Abstract
In recent clinical trials in patients with metastatic melanoma, adoptive transfer of tumor-reactive lymphocytes mediated the regression of metastatic tumor deposits. To better understand the role of individual T cell clones in mediating tumor regression, a 5′ RACE technique was used to determine the distribution of TCR β-chain V region sequences expressed in the transferred cells as well as in tumor samples and circulating lymphocytes from melanoma patients following adoptive cell transfer. We found that dominant T cell clones were present in the in vitro-expanded and transferred tumor-infiltrating lymphocyte samples and certain T cell clones including the dominant T cell clones persisted at relatively high levels in the peripheral blood of the patients that demonstrated clinical responses to adoptive immunotherapy. However, these dominant clones were either undetected or present at a very low level in the resected tumor samples used for tumor-infiltrating lymphocyte generation. These data demonstrated that there was selective growth and survival, both in vitro and in vivo, of individual T cell clones from a relatively small number of T cells in the original tumor samples. These results suggest that the persistent T cell clones played an active role in mediating tumor regression and that 5′ RACE analysis may provide an important tool for the analysis of the role of individual T cell clones in mediating tumor regression. A similar analysis may also be useful for monitoring autoimmune responses.
The adoptive transfer of bulk populations of tumor-infiltrating lymphocytes (TIL)2 with antitumor reactivity can mediate the regression of cancer in patients with metastatic melanoma (1–3). Although objective responses were observed in many of the recipients, the lack of response observed in some patients, as well as the limited duration of many of these responses, led to attempts to further enhance these treatments. Many factors such as the heterogeneity of the infused TILs, the phenotype of the growing tumors, and variations of in vivo inflammatory environments may be responsible for the differences in clinical responses (4 – 6). We have thus investigated the fate of the transferred T cell populations to better understand the mechanism of tumor regression in melanoma patients after adoptive cell transfer.
The autologous cells used in adoptive cell transfer immunotherapy are derived from pre-existing T cell responses in vivo. The analysis of the in vivo presence of effector T cells has been primarily conducted using Abs directed against families of the TCR β-chain V region (TRBV) gene products and have provided evidence that a limited number of clones specifically expand in vivo in response to tumor (7). Additional studies using panels of oligonucleotide primers designed to specifically amplify sequences derived from individual TRBV families (8) indicated that particular TRBV gene families were preferentially amplified from melanoma metastases obtained following IL-2-based immunotherapy (9). Regressing lesions obtained from four melanoma patients isolated following IL-2 treatment contained one or two dominant T cell clones (10). TRBV rearrangement studies indicated that DNP vaccine induced the expansion of particular T cell clones in melanoma metastases (11). CDR3 spectratyping analyses showed that a high frequency of T cell clonal expansions occurred in primary human melanoma (12).
Techniques based on the use of TRBV primers have also been used to follow the fate of adoptively transferred T cell clones. In one trial conducted using T cell clones that recognized the melanoma Ag, MART-1, adoptively transferred T cells were monitored using a highly sensitive PCR-based technique that used an oligonucleotide primer based on the CDR3 region sequence of the expressed TRBV gene product. This method, which was capable of detecting one cell in between 103 and 105 total lymphocytes, detected transferred cells in the peripheral blood of two of the treated five patients tested between 2 and 5 days following transfer (4). These clones were not, however, detected between 2 and 3 wk following transfer, and no clinical responses were observed in this trial. In a separate clinical trial conducted with antitumor T cell clones, evidence was presented indicating that T cell clones reactive with either MART-1 or gp100 peptides persisted at levels of between 1 and 2% of the peripheral blood CD8+ T cells for up to 14 days following adoptive transfer to melanoma patients, but no objective clinical responses were observed (13). It is not clear whether the levels of T cell engraftment were insufficient to mediate tumor regression, or if the function of these cells was inhibited in vivo.
The expressed TRBV genes were used to characterize lymphocyte persistence in our most recent clinical trial that used transfer of autologous antitumor T cells into melanoma patients following a nonmyeloablative chemotherapy designed to enhance the engraftment of adoptively transferred T cells. Six of the 13 patients that were treated with this regimen demonstrated objective clinical responses, and four other patients demonstrated mixed responses (1). Analysis of the TRBV sequences by FACS using a panel of TRBV-specific Abs revealed that distinct HLA-A2-restricted MART-1-reactive T cell clones also underwent significant expansion following adoptive transfer in two of the patients in this trial who demonstrated nearly complete regression of multiple metastases. Each of the two clones represented over 50% of the T cells in the peripheral circulation of the two treated patients for periods greater than 4 mo. The antitumor response observed in these patients provided preliminary evidence that the persistence of adoptively transferred tumor-reactive T cells was associated with tumor regression.
Because Abs are available that can recognize only about half of TRBV families, this approach provided an incomplete analysis of the presence and persistence of the T cells in TIL and PBL. This report describes studies that were conducted using a 5′ RACE technique to amplify all TRBV families and thus analyzed the presence of all T cells in the original tumor tissues as well as the persistence of T cells following adoptive transfer to patients following nonmyeloablative chemotherapy. The results demonstrated that dominant T cell clones were present in cultured TIL samples and persisted in the patients following adoptive cell transfer. Unexpectedly, these persistent T cell clonotypes were either undetected or present at a very low level in the original tumor samples used for TIL generation. The results provide further evidence that tumor regression may be associated with the in vivo persistence of individual tumor-reactive T cell clones that are suppressed in the native tumor and can be activated and expanded both by in vitro culture and in vivo growth following cell transfer.
Materials and Methods
Patient samples
Samples obtained from patients 1803 and 2023 include uncultured resected tumors (1803Tu and 2023Tu), TIL that were derived from these tumors by culturing in 6000 IU/ml IL-2 (TIL 1803 and TIL 2023) (5), and PBL samples obtained 1, 2, 4, and 8 wk following adoptive transfer of autologous TIL, designated 1803 or 2023PBL1w, 1803 or 2023PBL2w, 1803 or 2023PBL4w, and 1803 or 2023PBL8w, respectively.
RNA isolation
Total RNA was prepared from the fresh resected tumor samples, TIL samples, and PBL samples using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Total RNA solutions were quantitated by spectrophotometer and the quality was verified by gel electrophoresis.
