Abstract
The plant homologs of the archaeal DNA topoisomerase VI complex are required for the progression of endoreduplication cycles. Here, we describe the identification of MIDGET (MID) as a novel component of topoisomerase VI. We show that mid mutants show the same phenotype as rhl1, rhl2, and top6B mutants and that MID protein physically interacts with RHL1. The phenotypic analysis revealed new phenotypes, indicating that topoisomerase VI is involved in chromatin organization and transcriptional silencing. In addition, genetic evidence is provided suggesting that the ATR-dependent DNA damage repair checkpoint is activated in mid mutants, and CYCB1;1 is ectopically activated. Finally, we demonstrate that overexpression of CYCB1;2 can rescue the endoreduplication defects in mid mutants, suggesting that in mid mutants, a specific checkpoint is activated preventing further progression of endoreduplication cycles.
INTRODUCTION
Type II topoisomerases play an important role in disentangling DNA duplexes by passing one duplex through another. This process is essential to overcome topological problems, such as supercoiling associated with DNA replication, transcription, or formation of knots and tangles during recombination and replication (Liu and Wang, 1987; Wang, 2002). Decatenation by type II topoisomerases is achieved in three steps (Champoux, 2001; Wang, 2002). The first step is the cleavage of a DNA duplex by creating a covalent phosphotyrosyl protein–DNA bond. A second DNA duplex is captured and passed through the break of the first duplex. Finally, the first DNA duplex is relegated. The process is driven by ATP binding and hydrolysis.
Type II topoisomerases are found in virtually all organisms, including bacteria and all eukaryotes. Eukaryotic type II topoisomerases form homodimers. The monomers contain separate domains for ATP binding and hydrolysis, DNA binding, and cleavage and a metal binding TOPRIM domain. In bacteria, heterodimeric complexes are formed with each monomer containing a subset of functions. Archaea do not possess a typical type II topoisomerase (Bult et al., 1996). Instead, a topoisomerase VI is found, which forms a heterotetramer containing two A subunits required for DNA cleavage and two B subunits for ATP hydrolysis (Bergerat et al., 1994, 1997). While the B subunit is similar to the ATPase domains of type II topoisomerases, the topoisomerase A subunit is similar to another eukaryotic protein, Spo11, which mediates double-strand breaks during recombination (Bergerat et al., 1997; Nichols et al., 1999; Corbett and Berger, 2003a). It is therefore assumed that topoisomerases VI employ a different mechanism than classical type II topoisomerases.
For a long time it was believed that topoisomerases VI are specific for archae. However, a few years ago homologs were also found in the model plant Arabidopsis thaliana (Corbett and Berger, 2003b). In the Arabidopsis genome, one B subunit called At TOP6B, and three topoVI-A/Spo11 homologs (At SPO11-1, At SPO11-2, and At SPO11-3) were found (Hartung and Puchta, 2000, 2001). Genetic and biochemical data suggest that At SPO11-1 and At SPO11-2 represent the true SPO11 homologs required during meiosis, whereas At SPO11-3 function is linked to topoisomerase VI (Grelon et al., 2001; Hartung and Puchta, 2001; Stacey et al., 2006). It was demonstrated that At SPO11-3 and At TOP6B interact with each other and can form a functional topoisomerase VI (Sugimoto-Shirasu et al., 2002). Several groups have identified mutants for SPO11-3 (also called bin5 and rhl2) and At TOP6B (also called bin3 and hyp6) in different biological contexts (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002; Yin et al., 2002). Mutations in At TOP6B and At SPO11-3 result in an overall dwarf phenotype that was attributed to reduced endoreduplication levels (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002; Yin et al., 2002). Another link to growth regulation is suggested by the finding that the mutants exhibit reduced sensitivity to the plant hormone brassinosteroid (Yin et al., 2002). Microarray experiments showed that a significant number of genes is up- or downregulated in the mutants, suggesting a role in transcriptional regulation (Sugimoto-Shirasu et al., 2002; Yin et al., 2002).
In addition to canonical components known from archae, ROOT HAIRLESS1 (RHL1) was identified as a new component of the topoisomerase VI complex. RHL1 shares sequence similarity to the mammalian topoisomerase IIα and is proposed to function as a regulator of topoisomerase VI activity (Sugimoto-Shirasu et al., 2005).
In this work, we identified the MIDGET (MID) gene as a novel component of topoisomerase VI. MID protein physically interacts with RHL1, and mid mutants show the same phenotype as rhl1, rhl2, and top6B mutants. The DNA endoreduplication defect can be overcome by ectopic expression of cyclin B1;2. In addition to the already known phenotypes, we provide evidence that topoisomerase VI is also involved in chromatin organization and transcriptional silencing. At the genetic level, we show that the absence of MID function results in the activation of the DNA damage checkpoint response.
RESULTS
Phenotypic Analysis of mid Mutants: Reduced Cell Size and Endoreduplication Levels
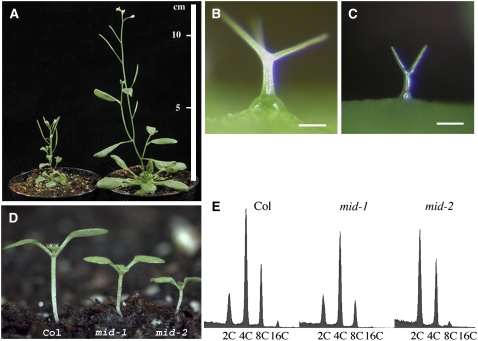
The mid-1 mutant was initially isolated as a recessive dwarf mutant from a population of 8000 activation-tagged T-DNA lines (Weigel et al., 2000). A second mid allele, mid-2, was obtained from the SALK T-DNA mutant collection (SALK_110705). Both alleles show a strong general growth reduction (Figures 1A and 1D) and carry T-DNA insertions in the At5g24630 gene. The mid-1 allele has a T-DNA insertion in exon V, and the mid-2 allele a T-DNA insertion in intron III. A 3596-bp genomic fragment containing the At5g24603 gene rescued the mid-1 mutant phenotype completely, indicating that At5g24630 encodes the MID gene.
