Abstract
The maize smut fungus Ustilago maydis switches from yeast to hyphal growth to infect maize (Zea mays) plants. This switching is promoted by mating of compatible cells and seems to be required for plant penetration. Although many genes distinctively expressed during this dimorphic switch have been identified and shown to be essential for the infection process, none seems to be explicitly required for polar growth control. Here, we report the characterization of pcl12, encoding a cyclin that interacts specifically with Cdk5, an essential cyclin-dependent kinase with regulatory roles in morphogenesis in U. maydis. Pcl12 fulfills the requirements to be a virulence-specific regulator of polar growth in U. maydis. First, pcl12 expression is induced during the pathogenic development. Secondly, Pcl12 is sufficient to induce hyperpolarized growth in U. maydis cells, as haploid cells overexpressing pcl12 in axenic conditions produce filaments that were morphologically indistinguishable from those produced during the infection process. Finally, cells defective in pcl12 showed impaired polar growth during the formation of the b-dependent filament, the induction of the conjugation tubes, or the formation of a promycelium in spore germination. However, in spite of this pivotal role during morphogenesis, pcl12 mutants were virulent. We discuss the implications of these results for the role of polar growth during the infection process.
INTRODUCTION
Many fungal pathogens undergo morphological transformations during host invasion, and frequently these morphological transitions correlate with the ability to switch between isotropic and polar growth or with changes in the direction of polarity axes (Gow et al., 2002). For instance, plant diseases caused by foliar fungal pathogens are initiated when spores attach to host surface and germinate. The process of germination implies the activation of a polarity axis and the emergence of a germ tube. For some fungi, the resulting germ tube has evolved to locate natural openings, such as stomata, or alternatively has also evolved the ability to elaborate specialized infection structures, such as appressoria, that enable direct penetration of plant cuticle (for a review, see Tucker and Talbot, 2001). It is clear that the ability to form polarized hyphae may represent an Achilles' heel that can be exploited to limit fungal invasion of the plant tissue (Harris, 2006). Paradoxically, how these processes are regulated in conjunction to the induction of the virulence program in phytopathogenic fungi is largely unknown.
The maize smut fungus Ustilago maydis is an excellent model system for the analysis of the molecular basis of fungal plant pathogenicity (Bölker, 2001; García-Pedrajas and Gold, 2004; Kahmann and Kämper, 2004). This basidiomycete fungus belongs to an important group of plant pathogens, the smut fungi, which can cause considerable grain yield loss and economic damage (Agrios, 1997). In the field, maize smut infections are dispersed by air-borne diploid teliospores (Christensen, 1963; Brown and Hovmoller, 2002). Germination of the teliospore on the plant surface is the first step in the infection process. Upon germination, meiosis takes place, and pairs of compatible haploid cells are generated. Pathogenic development is mediated by two independent loci: the a-locus, encoding a pheromone receptor system, and the b-locus, encoding a pair of homeoproteins (bW and bE). The process initiates with the recognition of mating pheromone secreted by haploid cells of the opposite mating type on the plant surface (Bölker et al., 1992). This induces a cell cycle arrest (García-Muse et al., 2003; Pérez-Martín et al., 2006) and triggers cells to the formation of long conjugation tubes. These filaments grow toward each other and fuse at their tips (Snetselaar et al., 1996). After cell fusion, the combination of the cytoplasm of compatible cells leads to the formation of the bE/bW transcription factor that triggers growth of the b-dependent hyphae on the surface of plant epidermis (Kahmann and Kämper, 2004). On the plant surface, infectious hyphae grow in a polar manner by expanding at the apical tip and inserting regularly spaced septa at the distal pole, resulting in the formation of characteristic empty sections (Steinberg et al., 1998). This growth mode enables the fungus to progress along the plant surface most likely to find an appropriate point of entry. Eventually, the filaments differentiate appressoria and penetrate the cuticule (Snetselaar and Mims, 1992, 1993). Because polarized growth of hyphae plays a key role in all of these steps, it is not surprising that cytoskeleton regulators, like Rac1, or molecular motors, such as myosin V, are required for pathogenic development in U. maydis (Weber et al., 2003; Mahlert et al., 2006). However, how these housekeeping elements are differentially regulated during the pathogenic development is not currently understood because no virulence-specific polarity regulators have been identified so far in U. maydis.
Recently, we described a kinase, Cdk5, that is required for sustained polar growth in U. maydis (Castillo-Lluva et al., 2007). Cdk5 belongs to a family of cyclin-dependent kinases (CDKs) implicated in the regulation of morphogenesis in organisms ranging from yeast to human (Xie et al., 2006). CDK activity requires the interaction with proteins known as cyclins (Morgan, 1997), which target the catalytic subunit to correct substrates. This idea is supported by the observation that a single catalytic subunit, in complex with different cyclins, can phosphorylate a different set of substrates (Roberts, 1999). The ortholog of U. maydis Cdk5 in Saccharomyces cerevisiae, Pho85, associates with 10 cyclins that can be divide into two subfamilies based on sequence similarity: the Pho80 cyclin subfamily (Pho80, Pcl6, Pcl7, Pcl8, and Pcl10) and the Pcl1,2 subfamily (Pcl1, Pcl2, Cgl1, Pcl5, and Pcl9; for a review, see Andrews and Measday, 1998; Carroll and O'Shea, 2002). It is thought that each cyclin directs Pho85 to phosphorylate distinct substrates involved in different cellular processes (Dephoure et al., 2005). Therefore, mutations in the different cyclins produce differing phenotypes, indicating that, in spite of some degree of functional overlapping, Pho85 complexes containing different cyclin subunits can have different functions (Carroll and O'Shea, 2002; Huang et al., 2002).
U. maydis cells carrying a cdk5 conditional mutation showed drastically reduced virulence (Castillo-Lluva et al., 2007). However, because of the essential role of Cdk5 for growth, it is not clear whether this requirement for virulence reflects specific roles of Cdk5 during the pathogenic development or whether it is an indirect effect of the various cellular abnormalities associated with the cdk5 conditional mutation. To address this issue, we reasoned that as it happens in budding yeast, it could be possible that distinct Pcl-like cyclins associate with Cdk5 and that some of these putative cyclins could be specifically required during the induction of the virulence program. Here, we report the identification of a Cdk5-associated cyclin, Pcl12, that plays a specific role in the hyphal morphogenesis of U. maydis during the pathogenic development. We describe the characterization of its properties and regulation, its role in controlling hyphal morphogenesis, and its importance for virulence.
RESULTS
The U. maydis Genome Encodes Seven Putative Pcl Cyclins
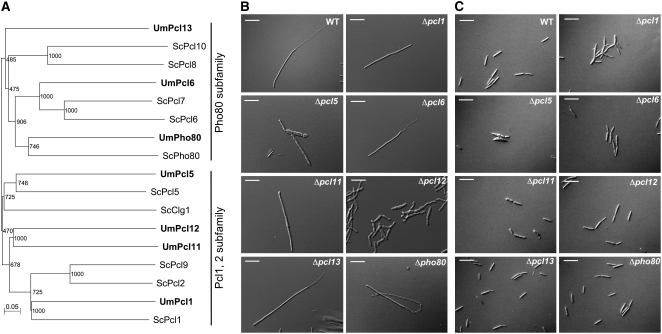
To explore the possibility that U. maydis may contain Pcl-related cyclins, we used the sequences of the described S. cerevisiae Pcl proteins to conduct a tBLASTn search against U. maydis genomic sequence data. We found seven significant matches (GenBank accession numbers EF494633 to EF494639). A multiple sequence alignment was used to construct a phylogenetic tree that confirmed the relatedness of the protein sequences to S. cerevisiae Pcl cyclins. In this tree, U. maydis Pcl cyclins grouped into the two major subfamilies described for S. cerevisiae Pcl cyclins: the Pho80 and the Pcl1,2 subfamilies (Figure 1A). We named these U. maydis cyclins with the name of the more related S. cerevisiae Pcl cyclin. In three cases, as no clear counterpart was present in S. cerevisiae, we followed the numeration of the cyclins described in S. cerevisiae; therefore, we named these new cyclins Pcl11, Pcl12, and Pcl13.
Figure 1.
Pcl-Like Cyclins in U. maydis.
(A) The genome of U. maydis encodes for seven Pcl-like cyclins. Dendrogram of Pcl cyclins from S. cerevisiae and U. maydis. Branch lengths are proportional to the estimated number of amino acid substitutions; the scale bar indicates amino acid substitutions per site. Bootstrap support values are given at branch nodes. S. cerevisiae cyclin sequences were downloaded from PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). Note that U. maydis Pcl cyclins group into the two S. cerevisiae subfamilies: Pho80 and Pcl1,2. Alignment data are presented in Supplemental Table 1 online.
(B) Morphology phenotype of pcl null mutants in AB33 background. AB33 strain carries compatible bE and bW alleles under the control of the nar1 promoter and forms b-dependent filaments upon shift to minimal medium with nitrate as nitrogen source. We observed that most pcl null mutants do not show impaired hyphal growth after 8 h of incubation in inducing conditions. Δpcl5 and Δpcl11 showed defects in filament formation, and Δpcl12 was severely impaired. Bars = 20 μm.