5′ RACE analysis of TRBV gene expression
The IMGT TRBV gene nomenclature was used in this research (available at http://imgt.cines.fr/textes/IMGTrepertoire/LocusGenes/nomenclatures/human/TRB/TRBV/Hu_TRBVnom.html). 5′ RACE analysis of TRBV gene expression was conducted by SMART RACE cDNA Amplification kit (BD Biosciences/Clontech, Palo Alto, CA) according to the manufacturer’s instructions. Briefly, 3 μl of total RNA solution (0.5 μg of RNA) for each sample was used in 10 μl of reverse transcription mixture containing 1 mM dNTP mix, 1 μM SMART II A oligonucleotide, 1 μM 5′-RACE CDS primer, 2 μM DTT, 1× first-strand buffer, and 100 U of PowerScript reverse transcriptase for cDNA synthesis. After incubation at 42°C for 1.5 h in a PTC-200 Peltier Thermal Cycler (MJ Research, Watertown, MA), the reverse transcription mixture was diluted with 100 μl of 10 mM tricine-EDTA buffer, heated at 72°C for 7 min, and stored at −20°C. A total of 2.5 μl of diluted reverse transcription mixture was used in 50 μl of PCR mixture containing 1 mM dNTP mixture, 1 × Advantage 2 PCR buffer, 1 × Advantage 2 polymerase mix (Advantage 2 PCR kit; BD Biosciences/Clontech), 1 × universal primer A mix (UPM), and 0.4 μM 3′TCRBCN primer. 3′ TCRBCN primer anneals the C region of the TCR β-chain, and its sequence is 5′-CGA GGT AAA GCC ACA GTC T-3′. PCR was performed by the initial five cycles of 30 s at 94°C and 2 min at 72°C, 25 cycles of 30 s at 94°C, 30 s at 65°C, and 2 min at 72°C, and the final cycle of 10 min at 72°C in a PTC-200 Peltier Thermal Cycler.
A total of 20 μl of PCR product for each sample was separated by electrophoresis using 2% agarose E-Gel (Invitrogen Life Technologies, Carlsbad, CA). Amplified bands of ~700 bp were purified and eluted with 8 μl of water using a Zymoclean Gel DNA Recovery kit (Zymo Research, Orange, CA). A total of 4 μl of the eluted sample was used for cloning and transformation with the pcDNA3.1/V5-His TOPO TA Expression kit (Invitrogen Life Technologies). After overnight culture of bacterial plates at 37°C, at least 96 separate bacterial colonies were transferred into 96-well deep plates with 1 ml of Super Broth (BioSource International, Camarillo, CA) and 100 μg/ml ampicillin for each well. After overnight culture at 37°C under 300 rpm shaking, plasmid DNA samples were prepared from 96-well deep plates using a QIAprep 96 Turbo Miniprep kit (Qiagen).
A total of 2 μl of plasmid DNA sample (0.5 μg DNA) was added in 20 μl of PCR containing 1 × BigDye Terminator version 1.1 (Applied Bio-systems, Foster City, CA) and 0.32 μM TCR-CB-50R primer. TCR-CB-50R primer is a nested primer annealing the constant region of TCR β-chain, and its sequence is 5′-TC TGA TGG CTC AAA CAC AG-3′. PCR was performed by the first cycle of 1 min at 96°C, and 25 cycles of 10 s at 94°C, 5 s at 50°C, and 4 min at 60°C. PCR products were cleaned using a DyeEx 96 kit (Qiagen). After drying of cleaned PCR products at 72°C for 1 h, the PCR products were dissolved in 10 μl of formamide and analyzed by an automated DNA sequencer (ABI PRISM 3100-Avant Genetic Analyze; Applied Biosystems). The sequence data were analyzed by comparison with known TRBV sequences using Vector NTI Suite 8 (InforMax, Frederick, MD).
RT-PCR of TRBV20-1 gene expression
A total of 5 μl of diluted reverse transcription mixture from the 1803Tu sample, which was the same sample for 5′ RACE analysis, was used in 50 μl of PCR mixture containing 1 mM dNTP mixture, 1 × Advantage 2 PCR buffer, 1 × Advantage 2 polymerase mix (Advantage 2 PCR kit; BD Biosciences/Clontech), 0.4 μM 3′TCRBCN primer, and 0.4 μM TRBV20-1-specific primer. TRBV20-1-specific primer anneals the V region of the TRBV20-1 β-chain, and its sequence is 5′-GCT GAT GGC AAC TTC CAA TG-3′. PCR was performed by the initial five cycles of 30 s at 94°C and 2 min at 72°C, 30 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 72°C, and the final cycle of 10 min at 72°C in a PTC-200 Peltier Thermal Cycler. PCR product was purified, cloned, and sequenced as 5′ RACE analysis. The sequences of the D region of TRBV20-1 were analyzed using Vector NTI Suite 8 (InforMax).
IFN-γ release assay
A total of 1 × 105 T cells were cocultured with 1 × 105 tumor cells in a 96-well flat plate with 200 μl of T cell assay medium (RPMI 1640 supplemented with 2% human serum, 2 mM glutamine, 10 mM HEPES buffer, 0.55 mM 2-ME, and 20 cetus unit (CU) IL-2) in a humidified incubator at 37°C and 5% CO2 for 20 h. The concentration of IFN-γ in the supernatant was determined by ELISA using a mAb against human IFN-γ (Endogen, Woburn, MA).
1803BV7-2 and 1803BV20-1 T cell sorting
1803BV7-2 and 1803BV20-1 T cell clones were sorted from the 1803TIL sample by the CELLection Pan Mouse IgG kit (Dynal, Lake Success, NY) using anti-human TCR Vβ6.7-FITC mAb (Endogen) and anti-human TCR Vβ2-PE mAb (Beckman Coulter, Miami, FL) respectively, as the TRBV7-2 gene product is recognized by the TCR Vβ6.7 Ab, whereas TRBV20-1 gene product is recognized by the TCR Vβ2 Ab. After sorting, the purity of the T cells was confirmed by FACS analysis and 5′ RACE analysis of TRBV gene expression. Sorted 1803BV7-2 and 1803BV20-1 T cells as well as 1803BV7-2 and 1803BV20-1 depleted T cells were rested in T cell growth medium (RPMI 1640 supplemented with 10% FCS, 2 mM glutamine, 10 mM HEPES buffer, 0.55 mM 2-ME, and 50 CU IL-2) in a humidified incubator at 37°C and 5% CO2 for 1 wk, and then their activities against tumor cells were determined by IFN-γ release assay.
2023BV24-1 and 2023BV30 T cell cloning
2023BV24-1 and 2023BV30 T cells were cloned from the TIL 2023 sample by limiting dilution. Briefly, five cells per well were seeded in 96-well round-bottom plates with 200 μl of X-Vivo 15 medium (Cambrex, Baltimore, MD) supplemented with 10% FCS, 10 mM HEPES buffer, 0.55 mM 2-ME, 50 CU IL-2, 3 μg/ml OKT3, and 5 × 104 allogeneic PBMC. After 2 wk of culture, T cells from growth positive wells were transferred into new 96-well round-bottom plates, and tested for the ability to release IFN-γ in response to autologous 2023mel tumor cells. The TRBV gene products expressed by tumor-reactive T cell clones were identified using 5′ RACE analysis of TRBV gene expression.
Clinical response evaluation
All patients underwent computed axial tomography of the chest, abdomen, and pelvis before adoptive cell transfer and 2 mo after the treatment. For each patient, the sum of the longest diameters of all tumors (World Health Organization Response Evaluation Criteria in Solid Tumors) before and after therapy was calculated. A partial response was defined as a decrease of ≥50% (but <100%) in the sum of the longest diameters of all evaluable metastases lasting ≥1 mo with no new or enlarging tumors.