Figure 1.
Reduced Growth of the mid Mutant.
(A) Reduced growth of the mid-1 mutant (left) compared with a wild-type plant (right) at the same age.
(B) Wild-type trichome.
(C) Trichome on a mid-1 mutant rosette leaf.
(D) One-week-old seedlings of the wild type, mid-1, and mid-2.
(E) Flow cytometric histogram of leaf nuclear DNA.
Bars in (B) and (C) = 100 μm.
The full-length sequence of the MID cDNA was determined using 3′ and 5′ rapid amplification of cDNA ends (RACE) techniques (see Supplemental Figure 1 online). MID is a single-copy gene that shows significant amino acid sequence similarity only to other plant genes, including the rice (Oryza sativa) Os02g0147700 gene (32% sequence identity), the Jerusalem artichoke (Cynara scolymus) EL434975 gene (35% sequence identity), and the barley (Hordeum vulgare) AK250018 gene (33% sequence identity). Several domains can be recognized, including two direct repeats, a putative HMG DNA binding domain, and a bipartite nuclear localization signal (Robbins et al., 1991; Schultz et al., 1998; Letunic et al., 2006).
The general reduction in plant size correlates with a strong size reduction of various cell types, including trichomes (Figures 1B and 1C), hypocotyl cells, and leaf pavement cells (data not shown). The general reduction in cell size suggested to us that MID might be involved in DNA endoreduplication. To test this, we analyzed endoreduplication levels of leaves by flow cytometry. Both alleles show a strong shift toward lower endoreduplication levels. In mid-2 alleles, the population of 2C cells has become the most prominent fraction (Figure 1E). We confirmed this result for trichome cells by measuring the relative fluorescence intensity of the 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclear DNA. Wild-type trichomes typically undergo four rounds of endoreduplication and have a DNA content of on average 32C. By contrast, mid trichomes have a DNA content of ∼8 C (n = 56), which corresponds to two endoreduplication cycles. Together, these data suggest that MID function is required for DNA endoreduplication. To further explore whether MID assists the progression of endoreduplications, we used the trichome system for a genetic analysis. Here, various mutants are known that show higher or lower endoreduplication levels than the wild type (Hulskamp et al., 1994; Perazza et al., 1999). Double mutants of mid-1 with mutants forming trichomes with a DNA content of 64C, such as triptychon, kaktus, and rastafari, exhibit a small trichome phenotype indistinguishable from mid-1 (data not shown). This epistatic behavior of the mid-1 mutation suggests that the MID gene is essential for the progression of endoreduplication cycles.
MID Function Is Required for Differentiation of Root Hairs and Seed Columella Cells
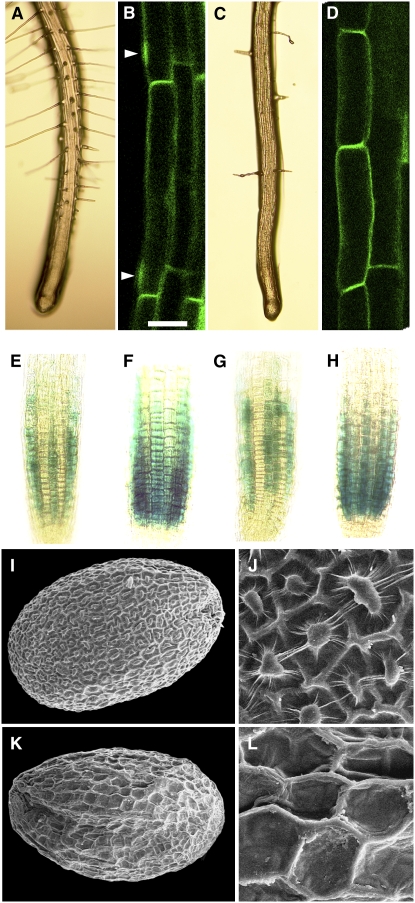
In addition to the cell size phenotype, the mid mutants produce fewer root hairs (Figures 2A and 2C). To decide whether this is because root hair cell fate is not properly determined or whether root hair cells do not differentiate, we analyzed the expression and localization of specific markers in the mid mutants. Early events during cell fate specification can be monitored using root hair patterning genes expressed in future root hair cell files (trichoblasts) and non-root hair cells (atrichoblasts). Here, we analyzed the expression of CAPRICE:β-glucuronidase (CPC:GUS) as an early marker for developing atrichoblasts (Lee and Schiefelbein, 2002) and ENHANCER OF GLABRA3 (EGL3):GUS (Bernhardt et al., 2005) as a marker for trichoblasts. Both markers showed an unchanged expression pattern in the mid mutant, indicating unperturbed position-dependent cell fate specification (Figures 2E to 2H).
Figure 2.
Differentiation Defects of Root Epidermal Cells and the Columella Cells in the Seed Coat of the mid Mutants.
(A), (B), (E), (F), (I), and (J) The wild type.
(C), (D), (G), (H), (K), and (L) mid-1.
(A) and (C) Light micrograph of roots.
(B) and (D) Laser scanning confocal micrograph showing GFP-ROP2 localization in epidermal root cells.
(E) and (G) CPC:GUS reporter.
(F) and (H) EGL3:GUS reporter.
(I) to (L) Scanning electron micrographs of the seed surface.
Arrowheads in (B) show accumulation of GFP-ROP2. Bar in (B) = 25 μm for (B) and (D), 50 μm for (E) to (H), 150 μm for (I) and (K), and 30 μm for (J) and (L).