(C) Morphology phenotype of pcl null mutants in FB2 background. FB2 is a wild-type haploid strain. Cells were grown as in (B). Note that Δpcl1 cells showed a defect in cell separation, while Δpcl5 and Δpcl11 showed morphogenetic defects. Δpcl12 cells showed a wild-type appearance. Bars = 20 μm.
To address whether some of these putative Pcl cyclins play specific roles during the formation of the infective filament in U. maydis, we took advantage of the AB33 strain (Brachmann et al., 2001). This haploid laboratory strain harbors the compatible bW1/bE2 genes under control of the nitrate-inducible nar1 promoter (Brachmann et al., 2001). Thus, b-dependent infective filament formation can be elicited in AB33 by changing the nitrogen source. The resulting monokaryotic filament mimics its dikaryotic counterpart in all aspects of filamentous growth: apical tip extension, nuclear migration, and formation of septa and empty sections at the distal pole (Steinberg et al., 1998; Brachmann et al., 2001; Schuchardt et al., 2005). We reasoned that for those pcl genes acting in the formation of the infective filament, their deletion in AB33 cells would result in some impairment in the formation of the infective filament under growth in induction conditions (nitrate-containing minimal medium). To discard effects of the nutritional conditions after the shifting to nitrate as nitrogen source, as well as to discriminate a general role in the cellular morphogenesis versus specific roles in the formation of the infective filament, we deleted pcl genes also in FB2 cells, a haploid wild-type strain with the same genetic background as AB33. From our analysis we found that mutants in pcl5 and pcl11 produced filaments that were slightly thicker than wild-type filaments and grew irregularly. However, these mutants also showed morphological defects in yeast-like cells (FB2 background), suggesting a general role of these genes in morphogenesis (Figures 1B and 1C). Interestingly, we found that mutants in pcl12 showed a wild-type appearance in FB2 cells, but formation of the b-dependent filament (i.e., AB33Δpcl12 cells) was clearly impaired (Figures 1B and 1C).
pcl12 Mutants Are Unable to Focalize the Growth in the Filament Apex
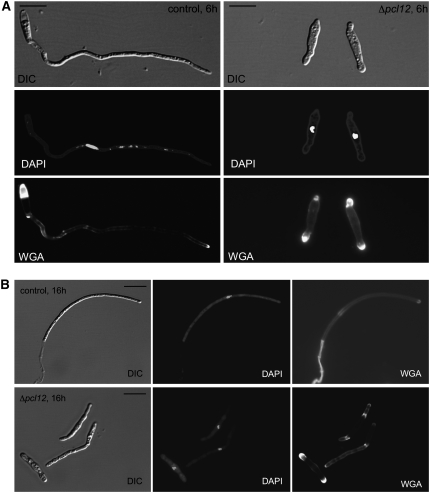
We decided to analyze in more detail the defects in filamentation associated to the lack of pcl12. Induction of bW2 and bE1 expression in AB33 cells for 6 h resulted in the formation of a filament (∼90 μm long), containing a single nucleus and accumulating chitin (stained with fluorescein isothiocyanate [FITC]-labeled wheat germ agglutinin [WGA]; Figure 2A, left panels) at the growing hyphal tip (Brachmann et al., 2001). In this filament, it was possible to observe the formation of empty cell compartments (brightly stained with FITC-WGA) at the basal cell end. Longer incubations (16 h) resulted in a filament consisting of a tip cell, which is cell cycle arrested, leaving basal compartments bounded by the cell wall and regularly separated by septa (Figure 2B, top panels). By contrast, AB33Δpcl12 cells shifted to inductive conditions for 6 h were unable to form the filament, and they showed swollen tips, which accumulated WGA staining (Figure 2A, right panels). Longer incubations in inducing conditions resulted in the formation of elongated cells (46 ± 20 μm, n = 20) that were irregular in shape. In contrast with wild-type hyphae, WGA staining was not restricted to the growing apex but was found along the length of the hypha with strong accumulations at swollen areas (Figure 2B, bottom panels). Only scarcely was it possible to find cells carrying empty basal compartments. The mutant cells were cell cycle arrested as wild-type cells and accumulated with a 2C DNA content and single nucleus, in agreement with a G2 cell cycle arrest (N. Mielnichuk, C. Sgarlata, and J. Pérez-Martín, unpublished data; see Supplemental Figure 1 online).
Figure 2.
Morphology of b-Dependent Filament in the Absence of Pcl12.
(A) Morphology after 6 h of induction. Control cells (AB33, left column) showed a typical filament, ∼90 μm, carrying a single nucleus (4′,6-diamidino-2-phenylindole [DAPI] staining, middle row), and the chitin accumulation (stained with FITC-labeled WGA, bottom row) was restricted to the growing apex and the basal septa; mutant cells do not show elongated growth, and WGA staining was located at both cell poles as a diffuse swelled area. DIC, differential interference contrast. Bars = 15 μm.
(B) Morphology after 16 h of induction. Bars = 10 μm.
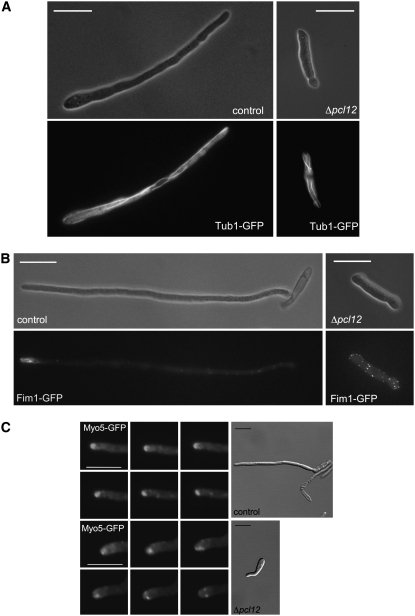
We analyzed the distribution of the actin and microtubule cytoskeletons using fimbrin-green fluorescent protein (GFP) and α-tubulin-GFP protein fusions, respectively. The microtubule cytoskeleton showed no difference between the wild-type and mutant cells (Figure 3A), and in both cases, it was compatible with a G2 cytoplasmic array, which is characterized by several bundles of microtubules that point toward the growth region (Steinberg et al., 2001; Pérez-Martín et al., 2006). By contrast, we found that while in AB33 cells fimbrin-GFP signal is concentrated at the growth tip (Figure 3B, left panels), in AB33Δpcl12 cells, actin patches were randomly distributed throughout the cell length (Figure 3B, right panels).
Figure 3.
pcl12 Mutants Are Unable to Focalize the Growth in the Filament Apex.
(A) The microtubule cytoskeleton was not affected by the absence of Pcl12. Control and Δpcl12 cells carrying a tub1-gfp allele were incubated for 6 h in nitrate. Note that in both cases the GFP fluorescence was compatible with a typical G2 microtubule array. Bars = 15 μm.
(B) Fimbrin-GFP localizes to the tip of b-dependent filament. Control and Δpcl12 cells carrying the fim1-gfp allele were incubated for 6 h in nitrate-containing minimal medium. Note that in control cells, fimbrin patches are accumulated at the tip of the filament, in mutant cells, the GFP signal is dispersed throughout the cell length. Bars = 15 μm.
(C) Myo5-GFP localization. Time series of cell poles of control (AB33) and Δpcl12 cells carrying a Myo5-GFP fusion grown for 6 h in minimal medium with nitrate. Images (taken every 2 s) were magnified from the cell shown in the DIC image and run left to right from the top to the bottom row. Note that in control cells, the GFP-Myo5 accumulation at the growth region remains stationary, while in Δpcl12, cells undergo rapid rearrangement and diffuse at the tip surface. Bars in DIC panels = 10 μm; bars in GFP panels = 5 μm.
The loss of polarization of actin patches observed in AB33Δpcl12 cells could be explained either by a defect in the ability to establish polar growth or by a defect in the maintenance of such polar growth. To distinguish between these possibilities, we introduced a fusion protein of GFP and Myo5, a class V myosin from U. maydis that is transported via actin cables toward the growing end of the cell (Weber et al., 2003). GFP-Myo5 signal appeared at the filament tip in control and mutant cells (Figure 3C), indicating that the ability to detect and direct transport toward the hyphal tip was not impaired. However, while in cells carrying a wild-type pcl12 allele, the GFP-Myo5 signal remained stationary associated with the tip (Figure 3C, top panels), and in AB33Δpcl12 cells, the fluorescent signal localization was unstable, and once it reached the hyphal apex, it diffused toward distal tip areas (Figure 3C, bottom panels; see Supplemental Movies 1 and 2 online).
From these observations it seems that in the absence of Pcl12, the ability to direct the transport of material to the hyphal apex was not impaired, but the ability to focalize the deposition of cell wall material in the hyphal apex was defective, resulting in a suboptimal ability to induce a strong polar growth.
pcl12 Encodes a Cdk5-Interacting Cyclin
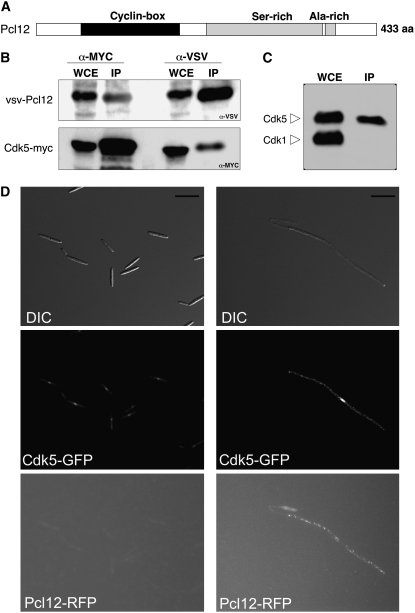
Pcl12 corresponds to the predicted protein UM10529 in the annotated U. maydis genome database (http://mips.gsf.de/genre/proj/ustilago/). The gene encoding protein UM10529 is predicted to have two introns by automated annotation. However, rapid amplification of cDNA ends analysis showed that pcl12 had a single intron (data not shown; GenBank accession number EF494639). Analysis of the amino acid sequence of Pcl12 indicated the presence of a cyclin box located between residues 59 and 184, a Ser-rich region located between residues 218 and 366, and an Ala-rich region between residues 367 and 379 (Figure 4A).
