Abstract
In rice (Oryza sativa) and Arabidopsis thaliana, gibberellin (GA) signaling is mediated by GIBBERELLIN-INSENSITIVE DWARF1 (GID1) and DELLA proteins in collaboration with a GA-specific F-box protein. To explore when plants evolved the ability to perceive GA by the GID1/DELLA pathway, we examined these GA signaling components in the lycophyte Selaginella moellendorffii and the bryophyte Physcomitrella patens. An in silico search identified several homologs of GID1, DELLA, and GID2, a GA-specific F-box protein in rice, in both species. Sm GID1a and Sm GID1b, GID1 proteins from S. moellendorffii, showed GA binding activity in vitro and interacted with DELLA proteins from S. moellendorffii in a GA-dependent manner in yeast. Introduction of constitutively expressed Sm GID1a, Sm G1D1b, and Sm GID2a transgenes rescued the dwarf phenotype of rice gid1 and gid2 mutants. Furthermore, treatment with GA4, a major GA in S. moellendorffii, caused downregulation of Sm GID1b, Sm GA20 oxidase, and Sm GA3 oxidase and degradation of the Sm DELLA1 protein. These results demonstrate that the homologs of GID1, DELLA, and GID2 work in a similar manner in S. moellendorffii and in flowering plants. Biochemical studies revealed that Sm GID1s have different GA binding properties from GID1s in flowering plants. No evidence was found for the functional conservation of these genes in P. patens, indicating that GID1/DELLA-mediated GA signaling, if present, differs from that in vascular plants. Our results suggest that GID1/DELLA-mediated GA signaling appeared after the divergence of vascular plants from the moss lineage.
INTRODUCTION
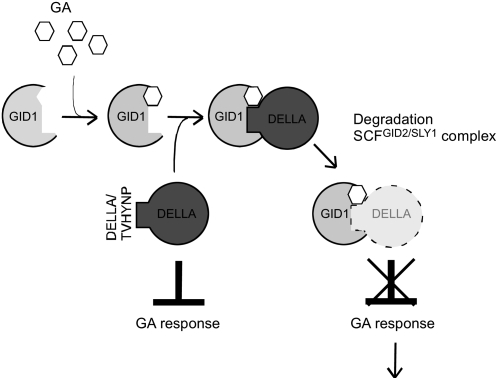
Gibberellins (GAs) comprise a large family of tetracyclic, diterpenoid plant hormones that play diverse biological roles in plant growth, including seed germination, stem elongation, leaf expansion, pollen maturation, and induction of flowering (Olszewski et al., 2002). Through molecular genetic studies on GA-insensitive mutants of rice (Oryza sativa) and Arabidopsis thaliana, the mechanism underlying GA perception has been revealed (reviewed in Ueguchi-Tanaka et al., 2007a). A well-characterized factor involved in the GA signaling pathway is the DELLA protein, which belongs to the GRAS superfamily of putative transcription factors. The DELLA protein functions as a negative regulator of GA signaling (Peng et al., 1997; Ikeda et al., 2001; Chandler et al., 2002) and is rapidly degraded when plants are treated with GA (Dill et al., 2001; Silverstone et al., 2001; Gubler et al., 2002; Itoh et al., 2002). Recent studies have identified rice GIBBERELLIN-INSENSITIVE DWARF2 (GID2) and Arabidopsis SLEEPY1 (SLY1), candidate F-box components of Skp1-Cullin-F box protein (SCF) E3 ubiquitin ligases, as apparently responsible for targeting DELLA proteins to the proteasome (McGinnis et al., 2003; Sasaki et al., 2003). More recently, GIBBERELLIN-INSENSITIVE DWARF1 (GID1) was identified as a soluble GA receptor in rice and Arabidopsis (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006). Based on these observations, the current model of GA signaling is as follows. In the absence of GA, the DELLA protein represses GA action. In the presence of GA, the GID1 receptor binds GA. The GID1/GA complex then interacts with the DELLA protein. This interaction results in DELLA protein degradation through the SCFGID2/SLY1 proteasome pathway and, consequently, allows GA action to occur (Ueguchi-Tanaka et al., 2007a) (Figure 1).
Figure 1.
Current Model of GID1-Mediated GA Signaling in Flowering Plants.
In the presence of GA, GID1 binds to DELLA, a negative regulator of GA action, to form the GA-GID1-DELLA complex. Subsequently, DELLA protein is degraded through the SCFGID2/SLY1 proteasome pathway, and as a consequence, GA actions occur.
When the GID1/DELLA-mediated GA perception system was established is an interesting question in the evolution of plant growth and development. Since the late 1950s, the presence of GAs has been reported in both seed plants and nonseed plants, including unicellular and multicellular algae, mosses, and ferns (Radley, 1961; Kato et al., 1962; Ergün et al., 2002). Although the presence of GAs in nonseed plants suggested that these species might use GA as a bioactive substance, unequivocal GA identification was not possible due to the lack of reliable chemical methods. To clarify this issue, MacMillan (2002) proposed three criteria for GA identification: (1) isolation of the pure product and comparison of its physical properties with an authentic specimen; (2) gas chromatography–mass spectrometry (GC-MS) comparing Kovats retention indices and m/z (and relative intensities) of at least six significant ions with those of standards; and (3) gas chromatography–single ion monitoring comparing retention times and m/z (and relative intensities) of at least six significant ions with those of standards (http://www.plant-hormones.info/occurrence_of_gas_in_plants.htm). When these criteria are applied to GA identification of GAs in nonseed plants, only a few GAs from fern are identified (Yamane et al., 1985, 1988; Yamauchi et al., 1996). Consequently, with the exception of antheridiogen, a pheromone promoting antheridium formation in tree ferns, it is still unclear whether nonseed plants use GAs as bioactive substances (Yamauchi et al., 1996; Banks, 1999; Menéndez et al., 2006).
Recent progress in genome research provides an alternative approach to determine when the GA perception system evolved. When genes encoding proteins homologous with the GA-related proteins in seed plants are found in the genomes of nonseed plants, these genes can be cloned and characterized functionally using biochemical and genetic techniques. Two model plants, the lycophyte Selaginella moellendorffii and the moss Physcomitrella patens, are well suited for in silico searches for GA-related genes because their whole-genome shotgun sequences and EST sequences are available to the public (http://selaginella.genomics.purdue.edu/ and http://moss.nibb.ac.jp/).
These plants are also suitable model plants for evolutionary studies. The lycophyte S. moellendorffii has a small genome (∼100 Mb) (Weng et al., 2005), about two-thirds the size of the Arabidopsis genome. The lycophytes form a basal group within the vascular plants and diverged from other vascular plants ∼400 million years ago (Weng et al., 2005). Leaves of the lycophytes and other vascular plants evolved in parallel (Gifford and Foster, 1989), so the study of S. moellendorffii will provide insights into vascular plant evolution, particularly the development of the plant body. The moss P. patens has been used as an experimental organism because it exhibits a high efficiency of homologous recombination (Schaefer and Zrÿd, 1997). It is believed that mosses and vascular plants diverged ∼430 million years ago (Kenrick and Crane, 1997), and, despite large differences in morphology and life cycle between flowering plants and mosses, P. patens contains some signal transduction systems similar to those found in flowering plants. For example, auxins and cytokinins are important developmental regulators for both flowering plants and mosses (Cove et al., 2006), and the desiccation stress response network mediated by abscisic acid is probably conserved in both (Knight et al., 1995). However, the GA signaling pathway has not been studied in these model nonseed plants. Very recently, Hayashi et al. (2006) reported the presence of ent-kaurene synthase, an enzyme involved in GA biosynthesis in seed plants, in P. patens. However, it remains to be determined whether P. patens contains GAs and uses them as growth substances.
We performed in silico screening of GA-related genes in the genomes of S. moellendorffii and P. patens using rice genes for the GA receptor GID1, DELLA protein genes, SLR1, and the F-box gene GID2 as queries. Several candidate sequences were found in both genomes, so we examined the GID1 homologs in an in vitro GA binding experiment and studied the interaction between GID1 and the DELLA homologs in a yeast two-hybrid assay. We also performed complementation experiments by introducing the candidate genes into rice plants. Our results demonstrate that S. moellendorffii has functional homologs of GID1, DELLA protein, and GID2, whereas P. patens does not.
RESULTS
Isolation of Putative GA Signaling Genes in S. moellendorffii and P. patens
The amino acid sequences of rice GID1, SLR1, and GID2 were used as queries to screen the available S. moellendorffii and P. patens databases (http://selaginella.genomics.purdue.edu/cgi-bin/blast_tmpl_s.cgi and http://moss.nibb.ac.jp/). Using a TBLASTN search, candidate genes were selected for preliminary phylogenetic analyses. In the S. moellendorffii genome database, two GID1-like sequences (Sm GID1a and Sm GID1b), two DELLA-like sequences (Sm DELLA1 and Sm DELLA2), and three GID2-like sequences (Sm GID2a, Sm GID2b, and Sm GID2c) were found. In the P. patens genome database, two GID1-like sequences (Pp GID1L1 and Pp GID1L2), two DELLA-like sequences (Pp DELLAL1 and Pp DELLAL2), and three GID2-like sequences (Pp GID2L1, Pp GID2L2, and Pp GID2L3) were found.
GID1-Like Genes
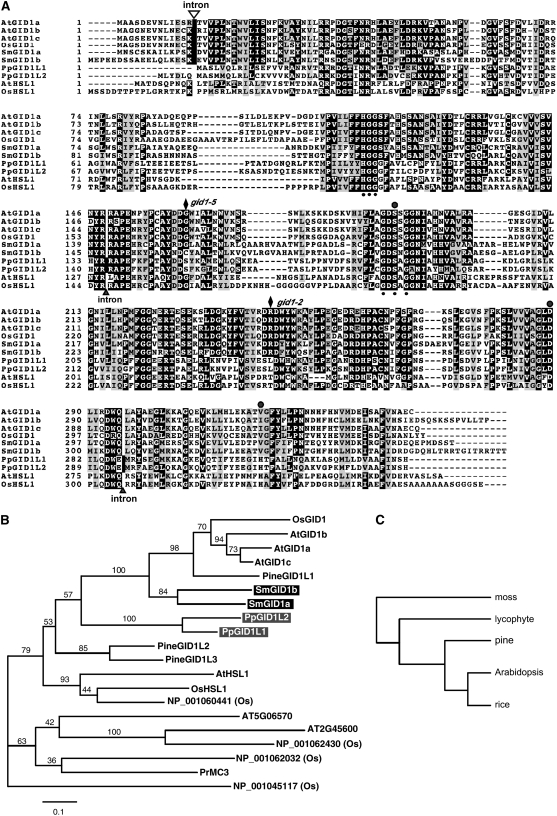
In the Sm GID1a and Sm GID1b genes, a single intron was predicted at the same site in the N-terminal portion of the predicted protein sequences. This predicted intron position is the same as that found in rice (Os GID1) and three Arabidopsis GID1 genes (At GID1a, -b, and -c) (Figure 2A, open triangle at the top of the first block). On the other hand, Pp GID1L1 and Pp GID1L2 contained two introns at the same site in each gene, localized at the middle and C terminus of the predicted protein sequences (Figure 2A, closed triangles at the bottom of the third and fifth blocks). These sites do not correspond to the intron position of GID1 genes in the flowering plants analyzed. The locations and lengths of these predicted introns were confirmed by sequencing the corresponding cDNA for each gene.
Figure 2.
Comparison of Deduced Amino Acid Sequences of GID1 Homologs in Land Plants.
(A) Amino acid sequences of GID1-like proteins from Arabidopsis and rice (angiosperms), S. moellendorffii (lycophyte), and P. patens (bryophyte) were aligned with ClustalW (http://align.genome.jp/). The open triangle indicates the position of an intron shared by the true GID1 receptors (Os GID1 and At GID1s) and Sm GID1s. Closed triangles at the bottom of the alignment are intron positions unique to Pp GID1L proteins. Small and large circles represent the conserved residues and the catalytic triad in the HSL family, respectively. The positions of gid1-2 and gid1-5 mutations in rice mutants are indicated by black diamonds at the top of the alignment. The amino acid residues are numbered from the first Met, and gaps (dashes) were introduced to achieve maximum similarity. Black and gray boxes indicate identical and similar residues, respectively. At, Arabidopsis thaliana; Os, Oryza sativa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
(B) Phylogenetic analysis of GID1 homologs. A maximum likelihood tree based on the JTT model (Jones et al., 1992) was obtained. The horizontal branch lengths are proportional to the estimated number of amino acid substitutions per residue. Bootstrap values were obtained by 1000 bootstrap replicates. Pr, Pinus radiata; Pine, Pinus taeda.