Results
Comparison of TRBV gene expression in TIL and tumor samples
Initial studies were focused on the analysis of the TRBV gene products expressed by populations of TIL that were administered to patients following treatment with a nonmyeloablative chemotherapy regimen. A 5′ RACE protocol was devised that uses a TRBV C region primer to amplify gene products expressed in polyclonal T cell populations in an attempt to obtain an unbiased, quantitative measurement of the representation of individual clones present in these cells. Studies conducted on the in vitro-cultured TIL 1803 indicated that a single dominant rearranged TRBV sequence derived from the TRBV20-1 germline gene represented ~65% of the sequences amplified from this TIL. The dominant TRBV20-1 sequence-corresponding T cell clone was designated as 1803BV20-1. Cell surface staining conducted using an Ab directed against the TCR Vb2 gene product, which corresponds to TRBV20-1, revealed that this gene product was expressed by ~68% of TIL 1803 T cells, similar to the results of the 5′ RACE analysis. Sequences derived from each of the individual TRBV germline genes represented between 5 and 10% of the 5′ RACE amplified products that were derived from the uncultured 1803 tumor sample (Fig. 1). A comparison of the CDR3 unique junctional region sequences revealed that none of the sequences present in the cultured TIL 1803 were identical to those found in the uncultured tumor sample. For example, three different rearranged sequences derived from the TRBV20-1 germline gene were isolated from the 1803 tumor sample, but these sequences were distinct from the dominant sequence present in TIL 1803 (Fig. 2).
FIGURE 1.
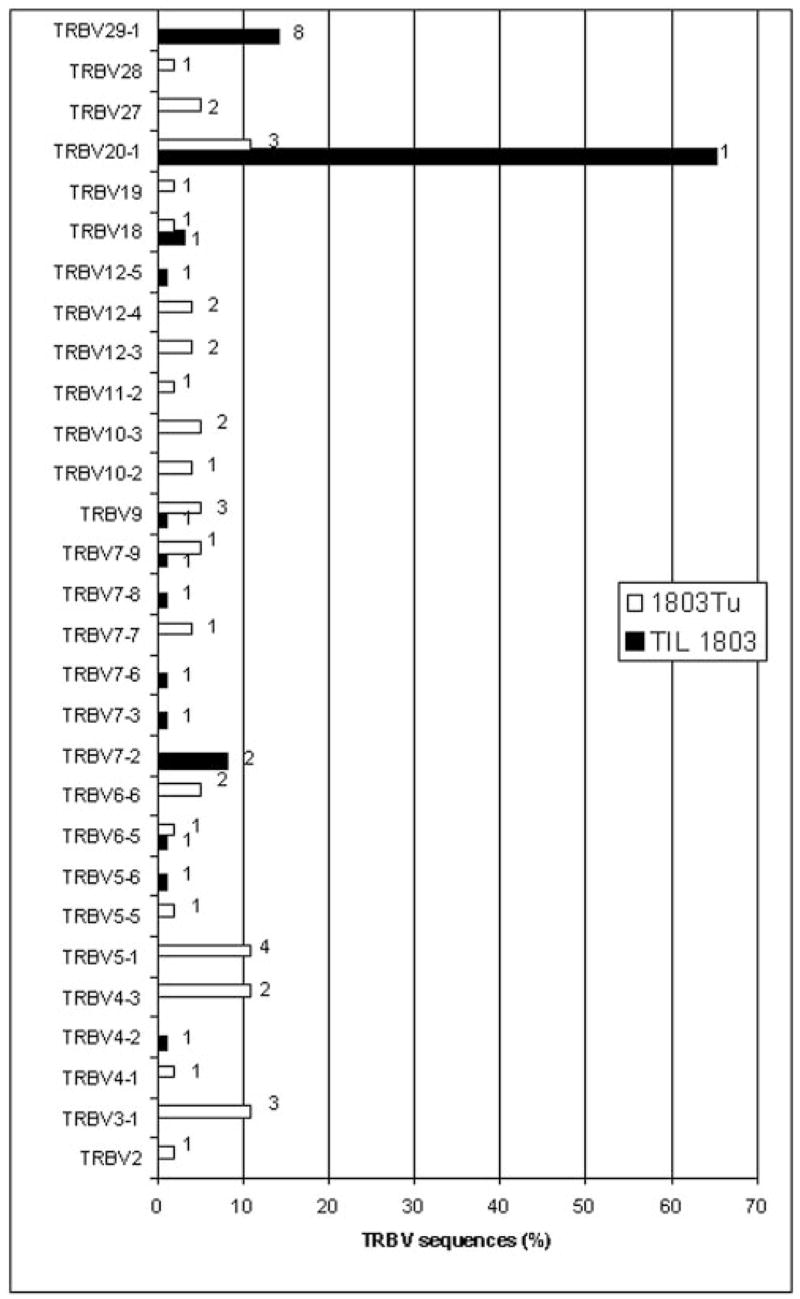
Diversity of TRBV sequences expressed in the TIL and uncultured tumor samples from melanoma patient 1803. TRBV sequences amplified from TIL 1803 and the uncultured 1803 tumor (1803Tu) were aligned to germline TRBV sequences and the percentage of sequences corresponding to each other was determined. The absence of a bar indicates that none of the sequences corresponded to that particular TRBV gene. The datum next to each bar represents the number of distinct sequences with the same TRBV sequence that were identified by comparison of CDR3 junctional region sequences. A total of 96 clones were sequenced from TIL 1803 and 96 clones were sequenced from 1803Tu.
FIGURE 2.
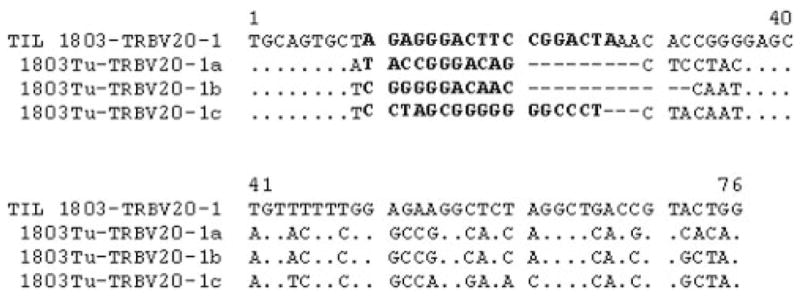
Alignment of the VDJ junctional region of TRBV20-1 sequences amplified from the 1803Tu sample and the TIL 1803 sample. Dots represent identical bases and dashes represent gaps introduced to align the D and J region sequences. The sequences in D region are noted in bold. The dominant TRBV20-1 sequence from the TIL 1803 sample was not detected in the uncultured 1803 tumor sample.
