Summary
Genetic modification of dendritic cells (DCs) with recombinant vectors encoding tumor antigens may aid in developing new immunotherapeutic treatments for patients with cancer. Here, we characterized antigen presentation by human DCs genetically modified with plasmid cDNAs, RNAs, adenoviruses, or retroviruses, encoding the melanoma antigen gp100 or the tumor-testis antigen NY-ESO-1. Monocyte-derived DCs were electroporated with cDNAs or RNAs, or transduced with adenoviruses. CD34+ hematopoietic stem cell-derived DCs were used for retroviral transduction. Genetically modified DCs were coincubated with CD8+ and CD4+ T cells that recognized major histocompatibility complex class I- and class II-restricted epitopes from gp100 and NY-ESO-1, and specific recognition was evaluated by interferonγ secretion. Cytokine release by both CD8+ and CD4+ T cells was consistently higher in response to DCs modified with adenoviruses than cDNAs or RNAs, and maturation of DCs after genetic modification did not consistently alter patterns of recognition. Also, retrovirally transduced DCs encoding gp100 were well recognized by both CD8+ and CD4+ T cells. These data suggest that DCs transduced with viral vectors may be more efficient than DCs transfected with cDNAs or RNAs for the induction of tumor reactive CD8+ and CD4+ T cells in vitro and in human vaccination trials.
Keywords: dendritic cells, electroporation, nonviral vectors, adenovirus, retrovirus
Dendritic cells (DCs) are potent antigen-presenting cells capable of stimulating both naive CD4+ T-helper cells and CD8+ cytotoxic T lymphocytes (CTLs).1 Therefore, DCs loaded with tumor-associated antigens may facilitate the development of new immunotherapies for the treatment of patients with cancer. DCs can readily be loaded with synthetic peptides, but this strategy is limited to the repertoire of known tumor-associated epitopes and to patients who express particular major histocompatibility complex (MHC) restriction elements. An alternate method for loading DCs with antigen is direct incubation with full-length recombinant protein. However, exogenously loaded protein is predominantly processed by DC via the endocytic compartment, and epitopes are most efficiently presented in the context of MHC class II molecules, eliciting CD4+ T-cell responses. Through cross-presentation, an exogenously loaded protein may be diverted to the endogenous processing pathway, resulting in presentation of MHC class I-binding epitopes recognized by CD8+ T cells.2,3 However, the extent to which cross-presentation truly results in CTL activation to exogenous self-antigens is unclear.4,5 In a previous report, we did not detect significant cross-presentation when the melanoma differentiation antigen gp100 was pulsed on DCs as an intact protein or from melanoma cell lysates.6 Alternatively, in several other reports, efficient cross-presentation was induced using full-length proteins formulated as antigen-antibody immune complexes,7 with ISCOMATRIX adjuvant,7,8 or with antibodies against DEC-205.9
Insertion of full-length antigens into DC by genetic modification has the potential to overcome the dependence of MHC class I processing on cross-presentation after loading with exogenous protein. A vector-transferred recombinant antigen synthesized in the cytosol may enter the degradation process of intracellular molecules, eventually yielding epitope presentation in the context of MHC class I molecules.10 Several gene delivery methods have been employed for genetic modification of DC, and these can be divided into viral and nonviral vectors.
The nonviral vectors, including plasmid cDNAs and in vitro transcribed RNAs (IVT RNAs), exclusively confer the antigen of interest and are relatively easy to produce. IVT RNA is an especially attractive vector because it does not integrate into the genome and is not controlled by promoter sequences. RNA transfections are transient, but a short-lived display of peptide on RNA-transfected DCs may be sufficient to prime antigen-specific T cells.11 DNA and RNA transfections are most efficiently carried out using electroporation.12 Briefly, high-voltage electrical pulses are applied to cells to induce the formation of transient pores in the cell membrane. DNA requires higher voltages to enter the nuclear membrane and is therefore associated with higher rates of cell damage in comparison to RNA, which only requires passage through the cytoplasmic membrane. DCs transfected with RNA were previously reported to be more efficient than DNA-transfected DCs for antigen-specific T-cell stimulation.13
Several viral vectors have been used to transduce DCs, including recombinant adenoviruses,14 pox viruses,15 retroviruses,16 and lentiviruses.17 In general, viral transductions induce higher levels of transgene expression than their nonviral counterparts. Adenoviral vectors transduce DCs with high efficiency and do not significantly reduce cell viability.18,19 However, proteins from the adenoviral backbone may dominate the immune response, and attempts to reduce the expression of adenoviral proteins in these vectors have not been successful.19,20 Retroviruses have the advantage that viral proteins are not expressed after integration of the transgene into the genome, and retrovirally transduced DCs can elicit both CD8+ and CD4+ T-cell responses to tumor antigens.16,21 However, retroviruses can only transduce dividing cells, and therefore, cannot mediate gene transfer into monocyte-derived DC populations. Instead, CD34+ hematopoietic stem cells (HSCs) must be mobilized and stimulated to proliferate in vitro using a cocktail of cytokines. After retroviral transduction, these cells can then be stimulated to differentiate into DCs expressing the transgene. Lentiviruses are thought to be able to transduce nondividing cells, but levels of transgene expression in DCs is generally not as high as with other viral vectors, and the production of stable high-titer viral supernatants is technically challenging.17
In this report, we compared patterns of antigen presentation by DCs genetically modified with viral and nonviral vectors. The use of antigen-specific T-cell lines provided a biologic assay for assessing presentation of specific epitopes because binding of T-cell receptor to relevant peptide-MHC class I or class II complexes on DC surfaces induced measurable interferon (IFN)γ secretion. We selected the human tumor-associated antigens gp100 and NY-ESO-1 as model proteins due to the availability of tumor reactive CD8+ and CD4+ T-cell lines with specificities for a variety of antigen-derived epitopes. DCs transduced with viral vectors were consistently recognized more efficiently than those transfected with cDNAs or IVT RNAs by both MHC class I and class II restricted T cells suggesting that viral vectors may be more efficient for the induction of tumor reactive CD8+ and CD4+ T cells in vitro and in human vaccination trials.
MATERIALS AND METHODS
Cell Lines and Reagents
Human melanoma cell lines, Epstein-Barr virus-transformed B cells, and T2 cells were routinely cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 2 mM L-glutamine (Invitrogen, Carlsbad, CA). DC cultures were initiated by plating human peripheral blood mononuclear cells (PBMC) in X-Vivo-20 (BioWhittaker, Walkersville, MD). Human lymphocytes and DCs were cultured in complete medium (CM) consisting of RPMI 1640, 2 mM L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin (Invitrogen), and 10% heat-inactivated human AB serum (Gemini Bio-Products, Woodland, CA; Valley Biomedical, Winchester, VA). Multiple melanoma-reactive T-cell lines and clones were used to evaluate the presentation of MHC-restricted epitopes by genetically modified DCs16,22–29 as presented in Table 1.
TABLE 1.