(C) A consensus phylogenetic species relationship among moss, lycophyte, pine, Arabidopsis, and rice. Note that node lengths do not reflect the accurate divergence time of each species.
We compared the amino acid sequences of the Sm GID1 and Pp GID1L proteins with those of Os GID1 and At GID1a, -b, and -c. We also included the amino acid sequences of hormone-sensitive lipases (HSLs) from Arabidopsis and rice (Figure 2A) in the comparison, because GID1 shares homology with the consensus sequence of the HSL family (Ueguchi-Tanaka et al., 2005). The Sm GID1 and Pp GID1L proteins contain the conserved HSL motifs HGG and GXSXG found in other GID1 proteins (Figure 2A, small closed circles at the bottom of the second and third blocks). Of the three conserved amino acids (S, D, and H) that form the catalytic site in the HSL family (Figure 2A, large closed circles at the top of the third, fourth, and fifth blocks), S and D are found at the corresponding positions in the GID1 proteins, but H is replaced with V or I in the rice and Arabidopsis GID1 proteins (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). The predicted amino acid sequences for the Sm GID1 and Pp GID1L proteins also contain S and D in the same relative positions. At the third amino acid residue, however, the Sm GID1 proteins contained V, as in other GID1 proteins, while Pp GID1L proteins contained H, as in HSLs. A rice GID1 loss-of-function allele, gid1-2, contains one amino acid substitution (R251T), indicating that R251 is important for GA receptor function (Ueguchi-Tanaka et al., 2005). This R residue is conserved in Sm GID1a and Sm GID1b, but not in Pp GID1L1 or Pp GID1L2, in which R is replaced by L (Figure 2A, closed diamond at the top of the fourth block). Another rice loss-of-function allele, gid1-5, contains E instead of G at position 169 (Ueguchi-Tanaka et al., 2007b), whereas Sm GID1b contains C and Pp GID1L1 and -L2 contain S at that position (Figure 2A, closed diamond at the top of the third block).
A phylogenetic analysis illustrated the relationships among the Sm GID1, Pp GID1L, and GID1 proteins from seed plants (Figure 2B). Sm GID1a and -b form a monophyletic clade that is sister to the clade of GID1 proteins from seed plants. The clade containing the S. moellendorffii and seed plant GID1 proteins is supported by a high bootstrap value and is distinct from other proteins. This clade combined with the Pp GID1L proteins forms another clade with weak bootstrap support. A phylogenetic species relationship among moss, lycophyte, pine (Pinus sp), Arabidopsis, and rice is shown in Figure 2C as a reference (Nishiyama, 2007).
DELLA-Like Genes
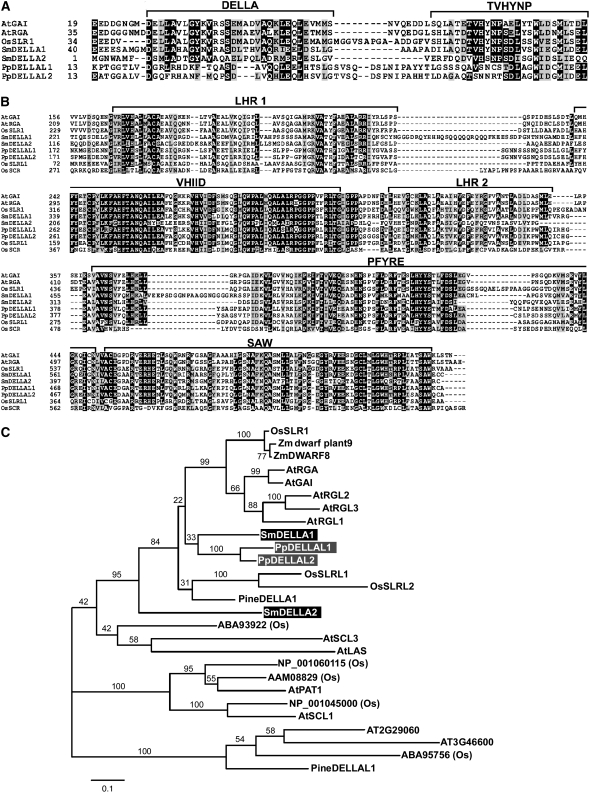
DELLA proteins can be divided into two domains based on their primary structure. DELLA/TVHYNP domains, found in the N-terminal portion of the proteins, are unique to the DELLA protein family and are involved in the interaction with GID1s in a GA-dependent manner (Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007b; Willige et al., 2007). The second domain, shared among GRAS proteins, is located in the C-terminal portion (Bolle, 2004; Itoh et al., 2005). These DELLA/TVHYNP and GRAS domains are interrupted by highly variable sequences of various lengths (Itoh et al., 2002). We compared the structures of the DELLA/TVHYNP and GRAS domains separately. Comparison of the DELLA/TVHYNP domains revealed that Sm DELLA1 contains a sequence that is very similar to those of DELLA proteins of Arabidopsis and rice, whereas Sm DELLA2 was less similar to flowering plant DELLA proteins (Figure 3A). Although we could align the amino acid sequences at the N-terminal portion of Pp DELLAL proteins with the DELLA/TVHYNP sequences of flowering plant DELLA proteins, the corresponding sequences of Pp DELLAL1 and -L2 only partially shared the conserved motifs of DELLA or TVHYNP (Figure 3A).
Figure 3.
Comparison of Deduced Amino Acid Sequences of DELLA Homologs in Land Plants.
(A) and (B) Amino acid sequence alignments of the N-terminal DELLA/TVHYNP domains (A) and C-terminal GRAS domains (B) of DELLA-like proteins from Arabidopsis and rice (angiosperms), S. moellendorffii (lycophyte), and P. patens (bryophyte) obtained using ClustalW (http://align.genome.jp/). The conserved regions or domains are presented at the top. The amino acid residues are numbered from the first Met, and gaps (dashes) were introduced to achieve higher similarity scores. Black and gray boxes indicate identical and similar residues, respectively. Abbreviations are as in Figure 1.
(C) Phylogenetic analysis of DELLA protein homologs. A maximum likelihood tree based on the JTT model (Jones et al., 1992) was obtained. The horizontal branch lengths are proportional to the estimated number of amino acid substitutions per residue. Bootstrap values were obtained by 1000 bootstrap replicates. Zm, Zea mays.
We also compared the GRAS domains of Sm DELLA and Pp DELLAL proteins with those of Arabidopsis and rice DELLA proteins and also with those of Os SCR, a protein in another GRAS subfamily (Kamiya et al., 2003). The Sm DELLA and Pp DELLAL proteins share GRAS domains, such as the leucine heptad repeat I (LHR 1), VHIID, LHR 2, PFYRE, and SAW domains, with high levels of similarity (Figure 3B). Recently, Itoh et al. (2005) reported unique DELLA proteins, Os SLRL1 and Os SLRL2, which contain regions shared with GRAS domains of typical DELLA proteins but lack the N-terminal DELLA/TVHYNP domains. These DELLA-like proteins lacking DELLA/TVHNYP domains can suppress GA signaling in a GA-independent manner in rice, indicating that these proteins may function as constitutive suppressors in GA signaling. Phylogenetic analysis demonstrated that Sm DELLA1, Pp DELLAL1, and Pp DELLAL2 form a clade with the DELLA proteins of flowering plants (Figure 3C). Together, these comparative analyses suggest that Sm DELLA1 might be involved in GA signaling in a manner similar to that of other typical DELLA proteins, since this protein contains both DELLA/TVHYNP and GRAS domains with high similarity to the flowering plant DELLA proteins analyzed. This hypothesis was tested using a number of biochemical and genetic approaches, as described below.
GID2-Like Genes
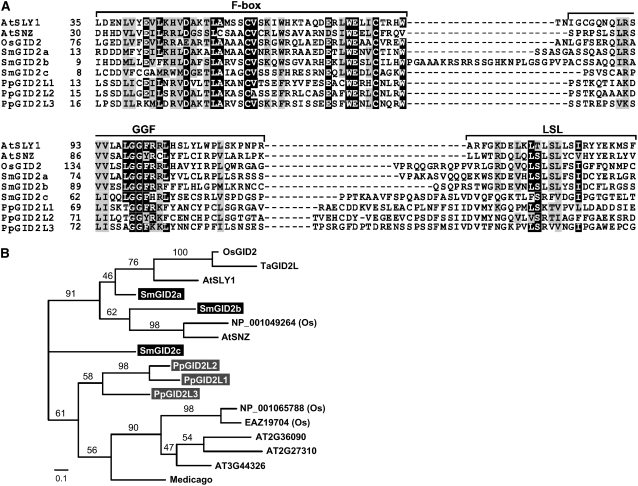
In a comparative study of Arabidopsis At SLY1 and its homologs, including Os GID2, McGinnis et al. (2003) proposed that the At SLY1/Os GID2 proteins contain three conserved domains: F-box, GGF, and LSL. Gomi et al. (2004) demonstrated that these three conserved domains are essential for the function of Os GID2. Noting these important domains, we aligned the predicted amino acid sequences of the Sm GID2 and Pp GID2L proteins with those of At SLY1 and Os GID2 (Figure 4A). At SNZ, a homolog of At SLY1 that also functions in GA signaling in Arabidopsis (Strader et al., 2004), was included in the analysis. In the phylogenetic analysis, Sm GID2a and Sm GID2b formed a clade with Os GID2, At SLY1, At SNZ, Ta GID2L (a Triticum aestivum GID2 homolog), and another rice gene with 91% bootstrap support (Figure 4B). Sm GID2c and Pp GID2L proteins were placed outside this clade. Although Sm GID2c and Pp GID2L proteins contain an F-box similar to that of At SLY1 and Os GID2, their sequences corresponding to the GGF and LSL regions showed low similarity to those in At SLY1 and Os GID2 (Figure 4A). These comparative studies suggest that Sm GID2a and Sm GID2b might function as F-box proteins in GA signaling, whereas the other GID2-like proteins might not.
Figure 4.
Comparison of Deduced Amino Acid Sequences of GID2 Homologs in Land Plants.
(A) Amino acid sequence alignment of GID2-like proteins from Arabidopsis and rice (angiosperms), S. moellendorffii (lycophyte), and P. patens (bryophyte) calculated using ClustalW (http://align.genome.jp/). The conserved regions or domains are presented at the top. The amino acid residues are numbered from the first Met, and gaps (dashes) were introduced to achieve maximum similarity. Black and gray boxes indicate identical and similar residues, respectively. Abbreviations are as in Figure 1.
(B) Phylogenetic analysis of GID2 homologs. A maximum likelihood tree based on the amino acid alignment was obtained using the JTT model (Jones et al., 1992). The horizontal branch lengths are proportional to the estimated number of amino acid substitutions per residue. Bootstrap values were obtained by 1000 bootstrap replicates. Ta, Triticum aestivum.
GA Binding Properties of Sm GID1 and Pp GID1L Proteins
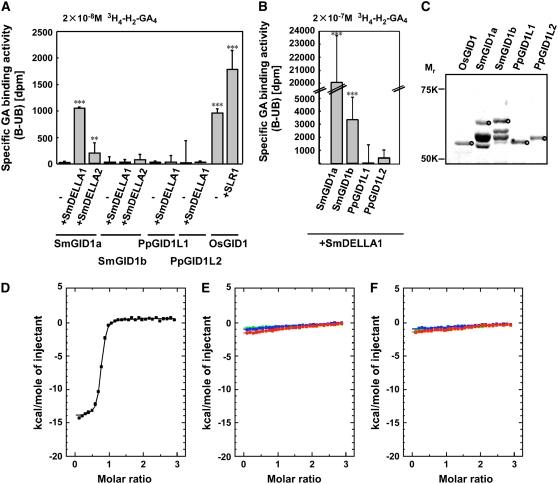
To examine the function of the Sm GID1 and Pp GID1L proteins, we studied the interaction between recombinant thioredoxin·histidine (Trx·His)-tagged GID1-like proteins and radioactive GA using a nonequilibrium gel permeation technique (Ueguchi-Tanaka et al., 2005). Because the presence of DELLA protein stabilizes the interaction between Os GID1 and GA (Ueguchi-Tanaka et al., 2007b), we performed the experiment in the presence and absence of DELLA proteins. Trx·His-Sm GID1a bound to [1,2,16,17-3H4]16,17-dihydro-GA4 (3H4-H2-GA4) in the presence of glutathione S-transferase (GST)–tagged Sm DELLA1 and Sm DELLA2, although the binding activity of Sm GID1a–Sm DELLA1 was much higher than that of Sm GID1a–Sm DELLA2 (Figure 5A). No binding activity was seen in the absence of Sm DELLA protein. We barely detected any GA binding activity of Sm GID1b or Pp GID1L proteins, regardless of whether Sm DELLA1 was included in the reaction. However, since a yeast two-hybrid experiment demonstrated that Sm GID1b interacted with GA4 with ∼6-fold lower affinity than Sm GID1a (see below), we reexamined the interaction between Sm GID1b and GA4 under a 10-fold higher level of 3H4-H2-GA4. As expected, Sm GID1b showed low but detectable GA binding activity under the high 3H4-H2-GA4 conditions, while no or very low activity of Pp GID1L proteins was observed (Figure 5B). In each experiment, approximately the same amount of Escherichia coli–produced Trx·His-GID1 protein was used (Figure 5C). These in vitro GA binding analyses demonstrate that Sm GID1a and Sm GID1b have GA4 binding activity only in the presence of Sm DELLA1. GA4 has a much higher affinity for Sm GID1a than for Sm GID1b, while Pp GID1L proteins have no or very low 3H4-H2-GA4 or GA4 binding activity.
