Abstract
RCC1, the only known guanine-nucleotide exchange factor for the Ran GTPase, is an ∼45-kD nuclear protein that can bind chromatin. An important question concerns how RCC1 traverses the nuclear envelope. We now show that nuclear RCC1 is not exported readily in interphase cells and that the import of RCC1 into the nucleoplasm is extremely rapid. Import can proceed by at least two distinct mechanisms. The first is a classic import pathway mediated by basic residues within the NH2-terminal domain (NTD) of RCC1. This pathway is dependent upon both a preexisting Ran gradient and energy, and preferentially uses the importin-α3 isoform of importin-α. The second pathway is not mediated by the NTD of RCC1. This novel pathway does not require importin-α or importin-β or the addition of any other soluble factor in vitro; however, this pathway is saturable and sensitive only to a subset of inhibitors of classical import pathways. Furthermore, the nuclear import of RCC1 does not require a preexisting Ran gradient or energy. We speculate that this second import pathway evolved to ensure that RCC1 never accumulates in the cytoplasm.
Keywords: nuclear transport, nuclear pore complex, importin, DNA-binding protein, permeabilized cells
Introduction
The exchange of macromolecules between the nucleus and cytoplasm is an essential biological activity in all eukaryotic cells (reviewed in Mattaj and Englmeier 1998; Melchior and Gerace 1998; Pemberton et al. 1998; Görlich and Kutay 1999; Nakielny and Dreyfuss 1999). Unlike the largely unidirectional transport processes of other organelles, nuclear transport is a bidirectional process. All nuclear transport is believed to occur through the nuclear pore complex, a large (∼125 MD) glycoprotein complex that contains ∼50–100 different proteins in mammals (reviewed in Davis 1995). The nuclear pore contains an ∼9-nm central channel that is believed to permit the passive diffusion of ions, small molecules, and proteins <∼50 kD (reviewed in Stoffler et al. 1999). Molecules above this diffusion limit require facilitated mechanisms to traverse the nuclear envelope.
Facilitated nuclear import and export are signal-mediated processes. The first such signal to be identified was a nuclear localization signal (NLS), which consists of a short string of basic amino acids, designated as either monopartite or bipartite by the number of basic clusters (Kalderon et al. 1984; Dingwall and Laskey 1991). Examples of NLS sequences are widespread in nature. The monopartite NLS from the SV-40 large T antigen and the bipartite NLS from nucleoplasmin are two of the most thoroughly studied. Importin-α (also called karyopherin-α, Srp1p, p56, PTAC 58, and pendulin) was found to bind directly to both types of NLS sequences and mediate their import (Adam and Gerace 1991; Cortes et al. 1994; Cuomo et al. 1994; Görlich et al. 1994; Kussel and Frasch 1995; Moroianu et al. 1995; Weis et al. 1995). In the cytoplasm, importin-α forms a stable complex with the transport receptor importin-β (also called p97, karyopherin-β, and Kap95; Chi et al. 1995; Enenkel et al. 1995; Radu et al. 1995; Görlich et al. 1996a; Weis et al. 1996). Importin-β is able to associate with the nuclear pore complex and transport NLS/importin-α complexes into the nucleoplasm (Görlich et al. 1995; Moroianu et al. 1995), where the trimeric import complex is dissociated (Moore and Blobel 1993; Rexach and Blobel 1995; Görlich et al., 1996; Moroianu et al. 1996).
Although the genome of the budding yeast, Saccharomyces cerevisiae, contains only a single importin-α gene (SRP1) (Kussel and Frasch 1995), duplication has occurred in metazoans, and mammals possess at least six importin-α isoforms (Cortes et al. 1994; Cuomo et al. 1994; Köhler et al. 1997; Seki et al. 1997; Takeda et al. 1997; Nachury et al. 1998). All of the isoforms bind the SV-40 NLS with similar affinity but show distinct preferences to other basic NLS sequences (Nadler et al. 1997; Seki et al. 1997; Prieve et al. 1998; Köhler et al. 1999; Welch et al. 1999). The structural basis for this preference remains obscure. In contrast to importin-α, the budding yeast genome contains 14 members of the importin-β (karyopherin-β) family, of which nine are known to mediate import and five export; mammals express at least 23 members of this family. (Görlich 1997; Wozniak et al. 1998; Görlich and Kutay 1999).
Importin-β family members are also able to transport proteins into the nucleus independently of importin-α (Siomi and Dreyfuss 1995; Weighardt et al. 1995; Görlich et al. 1997; Pollard et al. 1996; Weis et al. 1996; Jakel and Görlich 1998; Jakel et al. 1999). In these cases, importin-β binds directly to the import substrate and transports it into the nucleoplasm as a binary importin-β/import substrate complex.
Over the past decade, a wealth of experimental evidence has shown that much of the traffic across the nuclear envelope is governed by the small GTPase Ran (reviewed Mattaj and Englmeier 1998; Melchior and Gerace 1998; Pemberton et al. 1998; Görlich and Kutay 1999; Macara et al. 2000). Ran is a highly abundant (∼107 copies/cell), ∼25-kD protein that, at steady state, is ∼80% nuclear (Bischoff and Ponstingl 1991b). Like other small GTPases, Ran cycles between GTP- and GDP-bound states. Unlike other GTPases, however, Ran possesses only a single known guanine-nucleotide exchange factor, RCC1 (Bischoff and Ponstingl 1991a), and a single known GTPase-activating protein, RanGAP (Bischoff et al. 1994). RCC1 and RanGAP possess distinct subcellular localizations. While RCC1 is a nuclear protein (Ohtsubo et al. 1989), RanGAP is found both free in the cytoplasm and attached to the cytoplasmic face of the nuclear pore (Hopper et al. 1990; Matunis et al. 1996; Mahajan et al. 1997; Saitoh et al. 1997). The disparate localizations of these two proteins are believed to create a nuclear compartment with a high concentration of Ran-GTP, while Ran within the cytoplasm is predominantly in the GDP-bound state. This gradient of Ran-GTP across the nuclear envelope is crucial for most forms of nuclear transport (Mattaj and Englmeier 1998; Pemberton et al. 1998; Görlich and Kutay 1999; Macara et al. 2000).
The prevailing model for the role of Ran in nuclear transport asserts that eukaryotic cells use Ran to control the loading and unloading of cargo in a compartment-specific manner. Biochemically, import complexes can only be formed in the absence of Ran-GTP (Rexach and Blobel 1995; Chi et al. 1996; Görlich et al. 1996b). Thus, the relative absence of Ran-GTP in the cytoplasm allows for the formation of a stable import complex. These complexes can then translocate into the nucleus where they encounter high levels of Ran-GTP. The binding of Ran-GTP to the import receptor dissociates the receptor–cargo complex, thereby unloading cargo into the nucleoplasm (Moore and Blobel 1993; Görlich et al. 1996b). This model proposes that the gradient of Ran-GTP across the nuclear envelope is the primary driving force for the directionality of Ran-dependent transport.
Since nuclear transport is an essential biological activity in eukaryotic cells, understanding how the Ran gradient is established and maintained is of obvious interest. One intriguing question is how cells preserve the nuclear localization of RCC1. If nuclear RCC1 is required to establish the Ran gradient, can RCC1 be imported solely by a Ran-dependent mechanism? A recent study has shown that importin-α3 is able to stimulate the import of RCC1 into digitonin-permeabilized cells in vitro (Köhler et al. 1999). It is not known, however, whether there are other, additional transport mechanisms for RCC1.
RCC1 is a nuclear protein of ∼45 kD that can bind chromatin (Ohtsubo et al. 1989). The crystal structure for RCC1 has been solved and revealed a seven-bladed propeller (Renault et al. 1998), reminiscent of the structure for the β subunit of heterotrimeric G-proteins (Wall et al. 1995). Mutational analysis suggests that RCC1 has discrete Ran- and DNA-binding surfaces (Seino et al. 1992; Azuma et al. 1996; Renault et al. 1998). Mutation of a single aspartic acid residue to alanine (D182A) renders the protein unable to interact stably with Ran and catalyze nucleotide exchange (Azuma et al. 1996, Azuma et al. 1999). The first 25 amino acids of RCC1 comprise its NH2-terminal domain (NTD). The NTD is primarily unstructured, but it is located on the putative DNA-binding surface (Renault et al. 1998). The NTD harbors two clusters of basic amino acids that each resemble a monopartite NLS (see Fig. 1 A). Mutational analysis of this domain has implicated the NTD in both nuclear transport and DNA binding (Seino et al. 1992); however, a definitive role for this domain remains to be defined.
Figure 1.