Characteristics of Antigen Reactive T-cell Lines and Clones
| T-cell Clone/Line | CD4/CD8 | HLA Restriction Element | Peptide Specificity |
|---|---|---|---|
| TIL 1200 (line) | CD8 | HLA-A*0201 | gp100:154–162 |
| CL70 (clone) | CD8 | HLA-A*0201 | gp100:154–162 |
| RB154 (line) | CD8 | HLA-A*0201 | gp100:154–162 |
| CK3H6 (clone) | CD8 | HLA-A*0201 | gp100:209–217 |
| L2D8 (clone) | CD8 | HLA-A*0201 | gp100:209–217 |
| JR1E2 (clone) | CD8 | HLA-A*0201 | gp100:280–288 |
| JR1A4 (clone) | CD8 | HLA-A*0201 | gp100:280–288 |
| HT1D11 (clone) | CD8 | HLA-A*0201 | gp100:280–288 |
| BR-B8 (line) | CD4 | HLA-DRβ1*0401 | gp100:44–59 |
| B104 (line) | CD4 | HLA-DRβ1*0701 | gp100:170–190 |
| JE-D2 (line) | CD4 | HLA-DRβ1*0701 | gp100:420–435 |
| TH1F2L (line) | CD8 | HLA-A*0201 | NY-ESO-1:157–165 |
| J H1 (line) | CD8 | HLA-A*0201 | NY-ESO-1:157–165 |
| J H6 (line) | CD8 | HLA-A*0201 | NY-ESO-1:157–165 |
| M-8 (line) | CD8 | HLA-A*0201 | NY-ESO-1:157–165 |
| SG6 (line) | CD4 | HLA-DPβ1*0401 | NY-ESO-1:161–180 |
Expression of gp100 in melanoma cell lines was assessed by fluorescence-activated cell sorting (FACS) using a monoclonal antibody (mAb) (HMB45; Enzo Diagnostics, Farmingdale, NY), and expression of NY-ESO-1 was previously evaluated on the basis of specific IFNγ secretion by two T-cell clones, MB4 and M8, respectively, that specifically recognize peptides from these proteins in the context of human leukocyte antigen (HLA)-A*0201.26 The F002R melanoma cell line (F002Rmel) did not naturally express gp100, but it had previously been transduced with a vesicular stomatitis virus (VSV)-pseudotyped retroviral vector encoding either gp100 or green fluorescence protein (GFP).16 Expression of HLA class II molecules on the surfaces of melanoma cells was upregulated by transduction with a retroviral contstruct encoding the HLA class II transacti-vator gene (CIITA) as previously described.30 The expression of cell surface HLA class I and class II molecules on melanoma cells was confirmed by FACS using mAbs against HLA-A2 (One Lambda, Conestoga, CA), HLA-DR (L243; Becton Dickinson Biosciences, Franklin Lakes, NJ), HLA-DR4 (Accurate Chemical and Scientific Corp, Westbury, NY), and HLA-DR7 (Pel-Freez Biologicals, Rogers, AR). In addition, HLA haplotypes of cell lines were determined by DNA sequencing in the HLA Laboratory at the National Institutes of Health. The expression of gp100 and NY-ESO-1, HLA-A*0201, HLA-DRβ1*0401, and HLA-DRβ1*0701 in melanoma cell lines was as follows: 888mel (gp100+, NY-ESO-1−, HLA-A*0201−, HLA-DRβ1*0401−, HLA-DRβ1*0701−), 624mel CIITA (gp100+, NY-ESO-1+, HLA-A*0201+, HLA-DRβ1*0401+, HLA-DRβ1*0701+), 526mel CIITA (gp100+, NY-ESO-1−, HLA-A*0201+, HLA-DRβ1*0401+, HLA-DRβ1*0701−), F002Rmel CIITA-gp100 (gp100+, HLA-A*0201+, HLA-DRβ1 *0401−, HLA-DRβ1*0701+), F002Rmel CIITA-GFP (gp100−, HLA-A*0201+, HLA-DRβ1*0401−, HLA-DRβ1*0701+), and 1088mel CIITA (gp100+, HLA-DRβ1*0401+, HLA-DRβ1*0701−).
Generation of DC
PBMC and CD34+ HSCs were obtained from patients with metastatic melanoma who were enrolled in clinical trials in the Surgery Branch of the National Cancer Institute, as part of IRB approved protocols. Immature DCs derived from peripheral blood mononuclear cells were prepared as previously described.31 Briefly, PBMC were plated in X-Vivo-20 (BioWhittaker), and adherent cells were subsequently cultured in CM containing 1000 IU/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) (Peprotech, Rocky Hill, NJ) and 1000 IU/mL interleukin (IL)-4 (Peprotech). DCs were electroporated with cDNAs or IVT RNAs or were transduced with recombinant adenoviral vectors on day 6. Maturation cytokines were added where indicated as follows: 1 μg/mL prostaglandin E2 (PGE2) (Sigma-Aldrich); 10 ng/mL tumor necrosis factor (TNF)α (Sigma-Aldrich, St Louis, MO); 1 μg/mL soluble trimeric CD40L (Amgen-Immunex, Seattle, WA); 5 μg/mL lipo-polysaccharide (LPS) (Sigma-Aldrich); or 50 μg/mL IFNa∼(Roche).
For the preparation of HSC-derived DCs, CD34+ cells were mobilized in peripheral blood of patients by 5 daily subcutaneous injections of 10 μg/kg rhG-CSF (Filgrastim; Neupogen; Amgen, Thousand Oaks, CA), followed by lymphocytopheresis to obtain PBMCs on day 6. Mobilized CD34+ HSCs were then isolated using StemSep CD34-positive selection cocktail and magnetic colloid (StemCell Technologies, Vancouver, Canada). Cells were washed and plated in 6-well plates in CM containing 10 ng/mL TNFα (Sigma-Aldrich, St Louis, MO), 40 ng/mL stem cell factor (SCF) (R&D Systems, Minneapolis, MN), and 1000 IU/mL GM-CSF (Peprotech) as previously described.32
Plasmids
pcDNA3 plasmids containing full-length cDNAs encoding gp100,33 NY-ESO-1,34 and eGFP were used for cDNA electroporation. For in vitro transcription of mRNAs, a modified pGEM-4Z plasmid was constructed, to which a 64 bp length poly A tail had been added downstream of T7 promoter and multiple cloning site, as previously described.35 The full-length NY-ESO-1 was inserted into the pGEM-4Z-64A vector using HindIII and HpaI restriction enzymes. The full length gp100 was inserted using NotI and HindIII restriction digestion and ligation. Plasmids were linearized with Spel for use as templates for in vitro transcription of RNA with mMESSAGE mMACHINE High Yield Capped RNA Transcription Kit (Ambion) according to the manufacturer’s instructions.
DNA Electroporation
DNA electroporation was performed with the AMAXA electroporation system using the Human Dendritic Cell Nucleofector Kit I (AMAXA Biosystems, Cologne, Germany). Briefly, 2.5 × 106 monocyte-derived DCs were resuspended in 100 μL Nucleofector solution and were electroporated in an electroporation cuvette with an electrode gap of 2 mm. DNA was added immediately before electroporation at 5 μg/sample. DCs were recovered in prewarmed CM containing GM-CSF and IL-4 at a final concentration of 1 × 106 cells/mL. Cell viability ranged from 45% to 70% 24 hours posttransfection, as detected by propidium iodide staining. In some experiments, transfected cells were analyzed by flow cytometry or immunohistochemical staining 24 hours posttransfection.
RNA Electroporation
RNA electroporation was performed as previously described.36 Briefly, DCs were washed and gently resuspended in Opti-MEM media (Invitrogen, Grand Island, NY) at 2.5 × 107/mL. Cells were electroporated in 2-mm cuvettes at 300 V for 500 μs using an Electro Square Porator ECM 830 (BTX, San Diego, CA). The amount of IVT RNA used was 2 μg per 106 DCs. Cells were immediately transferred to media containing 50% fresh CM and 50% conditioned CM (collected from day 7 cultured DC), supplemented with GM-CSF and IL-4.
Adenoviral Vectors
Ad2 viruses encoding gp100 and eGFP were commercially supplied (Genzyme, Cambridge, MA) and were used to transduce monocyte-derived DCs as previously described.14 Briefly, DCs were resuspended in X-Vivo-15 (BioWhittaker) at 107/mL and incubated with Ad2 stocks at 37°C at a multiplicity of infection of 300. Transduced DCs were resuspended in CM containing GM-CSF and IL-4 at a final concentration of 1 × 106 cells/mL. Coincubation with responding T cells was performed after a 24 hour rest period.
Retroviral Vectors
The VSV-pseudotyped retroviral system was used as previously described.16,32 Briefly, the vectors were prepared by first inserting the complete gp100 and NY ESO-1 sequences into the pCLNC retroviral plasmid. The pCLNC-gp100 and pMDG-VSV plasmids were cotransfected into 293-gag-pol packaging cells using Lipofectamine Plus (Invitrogen). The 293-gag-pol packaging cells, provided by I. Verma (The Salk Institute, La Jolla, CA), were cultured in Dulbecco’s modified Eagles’s medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. Media was changed 16 and 48 hours after transfection, and culture supernatants were harvested on days 3 and 4. Producer cells were removed from retroviral supernatants by filtration through a 0.2-μm filter (Nalgene, Rochester, NY). Supernatants were immediately frozen at 70°C for future use.
Retroviral supernatants were added to cultured CD34+ cells on days 2 and 3 at a ratio of 1:1 CM. GM-CSF, SCF, TNFα, and polybrene were added, and cells were spun in the plate at 1000 g for 1 hour. On day 4 (or immediately after the second spinfection), transduced DCs were resuspended in fresh CM supplemented with GM-CSF and IL-4 as described above for monocyte-derived DCs. In some experiments, maturation was induced 16 hours before coculture of DCs and T cells with various cytokine cocktails as indicated.
FACS Analyses
DCs were characterized for expression of cell surface markers including CD11c, CD14, CD40, CD80, CD86, CD83, HLA-A, B, C, and HLA-DR using PE or fluorescein isothiocynate-conjugated mAbs (BD PharMingen, San Diego, CA). FACS analyses were performed on a FACSCalibur flow cytometer (BD Biosciences) using CellQuest software (BD Biosciences).