We used the localization of ROP2 (for Rho-related GTPase from plants-2) as a marker for the earliest indication of actual root hair initiation. In the wild type, ROP2 protein becomes localized at the future root hair outgrowth site (Jones et al., 2002). In the wild type, we found ROP2-green fluorescent protein (GFP) accumulation close to the apical end of trichoblasts marking the future root hair outgrowth site. By contrast, in the mid mutants, ROP2-GFP was not detectable in most of the root epidermal cells of the root hair initiation zone (Figures 2B and 2D).
The outer cell layer of the Arabidopsis seed coat is characterized by hexagonally shaped cells with thick radial cell walls and a spindle (columella) centrally positioned in the cell. This distinctive cell shape is a result of the massive mucilage secretion in a ring between the plasma membrane and the outer cell wall that forces the cytoplasm in a columnar shape in the cell center. Seed hydration releases mucilage, providing the seed with a gelatin-like coating. The outer cell layer of the mid mutant seeds does not develop columella (Figures 2I to 2K), and seeds of the mid mutant fail to release seed coat mucilage after hydration (data not shown). Thus, MID is required for seed epidermal differentiation.
MID:GUS Expression Is Predominantly Found in Young Developing Tissues
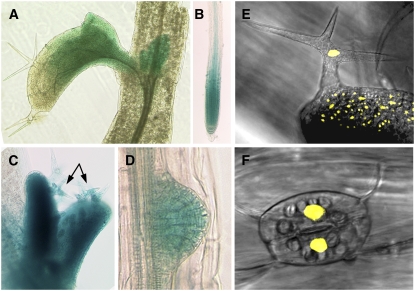
To study the expression of the MID gene, a MID:GUS fusion was created that contained the 5′ upstream region used in the rescue experiments. All 10 transformants analyzed showed the same expression pattern. Expression was found in the actively proliferating tissues of the shoot and root (Figures 3A to 3D). We also found expression in elongating hypocotyl cells and developing trichomes (Figure 3C) at a phase when their DNA undergoes endoreduplication; however, because of the stability of the used GUS reporter protein, it is not possible to decide whether this GUS staining is truly reflecting an expression in endoreduplicating tissues.
Figure 3.
Expression Analysis of MID and Localization of the MID-YFP Fusion Protein.
(A) to (D) Expression of the MID:GUS reporter.
(A) Rosette leaves.
(B) Meristematic region of the root.
(C) Young leaves with developing trichomes (arrows).
(D) Lateral root primordium.
(E) and (F) Nuclear localization of MID-YFP in trichomes (E) and stomata (F).
MID Is a Nuclear Protein
The MID protein contains a putative nuclear localization signal (predicted by PSORT algorithm) and AT hook-type DNA binding motives (predicted by SMART algorithm), suggesting that MID is located in the nucleus. To test the cellular localization of the MID protein, a translational fusion of MID and yellow fluorescent protein (YFP) was created and expressed from the 35S promoter. This 35S:MID-YFP construct rescued the mid phenotype completely. It has been previously reported that YFP alone is equally distributed in the cytoplasm and the nucleus and that protein fusions larger than 50 kD are excluded from the nucleus (Grebenok et al., 1997). In 35S:MID-YFP lines, a strong YFP fluorescence signal was found in the nuclei in different cell types, indicating that MID is a nuclear protein (Figures 3E and 3F).
MID Protein Is an Essential Component of the Arabidopsis Topoisomerase VI Complex
Except for the predicted nuclear localization and DNA binding motives, no further sequence similarity pointing to a potential function of MID was found. We therefore performed a yeast two-hybrid screen with MID as bait to identify interacting proteins. One of the found putative interactors, RHL1, was particularly interesting as the mid and rhl1 mutants show an identical phenotype. As RHL1 interacts with the A and B subunits of the Arabidopsis topoisomerase VI complex, RHL2 and TOP6B (Sugimoto-Shirasu et al., 2005), we speculated that MID is a new component of this complex. In support of this, the rhl2 and top6b mutant phenotypes are indistinguishable from mid and rhl1 (Sugimoto-Shirasu et al., 2002). To test this hypothesis, we created the mid rhl2 and mid topb6 double mutants. The double mutants showed the same phenotype as the single mutants (data not shown), indicating that the corresponding genes act in the same pathway.
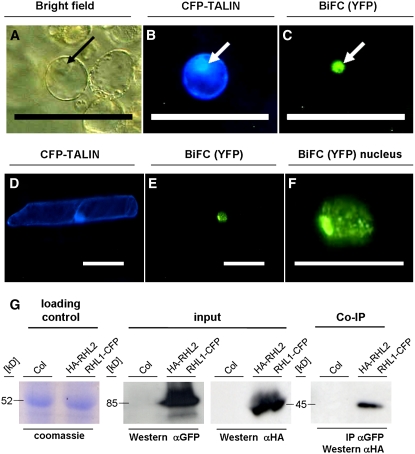
To test further the hypothesis that MID is a component of the topoisomerase VI complex, we tested interactions between MID and RHL1 by directed yeast two-hybrid assay and the bimolecular fluorescence complementation assay (BiFC; Walter et al., 2004). Directed yeast two-hybrid assays confirmed the MID–RHL1 interaction found in the yeast two-hybrid screen (data not shown). To independently verify MID–RHL1 interactions by BiFC, we cotransformed plasmids encoding BiFC fusion constructs of the two proteins with the N- or C-terminal part of YFP into Arabidopsis protoplasts. A restored YFP fluorescence was localized in the nuclei of transfected protoplasts, suggesting that MID and RHL1 can interact in planta (Figures 4A to 4C). Previous data on the interactions between RHL1 and RHL2 (Sugimoto-Shirasu et al., 2005) could also be confirmed by BiFC (Figures 4D to 4F).
Figure 4.
MID Protein Is a Component of the Arabidopsis Topoisomerase VI Complex.