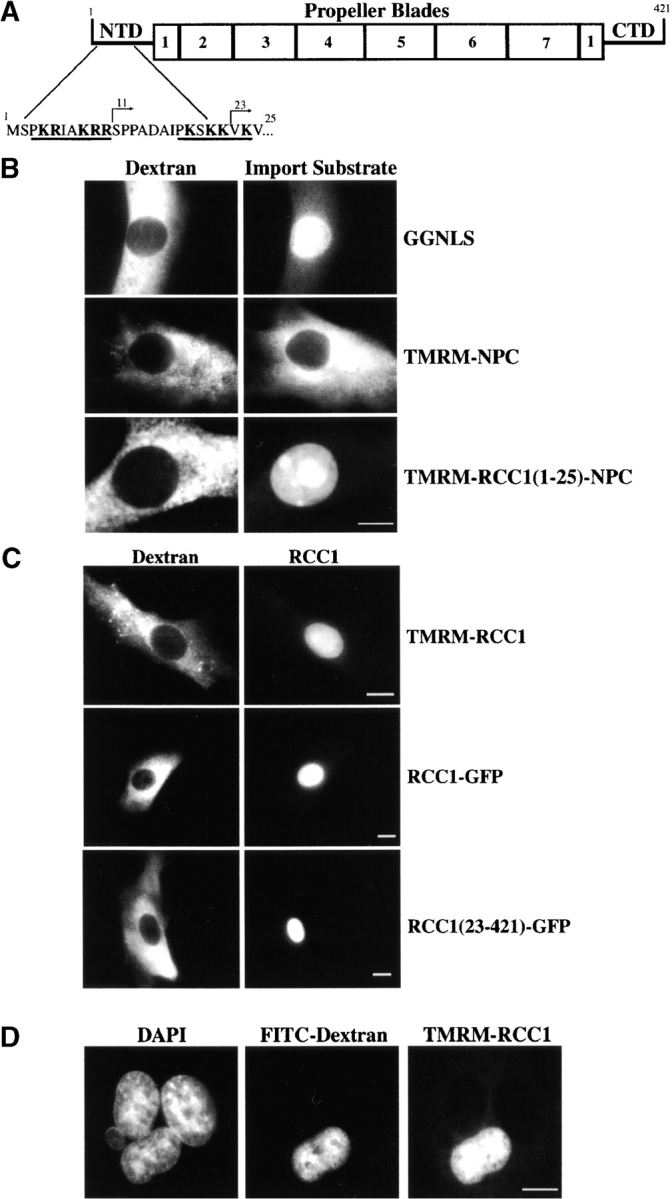
The NTD is a functional NLS but is not required for transport of RCC1. (A) The crystal structure of RCC1 displays a seven-bladed propeller with the NH2- and COOH-terminal domains extended beneath the putative DNA-binding surface. The propeller regions are numbered (1–7) and the sequence of the NTD is listed. Basic residues within the NTD are in bold; the two putative NLS sequences are underlined. (B) NPC and RCC1(1-25)-NPC were labeled with tetramethylrhodamine-5-maleimide (TMRM). These proteins, along with GGNLS as a positive control, were injected (1 mg/ml) into the cytoplasm using a fluorescent dextran as an injection site marker (left panels). Samples were incubated for 15 min before fixation and visualization of the injected import substrate (right panels). (C) TMRM-RCC1, RCC1-GFP, and RCC1(23-421)-GFP (∼18 μM) were injected into the cytoplasm of BHK21 cells as described above. In all experiments, a fluorescent dextran (left panels) was used at 1 mg/ml to mark the injection site. The cells were incubated for 15 min before analysis of the injected protein substrate (right panels). (D) Multinucleate BHK21 cells were injected with TMRM-RCC1 (right panel) and FITC-dextran (middle panel) as an injection site marker. Samples were incubated for ∼35–40 min total before fixation. Nuclei were visualized by staining DNA with DAPI (left panel). The TMRM-RCC1 (right panel) was intentionally overexposed so as to allow visualization of the cytoplasm of the multinucleate cell. Bars, 10 μm.
In this study, we show that the NTD is a bona fide transport sequence for RCC1. Furthermore, we demonstrate that the NTD confers high affinity binding to importin-α3 and access to a Ran-dependent import pathway. Surprisingly, deletion of the NTD from RCC1 abrogated its ability to bind importin-α, but it did not prevent the protein from entering the nucleoplasm. In addition, many treatments that inhibit the transport of Ran-dependent import substrates do not prevent the import of RCC1 constructs that are above the passive-diffusion limit of the nuclear pore. We conclude that RCC1 is imported by at least two different mechanisms. The first is a classic, Ran-dependent import pathway mediated by the NTD of RCC1. The second is a previously uncharacterized pathway that is saturable and temperature-sensitive but requires neither energy, soluble factors, nor a preexisting Ran gradient. We speculate that this second pathway evolved as a mechanism to scavenge RCC1 molecules and prevent any accumulation of Ran-GTP in the cytoplasm.
Materials and Methods
DNA Constructs
The cDNA encoding the full-length RCC1 protein was inserted into pGEX-2T (Pharmacia) as a BamHI-BamHI fragment. Creation of the RCC1 deletion constructs was accomplished by PCR amplification of the region encoding residues 23–421 and ligation into both pGEX-2T and pGEX-GFP as a BamHI-BamHI fragment. Creation of the RCC1(D182A) mutant was accomplished by megaprimer PCR using a mismatched primer for the initial amplification process and pGEX-RCC1 as the template. Subsequent amplification of the entire RCC1 cDNA was accomplished and the digested product was ligated into pGEX-2T as a BamHI-BamHI fragment. To fuse RCC1 to two IgG-binding domains from Protein A (z-domains), the zz domain from pQEzzBIB (provided by Dirk Görlich) was amplified by PCR and ligated as a BamHI-BamHI fragment into pGEX-GFP that had been digested BamHI-BglII, thereby removing the GFP and destroying the 3′ restriction site. RCC1 was then ligated into pGEX-zz as a BamHI-BamHI fragment. The cDNA encoding the first 25 amino acids of RCC1 was PCR-amplified and ligated into pQE70–nucleoplasmin core-domain (NPC) (Dirk Görlich) as a SphI-SphI fragment. All constructs were checked by automated sequencing.
Protein Expression, Purification, and Labeling
GST-fusion proteins were expressed in Escherichia coli XL1-Blue or BL21 essentially as described previously (Seino, 1992, Welch, 1999). The proteins were bound to glutathione-Sepharose beads (Pharmacia), eluted with glutathione, and exchanged into thrombin cleavage buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 2.5 mM CaCl2) using a PD10 column (Pharmacia). The protein was cleaved with 4U thrombin; the GST and thrombin were then removed by binding to glutathione-Sepharose and p-aminobenzamidine beads (Sigma), respectively. The recombinant protein was concentrated using a Centricon 30 (Amicon) and analyzed by SDS-PAGE.
RCC1(1-25)-NPC-His6 was expressed in E. coli BL21 and purified by binding to nickel-agarose beads (Qiagen; Görlich, 1996). The protein was then eluted with 200 mM imidazole, exchanged into thrombin cleavage buffer using a PD10 column (Pharmacia), concentrated using a Centricon 30 (Amicon), and analyzed by SDS-PAGE.
All labeling reactions occurred in thrombin-cleavage buffer. Protein samples were labeled with tetramethylrhodamine-5-maleimide (TMRM; Molecular Probes) at a 1:1 molar ratio for 60 min on ice. After quenching unreacted TMRM with 50 mM 2-mercaptoethanol (Sigma), the proteins were separated from unreacted label and exchanged into microinjection buffer (10 mM NaHPO4, pH 7.2, 70 mM KCl, and 1 mM MgCl2) using a Centrisep column (Princeton Separations).
Cell Culture
All cells were passaged in Dulbecco's modified eagle medium (supplemented with 10% fetal calf serum [vol/vol] and penicillin/streptomycin). Baby hamster kidney cells (BHK21) and HeLa cells were cultured in a humidified, 37°C/5% CO2 incubator. tsBN2 cells were grown at 33.5°C and, where indicated, were temperature-shifted to 39.5°C for 3 h. To energy-deplete BHK21 cells, cultures were washed twice in PBS, then placed into serum-free, glucose-free Dulbecco's minimal essential medium containing 10 mM sodium azide (Sigma) and 6 mM 2′deoxy-d-glucose (Sigma) for 3 h before experimentation.
Microinjection Studies
Cells were cultured on CELLocate gridded coverslips (Eppendorf). The samples were placed in Ringer's solution (25 mM Hepes, pH 7.2, 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 1 mM KH2PO4, 1 mg/ml BSA, and 0.2% glucose) before microinjection. When injecting energy-depleted cells, glucose-free Ringer's solution was used that was supplemented with metabolic inhibitors. When injecting temperature-shifted tsBN2 cells, the Ringer's media was supplemented with 10 μg/ml cycloheximide to prevent the resynthesis of endogenous RCC1. GGNLS, RCC1(1-25)-NPC, and NPC were injected at ∼1 mg/ml; Ran(G19V) was injected at 40 μM. All other samples were injected at ∼20 μM. Recombinant protein stocks were injected using the Eppendorf 5242 apparatus, and either a FITC- or TRITC-labeled dextran (Sigma) was used as an injection site marker (1 mg/ml). Samples were incubated for 15–20 min before fixation in 4% paraformaldehyde (PFA)/2% sucrose/PBS. The samples were then permeabilized in methanol and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The coverslips were mounted on glass slides using Gel Mount (Biomeda). Samples were visualized by fluorescence microscopy using a Nikon microscope with a 60× water immersion lens. Images were captured with a Hamamatsu CCD camera using Openlab software (Improvision).
When the import of proteins was assayed by time-lapse photography, cells were injected in ice-cold Ringer's solution as described above. The plate was washed quickly with 33.5°C Ringers before initiating the time-lapse program (∼1 min) and images were captured at 1-min intervals. The sample was maintained at the appropriate temperature using a heated-stage apparatus. The change in mean nuclear fluorescence intensity was measured using the Openlab software. All time-lapse images were captured as 20× images using the apparatus described above.
To determine whether RCC1 shuttles between the nucleus and cytoplasm, BHK21 cells were cultured on CELLocate coverslips as described above. Single nuclei of multinucleate cells were injected with TMRM-RCC1 (18 μM) using FITC-dextran (1 mg/ml) as an injection site marker (Herold et al. 1998). Cells were then placed in the 37°C tissue culture incubator for ∼25 min before fixation and visualization. Given the time required to inject cells as well as that required to find subsequent multinucleate BHK21 cells, we estimate that the shuttling assay proceeded for ∼35–40 min total.