Cytokine Release Assay
Recognition of target cells by melanoma reactive CD8+ and CD4+ T-cell lines and clones was evaluated on the basis of specific IFNγ secretion. Responder T cells (105) were coincubated with 0.5 to 1 × 105 genetically modified DCs (250 μL total) ~20 hours at 37°C, and the concentration of human IFNγ in coculture supernatants was measured by ELISA (Pierce-Endogen, Cambridge, MA). As positive controls for T-cell function, specific IFNγ secretion was measured in response to peptide-loaded target cells and melanomas. For HLA-A*0201 restricted CD8+ T-cell populations, T2 cells were incubated with 1 μM of the appropriate peptide 1 to 3 hours at 37°C. For class II HLA-restricted CD4+ T cells, Epstein-Barr virus-transformed B cells expressing HLA-DRβ1*0401 or HLA-DRβ1*0701 were incubated with 50 μM of the appropriate peptide approximately 3 hours at 37°C. In addition, melanoma cell lines expressing various combinations of HLA-A*0201, HLA-DRβ1*0401, and HLA-DRβ1*0701 were harvested. Responder T cells (105) were coincubated with 105 stimulator cells, and the concentration of human IFNγ in coculture supernatants was measured by ELISA (Pierce-Endogen).
RESULTS
Efficiencies of Gene Transfer Methods
To estimate transfection or transduction efficiencies, DCs genetically modified with viral and nonviral vectors encoding eGFP were analyzed by FACS (Fig. 1). Using the AMAXA electroporation system for cDNA plasmids, 24 hours after transfection, GFP expression ranged from 18% to 36%, and cell viability was 40% to 75% (data not shown). Transfection efficiencies with IVT RNAs were generally higher than those with cDNAs, and DCs electroporated with IVT RNA expressed detectable levels of GFP in 60% to 96% of transfected cells. Cell viabilities were also generally higher using IVT RNAs in comparison to cDNAs (ie, 80% to 90% vs. 40% to 75%, data not shown). Adenoviral vectors were comparable to IVT RNAs in terms of transduction/transfection efficiencies which generally exceeded 90%. However, expression levels, as evaluated by mean fluorescence intensities (MFIs), were usually higher for adenovirally transduced DCs. The transduction efficiency of the retroviral vector was comparable to plasmid cDNA electroporation (ie, ~23%). However, like the recombinant adenovirus, MFIs for retrovirally transduced DCs were usually higher than those for DCs transfected with cDNA or IVT RNA.
FIGURE 1.

Gene transfer efficiencies in DCs transfected or transduced with vectors encoding eGFP. For each panel, gene expression was evaluated by FACS ~24 hours after genetic modification. Percentages indicate the percent of cells gated through M1, and reported MFI values are for M1-gated cells. A, Monocyte-derived DCs from 2 different donors were electroporated with IVT RNA or cDNA encoding eGFP. B, Monocyte-derived DCs from 2 different donors were electroporated with IVT RNA or cDNA encoding eGFP or were transduced with an Ad2 vector encoding this protein. C, Monocyte-derived DCs from 1 donor were transduced with an Ad2 vector encoding eGFP, and CD34+ HSC-derived DCs from the same donor were transduced with a retroviral vector encoding this protein.
Expression of Phenotypic Markers on Genetically Modified DCs
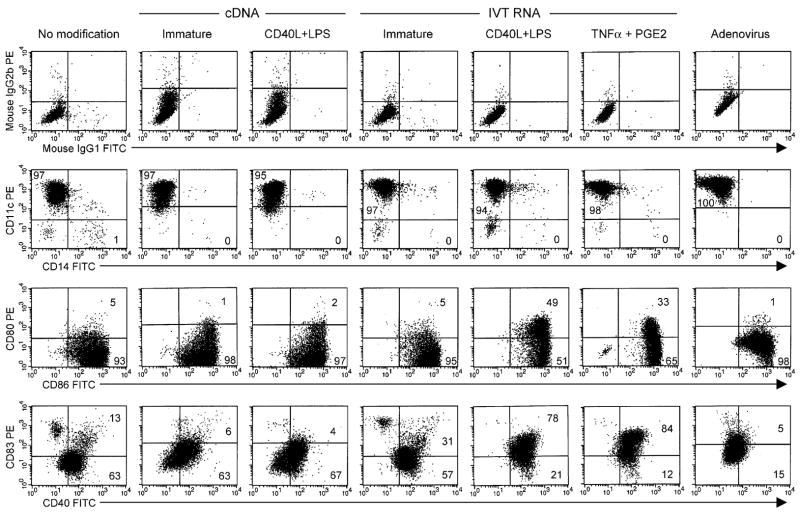
In the absence of any maturation cytokines, expression of various phenotypic markers was not significantly different on DCs transfected or transduced with cDNA, IVT RNA, or Ad2 (Fig. 2). All immature DC populations were essentially 100% positive for CD11c and negative for CD14, as is characteristic of monocyte-derived DCs. High levels of the costimulatory molecule CD86 were consistently observed, with much lower levels of CD80. Expression of CD40 on immature DCs was comparable after transfection with cDNA or IVT RNA but was lower after Ad2 transduction. Also, there was a trend for higher levels of the maturation marker CD83 on immature DCs electroporated with IVT RNA. In addition, genetic modification of immature DCs did not induce different patterns of cytokine secretion including IL-12p70, IL-1β, and IL-15 (data not shown). In contrast, significant upregulation of CD80, CD86, and CD40 was observed on RNA-transfected DCs after maturation with CD40L and LPS or TNFα and PGE2. However, this was not the case for DCs matured with CD40L and LPS after electroporation with cDNA.
FIGURE 2.
DC phenotype after genetic modification. Monocyte-derived DCs were electroporated with IVT RNA or cDNA encoding gp100 or were transduced with an adenoviral vector encoding this protein. Approximately, 16 hours after gene modification, maturation cytokines were added as indicated and described in the Materials and Methods section, and approximately 24 hours later, DCs were stained with fluorescein isothiocynate- (x-axis; FL1-H) and PE- (y-axis; FL2-H) conjugated mAbs as indicated and analyzed by FACS.
Presentation of gp100 Epitopes by DCs Genetically Modified With cDNA, IVT RNA, and Ad2
To determine if genetically modified DCs presented relevant MHC class I and class II restricted tumor-associated epitopes, IFNγ secretion by multiple CD8+ and CD4+ melanoma-reactive T-cell populations (Table 1) was measured in response to transfected or transduced DCs. The melanocyte differentiation antigen gp100 was initially selected as a model system because multiple MHC class I and class II restricted T-cell lines were available that specifically recognized epitopes from this protein.
In multiple experiments, recognition of DCs genetically modified with cDNA, IVT RNA, and Ad2 encoding gp100 by a variety of different MHC class I and class II restricted T-cell lines was directly compared. Data from 2 representative experiments are presented in Tables 2 and 3. In some experiments (Table 3), low amounts of IFNγ were secreted by HLA-A*0201 restricted gp100 reactive CTL in response to DCs transfected with gp100 cDNA. However, in 22 of 26 electroporation experiments (85%), no specific cytokine secretion was detected by CD8+ T-cell lines in response to immature DCs electro-porated with either gp100 cDNA or IVT RNA (Table 2). MHC class II restricted recognition of gp100 epitopes on electroporated DCs was also inconsistent. In some experiments (Table 3), low amounts of IFNγ were secreted by HLA-DR restricted gp100 reactive CD4+ T cells in response to DCs transfected with gp100 cDNA. In 3 of 4 additional electroporation experiments, DCs transfected with gp100 cDNA or IVT RNA were not well-recognized by HLA-DRβ1*0701 restricted gp100 reactive CD4+ T cells that recognized either gp100:170–190 or gp100:420–435. However, in most other experiments, the BR-B8 CD4+ T-cell line clearly recognized gp100:44–59 on electroporated DCs in the context of HLA-DRβ1*0401 (Table 2). In contrast, DCs transduced with Ad2 encoding gp100 were consistently well recognized by both MHC class I and class II restricted T-cell lines as evaluated by the high amounts of specific IFNγ secretion observed in multiple experiments (Tables 2 and 3).
TABLE 2.