Figure 5.
GA Binding Properties of GID1 Homologs.
(A) Affinity-purified Trx·His-Os GID1 or its homologs were incubated with 2 × 10−8 M 3H4-H2-GA4 in the presence or absence of GST-DELLA protein. The specific binding activity (B-UB) was calculated by subtracting the nonspecific binding activity (UB), which was evaluated by the addition of 0.125 mM GA4 to the assay solution, from the total binding activity (B).
(B) GA binding activity of affinity-purified Trx·His-GID1 homologs under a higher concentration of 3H4-H2-GA4 (2 × 10−7 M).
For (A) and (B), sd values were determined from more than three measurements. ** P < 0.01, *** P < 0.001 against vector control (data not shown), determined using the unpaired t test.
(C) SDS-PAGE profile of affinity-purified Trx·His-Os GID1 and its homologs. Circles indicate the recombinant proteins with approximately the expected molecular sizes (Os GID1, 57.0 kD; Sm GID1a, 57.7 kD; Sm GID1b, 59.2 kD; Pp GID1L1, 55.1 kD; and Pp GID1L2, 56.1 kD). The identification of protein corresponding to Os GID1 and its homologs was confirmed by immunoblot analysis using an Os GID1 antibody (data not shown). Although several bands were observed in each of the Sm GID1 lanes, the immunoreacting band was considered to be Sm GID1 protein, and it was concluded that the other bands were the result of degradation, immaturely transcribed protein, or protein unrelated to Sm GID1s. Approximately equal amounts (∼16 μg) of protein were used for the GA binding assay.
(D) to (F) Isothermal titration calorimetry analysis of the Pp GID1L–GA interaction. The heat exchange versus molar ratio for Trx·His-GID1 with titration of GA is shown.
(D) Integrated titration curve of Trx·His-Os GID1 with GA4. The line represents the best fitting curve calculated from a single-site binding model.
(E) Integrated titration curve of Trx·His-Pp GID1L1 with epi-GA4 (red line), GA9 (blue line), and GA12 (green line).
(F) Integrated titration curve of Trx·His-Pp GID1L2 with epi-GA4 (red line), GA9 (blue line), and GA12 (green line).
To further elucidate whether Pp GID1L1 and -L2 can bind to other GAs, we performed a thermodynamic analysis using isothermal titration calorimetry (ITC). ITC is the most direct method to measure the change of heat upon the formation of a complex. Figure 5D shows the heat exchange versus molar ratio for Trx·His-Os GID1 (40 μM) with titration of GA (final content of 500 μM) at 30°C. A clear thermodynamic change was observed in the Trx·His-Os GID1-GA4 titration experiments: the number of binding sites per protein molecule was approximately one (n = 1), and base dissociation constant was 5.16 × 106 M (Figure 5D). On the other hand, Trx·His-Pp GID1L1 (Figure 5E) and Trx·His-Pp GID1L2 (Figure 5F) did not show any heat change with any of the GAs tested (GA9, GA12, and 3-epi-GA4).
Properties of the GID1–DELLA Interaction in Yeast Cells
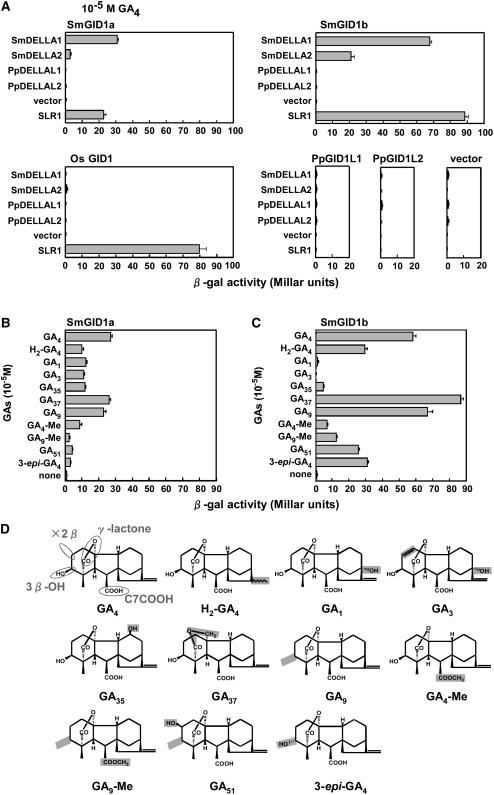
GID1 proteins in rice and Arabidopsis can interact with DELLA proteins in a GA-dependent manner in yeast cells (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006). Using a yeast two-hybrid assay, we examined the interaction preference between GID1 and DELLA proteins from rice, S. moellendorffii, and P. patens by measuring β-galactosidase (β-gal) activity (Figure 6A). We performed the experiment in the presence of 10−5 M GA4, which was an excess condition for the Os GID1–SLR1 interaction in yeast (Ueguchi-Tanaka et al., 2007b). Sm GID1a interacted with Sm DELLA1 and Os SLR1 with similar effectiveness and also interacted very weakly with Sm DELLA2. Sm GID1b interacted actively with Sm DELLA1 and Os SLR1 but less actively with Sm DELLA2. Os GID1 interacted with Os SLR1 but not with Sm DELLA1 or Sm DELLA2. Pp GID1L1 and Pp GID1L2 did not interact with any DELLA proteins. Similarly, the Pp DELLAL proteins did not interact with any of the GID1 proteins. We confirmed the expression of GID1 and DELLA proteins in yeast cells by an immunoblot analysis (data not shown), indicating that the lack of β-gal activity was caused not by a failure in protein production in yeast cells but by a lack of protein–protein interaction.
Figure 6.
Interaction between GID1 and DELLA Homologs in Yeast Cells.
(A) Interaction of various combinations of GID1 and DELLA proteins from rice, S. moellendorffii, and P. patens. GID1 proteins and DELLA proteins were used as bait and prey, respectively. β-Gal activity was detected in a liquid assay with Y187 transformants (means ± sd; n = 3). Only results in the presence of 10−5 M GA4 are presented, since no activity > 1.4 Millar units was detected in the absence of GA4 in any combination.
(B) and (C) Effects of various GAs on the Sm GID1s–Sm DELLA1 interaction in yeast cells. A two-hybrid assay was performed using Sm GID1s as bait and Sm DELLA1 as prey in the presence of 10−5 M of various GAs. β-Gal activity was determined as in (A) (means ± sd; n = 3).
(D) Chemical structures of GAs used in this study. Structures essential for bioactive GAs are circled in gray (free 2β- and 3β-hydroxylation of the A-ring, γ-lactone structure in the A-ring, and carboxylation of C7). The characteristic structure of each GA compared with GA4 is highlighted in gray. H2-GA4, 16,17-dihydro-GA4; GA4-Me, GA4 methyl ester; GA9-Me, GA9 methyl ester.
We then examined the effect of various GAs at 10−5 M on Sm GID1–Sm DELLA1 interactions in yeast cells (Figures 6B and 6C). These interactions were used as an indirect measure of the binding preferences of Sm GID1 proteins for various kinds of GAs. The structural features of GAs that affect the Sm GID1–Sm DELLA1 interaction do not fit the rules established for GA bioactivity in flowering plants. According to these rules, bioactive GAs should contain 3β-hydroxylation of the A-ring, γ-lactone structure in the A-ring, and carboxylation of C7, whereas 2β-hydroxylation of the A-ring causes inactivation of GA activity (Davis, 2004). By contrast, the effect of GA37 and GA9 (10−5 M) on the Sm GID1–Sm DELLA1 interactions was similar to that of GA4 for Sm GID1a–Sm DELLA1 and was even greater for Sm GID1b–Sm DELLA1 (Figures 6B and 6C). Both GA37 and GA9 are classified as inactive types because GA37 contains a δ-lactone structure in the A-ring and GA9 does not contain 3β-hydroxylation of the A-ring (Figure 6D). Furthermore, for the Sm GID1b–Sm DELLA1 interaction, GA51 and 3-epi-GA4 showed much higher activity than did GA1 or GA3, both of which are bioactive GAs in flowering plants. GA51 contains 2β-hydroxylation rather than 3β-hydroxylation in the A-ring, and 3-epi-GA4 contains 3α-hydroxylation instead of 3β-hydroxylation. The unique effectiveness of the Sm GID1–Sm DELLA1 interaction suggests that the GA selectivity of Sm GID1 proteins differs from that of GID1 proteins in flowering plants (see Discussion). By contrast, interactions between Pp GID1L and Pp DELLAL proteins were not observed in the presence of any of the GAs tested (see Supplemental Figure 1 online).
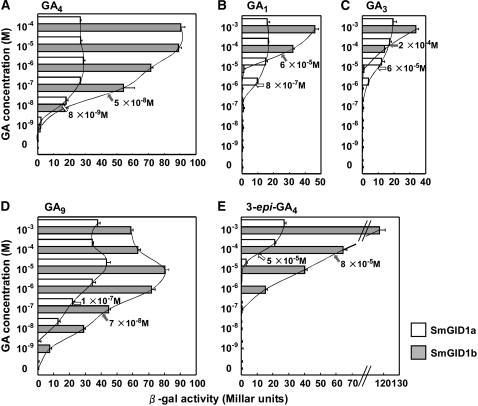
To further study the GA binding properties of Sm GID1 proteins, we examined the dose-dependence of the Sm GID1a– and Sm GID1b–Sm DELLA1 interactions in yeast cells in the presence of GA4, GA1, GA3, GA9, and 3-epi-GA4 (Figure 7). For both interactions, GA4 showed the highest affinity. The Sm GID1b–Sm DELLA1 interaction increased as the GA4 concentration increased from 10−9 to 10−5 M and finally reached a plateau, while the saturation point for Sm GID1a–Sm DELLA1 was almost 10−7 M (Figure 7A). Consequently, the 50% saturation points of the interactions were estimated as 8 × 10−9 M and 5 × 10−8 M, respectively. This indicates that Sm GID1a has higher affinity for GA4 than does Sm GID1b.
Figure 7.
Dose-Dependence of GA4, GA1, GA3, GA9, and 3-epi-GA4 in the Sm GID1s–Sm DELLA1 Interaction in Yeast Cells.
Two-hybrid assay using Sm GID1 proteins as bait and Sm DELLA1 as prey in the presence of various concentrations of GA4 (A), GA1 (B), GA3 (C), GA9 (D), and 3-epi-GA4 (E). β-Gal activity was determined as in Figure 6A (means ± sd; n = 3). The 50% saturation points are indicated by arrows.
The higher GA affinity of Sm GID1a was more obvious when we used GA1 or GA3 as a ligand (Figures 7B and 7C), whereas GA9 and 3-epi-GA4 had much stronger effectiveness in the Sm GID1b–Sm DELLA1 interaction than did GA1 or GA3 (Figures 7D and 7E). These results demonstrate the unique GA selectivity of Sm GID1 proteins, although the interacting affinity between Sm GID1a and GA4 was highest (8 × 10−9 M) among any interactions between Sm GID1 proteins and GAs, as with the flowering plant GID1 proteins (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). The β-gal activity was always higher in the Sm GID1b–Sm DELLA1 interaction than in Sm GID1a–Sm DELLA1 at high concentrations of various GAs, even in the case of low-affinity GAs such as GA1 and GA3. Although it is not clear why the Sm GID1b–Sm DELLA1 interaction always causes higher β-gal activity than the Sm GID1a–Sm DELLA1 interaction, it is possible that the Vmax of the Sm GID1b–Sm DELLA1 interaction may be higher than that of the Sm GID1a–Sm DELLA1 interaction.