Binding Assays
These assays were done using a protocol similar to that described by Welch et al. Importin-α was expressed in E. coli BL21 with a COOH-terminal His6 tag (Köhler et al. 1999; Welch et al. 1999). The importin-α-lysate was prepared by resuspension in lysis buffer (50 mM Tris, pH 8, 1 mM EDTA, pH 8, 1% Triton X-100, 150 mM NaCl, and 1 mg/ml lysozyme), sonication, and clearance by centrifugation. Equal amounts of the GST-fusion proteins were prebound to glutathione-Sepharose beads (Amersham Pharmacia) and the bacterial lysate was added in binding buffer (20 mM MOPS, pH 7.1, 100 mM KOAc, 5 mM MgOAc, 0.1% Tween-20, 5 mM DTT, and 0.5 mM PMSF). Binding reactions were incubated for 2.5 h at 4°C in an Eppendorf thermomixer. The supernatant was sampled to ensure that the beads were incubated with equal concentrations of importin-α. After washing (3×) in binding buffer, the beads were resuspended in Laemmli sample buffer and boiled. The proteins were subjected to SDS-PAGE and transferred to nitrocellulose. Importin-α was then visualized using an anti-His6 antibody (1:1,000; Babco), a HRP-coupled secondary antibody (1:20,000; The Jackson Laboratory), and a chemiluminescence reagent (KPL).
In Vitro Import Assays
We used a modified protocol from that described originally by Adam et al. 1990. HeLa cells were cultured on poly-l-lysine–coated coverslips to 50–70% confluence. After washing in ice-cold PBS, the cells were permeabilized with digitonin (0.005%; Calbiochem) in import assay buffer (20 mM Hepes-KOH, pH 7.5, 80 mM KOAc, 4 mM MgOAc, 2 mM DTT, and 250 mM sucrose) for five min on ice. Permeabilization was stopped by washing the samples in ice-cold import assay buffer and adding 1% BSA/assay buffer for 5 min at 25°C. A standard import assay consisted of the import substrate (0.6–3 μM), a rabbit reticulocyte lysate (50% vol/vol), and an energy-regenerating system (20 mM phosphocreatine, 1 mM ATP, 1 mM GTP, 20 U/ml creatine phosphokinase) in 1% BSA/import assay buffer. Import reactions were carried out for ∼25 min for GGNLS and 5–10 min for the RCC1 constructs. Where indicated, inhibitors were used at the following concentrations: 0.2 mg/ml wheat-germ agglutinin, 100 μM β-like import receptor binding domain (BIB; Jakel and Görlich 1998; Welch et al. 1999), 6 μM GST-importin-β(45-462), and 36 μM RCC1. Each import reaction was terminated by washing (3×) in import assay buffer. The cells were then fixed in 4% PFA/2% sucrose/PBS, permeabilized in methanol (−20°C), stained with DAPI, and visualized by fluorescence microscopy.
To assess the integrity of the nuclear envelope, import assays were done on digitonin-permeabilized HeLa cells using RCC1zz as an import substrate, as described above. All samples were fixed in 4% PFA/2% sucrose/PBS; however, the methanol permeabilization step was omitted in some samples. All samples were then blocked in 5% BSA/PBS before visualization of the RCC1zz by immunofluorescence.
Chromatin-binding Assay
HeLa cells were cultured on poly-l-lysine coverslips and permeabilized in 0.2% Triton X-100/import assay buffer (vol/vol) for six min at 25°C. RCC1, RCC1-GFP, or RCC1zz were added directly to the coverslips for 5 min. The cells were then washed, fixed, stained, and visualized as described previously.
Immunofluorescence
HeLa cells were fixed in 4% PFA/2% sucrose/PBS for 15 min at room temperature and, where indicated, permeabilized in methanol (−20°C) for 2 min. Samples were blocked in 5% BSA/PBS overnight at 4°C. RCC1zz was visualized directly using Texas red–conjugated rabbit IgG (The Jackson Laboratory; 1:1,000) for 60 min at 25°C. Nuclei were stained concomitantly with DAPI. Samples were then washed (5×) and analyzed by fluorescence microscopy.
To visualize endogenous Ran, tsBN2 cells were incubated at 33.5°C or 39.5°C for 3 h and then fixed, permeabilized, blocked, and incubated with a monoclonal anti-Ran antibody (Transduction Laboratories; 1:250) for 45 min at 25°C. A secondary, Texas red–conjugated, anti–mouse IgG (The Jackson Laboratory; 1:1000) was used for 35 min at 25°C. Nuclei were stained concomitantly with DAPI.
Results
The NTD of RCC1 Contains a Functional NLS but Is Not Required for Transport
RCC1, the only known guanine-nucleotide exchange factor for the Ran GTPase (Bischoff and Ponstingl 1991a), is a nuclear, chromatin-bound protein (Ohtsubo et al. 1989). The NTD of RCC1 is primarily unstructured, but it is located on the putative DNA-binding surface of RCC1 (Renault et al. 1998). It possesses two clusters of basic amino acids that each resemble a monopartite NLS (Fig. 1 A) and has been implicated in both nuclear transport and DNA binding (Seino et al. 1992). To test the functionality of the NTD in nuclear transport, a fusion protein was generated using the NTD of RCC1 and the nucleoplasmin core domain (RCC1(1-25)-NPC); this protein, labeled with TMRM, spontaneously forms pentamers (Görlich et al. 1996a; ∼100 kD) that are well above the passive diffusion limit of the nuclear pore (∼50 kD; Stoffler et al. 1999). GST-GFP-NLS (GGNLS), a fluorescent protein substrate containing a classic NLS from the SV-40 large T antigen (Kalderon et al. 1984; Welch et al. 1999), was used as a positive control; the nucleoplasmin core protein itself was used as a negative control (Fig. 1 B). When injected into the cytoplasm of BHK21 cells, RCC1(1-25)-NPC, but not NPC alone, concentrated rapidly within the nucleoplasm, demonstrating that the NTD of RCC1 contains a functional NLS.
These data showed that the NTD was sufficient to promote the nuclear localization of a fluorescent reporter protein. To test whether the NTD was necessary for nuclear translocation of RCC1, we created RCC1-GFP fusion proteins in which portions of the NTD had been deleted. Since the removal of the first 10 amino acid residues of RCC1 had no detectable effect on the ability of RCC1 to traverse the nuclear envelope (data not shown), we constructed a GFP-fusion protein in which almost the entire NTD was deleted (RCC1(23-421)-GFP). RCC1 and RCC1-GFP were used as controls. When injected into the cytoplasm, full-length RCC1, RCC1-GFP, and RCC1(23-421)-GFP were all imported rapidly into the nucleus (Fig. 1 C). Therefore, the nuclear import of RCC1 does not require its NTD. Since RCC1(23-421)-GFP (∼70 kD) is above the diffusion limit of the nuclear pore complex, it seemed unlikely that import of this molecule was due to passive diffusion. Taken together, these data imply that RCC1 possesses at least two independent import pathways.
To explore the transport dynamics of RCC1, we next asked whether RCC1 shuttles continuously between the nucleus and cytoplasm. To investigate this possibility, we injected single nuclei of multinucleate BHK21 cells (Herold et al. 1998). RCC1 did not equilibrate between the nuclei within the cells over a period of 35–40 min (Fig. 1 D), suggesting that RCC1 is not exported readily from the nucleus in interphase cells. Use of heterokaryon fusion assays also demonstrated that RCC1 does not shuttle (Lindsay, M., personal communication). Since RCC1 (∼45 kD) is below the theoretical diffusion limit of the nuclear pore (∼50 kD), these data suggest that RCC1 is efficiently retained within the nucleoplasm, presumably by binding to chromatin.
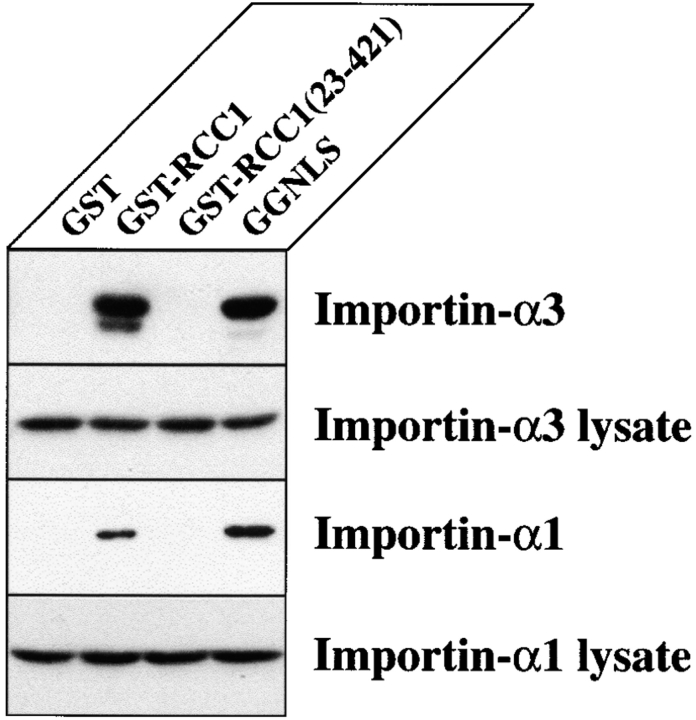
The nuclear import of RCC1 in vitro was recently reported to be stimulated by importin-α, and preferentially by the α3 isoform (Köhler et al. 1999). Since the NTD of RCC1 contains a NLS (Fig. 1 B), it seemed likely that this sequence would be necessary to confer binding to importin-α. To determine which RCC1 constructs could bind members of the importin-α family, we immobilized recombinant GST-RCC1 fusion proteins onto glutathione-Sepharose beads. We then tested their ability to bind individual importin-α family members from an E. coli lysate. Bound and unbound fractions were visualized by Coomassie staining (data not shown) and immunoblotting against the His6-tag contained on the COOH terminus of each importin-α family member (Fig. 2). GGNLS, a protein that binds to all known importin-α members tested to date, was used as a positive control (Welch, 1999). While GST-RCC1 was able to bind both importin-α3 and importin-α1, binding to importin-α3 was much more robust (Fig. 2). By Coomassie staining, GST-RCC1 was able to bind nearly equimolar amounts of importin-α3 while binding to importin-α1 was undetectable (data not shown). By immunoblotting, visualization of importin-α1 required an ∼12-fold longer exposure relative to that of importin-α3. These data are consistent with the specificity imparted by importin-α3 on the import of RCC1 in vitro (Köhler et al. 1999). The affinity of GST-RCC1(23-421) for these importin-α family members was too low to be detectable (Fig. 2). However, as described above, RCC1(23-421)-GFP is imported into nuclei in intact cells. These data therefore suggest that RCC1 can use both importin-α–dependent and –independent import pathways in vivo.
