Abstract
Seasonal control of flowering through responsiveness to daylength shows extreme variation. Different species flower in response to long days or short days (SDs), and this difference evolved several times. The molecular mechanisms conferring these responses have been compared in detail only in Arabidopsis thaliana and rice (Oryza sativa) and suggest that a conserved pathway confers daylength responses through regulation of FLOWERING LOCUS T (FT) transcription by CONSTANS (CO). We studied Pharbitis (Ipomoea nil; formerly, Pharbitis nil), a widely used SD model species and a member of the Convolvulaceae, and showed using transgenic plants together with detailed expression analysis that two putative orthologs of FT (Pn FT1 and Pn FT2) promote flowering specifically under SDs. These genes are expressed only under SDs, and light flashes given during the night reduce their expression and prevent flowering. We demonstrate that in Pharbitis a circadian rhythm set by the light-to-dark transition at dusk regulates Pn FT expression, which rises only when the night is longer than 11 h. Furthermore, Pharbitis accessions that differ in their critical night-length responses express Pn FT at different times after dusk, demonstrating that natural genetic variation influencing the clock regulating Pn FT expression alters the flowering response. In these assays, Pn FT mRNA abundance was not related to Pn CO expression, suggesting that Pn FT may be regulated by a different transcription factor in Pharbitis. We conclude that SD response in Pharbitis is controlled by a dedicated light sensitive clock, set by dusk, that activates Pn FT transcription in darkness, a different mechanism for measuring daylength than described for Arabidopsis and rice.
INTRODUCTION
Many plant species flower at characteristic times of the year. These seasonal patterns in flowering are created by responsiveness to environmental cues, such as changes in daylength or temperature. Plants can be classified based on their responsiveness to daylength (Thomas and Vince-Prue, 1997). Long-day (LD) plants flower when daylength exceeds a critical duration, while short-day (SD) plants show the reverse response, flowering when daylength is shorter than a critical length, and day-neutral plants do not respond to daylength. The precise daylength that induces flowering often differs between individuals of a species (Ray and Alexander, 1966; Koornneef et al., 2004). Such genetic differences were exploited during the domestication of plants to allow growth of crops at different latitudes and in nature may adapt accessions of a single species to particular ecological niches (Yano et al., 2000; Turner et al., 2005). Here, we characterize at the molecular level the induction of flowering of Pharbitis (Ipomoea nil; formerly, Pharbitis nil) by exposure to SDs and compare the mechanism by which this species measures daylength with those previously described for Arabidopsis thaliana, an LD plant, and rice (Oryza sativa), an SD plant.
The mechanisms by which plants detect and respond to daylength are best understood in Arabidopsis (Searle and Coupland, 2004; Imaizumi and Kay, 2006). Mutants disrupted in the response to daylength were defined because they flower later under LDs than wild-type plants but are not delayed in flowering under SDs. Molecular-genetic approaches defined the photoperiodic flowering pathway, which at its core comprises the GIGANTEA (GI), CONSTANS (CO), and FLOWERING LOCUS T (FT) genes (Searle and Coupland, 2004; Imaizumi and Kay, 2006). CO activates the transcription of FT in the vascular tissue (An et al., 2004), and in turn FT strongly promotes flowering (Kardailsky et al., 1999; Kobayashi et al., 1999). FT is required for activation in the meristem of transcription of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Schmid et al., 2003; Searle et al., 2006), a MADS box transcription factor that promotes flowering. The mechanism by which FT activates SOC1 likely involves movement of the FT protein from the leaf to the meristem (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007) where it interacts with the basic domain/leucine zipper transcription factor FD, as demonstrated for the activation of APETALA1 transcription (Abe et al., 2005; Wigge et al., 2005). Photoperiodic regulation of this pathway results in activation of FT transcription by CO under LDs but not SDs. The regulation of CO activity by LDs occurs at both the transcriptional and posttranscriptional levels. CO transcription is regulated by the circadian clock, with a diurnal peak in expression during the evening and night (Suarez-Lopez et al., 2001). In LDs, this peak is broader due to the activity of the ubiquitin ligase FLAVIN binding KELCH DOMAIN F BOX PROTEIN1, which degrades CYCLING DOF1, a repressor of CO transcription (Imaizumi et al., 2003, 2005). This combination of circadian clock control and responsiveness to light causes CO mRNA expression to coincide with exposure of plants to light under LDs but not SDs. At the posttranscriptional level, CO protein is stabilized when plants are exposed to light but degraded in darkness (Valverde et al., 2004). The degradation in darkness is due to ubiquitination and involves the SUPPRESSOR OF PHYA-105 (SPA1) protein (Laubinger et al., 2006). This posttranscriptional regulation ensures that CO protein only accumulates in the nucleus under LDs and, therefore, that FT transcription occurs under LDs but not SDs.