(A) to (F) BiFC of MID and RHL1, and of RHL1 and RHL2. Plasmids encoding BiFC fusion constructs of MID, RHL1, and RHL2 with the N- or C-terminal part of YFP were cotransformed into Arabidopsis protoplasts (MID and RHL1 in [A] to [C]) or cobombarded into leak (Allium porrum) cells (RHL1 and RHL2 in [D] to [F]). CFP-TALIN served as a transformation control ([B] and [D]). Bar = 100 μm in (A) to (E) and 50 μm in (F).
(A) Bright-field image of the transformed protoplast.
(C) YFP fluorescence indicates interaction between RHL1 and MID. The interaction was localized to the nucleus (arrows in [A] to [C]).
(E) and (F) YFP fluorescence indicates interaction between RHL1 and RHL2.
(F) Magnification of the nucleus shown in (E).
(G) Co-IP of HA-RHL2 with RHL1-CFP. Immunoprecipitation (IP) of proteins from the plant extract was done with anti-GFP beads, and the precipitate was analyzed with anti-HA antibody after protein gel blotting (Co-IP). The input for both assays is displayed after anti-GFP and anti-HA detection. Loading control (Coomassie staining) shows that equal amounts of total protein were loaded. Columbia plants (Col) served as a negative control. Western, protein gel blot.
To substantiate that MID and RHL1 are components of the topoisomerase VI complex in planta, coimmunoprecipitation (Co-IP) experiments were performed. We created transgenic plants harboring the 35S:RHL1-cyan fluorescent protein (CFP) and 35S:HA-RHL2 constructs and transgenic plants harboring the 35S:RHL1-CFP and 35S:HA-MID constructs. In these experiments, we could not detect the HA-MID protein in Co-IP experiments with the MID and RHL1 proteins with anti-HA beads (data not shown). However, we found that HA-RHL2 protein was immunoprecipitated with anti-GFP beads and RHL1-CFP with anti-HA beads from the plants harboring the 35S:RHL1-CFP and 35S:HA-RHL2 constructs (Figure 4G).
Taken together, the results of the yeast two-hybrid screen, genetic interactions, BiFC, and Co-IP suggest that MID protein is part of the RHL1/RHL2/TOP6B complex and its function is essential for activity of Arabidopsis topoisomerase VI.
MID Function Is Required for Chromatin Organization
Recent studies provide evidence for a role of Topo II in chromosome condensation during mitosis (Cuvier and Hirano, 2003) and implicated it in chromatin structure and organization in nonmitotic cells (Varga-Weisz et al., 1997; LeRoy et al., 2000).
To test genetically whether MID is involved in chromatin organization, we created a double mutant of mid-1 with a mutant defective in the p150 subunit (fasciata1 [fas1]) of chromatin assembly factor1 that was shown to affect the heterochromatin formation in nonmitotic cells in Arabidopsis (Kirik et al., 2006). The double mutant displayed dramatically reduced seedlings size and died shortly after germination (Figure 5A). This synergistic phenotype suggests the involvement of the Arabidopsis topoisomerase VI in chromatin organization.
Figure 5.
MID Function Is Required for Chromatin Condensation and Transcriptional Gene Silencing.
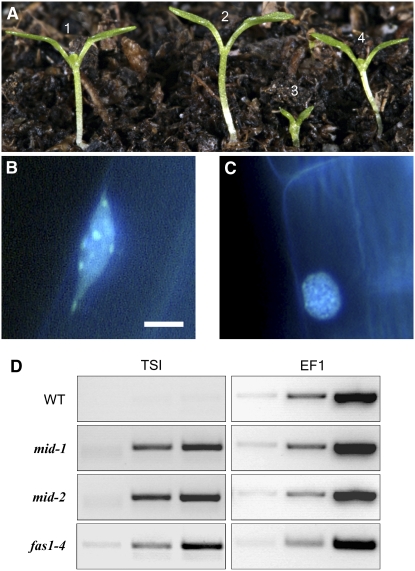
(A) Genetic interaction between the mid and fas mutations: 7-d-old seedling of (1) the wild type; (2) fas1-4; (3) fas1-4 mid-1; and (4) mid-1.
(B) and (C) DAPI-stained nuclear DNA of root epidermal cells in the wild type (B) and mid-1 (C). Note that mid mutant nuclei show smaller and less-defined chromocenters than the wild type. Bar = 5 mm.
(D) RT-PCR analysis of the TSI loci in the mid and fas1 mutants. RT-PCR with the TSI primers was performed for 30, 35, and 40 cycles (columns from left to right). As a cDNA control, RT-PCR was done with primers specific for the translation elongation factor EF1αA4 using 20, 22, and 24 cycles (columns from left to right).
To further address the role of the Arabidopsis topoisomerease VI complex in heterochromatin formation, we analyzed DAPI-stained nuclei of several cell types in the mid mutant. Mitotic cells showed no detectable defects in chromosome condensation and ploidy levels (data not shown), suggesting that mitotic chromosome assembly and segregation are not dependent on Topo VI in Arabidopsis. However, nuclei of endoreduplicating cells, such as trichomes, elongated root epidermal cells, and occasionally root hairs, exhibited aberrant staining patterns (Figures 5B and 5C). In the wild type, highly condensed heterochromatic DNA consisting of centromeric and pericentromeric repeats and rDNA genes (Fransz et al., 2002) was visible as brightly stained distinct chromocenters in 81% of the nuclei (n = 31; Figure 5B). In the mid mutants, chromocenters are smaller and less defined (Figure 5C). Only 12% of the analyzed nuclei showed clearly defined chromocenters (n = 52), suggesting that heterochromatin organization is affected by the mid mutation.
Generally, heterochromatic regions correlate with transcriptional gene silencing, whereas euchromatic regions show more transcriptional activity. Thus, defective heterochromatin formation in the mid mutant could result in a release of transcriptional silencing within heterochromatin regions. We used the TRANSCRIPTIONAL SILENCING INFORMATION A (TSI) gene as a marker to test this assumption. The TSI gene is located in centromeric regions and serves as a specific marker for disturbed transcriptional gene silencing (Steimer et al., 2000). As shown in Figure 5D, TSI expression is activated in the mid and fas1 mutants. Taken together, these data illustrate a role of Arabidopsis topoisomerase VI in heterochromatin-dependent transcriptional gene silencing.