Figure 2.
The NTD of RCC1 is required to bind importin-α. Equal amounts of the indicated GST-fusion proteins were immobilized on glutathione-Sepharose beads. The beads were incubated with a bacterial lysate expressing the indicated importin-α isoform. An aliquot of the supernatant was analyzed to ensure that the beads were incubated with equal concentrations of importin-α. The bound and unbound fractions were analyzed by SDS-PAGE and western blotting. An anti-His6 antibody was used to visualize the importin-α isoforms, each of which possess a COOH-terminal His6 tag. All lanes were exposed equally to film except the bound importin-α1 panel which required a 12-fold longer exposure.
Transport Kinetics of RCC1
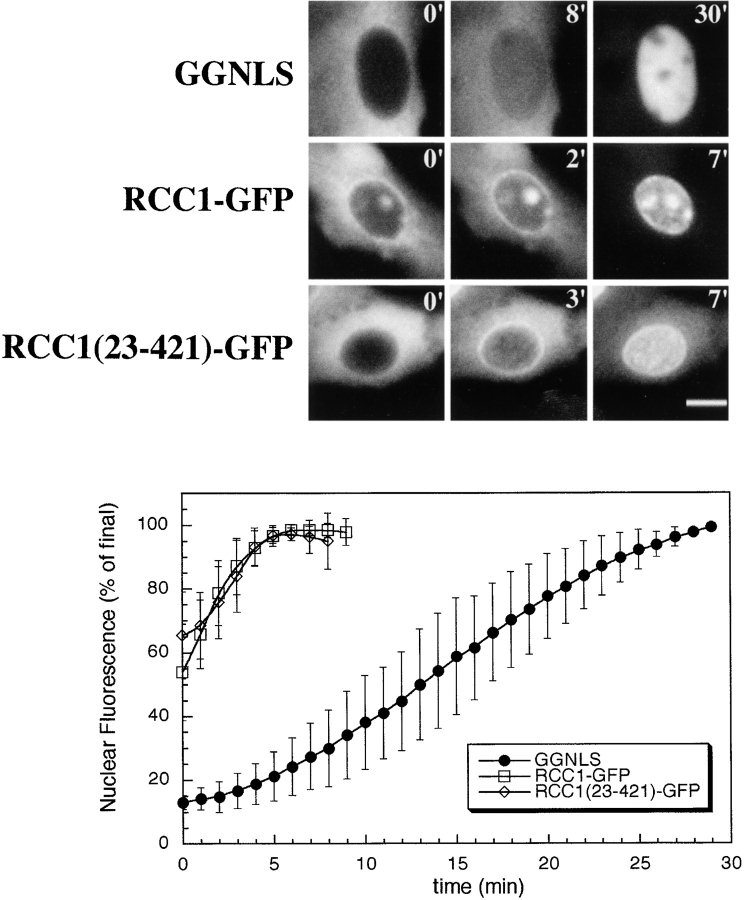
To estimate the rate of entry of RCC1 into the nucleus in vivo, we injected the cytoplasm of tsBN2 cells with the RCC1 constructs and visualized entry into the nucleus by time-lapse photography. GGNLS, a substrate known to be imported in a Ran- and energy-dependent manner, was used as a control (Welch et al. 1999). The rate of import is plotted in Fig. 3, shown below time-lapse stills from individual cells. When GGNLS was injected, we observed a steady increase in nuclear fluorescence that continued for ∼30 min. Nuclear import of the RCC1 constructs, however, was significantly different. When RCC1 was injected, almost all of the cells had concentrated the protein within their nucleus before the time-lapse program could be initiated (<∼1 min, data not shown). RCC1-GFP and RCC1(23-421)-GFP both displayed rapid import rates. In cells injected with RCC1-GFP, ∼50% of the cells had concentrated the protein within their nuclei before the time-lapse program was initiated (data not shown); the remaining cells imported RCC1-GFP within ∼5 min (Fig. 3). Import of RCC1(23-421)-GFP also occurred rapidly (see Discussion). Furthermore, during the early phase of import of all RCC1 constructs tested, the injected protein concentrated at the periphery of the nucleus (Fig. 3, middle panels), suggesting that RCC1 may dock at the nuclear pore more rapidly that it is translocated into the nucleoplasm. Another possibility is that these rings represent the binding of RCC1 to chromatin just under the surface of the nuclear envelope (see Discussion). Shortly after this peripheral accumulation, the protein became almost entirely nuclear within ∼2 min. GGNLS did not display this phenotype.
Figure 3.
Microinjected RCC1 accumulates at the nuclear rim and is imported rapidly into nuclei. GGNLS (1 mg/ml; top panels), RCC1-GFP (middle panels), and RCC1(23-421)-GFP (bottom panels) were injected at ∼18 μM into the cytoplasm of tsBN2 cells. Images were captured by time-lapse photography at 1-min intervals. Sample time-lapse stills are shown (top) and the elapsed time (min) after injection is shown in the upper right corner of each image. The change in the mean fluorescence intensity of the nucleus with time was plotted. The mean of four separate cells is shown and the standard deviation from the mean is shown as error bars. Bar, 10 μm.
Nuclear Import of RCC1 Does Not Require a Ran Gradient In Vivo
The nuclear import of many proteins has been shown to require a gradient of Ran-GTP across the nuclear envelope. Since RCC1(23-421)-GFP is unable to bind importin-α but is still capable of nuclear import (Fig. 2 and Fig. 3), an intriguing question was whether the import of RCC1 was a Ran-dependent process. To address this issue in vivo, we artificially collapsed the Ran gradient by coinjecting the various RCC1 constructs with Ran(G19V). This mutant protein, shown previously to be predominantly in the GTP-bound state, harbors a single amino acid substitution that renders the protein GTPase-deficient and insensitive to both RanGAP and RCC1-catalyzed nucleotide exchange (Lounsbury, 1996). Mammalian cells harbor ∼10 million Ran molecules, and ∼10–20% are in the cytoplasm at steady state (Bischoff and Ponstingl 1991b). By injecting the cytoplasm with one-tenth the cellular volume (∼5 × 10−13 L) of 40 μM Ran(G19V), ∼12 million G19V molecules are injected, an amount that should be sufficient to collapse the Ran gradient in vivo. GGNLS, a protein known to require a Ran gradient for import, was used as a control for Ran gradient collapse (Welch et al. 1999). As shown in Fig. 4, the coinjection of Ran(G19V) completely prevented the import of both GGNLS and RCC1(1-25)-NPC. Since RCC1(1-25)-NPC contains a NLS (Fig. 2) that is necessary to confer binding to importin-α (Fig. 3), these data suggest strongly that the NTD of RCC1 is a classic, Ran-dependent NLS imported specifically by the importin-α/β heterodimer. In contrast, the import of RCC1 and RCC1-GFP was not prevented by coinjection of Ran (G19V). Moreover, a mutant form of RCC1, D182A, which can neither interact with Ran stably nor catalyze nucleotide exchange (Azuma et al. 1996, Azuma et al. 1999), was imported efficiently in the presence of Ran(G19V). Taken together, these data suggest that RCC1 can be imported by at least two pathways: (a) a classic Ran-dependent pathway and (b) a Ran-independent pathway that is not passive diffusion.
Figure 4.
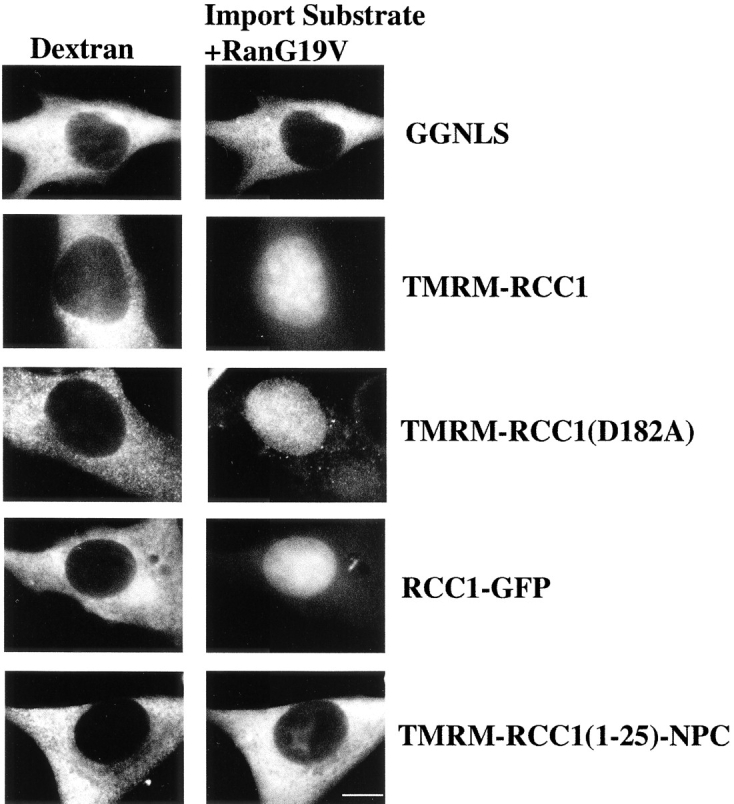
The import of RCC1 is not prevented by collapse of the Ran gradient in vivo. The indicated protein substrates were coinjected into the cytoplasm of BHK21 cells with Ran(G19V) (∼40 μM), to collapse the Ran gradient, and a fluorescent dextran (1 mg/ml) as an injection site marker (left panels). The cells were incubated for 15 min before fixation and visualization of the injected protein substrate (right panels). Bar, 10 μm.