Functional Analysis of GA Signaling Genes of S. moellendorffii and P. patens in Transgenic Rice Plants
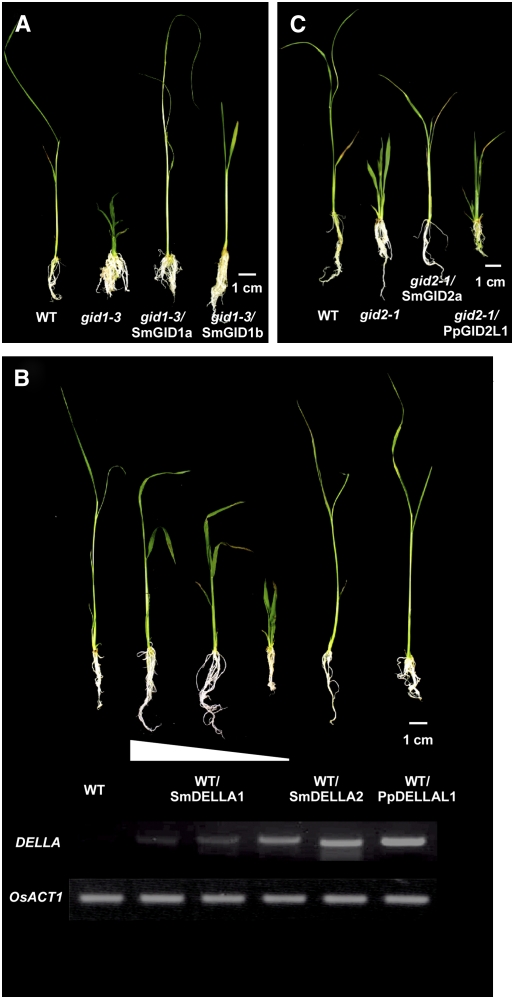
To test whether the putative GA signaling genes in S. moellendorffii possess similar functions to their flowering plant counterparts in vivo, we produced transgenic rice plants overexpressing each gene under the control of the constitutive Act1 promoter (proAct1) (McElroy et al., 1990). When we introduced Sm GID1a and Sm GID1b into the gid1-3 background (Ueguchi-Tanaka et al., 2005), all transgenic plants carrying the Sm GID1a (n = 6) or Sm GID1b (n = 9) DNA sequences were taller than nontransgenic gid1 dwarf plants (Figure 8A; see Supplemental Figure 2 online). Sm GID1b plants were shorter than Sm GID1a or wild-type plants (Figure 8A; see Supplemental Figure 2 online), even though the levels of Sm GID1 mRNA were similar in both Sm GID1a and Sm GID1b transgenic plants (data not shown). This suggests that the GA receptor function of Sm GID1b is weaker than that of Sm GID1a in rice cells. This is reasonable when we consider the GA preference of Sm GID1b and that the dominant GA in the rice plant at the vegetative stage is GA1 (Kobayashi et al., 1989): Sm GID1b is much less sensitive than Sm GID1a to GA1 under physiological conditions (<10−5 M) (Figure 7B).
Figure 8.
Complementation of the Dwarf Phenotype of Rice gid1 and gid2 Mutants by Expression of Sm GID1s, Sm GID2a, and Pp GID2L1, and Phenotypic Analysis of Rice Overproducers of Sm DELLAs and Pp DELLAL1.
(A) Gross morphology of the wild type, gid1-3, and Sm GID1a and Sm GID1b overproducers in gid1-3 mutants at the young seedling stage. Expression of Sm GID1a and Sm GID1b completely and partially complemented the gid1-3 dwarf phenotype, respectively.
(B) Gross morphology of the wild type and Sm DELLA1, Sm DELLA2, and Pp DELLAL1 overproducers in wild-type T65 plants. The panels at bottom present the results of RT-PCR analysis of each transcript of the transgene. Higher expression of Sm DELLA1 was associated with more severe dwarfism of transformants, while there was no dwarfism in transformants highly expressing Sm DELLA2 or Pp DELLAL1. The Os ACT1 gene was used as an internal standard to ensure that the same amount of cDNA was used as the DNA template in each PCR. Results presented are representative of three independent experiments.
(C) Gross morphology of the wild type, gid2-1, and Sm GID2a and Pp GID2L1 overproducers in the gid2-1 mutant at the young seedling stage. Only Sm GID2a partially complemented the gid2-1 dwarf phenotype.
To examine DELLA-like protein function, we introduced Sm DELLA1, Sm DELLA2, and Pp DELLAL1 clones into wild-type Taichung 65 (T65) rice. We used a wild-type background because the regeneration frequency of rice slr1 callus is quite low, and if the introduced proteins had little or no DELLA function, it would be difficult to obtain any transgenic plants. On the other hand, transgenic plants overproducing functional DELLA proteins would be expected to have a semidwarf phenotype because of the suppression of GA action by accumulated DELLA proteins (Itoh et al., 2002). As expected, the plants overexpressing Sm DELLA1 showed a dwarf phenotype; the severity of dwarfism appeared to be associated with the level of Sm DELLA1 expression (Figure 8B; see Supplemental Figure 2 online). On the other hand, we did not see dwarf plants among >30 independent plants transformed with constructs for Sm DELLA2 or Pp DELLAL1 overexpression. These observations demonstrate that Sm DELLA2 and Pp DELLAL1 do not suppress GA signaling in rice cells or that their effects are at an undetectable level.
Introduction of Sm GID2a partially rescued the dwarf phenotype of gid2-1 (Figure 8C; see Supplemental Figure 2 online), indicating that Sm GID2a functions at least in part as a GA-related F-box protein in rice. The lack of complete rescue of the gid2 dwarf phenotype by transformation with Sm GID2a may be caused by an incomplete degradation of rice DELLA protein SLR1 by Sm GID2a, possibly because of the low interaction between SLR1 and Sm GID2a. On the other hand, the dwarf phenotype of gid2-1 was not rescued by overexpression of Pp GID2L1; this result is not surprising given the sequence differences observed (Figure 4). Again, the homologous gene in P. patens was unable to substitute for its counterpart gene in rice.
Expression Analysis of GA-Related Genes in S. moellendorffii
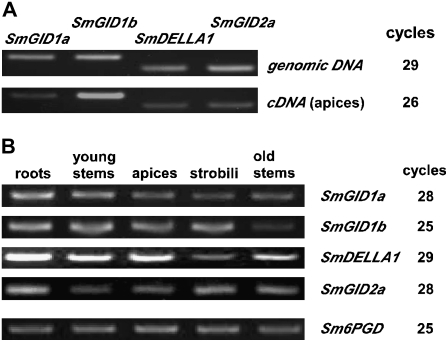
The above results demonstrate that S. moellendorffii has all the components of GA signaling we have examined: GID1, DELLA, and GID2 proteins. To confirm the expression of these genes in S. moellendorffii, we performed RT-PCR using total RNA isolated from various organs (Figure 9). For this experiment, we prepared primers specific for Sm GID1a, Sm GID1b, Sm DELLA1, and Sm GID2a. The primers produced almost the same amount of PCR product for each gene when used with S. moellendorffii genomic DNA as the template (Figure 9A), indicating that they amplify each product at a similar efficiency. We then compared the expression levels of these genes using cDNA produced from total RNA of the vegetative shoots of S. moellendorffii (Figure 9A). There was more PCR product from Sm GID1b than from the other genes, indicating that Sm GID1b is more actively transcribed in the shoots than the other genes. As the expression levels of each gene differed, we selected a suitable cycle number for PCR amplification of each gene to compare the amounts of mRNA in various organs (Figure 9B). The products of each gene were observed in various organs, and the levels did not appear to differ much among organs. This lack of organ-specific expression is similar to the expression patterns of GID1, DELLA, and GID2/SLY1 in rice and Arabidopsis (Kaneko et al., 2003; McGinnis et al., 2003; Gomi et al., 2004; Nakajima et al., 2006).
Figure 9.
Expression of GA Signaling Genes in S. moellendorffii.
(A) PCR was performed using genomic DNA from S. moellendorffii or cDNA produced from the apical part of vegetative shoots as a DNA template. The results of genomic PCR indicate that the primers for each gene work similarly. Compared with other genes, Sm GID1b is preferentially expressed in the apical part of vegetative shoots. The number of PCR cycles used is shown at right.
(B) RT-PCR of GA signaling genes in various organs of S. moellendorffii. Total RNA was isolated from the organs indicated at the top, and 2 μg was used for the RT reaction. “Roots” indicates a mixture of roots and rhizophores, and “apices” indicates apical parts of the vegetative shoot. Expression of the Sm 6PGD gene, an ortholog of the Sr 6PGD gene (Tanabe et al., 2003), was used as a control. The number of PCR cycles used is shown at right. Results presented are representative of at least three independent experiments.
GA Signaling and Synthesis in S. moellendorffii
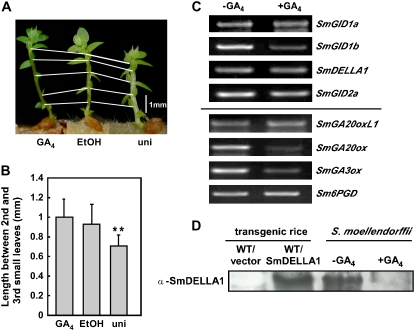
The presence of functional homologs of GID1, DELLA, and GID2 in S. moellendorffii strongly indicates that GA functions as a bioactive substance in this species. To confirm this, we treated S. moellendorffii bulbils with 10−5 M GA4 or 10−6 M uniconazole, a GA biosynthesis inhibitor, and observed the effect on plant growth at ∼2 weeks after germination by measuring the length between adjacent small leaves. The stem length of GA4-treated plants was increased slightly compared with that of the untreated plants, although the unpaired t test did not indicate a significant difference (P > 0.05) (Figures 10A and 10B). By contrast, the uniconazole treatment caused a significant dwarf phenotype compared with the untreated plants (P < 0.01). However, treatment with 10−5 M GA4 at 10 d after uniconazole treatment did not restore its inhibitory growth effect on the plant (data not shown).
Figure 10.
Effects on Growth, Feedback Regulation of GA-Related Genes, and Degradation of Sm DELLA1 of GA4 Treatment in S. moellendorffii.
(A) Gross morphology of 10−5 M GA4-treated and 10−6 M uniconazole-treated plants. Ethanol (0.01%) solution was used as a control. The positions of corresponding small leaves on each plant are connected with white lines. EtOH, ethanol; uni, uniconazole.
(B) Length of stem between the second and third small leaves from the bottom (±se; n = 12, 20, and 10 for GA4, ethanol, and uniconazole treatments, respectively). ** Significant difference (P < 0.01) compared with control (ethanol) treatment from the unpaired t test analysis.
(C) Downregulation of GA-related genes in S. moellendorffii after GA4 treatment. Plants were treated for 3 d with or without GA4 at a concentration of 10−4 M, total RNA was isolated from young shoots, and RT-PCR was performed. The Sm 6PGD gene was used as a control. Results presented are representative of at least three biological replicates.
(D) Disappearance of Sm DELLA1 protein after GA4 treatment. S. moellendorffii plants were treated with either buffer only or 10−4 M GA4 for 12 h. Sm DELLA1 protein was detected by immunoblot analysis of crude protein extract using the anti-Sm DELLA1 antibody. The specificity of the antibody was confirmed using transgenic rice overexpressing Sm DELLA1 (positive control) and transgenic rice possessing vector only (negative control).
We then examined the GA signaling pathway in S. moellendorffii using a molecular biological approach. GA negatively regulates the expression of genes encoding GA-synthesizing enzymes such as GA20 oxidase and GA3 oxidase (Chiang et al., 1995; Phillips et al., 1995). To examine the downregulation of GA20 and GA3 oxidase genes by GA in S. moellendorffii, we screened and isolated genes homologous with the GA20 and GA3 oxidases. We found three GA20 oxidase–like genes and one GA3 oxidase–like gene in the S. moellendorffii genome (see Supplemental Figure 3 online). We examined the GA20 or GA3 oxidase activity of each gene's protein product and confirmed that one of the GA20 oxidase candidates, Sm GA20ox, and one GA3 oxidase–like gene, Sm GA3ox, encode functional enzymes (see below).
We examined the expression of GA-related genes in shoots of S. moellendorffii with or without GA4 treatment (Figure 10C). With GA4 treatment, the expression of Sm GID1b, Sm GA20ox, and Sm GA3ox decreased markedly, while the expression of Sm GID1a, Sm DELLA1, Sm GID2a, and Sm GA20oxL1 was unchanged. Downregulation of the GID1 gene by GA treatment also occurs in rice and Arabidopsis seedlings (Griffiths et al., 2006, M. Ueguchi-Tanaka, unpublished data), indicating that suppression of the GID1 gene by GA is a common phenomenon in vascular plants. Furthermore, the downregulation of Sm GA20ox and Sm GA3ox by GA strongly suggests that the negative feedback regulation of GA synthesis genes mediated by the GA signaling pathway functions in S. moellendorffii as in flowering plants.