Figure 4.
Pcl12 Is a Cyclin of Cdk5.
(A) Domain organization of Pcl12. Domain analysis was done using the SMART server (http://smart.embl-heidelberg.de). aa, amino acids.
(B) Pcl12 associates with Cdk5. Lysates prepared from SONU167 cells, expressing a VSV-tagged version of Pcl12 and a MYC-tagged Cdk5 allele, were immunoprecipitated with anti-MYC or anti-VSV antibodies to pull down Cdk5 or Pcl12, respectively. The whole-cell extracts (WCE) and the immunoprecipitates (IP) were separated by SDS-PAGE and immunoblotted with anti-MYC and anti-VSV to detect Cdk5 and Pcl12, respectively.
(C) Pcl12 is a Cdk5-specific cyclin. A lysate prepared from SONU160 cells, expressing a VSV-tagged version of Pcl12, was immunoprecipitated with anti-VSV antibodies and assayed by protein gel blots using anti-PSTAIRE antibodies that detect both Cdk5 and the mitotic CDK, Cdk1. Note that only Cdk5 was found in the immunoprecipitate fraction, indicating that Pcl12 specifically interacts with Cdk5.
(D) Localization of Pcl12-RFP and Cdk5-GFP in the b-dependent filament. AB33 derivate cells carrying pcl12-rfp and cdk5-gfp alleles were grown for 8 h in ammonium-containing (left column) or nitrate-containing (right column) minimal medium. Bars = 15 μm.
In S. cerevisiae, Pcl cyclins specifically interact with the Pho85 kinase, whose ortholog in U. maydis is Cdk5 (Castillo-Lluva et al., 2007). To test physical interaction between the Cdk5 kinase and Pcl12 in U. maydis, we expressed an N-terminal VSV-tagged Pcl12 allele in a strain carrying a C-terminal myc-tagged functional cdk5 allele (Castillo-Lluva et al., 2007), and we performed a coimmunoprecipitation assay (Figure 4B). We found that VSV-Pcl12 associated with Cdk5-myc, supporting the notion of Pcl12 as a Pcl-like cyclin. To determine whether the interaction between Pcl12 and Cdk5 is specific (i.e., Pcl12 is not able to interact in U. maydis with other CDKs, such as the mitotic kinase Cdk1) (García-Muse et al., 2004), VSV-Pcl12 immunoprecipitates were also probed using anti-PSTAIRE antibodies that detect both Cdk5 and Cdk1 (Castillo-Lluva et al., 2007). No interaction was detected between Cdk1 and Pcl12 (Figure 4C).
We also sought to analyze to what extent the localization patterns of Pcl12 and Cdk5 overlap. To analyze the subcellular location of Pcl12, we fused a red fluorescent protein (RFP) tag to the C terminus of the endogenous pcl12 gene in AB33-derived cells carrying a cdk5-gfp allele. The resulting strain was indistinguishable from AB33 cells with respect to the ability to produce the b-dependent filament upon induction (data not shown), indicating that the fusion proteins were functional. We found that in noninduction conditions for b genes (i.e., cells growing in ammonium), the Cdk5-GFP signal was mainly located at the nucleus, as we previously reported in yeast-like cells (Castillo-Lluva et al., 2007). No signal was apparent for Pcl12-RFP, suggesting a low level of expression of this gene under this condition (Figure 4D, left column). Interestingly, in cells growing in nitrate (i.e., b-expression was induced), we observed RFP single spots that were localized through the filament (Figure 4D, right column) but excluded from the nucleus. In these conditions, GFP-Cdk5 can be detected in the nucleus but also in the cytoplasm in the form of dots that were coincident with the RFP dots observed.
Together, these data indicate that the Pcl12 cyclin specifically interacts with its partner kinase Cdk5 and is distributed throughout the entire b-dependent filament.
Pcl12 Is Highly Expressed during the b-Dependent Filament Induction, and Its Ectopic Expression Resulted in Filament Formation
As the data above suggest that pcl12 could be expressed in a b-dependent manner, we analyzed the expression pattern of pcl12 under b-induced conditions, using the pair of strains AB33 and AB34, in which the combination of either bW2 and bE1 (active b complex) or bW2 and bE2 (nonactive b complex) genes, respectively, was induced by shifting the cells to nitrate-containing medium (Brachmann et al., 2001). In agreement with its role during b-dependent filament formation, we found that pcl12 transcription was strongly induced in the presence of an active b complex (Figure 5A).
Figure 5.
Pcl12 Is Sufficient to Induce Filamentation.
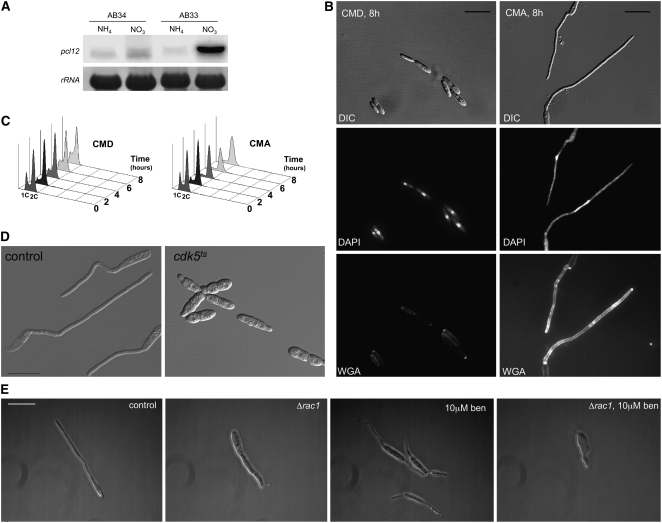
(A) RNA gel blot analysis of pcl12 expression. Total RNA was extracted from AB33 (active b) and AB34 (non-active b) cells growing in minimal medium with ammonium (noninducing conditions; NH4) or nitrate (inducing conditions; NO3) as nitrogen source. In all cases, 10 μg of total RNA was loaded per lane. The same filter was hybridized in succession with probes for pcl12 and 18s rRNA as loading control.
(B) Morphology of cells expressing an extra copy of pcl12 under the control of the crg1 promoter grown for 8 h in induction (complete medium with arabinose [CMA]) or repression conditions (complete medium with glucose [CMD]). Observe the production of a filament carrying a single nucleus (DAPI) and supporting a focalized apex growth (WGA staining). Bar = 15 μm.
(C) pcl12 overexpression induces a G2 cell cycle arrest. Fluorescence-activated cell sorter (FACS) analysis of SONU147 cells in repressive and induction conditions. Note the accumulation of cells carrying a 2C DNA content in CMA.
(D) Pcl12-induced filamentation is dependent on cdk5. Overexpression of pcl12 in cells carrying a wild-type cdk5 allele (control) or carrying a cdk5ts allele. Cells were grown in CMD and then shifted to induction conditions (CMA) and restrictive temperature (34°C) for 6 h. Bar: 20 μm.
(E) Pcl12-induced filamentation is dependent on rac1 and the microtubule cytoskeleton. Overexpression of pcl12 in cells carrying a wild-type rac1 allele (control) or carrying a deletion of rac1 (Δrac1) in the presence or not of 10 μM benomyl. Cells were grown in CMD and then shifted to induction conditions (CMA) in the presence or absence of benomyl (ben) for 6 h. Bar = 15 μm.
The above results suggest a correlation between pcl12 expression and filament formation. Therefore, we wondered about the consequence of the forced expression of pcl12 in haploid cells during axenic growth conditions. For this, we introduced an extra copy of pcl12 into haploid U. maydis cells under the control of the crg1 promoter, which can be induced by arabinose (Bottin et al., 1996). Encouragingly, haploid cells carrying this extra copy of pcl12 displayed hyperpolarized growth after the shift to inducing conditions (Figure 5B, right panel). Furthermore, these filaments were indistinguishable from those formed after expression of an active b complex (i.e., those observed in AB33 cells), and extensive incubations resulted in filaments that left empty sections behind (data not shown). These filaments were composed of a single apical cell carrying a single nucleus that accumulates with a 2C DNA content (Figures 5B and 5C). The highly polarized growth triggered by induced expression of pcl12 in yeast cells was dependent on Cdk5, as it was abrogated in a cdk5ts background at restrictive temperature (Figure 5D).