To further test the import of RCC1 in vivo, we conducted microinjection experiments in tsBN2 cells. These cells possess a temperature-sensitive RCC1 allele and, when cultured at 39.5°C, endogenous RCC1 protein is degraded rapidly, resulting in the loss of the Ran gradient (Fig. 5 b; Nishimoto et al. 1978; Uchida et al. 1990; Ohtsubo et al. 1991; Tachibana et al. 1994; Richards et al. 1997). As a result, it has been shown that the nuclear import of Ran-dependent cargo is severely compromised in temperature-shifted tsBN2 cells (Uchida et al. 1990; Tachibana et al. 1994; Richards et al. 1997). When tsBN2 cells cultured at 33.5°C were injected, GGNLS and all RCC1 constructs tested accumulated within the nucleoplasm (Fig. 5 A). In contrast, when temperature-shifted tsBN2 cells were injected, the import of GGNLS and RCC1(1-25)-NPC was prevented completely. However, RCC1, RCC1(D182A), and RCC1-GFP all entered nuclei rapidly, further suggesting that RCC1 possesses a Ran-independent import pathway.
Figure 5.
The import of RCC1 does not require a Ran gradient in vivo. (A) tsBN2 cells were cultured at 33°C (left) or 39.5°C (right) for 3 h before experimentation. The indicated protein substrates were coinjected with a fluorescent dextran to mark the site of injection, as described previously. Samples were incubated for 15 min before fixation. (B) Temperature-shifted (top right) and unshifted (top left) tsBN2 cells were fixed and endogenous Ran was stained with an anti-Ran antibody. Nuclei were visualized by staining DNA with DAPI. Unlabeled RCC1 and the mutant RCC1 (D182A) (18 μM) were coinjected with FITC-dextran as an injection site marker (bottom, left panels). Cells were incubated for 30 min before fixation and visualization of endogenous Ran (bottom, right panels) by immunofluorescence. Bars, 10 μm.
Since RCC1 is slightly below the diffusion limit of the nuclear pore, one possible explanation for the data described above is that small amounts of RCC1 were able to enter nuclei by passive diffusion, which might reestablish the Ran gradient and prime the import of RCC1 by a Ran-dependent pathway. The fact that RCC1-GFP, a molecule above the diffusion limit of the nuclear pore, accumulated rapidly within nuclei argues against this possibility. In addition, RCC1(D182A), a mutant that is inactive catalytically (Azuma et al. 1996), was able to accumulate within the nuclei of temperature-shifted tsBN2 cells at an efficiency comparable to the wild-type protein. One could speculate, however, that although the ability of the RCC1(D182A) mutant to catalyze nucleotide exchange is severely compromised in vitro, there may be sufficient residual activity to reestablish the Ran gradient in vivo. To test this possibility experimentally, tsBN2 cells were cultured at 39.5°C for 3 h and stained for endogenous Ran. In tsBN2 cells cultured at 33.5°C, Ran is ∼80% nuclear while in temperature-shifted tsBN2 cells the Ran is distributed diffusely throughout the cell (Fig. 5 B, top). The Ran gradient could be reestablished by a cytoplasmic injection of wild-type RCC1 but not the mutant RCC1(D182A). The fact that RCC1(D182A) did not reestablish the Ran gradient cannot be explained by a lack of import, because it accumulates within the nuclei of temperature-shifted tsBN2 cells (Fig. 5 A). Thus, RCC1(D182A) can enter nuclei in the absence of a Ran gradient. These data demonstrate that (a) a cytoplasmic injection of RCC1 can quickly reestablish the Ran gradient, (b) the ability to reestablish the Ran gradient is dependent upon the catalytic activity of RCC1, and (c) the ability of RCC1 to be imported is independent of both the catalytic activity of RCC1 and a preexisting Ran gradient.
Import of RCC1 into Digitonin-permeabilized Cells
We have demonstrated that, in intact cells, import of RCC1 is mediated by both Ran-dependent and -independent import pathways. To determine whether RCC1 requires specific, soluble transport factors in vitro, HeLa cells were permeabilized with digitonin (Adam et al. 1990) and the import of TMRM-RCC1 and RCC1-GFP was observed using GGNLS as a control. GGNLS is imported by a classical, temperature-dependent and factor-dependent import pathway (Welch et al. 1999). This process was inhibited by: (a) removal of the soluble factors (reticulocyte lysate); (b) addition of the pore-binding protein WGA; (c) addition of a dominant-negative fragment of importin-β (β(45-462)); (d) addition of an importin-β–binding protein fragment (BIB; Jakel and Görlich 1998); and (e) carrying out the import reaction at low temperature (Fig. 6 A). In contrast to this classic import substrate, RCC1 and RCC1-GFP were imported rapidly under every condition tested except when the reaction was carried out at 4°C, or when β(45-462) was added. However, since the reaction did not require the addition of importin-β, and the BIB domain did not inhibit the import of RCC1, these data suggest that the importin-α–independent pathway for RCC1 is not mediated by direct binding to importin-β. The β(45-462) fragment is known to associate almost irreversibly with nucleoporins and to inhibit a variety of different nucleocytoplasmic transport pathways (Kutay et al. 1997). The fact that β(45-462) inhibited the import of the RCC1 constructs confirms that RCC1 uses nuclear pores for transport and that its import does not occur by passive diffusion (see Discussion). To address whether treatment at 4°C was inhibiting nuclear import or retention, HeLa cells were permeabilized with Triton X-100 and the ability of RCC1 to bind chromatin was assayed directly. In Triton X-100–permeabilized cells, the binding of RCC1 and RCC1-GFP to chromatin was a temperature-insensitive event (Fig. 6 B). Thus, low temperature prevented the translocation of RCC1 across the nuclear envelope, and not the nuclear retention by binding to chromatin.
Figure 6.
RCC1 is imported by a temperature-sensitive and factor-independent pathway in vitro. (A) The import reactions using GGNLS (0.2 mg/ml; top panels), TMRM-RCC1 (0.7 μM; middle panels), and RCC1-GFP (0.7 μM; lower panels) were accomplished as described in Materials and Methods. In brief, digitonin-permeabilized HeLa cells were incubated with an import substrate and a reticulocyte lysate (+RL) or with buffer alone (−RL). All reactions contained an energy regenerating system. Where indicated, reactions containing a reticulocyte lysate were supplemented with the BIB domain (100 μM), WGA (0.2 mg/ml), or GST-importin-β(45-462) (6 μM). A reaction carried out at 4°C is also shown. (B) HeLa cells were permeabilized with Triton X-100 and RCC1 (0.7 μM; top panels) or RCC1-GFP (0.7 μM; bottom panels) were added either at 25 or 4°C. Samples were incubated for 5 min before fixation. Nuclei were visualized by staining DNA with DAPI (left panels).
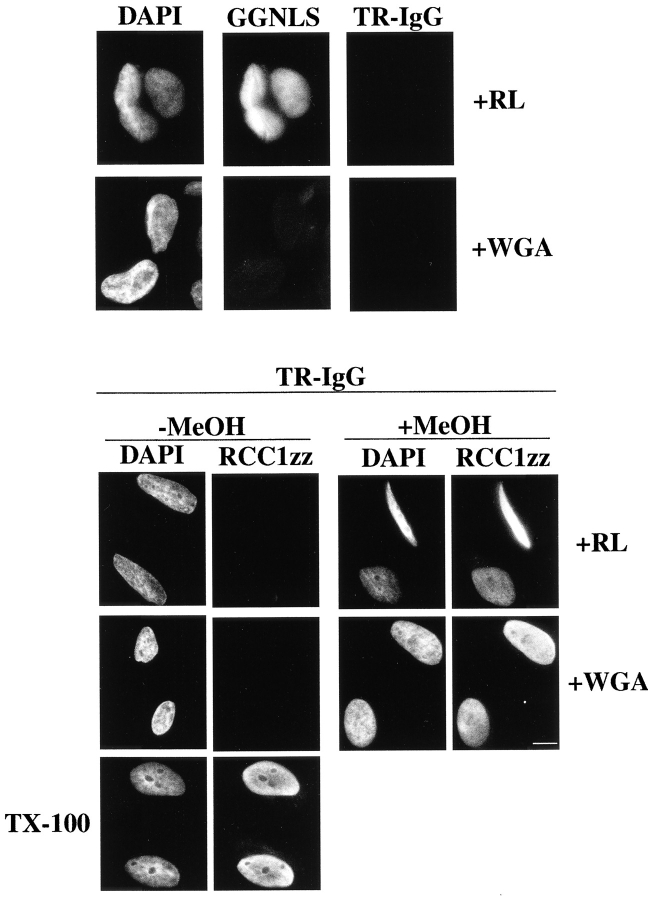
Although WGA inhibited the import of GGNLS completely, the transport of RCC1 and RCC1-GFP appeared much less sensitive. The fact that WGA did not prevent the import of RCC1 was surprising, and raised the possibility that digitonin was perturbing the integrity of the nuclear envelope. Although digitonin is not expected to disrupt the nuclear envelope at the concentrations used in these experiments (0.005%), we wished to test this possibility empirically. If the nuclear envelope was disrupted, then it would allow access of an antibody into the nuclear compartment (Adam et al. 1990). Initially, we observed that the digitonin permeabilization of HeLa cells did not permit the access of an antibody targeted to the nuclear antigen C23 (data not shown). As a more direct approach, we designed an assay that would allow recognition of damaged nuclei using identification of the imported RCC1 itself as a marker. An RCC1 fusion protein was constructed with a COOH-terminal zz tag, which binds efficiently to the constant domain from IgG (Görlich et al. 1997). Import assays were performed on digitonin-permeabilized cells using RCC1zz (∼60 kD) and GGNLS as import substrates in the presence or absence of WGA. All samples were then fixed in paraformaldehyde; however, only half of the RCC1zz samples were permeabilized with methanol. GGNLS was visualized directly; RCC1zz was stained using a Texas red–conjugated rabbit IgG (TR-IgG). As shown in Fig. 7, when cells were permeabilized with methanol, RCC1zz was detectable in nuclei regardless of whether or not WGA was included in the import reaction. If the cells were not permeabilized with methanol, only background staining was observed. Thus, the ability of TR-IgG to gain access to the nuclear compartment in digitonin-permeabilized cells was entirely dependent on additional permeabilization with methanol. As a positive control, the nuclear envelope was intentionally disrupted with Triton X-100 before incubation with RCC1zz. In these cells, RCC1zz was readily detectable in the absence of methanol permeabilization (Fig. 7, bottom). Since WGA inhibited the import of GGNLS in this assay, these data can not be explained by a defect in the activity of the WGA. Furthermore, the staining of nuclei was dependent upon the addition of RCC1zz; thus, TR-IgG recognized RCC1zz specifically and did not cross-react with endogenous RCC1 (Fig. 7, top). Taken together, these data demonstrate that RCC1 can be imported by a factor-independent pathway that it is resistant to all import inhibitors tested except low temperature and β(45-462).