We further examined the GA-dependent degradation of Sm DELLA1 in young shoots of S. moellendorffii because this phenomenon is one of the most direct and sensitive events under the control of the GA signaling pathway mediated by the GID1/DELLA system. First, we performed a protein blot analysis of the crude extract of young shoots of S. moellendorffii with the antibody to the rice DELLA protein, SLR1 (Itoh et al., 2002), but this antibody did not detect Sm DELLA1 (data not shown). We then produced a specific antibody to Sm DELLA1 and used it for protein gel blot analysis. The antibody recognized a single band, which migrated at the estimated molecular weight of Sm DELLA1. This band was detected in a transgenic rice plant carrying the proAct1-Sm DELLA1 construct but not in a control plant (Figure 10D). An immunoreactive band with the same mobility was detected in the extract of young shoots of S. moellendorffii; this band almost disappeared within 12 h of the application of 10−4 M GA4 (Figure 10D). This result clearly demonstrates that the GA perception pathway mediated by the GID1/DELLA system occurs in S. moellendorffii.
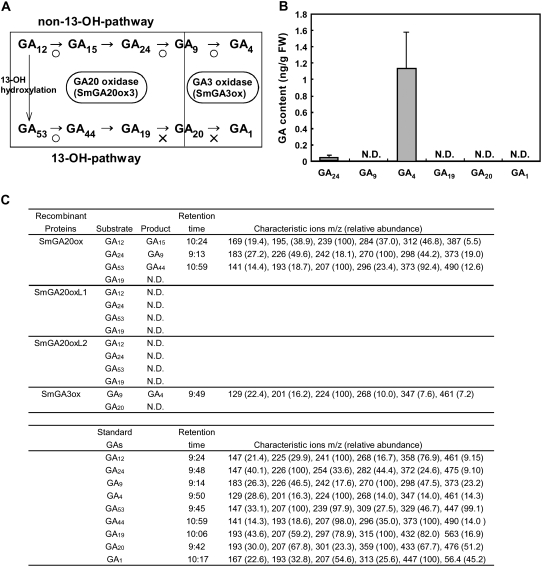
Next, we directly examined the amount of bioactive GA in S. moellendorffii (Figure 11B). Some level of GA4 and a very low level of GA24 were detected in young shoots, while no GA1 or GA19 was found. Identification of GA4 was confirmed by GC-MS (see Supplemental Figure 4 online). Detection of non-13-OH (hydroxy)-GAs (such as GA4 and GA24) but not 13-OH-GAs (such as GA1, GA19, and GA20) suggests that S. moellendorffii may preferentially produce non-13-OH-GAs and use GA4 as a bioactive GA. This is reasonable given that Sm GID1a and -1b preferentially interact with GA4 but not with GA1 (Figures 6B, 6C, 7A, and 7B). To test this possibility, we examined the enzymatic activity of Sm GA20ox, Sm GA20oxL1, Sm GA20oxL2, and Sm GA3ox produced in E. coli. The recombinant Sm GA20ox catalyzes the conversion of GA12 to GA15, GA24 to GA9, and GA53 to GA44, while the conversion of GA19 to GA20 was not observed (Figures 11A and 11C). This suggests that Sm GA20ox catalyzes the conversion of GA12 to GA9 but converts GA53 to GA20 inefficiently if at all. Similarly, Sm GA3ox catalyzed the conversion of GA9 to GA4 but not GA20 to GA1 (Figures 11A and 11C). Two other GA20 oxidase homologs in S. moellendorffii, Sm GA20oxL1 and Sm GA20oxL2, did not catalyze any conversions. These results demonstrate that Sm GA20ox and Sm GA3ox catalyze the GA20 oxidation and the GA3 oxidation of non-13-OH-GAs but do not efficiently use 13-OH-GAs as substrates. This observation supports the idea that S. moellendorffii preferentially produces non-13-OH-GAs and uses GA4 as a bioactive GA. We also examined the endogenous GA content (see Supplemental Table 1 online) and enzymatic activity of homolog proteins of GA20 oxidase and GA3 oxidase in P. patens (see Supplemental Table 2 online), but we did not detect either endogenous GA or GA oxidase activity in this species.
Figure 11.
GA Content and in Vitro Activity of GA20 Oxidase and GA3 Oxidase Homologs in S. moellendorffii.
(A) The late stage of GA biosynthesis. In many flowering plants, GA12 is often converted to GA53 by hydroxylation at C-13. GA12 or GA53 is converted, via parallel pathways, to other GAs through a series of oxidations at C-20 to finally form GA9 or GA20 by GA20 oxidase. GA9 or GA20 is oxidized to the bioactive GA4 or GA1 by GA3 oxidase. Sm GA20ox can catalyze the steps from GA12 to GA15, from GA24 to GA9, and from GA53 to GA44 (circles), but not from GA19 to GA20 (crosses). Sm GA3ox catalyzes the step from GA9 to GA4 but not the step from GA20 to GA1.
(B) GA content of S. moellendorffii shoots. One gram of S. moellendorffii shoots was used for GA content measurement by liquid chromatography–mass spectrometry analysis (see Methods). Tests were performed on four independent plants. Error bars indicate sd. FW, fresh weight; N.D., no expected product was detected.
(C) In vitro enzymatic activity of GA20 oxidase and GA3 oxidase homologs in S. moellendorffii. N.D., no expected product was detected.
DISCUSSION
Evolution of GA Perception Mediated by the GID1/DELLA System
In silico screening of the genomic DNA of S. moellendorffii and P. patens was used to identify candidate genes for each of three GA signal-related genes: GID1, DELLA, and GID2. To determine whether any of these candidates are actually involved in GA signaling, we studied their biochemical and biological properties. We used an in vitro GA binding assay and a yeast two-hybrid assay to examine the GA binding activity of GID1 candidate proteins and their GA-dependent interaction with DELLA proteins. We examined the ability of the GID2 candidate genes to complement a rice gid2 mutant. We also used transgenic experiments with rice mutants and wild types to study the GID1 and DELLA candidates, respectively.
All results from these experiments demonstrate that S. moellendorffii contains bioactive GID1, DELLA, and GID2 counterpart genes. Furthermore, the expression of GA biosynthesis genes, such as Sm GA20ox and Sm GA3ox, and the GA receptor Sm GID1b in S. moellendorffii is downregulated by GA treatment, similar to the GA feedback regulation in flowering plants (Chiang et al., 1995; Phillips et al., 1995; Griffiths et al., 2006). The GA-dependent degradation of DELLA-like protein also strongly supports the presence of a similar GA perception system in S. moellendorffii. However, treatment with GA4 failed to restore the dwarf phenotype of S. moellendorffii caused by uniconazole, suggesting either that the effect of uniconazole was not due to the inhibition of GA biosynthesis or that the timing of our treatment was inappropriate. Although we did not observe clear changes in strobilus formation or sporulation when S. moellendorffii was treated with GA4 or uniconazole (data not shown), it is still possible that unusual GAs in flowering plants may be involved in reproduction, especially in the development of sexual organs, because some GAs (antheridiogens) are involved in the formation of sexual organs in fern gametophytes (Yamauchi et al., 1996; Banks, 1999; Menéndez et al., 2006). Taken together, we conclude that S. moellendorffii has a GA perception mechanism mediated by the GID1/DELLA system similar to that found in flowering plants. Further studies on the biological function of GA in S. moellendorffii should expand our knowledge of GA function.
In contrast with S. moellendorffii, we found no functional homologs in the moss P. patens, although there are some genes encoding proteins homologous with rice GID1, SLR1, and GID2. The GID1 homologs in P. patens, Pp GID1L1 and Pp GID1L2, did not show in vitro GA binding activity in two different analyses, nor did they interact with any DELLA proteins in the presence of various kinds of GAs in yeast cells. Transgenic rice expressing the gene encoding Pp DELLAL1, a protein that does not contain typical DELLA/TVHYNP domains but contains conserved GRAS domains similar to those in seed plant DELLA proteins, did not show any GA-insensitive phenotypes. Furthermore, the overexpression of Pp GID2L1 in rice gid2-1 did not rescue its dwarf phenotype. These results suggest that P. patens does not contain a GA perception mechanism mediated by the GID1/DELLA system, although it is possible that GAs other than those that we tested may initiate the GID1/DELLA pathway in this organism. Another possibility is that we did not find functional homologs of GID1, DELLA, and GID2 due to the incomplete sequence of the P. patens genome. However, we found several homologs of the auxin receptor TIR1 and auxin signal components such as Aux/IAA and ARF (Dharmasiri and Estelle, 2002) in the P. patens genome (data not shown), suggesting that we would have found a functional homolog to at least one of the three GA signal–related genes if it were present in the genome.
Very recently, a similar study on the evolution of the GA/DELLA mechanism in Arabidopsis, Selaginella kraussiana, and P. patens was reported (Yasumura et al., 2007). Although the authors suggest that the GA perception mechanism emerged after bryophytes and vascular plants diverged, which is consistent with our observations, some intriguing differences were also noted. First, Pp GLP1 (corresponding to Pp GID1L1) and Sk DELLA (S. kraussiana DELLA protein) interacted in a GA-independent manner in the yeast two-hybrid assay, whereas Pp DELLAa (corresponding to Pp DELLAL1) did not bind to GID1 of any species tested. Second, introducing GFP-Pp DELLAa into an Arabidopsis gai-t6 rga-24 ga1-3 triple mutant strain (null for the DELLA proteins GAI and RGA and containing almost no GA) resulted in dwarfism, whereas the triple mutant normally confers a tall mutant phenotype. From these observations, the authors suggest that the ability of GID1 to interact with DELLA, and the growth-restraining ability of DELLA, evolved before bryophyte and lycophyte divergence, whereas the ability of DELLA to bind to GID1 evolved at a later stage, between the divergence of bryophytes and lycophytes.
These results are clearly different from our findings. We did not detect an interaction of Pp GID1L proteins with any DELLA protein tested (Figure 6A), and overproduction of Pp DELLAL1 in our rice plants did not cause dwarfing (Figure 8B; see Supplemental Figure 2 online). In addition, the unique GA preference observed for Sm GID1 proteins (Figures 6B and 6C) was not observed for the S. kraussiana GID1. The use of different plant species (Arabidopsis versus rice, S. kraussiana versus S. moellendorffii) could be the major cause of these differences. For Pp DELLAL1, differences in the growth-restraining activity observed for rice and Arabidopsis may be caused not merely by differences in plant species but may also depend on whether mutant or wild-type plants were used (gai-t6 rga-24 ga1-3 for Arabidopsis, wild-type T65 for rice).
Green plants first colonized land around the mid-Ordovician period (470 million years ago) and subsequently diverged into various lineages (Kenrick and Crane, 1997). The mosses and vascular plants diverged early in the Silurian period (430 million year ago) (Kenrick and Crane, 1997). The presence of a GID1/DELLA-mediated GA perception mechanism in S. moellendorffii, which diverged early from the lineage of ferns and seed plants, strongly suggests that the last common ancestor of vascular plants used a GID1/DELLA-mediated GA perception system. By contrast, failure to identify functional homologs of GID1, DELLA, GID2, GA20 oxidase, or GA3 oxidase in P. patens suggests that bryophytes may not use GA as a bioactive substance, although the possibility that P. patens utilizes GAs that were not tested in this study, or that it contains an alternative GA signaling pathway, cannot be excluded. It is also necessary to examine whether other bryophyte species (mosses, hornworts, and liverworts) lack the GID1/DELLA-mediated perception system and functional homologs of GA20 oxidase and GA3 oxidase. Based on our current data, we hypothesize that the usage of GA as a bioactive substance was a key event in the evolution of the body plans of vascular plants. However, further studies of GA perception, response, and biosynthesis in nonflowering land plants are necessary to evaluate this hypothesis.
Properties of Sm GID1s
The ligand selectivity of Sm GID1 proteins, especially Sm GID1b, differed from that of GID1 proteins in flowering plants (Figures 6 and 7). For example, the effectiveness of GA37 and GA9 at 10−5 M was similar to that of GA4, the most effective GA in flowering plants, for the Sm GID1a–Sm DELLA1 interaction and higher than that of GA4 for the Sm GID1b–Sm DELLA1 interaction. By contrast, bioactive GAs such as GA1 and GA3 had a very low effect on the Sm GID1b–Sm DELLA1 interaction and an intermediate effect on the Sm GID1a–Sm DELLA1 interaction. Furthermore, GAs known to be inactive in flowering plants, such as GA51 and 3-epi-GA4, had an apparent effect on the Sm GID1b–Sm DELLA1 interaction.