Formation of the b-dependent filament is dependent on rac1 (Mahlert et al., 2006). Therefore, we analyzed the pcl12-induced filament in the absence of rac1 and found that cells expressing high levels of pcl12 in the absence of rac1 were still able to undergo a certain degree of polar growth, although the filament produced was shorter and thicker than control filament (Figure 5E). As both actin- and microtubules-based transport collaborates in b-dependent hyphal growth (Fuchs et al., 2005; Schuchardt et al., 2005), we wondered whether microtubules were responsible for this residual polar growth found in rac1 mutants. Treatment of pcl12-overexpressing cells with benomyl, a microtubule destabilizer, resulted in bipolar growth and the formation of very short hyphae, in striking similarity to what has been reported after treatment of b-dependent hyphae with benomyl (Fuchs et al., 2005). Furthermore, disruption of microtubules abolished the residual growth of Δrac1 hyphae and led to rounded cells (Figure 5E). In conclusion, both rac1 and microtubule activity are needed to maintain pcl12-dependent polarized hyphal growth, as it has been showed for b-dependent filaments (Fuchs et al., 2005; Schuchardt et al., 2005).
Pcl12 Is Required for the Formation of b-Dependent Filament
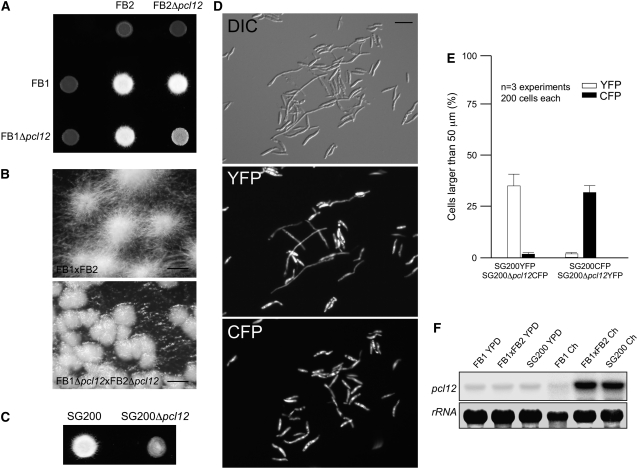
Even when the b-dependent filament produced in AB33 cells mimics its dikaryotic counterpart in all aspects of filamentous growth, we sought to analyze the consequences of the absence of Pcl12 in the formation of the infective filament in more native conditions. Crosses of compatible haploid wild-type cells on charcoal-containing plates resulted in the formation of dikaryotic hyphae consisting of a tip cell, ∼150 μm long, which formed basal empty compartments as it grew (Holliday, 1974; Steinberg et al., 1998). We crossed compatible wild-type and Δpcl12 strains on charcoal-containing plates. Wild-type crosses led to a typical fuzzy colony appearance, which was attributable to the massive formation of dikaryotic hyphae (Banuett and Herskowitz, 1989). By contrast, Δpcl12 mutants were strongly attenuated in filament formation (Figures 6A and 6B).
Figure 6.
Pcl12 Is Required for the Proper Formation of b-Dependent Filament.
(A) Crosses of control strains FB1 × FB2 and Δpcl12 mutant strains in charcoal-containing agar plates. Note the gray appearance of mutant cross, indicating impairment in filament formation.
(B) Magnification of the colony border of (A). Note that while the control cross generates a mat of filamentous cells, in the mutant cross, this appearance is impaired. Bars = 0.5 mm.
(C) Solopathogenic strain SG200Δpcl12 growing on charcoal-containing plates also showed defective filamentation.
(D) Analysis of ability to produce filaments by Δpcl12 cells versus control cells. Expression of yellow- and cyan-shifted derivatives of GFP (YFP and CFP) allows the identification of SG200 and SG200Δpcl12 strains when spotted together on charcoal-containing plates. Cells were scrapped from agar surface, mounted on microscopy slides, and epifluorescence observed. The top image shows a DIC image of cells on charcoal surface. Middle image shows fluorescence in CFP channel (SG200-YFP cells), and the bottom image shows fluorescence in YFP channel (SG200Δpcl12-YFP mutant). Bar = 20 μm.
(E) Quantification of (D).
(F) Expression of pcl12 during crosses on charcoal plates. Cultures of haploid wild-type (FB1), solopathogenic (SG200), and crosses of two compatible haploid strains (FB1 × FB2) were plated on solid YPD and charcoal-containing plates (Ch). Approximately equal amounts of cell material were scraped from the plate surface, and total RNA was extracted and submitted to RNA gel blot analysis. In all cases, 10 μg of total RNA was loaded per lane. The same filter was hybridized in succession with probes for pcl12 and 18s rRNA as loading control.
To compare the efficiency of filament formation quantitatively in charcoal plates, we took advantage of the SG200 strain, which harbors a compatible combination of the bE1 and bW2 genes and the mfa2 gene inserted in the a1 locus and is thus able to form the infective filament without the need for a mating partner (Bölker et al., 1995). SG200 cells lacking pcl12 were also severely impaired in the formation of long filaments (Figure 6C). To differentiate wild-type from Δpcl12 cells, we used combinations of SG200 and SG200Δpcl12 strains that were tagged with constitutively expressed genes for cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) (Figure 6D). We cospotted into charcoal plates a 1:1 mixture of wild-type and mutant cells, and we counted the number and length of the filaments produced. We found that irrespective of the combination used (i.e., SG200-CFP and SG200Δpcl12-YFP or SG200-YFP and SG200Δpcl12-CFP), long filament formation was reduced in Δpcl12 cells compared with wild-type cells (Figure 6E).
We also analyzed the expression of pcl12 in conditions that produce dikaryotic hyphae (combination of compatible haploid cells) as well as in solopathogenic SG200 cells. For this, we scrapped different cell combinations from charcoal-containing plates. As a control we used cells growing in solid rich medium (YPD), a condition in which the expression of the pathogenicity program is repressed (Spellig et al., 1994; Castillo-Lluva and Pérez-Martín, 2005). In agreement with the data obtained for AB33 cells, pcl12 expression was highly induced in conditions that allow the formation of the filament (i.e., cells carrying a compatible combination of b genes) (Figure 6F).
Absence of Pcl12 Delays the Formation of the Conjugation Tube
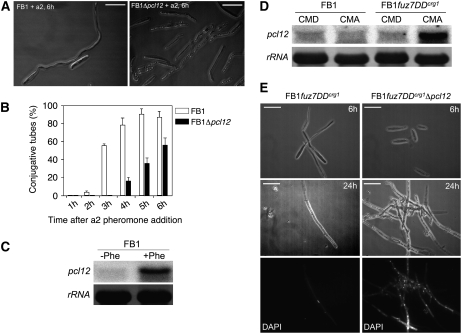
In response to pheromone, U. maydis cells undergo a cell cycle arrest and form long conjugation tubes that, under normal conditions, search for a compatible mating partner (Spellig et al., 1994; García-Muse et al., 2003). As formation of conjugation tubes implies the induction of a strong polar growth, we sought to analyze whether this response was also altered in Δpcl12 mutants. Pheromone treatment of cells in liquid culture results in the formation of long conjugation tubes (Szabo et al., 2002), and this allowed us to quantify tube formation in pcl12 null mutant cells. Our analysis revealed that Δpcl12 cells produced significantly fewer tubes (Figure 7B). In addition, the tubes were shorter than those produced by wild-type cells and they showed a high vacuole content (Figure 7A). We confirmed these results using confrontation assays of compatible haploid cells (see Supplemental Figure 2 online). In agreement with some role of Pcl12 in the formation of the conjugation tubes, we found that pcl12 was induced after pheromone treatment (Figure 7C). These data suggest that Pcl12 plays a role in the polar growth of the conjugation tube, although we cannot exclude some indirect effect such as the possibility that Pcl12 participate in pheromone sensing as it has been recently proposed for Myo5 and Yup1 (Weber et al., 2003; Fuchs et al., 2006). To address this issue, we took advantage of the fuz7DD allele, which encodes an activated version of the mitogen-activated protein kinase (MAPK) kinase involved in pheromone response (Banuett and Herskowitz, 1994; Müller et al., 2003), since cells expressing this allele under the arabinose-induced crg1 promoter were largely independent on the upstream components (i.e., pheromone and receptors) for conjugation tube formation when grown in arabinose-containing medium (Müller et al., 2003). As we observed previously after pheromone treatment, pcl12 expression was increased after fuz7DD induction (Figure 7D). We found that cells lacking pcl12 and expressing fuz7DD were severely delayed in the formation of conjugation tube with respect to control cells. For instance, 6 h after shift to arabinose-containing medium, almost the totality of control cells produced conjugation tubes, whereas Δpcl12 cells only showed a slight tip elongation (Figure 7E, top row). These results indicate that the delay observed in conjugation formation after pheromone treatment was not due to problems in pheromone sensing, supporting a direct role of Pcl12 during the polar growth of the conjugation tube. Strikingly, we observed a bizarre behavior of pcl12 mutants after a prolonged fuz7DD induction. Incubation of more than 24 h of control cells under inducing conditions resulted in long filaments in which only the tip cell was filled with cytoplasm carrying a single nucleus, and the remaining part of the hyphae consisted of empty sections separated by septa (Figure 7E). This behavior has been previously noted as a consequence of a permanent G2 cell cycle arrest due to a sustained MAPK activation (Flor-Parra et al., 2006). However, in Δpcl12 cells, the filaments seemed to be not cell cycle arrested as they were formed by compartments carrying each a single nucleus (Figure 7E). Currently, we do not have an explanation for this behavior, but it links Pcl12 with the ability to arrest cell cycle progression in response to a sustained MAPK activation (García-Muse et al., 2003).