Figure 7.
Import assays were accomplished on digitonin-permeabilized HeLa cells. Reactions consisted of GGNLS (0.2 mg/ml; top) or RCC1zz (0.7 μM; bottom) as import substrates, an energy regenerating system, and a reticulocyte lysate. Where indicated, 0.2 mg/ml WGA was added. All samples that used GGNLS as an import substrate were permeabilized with methanol and stained with a Texas red–conjugated rabbit IgG (TR-IgG; top, right panels). When RCC1zz was used as the import substrate, half of the samples were permeabilized with methanol (+MeOH) while half were not (−MeOH). RCC1zz was then visualized in all samples using a Texas red–conjugated rabbit IgG. When Triton-permeabilized cells were used (bottom, left panels), the permeabilization with methanol was omitted. All nuclei were visualized with DAPI. Bar, 10 μm.
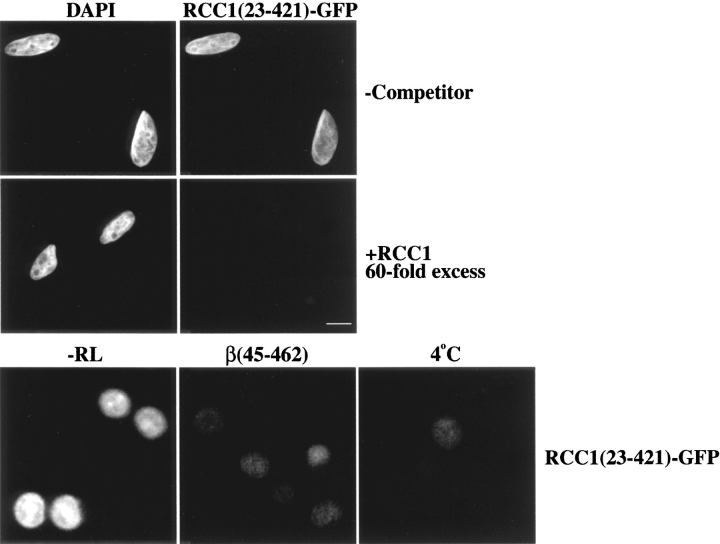
To determine whether the Ran-independent accumulation of RCC1 in the nucleus was saturable, we performed a competition experiment in vitro using RCC1(23-421)-GFP as an import substrate and unlabeled RCC1 as a competitor. Since RCC1(23-421)-GFP does not possess the NTD, it can only access the Ran/importin-α–independent import pathway. Furthermore, import reactions were carried out in the absence of a reticulocyte lysate so that only the Ran-independent pathway could be observed. Under these conditions, RCC1(23-421)-GFP rapidly entered the nuclei of digitonin-permeabilized cells in the absence of any competitor (Fig. 8, top). In contrast, the nuclear accumulation of RCC1(23-421)-GFP was prevented by the addition of a 60-fold molar excess of RCC1. Significantly, import of RCC1(23-421)-GFP was also inhibited by low temperature or by the addition of importin-β(45-462) (Fig. 8, bottom). These data confirm that importin-β(45-462) inhibits both the Ran-dependent and -independent import pathways of RCC1 and suggest that, in the absence of soluble factors, RCC1, RCC1-GFP, and RCC1(23-421)-GFP enter nuclei by the same saturable pathway.
Figure 8.
Import of RCC1 into nuclei is saturable. Digitonin-permeabilized HeLa cells were incubated with RCC1(23-421)-GFP as an import substrate (0.6 μM). All reactions were done in the absence of a reticulocyte lysate (−RL) but in the presence of an energy regenerating system. Import was accomplished in the presence or absence of a 60-fold molar excess of unlabeled RCC1. Nuclei were visualized with DAPI (top left panels). Where indicated, GST-importin-β(45-462) (6 μM) was added; a reaction incubated on ice is also shown. Bar, 10 μm.
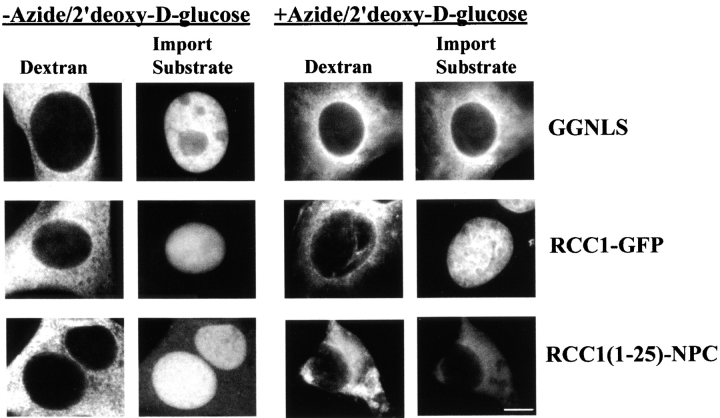
Import of RCC1 in Energy-depleted Cells
To determine the energy requirements for the import of RCC1 in vivo, BHK cells were treated with 10 mM sodium azide and 6 mM 2′deoxy-d-glucose to inhibit both mitochondrial and glycolytic mechanisms for ATP production. Under these conditions, it has been shown that ATP pools are depleted rapidly and reversibly (Breeuwer and Goldfarb 1990; Weldon et al. 1998). Energy-depleted cells were microinjected with the RCC1 constructs and import of the proteins was monitored by fluorescence microscopy. In the absence of metabolic inhibitors, GGNLS, RCC1-GFP, and RCC1(1-25)-NPC were all transported rapidly into nuclei (Fig. 9). In energy-depleted cells, however, RCC1-GFP was still imported rapidly while GGNLS and RCC1 (1-25)-NPC remained cytosolic. These data demonstrate that RCC1 is imported by at least two pathways. The first is mediated by the NTD of RCC1 and is a classic Ran (Fig. 4 and Fig. 5) and energy-dependent (Fig. 9) pathway. The second pathway is both saturable and sensitive to temperature, but requires neither energy, soluble factors, nor a Ran gradient.
Figure 9.
Import of RCC1 into nuclei does not require energy in vivo. BHK21 cells were incubated in serum- and glucose-free media with (right panels) or without (left panels) sodium azide and 2′deoxy-d-glucose for 3 h. The indicated protein substrates were coinjected with a fluorescent dextran as an injection site marker at concentrations identical to that described previously. Samples were incubated 15 min before fixation. Bar, 10 μm.
Discussion
The protein RCC1 possesses at least two distinct biological functions. The first and best-established function is to generate a Ran-GTP gradient across the nuclear pores (Bischoff and Ponstingl, 1991), which is used to drive the transport of a wide variety of cargo molecules against their concentration gradients (Mattaj and Englmeier 1998; Melchior and Gerace 1998; Pemberton et al. 1998; Görlich and Kutay 1999; Nakielny and Dreyfuss 1999; Macara, 2000). A second, more recently characterized function is to create a Ran-GTP gradient near the chromatin surface, which is required for mitotic spindle formation (Carazo-Salas, 1999; Ohba et al. 1999; Wilde and Zheng 1999). In both cases, RCC1 behaves as a sort of identification tag for chromatin and hence, in interphase cells, as a marker for the nuclear compartment. It operates by efficiently catalyzing guanine-nucleotide exchange on the Ran GTPase (Bischoff and Ponstingl, 1991). The amount of RCC1 associated with the chromatin must double with each cell cycle. One can imagine two extreme mechanisms to achieve this result. In the first mechanism, RCC1 is only synthesized in S-phase, and must await breakdown of the nuclear envelope in prophase to associate with the chromatin. This mechanism would be problematic for cells such as those of the budding yeast, Saccharomyces cerevisiae, that use a closed mitotic cycle. In the second mechanism, RCC1 is synthesized throughout the cell cycle and is imported continuously into the nucleus, either by passive diffusion or by a facilitated process.