The relatively high effectiveness of 3-epi-GA4 indicates that a stereoscopic hydroxylation structure at the C3 site is much less important for Sm GID1s, especially for Sm GID1b, than for GID1s in flowering plants. The intermediate effectiveness of GA51 also indicates that the absence of 3β-hydroxylation and the presence of 2β-hydroxylation does not prevent the interaction of the GA molecule with Sm GID1 proteins. Moreover, the high effectiveness of GA37 demonstrates that the γ-lactone structure in the A-ring can be replaced with the δ-lactone structure. These results suggest that Sm GID1 proteins, especially Sm GID1b, do not recognize the A-ring of GA as strictly as do GID1 proteins in flowering plants. By contrast, the low or almost nonexistent effect of GA1 and GA3 at the concentration of 10−5 M on the Sm GID1a–Sm DELLA1 or Sm GID1b–Sm DELLA1 interaction suggests that Sm GID1 proteins more strictly recognize the C-ring structure of GA than do GID1s in flowering plants. Specifically, Sm GID1b discriminates between GAs based on the presence or absence of 13-hydroxylation: GA4 is active, whereas GA1 is inactive.
Such discrimination was also observed in the reaction of GA3 oxidation by Sm GA3 oxidase; that is, Sm GA3 oxidase actively catalyzed the conversion of GA9 to GA4 but not that of GA20 to GA1 (Figure 11C). These results indicate that S. moellendorffii specifically metabolizes non-13-OH-GAs to produce GA4 as an active GA and specifically perceives GA4 with Sm GID1 proteins. In flowering plants, 13-OH- and non-13-OH-pathways are used differentially from species to species. For example, GA1 predominates in cereals and legumes and GA4 predominates in Arabidopsis and cucurbits (Kobayashi et al., 1989; Fleet et al., 2003; Davis, 2004; Lange et al., 2005). Moreover, the two pathways are also used differentially among organs within a species or in response to different environmental conditions (Davis, 2004). It is possible that the non-13-OH-pathway is the default state of GA synthesis, and the end product of this pathway, GA4, is the most active GA in terms of affinity to the GID1/GA receptor. In this context, the 13-OH-GA synthetic pathway may have come into existence at later stage(s) during the evolution of vascular plants. If so, it is interesting to speculate why vascular plants developed the 13-OH-GA synthetic pathway during their evolution, given the biological significance of 13-OH–type GAs such as GA1 and GA3.
METHODS
Plant Materials and Growth Conditions
Physcomitrella patens subsp patens, originally collected in Gransden Wood (Ashton and Cove, 1977), was grown on BCDATG medium at 25°C under continuous light (Nishiyama et al., 2000). Vegetatively propagated protonemata containing young gametophores at 13 d after inoculation were used for RNA extraction and analysis of endogenous GA content. Selaginella moellendorffii was grown in the laboratory at room temperature. A japonica-type rice cultivar (Oryza sativa cv Taichung 65) and its irradiation-induced mutants, gid1-3 (Ueguchi-Tanaka et al., 2005) and gid2-1 (Sasaki et al., 2003), were used to create transgenic rice plants. Rice plants were grown in a growth chamber at 30°C under continuous light.
Screening of GA-Related Genes in S. moellendorffii and P. patens, and Phylogenetic Analysis
The amino acid sequences of rice GID1, SLR1, GID2, GA20ox2, and GA3ox1 were used as queries to screen the available S. moellendorffii and P. patens genomic databases (http://selaginella.genomics.purdue.edu/cgi-bin/blast_tmpl_s.cgi and http://moss.nibb.ac.jp/) by TBLASTN, and candidate genes were selected for preliminary phylogenetic analyses. The genes used in the preliminary analyses along with their e-values and bit thresholds are as follows: Sm GID1 (〈e-50, bit 〉 200), Sm DELLA (〈e-70, bit 〉 250), Sm GID2 (〈e-03, bit 〉 30), Sm GA20ox (〈e-30, bit 〉 100), Sm GA3ox (〈e-20, bit 〉 70), Pp GID1L (〈e-20, bit 〉 100), Pp DELLAL (〈e-100, bit 〉 300), Pp GID2L (〈e-04, bit 〉 30), Pp GA20ox (〈e-26, bit 〉 100), and Pp GA3ox (〈e-20, bit 〉 100). For GA oxidase genes, candidate sequences were also manually checked and selected for further analyses based on the presence of conserved amino acids important for their function.
After obtaining candidate sequences, the deduced full coding region for each gene was PCR-amplified using cDNA of S. moellendorffii or P. patens as the template. Exon and intron regions were confirmed by sequencing the PCR products. Highly similar sequences from other species were identified with PSI-BLAST (Altschul et al., 1997) from a combined data set including the National Center for Biotechnology Information nonredundant data set, the poplar v1.1 proteins (proteins.Poptr1_1.JamboreeModels.fasta) data set, and the Computational Biology and Functional Genomics Laboratory (http://compbio.dfci.harvard.edu/tgi/plant.html). The queries, inclusion limits, and e-value thresholds were set differently for different families (see Supplemental Table 3 online).
Tentative clusters of expressed sequence tags in pine (Pinus taeda) were searched with TBLASTN using SLR1, GID1, and GID2 as queries, and the translated sequences were obtained. Because GA3 oxidases were identified as distantly related in the data set of GA20 oxidases and the outgroup found in the search had overlaps, the GA3 oxidase and GA20 oxidase sequences were analyzed together. The sequences of each data set were aligned with P. patens and S. moellendorffii sequences determined in this study using the einsi algorithm of MAFFT version 6.2 (Katoh et al., 2005). The unambiguously aligned regions were manually selected, and partial sequences lacking those regions were removed with MacClade version 4.08 (http://macclade.org/index.html). A neighbor-joining (NJ) tree (Saitou and Nei, 1987) was obtained with PROTDIST and NEIGHBOR in the PHYLIP version 3.65 package (http://evolution.genetics.washington.edu/phylip.html). Bootstrap analyses were performed by repeating the procedure on 100 data sets prepared with SEQBOOT.
To find a maximum likelihood tree, distantly related outgroup sequences were reduced and ingroup sequences from rice, Arabidopsis, pine, S. moellendorffii, and P. patens plus some well-characterized genes were selected. A distance matrix was obtained with ProtML in the MOLPHY-2.3 package (Adachi and Hasegawa, 1996) under the JTT model (Jones et al., 1992), and NJ trees were obtained with NJDist. Maximum likelihood trees were searched with the nearest neighbor interchange algorithm implemented in ProtML as a local rearrangement search starting with the NJ tree. Bootstrap analyses were performed by repeating the procedure on 1000 data sets prepared with SEQBOOT. For the alignments shown in Figures 2 to 4, sequences were aligned with ClustalW version 1.81 with default parameters (Thompson et al., 1994; http://align.genome.jp/), followed by manual alignment. Boxshade (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html) was used to draw the alignments with default parameters.
Genomic DNA Isolation, RNA Isolation, and cDNA Synthesis
For isolation of the genomic DNA from S. moellendorffii and total RNA of S. moellendorffii and rice, plants were ground with liquid nitrogen in a mortar and pestle. Genomic DNA was isolated using ISOPLANT (Nippongene) according to the instruction manual. Total RNA of S. moellendorffii and rice was isolated using Trizol reagent (Invitrogen). Total RNA from P. patens was extracted according to Hasebe et al. (1998) and further purified with ISOGEN-LS (Wako Pure Chemical). Total RNA was treated with RNase-free DNase for 30 min at 37°C, followed by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. Single-stranded cDNA was synthesized from 2 μg of total RNA using the OmniScript reverse transcriptase kit (Qiagen), according to the instruction manual.
Plasmid Construction
The deduced full coding region of each gene was obtained by PCR using cDNA of S. moellendorffii or P. patens as the template. For each product, the sequence was confirmed to ensure that no mutations were introduced. The primers used in this study are listed in Supplemental Table 4 online. For the GID1 genes, Sm DELLA genes, Pp DELLAL1, and Sm GID2a, each PCR product was first cloned into the pCR4 Blunt-TOPO vector (Invitrogen) and the plasmid was further used for subcloning into various vectors. Pp GID2L1 and several GA20ox- and GA3ox-like genes were cloned directly into the vector of purpose.
For the GA binding assay, GID1 genes each containing suitable restriction enzyme sites at both ends were cloned into the pET32a vector (Novagen/Merck Biosciences) to produce the Trx·His-GID1 plasmids. Cloning was performed using the BamHI-HindIII site for Os GID1, Sm GID1b, and Pp GID1L2; using SalI-NotI for Sm GID1a; and using BamHI-XhoI for Pp GID1L1. DELLA genes with suitable restriction enzyme sites at both ends were cloned into the pGEX-4T-1 vector (GE Healthcare), using the EcoRI site for Os SLR1 and EcoRI-NotI for the Sm DELLA genes, to produce the GST-DELLA plasmids.
For the yeast two-hybrid assay, Sm GID1 and Pp GID1L genes containing appropriate restriction sites at both ends were cloned into a pGBKT7 DNA-BD shuttle vector (Clontech), using the NdeI-SmaI site for Sm GID1a and Pp GID1L1, NcoI-SmaI for Sm GID1b, and NdeI-EcoRI for Pp GID1L2, to produce pGBKT7 DNA-BD-GID1 bait plasmids. Similarly, the entire coding region of each Sm DELLA and Pp DELLAL sequence containing appropriate restriction sites at both ends was cloned into the pGADT7 AD vector (Clontech) using the NdeI-SmaI site to produce pGADT7 AD-DELLA prey plasmids. The Os GID1 bait plasmid and the Os SLR1 prey plasmid were constructed as described previously (Ueguchi-Tanaka et al., 2005).
To construct vectors for the production of transgenic rice, Sm GID1, Sm DELLA, and Sm GID2a genes containing appropriate restriction sites were introduced at the site between proAct1 (McElroy et al., 1990) and the NOS terminator of the binary vector pActNos/Hm2 (Sentoku et al., 2000) to produce proAct1-Sm GID1, -Sm DELLA, and -Sm GID2 transformation vectors. The XbaI-SmaI site was used for cloning the Sm GID1, Sm DELLA, and Sm GID2a genes, and the XbaI site was used for Pp GID2L1. For Pp DELLAL1, the PCR product cloned in pCR4-TOPO (Invitrogen) was digested with NotI/SmaI, blunt-ended using a DNA blunting kit (Takara), and cloned into the SmaI site of the vector.
The full-length coding regions for the GA20 oxidases and GA3 oxidases containing appropriate restriction sites were cloned into pMAL-c2x (New England Biolabs), using BamHI-HindIII sites for Sm GA20oxL1, BamHI-XhoI for Sm GA20oxL2 and Pp GA20oxL1, HindIII for Sm GA20ox, and NcoI-BamHI for Sm GA3ox and Pp GA3oxL, to produce MBP fusion plasmids.
Production of Recombinant Proteins
Escherichia coli BL21 (DE3) pLysS Rosseta-gami 2 (Novagen) was used as a host strain for recombinant protein production. To produce recombinant Trx·His-GID1 proteins for use in the GA binding assay, 10 mL of precultured cells was added to 500 mL of Luria-Bertani medium in a 2-liter flask and cultured at 37°C until the OD600 was 0.4 to 0.6. Induction of recombinant proteins was performed by the addition of 0.01 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and further incubated at 16°C for 18 h. Cells were harvested and resuspended with buffer A (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM imidazole, and 0.1% Triton X-100). The cells were lysed by sonication (20 kHz, 10 s × 20 times). Each lysate was centrifuged at 16,000g for 30 min, and the supernatants were mixed with 400 μL of TALON Metal Affinity Resin (Clontech) and rotated for 2 h at 4°C. The resin was washed five times with buffer A and eluted five times with 400 μL of 500 mM imidazole in buffer A. Five portions of eluate were gathered and desalted using a PD-10 column (GE Healthcare) equilibrated with buffer B (20 mM Tris-HCl, pH 7.5, 0.15 M NaCl, and 2 mM 2-mercaptoethanol [2-ME]). Preparation of the Trx·His-GID1 proteins for the ITC assay was identical except that buffer A was adjusted to pH 9.4 and ITC buffer containing 20 mM PBS, pH 9.4, 100 mM NaCl, and 2.5 mM 2-ME was used in the PD-10 column desalting procedure.
For the production of recombinant GST-DELLA proteins, the cell culture and induction were performed as for Trx·His-GID1, except that the induction temperature was 24°C instead of 16°C. Cells were harvested and resuspended with buffer C (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, and 1 mM DTT). The cells were lysed by sonication (20 kHz, 10 s × 20 times) and 1% Triton X-100 was added. The lysates were centrifuged at 16,000g for 30 min, and the supernatants were mixed with 2 mL of glutathione Sepharose 4B beads (GE Healthcare) and rotated for 2 h at 4°C. The beads were washed five times with PBS containing 1% Triton X-100 and eluted five times with 400 μL of 20 mM glutathione in buffer C. Five portions of the eluate were gathered and desalted using a PD-10 column (GE Healthcare) equilibrated with buffer B. The production of recombinant proteins was confirmed by 7.5% SDS-PAGE. Furthermore, GID1 proteins were confirmed by immunoblot analysis using the Os GID1 antibody as described previously (Ueguchi-Tanaka et al., 2005).