Recognition of DCs Genetically Modified With gp100 cDNA, IVT RNA, or Ad2 by HLA-A*0201 and HLA-DRβ1*0401 Restricted, gp100-reactive T-cell Populations*
| Target Cells | HLA Expression | Preloaded Peptide† | Genetic Modification | % GFP Positive‡ | MFI§ | CK3H6 (A2) | BR-B8 (DR4) |
|---|---|---|---|---|---|---|---|
| Donor 3 DC | A2+DR4+ | — | gp100 cDNA | — | — | 48 | 1814|| |
| Donor 3 DC | A2+DR4+ | — | GFP cDNA | 36 | 586 | 42 | 37 |
| Donor 3 DC | A2+DR4+ | — | gp100 IVT RNA | — | — | 57 | > 2000 |
| Donor 3 DC | A2+DR4+ | — | GFP IVT RNA | 96 | 130 | 48 | 62 |
| Donor 3 DC | A2+DR4+ | — | gp100 Ad2 | — | — | 1080 | > 2000 |
| Donor 3 DC | A2+DR4+ | — | GFP Ad2 | 96 | 1675 | 28 | 27 |
| Donor 4 DC | A2+DR4+ | — | gp100 cDNA | — | — | 34 | 569 |
| Donor 4 DC | A2+DR4+ | — | gp100 IVT RNA | — | — | 34 | > 2000 |
| Donor 4 DC | A2+DR4+ | — | GFP IVT RNA | 61 | 50 | 51 | 70 |
| Donor 4 DC | A2+DR4+ | — | gp100 Ad2 | — | — | > 2000 | > 2000 |
| Donor 4 DC | A2+DR4+ | — | GFP Ad2 | 90 | 556 | 35 | 53 |
| T2 cells | A2+ | HBVc:18–26(23Y) | — | — | — | 44 | — |
| T2 cells | A2+ | gp100:209–217 | — | — | — | > 2000 | — |
| EBV-B | DR4+ | IgK:188–201 | — | — | — | — | 35 |
| EBV-B | DR4+ | gp100:170–190 | — | — | — | — | 328 |
IFNγ secretion (pg/mL) in 20 h coculture supernatants of target cells expressing HLA-A*0201 and/or HLA-DRβ1*0401 with T lymphocytes.
EBV-B cells were preincubated with 50 mg/mL gp100:44–59 or Influenza HA:306–324, and T2 cells were preincubated with 1 μg/mL gp100:209–217 or HBVc:18–26(23Y).
Percentage of cells expressing GFP by FACS (Fig. 1B).
MFIs of GFP-positive cells by FACS (Fig. 1B).
Underlined values indicate that IFNg secretion in response to DCs genetically modified with gp100 vectors, or peptide-loaded T2 cells or EBV-B cells, was ≥ 50 pg/mL and at least twice background with relevant control target cells.
TABLE 3.
Recognition of DCs Genetically Modified With gp100 cDNA or Ad2 by HLA-A*0201 (A2), HLA-DRβ1*0401 (DR4), and HLA-DRβ1*0701 (DR7) restricted, gp100-reactive T-cell Populations*
| Target Cells | HLA Expression | Preloaded Peptide† | Genetic Modification | TIL1200 (A2) | CK3H6 (A2) | JR1E2 (A2) | BR-B8 (DR4) | B104 (DR7) | JE-D2 (DR7) |
|---|---|---|---|---|---|---|---|---|---|
| Donor 3 DC | A2+DR4+DR7+ | — | GFP Ad2 | 68 | 25 | 25 | 22 | 23 | 29 |
| Donor 3 DC | A2+DR4+DR7+ | — | gp100 Ad2 | 3677‡ | 6779 | 6445 | 2033 | > 10000 | 4476 |
| Donor 3 DC | A2+DR4+DR7+ | — | GFP cDNA | 105 | 28 | 24 | 22 | 25 | 28 |
| Donor 3 DC | A2+DR4+DR7+ | — | gp100 cDNA | 177 | 92 | 29 | 62 | 486 | 34 |
| Donor 4 DC | A2+DR4+DR7+ | — | GFP Ad2 | 84 | 23 | 17 | 17 | 21 | 17 |
| Donor 4 DC | A2+DR4+DR7+ | — | gp100 Ad2 | 1162 | 2317 | 1218 | 1814 | > 10000 | 847 |
| Donor 4 DC | A2+DR4+DR7+ | — | GFP cDNA | 67 | 24 | 21 | 18 | 24 | 18 |
| Donor 4 DC | A2+DR4+DR7+ | — | gp100 cDNA | 124 | 201 | 48 | 74 | 238 | 268 |
| media | — | — | — | 112 | 21 | 19 | 18 | 22 | 19 |
| T2 cells | A2+ | HBVc:18–26(23Y) | — | 162 | 37 | 28 | — | — | — |
| T2 cells | A2+ | gp100:154–162 | — | 7386 | — | — | — | — | — |
| T2 cells | A2+ | gp100:209–217 | — | — | > 10000 | — | — | — | — |
| T2 cells | A2+ | gp100:280–288 | — | — | — | 9214 | — | — | — |
| EBV-B | DR4+ | HA:306–324 | — | — | — | — | 15 | — | — |
| EBV-B | DR4+ | gp100:44–59 | — | — | — | — | 1279 | — | — |
| EBV-B | DR7+ | IgK:188–201 | — | — | — | — | — | 65 | 19 |
| EBV-B | DR7+ | gp100:170–190 | — | — | — | — | 8583 | ||
| EBV-B | DR7+ | gp100:420–435 | — | — | — | — | — | — | 9190 |
| F002Rmel CIITA | A2+DR4−DR7+ | — | VSV-GFP | 81 | 21 | 27 | 17 | 17 | 22 |
| F002Rmel CIITA | A2+DR4−DR7+ | — | VSV-gp100 | 7321 | 8668 | > 10000 | 17 | > 10000 | > 10000 |
| 1088mel CIITA | A2+DR4+DR7− | — | — | 7640 | 9943 | > 10000 | 824 | 16 | 20 |
| 888mel | A2−DR4−DR7− | — | gp100 cDNA | 147 | 16 | 18 | 20 | 20 | 22 |
IFNγ secretion (pg/mL) in 20 h coculture supernatants of target cells expressing HLA-A*0201, HLA-DRβ1*0401, and/or HLA-DRβ1*0701 with T lymphocytes.
EBV-B cells were preincubated with 50 μg/mL MHC class II-restricted peptides, and T2 cells were preincubated with 1 μg/mL MHC class I-restricted peptides.
Underlined values indicate that IFNγ secretion in response to DCs genetically modified with gp100 vectors, peptide-loaded T2 cells or EBV-B cells, or melanoma cell lines was ≥ 50 pg/mL and at least twice background with relevant control target cells.
Effect of Maturation on Presentation of gp100 Epitopes by Electroporated DCs
To determine if maturation of DCs after transfection with nonviral vectors encoding gp100 would enhance presentation of MHC class I and class II restricted epitopes, IFNγ secretion by gp100 reactive T cells was measured in response to electroporated DCs subsequently stimulated with various cytokine cocktails (Tables 4 and 5). Addition of maturation cytokines did not consistently or significantly effect MHC class I or class II presentation of gp100 epitopes. However, there may have been a slight trend toward enhanced recognition of gp100:209–217 by the L2D8T cell clone upon maturation of DCs with soluble CD40L and LPS. Also, IFNγ secretion by the MHC class II restricted T-cell lines BR-B8 and B104 was usually lower after DC maturation with TNFα and PGE2.
TABLE 4.