Although the size of RCC1 (∼45 kD) is such that it could, in principle, diffuse passively through the nuclear pores, the rate of diffusion, based on comparison with ovalbumin, which is of similar size (Featherstone et al. 1988), is likely to be too low to be practicable. Also, accumulation of newly synthesized RCC1 within the cytoplasm would likely reduce the efficiency of nucleocytoplasmic transport and disrupt microtubule dynamics formation. Therefore, one might predict that RCC1 is imported by a facilitated mechanism throughout interphase. In support of this prediction, a conserved polybasic sequence that resembles a classic NLS is found near the NH2 terminus of RCC1 from many species (Fig. 1 A). Attachment of this NTD to β-galactosidase permits accumulation of the fusion protein within nuclei of transiently transfected COS 7 cells (Seino et al. 1992). Also, a study of multiple importin-α isoforms showed that RCC1 can accumulate within the nuclei of digitonin-permeabilized cells in the presence of energy and soluble transport factors, and that accumulation was enhanced specifically by the importin-α3 isoform (Köhler et al. 1999). These studies suggest that the import of RCC1 can be mediated by the classical importin-α/β pathway through a conventional NLS. However, these studies did not define the NH2-terminal sequence as the signal recognized by importin-α3. The NTD has also been reported to bind DNA (Seino et al. 1992), thus complicating analysis of the transfection data for the NTD-β-galactosidase construct described above. Because the transfected cells had undergone at least one cell cycle (Seino et al. 1992), one could imagine that the NTD caused binding of the fusion protein to exposed chromatin during mitosis, and hence mediated trapping within nuclei rather than import through the nuclear pores. Moreover, an NH2-terminal deletion mutant of RCC1 was not excluded from the nuclei of transfected tsBN2 cells and was capable of rescuing these cells at the nonpermissive temperature (Seino et al. 1992). Either the deletion mutant could diffuse into the nuclei, where it would be retained by binding to chromatin, or there exists a second transport system that is independent of the putative NH2-terminal NLS.
Given the physiological importance of RCC1 import, we examined the mechanism by which it is translocated into the nucleus using a combination of intact and permeabilized cell assays. We find that the NTD of RCC1 is necessary for binding importin-α and that RCC1 preferentially recognizes the α3 isoform. Attachment of the first 25 amino acids from RCC1 are also sufficient to target a fluorescent reporter protein to the nucleus upon microinjection into the cytoplasm. The NTD of RCC1 is not sufficient, however, to target proteins to chromatin in vitro (data not shown). Furthermore, we show that NTD-mediated transport is dependent upon both a preexisting Ran gradient and energy in vivo. These data confirm that the NTD of RCC1 is a NLS and not a DNA-binding domain.
By time-lapse photography, we found that almost all of the cells concentrated the injected RCC1 protein within their nucleus before we could initiate the time lapse program (<1 min). Assuming that (a) each cell was injected with ∼5 × 10−13 L of substrate, (b) there are ∼3,000 nuclear pores per nucleus, and (c) ∼80% of the injected protein became nuclear within one minute, we can estimate that ∼72,000 RCC1 molecules were imported per second (∼24 events/pore/s). To our knowledge, this is one of the most rapid nuclear import process recorded to date. In fact, kinetic analysis of RCC1 demonstrated that its import was at least 30-fold greater than the import of a classic NLS-containing import substrate (GGNLS) under similar conditions. Since RCC1 and GGNLS can bind to importin-α3 with similar affinity (Fig. 3), each can access the importin-α/β import pathway; however, GGNLS can both bind and be imported by every importin-α molecule tested to date while RCC1 can not (Köhler et al. 1999; Welch et al. 1999). The fact that RCC1 is imported much more rapidly than GGNLS thus suggests that RCC1 possesses another import pathway that is independent of that mediated by importin-α3.
Removal of the NTD from RCC1 abolished binding to importin-α but did not prevent the RCC1 deletion mutant from entering the nucleoplasm (Fig. 1 C). Kinetic analysis of RCC1(23-421)-GFP in vivo is complicated by the fact that a subset of cells did not import the protein within the time frame of the time-lapse experiment; analysis of those cells that imported, however, showed that the rate was indistinguishable from that of RCC1-GFP. The fact that not all of the cells import RCC1(23-421)-GFP within 25 min suggests that the import might be a regulated process. Efforts to either increase or decrease the import of RCC1 (23-421)-GFP have only been successful by treatment at low temperature, the addition of importin-β(45-462), and by competition with excess, unlabeled RCC1 in vitro (Fig. 8, data not shown). The heterogeneity of import may also reflect differences in individual cells to import under time-lapse conditions; however, since cells that did not import RCC1(23-421)-GFP were excluded from kinetic analysis, the overall mean import rate for RCC1-GFP in vivo must be more rapid than for RCC1(23-421)-GFP. This observation, plus the fact that the NTD of RCC1 confers high-affinity binding to importin-α3, suggests that the NTD can facilitate the import of RCC1 in vivo.
Interestingly, all of the RCC1 constructs that were tested accumulated at the nuclear periphery before import (Fig. 3). In our hands, the import of molecules by classic transport pathways, such as GGNLS (Fig. 3) and RCC1 (1-25)-NPC (data not shown), do not show this phenotype. A similar accumulation at the nuclear envelope was observed during the import of the RCC1 constructs into the nuclei of temperature-shifted tsBN2 cells (data not shown); therefore, this phenotype is not due to import by a Ran-dependent process. This phenomenon could represent either RCC1 that is bound either directly or indirectly to the nuclear pore, or RCC1 that is binding to chromatin just as it enters the nucleus. The latter hypothesis, however, predicts that accumulation of RCC1 in the nucleus would proceed from the nuclear periphery to the nuclear center, i.e., that the ring of RCC1 would thicken over time, and this process is not observed. If RCC1 is docking at the nuclear pore, then this may be the rate-limiting step in the import of RCC1. The fact that the import of RCC1 was inhibited by importin-β(45-462) suggests that either RCC1 binds to some of the same nucleoporins as importin-β or that access to RCC1-binding sites on nuclear pores is obscured by importin-β. The identity of the putative nucleoporin docking sites for RCC1 remains to be investigated. It is noteworthy that staining of RCC1 at the nuclear periphery was not prominent in digitonin-permeabilized cells incubated with RCC1 at 4°C. Therefore, either binding is of very low affinity or access to the docking site is reduced at low temperatures. Low-affinity binding to nucleoporins is also a characteristic of NTF2, which has an ∼25 μM affinity for nucleoporin-FxFG repeats (Chaillan-Huntington et al. 2000).
A surprising feature of the factor-independent RCC1 import pathway is that, compared with GGNLS, it is much less sensitive to inhibition by WGA. WGA binds with high affinity to O-linked glycoproteins of the nuclear pore (Stoffler et al. 1999), including the p62 complex which is localized near the central gated channel of the pore (Guan et al. 1995). WGA is presumed not to occlude the channel because it does not abrogate the passive diffusion of small dextrans (Finlay et al. 1987). For this reason, WGA has been regarded as a marker to distinguish active from passive transport. However, different transport pathways use different nucleoporins (Shah and Forbes 1998; Shah et al. 1998; Moy and Silver 1999; Seedorf et al. 1999), and WGA may inhibit selectively those pathways that require the p62 complex. RCC1 may represent the first member of a class of proteins that are translocated across the nuclear pore complex independently of interactions with O-linked glycoproteins.
In digitonin-permeabilized cells, the import of RCC1 could only be inhibited by importin-β(45-462), low temperature, or competition with excess, unlabeled RCC1. The fact that the import reaction was inhibited by low temperature suggests strongly that import does not proceed by passive diffusion, and that the nuclear envelope was not disrupted during permeabilization of the plasma membrane (Fig. 6 B). The inhibition due to excess RCC1 could be explained by competition for a saturable transport process and/or competition for chromatin-binding sites. Evidence from time-lapse photography, however, suggests that the rate-limiting step for the import of RCC1 in vivo is transit across the nuclear pore.
Interestingly, the process by which RCC1 traverses the nuclear envelope does not require energy in vivo (Fig. 9). These data cannot be explained by the assertion that energy depletion alters the permeability of the nuclear envelope, because neither the injection site marker, GGNLS, nor RCC1(1-25)-NPC entered the nucleoplasm. Furthermore, since RCC1-GFP is beyond the diffusion limit of the nuclear pore, import in energy-depleted cells can not proceed by passive diffusion. Energy-independent translocation of this sort is not without precedent. Evidence from in vitro import assays suggest that single rounds of transport mediated by transportin, Crm1, and importin-β are energy independent (Schwoebel et al. 1998; Englmeier et al. 1999; Ribbeck et al. 1999). In addition, proteins such as β-catenin (Yokoya et al. 1999) and hnRNP K (Michael et al. 1997) are able to translocate across the nuclear envelope in the absence of soluble factors. It is noteworthy that transport of these proteins is inhibited by the dominant-negative importin-β(45-462), as is the case for RCC1. It seems likely that the translocation of all these proteins occurs by a process of facilitated diffusion that requires specific, low-affinity interactions with nucleoporins that line the central channel of the pores. Facilitated diffusion cannot occur against a concentration gradient. Thus, nuclear pores must import and export RCC1 at the same rate and equilibrate the levels of free RCC1 in the cytoplasm to a concentration identical to that free in the nucleoplasm. The fact that RCC1 is >90% nuclear at steady state and does not shuttle detectably in vivo implies that the binding of RCC1 to chromatin is much more rapid than translocation across the nuclear envelope. In addition, it suggests that binding to chromatin can be sufficient to establish and maintain the nuclear localization of RCC1 and, hence, the Ran gradient. In effect, RCC1 acts as a chromatin marker, an activity key for the evolution of the nuclear compartment and the open mitotic cycle.