Analysis of the mechanisms underlying photoperiodic response in rice enabled comparison of how LD and SD responses are controlled. The response of rice was studied both by exploiting natural genetic variation to identify quantitative trait loci that impair photoperiodic flowering and by isolating mutations that alter the response (Izawa et al., 2000; Yano et al., 2000; Kojima et al., 2002). These experiments led to the demonstration that rice homologs of GI (Os GI), CO (Heading Date1 [Hd1]), and FT (Hd3a) confer the response to SDs (Hayama et al., 2003). However, the effect of daylength on Hd3a mRNA level is reversed compared with its effect on FT mRNA in Arabidopsis. In rice, Hd3a mRNA accumulates under SDs but not LDs (Kojima et al., 2002). Reversal of photoperiodic response type between Arabidopsis and rice appears to be due to Hd1 acting as a light-mediated repressor of Hd3a transcription under LDs, whereas in Arabidopsis, CO is a light-mediated activator of FT transcription (Izawa et al., 2002; Hayama et al., 2003; Hayama and Coupland, 2004).
Rice and Arabidopsis are distantly related among the angiosperms. Arabidopsis is a member of the clade of Rosids within the Eudicots, whereas rice is a monocot, and these clades diverged 150 to 300 million years ago (Soltis et al., 1999). However, LD and SD photoperiodic response types occur within single families in both the monocots and dicots, indicating that this difference evolved independently several times. For example, LD and SD response types are present within the Solanaceae (Smykal et al., 2004), which is within the clade of Asterids in the Eudicots. Similarly, the monocot grasses include both LD and SD species (Yano et al., 2000; Turner et al., 2005). Studying the mechanistic basis of photoperiod response in a wider range of plant species will help determine whether the distinction between LD and SD responses evolved in different families through similar changes to the photoperiodic flowering pathway as have been described by comparing Arabidopsis and rice. We have studied the molecular basis of photoperiod response in I. nil (here called Pharbitis and also known as Japanese morning glory), a member of the Convolvulaceae in the order Solanales.