Similar studies conducted with TIL 2023 indicated that ~80% of the TRBV sequences amplified from this TIL corresponded to a single rearranged transcript derived from the TRBV24-1 germline gene (Fig. 3). The dominant TRBV24-1 sequence-corresponding T cell clone was designated as 2023BV24-1. Abs directed against the TRBV24-1 gene product are not available, however, and thus a correlation could not be established between the 5′ RACE results and FACS analysis in TIL 2023. As observed for the samples obtained from patient 1803, none of the sequences amplified from the uncultured 2023 tumor sample corresponded to the sequences amplified from cultured TIL 2023 (data not shown). Taken together, these results suggest that T cells that predominate in populations of TIL are present at only a very low frequency in the uncultured tumor samples that were used to derive the TIL and were thus not seen by sequencing 96 clones from the fresh tumor.
FIGURE 3.

Diversity of TRBV sequences expressed in the TIL and uncultured tumor samples from melanoma patient 2023. TRBV sequences amplified from TIL 2023 and the uncultured 2023 tumor (2023Tu) were aligned to germline TRBV sequences and the percentage of sequences corresponding to each other was determined. The absence of a bar indicates that none of the sequences corresponded to that particular TRBV gene. The datum next to each bar represents the number of distinct sequences with the same TRBV sequence that were identified by comparison of CDR3 junctional region sequences. A total of 192 clones were sequenced from TIL 2023 and 96 clones were sequenced from 2023Tu.
Further studies were then conducted to determine the frequency of T cells in the uncultured tumor samples, which corresponded to the dominant clone in TIL 1803. The TRBV20-1 sequences were amplified from 1803Tu using a primer specific for this gene product by RT-PCR and 64 amplified products of the TRBV20-1 family were sequenced. These results indicate that 1 of the total of 64 cDNA products sequenced corresponded to the sequence present in TIL 1803 (Table I). As 11% of the sequences amplified from 1803Tu were derived from the TRBV20-1 gene, ~0.2% of the sequences from the uncultured 1803 tumor corresponded to the dominant sequence isolated from TIL 1803. The results demonstrate that certain T cell clones that are represented at relatively low levels in tumor samples preferentially expand during in vitro culture, in some cases actually becoming the dominant clone.
Table I.
Frequencies of TRBV20-1 sequences with unique D region in the 1803Tu samplea
| TRBV20-1 Sequence | Sequence of Unique D Region | Frequency |
|---|---|---|
| TRBV20-1-TIL | AGA GGG ACT TCC GGA CT | N/A |
| TRBV20-1-1 | AGA GAT AAC CCC GGG ACT AG | 1/64 |
| TRBV20-1-2 | TGG GGG ACA GG | 2/64 |
| TRBV20-1-3 | AGA GCC GAC AGG GGG AA | 6/64 |
| TRBV20-1-4 | AGT CGG GGG ACA GG | 1/64 |
| TRBV20-1-5 | AGA GGT TGG GAC GAG | 5/64 |
| TRBV20-1-6 | TGG ACA GCG AG | 2/64 |
| TRBV20-1-7 | AGA CGA CAG AAC GG | 1/64 |
| TRBV20-1-8 | AGA GTT GAT AGG A | 1/64 |
| TRBV20-1-9 | AGG ACC GGG ACA GGG | 2/64 |
| TRBV20-1-10 | AGA GGG CCA GGG GC | 2/64 |
| TRBV20-1-11 | CAG CAA CTA GCG GGG | 1/64 |
| TRBV20-1-12 | GAA GAG CCA AAC AGG AGG | 1/64 |
| TRBV20-1-13 | AGA GGG ACT TCC GGA CT | 1/64 |
| TRBV20-1-14 | AGA GAT CGA CAG | 2/64 |
| TRBV20-1-15 | AGA GGC GTT CAG GGG | 1/64 |
| TRBV20-1-16 | AGA GCC GAC AGG GGG | 1/64 |
| TRBV20-1-17 | AGA GAT GGT CCC GGG | 1/64 |
| TRBV20-1-18 | AGT TTC TGG GAC GT | 1/64 |
| TRBV20-1-19 | AGA GAC TCT GGG | 1/64 |
| TRBV20-1-20 | CAG CAC ACT AGC GGG | 1/64 |
| TRBV20-1-21 | AGA GGG CTC AGG GG | 1/64 |
| TRBV20-1-22 | AGA GGG CCA GGG | 1/64 |
| TRBV20-1-23 | AGA GAT TGG TGA GG | 1/64 |
| TRBV20-1-24 | CCT GGC TGG GGG | 1/64 |
| TRBV20-1-25 | AGA GGG GGA CAG GGG | 1/64 |
| TRBV20-1-26 | AGA GAG GGT TCG TG | 1/64 |
| TRBV20-1-27 | AGA GAT TGG GGC GGT TGC C | 1/64 |
| TRBV20-1-28 | TAC TGA CGA GGG ATT GG | 1/64 |
| TRBV20-1-29 | GGT TGG TTG GG | 1/64 |
| TRBV20-1-30 | GGA GGT TGG GA | 1/64 |
| TRBV20-1-31 | GGG GAG GTT GGG | 1/64 |
| TRBV20-1-32 | GCT ACA GGC CCC CT | 1/64 |
| TRBV20-1-33 | AGG CAG GCA TT | 1/64 |
| TRBV20-1-34 | CGG GAC A | 1/64 |
| TRBV20-1-35 | AGA AGT GAT G | 3/64 |
| TRBV20-1-36 | AGA GAT TGG AGC GGA GCA AGC A | 2/64 |
| TRBV20-1-37 | AGA GAT TGG TCG AGA CAA CAG | 1/64 |
| TRBV20-1-38 | GGG ATG GAC ACA T | 1/64 |
| TRBV20-1-39 | CCA GGG GGA AT | 1/64 |
| TRBV20-1-40 | AGA CGG ACA GGG GAG CC | 1/64 |
| TRBV20-1-41 | CCT AGC GGG GGG GC | 3/64 |
| TRBV20-1-42 | AGA GAC TCA GGG TTG AG | 2/64 |
| TRBV20-1-43 | AGA GAC GTG TGG GGG GG | 1/64 |
| TRBV20-1-44 | CCG GGA CAG GG | 1/64 |
Total RNA was isolated from the 1803Tu sample and reverse-transcribed into cDNA. TRBV20-1 sequences were amplified with the TRBV20-1-specific primer and sequenced. The D region sequences were compared. The TRBV20-1-13 sequence from the 1803FrTu sample has the same D region as the TRBV20-1 sequence from the 1803TIL sample, which is noted in bold.