Figure 7.
Pheromone Response in Δpcl12 Cells.
(A) Formation of conjugation tubes in wild-type and Δpcl12 cells after 6 h of a2 pheromone addition. Note that conjugation tubes in Δpcl12 cells were shorter and showed a dramatic vacuolization. Bars = 15 μm.
(B) Quantitative analysis of conjugative tube formation.
(C) pcl12 expression is enhanced after pheromone treatment. FB1 cells were treated with a2 pheromone or DMSO as a solvent control, and after 6 h, RNA was extracted and submitted to RNA gel blot analysis using pcl12 and 18s RNA probes.
(D) pcl12 is induced by the MAPK cascade. UMN4 cells carrying fuz7DD, a constitutive allele of the Fuz7 MAPK kinase, under the control of Pcrg1 promoter were grown to an OD600 of 0.2 in noninducing conditions (CMD) and then shifted to noninducing conditions (CMD) and inducing conditions (CMA) for 6 h.
(E) Pcl12 is required for sustained MAPK-induced cell cycle arrest. Cells carrying fuz7DD under Pcrg1 control and grown for 6 h in arabinose produce structures resembling conjugation tubes (left column, top). Prolonged incubation (24 h, left column, middle and bottom images) resulted in a single-celled filament that is cell cycle arrested and leaves septa behind. However, in pcl12 defective cells (right column), there is a clear delay in the production of conjugation tubes (top image), and prolonged incubation resulted in a filament that is not cell cycle arrested (see the multiple nuclei in DAPI image, bottom). Bars = 15 μm.
Analysis of the Virulence of Δpcl12 Mutants
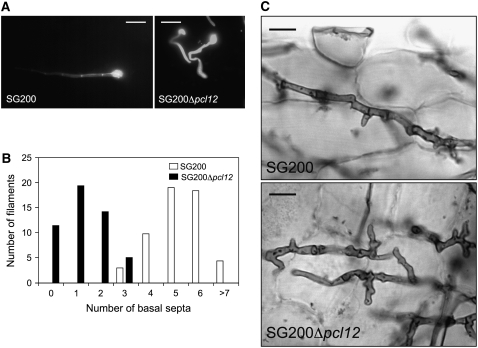
U. maydis infection of maize (Zea mays) results in anthocyanin pigment production by the plant and the formation of tumors that are filled with proliferating fungal cells that eventually differentiate into black teliospores (Banuett and Herskowitz, 1996). To assess the ability of Δpcl12 strains to infect and cause symptoms in plants, we inoculated by stem injection maize plants with mixtures of wild-type and mutant strains. We were surprised to observe disease symptoms with compatible Δpcl12 strains that were as severe as symptoms induced by wild-type strains (data not shown). Because of the dramatic defects observed during filament formation on charcoal plates, we wondered whether these defects were apparent on the plant surface. To address this issue, we examined the role of pcl12 during the morphological changes that U. maydis cells undergo during plant infection. For this, we inoculated plants with the solopathogenic strain SG200 and a Δpcl12 derivative and observed fungal cells onto the leaf surface after staining with Calcofluor. In SG200 cells, we observed hyphae formation that showed regular basal septation up to the appressorial tip, which is strongly stained by calcofluor (Figure 8A). By contrast, SG200Δpcl12 cells formed short filaments that were swollen, misshapen, and that had a limited number of basal septa and an abnormal chitin distribution (Figures 8A and 8B). Nevertheless, in spite of these defects, mutant cells were able to produce appressoria and entered the plant. Further time-course experiments 1 week after infection did not reveal significant differences between wild-type and mutant cells proliferating inside the plant tissue. Only a slight different morphology is apparent since mutant hyphae appear slightly curved or curled (Figure 8C). However, tumors were observed, and they had a wild-type appearance and were filled with black teliospores (data not shown).
Figure 8.
Analysis of Δpcl12 in Planta.
(A) Control and Δpcl12 hyphae stained with calcofluor after incubation for 24 h. Note that both hyphae are able to form appressorium, but in SG200 Δpcl12 hyphae, the appressorium was formed shortly with no basal septa, in contrast with the control strain that showed regular septation up to the appressorial tip. Bars = 10 μm.
(B) Quantification of the number of basal septa in appressorium-forming filaments.
(C) Images of Chlorazole Black E–stained control and Δpcl12 hyphae growing inside the plant tissue 1 week after infection. Bars = 10 μm.
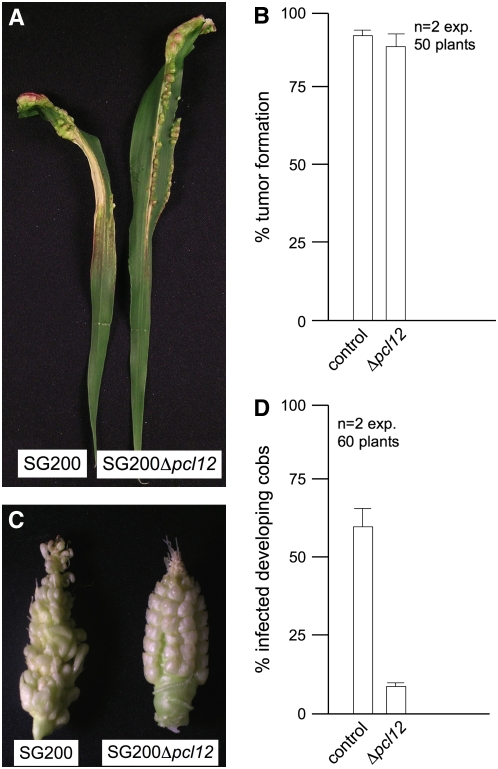
In the field, smut infections are frequently found in cobs, where kernels may be infected through the stigma (Christensen, 1963). Rapid directional growth of infective filament through the stigma frequently precedes the plant infection (Snetselaar and Mims, 1993). Consequently, sustained polar growth may be an important determinant of pathogenicity for U. maydis in these conditions because hyphae that fail to reach the ovary will not produce tumors. Since the morphological defects observed on charcoal plates were also apparent on plant surfaces, we reasoned that infection of developing cobs might uncover defects in the pcl12 mutants. Cultures of the solopathogenic strain SG200 and a Δpcl12 derivative were used to infect stems and developing cobs of maize plants. Encouragingly, while infection after stem injection gave no differences between wild-type and mutant strains, we observed that the ability to infect cobs was impaired in the Δpcl12 strain (Figure 9).
Figure 9.
Virulence of Δpcl12 cells
(A) Leaves of plants inoculated either with SG200 or SG200Dpcl12. Note the presence of anthocyanin stains and tumors
(B) Quantification of tumor formation on infected maize plants.
(C) Cobs of plants inoculated either with SG200 or SG200Dpcl12. Note the anomalous development of infected kernels.
(D) Quantification of tumor formation on infected cobs.
Teliospore Germination Requires Pcl12
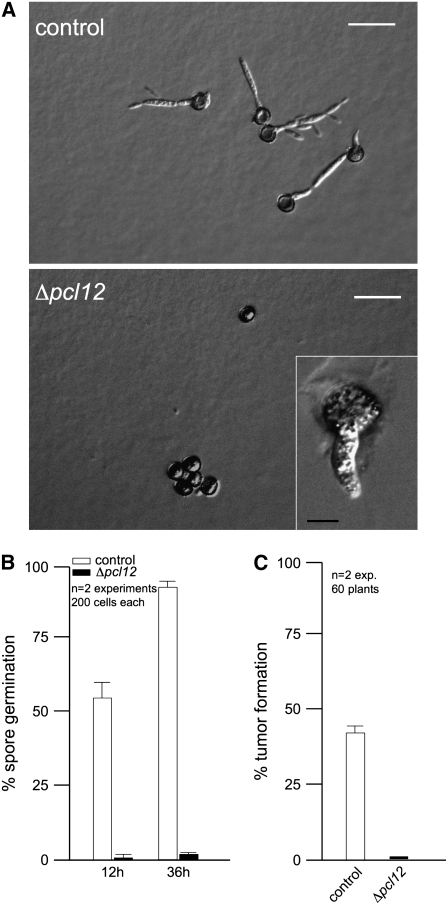
U. maydis teliospore germination has been studied at the morphological level by electron and light microscopy (O'Donnell and McLaughlin, 1984). It is a complex process that includes a switch from dormancy to physiological activity, the rupture of the thick cell wall, extension of a tubular promycelium, and the completion of meiosis to produce haploid cells. Since emergence of the promycelium implies the establishment of a new polarity axis, we wondered whether this process could be affected in Δpcl12 mutants. Tumors from infected plants with wild-type or Δpcl12 mutant cells were ground and teliospores isolated. Teliospore preparations were plated onto complete medium agar-coated slides and incubated for 12 and 36 h to observe and quantify teliospore germination (Figures 10A and 10B). Wild-type teliospores germinate by extending a promycelium, with subsequent meiosis and the formation of haploid progeny as buds from the promycelium (Figure 10A, top panel). However, the majority of Δpcl12 teliospores were unable to germinate, and only scarcely was it possible to observe in some teliospores a short and thick promycelium (Figure 10A, bottom panel and inset). In those few cases in which Δpcl12 teliospores managed to germinate, promycelium eventually rise wild-type-looking cells (data not shown).
Figure 10.
Pcl12 Is Required for Teliospore Germination.