Pharbitis is widely used as a model species to study the physiology of the SD flowering response (Imamura, 1967; Thomas and Vince-Prue, 1997), and these studies provide a strong basis for a molecular analysis. Physiological experiments established that the flowering response of Pharbitis is determined by night length rather than the length of the day (Saji et al., 1984), as previously shown for other SD species (Hamner and Bonner, 1938). Pharbitis plants grown under continuous light remain vegetative, but exposure to a single dark period is sufficient to induce flowering (Takimoto and Hamner, 1965; Vince-Prue and Gressel, 1985). The precise length of the dark period required to induce flowering differs between Pharbitis accessions, and this appears to be correlated with the latitude at which individual accessions are found (Imamura et al., 1966). Disruption of the dark period by exposure to red light for 5 min prevents flowering if given ∼8 to 9 h after the onset of darkness (Takimoto and Hamner, 1965). Maximum sensitivity to these night breaks shows a circadian rhythm, suggesting that they disrupt a circadian clock–regulated process required for flowering (Lumsden and Furuya, 1986). The timing of maximum sensitivity to night breaks always occurs at a constant time in darkness if the length of the preceding light period is longer than 6 h (Lumsden et al., 1982). Based on these observations, a model for photoperiodic flowering was proposed for Pharbitis (Lumsden, 1998). According to this model, a circadian rhythm, called a photoperiodic response rhythm, is initiated by the transition from light to dark at dusk and continues in darkness. This rhythm is suspended under continuous light. If plants are held in darkness for at least 9 h, processes controlled by this rhythm initiate flowering. However, exposure to light in the form of night breaks at crucial phases of the photoperiodic response rhythm inhibits this floral inductive activity and suppresses flowering. This model was proposed on the basis of physiological experiments, and although homologs of CO are present in Pharbitis (Liu et al., 2001; Kim et al., 2003), the model has not been tested at the molecular level. However, the physiological model established for Pharbitis differs in important respects from that established in rice using molecular-genetic approaches. Here, we test the Pharbitis model at the molecular level and compare this system with those of Arabidopsis and rice. We show that expression of Pharbitis FT homologs is activated under SDs but not LDs and that this is regulated by a circadian clock set by dusk and suppressed by light. We compare the mechanisms regulating photoperiodism in Pharbitis and rice and propose that despite the phenotypic similarities and that transcription of FT homologs is regulated by daylength in both species, the mechanisms by which they measure daylength differ.
RESULTS
Isolation of FT Homologs from Pharbitis
PCR primers were designed to regions of the Arabidopsis FT gene that are conserved in other members of the phosphatidylethanolamine binding protein/RAF kinase inhibitor protein (PEBP/RKIP) family. These primers were then used to amplify FT-like genes from cDNA made from Pharbitis cotyledon RNA. Two partial cDNA clones showing sequence similarity to FT were isolated. Full-length cDNAs of these genes, which were called Pn FT1 and Pn FT2, were obtained by conducting rapid amplification of cDNA ends (RACE) from the 5′ and 3′ termini. The predicted amino acid sequences of Pn FT1 and Pn FT2 were derived from the cDNA sequences and are 78% identical to each other and 70 to 75% identical to Arabidopsis FT (see Supplemental Figure 1 online). Gel blot hybridizations suggest that there are likely to be at least two additional Pharbitis genes with homology to Pn FT1 and Pn FT2 (see Supplemental Figure 2 online), but the hybridization signal is relatively weak, and it remains unclear whether these genes encode proteins closely related to FT or to other proteins within the FT-like protein family (Figure 1A). However, a phylogenetic tree comprising PEBP/RKIP members from a wide range of monocotyledonous and dicotyledonous plant species showed that Pn FT1 and Pn FT2 proteins cluster within a clade containing the FT proteins of Arabidopsis, tomato (Solanum lycopersicum), and poplar (Populus spp) (Figure 1A), all of which are implicated in flowering-time control (Kardailsky et al., 1999; Kobayashi et al., 1999; Bohlenius et al., 2006; Hsu et al., 2006; Lifschitz et al., 2006). These analyses suggested that Pn FT1 and Pn FT2 are closely related to FT and may play a similar role in the promotion of flowering.
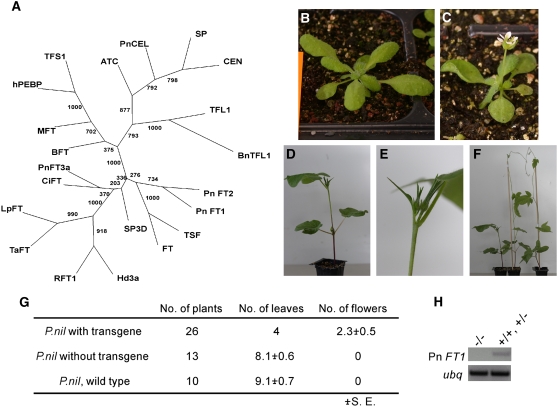
Figure 1.
Cloning of Pn FT1 and Pn FT2 from Pharbitis and the Effect of the Overexpression of Pn FT1 on the Flowering Time in Arabidopsis and Pharbitis.