In vivo persistence of T cell clones derived from TIL following adoptive transfer
The presence and persistence of T cells in the peripheral blood of patients following adoptive TIL transfer were then evaluated using the 5′ RACE technique. The analysis of peripheral blood samples obtained from patient 1803 indicated that two rearranged sequences derived from the TRBV7-2 and 20-1 germline genes were identical to sequences present in the administered TIL 1803 (Fig. 4), indicating that a minimum of two T cell clones that were derived from this TIL persisted in vivo following adoptive transfer. The relative frequency of the TRBV7-2-expressing clone, designated 1803BV7-2, which comprised only 8% of the TRBV sequences in the administered TIL, increased in the peripheral blood to ~60% of the total, and was maintained at relatively high levels for 8 wk, at which time it represented 40% of the peripheral T cells. Representation of the TRBV20-1-expressing T cells (1803BV20-1), which comprised ~65% of the T cells in TIL 1803, appeared to dramatically decline 1 wk following transfer, at which time it only represented ~4% of the total T cell repertoire. The relative levels of this clone then appeared to increase in vivo, and at 8 wk represented ~20% of the T cells in the peripheral blood of patient 1803. These results provide evidence for the selective survival and in vivo growth of individual clones present in the infused TIL samples following adoptive transfer.
FIGURE 4.
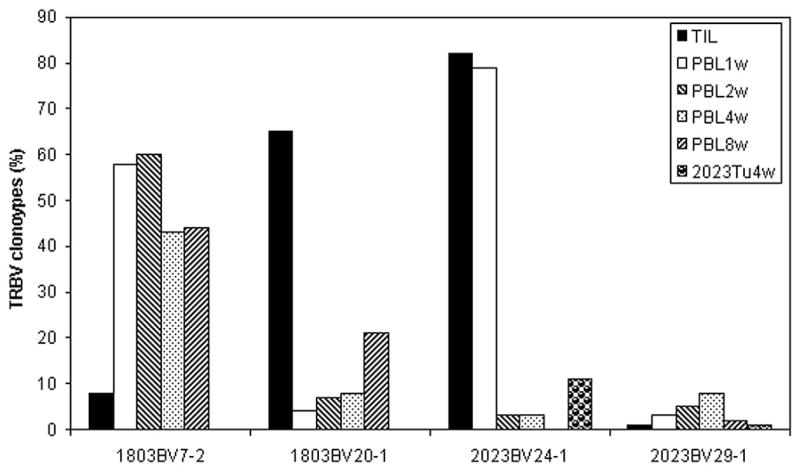
Persistence of T cell clones in melanoma patients 1803 and 2023 after adoptive cell transfer. The TRBV sequences expressed in TIL as well as PBL samples obtained at 1, 2, 4, and 8 wk following adoptive TIL transfer were analyzed and compared. 2023Tu4w was the tumor sample obtained from patient 2023 at 4 wk following adoptive transfer and analyzed by 5′ RACE analysis of TRBV gene expression, and the TRBV sequences expressed in 2023Tu4w were also compared with those in TIL 2023 and PBL samples. Two T cell clones, 1803BV7-2 and 1803BV20-1, derived from the TRBV7-2 and TRBV20-1 germline genes persisted in patient 1803. Two T cell clones, 2023BV24-1 and 2023BV29-1, derived from the TRBV24-1 and TRBV29-1 germline genes persisted in patient 2023.
Analysis of samples from patient 2023 indicated that the dominant administered clone expressing TRBV24-1 (2023BV24-1) comprised ~80% of the T cells present in TIL 2023 as well as 80% of the T cells 1 wk following transfer (Fig. 4). The relative representation of this clone in the peripheral blood of patient 2023 then declined to a level of 3% at 2 and 4 wk following adoptive transfer, and was not detected 8 wk following transfer. Another clone expressing TRBV29-1 (2023BV29-1) represented 1% of the sequences present in TIL 2023 and appeared to increase up to 8% of the sequences obtained from PBL 4 wk following transfer and decrease to 2% of the T cells present in PBL 8 wk following transfer. Approximately 11% and 1% of the sequences amplified from the resected tumor sample obtained 4 wk following adoptive transfer corresponded to 2023BV24-1 and 2023BV29-1 TRBV sequences, respectively (Fig. 4). T cells expressing the 2023BV24-1 TRBV sequence comprised only 3% of the T cells in the peripheral blood at this time, suggesting that they may have selectively accumulated in tumor tissue and played a role in mediating tumor regression.
The sequences obtained from TIL were compared with those derived from PBL samples obtained before adoptive transfer in an attempt to determine whether they were present at detectable levels in peripheral blood before therapy. The result indicated that the persistent clone 1803BV20-1 comprised ~2% of the total T cells present in the peripheral blood of patient 1803 8 days before adoptive transfer. In addition, the persistent 2023BV29-1 clone was present at a level of ~4% in the peripheral blood of patient 2023 7 days before adoptive transfer. Thus, while the ablation may have eliminated or significantly reduced the number of cells in the peripheral blood of patients 1803 and 2023 corresponding to the 1803BV20-1 and 2023BV29-1 clones, respectively, it is formally possible that some or all of the cells detected in the peripheral blood of these patients following adoptive transfer represented cells that were relatively resistant to the nonmyeloablative chemotherapy and that repopulated the peripheral blood and not T cells that were derived from the transferred TIL.
Tumor reactivity of persistent T cell clones
Attempts were then made to determine whether or not the T cell clones that persisted following adoptive transfer were tumor reactive. An autologous tumor cell line was not available from the 1803 tumor, however, TIL 1803 T cells recognized uncultured 1803 tumor as well as the allogeneic 888mel cell line that shared expression of HLA-A1 with 1803 tumor cells but not 888EBV-B cells (Fig. 5A). Ab blocking assays confirmed that the recognition of uncultured 1803 tumor cells and 888mel tumor cells by TIL 1803 T cells were restricted by HLA-A1 (data not shown). Further studies were then conducted using the 888mel cell line due to the limited availability of uncultured 1803 tumor cells. The 1803BV7-2 and 1803BV20-1 T cells were then isolated from TIL 1803 by positive selection using anti-Vβ6.7 and anti-Vβ2 Abs, respectively, and tested for their ability to respond to 888mel. The results indicated that the predominantly persistent 1803BV7-2 T cells strongly recognized 888mel cells, whereas 1803BV20-1 T cells failed to recognize this cell line. In addition, the T cells that were depleted of 1803BV7-2 T cells only weakly recognized 888mel, whereas the T cells that were depleted of 1803BV20-1 T cells responded similarly to the unseparated TIL 1803 (Fig. 5A). 1803BV20-1 T cells were sorted by FACS to a purity of 99.34% and showed no recognition of 888mel when tested using an IFN-γ release assay, which was also confirmed by an intracellular IFN-γ staining assay of TIL 1803 (data not shown). These results indicate that the 1803BV7-2 T cell clone that persisted at a high level in vivo in patient 1803 following adoptive transfer was tumor-reactive. The 1803BV20-1 clone may be reactive to a tumor Ag exclusive to the 1803 tumor though insufficient 1803 tumor cells were available for testing this.
FIGURE 5.