(A) Images of control and Δpcl12 teliospores germinated on CM glucose–containing agar slides after 1 d of incubation. Note that wild-type teliospores extend a promycelium, from which haploid progeny are generated. In mutant teliospores, no germination is observed and, scarcely, a short and thick filament is observed in some teliospores (inset). Bars = 20 μm. Inset bar = 5 μm.
(B) Quantification of teliospore germination after 12 and 36 h. Error bars represent values ± sd.
(C) Quantitative analysis of tumor formation in plants sprayed with wild-type and Δpcl12 teliospores and incubated for 14 d after infection. Error bars represent values ± sd.
In the field, germination of the air-borne diploid teliospores is the first step in the infection process (Christensen, 1963; Brown and Hovmoller, 2002), and we wondered whether the requirement of Pcl12 for spore germination might have consequences in the ability of teliospores to infect plants. Therefore, we prepared teliospore suspensions from wild-type and mutant strains, and maize seedlings were then sprayed with these suspensions. Infection with wild-type spores led to tumor production in ∼40% of the inoculated plants. By contrast, no infection symptoms were observed in the plants sprayed with Δpcl12 teliospores (Figure 10C).
DISCUSSION
Pathogenic fungi undergo strong morphological changes during the process of plant infection. In maize smut fungus, cells switch to hyphal growth after mating, and this dimorphic switch is required for plant penetration. Hyphal morphogenesis in U. maydis is mediated by several signaling pathways, from which we currently have a fair amount of knowledge (for a review, see Feldbrügge et al., 2004). On the other hand, the knowledge of the machinery that executes morphogenesis in this fungus is increasing day to day, and the role of cytoskeleton components during the pathogenic development is an active area of research (Steinberg, 2006, 2007). However, a knowledge gap exists between the signal transmission pathways and the machinery involved in morphogenesis since how the housekeeping cytoskeleton components are differentially regulated during the pathogenic development is not currently understood, as no virulence-specific polarity regulators have been identified so far in U. maydis.
In this study, we have identified a gene, pcl12, which fulfills the requirements to be a virulence-specific regulator of polar growth in U. maydis. This gene encodes a cyclin that interacts specifically with Cdk5, an essential CDK with regulatory roles in morphogenesis in U. maydis (Castillo-Lluva et al., 2007). Cells defective in pcl12 showed drastic morphological changes that affect specifically to situations that demand a strong polar growth associated with the infection process, such as the formation of the b-dependent filament, the induction of the conjugation tubes, or the formation of a promycelium during spore germination. The outstanding feature of hyphal growth is its extreme directionality. Briefly, a fungal hypha is an intensely polarized secretory system. Vesicles, laden with precursors and/or enzymes for the construction of wall and plasma membrane, are brought forward, fill the hyphal apex, and undergo exocytosis there. Deposition of a new cell wall is confined to that apex and is so organized as to generate a tube of constant diameter capped by a tapered tip that is continuously made afresh (Harold, 2002). In the absence of Pcl12, the cell machinery is able to localize and to transport cell material to the hyphal apex, as we showed using a Myo5-GFP fusion. However, the deposition of this material is not focalized in a single point (i.e., the hyphal apex, compare the localized WGA staining in wild-type filament tips versus the wide distribution of WGA staining at the tips in Δpcl12 mutants) but is redistributed along the surface of the hyphal tip, resulting in the formation of a thicker and irregular filament that is unable to reach wild-type length. Thus, it seems that one of the roles of Pcl12 is to prevent passive diffusion of cortical factors that reach the hyphal apex (for instance, compare Supplemental Movies 1 and 2 online). Therefore, we believe that Pcl12 may function to define the shape of the hyphae by allowing the formation of a boundary domain that helps to restrict the area of the growing tip.
Our transcriptional analysis showed that pcl12 expression is induced during pathogenic development. The increase in the expression of pcl12 was dependent on both a-locus (i.e., pheromone signaling) and b-locus signaling (i.e., the presence of an active b heterodimer). A close inspection of the pcl12 promoter has not revealed the presence of sites related to the two known binding sites for the bW/bE complex (Romeis et al., 2000; Brachmann et al., 2001) nor PRE sites (the binding sites for Prf1, the pheromone response factor; Hartmann et al., 1996; Urban et al., 1996). Although the possibility that the bW/bE or Prf1 binds to the pcl12 regulatory region cannot be excluded, we consider it more likely that pcl12 is not a direct target for either the b proteins or Prf1, but it could be regulated by an uncharacterized yet transcriptional factor(s) in the MAPK cascade and the b regulatory cascade.
Disappointingly, we found that, using standard pathogenicity tests, Δpcl12 cells were able to infect plants at similar ratios than wild-type cells. Therefore, it is fair to conclude from these results that a precise control of polarity is not so necessary for virulence. However, we believe that this is a naive conclusion that deserves some clarifications. In the first place, it is worthwhile to note that while in wild-type infective filaments the appressoria seemed to differentiate after an extensive period of polar growth (that can be followed by the number of basal septa), in pcl12 mutants, appressoria were produced shortly, and mutant filaments that developed appressoria without previous production of basal septation were frequently found. It is assumed that appressorium formation is the response to plant surface clues and that sustained polar growth enables the infective filaments to navigate through the plant surface to locate points of weakened surface integrity, thereby gaining access to vulnerable sites for infection. We believe that the way pathogenicity tests are performed in U. maydis, by stem injection with ∼105 cells, precludes the observation of defects related with the ability to find places to penetrate the plant, as probably all the surface susceptible to be infected is saturated by fungal cells. Consistent with this explanation, we found that in conditions in which the ability to produce a long infective filament could be determinant, such as the infection of developing cobs, pcl12 mutants showed a lower ratio of infectivity than wild-type cells. A second line of evidence to support the importance of a proper regulation of the polarity during the infection is the requirement of Pcl12 for teliospore germination. In U. maydis, the germination of teliospores requires the teliospore to break dormancy, extend a promycelium, and undergo meiosis. We did not trace at which of these steps the teliospore germination is defective in Δpcl12 mutants. However, spore germination in other fungi requires first the establishment of a polarity axis and then the restriction of the growth to the immediate tip to permit the emergence of a germ tube. We believe that in Δpcl12 mutants, the lack of spore germination is the result of the inability of these cells to restrict the growth at the polarity axis during germ tube emergence. In any case, the dramatic defect observed in germination supports a role of Pcl12 in virulence, as the field germination of the air-borne diploid teliospores is the first step in the infection process (Christensen, 1963; Brown and Hovmoller, 2002). In agreement with this view, we have been unable to infect plants spraying maize seedlings with Δpcl12 teliospores.
In conclusion, our experiments indicated that Pcl12 represents a polarity- and virulence-specific regulator in maize smut fungus. As genes encoding homologs of the Pcl12-Cdk5 complex can be found in the genome of other phytopathogenic fungi (our unpublished data), it is tempting to speculate that these regulators may play a broad role in phytopathogenic fungi. Therefore, our results provide valuable insights into the molecular mechanisms that coordinate the induction of polar growth with the pathogencity program in phytopathogenic fungi. However, a cautionary note is required, as our results also underpin that fungal morphogenesis could be a component, but not a complete explanation, of host invasion.
METHODS
Strains and Growth Conditions
Ustilago maydis strains are listed in Table 1. Growth conditions for U. maydis strains and source of antibiotics were described previously (Brachmann et al., 2004). Filamentous growth of AB33 derivatives was induced by shifting cells of an exponential growing culture (OD600 = 0.4 to 0.5) from liquid complete medium to nitrate minimal medium. Controlled expression of genes under the crg1 promoter was performed as described previously (Brachmann et al., 2001; García-Muse et al., 2004). FACS analysis was described previously (García-Muse et al., 2003).
Table 1.