(A) Sequence comparison of Pharbitis FT and related proteins from other flowering plants. The predicted sequences of FT-like proteins were aligned using ClustalW. An unrooted phylogenetic tree was constructed by the neighbor-joining method using Phylip 3.65 (Felsenstein, 1989). Bootstrap values of 1000 samplings are shown on each branch. Arabidopsis FT (BAA77838), TSF (BAD93590), MFT (Q9XFK7), BFT (Q9FIT4), TFL1 (AAB41624), ATC (BAA75931); Oryza FT-Hd3a (BAB61030), RFT1 (BAB78480), tomato SP (AAC26161), SFT/SP3D (AAO31792), Lolium FT 3-LpFT3 (ABC33722), Triticum FT (TaFT; AAW23034), citrus FT (CiFT; BAA77836), Brassica napus TFL (Bn TFL1; BAA33415), Antirrhinum CEN (AAB36112), Pharbitis Pn CEL (BAE44112), human hPEBP (P30086), Populus nigra Pn FT3a (BAD08336), S. cerevisiae TFS1 (CAA44015), and Pharbitis FT (Pn FT1 and Pn FT2).
(B) and (C) The effect of overexpression of Pn FT1 on flowering time of Arabidopsis. co-2 (B) and 35S:Pn FT1 in co-2 (C) were grown under SDs.
(D) to (H) The effect of overexpression of Pn FT1 on flowering time of Pharbitis cv Violet. Thirty-nine T2 plants, generated from two T1 transgenic plants heterozygous for the transgene, were grown under LDs (16 h light/8 h dark) for 21 d.
(D) and (E) A Pharbitis 35S:Pn FT1 transgenic plant (D) and the primary shoot apex illustrating the bud of the terminal flower produced in a transgenic plant (E) are shown.
(F) and (G) Flowering time of Pharbitis 35S:Pn FT1 plants. In (F), from left to right, a plant carrying the transgene, a plant without the transgene, and the wild-type plant are shown. In (G), a comparison of the flowering times of the plants shown in (F) is shown. In the transgenic plants, no more than two or three buds are formed before the shoot apical meristem is converted to a terminal flower.
(H) Comparison of Pn FT1 mRNA levels in T2 plants that have inherited the 35S:Pn FT1 transgene (+/+ and +/−) with those that have not inherited the transgene (−/−). RNA levels were tested by RT-PCR.
Overexpression of Pn FT1 Accelerates Flowering in Arabidopsis and Pharbitis
Whether Pn FT1 can promote flowering in Arabidopsis and Pharbitis transgenic plants was then tested. In Arabidopsis, FT transcription is activated by CO, and the late-flowering phenotype of co mutants can be complemented by overexpression of FT (Kardailsky et al., 1999; Kobayashi et al., 1999; Suarez-Lopez et al., 2001; An et al., 2004). Twelve independent transgenic co mutant lines carrying 35S:Pn FT1 flowered much earlier than co-2 mutants. Two lines that contained the transgene at a single locus were scored in the T2 generation and flowered much earlier than the co-2 mutant under LDs and SDs. These plants also displayed a terminal flower at the apex of the shoot (Figures 1B and 1C). This experiment suggests that Pn FT1 can promote flowering and induce formation of a terminal flower in Arabidopsis in a similar way to previously described for FT (Kardailsky et al., 1999; Kobayashi et al., 1999).
In addition, transgenic Pharbitis plants carrying 35S:Pn FT1 or UBQ:Pn FT1 were examined. The 35S:Pn FT1 plants expressed Pn FT1 mRNA at a higher level than wild-type plants under LDs (Figure 1H). Furthermore, in a segregating T2 population grown under LDs (16 h light/8 h dark), all the transgenic plants produced flowers by 21 d after germination, while none of the nontransgenic plants were flowering 40 d after germination (Figure 1G). The transgenic plants produced four leaves under these conditions, and their primary shoot apex ended in a terminal flower (Figures 1D and 1E), whereas sibling plants that had not inherited the transgene and wild-type control plants continued to grow vegetatively, developing eight or nine leaves on the primary shoot by the same stage (Figures 1F and 1G). Similar early flowering and terminal flower phenotypes were observed for the UBQ:Pn FT1 transformants (data not shown). These results indicate that Pn FT1 promotes flowering in Pharbitis and therefore plays a similar role in the promotion of flowering as FT-like genes in other plant species (Kardailsky et al., 1999; Kobayashi et al., 1999; Bohlenius et al., 2006; Hsu et al., 2006; Lifschitz et al., 2006).