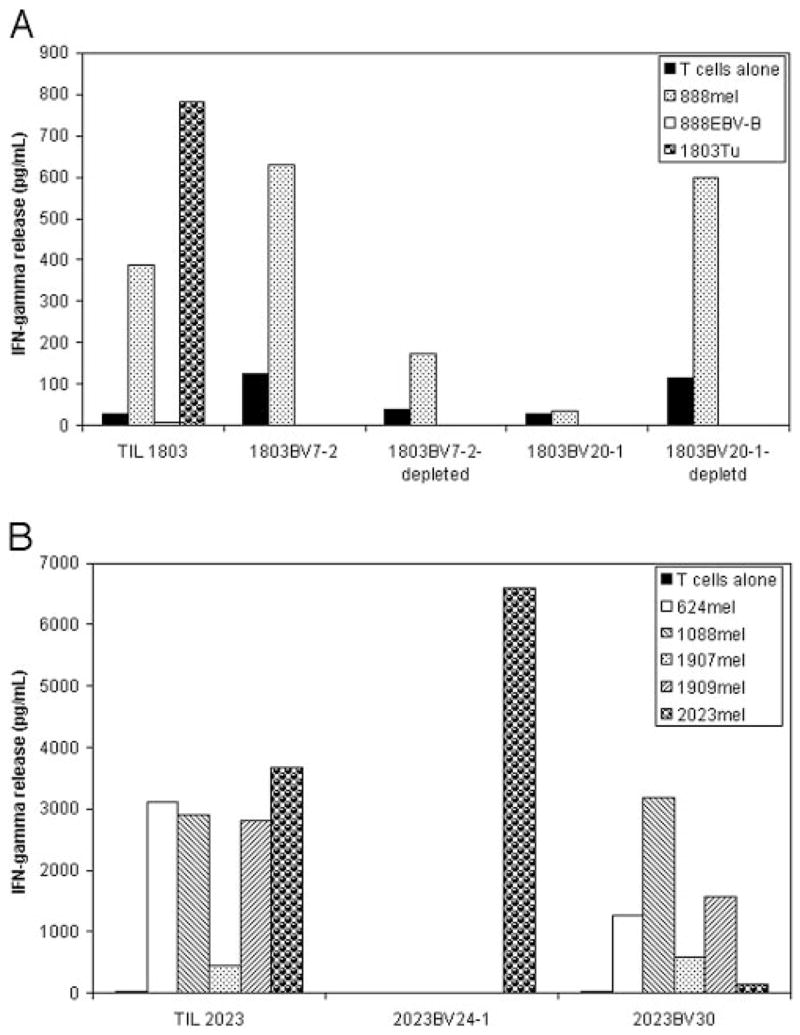
Tumor reactivity of TIL and T cell clones derived from TIL 1803 (A) and TIL 2023 (B). 1803BV7-2 and 1803BV20-1 T cells were sorted from TIL 1803 using corresponding Abs. Tumor reactivity of TIL 1803 cells was tested against uncultured 1803 tumor cells and HLA-A1-matched allogeneic 888mel as well as 888EBV-B cells. Tumor reactivity of 1803BV7-2, 1803BV20-1, and their depleted T cell samples were tested against 888mel only because of limited 1803Tu cells. 2023BV24-1 and 2023BV30 T cells were cloned from TIL 2023 by limiting dilution, and their tumor reactivity was tested against autologous 2034mel as well as HLA-A2-matched allogeneic 624mel, 1088me, 1907mel, and 1909mel.
TIL 2023 T cells recognized the autologous 2023mel cell line as well as the allogeneic 624mel, 1088mel, 1907mel, and 1909mel cell lines that shared expression of HLA-A2 with autologous tumor cells (Fig. 5B). Limiting dilution studies resulted in the isolation of a 2023BV24-1 clone as well as a TRBV30 clone (2023BV30) that was not a persistent T cell clone, but a 2023BV29-1 clone was not obtained, presumably because it was represented at a relatively low frequency in this TIL. The 2023BV24-1 clone recognized the autologous melanoma cell line but failed to recognize any of the allogeneic melanoma cell lines that were tested, whereas the 2023BV30 clone recognized the allogeneic cell lines but only weakly recognized the autologous melanoma cell line. Thus, the 2023BV24-1 clone that persisted in vivo in patient 2023 following adoptive transfer was tumor-reactive.
Clinical responses observed following adoptive transfer
An objective partial response of metastatic lesions in the lung, lymph nodes, and intra-abdominal and i.m. sites was observed in patient 1803 (Fig. 6A). A partial response of multiple s.c. and lymph node metastases was observed following treatment of patient 2023 (Fig. 6, B and C). The observation that at least one tumor-reactive T cell clone, clone 1803BV7-2, persisted in patient 1803 following adoptive transfer, and a tumor-reactive clone, clone 2023BV24-1, persisted in patient 2023 following adoptive transfer is compatible with the hypothesis that these persistent T cells played an active role in mediating the tumor regression that was observed in these patients (Table II).
FIGURE 6.
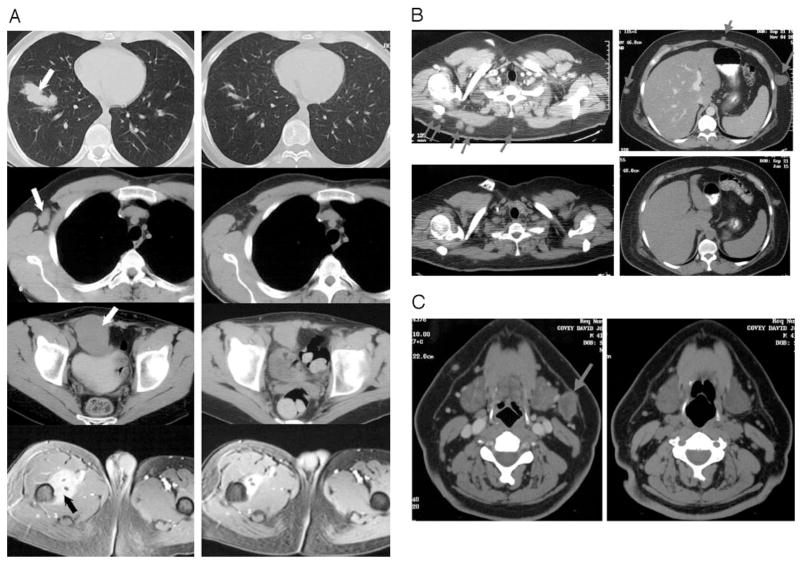
Clinical response in melanoma patients after adoptive cell transfer. A, Computed axial tomography scans in patient 1803. Left panels, Disease status before adoptive cell transfer. Right panels, Disease status after treatment. Panels in the top row show the metastases in the lung. Panels in the second row show a metastasis in right axillary lymph node. Panels in the third row show a metastasis in the intra-abdominal site. Panels in the last row show a metastasis in the muscles of the thigh site. B, Computed axial tomography scans illustrating the s.c. metastases in patient 2023. Top panels, Disease status before treatment. Bottom panels, Disease status after the treatment. C, Computed axial tomography scans illustrating a metastasis in the left cervical lymph node in patient 2023. Left panels, Disease status before the treatment. Right panels, Disease status after the treatment. Arrows show sites of metastases.
Table II.