Recognition of DCs Transfected with gp100 cDNA in the Presence or Absence of Maturation Cytokines by HLA-A*0201 (A2), HLA-DRβ1*0401 (DR4), and HLA-DRβ1*0701 (DR7) Restricted, gp100-reactive T-cell Populations*
| Target Cells | HLA Expression | Preloaded Peptide† | Genetic Modification | Maturation Cytokines‡ | CK3H6 (A2) Exp. 1 | JR1E2 (A2) Exp. 1 | B104 (DR7) Exp. 1 | CL70 (A2) Exp. 2 | L2D8 (A2) Exp. 2 | BR-B8 (DR4) Exp. 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Donor 5 DC | A2+ DR7+ | — | gp100 cDNA | None | 72 | 76 | 81 | — | — | — |
| Donor 5 DC | A2+ DR7+ | — | GFP cDNA | None | 104 | 71 | 834§ | — | — | — |
| Donor 5 DC | A2+ DR7+ | — | gp100 cDNA | TNFα + PGE2 | 75 | 67 | 76 | — | — | — |
| Donor 5 DC | A2+ DR7+ | — | GFP cDNA | TNFα + PGE2 | 76 | 76 | 71 | — | — | — |
| Donor 6 DC | A2+ DR7+ | — | gp100 cDNA | None | 72 | 71 | 78 | — | — | — |
| Donor 6 DC | A2+ DR7+ | — | GFP cDNA | None | 212 | 69 | > 2000 | — | — | — |
| Donor 6 DC | A2+ DR7+ | — | gp100 cDNA | TNFα + PGE2 | 67 | 68 | 70 | — | — | — |
| Donor 6 DC | A2+ DR7+ | — | GFP cDNA | TNFα + PGE2 | 78 | 68 | 77 | — | — | — |
| Donor 7 DC | A2+DR4+ | — | gp100 cDNA | None | — | — | — | 20 | 84 | 4605 |
| Donor 7 DC | A2+DR4+ | — | GFP cDNA | None | — | — | — | 88 | 88 | 250 |
| Donor 7 DC | A2+DR4+ | — | gp100 cDNA | TNFα + PGE2 | — | — | — | 19 | 58 | 534 |
| Donor 7 DC | A2+DR4+ | — | GFP cDNA | TNFα + PGE2 | — | — | — | 130 | 98 | 238 |
| Donor 7 DC | A2+DR4+ | — | gp100 cDNA | CD40L+LPS | — | — | — | 205 | 1285 | 3527 |
| Donor 7 DC | A2+DR4+ | — | GFP cDNA | CD40L+LPS | — | — | — | 251 | 390 | 524 |
| Donor 8 DC | A2+DR4+ | — | gp100 cDNA | None | — | — | — | 22 | 263 | 11737 |
| Donor 8 DC | A2+DR4+ | — | GFP cDNA | None | — | — | — | 33 | 162 | 341 |
| Donor 8 DC | A2+DR4+ | — | gp100 cDNA | TNFα + PGE2 | — | — | — | 22 | 246 | 2324 |
| Donor 8 DC | A2+DR4+ | — | GFP cDNA | TNFα + PGE2 | — | — | — | 69 | 158 | 313 |
| Donor 8 DC | A2+DR4+ | — | gp100 cDNA | CD40L+LPS | — | — | — | 54 | 603 | 4973 |
| Donor 8 DC | A2+DR4+ | — | GFP cDNA | CD40L+LPS | — | — | — | 407 | 309 | 381 |
| T2 or EBV-B | T cell matched | control | — | — | 65 | 65 | 70 | 18 | 22 | 144 |
| T2 or EBV-B | T cell matched | relevant | — | — | > 2000 | > 2000 | > 2000 | 3981 | 11662 | 6714 |
| F002Rmel CIITA | A2+DR4−DR7+ | — | VSV-GFP | — | 70 | 76 | 93 | — | — | — |
| F002Rmel CIITA | A2+DR4−DR7+ | — | VSV-gp100 | — | 1676 | > 2000 | > 2000 | — | — | — |
| 526mel CIITA | A2+DR4+DR7− | — | — | — | 1403 | > 2000 | 68 | — | — | — |
IFNγ secretion (pg/mL) in 20 h coculture supernatants of target cells expressing HLA-A*0201, HLA-DRβ1*0401, and/or HLA-DRβ1*0701 with T lymphocytes.
EBV-B cells were preincubated with 50 μg/mL MHC class II-restricted peptides, and T2 cells were preincubated with 1 μg/mL MHC class I-restricted peptides. Control peptides were the same as those in Tables 2 and 3, and relevant peptides were those indicated in Table 1 for each specific T-cell line.
DCs were matured with the indicated cytokines as described in the Materials and Methods section.
Underlined values indicate that IFNγ secretion in response to DCs genetically modified with gp100 vectors, peptide-loaded T2 cells or EBV-B cells, or melanoma cell lines was ≥ 50 pg/mL and at least twice background with relevant control target cells.
TABLE 5.
Recognition of DCs Transfected With gp100 IVT RNA in the Presence or Absence of Maturation cytokines by HLA-A*0201 (A2) and HLA-DRβ1*0401 (DR4) Restricted, gp100-reactive T-cell Populations*
| Target Cells | HLA Expression | Preloaded Peptide† | Genetic Modification | Maturation Cytokines‡ | CL70 (A2) | L2D8 (A2) | HT1E11 (A2) | BR-B8 (DR4) |
|---|---|---|---|---|---|---|---|---|
| Donor 9 DC | A2+DR4+ | — | gp100 RNA | None | 12 | 38 | 12 | 49904§ |
| Donor 9 DC | A2+DR4+ | — | GFP RNA | None | 9 | 27 | 17 | 147 |
| Donor 9 DC | A2+DR4+ | — | gp100 RNA | TNFα + PGE2 | 25 | 51 | 17 | 24572 |
| Donor 9 DC | A2+DR4+ | — | GFP RNA | TNFα + PGE2 | 16 | 21 | 18 | 51 |
| Donor 9 DC | A2+DR4+ | — | gp100 RNA | CD40L+LPS | 15 | 144 | 14 | 19725 |
| Donor 9 DC | A2+DR4+ | — | GFP RNA | CD40L+LPS | 24 | 41 | 28 | 72 |
| Donor 10 DC | A2+DR4+ | — | gp100 RNA | None | 36 | 46 | 28 | 57778 |
| Donor 10 DC | A2+DR4+ | — | GFP RNA | None | 26 | 29 | 27 | 209 |
| Donor 10 DC | A2+DR4+ | — | gp100 RNA | TNFα + PGE2 | 23 | 57 | 33 | 24617 |
| Donor 10 DC | A2+DR4+ | — | GFP RNA | TNFα + PGE2 | 23 | 22 | 28 | 57 |
| Donor 10 DC | A2+DR4+ | — | gp100 RNA | CD40L+LPS | 38 | 105 | 34 | 22456 |
| Donor 10 DC | A2+DR4+ | — | GFP RNA | CD40L+LPS | 25 | 26 | 29 | 132 |
| Donor 11 DC | A2+DR4+ | — | gp100 RNA | None | 18 | 191 | 9 | 39000 |
| Donor 11 DC | A2+DR4+ | — | GFP RNA | None | 12 | 26 | 15 | 48 |
| Donor 11 DC | A2+DR4+ | — | gp100 RNA | TNFα + PGE2 | 28 | 89 | 13 | 12721 |
| Donor 11 DC | A2+DR4+ | — | GFP RNA | TNFα + PGE2 | 12 | 21 | 21 | 53 |
| Donor 11 DC | A2+DR4+ | — | gp100 RNA | CD40L+LPS | 8 | 42 | 15 | 36278 |
| Donor 11 DC | A2+DR4+ | — | GFP RNA | CD40L+LPS | 17 | 22 | 28 | 39 |
| T2 or EBV-B | T cell matched | control | — | — | 36 | 84 | 791 | 82 |
| T2 or EBV-B | T cell matched | relevant | — | — | 7450 | 45424 | 9156 | 5604 |
IFNγ secretion (pg/mL) in 20 h coculture supernatants of target cells expressing HLA-A*0201 and/or HLA-DRβ1*0401, with T lymphocytes.
EBV-B cells were preincubated with 50 μg/mL MHC class II-restricted peptides, and T2 cells were preincubated with 1 μg/mL MHC class I-restricted peptides. Control peptides were the same as those in Tables 2 and 3, and relevant peptides were those indicated in Table 1 for each specific T-cell line.
DCs were matured with the indicated cytokines as described in the Materials and Methods section.
Underlined values indicate that IFNγ secretion in response to DCs genetically modified with gp100 vectors, peptide-loaded T2 cells or EBV-B cells, or melanoma cell lines was ≥ 50 pg/mL and at least twice background with relevant control target cells.
Presentation of NY-ESO-1 Epitopes by DCs Transfected With IVT RNA
Because of improved DC viability after electroporation with IVT RNA as compared with cDNA in previous experiments, nonviral-mediated transfections of DCs with NY-ESO-1 were performed solely with IVT RNA (Table 6). Also, because of the previously described trends for recognition of gp100 epitopes after maturation of genetically modified DCs, only the combination of CD40L and LPS was evaluated as a maturation reagent for DCs transfected with cDNA or IVT RNA encoding NY-ESO-1. MHC class I restricted presentation of the NY-ESO-1:157-165 epitope was observed by DCs from 4 of 5 patients evaluated using 3 different HLA-A*0201-restricted T-cell lines. The addition of the maturation cocktail containing CD40L and LPS was necessary for antigen detection by the T-cell clones JH1 and M8 for 1 population of DCs (Donor 15), but no impact was observed for DCs from a second patient (Donor 16). However, DC maturation was not required for the highly avid T-cell line TH1F2L. MHC class II-restricted presentation of the NY-ESO-1:161–180 epitope was also observed using the HLA-DPβ1*0401-restricted T-cell line SG6. However, DC maturation was required for this presentation for both DC populations evaluated.