For the production of recombinant MBP-GA20 oxidases and MBP-GA3 oxidases, cell culture and induction were performed as for Trx·His-GID1, except that 0.4 mM IPTG was used instead of 0.01 mM IPTG. Cells were harvested and resuspended with buffer containing PBS, pH 7.4, 10 mM 2-ME, and 0.1 mg/mL lysozyme. After incubation for 30 min on ice, the suspension was frozen and kept at −80°C overnight, thawed on ice, and sonicated (20 kHz, 10 s × 20 times). The lysates were centrifuged at 16,000g for 30 min, and the supernatants were collected by centrifugation and used as crude extracts for each enzyme assay.
GA Binding Assay of GID1s
For the binding assay, 3H4-H2-GA4 was used as the labeled form of GA. GA binding was performed as reported previously (Nakajima et al., 1997) with the following modifications. One hundred microliters of purified Trx·His-GID1s (16 μg) was incubated at room temperature with 100 μL of 3H4-H2-GA4 (6 or 60 pmol), either with an excess of unlabeled GA4 (at a final concentration of 0.125 mM) for nonspecific binding or without unlabeled GA4 for total binding. After 20 min, 100 μL of GST-DELLA or GST alone (16 μg) was added to the solution and incubated for another 40 min. One hundred microliters of the mixture was then fractionated on a NAP-5 column (GE Healthcare). After discarding a void volume binding buffer eluate (600 μL), a 200-μL fraction was collected and its radioactivity was measured. The specific binding activity, which reflected the number of replaceable GA binding sites, was calculated by subtraction of nonspecific binding from total binding.
ITC experiments were performed with a VP-ITC microcalorimeter (MicroCal). The instrument design and its operation have been described in detail elsewhere (Wiseman et al., 1989). The instrument was allowed to equilibrate overnight. Since Pp GID1L proteins are stable only at basic pH, the ITC analysis of Trx·His-Os GID1 and Trx·His-Pp GID1L proteins with GAs was performed at pH 9.4. GAs were prepared by dissolving in ITC buffer (see above) and injected in 8-μL increments (final content of 500 μM) into the sample cell containing 1.4482 mL of GID1 solution (40 μM) at 30°C. The stirrer was kept rotating at 400 rpm during the experiments. The baseline was judged to have reached stability when root mean square noise was <5 ncal/s. The heat produced in the GA-GID1 binding experiment was subtracted from the heat produced in two control experiments. For the controls, injection of GA solution into buffer solution and injection of buffer solution into GID1 solution were performed under the same conditions used in the GA-GID1 binding experiment. Nonlinear fitting of the data was performed using MicroCal Origin 7.0 (Origin-Lab).
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed as described previously (Ueguchi-Tanaka et al., 2005) using the BD Matchmaker Two-Hybrid System 3 (Clontech). Vector cassettes for DNA-BD and DNA-AD were used as negative controls, and Saccharomyces cerevisiae strain Y187 was used as the host. GAs dissolved in ethanol, or ethanol only, were added to the culture medium at a dilution rate of 1:1000. Expression of DNA-BD and DNA-AD fusion proteins was confirmed by immunoblot analysis using anti-c-Myc (Clontech) and anti-HA (Sigma-Aldrich) antibodies. Details of the methods used for the yeast assays can be found in the manufacturer's instructions (Yeast Protocols Handbook PT3024-1; http://www.clontech.com/). Experiments were independently repeated at least three times.
Overexpression of GID1-Like, DELLA-Like, and GID2-Like Genes of S. moellendorffii and P. patens in Rice
proAct1-Sm GID1s, proAct1 DELLAs, and proAct1 GID2s were introduced into rice gid1-3 mutant plants (Ueguchi-Tanaka et al., 2005), wild-type T65 plants, and gid2-1 mutant plants (Sasaki et al., 2003), respectively, by Agrobacterium tumefaciens–mediated transformation (Hiei et al., 1994). Expression of these transgenes in rice shoots was confirmed by RT-PCR as described below. For each transgenic plant, the length of the second leaf sheath was measured.
RT-PCR
cDNAs prepared from various plant parts of S. moellendorffii were used for RT-PCR analysis. Young stems (excluding microphylls), apical parts of vegetative shoots (consisting mainly of microphylls), and a mixture of roots and rhizophores were obtained from plants <6 cm in height. Strobili and old stems were obtained from plants >15 cm in height. RT-PCR was performed in a 50-μL solution containing a 2.5-μL aliquot of cDNA as the DNA template, 0.2 μM gene-specific primers (see Supplemental Table 4 online), 10 mM deoxynucleotide triphosphates, 1 unit of ExTaq DNA polymerase (Takara), and reaction buffer. Amplifications of Sm 6PGD and Os ACT1 cDNAs were performed as controls for S. moellendorffii and rice, respectively, to ensure that equal amounts of cDNA were added to each PCR. The reaction included an initial 5-min denaturation at 94°C, followed by 25 to 31 cycles of PCR (94°C for 30 s, 56°C for 30 s, and 72°C for 30 s), and a final 10-min extension at 72°C. The number of cycles used for amplification with each primer pair was adjusted to be in the linear range. All RT-PCR data are representative of at least three independent experiments.
Antibody Production
GST-Sm DELLA1 recombinant protein was produced in E. coli and subsequently purified by glutathione beads by the same method described above. This protein was used for the production of antibodies after exchanging the buffer for PBS using a PD-10 desalting column (GE Healthcare). Sm DELLA1 polyclonal antibody was produced by immunization of a rabbit (Operon Biotechnologies).
Immunoblot Analysis of the Sm DELLA1 Protein
Crude protein extracts of young S. moellendorffii shoots and seedlings of transgenic rice were prepared by grinding with liquid nitrogen in the presence of sea sand (425 to 850 μm; Wako Pure Chemical) followed by an equal volume of 2× sample buffer (1× sample buffer is 67.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromphenol blue, and 0.1 M DTT) and boiling for 5 min. Protein samples were separated by 7.5% SDS-PAGE and transferred to a Hybond enhanced chemiluminescence nitrocellulose membrane (GE Healthcare) by semidry blotting. The blots were treated with 5% skim milk in TBST (0.1% Tween 20 in 2 mM Tris-HCl, pH 7.6, and 13.7 mM NaCl) for 1 h and subsequently incubated with anti-Os SLR1 antiserum (Itoh et al., 2002) or anti-Sm DELLA1 antiserum for 1 h. Blots were washed three times with TBST for 15 min each. Goat anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody was incubated for 45 min, and blots were washed following the same procedure described above. All reactions were conducted at room temperature. Detection of peroxidase activity was performed according to the instruction manual from Pierce.
GA Treatment of S. moellendorffii
Young S. moellendorffii plants were grown in pots to no more than 5 cm in height. Pots were submerged in water containing 0.1% ethanol, up to about one-third from the bottom, either with or without 10−4 M GA4. To ensure complete infiltration of GA4, plants were sprayed with 10−4 M GA4 solution (containing 0.1% ethanol and 0.02% Tween 20) at 24 and 72 h after the start of submergence. One hour after the second spray, total RNA was isolated from young shoots.
For the DELLA protein disappearance experiment, two different conditions were used, and both showed the disappearance of Sm DELLA1 protein in GA4-treated plants. Plants were either treated as described above for 1 month or dipped completely in a solution containing 10−4 M GA4, 0.1% ethanol, and 0.02% Tween 20 with continuous shaking using a seesaw shaker for 12 h. Plants not treated with GA solution were used as negative controls. Crude protein was extracted as described above.
To observe the effects of GA4 and uniconazole on plant growth, bulbils of S. moellendorffii were treated with either GA4 or uniconazole (each containing 0.01% ethanol) at concentrations of 10−5 M and 10−6 M, respectively. Ethanol solution (0.01%) was used as a negative control. Pots were filled with vermiculite, and Rockwool was placed on top. Pots were then submerged in each solution up to about one-third from the bottom, and the same solution was used to soak the Rockwool. Bulbils of S. moellendorffii were placed on the surface of the Rockwool to germinate. Growth effects were assessed by measuring the length between the second and third small leaves.
Enzyme Assay
Crude extracts were incubated at 30°C with GA substrates (1 μg each) in 200 μL of 100 mM Tris-HCl, pH 7.9, 4 mM ascorbic acid, 4 mM 2-oxoglutaric acid, 0.5 mM FeSO4, 4 mM DTT, 2 mg/mL BSA, and 1 mg/mL catalase. The reactions were stopped after overnight incubation by adding 25 μL of acetic acid. The solution was passed through a C18-HD high-performance extraction disk cartridge (1 mL; Empore). After the column was washed with 3 mL of water, substances retained on the column were eluted with 500 μL of methanol. The methanol eluate was evaporated with dry N2 gas. After trimethylsilyl (TMSi) ester–TMSi ether derivatization with N-methyl-N-trimethylsilyl-trifluoroacetamide, products were analyzed by full-scan GC-MS, and identical ion peaks were compared with standard GAs (GA12, GA24, GA9, GA4, GA53, GA44, GA19, GA20, and GA1) and published data (GA15) (Gaskin and MacMillan 1992).
GC-MS Analysis of the Enzyme Assay
Full-scan GC-MS analysis of GAs was performed using a mass spectrometer (JMS-K9; JEOL) connected to a gas chromatograph (6890N; Agilent Technology). The trimethylsilylated derivatives (TMSi ester–TMSi ether) were injected (250°C) into an HP-5 MS column (0.32 mm i.d. × 30 m, 0.25 μm film thickness; Agilent Technology). The column temperature was kept at 100°C for 2 min, then increased at a rate of 30°C/min to 260°C and held for 1 min, and then increased at a rate of 30°C/min to 300°C. The flow rate of the carrier He gas was 1.5 mL/min, and mass spectra were acquired by scanning from m/z 50 to 750 at 70 eV.
Endogenous GA Analysis
Tissue samples (∼1 g fresh weight) were ground to a fine powder under liquid nitrogen and then soaked in 5 mL of extraction solvent (methanol:formic acid:water, 15:1:4). For the internal standards, 5 pmol of stable isotope GAs (2H2-GA1, 2H2-GA4, 2H2-GA9, 2H2-GA19, 2H2-GA20, and 2H2-GA24) was added to the extract. To remove interfering compounds, the extract was first passed through a Sep-Pak Vac tC18 cartridge (Waters). The pass fraction was dried and reconstituted with 1 M formic acid. The pass fraction was then further fractionated using an MCX column (Waters), and the eluate was recovered by methanol in the solid-phase extraction (Dobrev and Kaminek, 2002). The fraction was dried and reconstituted with water. Subsequently, compounds contained in a fraction were further purified using DEAE-Sephadex (GE Healthcare). GAs were eluted from the DEAE-Sephadex with 1% acetic acid.
The GA contents were measured using a liquid chromatography–mass spectrometry system (UPLC/Quattro Ultima Pt; Waters) with an ODS column (AQUITY UPLC BEH C18, 1.7 μm, 2.1 × 50 mm; Waters) at a flow rate of 0.25 mL/min. The gradients of solvent A (0.05% formic acid) and solvent B (0.05% formic acid in acetonitrile) were applied at a flow rate of 0.25 mL/min according to the following profile: 0 min, 99% A + 1% B; 13 min, 66% A + 34% B; 15 min, 99% A + 1% B. Quantification was performed in the multiple reaction monitoring mode. The mother and daughter ions for the detection of each GA type were as follows: m/z 345 and 259 for GA1, m/z 331.2 and 243 for GA4, m/z 315.2 and 271 for GA9, m/z 361.1 and 243 for GA19, m/z 331.2 and 287 for GA20, and m/z 345.2 and 257 for GA24, respectively. Cone voltage and collision energy were 85 V and 18 eV for GA1, 88 V and 17 eV for GA4, 95 V and 18 eV for GA9, 85 V and 19 eV for GA19, 85 V and 19 eV for GA20, and 80 V and 23 eV for GA24, respectively. Capillary voltage was 3.12 kV. For the identification of GA4, mass spectra were obtained by daughter ion scanning of negative ions from m/z 50 to 400 with collision energy at 25 eV and compared with those of a GA4 standard.
GAs
3H4-H2-GA4 was custom ordered from DuPont–New England Nuclear. GA4, GA9, and 3-epi-GA4 used in the yeast two-hybrid assay and GAs used in the enzyme assay were purchased from L.N. Mander (Australian National University). 2H2-GA1, 2H2-GA4, 2H2-GA9, 2H2-GA19, 2H2-GA20, and 2H2-GA24 were purchased from OlChemIm.