Summary of tumor-reactive T cell persistence in melanoma patients after adoptive cell transfera
| Patient | Clonotype | Pre-PBL | TIL | PBL 1 wk | PBL 2 wk | PBL 4 wk | PBL 8 wk | Tumor 4 wk | Tumor Reactivity | Clinical Response |
|---|---|---|---|---|---|---|---|---|---|---|
| 1803 | TRBV7-2 | 0 | 8 | 58 | 60 | 42 | 44 | N/A | Yes | PR |
| TRBV20-1 | 2 | 65 | 4 | 7 | 8 | 21 | N/A | Unknown | ||
| 2023 | TRBV24-1 | 0 | 82 | 79 | 3 | 3 | 0 | 11 | Yes | PR |
| TRBV29-1 | 4 | 1 | 3 | 5 | 8 | 2 | 1 | Unknown |
Pre-PBL was the PBL sample obtained prior to adoptive cell transfer, and the other PBL samples were obtained at different time points after treatment. N/A, not available; PR, partial response.
Discussion
This represents the first report in which 5′ RACE analysis of TRBV gene expression was used to characterize the repertoire of T cells following adoptive cell transfer. Because the Ag-specific TCR repertoire exhibited a restricted Vα chain usage and usage of a variety of Vβ chains (14, 15), TRBV is a preferred marker to define the TCR repertoire in TIL samples. TCR α-chains result from the rearrangement of V genes and J genes, whereas TCR β-chains result from the rearrangement of V genes and J genes as well as D genes. Several reports showed that TCR α-chains played a dominant role in Ag recognition, and a specific TCR α-chain paired with multiple TCR β-chains maintained the same T cell function (14, 15). Therefore, TCR β-chains are more diversified and have been analyzed in this report. 5′ RACE analysis of TRBV gene expression demonstrated that dominant T cell clones, 1803BV20-1 and 2023BV24-1, were detected in the TIL samples from melanoma patients 1803 and 2023, respectively (Figs. 1 and 3). 5′ RACE analysis of TCR Vα gene expression showed similar results (data not shown). These dominant clones were either undetected or present at a very low level in the fresh tumor samples that were used to generate these TILs (Table I; Figs. 2 and 3). The data indicate that in vitro culture resulted in preferential expansion of certain clones from a relatively small number of T cells in the original tumor samples. In in vitro culture conditions, anti-CD3 Ab (OKT3) and IL-2 were used to drive nonspecific T cell proliferation (16). Therefore, T cell populations with a high capacity for cell division expanded preferentially during the TIL generation culture. In addition, selective growth and persistence of certain T cell clones appears to have occurred in vivo following adoptive cell transfer. For example, 1803BV7-2 T cells accounted for 8% of T cells in the administered TIL but ~60% of the total T cells in the peripheral blood obtained at 1 or 2 wk after adoptive transfer and then maintained at a high frequency for more than 2 mo (Fig. 4). Tumor Ag stimulation, cytokine regulation, and homeostatic expansion may be involved in the selective growth and persistence of T cell clones in vivo following adoptive cell transfer (17–19). Thus, selective growth both in vitro and in vivo of individual T cell clones in melanoma patients following adoptive cell transfer resulted in striking differences in clonotypes represented in the fresh tumor and in the cultured TIL.
Tumor-reactive T cell clones, 1803BV7-2 and 2023BV24-1, were present in the cultured TILs from patient 1803 and patient 2023, respectively (Figs. 1, 3, and 5). However, these tumor-reactive T cell clones were not detected in the original tumors by 5′ RACE analysis of TRBV gene expression, indicating that the tumor-reactive T cell clones in the TILs are only minimally present in the original tumors. In our studies, between 96 and 192 clones were sequenced each sample for TRBV gene expression analysis. Thus, T cell clonotypes present at a level <1% could have easily been missed. In a previous study, a TCR Vβ16 gene segment was strongly overexpressed at the site of a spontaneously regressive melanoma, suggesting that clonal expansion of TRBV16 T cells occurred locally and antitumor responses might take place in regressing melanoma (20). Results obtained from the analysis of 12 different autopsy specimens of tumor tissue from three patients with metastatic melanoma showed an overall predominance of a very limited number of TRBV transcripts at very high frequencies (21). In these reports, however, the tumor reactivity of dominant-specific TRBV-expressing T cells was not evaluated. Single-stranded conformational polymorphism analysis indicated that a TCR Vβ2-expressing antitumor cytotoxic T cell clone was dominant in the fresh tumor tissue, but these cells were incapable of preventing tumor cell progression (22). RT-PCR analysis conducted using TRBV primers capable of amplifying each of the individual TRBV genes indicated that relatively few TRBV-gene families were expressed at significant levels in both the regressive and progressive regions of the same primary human malignant melanoma (23). Moreover, comparison of the T cell clonotypes present in different melanoma lesions from individual patients demonstrated that multiple clonotypic TCR transcripts were present in all cases at low levels and the T cell infiltration of malignant melanoma was exceedingly heterogeneous (24). Therefore, it seems that tumor-reactive T cells are naturally present in melanoma tumor tissues at low levels, suggesting that in vivo accumulation of antitumor CTLs is usually not sufficient to eradicate growing melanomas.
The situation may be quite different, however, following adoptive cell transfer that may result in in vivo accumulation of anti-tumor T cells in the tumor sites at a level high enough to cause tumor regression. This is evidenced by the fact that in patient 2023 ~11% of the sequences amplified from the resected tumor sample obtained 4 wk following adoptive transfer corresponded to the tumor-reactive 2023BV24-1 TRBV sequence in the administered TIL 2023 (Fig. 4), however, 2023BV24-1 TRBV sequences were not detected in the original tumor resected before adoptive cell transfer (Fig. 3).
In this report, using 5′ RACE analysis of TRBV gene expression, we found that the tumor-reactive T cell clone, 1803BV7-2, persisted in patient 1803 for more than 2 mo at a high frequency after adoptive cell transfer, whereas the tumor-reactive T cell clone, 2023BV24-1, persisted in patient 2023 at a high frequency for 1 wk, decreased dramatically after 1 wk and was not detected in the peripheral blood in 2 mo after adoptive cell transfer (Fig. 4). Clinical observation indicated that patient 1803 had an objective partial response with progressive tumor regression after adoptive transfer (Fig. 6A), and patient 2023 had the partial response after adoptive transfer (Fig. 6, B and C). Thus, there was an association between the clinical outcomes and the persistence of tumor-reactive T cell clones after adoptive cell transfer. FACS analysis and tetramer staining demonstrated that the persistent clonal repopulation of MART-1-reactive T cells could be responsible for tumor destruction in melanoma patients after adoptive transfer of autologous tumor-reactive TILs (1). These results suggest that the persistence of tumor-reactive T cell clones is associated with tumor regression.
In summary, the results of this study demonstrate that 5′ RACE analysis of TRBV gene expression is a powerful tool for analyzing individual T cell subpopulations and monitoring the presence and persistence of adoptively transferred lymphocytes in patients with melanoma. This method may provide a means to analyze the TRBV diversity in patients bearing tumors of different histologies as well as in patients with autoimmune diseases. The results in this paper demonstrate that there is selective growth and survival, both in vitro and in vivo, of individual T cell clones from a relatively small number of T cells in the original tumor samples.
