cell lines and human colon cancers.
Acknowledgments
Work supported by NIH grants GM65113 (PCK), AI44629 (PCK) and DK070642 (PCK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Senger DR, Perruzzi CA, Gracey CF, Papadopoulos A, Tenen DG. Secreted phosphoproteins associated with neoplastic transformation: close homology with plasma proteins cleaved during blood coagulation. Cancer Res. 1988;48:5770–4. [PubMed] [Google Scholar]
- 2.Bellahcene A, Castronovo V. Increased expression of osteonectin and osteopontin, two bone matrix proteins, in human breast cancer. Am J Pathol. 1995;146:95–100. [PMC free article] [PubMed] [Google Scholar]
- 3.Singhal H, Bautista DS, Tonkin KS, O’Malley FP, Tuck AB, Chambers AF, et al. Elevated plasma osteopontin in metastatic breast cancer associated with increased tumor burden and decreased survival. Clin Cancer Res. 1997;3:605–11. [PubMed] [Google Scholar]
- 4.Casson AG, Wilson SM, McCart JA, O’Malley FP, Ozcelik H, Tsao MS, et al. ras mutation and expression of the ras-regulated genes osteopontin and cathepsin L in human esophageal cancer. Int J Cancer. 1997;72:739–45. doi: 10.1002/(sici)1097-0215(19970904)72:5<739::aid-ijc6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal D, Chen T, Irby R, Quackenbush J, Chambers AF, Szabo M, et al. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. J Natl Cancer Inst. 2002;94:513–21. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- 6.Coppola D, Szabo M, Boulware D, Muraca P, Alsarraj M, Chambers AF, et al. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10:184–90. doi: 10.1158/1078-0432.ccr-1405-2. [DOI] [PubMed] [Google Scholar]
- 7.Wai PY, Mi Z, Guo H, Sarraf-Yazdi S, Gao C, Wei J, et al. Osteopontin silencing by small interfering RNA suppresses in vitro and in vivo CT26 murine colon adenocarcinoma metastasis. Carcinogenesis. 2005;26:741–51. doi: 10.1093/carcin/bgi027. [DOI] [PubMed] [Google Scholar]
- 8.Wai PY, Mi Z, Gao C, Guo H, Marroquin C, Kuo PC. Ets-1 and runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells. J Biol Chem. 2006;281:18973–82. doi: 10.1074/jbc.M511962200. [DOI] [PubMed] [Google Scholar]
- 9.Parker SL, Tong T, Bolden S, Wingo PA. Cancer statistics, 1996. CA Cancer J Clin. 1996;46:5–27. doi: 10.3322/canjclin.46.1.5. [DOI] [PubMed] [Google Scholar]
- 10.Martin R, Paty P, Fong Y, Grace A, Cohen A, DeMatteo R, et al. Simultaneous liver and colorectal resections are safe for synchronous colorectal liver metastasis. J Am Coll Surg. 2003;197:233–41. doi: 10.1016/S1072-7515(03)00390-9. [DOI] [PubMed] [Google Scholar]
- 11.Scheele J, Stangl R, Altendorf-Hofmann A, Gall FP. Indicators of prognosis after hepatic resection for colorectal secondaries. Surgery. 1991;110:13–29. [PubMed] [Google Scholar]
- 12.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–56. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 13.Ahmad SA, Berman RS, Ellis LM. Biology of colorectal liver metastases. Surg Oncol Clin N Am. 2003;12:135–50. doi: 10.1016/s1055-3207(02)00078-9. [DOI] [PubMed] [Google Scholar]
- 14.Berman RS, Portera CA, Jr, Ellis LM. Biology of liver metastases. Cancer Treat Res. 2001;109:183–206. doi: 10.1007/978-1-4757-3371-6_10. [DOI] [PubMed] [Google Scholar]
- 15.Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, et al. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145:610–23. [PMC free article] [PubMed] [Google Scholar]
- 16.Gotoh M, Sakamoto M, Kanetaka K, Chuuma M, Hirohashi S. Overexpression of osteopontin in hepatocellular carcinoma. Pathol Int. 2002;52:19–24. doi: 10.1046/j.1440-1827.2002.01316.x. [DOI] [PubMed] [Google Scholar]
- 17.Smith JH, Denhardt DT. Molecular cloning of a tumor promoter-inducible mRNA found in JB6 mouse epidermal cells: induction is stable at high, but not at low, cell densities. J Cell Biochem. 1987;34:13–22. doi: 10.1002/jcb.240340103. [DOI] [PubMed] [Google Scholar]
- 18.Craig AM, Denhardt DT. The murine gene encoding secreted phosphoprotein 1 (osteopontin): promoter structure, activity, and induction in vivo by estrogen and progesterone. Gene. 1991;100:163–71. doi: 10.1016/0378-1119(91)90362-f. [DOI] [PubMed] [Google Scholar]
- 19.Denhardt DT, Guo XJ. Osteopontin: a protein with diverse functions. FASEB J. 1993;7:1475–82. [PubMed] [Google Scholar]
- 20.Hijiya N, Setoguchi M, Matsuura K, Higuchi Y, Akizuki S, Yamamoto S. Cloning and characterization of the human osteopontin gene and its promoter. Biochem J. 1994;303:255–62. doi: 10.1042/bj3030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malyankar UM, Hanson R, Schwartz SM, Ridall AL, Giachelli CM. Upstream stimulatory factor 1 regulates osteopontin expression in smooth muscle cells. Exp Cell Res. 1999;250:535–47. doi: 10.1006/excr.1999.4537. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Wrana JL, Sodek J. Characterization of the promoter region of the porcine opn (osteopontin, secreted phosphoprotein 1) gene. Identification of positive and negative regulatory elements and a ‘silent’ second promoter. Eur J Biochem. 1992;207:649–59. doi: 10.1111/j.1432-1033.1992.tb17092.x. [DOI] [PubMed] [Google Scholar]
- 23.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–48. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]