Key questions that remain to be explored include the identity of the RCC1-binding proteins on chromatin and at the nuclear pores. It will also be of interest to determine whether other import cargoes use the same pathway for transit across the nuclear envelope as RCC1.
Acknowledgments
We thank all members of the Macara lab for their continued advice throughout the experimental and preparatory phases of this manuscript. In particular, we thank Clark Peterson, Katie Welch, Amy Brownawell, Mark Lindsay, Alicia Smith, Kendra Plafker, and Scott Plafker for reagents and helpful suggestions. We also thank Takeharu Nishimoto, Dirk Görlich, and Matthias Köhler for their generosity in providing some of the expression plasmids used in this study (RCC1, nucleoplasmin, nucleoplasmin core, the BIBzz domains, and importin-α3).
This work was supported by a grant from the National Institutes of Health, DHHS (GM-50526).
Footnotes
Abbreviations used in this paper: BIB: β-like import receptor binding domain; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; GGNLS, GST-GFP-NLS; GST, glutathione-S-transferase; NLS, nuclear localization signal; NPC, nucleoplasmin core-domain; NTD, NH2-terminal domain; PFA, paraformaldehyde; RCC1, regulator of chromosome condensation protein; TMRM, tetramethylrhodamine-5-maleimide.
References
- Adam S.A., Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Adam S.A., Marr R.S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Seino H., Seki T., Uzawa S., Klebe C., Ohba T., Wittinghofer A., Hayashi N., Nishimoto T. Conserved histidine residues of RCC1 are essential for nucleotide exchange on Ran. J. Biochem. 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- Azuma Y., Renault L., Garcia-Ranea J.A., Valencia A., Nishimoto T., Wittinghofer A. Model of the Ran-RCC1 interaction using biochemical and docking experiments. J. Mol. Biol. 1999;289:1119–1130. doi: 10.1006/jmbi.1999.2820. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1 Nature 354 1991. 80 82a [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide Proc. Natl. Acad. Sci. USA. 88 1991. 10830 10834b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer M., Goldfarb D.S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990;60:999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chaillan-Huntington C., Braslavsky C.V., Kuhlmann J., Stewart M. Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J. Biol. Chem. 2000;275:5874–5879. doi: 10.1074/jbc.275.8.5874. [DOI] [PubMed] [Google Scholar]
- Chi N.C., Adam E.J., Adam S.A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N.C., Adam E.J.H., Visser G.D., Adam S.A. Ranbp1 stabilizes the interaction of ran with P97 in nuclear protein import. J. Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes P., Ye Z.S., Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1 Proc. Natl. Acad. Sci. USA. 91 1994. 7633 7637function as well as sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo C.A., Kirch S.A., Gyuris J., Brent R., Oettinger M.A. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I. The nuclear pore complex. Annu. Rev. Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R.A. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Enenkel C., Blobel G., Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo J.C., Mattaj I.W. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- Featherstone C., Darby M.K., Gerace L. A monoclonal antibody against the nuclear pore complex inhibits nucleocytoplasmic transport of protein and RNA in vivo. J. Cell Biol. 1988;107:1289–1297. doi: 10.1083/jcb.107.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D.R., Newmeyer D.D., Price T.M., Forbes D.J. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. Nuclear protein import. Curr. Opin. Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Görlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Görlich D., Prehn S., Laskey R.A., Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D., Vogel F., Mills A.D., Hartmann E., Laskey R.A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D., Henklein P., Laskey R.A., Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus EMBO (Eur. Mol. Biol. Organ.) J. 15 1996. 1810 1817a [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Pante N., Kutay U., Aebi U., Bischoff F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import EMBO (Eur. Mol. Biol. Organ.) J. 15 1996. 5584 5594b [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of Rangtp binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T., Muller S., Klier G., Pante N., Blevitt J.M., Haner M., Paschal B., Aebi U., Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A., Truant R., Wiegand H., Cullen B.R. Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 1998;143:309–318. doi: 10.1083/jcb.143.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A.K., Traglia H.M., Dunst R.W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J. Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Albig W., Kutay U., Bischoff F.R., Schwamborn K., Doenecke D., Görlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Köhler M., Ansieau S., Prehn S., Leutz A., Haller H., Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Köhler M., Speck C., Christiansen M., Bischoff F.R., Prehn S., Haller H., Görlich D., Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussel P., Frasch M. Yeast Srp1, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol. Gen. Genet. 1995;248:351–363. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde E., Bischoff F.R., Mattaj I.W., Görlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury K.M., Richards S.A., Carey K.L., Macara I.G. Mutations within the Ran/Tc4 GTPase—effects on regulatory factor interactions and subcellular localization. J. Biol. Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- Macara I., Brownawell A., Welch K. Ran. In: Hall A., editor. GTPases. Oxford University Press; New York: 2000. pp. 198–221. [Google Scholar]
- Mahajan R., Delphin C., Guan T.L., Gerace L., Melchior F. A small ubiquitin-related polypeptide involved in targeting Rangap1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W., Englmeier L. Nucleocytoplasmic transportthe soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein Rangap1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F., Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Eder P.S., Dreyfuss G. The K nuclear shuttling domain—a novel signal for nuclear import and nuclear export in the HnRNP K protein. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.S., Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moroianu J., Blobel G., Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J., Blobel G., Radu A. Nuclear protein importRan-GTP dissociates the karyopherin alpha/beta heterodimer by displacing alpha from an overlapping binding site on beta. Proc. Natl. Acad. Sci. USA. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy T.I., Silver P.A. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Ryder U.W., Lamond A.I., Weis K. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc. Natl. Acad. Sci. USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler S.G., Tritschler D., Haffar O.K., Blake J., Bruce A.G., Cleaveland J.S. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Nishimoto T., Eilen E., Basilico C. Premature of chromosome condensation in a ts DNA-mutant of BHK cells. Cell. 1978;15:475–483. doi: 10.1016/0092-8674(78)90017-x. [DOI] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki H., Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M., Yoshida T., Seino H., Nishitani H., Clark K.L., Sprague G.F., Jr., Frasch M., Nishimoto T. Mutation of the hamster cell cycle gene RCC1 is complemented by the homologous genes of Drosophila and S. cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:1265–1273. doi: 10.1002/j.1460-2075.1991.tb08068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel G., Rosenblum J.S. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Michael W.M., Nakielny S., Siomi M.C., Wang F., Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Prieve M.G., Guttridge K.L., Munguia J., Waterman M.L. Differential importin-alpha recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol. Cell. Biol. 1998;18:4819–4832. doi: 10.1128/mcb.18.8.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A., Blobel G., Moore M.S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L., Nassar N., Vetter I., Becker J., Klebe C., Roth M., Wittinghofer A. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nucleiassociation and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Kutay U., Paraskeva E., Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both ran and energy. Curr. Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- Richards S.A., Carey K.L., Macara I.G. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Sparrow D.B., Shiomi T., Pu R.T., Nishimoto T., Mohun T.J., Dasso M. Ubc9p and the conjugation of Sumo-1 to Rangap1 and RanBP2. Curr. Biol. 1997;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- Schwoebel E.D., Talcott B., Cushman I., Moore M.S. Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J. Biol. Chem. 1998;273:35170–35175. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- Seedorf M., Damelin M., Kahana J., Taura T., Silver P.A. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino H., Hisamoto N., Uzawa S., Sekiguchi T., Nishimoto T. DNA-binding domain of RCC1 protein is not essential for coupling mitosis with DNA replication. J. Cell Sci. 1992;102:393–400. doi: 10.1242/jcs.102.3.393. [DOI] [PubMed] [Google Scholar]
- Seki T., Tada S., Katada T., Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem. Biophys. Res. Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- Shah S., Forbes D.J. Separate nuclear import pathways converge on the nucleoporin nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich S., Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D., Fahrenkrog B., Aebi U. The nuclear pore complexfrom molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Tachibana T., Imamoto N., Seino H., Nishimoto T., Yoneda Y. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J. Biol. Chem. 1994;269:24542–24545. [PubMed] [Google Scholar]
- Takeda S., Fujiwara T., Shimizu F., Kawai A., Shinomiya K., Okuno S., Ozaki K., Katagiri T., Shimada Y., Nagata M. Isolation and mapping of karyopherin alpha 3 (KPNA3), a human gene that is highly homologous to genes encoding Xenopus importin, yeast SRP1 and human RCH1. Cytogenet. Cell Genet. 1997;76:87–93. doi: 10.1159/000134521. [DOI] [PubMed] [Google Scholar]
- Uchida S., Sekiguchi T., Nishitani H., Miyauchi K., Ohtsubo M., Nishimoto T. Premature chromosome condensation is induced by a point mutation in the hamster RCC1 gene. Mol. Cell. Biol. 1990;10:577–584. doi: 10.1128/mcb.10.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M.A., Coleman D.E., Lee E., Iniguez-Lluhi J.A., Posner B.A., Gilman A.G., Sprang S.R. The structure of the G protein heterotrimer Giα1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Weighardt F., Biamonti G., Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteinsa search for the targeting domains in hnRNP A1. J. Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- Weis K., Mattaj I.W., Lamond A.I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Weis K., Ryder U., Lamond A.I. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Welch K., Franke J., Köhler M., Macara I.G. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-alpha3. Mol. Cell. Biol. 1999;19:8400–8411. doi: 10.1128/mcb.19.12.8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon R.A., Jr., Parker W.B., Sakalian M., Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J. Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wozniak R.W., Rout M.P., Aitchison J.D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yokoya F., Imamoto N., Tachibana T., Yoneda Y. Beta-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]