The ATR-Dependent DNA Damage Repair Checkpoint Is Activated in mid Mutants
Cell cycle checkpoints detect various defects at different cell cycle phases and impose delays in the cell cycle progression. Two closely related protein kinases, ATM (ataxia telangiectasia-mutated) and ATR (A-T and red3 related), sense DNA abnormalities and send signals to regulate mitotic progression (Kurz and Lees-Miller, 2004; Shechter et al., 2004). To test whether defects in the Arabidopsis Topo VI complex activate a DNA damage response pathway, we created the respective double mutants mid atr and mid atm. Arabidopsis atm and atr mutants are viable and do not show any effect on plant growth and development under normal conditions except for a reduced fertility of the atm mutant (Culligan et al., 2004). atm mid double mutant plants were similar in size as the mid mutant and partially sterile (data not shown). By contrast, we could not isolate an atr mid double mutant. Among the 63 PCR-tested siblings of the mid-1/mid-1 atr/ATR progeny, 35 plants had mid-1/mid-1 atr/ATR and 18 plants had mid-1/mid-1 ATR/ATR genotype (close to a 2:1 ratio). The mid-1/mid-1 atr/ATR plants showed normal gametophytic transmission. Thus, genetic analysis suggests that the double homozygous mid atr plants are lethal.
In a recent study, Culligan et al. (2006) reported that genomic stress caused by γ-irradiation leads to a strong transcriptional activation of the Arabidopsis B-type cyclin (CYCB1;1). This upregulation depends on both the ATM and ATR kinases, with the ATM pathway inducing the CYCB1;1 transcription and the ATR pathway suppressing the degradation of the CYCB1;1 protein. We speculated that if the genetic link between MID and the ATM/ATR checkpoint is correct, mid mutants might also show an upregulation of CYCB1;1. We analyzed the expression of the pCYCB1;1:GUS reporter in the mid mutants (Colon-Carmona et al., 1999) and found expression not only in mitotic cells but also in differentiated cells, including cells that have endoreduplicated DNA-like trichomes and hypocotyl cells (Figures 6A and 6B). One explanation for this finding is that the mid mutation induces mitotic entry in these cell types causing an induction of the mitotic cyclinB1;1 similar as in siamese mutants (Walker et al., 2000). However, in >300 inspected trichomes, we found no multicellular situation as it is typical for siamese mutants. The data suggest that a loss of the topoisomerase VI function causes genomic stress that activates the ATR-dependent checkpoint and an ectopic induction of CYCB1;1:GUS.
Figure 6.
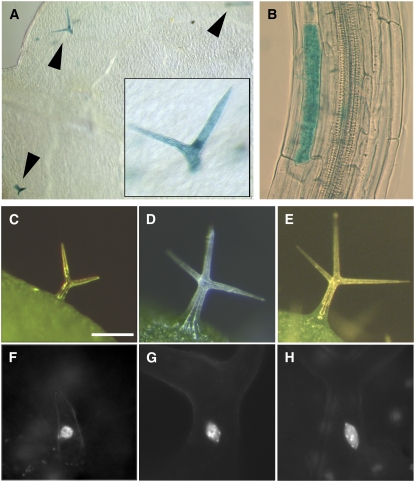
Ectopic Expression of CYCB1;1 in the mid Mutants and Rescue by CYCB1;2.
(A) Ectopic expression of CYCB1;1:GUS in mid mutant trichomes. Arrowheads indicate GUS-stained trichomes. Inset is an enlargement of a trichome on the same leaf.
(B) Ectopic expression of CYCB1;1:GUS in the mid elongated hypocotyl cell.
(C) to (H) Rescue of the trichome cell size and DNA endoreduplication defects by overexpression of the mitotic cyclin B1;2 in the mid mutant trichomes. Bar in (C) = 200 μm for (C) to (E) and 50 μm for (F) to (H).
(C) mid-2 trichome.
(D) Wild-type trichome.
(E) mid-2 GL2:CYCB1;2 trichome.
(F) to (H) DAPI-stained nuclei in trichomes.
(F) mid-2.
(G) The wild type.
(H) mid-2 GL2:CYCB1;2. Note restored nuclear size and brightly stained chromocenters in.
Endoreduplication Defects in the mid and rhl2 Mutants Can Be Rescued by B-Type Cyclin Activity
Our findings suggesting that a G2 damage repair checkpoint is activated in the mid mutants led us to the hypothesis that the reduced endoreduplication levels are caused by a checkpoint-driven cell cycle arrest in endoreduplicating cells. If this hypothesis is correct, one would expect that bypassing the DNA checkpoint by the expression of cell cycle regulators would restore DNA endoreduplication in the Topo VI mutants.
We tested this assumption by expressing a truncated version of the Arabidopsis CYCB1;2 gene under the control of the GL2 promoter in the mid mutants. This CYCB1,2 version carries a deletion of 133 amino acids at the C terminus, and it has been shown that its expression from the GL2 promoter leads to multiple nuclei in trichomes (Schnittger et al., 2002). We analyzed 20 transformants and found weak rescue in five lines and full rescue of the mid mutant trichome phenotype in 15 lines. In these transformants, trichomes were rescued to normal size and branching. Also, the nuclear size was like the wild type (mid-2: 275 ± 82 μm2, n = 17; GL2:CYCB1,2 mid-2: 562 ± 100 μm2, n = 10; Columbia: 597 ± 163 μm2, n = 14; Figures 6C to 6H). The dwarf phenotype was not rescued. The same results were obtained for rhl2 mutants in which the GL2:CYCB1;2 construct was introduced (data not shown).