Footnotes
Abbreviations used in this paper: TIL, tumor-infiltrating lymphocyte; TRBV, TCR β-chain V region; CU, cetus unit; MART, melanoma Ag recognized by T cells.
References
- 1.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Packard BS, Aebersold PM, Solomon D, Topalian SL, Toy ST, Simon P, Lotze MT, Yang JC, Seipp CA, et al. Use of tumor infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma: preliminary report. N Engl J Med. 1988;319:1676. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Yannelli JR, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR, Seipp CA, Einhorn JH, White DE. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J Natl Cancer Inst. 1994;86:1159. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 4.Dudley ME, Wunderlich J, Nishimura MI, Yu D, Yang JC, Topalian SL, Schwartzentruber DJ, Hwu P, Marincola FM, Sherry R, et al. Adoptive transfer of cloned melanoma-reactive T lymphocytes for the treatment of patients with metastatic melanoma. J Immunother. 2001;24:363. doi: 10.1097/00002371-200107000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormier JN, Hijazi YM, Abati A, Fetsch P, Bettinotti M, Steinberg SM, Rosenberg SA, Marincola FM. Heterogeneous expression of melanoma-associated antigens and HLA-A2 in metastatic melanoma in vivo. Int J Cancer. 1998;75:517. doi: 10.1002/(sici)1097-0215(19980209)75:4<517::aid-ijc5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 7.Sensi M, Farina C, Maccalli C, Anichini A, Berd D, Parmiani G. Intralesional selection of T cell clonotypes in the immune response to melanoma antigens occurring during vaccination. J Immunother. 1998;21:198. doi: 10.1097/00002371-199805000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Genevee C, Diu A, Nierat J, Caignard A, Dietrich PY, Ferradini L, Roman-Roman S, Triebel F, Hercend T. An experimentally validated panel of subfamily-specific oligonucleotide primers (Vα1-w29/Vβ1-w24) for the study of human T cell receptor variable V gene segment usage by polymerase chain reaction. Eur J Immunol. 1992;22:1261. doi: 10.1002/eji.1830220522. [DOI] [PubMed] [Google Scholar]
- 9.Willhauck M, Mohler T, Scheibenbogen C, Pawlita M, Brossart P, Schmier JW, Keilholz U. T-cell receptor β variable region diversity in melanoma metastases after interleukin 2-based immunotherapy. Clin Cancer Res. 1996;2:767. [PubMed] [Google Scholar]
- 10.Willhauck M, Scheibenbogen C, Pawlita M, Mohler T, Thiel E, Keilholz U. Restricted T-cell receptor repertoire in melanoma metastases regressing after cytokine therapy. Cancer Res. 2003;63:3483. [PubMed] [Google Scholar]
- 11.Manne J, Mastrangelo MJ, Sato T, Berd D. TCR rearrangement in lymphocytes infiltrating melanoma metastases after administration of autologous dinitrophenyl-modified vaccine. J Immunol. 2002;169:3407. doi: 10.4049/jimmunol.169.6.3407. [DOI] [PubMed] [Google Scholar]
- 12.Pisarra P, Mortarini R, Salvi S, Anichini A, Parmiani G, Sensi M. High frequency of T cell clonal expansions in primary human melanoma: involvement of a dominant clonotype in autologous tumor recognition. Cancer Immunol Immunother. 1999;48:39. doi: 10.1007/s002620050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, Greenberg PD. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci USA. 2002;99:16168. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich PY, Le Gal FA, Dutoit V, Pittet MJ, Trautman L, Zippelius A, Cognet I, Widmer V, Walker PR, Michielin O, et al. Prevalent role of TCR α-chain in the selection of the preimmune repertoire specific for a human tumor-associated self-antigen. J Immunol. 2003;170:5103. doi: 10.4049/jimmunol.170.10.5103. [DOI] [PubMed] [Google Scholar]
- 15.Yokosuka T, Takase K, Suzuki M, Nakagawa Y, Taki S, Takahashi H, Fujisawa T, Arase H, Saito T. Predominant role of T cell receptor (TCR)-α chain in forming preimmune TCR repertoire revealed by clonal TCR reconstitution system. J Exp Med. 2002;195:991. doi: 10.1084/jem.20010809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayan P, Pandey R, Yadav VS, Tandon R, Haq W, Dhar MM, Singh VK. Inhibition of anti-CD3 and interleukin-2 stimulated T lymphocyte proliferation by peptidomimetic opioid compound. Immunopharmacol Immunotoxicol. 2003;25:225. doi: 10.1081/iph-120020472. [DOI] [PubMed] [Google Scholar]
- 17.Powell DJ, Jr, Rosenberg SA. Phenotypic and functional maturation of tumor antigen-reactive CD8+ T lymphocytes in patients undergoing multiple course peptide vaccination. J Immunother. 2004;27:36. doi: 10.1097/00002371-200401000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schluns KS, Lefrancois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol. 2003;3:269. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 19.La Gruta NL, I, Driel R, Gleeson PA. Peripheral T cell expansion in lymphopenic mice results in a restricted T cell repertoire. Eur J Immunol. 2000;30:3380. doi: 10.1002/1521-4141(2000012)30:12<3380::AID-IMMU3380>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Ferradini L, Mackensen A, Genevee C, Bosq J, Duvillard P, Avril MF, Hercend T. Analysis of T cell receptor variability in tumor-infiltrating lymphocytes from a human regressive melanoma: evidence for in situ T cell clonal expansion. J Clin Invest. 1993;91:1183. doi: 10.1172/JCI116278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strohal R, Brna C, Mossbacher U, Fischer G, Pehamberger H, Stingl G. First comparative delineation of the T cell receptor repertoire in primary and multiple subsequent/coexisting metastatic melanoma sites. J Invest Dermatol. 1998;111:1085. doi: 10.1046/j.1523-1747.1998.00450.x. [DOI] [PubMed] [Google Scholar]
- 22.Hishii M, Andrews D, Boyle LA, Wong JT, Pandolfi F, van den Elsen PJ, Kurnick JT. In vivo accumulation of the same anti-melanoma T cell clone in two different metastatic sites. Proc Natl Acad Sci USA. 1997;94:1378. doi: 10.1073/pnas.94.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.thor Straten P, Becker JC, Seremet T, Bröcker EB, Zeuthen J. Clonal T cell responses in tumor infiltrating lymphocytes from both regressive and progressive regions of primary human malignant melanoma. J Clin Invest. 1996;98:279. doi: 10.1172/JCI118790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.thor Straten P, Guldberg P, Gronbaek K, Hansen MR, Kirkin AF, Seremet T, Zeuthen J, Becker JC. In situ T cell responses against melanoma comprise high numbers of locally expanded T cell clonotypes. J Immunol. 1999;163:443. [PubMed] [Google Scholar]