U. maydis Strains Used in This Study
| Strain | Relevant Genotype | Reference |
|---|---|---|
| FB1 | a1 b1 | Banuett and Herskowitz (1989) |
| FB2 | a2 b2 | Banuett and Herskowitz (1989) |
| SG200 | a1mfa2 b1bW2 | Bölker et al. (1995) |
| AB33 | a2 Pnar1:bW2 Pnar1:bE1 | Brachmann et al. (2001) |
| AB34 | a2 Pnar1:bW2 Pnar1:bE2 | Brachmann et al. (2001) |
| SONU82 | a2 b2 Δpcl1 | This work |
| SONU87 | a2 b2 Δpcl6 | This work |
| SONU92 | a2 b2 Δpcl5 | This work |
| SONU96 | a2 b2 Δpcl11 | This work |
| SONU97 | a2 b2 Δpho80 | This work |
| UMP94 | a2 b2 Δpcl13 | This work |
| UMN84 | a2 b2 Δpcl12 | This work |
| UMN70 | a2 Pnar1:bW2 Pnar1:bE1Δpcl1 | This work |
| UMP90 | a2 Pnar1:bW2 Pnar1:bE1Δpcl6 | This work |
| UMN71 | a2 Pnar1:bW2 Pnar1:bE1Δpcl5 | This work |
| SONU158 | a2 Pnar1:bW2 Pnar1:bE1Δpcl11 | This work |
| UMP91 | a2 Pnar1:bW2 Pnar1:bE1Δpho80 | This work |
| UMP92 | a2 Pnar1:bW2 Pnar1:bE1Δpcl13 | This work |
| UMP93 | a2 Pnar1:bW2 Pnar1:bE1Δpcl12 | This work |
| SONU160 | a1 b1 Pcrg1:VSV-pcl12 | This work |
| SONU167 | a1 b1 cdk5-myc Pcrg1:VSV-pcl12 | This work |
| UMP117 | a2 Pnar1:bW2 Pnar1:bE1 cdk5-GFP pcl12-RFP | This work |
| UMP100 | a2 Pnar1:bW2 Pnar1:bE1 fim1-GFP | This work |
| UMN92 | a2 Pnar1:bW2 Pnar1:bE1Δpcl12 fim1-GFP | This work |
| UMP99 | a2 Pnar1:bW2 Pnar1:bE1 tub1-GFP | This work |
| UMN81 | a2 Pnar1:bW2 Pnar1:bE1Δpcl12 tub1-GFP | This work |
| UMN94 | a2 Pnar1:bW2 Pnar1:bE1 myo5-GFP | This work |
| UMN95 | a2 Pnar1:bW2 Pnar1:bE1Δpcl12 myo5-GFP | This work |
| SONU149 | a1 b1 Δpcl12 | This work |
| SONU154 | a1mfa2 b1bW2 Δpcl12 | This work |
| SG200CFP | a1 mfa2 bW2 bE1 POMA:CFP | Flor-Parra et al. (2006) |
| SG200YFP | a1 mfa2 bW2 bE1 POMA:YFP | Flor-Parra et al. (2006) |
| UMN88 | a1mfa2 b1bW2 Δpcl12 POMA:CFP | This work |
| UMN89 | a1mfa2 b1bW2 Δpcl12 POMA:YFP | This work |
| SONU147 | a1 b1 Pcrg1:pcl12 | This work |
| SONU150 | a1 b1 Pcrg1:pcl12 cdk5ts | This work |
| SONU164 | a1 b1 Pcrg1:pcl12 Δrac1 | This work |
| UMN4 | a1 b1 Pcrg1:fuz7DD | Flor-Parra et al. (2006) |
| SONU155 | a1 b1 Pcrg1:fuz7DD Δpcl12 | This work |
DNA, RNA, and Protein Analysis
U. maydis DNA isolation was performed as previously described (Tsukuda et al., 1988). RNA isolation from liquid cultures and solid medium were performed as described (Flor-Parra et al., 2006). RNA gel blot analysis was performed as described previously (Garrido and Pérez-Martín, 2003). A 1420-bp DNA fragment spanning the coding sequence of pcl12 was used as probe. A 5′-end-labeled oligonucleotide complementary to the U. maydis 18S rRNA (Bottin et al., 1996) was used as loading control in RNA gel blot analyses. A phosphor imager (Molecular Imager FX; Bio-Rad) and the suitable program (Quantity One; Bio-Rad) were used for visualization and quantification of radioactive signals.
The procedures used here for obtaining protein extracts, for immunoprecipitation, and for protein gel blot analysis have all been described previously (Garrido et al., 2004). The anti-PSTAIRE (Santa Cruz Biotechnology), anti-myc 9E10, and anti-VSV (Roche Diagnostics) antibodies were diluted 1:10,000 in PBS + 0.1% Tween + 10% dry milk for use. Anti-mouse-Ig-horseradish peroxidase and anti-rabbit-Ig-horseradish peroxidase (Roche Diagnostics) were used as secondary antibodies at a dilution of 1:10,000. All protein gel blots were visualized using enhanced chemiluminescence (Renaissance; Perkin-Elmer).
Plasmid and Strain Constructions
Plasmid pGEM-T easy (Promega) was used for cloning, subcloning, and sequencing of genomic fragments and fragments generated by PCR. Plasmid pRU11 (Brachmann et al., 2001) was used to express genes under the control of the Pcrg1 promoter. Sequence analysis of fragments generated by PCR was performed with an automated sequencer (ABI 373A; Applied Biosystems) and standard bioinformatic tools. To construct the different strains, transformation of U. maydis protoplasts with the indicated plasmids was performed as described previously (Tsukuda et al., 1988). Homologous recombination of gene replacement into the corresponding loci was verified by diagnostic PCR and subsequent DNA gel blot analysis.
Plasmids for GFP tagging of fimbrin, myosin V, and α-tubulin were already described (Steinberg et al., 2001; Weber et al., 2003; Castillo-Lluva et al., 2007). Plasmids allowing the constitutive expression of cfp and yfp genes were described by Flor-Parra et al. (2006). Strains carrying the cdk5-myc and cdk5-gfp alleles were also described (Castillo-Lluva et al., 2007).
Deletion of the different pcl genes was done by homologous replacement following the protocols of Brachmann et al. (2004). Briefly, for each gene to be deleted, a pair of DNA fragments flanking the desired open reading frame were amplified and ligated to a hygromicin or carboxin resistance cassette via SfiI sites. The 5′ fragment spans from nucleotide −1000 to nucleotide −1 (considering the adenine in the ATG as nucleotide +1, and it was produced by PCR amplification using specific primers. The 3′ fragment spans from the end of the open reading frame in each gene and 1000 bp downstream, and they were produced by PCR amplification using specific primers.
To overexpress pcl12, the open reading frame was amplified using PCR and specific primers as an NdeI-EcoRI fragment spanning from initiation codon to 100 bp downstream the stop codon. This fragment was cloned into the corresponding sites of pRU11, an integrative U. maydis vector that contains the crg1 promoter (Brachmann et al., 2001), and the resulting plasmid (pRU11-pcl12) was linearized and integrated into the cbx1 locus by homologous recombination.
To produce a VSV-Pcl12 protein, we inserted the sequence encoding the VSV epitope at the NdeI site of pRU11-pcl12 in frame with the first ATG. This plasmid, pRU11-vsv-pcl12, was linearized and integrated into the cbx1 locus by homologous recombination.
For C-terminal fusion of Pcl12 with RFP, the adaptation of the SfiI-dependent gene replacement strategy for C-terminal RFP tag described by Becht et al. (2006) was used.
Plant Infection and Mating Assays
Pathogenic development of wild-type and mutant strains was assayed by stem injection of maize plants, as described (Gillissen et al., 1992), as well as by infection of silks as described (Snetselaar and Mims, 1993).The variety Early Golden Bantam (Olds Seeds) was used in these infections. Staining of infected plant samples with Calcofluor and Chlorazole Black E staining was done as described previously (Flor-Parra et al., 2006).
For charcoal mating assays, strains were crossed on charcoal-containing complete medium plates (Holliday, 1974) and incubated at 22°C.
To assay formation of conjugative tubes in response to pheromone, 0.5 μL of pheromone a2 of U. maydis (2.5 μg/μL stock in DMSO, final concentration 2.5 × 10−3 μg/μL) (Szabo et al., 2002) to 500 μL of cell suspension in a 2-mL reaction tube and incubated at 22°C and 200 rpm.
To assay the germination of teliospores, infected plants were incubated for 20 d. Tumors containing spores were dried at room temperature and minced using a mortar and pestle. The tumor material was incubated overnight in 1.5% copper (II) sulfate. After extensive washing in sterile distilled water, spores were plated on 2% complete medium–containing agar slides and incubated in a moist chamber at 22°C.
To assay teliospore virulence, suspensions of U. maydis teliospores (1 × 104 teliospores/mL), obtained as above, were prepared in complete medium plus 0.1% gelatin and 1% of activated charcoal and incubated with agitation at room temperature during 6 h. To infect the plants, the teliospore suspension was filtered through a cloth cheese to remove the charcoal. Two-week-old seedlings of Gaspar Flint maize cultivar were sprayed using an artists' airbrush. Plants were incubated in plastic bags for 18 h to maintain high humidity and then transferred to controlled environment chambers at 22°C and 90% humidity. Plants were incubated for 2 weeks for full disease symptoms to become apparent.
Light Microscopy and Image Processing
For in vivo observations, cells from logarithmically growing cultures were placed on a thin 1% agarose layer and immediately observed using a Nikon Eclipse 600 FN microscope coupled to a Hamamatsu ORCA-100 cooled CCD camera that was controlled by MetaMorph software (Universal Imaging). Nuclear staining was performed with DAPI as described previously (García-Muse et al., 2003), and WGA staining was performed as described by Castillo-Lluva et al. (2004). Epifluorescence was observed using filters for DAPI, FITC, rhodamine, and GFP. All measurements and image processing, including adjustment of brightness and contrast, were performed with MetaMorph and Photoshop (Adobe Systems).
Phylogenetic Analyses
Protein sequences of Saccharomyces cerevisiae Pcl cyclins were downloaded from PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). The neighbor-joining method (Saitou and Nei, 1987) was used to create phylogenies. A matrix of the data set was created using MOLPHY (Adachi and Hasegawa, 1996) and then used to create trees with the NEIGHBOR program of the PHYLIP package using random order of addition of taxa. Bootstrap values were obtained from 1000 replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Um Pcl1 (EF494633), Um Pcl5 (EF494635), Um Pcl6 (EF494634), Um Pcl11 (EF494636), Um Pcl12 (EF494639), Um Pcl13, (EF494638), and Um Pho80 (EF494637).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FACS Analysis of AB33 and AB33Δpcl12 Cells.
Supplemental Figure 2. Confrontation Assay.
Supplemental Table 1. PHYLIP File of Amino Acid Sequences Used in Construction of the Phylogenetic Tree in Figure 1.