Pn FT1 and Pn FT2 Expression Is Only Induced under Inductive SDs
Transcription of FT and Hd3a, a rice homolog of FT, responds to daylength, so that FT mRNA accumulates only under those conditions that promote flowering. Therefore, in Arabidopsis, FT mRNA accumulates only under LDs (Suarez-Lopez et al., 2001), whereas in rice, the mRNA of Hd3a accumulates under SDs (Izawa et al., 2002; Kojima et al., 2002). To determine whether accumulation of Pn FT1 and Pn FT2 mRNAs is also correlated with the photoperiodic response of flowering in Pharbitis, the abundance of these mRNAs was tested under LDs (16 h light/8 h dark) and SDs (10 h light/14 h dark).
Abundance of Pn FT1 and Pn FT2 mRNAs showed a diurnal rhythm in SD-grown plants, so that their levels started to rise during the night and reached their maximal level in the morning (Figure 2A). By contrast, the transcripts of both genes could not be detected under LDs (Figure 2A). To determine with more accuracy the conditions that induce expression of Pn FT, plants were grown under five different daylengths (see Supplemental Figures 3A and 3B online). Pn FT expression was induced under cycles of 12 h light/12 h dark but not under 13 h light/11 h dark. The conditions in which Pn FT expression was induced correlated precisely with those that induced flowering (see Supplemental Figure 3C online). The induction of Pn FT1 and Pn FT2 transcription precisely under those conditions that induce flowering combined with the ability of Pn FT1 to promote flowering when overexpressed in transgenic Pharbitis plants is consistent with these genes being involved in the promotion of flowering of Pharbitis under SDs.
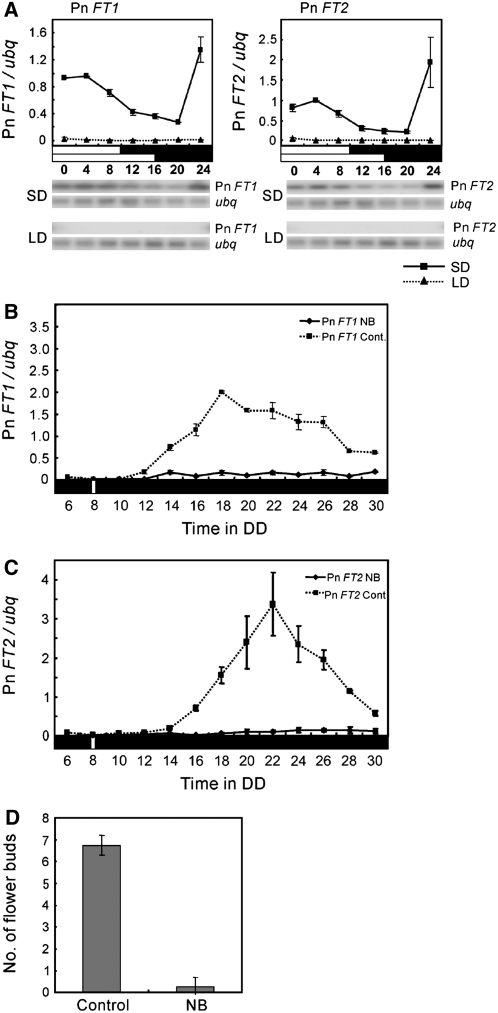
Figure 2.
Pn FT1 and Pn FT2 Transcript Abundance Correlates with the Flowering Behavior of Pharbitis cv Violet.