TABLE 6.
Recognition of DCs Transfected with NY-ESO-1 IVT RNA in the Presence or Absence of Maturation Cytokines by HLA-A*0201 (A2) and HLA-DPβ1*0401 (DP4) Restricted, NY-ESO-1-reactive T-cell Populations*
| Target Cells | HLA Expression | Preloaded Peptide† | Genetic Modification | Maturation Cytokines‡ | TH1F2L (A2) Exp. 1 | SG6 (DP4) Exp. 1 | J-H1 (A2) Exp. 2 | M8 (A2) Exp. 2 | SG6 (DP4) Exp. 2 |
|---|---|---|---|---|---|---|---|---|---|
| Donor 12 DC | A2+DP4+ | — | ESO RNA | None | 377§ | 14 | — | — | — |
| Donor 12 DC | A2+DP4+ | — | GFP RNA | None | 52 | 0 | — | — | — |
| Donor 13 DC | A2+DP4+ | — | ESO RNA | None | 279 | 13 | — | — | — |
| Donor 13 DC | A2+DP4+ | — | GFP RNA | None | 86 | 5 | — | — | — |
| Donor 14 DC | A2+DP4+ | — | ESO RNA | None | 897 | 13 | — | — | — |
| Donor 14 DC | A2+DP4+ | — | GFP RNA | None | 215 | 5 | — | — | — |
| Donor 15 DC | A2+DP4+ | — | ESO RNA | None | — | — | 14 | 28 | 17 |
| Donor 15 DC | A2+DP4+ | — | GFP RNA | None | — | — | 14 | 15 | 18 |
| Donor 15 DC | A2+DP4+ | — | ESO RNA | CD40L+LPS | — | — | 166 | 267 | 605 |
| Donor 15 DC | A2+DP4+ | — | GFP RNA | CD40L+LPS | — | — | 15 | 16 | 17 |
| Donor 16 DC | A2+DP4+ | — | ESO RNA | None | — | — | 16 | 63 | 19 |
| Donor 16 DC | A2+DP4+ | — | GFP RNA | None | — | — | 18 | 33 | 17 |
| Donor 16 DC | A2+DP4+ | — | ESO RNA | CD40L+LPS | — | — | 32 | 81 | 211 |
| Donor 16 DC | A2+DP4+ | — | GFP RNA | CD40L+LPS | — | — | 24 | 56 | 74 |
| T2 cells | A2+ | HBVc:18–26(23Y) | 388 | — | 17 | 14 | — | ||
| T2 cells | A2+ | ESO:157–165 | > 2000 | — | 1294 | 1264 | — | ||
| EBV-B | DP4+ | HA:306–324 | — | — | — | — | — | — | 62 |
| EBV-B | DP4+ | ESO:161–180 | — | — | — | — | — | — | 1366 |
| 624mel | A2+DP4− | — | — | — | 1766 | 21 | — | — | — |
| 888mel | A2−DP4− | — | — | — | 39 | 9 | — | — | — |
IFNγ secretion (pg/mL) in 20 h coculture supernatants of target cells expressing HLA-A*0201 and/or HLA-DPβ1*0401 with T lymphocytes.
EBV-B cells were preincubated with 50 μg/mL MHC class II-restricted peptides, and T2 cells were preincubated with 1 μg/mL MHC class I-restricted peptides.
DCs were matured with the indicated cytokines as described in the Materials and Methods section.
Underlined values indicate that IFNγ secretion in response to DCs genetically modified with NY-ESO-1 vectors, peptide-loaded T2 cells or EBV-B cells, or melanoma cell lines was ≥ 50 pg/mL and at least twice background with relevant control target cells.
Presentation of gp100 and NY-ESO-1 Epitopes by Retrovirally Transduced DCs
Data from the Ad2 vector transductions suggested that virally mediated gene transfer into DCs consistently enabled efficient presentation of both MHC class I and class II-restricted epitopes from tumor-associated antigens. However, adenoviral vectors also enable presentation of epitopes from the adenovirus backbone, and these may dominate the immune response when used to stimulate a heterogeneous population of T cells, such as peripheral blood leukocyte. To avoid this disadvantage, we evaluated epitope presentation by DCs after retroviral transduction because nonreplicating retroviral vectors only induce expression of the transgene after genomic integration into host cells. Retroviral transductions were performed using the Moloneymurine leukemia virus (MMLV) retrovirus produced by the 293-gag-pol packaging cell line as previously described.16,32 Since retroviruses can only transduce dividing cells, we first isolated CD34+ HSCs and stimulated them to proliferate with TNFα, SCF, and GM-CSF. These cells were then retrovirally transduced and were subsequently differentiated into DCs using GM-CSF and IL-4. DCs transduced with MMLV encoding both gp100 and NY-ESO-1 were well recognized by both MHC class I and class II-restricted T-cell lines (Table 7), and similar results were observed using CD34+ HSC-derived DCs from a second donor (data not shown). However, it is not possible to compare results directly between retrovirally transduced DCs and other means of genetic modification because of the differences in DC preparation.
TABLE 7.
Recognition of DCs Genetically Modified with Retroviral Vectors Encoding gp100 or NY-ESO-1 by HLA-A*0201 (A2) and HLA-DRβ1*0701 (DR7) Restricted, Antigen-reactive T-cell Populations*
| Target Cells | HLA Expression |
Preloaded Peptide† | MMLV Modification |
RB-154 (A2) | L2D8 (A2) | HT1E11 (A2) | B104 (DR7) | JE-D2 (DR7) | JH1 (A2) | M8 (A2) |
|---|---|---|---|---|---|---|---|---|---|---|
| Donor 17 HSC-DC | A2+DR7+ | — | gp100 | > 1000‡ | > 1000 | 783 | 745 | > 1000 | — | — |
| Donor 17 HSC-DC | A2+DR7+ | — | NY-ESO-1 | — | — | — | — | — | > 1000 | 287 |
| Donor 17 HSC-DC | A2+DR7+ | — | GFP | 29 | 19 | 272 | 30 | 24 | 19 | 30 |
| T2 cells | A2+ | HBVc:18–26(23Y) | — | 22 | 16 | 25 | — | 17 | 17 | |
| T2 cells | A2+ | gp100:154–162 | — | > 1000 | — | — | — | — | — | — |
| T2 cells | A2+ | gp100:209–217 | — | — | > 1000 | — | — | — | — | — |
| T2 cells | A2+ | gp100:280–288 | — | — | — | > 1000 | — | — | — | — |
| T2 cells | A2+ | ESO:157–165 | — | — | — | — | — | — | > 1000 | > 1000 |
| EBV-B | DR7+ | IgK:188–201 | — | — | — | — | 15 | 16 | — | — |
| EBV-B | DR7+ | gp100:170–190 | — | — | — | > 1000 | — | — | — | |
| EBV-B | DR7+ | gp100:420–435 | — | — | — | — | — | > 1000 | — | — |
| F002Rmel CIITA | A2+DR7+ | — | GFP | 16 | 15 | 18 | 14 | 60 | — | — |
| 624mel CIITA | A2+DR7+ | — | — | 588 | > 1000 | 26 | 445 | 534 | 76 | 198 |
| 526mel (ESO−) | A2+DR7− | — | — | > 1000 | > 1000 | 27 | 18 | 16 | 15 | 14 |
IFNγ secretion (pg/mL) in 20 h coculture supernatants of target cells expressing HLA-A*0201 and/or HLA-DRβ1*0701 with T lymphocytes.
EBV-B cells were preincubated with 50 μg/mL MHC class II-restricted peptides, and T2 cells were preincubated with 1 μg/mL MHC class I-restricted peptides.
Underlined values indicate that IFNγ secretion in response to DCs genetically modified with gp100 or NY-ESO-1 vectors, peptide-loaded T2 cells or EBV-B cells, or melanoma cell lines was ≥ 50 pg/mL and at least twice background with relevant control target cells.
DISCUSSION
Genetic modification of DCs with tumor-associated antigens may facilitate the development of new immunotherapies for the treatment of patients with cancer. The results presented here suggest that genetic engineering of DCs is feasible using both viral and nonviral gene delivery methods. Gene transfer efficiencies of greater than 18% were observed with all methods as evaluated by FACS after genetic manipulation with viral and nonviral vectors encoding eGFP. High transfection efficiencies did not correlate with efficient antigen processing and presentation of relevant T-cell epitopes. However, higher amounts of protein produced by DCs transduced with viral vectors in comparison with those transfected with cDNA or IVT RNA may have been related to the observation that these cells consistently presented both MHC class I and class II-restricted T-cell epitopes.