In summary, these experiments suggest that the ATR-dependent DNA checkpoint response is relevant in endoreduplication cycles and that the expression of mitotic B type cyclin can bypass this checkpoint in endoreduplicating cells.
DISCUSSION
In this study, we have identified a novel component of the topoisomerase VI complex in plants. Physical interactions with known topoisomerase VI proteins and the fact that the mid mutant phenotype is indistinguishable from mutants of the other components indicate that MID is either a direct regulator or functional component of the plant topoisomerase VI complex. Our analysis of mid and the other topoisomerase VI mutants revealed new insights in the function of plant topoisomerase VI.
The Role of Topoisomerase VI in Chromatin Organization
Earlier genetic and biochemical studies in yeast and animal cells have implicated DNA topoisomerase II in chromatin condensation during mitosis (Uemura et al., 1987; Wang, 2002; Cuvier and Hirano, 2003). In addition, a structural role of eukaryotic DNA topoisomerase II was postulated for the higher-order organization of chromosomes (Gasser et al., 1986), and an association with components of chromatin-remodeling complexes has been reported (Varga-Weisz et al., 1997; LeRoy et al., 2000). However, the functional relevance of topoisomerase II in chromatin structure and organization was not shown so far.
We did not find any defects in the chromosomal condensation in mitotic cells of mid mutants, indicating that Arabidopsis topoisomerase VI does not play a role in chromosome compaction during mitosis. This function could be performed by the Arabidopsis topoisomerase II gene (At TopII), which was shown to be expressed in mitotically active plant tissues (Xie and Lam, 1994) and thus can potentially carry out topoisomerase II functions in chromosome condensation.
Our evidence suggests that topoisomerase VI is important for heterochromatin formation during interphase. This is supported by three lines of evidence. First, interphase nuclei in various cell types lack the typical formation of chromocenters similar to that found in fas1 mutants, which were previously shown to be defective in heterochromatin formation (Kirik et al., 2006). Second, the synergistic phenotype of the fas1 mid-1 double mutants suggests a functional role of MID in chromatin organization. Third, the transcriptional activation of TSI that normally is silenced due to its position in heterochromatic regions of centromeres suggests that topoisomerase VI function contributes to heterochromatin formation and is involved in gene silencing.
Together, these results show that topoisomerase VI plays a functional role in chromatin organization and gene silencing.
The Role of Topoisomerase VI in Cell Cycle Checkpoint Control
Plants and animals possess a topoisomerase II–dependent G2 checkpoint that monitors the catenation status in the cell. This checkpoint can be bypassed by the overexpression of CYCB2 in plant cells (Gimenez-Abian et al., 2002) and by the overexpression of the nuclear-localized cyclin B1 in human cells (Deming et al., 2001). The ability to overcome the checkpoint by cyclin overexpression suggests that the topoisomerase II–dependent G2 checkpoint transiently inhibits mitotic CDK activation.
Our analysis of the MID gene suggests that the loss of the Topo VI function activates the ATR-dependent cell cycle checkpoint that senses persisting single-stranded DNA (Sancar et al., 2004). This is supported by genetic interaction between atr and mid mutants and the finding that CYCB1;1-GUS protein, whose level is specifically suppressed by ATR (Culligan et al., 2004), is ectopically activated in the mid mutants. To date, the ATR checkpoint was mostly analyzed in the context of the mitotic cell cycle. One possible explanation for the expression of CYCB1;1:GUS in endoreduplicating cell types in the mid mutant is that these cells remain diploid and are arrested in the G2/S phase as a consequence of the ATR-dependent cell cycle arrest in the G2/S phase. This scenario, however, is not likely, as we found a DNA content of 8C in trichomes corresponding to two endoreduplication cycles. In the second scenario, developing mid cells become arrested in the last mitotic cycle before DNA endoreduplication. Endoreduplication cycles would start but may be inefficient because of the perturbed cell cycle. Consistent with this hypothesis, DNA damage does not activate a checkpoint response in endoreduplicating cells (Hefner et al., 2006). In this scenario, CYCB1;1 expression may persist in endoreduplicating cells because the cells are halted in a mitotic phase before entering endoreduplication. Along the same lines, CYCB1;2 expression would overcome the mitotic arrest and allow normal endoreduplication cycles. Finally, the results could be explained by a scenario in which the loss of topoisomerase VI activity can trigger an ATR-dependent DNA endoreduplication checkpoint.
A Mixed Bag of Phenotypic Defects in Topoisomearase VI Mutants: What Are the Underlying Reasons?
The wide range of pleiotropic phenotypes is likely due to three main molecular defects recognized in topoisomerase VI mutants: the endoreduplication defect, the defects in chromatin structure, and the misexpression of many genes (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002; Yin et al., 2002). It is conceivable that most growth defects can be attributed to the endoreduplication defects. Differentiation defects, however, are more difficult to explain. Unbalanced transcription of genes important for epidermal cell differentiation may be responsible for the observed differentiation defects. Microarray analysis had shown that in the rhl2 and top6b mutants, ∼321 genes are downregulated (Yin et al., 2002). Our finding that chromatin organization is disturbed in the mid mutant suggests that the misregulation of at least some loci is due to disturbed gene silencing. This explanation is consistent with the finding that a correct balance between euchromatin and heterochromatin is important for root hair differentiation and that the chromatin assembly mutant fasciata2 displays a reduced number of root hairs (Costa and Shaw, 2006). The analysis and classification of up- and downregulated genes in topoisomerase VI mutants may provide a useful tool to recognize the molecular function of topoisomerase VI.