Accession Numbers
GenBank/EMBL accession numbers and Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: Os GID1 (Q6L545), Os HSL1 (ABA92266), Os SLR1 (BAE96289), Os SLRL1 (AAR31213), Os SLRL2 (AAT69589), Os SCR (ABA91267), Os GID2 (Q7XAK4), Os GA20ox1 (AAP21386), Os GA20ox2 (Q8RVF5), Os GA20ox3 (BAB90378), Os GA20ox4 (AAT44252), Os GA3ox1 (AAT77356), Os GA3ox2 (BAB17075), Os GA2ox1 (AAV43914), Os GA2ox2 (BAD53498), Os GA2ox3 (AAU03107), Os GA2ox4 (AAU03107), Os ACT1 (CT831215), Sr 6PGD (AB086022), At GID1a (AT3G05120), At GID1b (AT3G63010), At GID1c (AT5G27320), At HSL1 (AT5G23530), At GAI (AT1G14920), At RGA (AT2G01570), At SLY1 (AT4G24210), At SNZ (AT5G48170), At SCL3 (AT1G50420), At LAS (AT1G55580), At PAT (AT5G48150), At SCL1 (AT2G29060), At GA20ox1 (AT4G25420), At GA20ox2 (AT5G51810), At GA20ox3 (AT5G07200), At GA20ox4 (AT1G60980), At GA3ox1 (AT1G15550), At GA3ox2 (AT1G80340), At GA3ox3 (AT1G80330), At GA3ox4 (AT4G21690), At GA2ox1 (AT1G78440), At GA2ox2 (AT1G30040), At GA2ox3 (AT2G34555), At GA2ox4 (AT1G47990), At GA2ox5 (AT1G02400), Pr MC3 (AAD04946), Zm dwarf plant9 (ABI84225), Zm DWARF8 (Q9ST48), Ta GID2L (ABK79908), and Medicago truncatula DELLA (ABE77443). Sequence data obtained from the Computational Biology and Functional Genomics Laboratory (http://compbio.dfci.harvard.edu/tgi/plant.html) can be found under the following accession numbers: pine GID1L1 (TC73510), pine GID1L2 (TC57783), pine GID1L3 (TC76887) pine DELLA1 (TC59241), and pine DELLAL1 (TC60068).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effects of Various GAs on the Pp GID1L–Pp DELLAL Interactions in Yeast Cells.
Supplemental Figure 2. Length of the Second Leaf Sheath of Transgenic Rice Plants.
Supplemental Figure 3. Phylogenetic Analysis of GA Oxidase Homologs Using the Maximum Likelihood Method.
Supplemental Figure 4. GC-MS Spectrum of S. moellendorffii Endogeneous GA4.
Supplemental Figure 5. Phylogenetic Analysis of GID1 Homologs Using the NJ Method.
Supplemental Figure 6. Phylogenetic Analysis of DELLA Homologs Using the NJ Method.
Supplemental Figure 7. Phylogenetic Analysis of GID2 Homologs Using the NJ Method.
Supplemental Figure 8. Phylogenetic Analysis of GA Oxidase Homologs Using the NJ Method.
Supplemental Table 1. Endogenous GA Content in P. patens.
Supplemental Table 2. In Vitro Activity of GA20 Oxidase and GA3 Oxidase Homologs in P. patens.
Supplemental Table 3. Inclusion Limits and e-Value Thresholds Used for Phylogenetic Analyses Using the NJ Method.
Supplemental Table 4. Primers Used.
Supplemental Data Set 1. Text File of Amino Acid Alignments Presented in Figure 2A.
Supplemental Data Set 2. Text File of Amino Acid Alignments Presented in Figure 2B.
Supplemental Data Set 3. Text File of Amino Acid Alignments Presented in Figure 3A.
Supplemental Data Set 4. Text File of Amino Acid Alignments Presented in Figure 3B.
Supplemental Data Set 5. Text File of Amino Acid Alignments Presented in Figure 3C.
Supplemental Data Set 6. Text File of Amino Acid Alignments Presented in Figure 4A.
Supplemental Data Set 7. Text File of Amino Acid Alignments Presented in Figure 4B.
Supplemental Data Set 8. Text File of Amino Acid Alignments Used to Construct the Phylogenetic Tree Presented in Supplemental Figure 3 Online.
Supplemental Data Set 9. Text File of Amino Acid Alignments Used to Construct the Phylogenetic Tree Presented in Supplemental Figure 5 Online.
Supplemental Data Set 10. Text File of Amino Acid Alignments Used to Construct the Phylogenetic Tree Presented in Supplemental Figure 6 Online.
Supplemental Data Set 11. Text File of Amino Acid Alignments Used to Construct the Phylogenetic Tree Presented in Supplemental Figure 7 Online.
Supplemental Data Set 12. Text File of Amino Acid Alignments Used to Construct the Phylogenetic Tree Presented in Supplemental Figure 8 Online.
Supplementary Material
Acknowledgments
We thank Hitomi Kihara, Mayuko Kawamura, Kazuko Kigaku, Kozue Ohmae, and Hiroko Ohmiya for their technical assistance, Naomi Sumikawa for cultivation of S. moellendorffii, and Yuji Hiwatashi for P. patens RNA. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (M.M., M.N., M.U.-T., T.T., T.N., and M.H.), the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project IP1003; M.M. and M.A), and by Exploratory Research for Advanced Technology, Japan Science and Technology (T.N. and M.H.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Miyako Ueguchi-Tanaka (mueguchi@agr.nagoya-u.ac.jp) and Makoto Matsuoka (makoto@agr.nagoya-u.ac.jp).
Online version contains Web-only data.
References
- Adachi, J., and Hasegawa, M. (1996). MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 28 1–150. [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton, N.W., and Cove, D.J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants in the moss Physcomitrella patens. Mol. Gen. Genet. 154 87–95. [Google Scholar]
- Banks, J.A. (1999). Gametophyte development in ferns. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 163–186. [DOI] [PubMed] [Google Scholar]
- Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692. [DOI] [PubMed] [Google Scholar]
- Chandler, P., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, H., Hwang, I., and Goodman, H. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove, D., Benzanilla, M., Harries, P., and Quatrano, R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57 497–520. [DOI] [PubMed] [Google Scholar]
- Davis, P. J. (2004). Plant Hormones: Biosynthesis, Signal Transduction, Action! (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Dharmasiri, S., and Estelle, M. (2002). The role of regulated protein degradation in auxin response. Plant Mol. Biol. 49 401–409. [PubMed] [Google Scholar]
- Dill, A., Jung, H., and Sun, T. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev, P.I., and Kaminek, M. (2002). Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 950 21–29. [DOI] [PubMed] [Google Scholar]
- Ergün, N., Topcuoğlu, Ş.F., and Yildiz, A. (2002). Auxin (indole-3-acetic acid), gibberellic acid (GA3), abscisic acid (ABA) and cytokinin (zeatin) production by some species of mosses and lichens. Turk. J. Bot. 26 13–18. [Google Scholar]
- Fleet, C.M., Yamaguchi, S., Hanada, A., Kawaide, H., David, C.J., Kamiya, Y., and Sun, T.P. (2003). Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 132 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin, P., and MacMillan, J. (1992). GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra. (Bristol, UK: Cantocks Enterprises).
- Gifford, E.M., and Foster, A.S. (1989). Morphology and Evolution of Vascular Plants (New York: Freeman and Company).
- Gomi, K., Sasaki, A., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Kitano, H., and Matsuoka, M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37 626–634. [DOI] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P.M., White, R.G., Llewellyn, D.J., and Jacobsen, J.V. (2002). Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 129 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasebe, M., Wen, C.-K., Kato, M., and Banks, J.A. (1998). Characterization of MADS homeotic genes in the fern Ceratopteris richardii. Proc. Natl. Acad. Sci. USA 95 6222–6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K., Kawaide, H., Notomi, M., Sakigi, Y., Matsuo, A., and Nozaki, H. (2006). Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 580 6175–6181. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Shimada, A., Ueguchi-Tanaka, M., Kamiya, N., Hasegawa, Y., Ashikari, M., and Matsuoka, M. (2005). Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J. 44 669–679. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. [DOI] [PubMed] [Google Scholar]
- Kamiya, N., Itoh, J., Morikami, A., Nagato, Y., and Matsuoka, M. (2003). The SCARECROW gene's role in asymmetric cell divisions in rice plants. Plant J. 36 45–54. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., Itoh, H., Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Ashikari, M., and Matsuoka, M. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35 104–115. [DOI] [PubMed] [Google Scholar]
- Kato, J., Purves, W.K., and Phinney, B.O. (1962). Gibberellin-like substances in plants. Nature 196 687–688. [DOI] [PubMed] [Google Scholar]
- Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick, P., and Crane, P.R. (1997). The origin and early evolution of plants on land. Nature 389 33–39. [Google Scholar]
- Knight, C.D., Sehgal, A., Atwal, K., Wallace, J.C., Cove, D.J., Coates, D., Quatrano, R.S., Bahadur, S., Stockley, P.G., and Cuming, A.C. (1995). Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell 7 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, M., Sakurai, A., Saka, H., and Takahashi, N. (1989). Quantitative analysis of endogenous gibberellins in normal and dwarf cultivars of rice. Plant Cell Physiol. 30 963–969. [Google Scholar]
- Lange, T., Kappler, J., Fischer, A., Frisse, A., Padeffke, T., Schmidtke, S., and Lange, M.J. (2005). Gibberellin biosynthesis in developing pumpkin seedlings. Plant Physiol. 139 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan, J. (2002). Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20 387–442. [DOI] [PubMed] [Google Scholar]
- McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez, V., Revilla, M., Bernard, P., Gotor, V., and Fernández, H. (2006). Gibberellins and antheridiogen on sex in Blechnum spicant L. Plant Cell Rep. 25 1104–1110. [DOI] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Nakajima, M., Takita, K., Wada, H., Mihara, K., Hasegawa, M., Yamaguchi, I., and Murofushi, N. (1997). Partial purification and characterization of a gibberellin-binding protein from seedlings of Azukia angularis. Biochem. Biophys. Res. Commun. 241 782–786. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T. (2007). Evolutionary developmental biology of nonflowering land plants. Int. J. Plant Sci. 168 37–47. [Google Scholar]
- Nishiyama, T., Hiwatashi, Y., Sakakibara, K., Kato, M., and Hasebe, M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7 9–17. [DOI] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, A.L., Ward, D.A., Uknes, S., Appleford, N.E.J., Lange, T., Huttly, A.K., Gaskin, P., Graebe, J.E., and Hedden, P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley, M. (1961). Gibberellin-like substances in plants. Nature 191 684–685. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schaefer, D.G., and Zrÿd, J. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11 1195–1206. [DOI] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., and Matsuoka, M. (2000). Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220 358–364. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, L.C., Ritchie, S., Soule, J.D., McGinnis, K.M., and Steber, C.M. (2004). Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc. Natl. Acad. Sci. USA 101 12771–12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, Y., Uchida, M., Hasebe, M., and Ito, M. (2003). Characterization of the Selaginella remotifolia MADS-box gene. J. Plant Res. 116 71–75. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T., Hsing, Y., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Ashikari, M., and Matsuoka, M. (2007. a). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58 183–198. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Hongyu, X., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2007. b). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J.K., Tanurdzic, M., and Chapple, C. (2005). Functional analysis and comparative genomics of expressed sequence tags from the lycophyte Selaginella moellendorffii. BMC Plant Biol. 6 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B.C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E.M., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman, T., Williston, S., Brandts, J.F., and Lin, L.N. (1989). Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179 131–137. [DOI] [PubMed] [Google Scholar]
- Yamane, H., Fujioka, S., Spray, C.R., Phinney, B.O., Macmillan, J.M., Gaskin, P., and Takahashi, N. (1988). Endogenous gibberellins from sporophytes of two tree ferns, Cibotium glaucum and Dicksonia antarctica. Plant Physiol. 86 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, H., Yamaguchi, I., Kobayashi, M., Takahashi, M., Sato, Y., Takahashi, N., Iwatsuki, K., Phinney, B.O., Spray, C.R., Gaskin, P., and Macmillan, J. (1985). Identification of ten gibberellins from sporophytes of the tree fern, Cyathea australis. Plant Physiol. 78 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, T., Oyama, N., Yamane, H., Murofushi, N., Schraudolf, H., Pour, M., Furber, M., and Mander, L.N. (1996). Identification of antheridiogens in Lygodium circinnatum and Lygodium flexuosum. Plant Physiol. 111 741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura, Y., Crumpton-Taylor, M., Fuentes, S., and Harberd, N. (2007). Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr. Biol. 17 1225–1230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.