(A) Pn FT1 and Pn FT2 expression under LD and SD. Expression levels were quantified from DNA gel blots shown below and normalized to Ubiquitin expression. Plants were grown under LDs (16 h light/8 h dark) or SDs (10 h light/14 h dark) for 4 d, and cotyledons were harvested every 4 h in the last day–night cycle.
(B) and (C) The effect of night break on Pn FT1 and Pn FT2 mRNA levels in Pharbitis. Real-time PCR was performed to quantify Pn FT1 and Pn FT2 mRNA. All seedlings were grown under continuous light and then transferred to continuous darkness on day 5. One population of plants received 10 min of white light (a night break) 8 h after transfer to dark, whereas the control population did not receive the light treatment. Cotyledons from both populations were harvested every 2 h from 6 to 30 h in dark. Plants exposed to a night break (NB; solid line) or control plants (cont; dotted line). Error bars show se of three PCR experiments. A biological replicate provided a similar result.
(D) Flowering time of control plants and plants exposed to night break (NB). Plants were first grown under the same conditions as used in (B) and (C), and after 48 h in continuous darkness, they were transferred to continuous light. Flowering was measured 4 weeks after the start of the experiment as the total number of flower buds present on each of eight plants in both groups.
The Effect of Night Breaks on Pn FT1 and Pn FT2 Expression
Flowering of Pharbitis is inhibited by a short exposure to light given during the night. Such an exposure to light, called a night break, is most effective 8 to 9 h after dark (Takimoto and Hamner, 1965). In rice, night breaks suppress transcription of Hd3a and hence delay flowering (Ishikawa et al., 2005). To test the effect of night breaks on Pn FT1 and Pn FT2 expression, Pharbitis plants were grown under continuous light, shifted to constant darkness for 48 h, and then back to constant light. One population of plants was exposed to a night break of white light for 10 min and 8 h after the shift to darkness, whereas a control population was not exposed to the night break. The abundance of the mRNAs of Pn FT1 and Pn FT2 was examined every 2 h between 6 and 30 h after the shift to darkness. In the control plants, Pn FT1 and Pn FT2 mRNA levels peaked between 18 and 22 h in darkness (Figures 2B and 2C). By contrast, in those plants exposed to a night break, no clear peak in Pn FT1 or Pn FT2 expression was observed, and the abundance of the mRNAs was at least 10-fold lower at the times that they peaked in control plants. The flowering behavior of both populations of plants was also measured by scoring flowering 25 d after return to continuous light. The control plants that were not exposed to night breaks had produced at least six flower buds, whereas six of eight plants exposed to night breaks had produced no flower buds and the remaining plants had only produced a single flower bud (Figure 2D). Therefore, exposure to night breaks suppresses Pn FT1 and Pn FT2 mRNA levels, and this correlates with a strong reduction in flowering. Thus, in Pharbitis and rice, the two SD plants tested so far, night breaks reduce expression of FT homologs, suggesting that in both species a light-sensitive mechanism activates transcription of these genes.
Expression of Pn FT1 and Pn FT2 Is Regulated by the Circadian Clock and Cycles with Higher Amplitude under Constant Darkness Than under Constant Light
In Arabidopsis, the circadian pattern of CO mRNA expression sets the light-sensitive phase of the photoperiodic response, and CO activates FT transcription when the circadian rhythm in CO mRNA expression coincides with exposure of plants to light (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002). Thus, FT transcription shows a circadian rhythm in wild-type plants under constant light (Harmer et al., 2000; Imaizumi et al., 2005). To test how Pn FT mRNA expression in Pharbitis is influenced by light, Pn FT expression was examined every 3 to 4 h in Pharbitis plants grown under LD or SD and then transferred to continuous light or continuous darkness (Figures 3A and 3B; see Supplemental Figure 4A online).