METHODS
Plant Materials and Growth Conditions
The mid-1 mutant (Columbia ecotype) was isolated in a screen of 8000 activation tagging T-DNA lines obtained from the Arabidopsis Stock Center (Weigel et al., 2000). The mid-2 mutant allele was isolated from the SALK T-DNA collection (Columbia ecotype; SALK_110705). In addition, the following other mutants were used in this study: atr-4 (ecotype Columbia; SALK_054383), atm-3 (ecotype Columbia; SALK_089805), fas1-4 (in C24 background; Kirik et al., 2006), kaktus, rastafari, and triptychon (Landsberg erecta background; Perazza et al., 1999), and rhl2 and top6B (Columbia ecotype; Larkin et al., 1999). Lines carrying multiple mutations and/or transgenes were constructed by crossing single mutants or transgenic plants, examining the F2 progeny for putative mutant phenotypes and verifying the genotype of selected plants in subsequent generations by backcrossing to single mutants and/or PCR-based tests.
Cytological Analysis
The histochemical analysis of plants containing the GUS reporter constructs was performed essentially as described previously (Vroemen et al., 1996). Flow cytometry analysis was performed on propidium iodide–stained nuclei of the fully developed rosette leaves. The DISCUS software package (Carl H. Hilgers-Technisches Büro) was used to measure the fluorescence intensity of DAPI-stained nuclei. The C value was calculated using wild-type trichomes (32C) as a reference (Walker et al., 2000).
Molecular Biology Methods
For RT-PCR and 5′ RACE analysis RNA was isolated from rosettes of 2-week-old plants. The 5′ RACE was done according to Invitrogen recommendations to determine the open reading frame of the MID gene. Gene-specific TSI-A primers used in RT-PCR and TA-F1 and TA-R1 were described previously (Steimer et al., 2000). Amplifications of the translation elongation factor EF1αA4 cDNA (primers EF1α-UP, 5′-ATGCCCCAGGACATCGTGATTTCAT-3′, and EF1α-RP, 5′-TTGGCGGCACCCTTAGCTGGATCA-3′) were used as control. 35S:MID-YFP was constructed by fusing MID cDNA and EYFP and cloning the fusion into p35S-HPT vector (Becker et al., 1992).
For rescue experiments, a 3596-bp genomic fragment was used that included 433-bp upstream and 18-bp downstream sequences. This fragment was introduced into a modified pCAMBIA3000 vector carrying a transcriptional terminator sequence.
The MID:GUS reporter construct was designed as a translational fusion to GUS containing a 2115-bp MID genomic fragment comprising 433 bp upstream of the START codon and a part of the MID transcriptional unit including seven introns and coding sequence for the first 272 amino acids.
35S:MID-HA, 35S:RHL1-CFP, and 35S:RHL2-HA constructs were created using pEarleyGate plasmid vectors and Gateway-compatible vectors for plant functional genomics and proteomics (Earley et al., 2006). Details of the used constructs are available upon request.
Yeast Two-Hybrid Screen and Assay
Fusions with the GAL4 activation domain and GAL4 DNA binding domain were performed into the bait vector pCD2attR and into the pray vector pCACT2attR. All used constructs and empty vectors did not show self-activation in yeast. Yeast two-hybrid screens were performed as described before (Soellick and Uhrig, 2001). The Gal4BD-fused MID and RHL1 proteins were used as bait proteins to screen an Arabidopsis thaliana cDNA library (flowers, siliques, and green seeds).
BiFC
Polyethylene glycol–mediated transfection of Arabidopsis protoplasts was performed as previously described (Mathur and Koncz, 1998). Biolistic transformation was performed using a Helios particle bombardment device (Bio-Rad). The bombarded cells of leak (Allium porrum) were then placed at 22°C in the dark and analyzed after 24 h. For every assay, 80 to 200 different transformed cells of at least two independent transformations were analyzed.
Co-IP
Protein extracts were made from double transformed plants (Columbia ecotype) expressing HA-RHL2 and RHL1-CFP fusions under control of the cauliflower mosaic virus 35S promoter. Expression of RHL1-CFP was tested by fluorescence microscopy and expression of HA-RHL2 by protein gel blotting (see below). Young leaves (0.25 g) at the rosette stage were ground in 1 mL of lysis buffer complemented with 10 mM DTT, 0.1% SDS, and 50 μL protease inhibitor (Complete EDTAfree protease inhibitor). A 900-μL aliquot of this lysate was used with 50 μL of anti-GFP MicroBeads (Miltenyi Biotec) according to the manufacturer's instructions. HA and CFP fusion proteins were visualized by protein gel blotting using a monoclonal rat anti-HA antibody or a mouse anti-GFP antibody (Roche) and a horseradish peroxidase–conjugated goat anti-rat antibody or goat anti-mouse antibody (Jackson).
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library under the following accession number: At5g24630 (MID).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Sequence Information on the MIDGET Gene and MIDGET Protein.
Supplementary Material
Acknowledgments
We thank Arp Schnittger for providing the GL2:CYCB1;1 and GL2:CYCD3;2 constructs, Klaus Schmitz for help with scanning electron microscopy, Christoph Spitzer for help with the protein Co-IP method, and Irene Klinkhammer for excellent technical assistance. We also acknowledge the Signal Program and the ABRC for providing material used for the isolation of midget-1, midget-2, atr, and atm T-DNA insertion lines.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Viktor Kirik (vkirik@stanford.edu).
Online version of contains Web-only data.