Our hypothesis was that antigen present in the cytosol would be degraded by the proteasome, and resulting peptides would be assembled with MHC class I molecules in the endoplasmic reticulum. Peptide-MHC class I complexes would then migrate to the surfaces of the gene-modified DCs and activate CD8+ T cells as previously described.37 We also anticipated low levels of CD4+ T-cell stimulation, because most endogeneously produced proteins do not efficiently enter the endocytic pathway associated with presentation of MHC class II-restricted epitopes.38 However, we found that DCs transfected with nonviral vectors encoding gp100 did not consistently stimulate tumor reactive CTL, and there may have been a slight trend toward enhanced presentation of MHC class II-restricted T-cell epitopes. In contrast to nonviral vectors, transduction of DCs with recombinant Ad2 and MMLV vectors consistently induced potent presentation of both MHC class I and class II-restricted epitopes. The apparent difference in antigen processing between viral and nonviral vectors could not be attributed to transfection/transduction efficiencies or to differences in phenotypic markers expressed on DCs. The efficiencies of RNA electroporation and Ad2 transduction were similarly high, and both methods maintained high levels of viability of phenotypically similar DCs. However, in multiple experiments, much higher amounts of IFNγ were secreted by several different CD8+ and CD4+ T-cell lines in response to DCs transduced with Ad2 in comparison with those transfected with IVT RNA encoding gp100. A similar trend of enhanced MHC class I and class II-restricted epitope presentation with a virus compared with a nonviral vector was also observed for NY-ESO-1, although we only evaluated IVT RNA and MMLV for this particular antigen. In contrast, the overall amount of protein produced by DCs transduced with viral vectors was probably higher than those transfected with cDNA or IVT RNA as suggested by higher MFIs after genetic modification with eGFP (Fig. 1). Therefore, enhanced epitope presentation by virally transduced DCs may be related to increased quantities of proteins produced by these cells.
To determine if maturation of DCs after transfection with nonviral vectors would enhance presentation of MHC class I and class II-restricted epitopes, T-cell recognition of electroporated DCs subsequently stimulated with 2 different cytokine cocktails was evaluated. In particular, we evaluated recognition of gp100-transfected DCs after maturation with either CD40L and LPS or TNFα and PGE2. Addition of these maturation cytokines did not consistently or significantly effect MHC class I or class II presentation of gp100 epitopes. However, there may have been a slight trend toward enhanced recognition of gp100:209–217 by one particular T-cell clone upon DC maturation with CD40L and LPS and a trend toward decreased recognition of MHC class II-restricted epitopes upon DC maturation with TNFα and PGE2. On the basis of these trends for gp100, we only evaluated recognition of NY-ESO-1 epitopes after DC maturation with CD40L and LPS. Likewise, addition of these maturation cytokines did not consistently or significantly effect MHC class I or class II presentation of NY-ESO-1 epitopes. Despite these findings, we did not evaluate the effect of DC maturation on the priming of naïve T cells. Conclusions from such experiments might be very different than those presented here because IL-12 production is believed to be critical in the priming phase of the immune response.
In the experiments presented here, DC viability ranged from 40% to 90% depending on the method of transfection or transduction employed and was lowest using the cDNA electroporation protocol. Therefore, it is possible that antigen released by dead cells may have contributed, in part, to presentation of epitopes by surviving DCs. For the observed recognition of epitopes by CD4+ T cells, this phenomena may have contributed significantly because the exogeneous pathway is preferred for MHC class II-restricted antigen processing and presentation. However, it is unlikely that this phenomena contributed significantly to cross-presentation. In a previous report, we did not detect any cross-presentation when gp100 was pulsed on DCs as an intact protein or from melanoma cell lysates.6 Furthermore, in several other reports, efficient cross-presentation of NY-ESO-1 was only observed using full-length protein formulated as antigen-antibody immune complexes7 or with ISCOMA-TRIX adjuvant,7,8 not using soluble protein alone.
Presentation of gp100 epitopes in the context of MHC class II molecules after genetic modification of DCs may result from a unique structural characteristic. This glycoprotein, and the other melanosomal membrane glycoproteins, tyrosinase, TRP-1, and TRP-2, contain a dileucine-based sorting motif, the melanosomal transport signal (MTS).39 This hexapeptide sequence normally directs melanosomal proteins to the melanosome, but it may also facilitate entry of these proteins into the endocytic pathway, on the basis of structural similarities between the endosome and melanosome.40 The melanosomal transport signal-based targeting of intracellular gp100 to the endocytic pathway could explain its frequent presentation by MHC class II molecules, both in tumor cells and genetically modified DCs.
Lack of a clear distinction between viral and nonviral vectors in terms of gene transfer efficiency and expression of costimulatory molecules directed our focus to the role of the vector itself in determining how the cell degrades and processes the inserted antigen. Viral vectors seem to activate proteasomal processing and MHC class I presentation more efficiently than nonviral vectors. Although our results do not elucidate any cellular mechanisms involved, perhaps cofactors associated with cellular reactions to viral infection are crucial for effective antigen processing and presentation by DC. Also, it seems possible that rapid turnover of defective transgene products, prominent with viruses but not nonviral vectors, contributes to enhanced antigen processing.41 To address this hypothesis, in a preliminary experiment, we first transduced DCs with Ad2-GFP and subsequently transfected these antigen-presenting cells with IVT RNA encoding gp100 (data not shown). In that experiment, no enhanced recognition by gp100 reactive CD8+ or CD4+ T cells was observed, but similar experiments should be repeated in the future to address this issue more thoroughly.
Our results support the use of DCs transduced with recombinant retroviral vectors for the development of new cancer immunotherapies. Although adenoviral vectors are associated with higher transduction efficiencies, the application of these vectors is severely limited by preexisting immunity and by rapidly developing anti-vector immune responses.42 Despite lower transduction efficiencies with MMLV, this retroviral vector induced efficient presentation of both MHC class I and class II-restricted T-cell epitopes from both gp100 and NY-ESO-1. Since clinical evidence strongly suggests that CD8+ T cells can mediate tumor regression,43 MHC class I-restricted presentation of tumor-associated epitopes seems critical for any DC gene modification technique. In addition, MCH class II-restricted epitope presentation may be important for eliciting CD4+ T cell help which may, in turn, enhance CTL responses.11,15,36 Therefore, the broad spectrum of antigen presentation in retrovirally transduced DC and the low immunogenicity of the retroviral vector delineate this gene delivery method as a valuable tool both for expanding the spectrum of known tumor-associated antigens, and for potential clinical applications of DC-based anticancer immunotherapy.