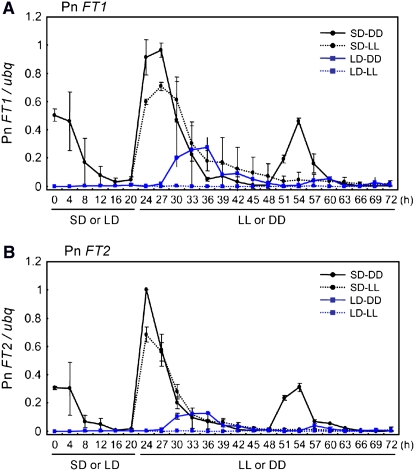
Figure 3.
Abundance of Pn FT1 and Pn FT2 Transcripts in Pharbitis cv Violet Shows a Circadian Rhythm, and Their Expression Requires Darkness.
Quantification of Pn FT1 and Pn FT2 from RT-PCR followed by DNA gel blotting. Plants were grown under LDs or SDs for 3 d and then shifted to continuous light (LL) or continuous darkness (DD). Cotyledons were harvested for RNA extraction every 4 h during the last LD or SD and every 3 h for 48 h in LL and DD. Solid black line indicates plants transferred from SD to DD. Dotted black line indicates plants transferred from SD to LL. Solid blue line indicates plants transferred from LD to DD. Dotted blue line indicates plants transferred from LD to LL. Error bars show se of three PCR experiments. A biological replicate provided a similar result.
In plants exposed to LDs, no expression of Pn FT1 or Pn FT2 mRNA was detected. However, transcript levels of both Pn FT1 and Pn FT2 were increased after the shift to continuous darkness and showed a circadian rhythm with peaks during the subjective mid day (Figures 3A and 3B). Therefore, robust cycling of Pn FT1 and Pn FT2 expression is initiated under constant darkness. By contrast, the transcripts of these genes were not reliably detected in continuous light following LD and showed no circadian rhythm. The circadian rhythm in Pn FT1 and Pn FT2 expression is therefore not induced in continuous light.
In plants exposed to SDs, Pn FT1 and Pn FT2 mRNAs were detected before transfer to continuous darkness or continuous light. After transfer from SD to continuous darkness, the transcripts of both genes peaked soon after the first subjective dawn and also at the second subjective dawn, demonstrating that Pn FT expression shows a circadian rhythm under continuous darkness. However, the phase of expression of Pn FT under continuous darkness is different after entrainment to SD compared with entrainment to LDs (Figures 3A and 3B), suggesting that the timing of dusk influences the phase of Pn FT expression. By contrast, those plants shifted from SDs to continuous light did not cycle after the first peak, indicating that under continuous light the circadian oscillations in Pn FT1 and Pn FT2 mRNAs dampened to a low level.
Taken together, these results demonstrate that unlike FT in Arabidopsis, Pn FT1 and Pn FT2 expression does not require exposure of plants to light but is induced in darkness. Furthermore, the rhythm in Pn FT1 and Pn FT2 expression occurs under continuous darkness but not continuous light, suggesting that a clock output gene that regulates Pn FT is activated in darkness.
The Circadian Rhythms in Pn FT1 and Pn FT2 Expression Are Set by Lights off at Dusk
In the photoperiodic response of Pharbitis, the length of the night rather than the length of the day determines whether flowering occurs (Imamura et al., 1966). Therefore, a timekeeping mechanism that specifically measures the length of the night appears to play an important role in regulating flowering of Pharbitis. To address whether a circadian rhythm set by dusk regulates Pn FT expression, we entrained Pharbitis plants to light/dark cycles of 14 h light/10 h dark, transferred them to continuous light, and then released populations of plants into darkness at 6-h intervals. If a timing mechanism set by dusk regulates Pn FT expression, then Pn FT mRNA abundance should rise at a constant time after transfer to dark, regardless of how long the plants were exposed to continuous light. By contrast, if the circadian rhythm in Pn FT expression is regulated by the transition from dark to light at dawn, then its peak in expression will occur at a constant time after the transfer to continuous light, as shown in Arabidopsis for the circadian rhythm in CAB2 expression (McWatters et al., 2000).