References
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bergerat, A., de Massy, B., Gadelle, D., Varoutas, P.C., Nicolas, A., and Forterre, P. (1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386 414–417. [DOI] [PubMed] [Google Scholar]
- Bergerat, A., Gadelle, D., and Forterre, P. (1994). Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 269 27663–27669. [PubMed] [Google Scholar]
- Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 291–298. [DOI] [PubMed] [Google Scholar]
- Bult, C.J., et al. (1996). Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273 1058–1073. [DOI] [PubMed] [Google Scholar]
- Champoux, J.J. (2001). DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70 369–413. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Corbett, K.D., and Berger, J.M. (2003. a). Structure of the topoisomerase VI-B subunit: Implications for type II topoisomerase mechanism and evolution. EMBO J. 22 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett, K.D., and Berger, J.M. (2003. b). Emerging roles for plant topoisomerase VI. Chem. Biol. 10 107–111. [DOI] [PubMed] [Google Scholar]
- Costa, S., and Shaw, P. (2006). Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439 493–496. [DOI] [PubMed] [Google Scholar]
- Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2 phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K.M., Robertson, C.E., Foreman, J., Doerner, P., and Britt, A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48 947–961. [DOI] [PubMed] [Google Scholar]
- Cuvier, O., and Hirano, T. (2003). A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 160 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming, P.B., Cistulli, C.A., Zhao, H., Graves, P.R., Piwnica-Worms, H., Paules, R.S., Downes, C.S., and Kaufmann, W.K. (2001). The human decatenation checkpoint. Proc. Natl. Acad. Sci. USA 98 12044–12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Fransz, P., De Jong, J.H., Lysak, M., Castiglione, M.R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, S.M., Laroche, T., Falquet, J., Boy de la Tour, E., and Laemmli, U.K. (1986). Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 188 613–629. [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian, J.F., Weingartner, M., Binarova, P., Clarke, D.J., Anthony, R.G., Calderini, O., Heberle-Bors, E., Moreno Diaz de la Espina, S., Bogre, L., and De la Torre, C. (2002). A topoisomerase II-dependent checkpoint in G2-phase plant cells can be bypassed by ectopic expression of mitotic cyclin B2. Cell Cycle 1 187–192. [PubMed] [Google Scholar]
- Grebenok, R.J., Pierson, E., Lambert, G.M., Gong, F.C., Afonso, C.L., Haldeman-Cahill, R., Carrington, J.C., and Galbraith, D.W. (1997). Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11 573–586. [DOI] [PubMed] [Google Scholar]
- Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, F., Angelis, K.J., Meister, A., Schubert, I., Melzer, M., and Puchta, H. (2002). An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 12 1787–1791. [DOI] [PubMed] [Google Scholar]
- Hartung, F., and Puchta, H. (2000). Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, F., and Puchta, H. (2001). Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene 271 81–86. [DOI] [PubMed] [Google Scholar]
- Hefner, E., Huefner, N., and Britt, A.B. (2006). Tissue-specific regulation of cell-cycle responses to DNA damage in Arabidopsis seedlings. DNA Repair (Amst.) 5 102–110. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Misera, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76 555–566. [DOI] [PubMed] [Google Scholar]
- Jones, M.A., Shen, J.J., Fu, Y., Yang, Z., and Grierson, C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, A., Pecinka, A., Wendeler, E., and Reiss, B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18 2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, E.U., and Lees-Miller, S.P. (2004). DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst.) 3 889–900. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Walker, J.D., Bolognesi-Winfield, A.C., Gray, J.C., and Walker, A.R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2002). Cell patterning in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy, G., Loyola, A., Lane, W.S., and Reinberg, D. (2000). Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275 14787–14790. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Pils, B., Pinkert, S., Schultz, J., and Bork, P. (2006). SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 34 D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L.F., and Wang, J.C. (1987). Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., and Koncz, C. (1998). PEG-mediated protoplast transformation with naked DNA. Methods Mol. Biol. 82 267–276. [DOI] [PubMed] [Google Scholar]
- Nichols, M.D., DeAngelis, K., Keck, J.L., and Berger, J.M. (1999). Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 18 6177–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza, D., Herzog, M., Hulskamp, M., Brown, S., Dorne, A., and Bonneville, J. (1999). Trichome cell growth in Arabidopsis thaliana can be depressed by mutations in at least five genes. Genetics 152 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, J., Dilworth, S.M., Laskey, R.A., and Dingwall, C. (1991). Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 64 615–623. [DOI] [PubMed] [Google Scholar]
- Sancar, A., Lindsey-Boltz, L.A., Unsal-Kacmaz, K., and Linn, S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73 39–85. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Stierhof, Y.D., and Hulskamp, M. (2002). Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 12 415–420. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter, D., Costanzo, V., and Gautier, J. (2004). Regulation of DNA replication by ATR: Signaling in response to DNA intermediates. DNA Repair (Amst.) 3 901–908. [DOI] [PubMed] [Google Scholar]
- Soellick, T.R., and Uhrig, J.F. (2001). Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2 RESEARCH0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, N.J., Kuromori, T., Azumi, Y., Roberts, G., Breuer, C., Wada, T., Maxwell, A., Roberts, K., and Sugimoto-Shirasu, K. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48 206–216. [DOI] [PubMed] [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Mittelsten Scheid, O., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Roberts, G.R., Stacey, N.J., McCann, M.C., Maxwell, A., and Roberts, K. (2005). RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl. Acad. Sci. USA 102 18736–18741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Stacey, N.J., Corsar, J., Roberts, K., and McCann, M.C. (2002). DNA Topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12 1782–1786. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Ohkura, H., Adachi, Y., Morino, K., Shiozaki, K., and Yanagida, M. (1987). DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50 917–925. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz, P.D., Wilm, M., Bonte, E., Dumas, K., Mann, M., and Becker, P.B. (1997). Chromatin remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388 598–602. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Langeveld, S., Mayer, U., Ripper, G., Jürgens, G., Kammen, A.V., and Vries, S.C.D. (1996). Pattern formation in the Arabidopsis embryo revealed by position-specific lipid transfer protein gene expression. Plant Cell 8 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.D., Oppenheimer, D.G., Concienne, J., and Larkin, J.C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127 3931–3940. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, J.C. (2002). Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 3 430–440. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, S., and Lam, E. (1994). Abundance of nuclear DNA topoisomerase II is correlated with proliferation in Arabidopsis thaliana. Nucleic Acids Res. 22 5729–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Cheong, H., Friedrichsen, D., Zhao, Y., Hu, J., Mora-Garcia, S., and Chory, J. (2002). A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc. Natl. Acad. Sci. USA 99 10191–10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.