References
- 1.Banchereau J, Briere F, Caux C, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Shen Z, Reznikoff G, Dranoff G, et al. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J Immunol. 1997;158:2723–2730. [PubMed] [Google Scholar]
- 3.Larsson M, Fonteneau JF, Bhardwaj N. Dendritic cells resurrect antigens from dead cells. Trends Immunol. 2001;22:141–148. doi: 10.1016/s1471-4906(01)01860-9. [DOI] [PubMed] [Google Scholar]
- 4.Ludewig B, McCoy K, Pericin M, et al. Rapid peptide turnover and inefficient presentation of exogenous antigen critically limit the activation of self-reactive CTL by dendritic cells. J Immunol. 2001;166:3678–3687. doi: 10.4049/jimmunol.166.6.3678. [DOI] [PubMed] [Google Scholar]
- 5.Melief CJ. Mini-review: regulation of cytotoxic T lymphocyte responses by dendritic cells: peaceful coexistence of cross-priming and direct priming? Eur J Immunol. 2003;33:2645–2654. doi: 10.1002/eji.200324341. [DOI] [PubMed] [Google Scholar]
- 6.Parkhurst MR, DePan C, Riley JP, et al. Hybrids of dendritic cells and tumor cells generated by electrofusion simultaneously present immunodominant epitopes from multiple human tumor-associated antigens in the context of MHC class I and class II molecules. J Immunol. 2003;170:5317–5325. doi: 10.4049/jimmunol.170.10.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnurr M, Chen Q, Shin A, et al. Tumor antigen processing and presentation depend critically on dendritic cell type and the mode of antigen delivery. Blood. 2005;105:2465–2472. doi: 10.1182/blood-2004-08-3105. [DOI] [PubMed] [Google Scholar]
- 8.Maraskovsky E, Sjolander S, Drane DP, et al. NY-ESO-1 protein formulated in ISCOMATRIX adjuvant is a potent anticancer vaccine inducing both humoral and CD8+ t-cell-mediated immunity and protection against NY-ESO-1+ tumors. Clin Cancer Res. 2004;10:2879–2890. doi: 10.1158/1078-0432.ccr-03-0245. [DOI] [PubMed] [Google Scholar]
- 9.Bonifaz L, Bonnyay D, Mahnke K, et al. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J Exp Med. 2002;196:1627–1638. doi: 10.1084/jem.20021598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.York IA, Goldberg AL, Mo XY, et al. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol Rev. 1999;172:49–66. doi: 10.1111/j.1600-065x.1999.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 11.Bonehill A, Heirman C, Tuyaerts S, et al. Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA class I and class II molecules. J Immunol. 2004;172:6649–6657. doi: 10.4049/jimmunol.172.11.6649. [DOI] [PubMed] [Google Scholar]
- 12.Van Tendeloo VF, Ponsaerts P, Lardon F, et al. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98:49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- 13.Strobel I, Berchtold S, Gotze A, et al. Human dendritic cells transfected with either RNA or DNA encoding influenza matrix protein M1 differ in their ability to stimulate cytotoxic T lymphocytes. Gene Ther. 2000;7:2028–2035. doi: 10.1038/sj.gt.3301326. [DOI] [PubMed] [Google Scholar]
- 14.Linette GP, Shankara S, Longerich S, et al. In vitro priming with adenovirus/gp100 antigen-transduced dendritic cells reveals the epitope specificity of HLA-A*0201-restricted CD8+ T cells in patients with melanoma. J Immunol. 2000;164:3402–3412. doi: 10.4049/jimmunol.164.6.3402. [DOI] [PubMed] [Google Scholar]
- 15.Bonini C, Lee SP, Riddell SR, et al. Targeting antigen in mature dendritic cells for simultaneous stimulation of CD4+ and CD8+ T cells. J Immunol. 2001;166:5250–5257. doi: 10.4049/jimmunol.166.8.5250. [DOI] [PubMed] [Google Scholar]
- 16.Lapointe R, Royal RE, Reeves ME, et al. Retrovirally transduced human dendritic cells can generate T cells recognizing multiple MHC class I and class II epitopes from the melanoma antigen glycoprotein 100. J Immunol. 2001;167:4758–4764. doi: 10.4049/jimmunol.167.8.4758. [DOI] [PubMed] [Google Scholar]
- 17.Lizee G, Gonzales MI, Topalian SL. Lentivirus vector-mediated expression of tumor-associated epitopes by human antigen presenting cells. Hum Gene Ther. 2004;15:393–404. doi: 10.1089/104303404322959542. [DOI] [PubMed] [Google Scholar]
- 18.Dietz AB, Vuk-Pavlovic S. High efficiency adenovirus-mediated gene transfer to human dendritic cells. Blood. 1998;91:392–398. [PubMed] [Google Scholar]
- 19.Zhong L, Granelli-Piperno A, Choi Y, et al. Recombinant adenovirus is an efficient and non-perturbing genetic vector for human dendritic cells. Eur J Immunol. 1999;29:964–972. doi: 10.1002/(SICI)1521-4141(199903)29:03<964::AID-IMMU964>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Roth MD, Cheng Q, Harui A, et al. Helper-dependent adenoviral vectors efficiently express transgenes in human dendritic cells but still stimulate antiviral immune responses. J Immunol. 2002;169:4651–4656. doi: 10.4049/jimmunol.169.8.4651. [DOI] [PubMed] [Google Scholar]
- 21.Batchu RB, Moreno AM, Szmania S, et al. High-level expression of cancer/testis antigen NY-ESO-1 and human granulocyte-macrophage colony-stimulating factor in dendritic cells with a bicistronic retroviral vector. Hum Gene Ther. 2003;14:1333–1345. doi: 10.1089/104303403322319417. [DOI] [PubMed] [Google Scholar]
- 22.Parkhurst MR, Riley JP, Robbins PF, et al. Induction of CD4+ Th1 lymphocytes that recognize known and novel class II MHC restricted epitopes from the melanoma antigen gp100 by stimulation with recombinant protein. J Immunother. 2004;27:79–91. doi: 10.1097/00002371-200403000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakker AB, Schreurs MW, Tafazzul G, et al. Identification of a novel peptide derived from the melanocyte-specific gp100 antigen as the dominant epitope recognized by an HLA-A2.1-restricted anti-melanoma CTL line. Int J Cancer. 1995;62:97–102. doi: 10.1002/ijc.2910620118. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami Y, Eliyahu S, Jennings C, et al. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 25.Dudley ME, Wunderlich JR, Yang JC, et al. A phase I study of nonmyeloablative chemotherapy and adoptive transfer of autologous tumor antigen-specific T lymphocytes in patients with metastatic melanoma. J Immunother. 2002;25:243–251. doi: 10.1097/01.CJI.0000016820.36510.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khong HT, Rosenberg SA. Pre-existing immunity to tyrosinase-related protein (TRP)-2, a new TRP-2 isoform, and the NY-ESO-1 melanoma antigen in a patient with a dramatic response to immunotherapy. J Immunol. 2002;168:951–956. doi: 10.4049/jimmunol.168.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khong HT, Wang QJ, Rosenberg SA. Identification of multiple antigens recognized by tumor-infiltrating lymphocytes from a single patient: tumor escape by antigen loss and loss of MHC expression. J Immunother. 2004;27:184–190. doi: 10.1097/00002371-200405000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touloukian CE, Leitner WW, Topalian SL, et al. Identification of a MHC class II-restricted human gp100 epitope using DR4-IE transgenic mice. J Immunol. 2000;164:3535–3542. doi: 10.4049/jimmunol.164.7.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bownds S, Tong-On P, Rosenberg SA, et al. Induction of tumor-reactive cytotoxic T-lymphocytes using a peptide from NY-ESO-1 modified at the carboxy-terminus to enhance HLA-A2.1 binding affinity and stability in solution. J Immunother. 2001;24:1–9. doi: 10.1097/00002371-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y, Boss JM, Hu SX, et al. Apoptosis-independent retinoblastoma protein rescue of HLA class II messenger RNA IFN-gamma inducibility in non-small cell lung carcinoma cells. Lack of surface class II expression associated with a specific defect in HLA-DRA induction. J Immunol. 1996;156:2495–2502. [PubMed] [Google Scholar]
- 31.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves ME, Royal RE, Lam JS, et al. Retroviral transduction of human dendritic cells with a tumor-associated antigen gene. Cancer Res. 1996;56:5672–5677. [PubMed] [Google Scholar]
- 33.Kawakami Y, Eliyahu S, Delgado CH, et al. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang RF, Johnston SL, Zeng G, et al. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J Immunol. 1998;161:3598–3606. [PubMed] [Google Scholar]
- 35.Boczkowski D, Nair SK, Snyder D, et al. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184:465–472. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Boczkowski D, Nair SK, et al. Inhibition of invariant chain expression in dendritic cells presenting endogenous antigens stimulates CD4+ T-cell responses and tumor immunity. Blood. 2003;102:4137–4142. doi: 10.1182/blood-2003-06-1867. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg AL, Cascio P, Saric T, et al. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol. 2002;39:147–164. doi: 10.1016/s0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 38.Brodsky FM. Antigen processing and presentation: close encounters in the endocytic pathway. Trends Cell Biol. 1992;2:109–115. doi: 10.1016/0962-8924(92)90015-f. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Bartido S, Yang G, et al. A role for a melanosome transport signal in accessing the MHC class II presentation pathway and in eliciting CD4+ T cell responses. J Immunol. 1999;163:5820–5826. [PubMed] [Google Scholar]
- 40.Calvo PA, Frank DW, Bieler BM, et al. A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. J Biol Chem. 1999;274:12780–12789. doi: 10.1074/jbc.274.18.12780. [DOI] [PubMed] [Google Scholar]
- 41.Yewdell J. To DRiP or not to DRiP: generating peptide ligands for MHC class I molecules from biosynthesized proteins. Mol Immunol. 2002;39:139–146. doi: 10.1016/s0161-5890(02)00097-4. [DOI] [PubMed] [Google Scholar]
- 42.Sumida SM, Truitt DM, Kishko MG, et al. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J Virol. 2004;78:2666–2673. doi: 10.1128/JVI.78.6.2666-2673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudley ME, Wunderlich JR, Robbins PF, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]