Supplemental Movie 1. Myo5-GFP Localization in AB33 Cells.
Supplemental Movie 2. Myo5-GFP Localization in AB33Δpcl12 Cells.
Supplementary Material
Acknowledgments
We thank I. Alvarez-Tabarés for critical reading of the manuscript. We also acknowledge the help of J.R. Valverde with the phylogenetic analysis. We also appreciate the suggestions of anonymous reviewers that clearly improved the work. This work was supported by Grants from Spanish government (BIO2005-02998) and the European Union (MRTN-CT-2005-019277). We declare no competing financial interests.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: José Pérez-Martín (jperez@cnb.uam.es).
Online version contains Web-only data.
References
- Adachi, J., and Hasegawa, M. (1996). MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 28 1–150. [Google Scholar]
- Agrios, G.N. (1997). Plant Pathology. (London: Academic Press).
- Andrews, B., and Measday, V. (1998). The cyclin family of budding yeast: Abundant use of a good idea. Trends Genet. 14 66–72. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1989). Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1994). Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8 1367–1378. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122 2965–2976. [DOI] [PubMed] [Google Scholar]
- Becht, P., König, J., and Feldbrügge, M. (2006). Thr RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 119 4964–4973. [DOI] [PubMed] [Google Scholar]
- Bölker, M. (2001). Ustilago maydis – A valuable model system for the study of fungal dimorphism and virulence. Microbiol. 147 1395–1401. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Genin, S., Lehmler, C., and Kahmann, R. (1995). Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73 S320–S325. [Google Scholar]
- Bölker, M., Urban, M., and Kahmann, R. (1992). The a mating type locus of U. maydis specifies cell signalling components. Cell 68 441–450. [DOI] [PubMed] [Google Scholar]
- Bottin, A., Kämper, J., and Kahmann, R. (1996). Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 25 342–352. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., König, J., Julius, C., and Feldbrügge, M. (2004). Reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272 216–226. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Weinzierl, G., Kämper, J., and Kahmann, R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42 1047–1063. [DOI] [PubMed] [Google Scholar]
- Brown, J.K.M., and Hovmoller, M.S. (2002). Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297 537–541. [DOI] [PubMed] [Google Scholar]
- Carroll, A.S., and O'Shea, E.K. (2002). Pho85 and signalling environmental conditions. Trends Biochem. Sci. 27 87–93. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva, S., Alvarez-Tabares, I., Weber, I., Steinberg, G., and Pérez-Martín, J. (2007). Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J. Cell Sci. 120 1584–1595. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva, S., García-Muse, T., and Pérez-Martín, J. (2004). A member of the Fizzy-related family of APC activators is required at different stages of plant infection by Ustilago maydis. J. Cell Sci. 117 4143–4156. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva, S., and Pérez-Martín, J. (2005). The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell 17 3544–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, J.J. (1963). Corn smut caused by Ustilago maydis. Am. Phytopathol. Soc. Monogr. 2 1–41. [Google Scholar]
- Dephoure, N., Howson, R.W., Blethrow, J.D., Shokat, K.M., and O'Shea, E.K. (2005). Combining chemical genetics and proteomics to identify protein kinase substrates. Proc. Natl. Acad. Sci. USA 102 17940–17945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge, M., Kämper, J., Steinberg, G., and Kahmann, R. (2004). Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7 666–672. [DOI] [PubMed] [Google Scholar]
- Flor-Parra, I., Vranes, M., Kämper, J., and Pérez-Martín, J. (2006). Biz1, a zinc finger protein that is required for plant invasion by Ustilago maydis regulates the levels of a mitotic cyclin. Plant Cell 18 2369–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, U., Hause, G., Schuchardt, I., and Steinberg, I. (2006). Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell 18 2066–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, U., Manns, I., and Steinberg, G. (2005). Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell 16 2746–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse, T., Steinberg, G., and Pérez-Martín, J. (2003). Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot. Cell 2 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse, T., Steinberg, G., and Pérez-Martín, J. (2004). Characterization of B-type cyclins in the smut fungus Ustilago maydis: Roles in morphogenesis and pathogenicity. J. Cell Sci. 117 487–506. [DOI] [PubMed] [Google Scholar]
- García-Pedrajas, M.D., and Gold, S.E. (2004). Kernel knowledge: Smut of corn. Adv. Appl. Microbiol. 56 263–290. [DOI] [PubMed] [Google Scholar]
- Garrido, E., and Pérez-Martín, J. (2003). The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47 729–743. [DOI] [PubMed] [Google Scholar]
- Garrido, E., Voss, U., Müller, P., Castillo-Lluva, S., Kahmann, R., and Pérez-Martín, J. (2004). The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev. 18 3117–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen, B., Bergemann, J., Sandmann, C., Schrör, B., Bölker, M., and Kahmann, R. (1992). A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68 647–657. [DOI] [PubMed] [Google Scholar]
- Gow, N.A., Brown, A.J., and Odds, F.C. (2002). Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5 366–371. [DOI] [PubMed] [Google Scholar]
- Harold, F.M. (2002). Force and compliance: Rethinking morphogenesis in walled cells. Fungal Genet. Biol. 37 271–282. [DOI] [PubMed] [Google Scholar]
- Harris, S.D. (2006). Cell polarity in filamentous fungi: Shaping the mold. Int. Rev. Cytol. 251 41–77. [DOI] [PubMed] [Google Scholar]
- Hartmann, H.A., Kahmann, R., and Bölker, M. (1996). The pheromone response factor coordinates filamentous growth and pathogenic development in Ustilago maydis. EMBO J. 15 1632–1641. [PMC free article] [PubMed] [Google Scholar]
- Holliday, R. (1974). Ustilago maydis. In Handbook of Genetics, R.C. King, ed (New York: Plenum Press), pp. 575–595.
- Huang, D., Moffat, J., and Andrews, B. (2002). Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell. Biol. 22 5076–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann, R., and Kämper, J. (2004). Ustilago maydis: How its biology relates to pathogenic development. New Phytol. 164 31–42. [DOI] [PubMed] [Google Scholar]
- Mahlert, M., Leveleki, L., Hlubek, A., Sandrock, B., and Bölker, M. (2006). Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 59 567–578. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microporocessors. Annu. Rev. Cell Dev. Biol. 13 261–291. [DOI] [PubMed] [Google Scholar]
- Müller, P., Weinzierl, G., Brachmann, A., Feldbrügge, M., and Kahmann, R. (2003). Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, K.L., and McLaughlin, D.J. (1984). Ultrastructure of meiosis in Ustilago maydis. Mycologia 76 468–485. [Google Scholar]
- Pérez-Martín, J., Castillo-Lluva, S., Sgarlata, C., Flor-Parra, I., Mielnichuk, N., Torreblanca, J., and Carbó, N. (2006). Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol. Genet. Genomics 276 211–229. [DOI] [PubMed] [Google Scholar]
- Roberts, J.M. (1999). Evolving ideas about cyclins. Cell 98 129–132. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Brachmann, A., Kahmann, R., and Kämper, J. (2000). Identification of a target gene for the be-bW homeodomain protein complex in Ustilago maydis. Mol. Microbiol. 37 54–66. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schuchardt, I., Assmann, D., Thines, E., Schuberth, C., and Steinberg, G. (2005). Myosin-V, Kinsein-1, and Kinesin-3 cooperate in long-distance transport in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 16 5191–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar, K.M., Bölker, M., and Kahmann, R. (1996). Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genet. Biol. 20 299–312. [DOI] [PubMed] [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1992). Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84 193–203. [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1993). Infection of maize stigmas by Ustilago maydis: Light and electron microscopy. Phytopathology 83 843–850. [Google Scholar]
- Spellig, T., Bölker, M., Lottspeich, F., Frank, R.W., and Kahmann, R. (1994). Pheromone trigger filamentous growth in Ustilago maydis. EMBO J. 13 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, G. (2006). Preparing the way: Fungal motors in microtubule organization. Trends Microbiol. 15 14–21. [DOI] [PubMed] [Google Scholar]
- Steinberg, G. (2007). On the move: Endosomes in fungal growth and pathogenicity. Nat. Rev. Microbiol. 5 309–316. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., Scliwa, M., Lehmler, C., Bölker, M., Kahmann, R., and McIntosh, J.R. (1998). Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111 2235–2246. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., Wedlich-Söldner, R., Brill, M., and Schulz, I. (2001). Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114 609–622. [DOI] [PubMed] [Google Scholar]
- Szabo, Z., Tonnis, M., Kessler, H., and Feldbrugge, M. (2002). Structure-function analysis of lipopeptide pheromones from the plant pathogen Ustilago maydis. Mol. Genet. Genomics 268 362–370. [DOI] [PubMed] [Google Scholar]
- Tsukuda, T., Carleton, S., Fotheringham, S., and Holloman, W.K. (1988). Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8 3703–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, S.L., and Talbot, N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39 385–417. [DOI] [PubMed] [Google Scholar]
- Urban, M., Kahmann, R., and Bölker, M. (1996). Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251 31–37. [DOI] [PubMed] [Google Scholar]
- Weber, I., Gruber, C., and Steinberg, G. (2003). A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15 2826–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Samuels, B.A., and Tsai, L.H. (2006). Cyclin-dependent kinase 5 permits efficient cytoskeletal remodeling -a hypothesis on neuronal migration. Cereb. Cortex 16 i64–i68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.