In all populations of plants, the abundance of Pn FT1 and Pn FT2 transcripts increased ∼14 h after transfer from light to dark, and the first peak in abundance of the transcripts occurred ∼16 to 18 h after the transition to dark (Figures 4A, 4B, and 4F). No effect of the timing of the transfer to light on the phase of Pn FT1 or Pn FT2 expression was observed. Furthermore, a second peak of Pn FT1 or Pn FT2 expression was clearly observed for those plants held in light for up to 16 h prior to transfer to dark, and the time of the second peak was also always correlated with the time of the light-to-dark transition (Figures 4A and 4B). The presence of the second peak suggests that rhythmic expression of Pn FT under these conditions is caused by circadian clock control. In addition, these results indicate that the phase of the circadian rhythms in both Pn FT1 and Pn FT2 transcripts is set by the transition from light to dark and, therefore, that a clock set by dusk regulates the rhythm of Pn FT expression in continuous darkness.
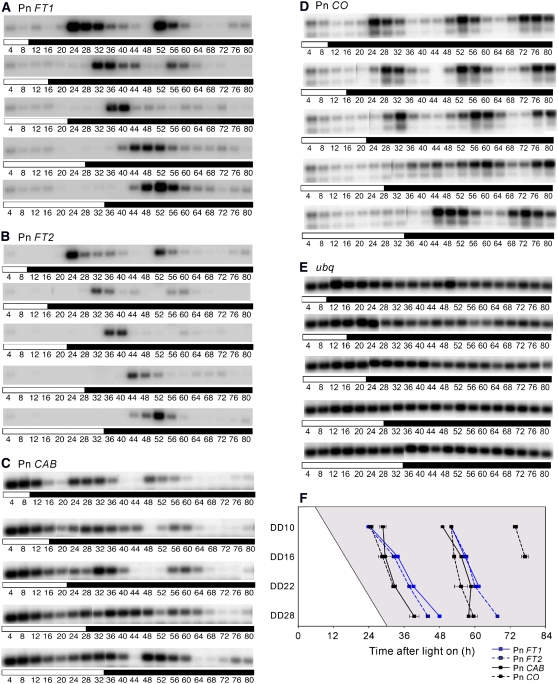
Figure 4.
Pn FT1 and Pn FT2 Transcript Rhythms Are Set by Light off in Pharbitis cv Violet.
(A) to (E) DNA gel blots of RT-PCR products detecting expression of Pn FT1 (A), Pn FT2 (B), Pn CAB (C), Pn CO (D), and ubq (E). Plants were grown under 14 h light/10 h dark and at dawn (Time 0) on the fifth day were divided into five populations, which were exposed to light for different lengths of time and then transferred to darkness. Cotyledons were harvested from the five populations every 4 h.
(F) A diagram illustrating the phase relationship between Pn FT1, Pn FT2, Pn CO, and Pn CAB in darkness. The peak times of expression of the genes relative to dawn is plotted. The horizontal axis represents time after dawn on the fifth day (see legend to [A] to [E]). The vertical axis represents the time after dawn at which each population was transferred to darkness and this time is indicated (e.g., DD10 spent 10 h in light before transfer to darkness). The results for the first four populations of plants shifted to darkness after 10, 16, 22, and 28 h are plotted, and the time that each population was transferred to darkness is illustrated in the diagram by the transition from light to shade. Error bars show se of three PCR experiments. A biological replicate provided a similar result.
In Arabidopsis, the circadian phase of CAB2 gene expression is predominantly set by the dark-to-light transition at dawn (McWatters et al., 2000). Therefore, we tested the entrainment of Pharbitis CAB (Pn CAB) in the same regime as used to analyze Pn FT expression. In contrast with Pn FT1 and Pn FT2, the phase of Pn CAB expression shifted gradually as the length of time spent in light was increased (Figures 4C and 4F). Therefore, the phase of Pn CAB expression is not controlled exclusively by the time of transition from light to dark but is strongly influenced by the timing of the transition from dark to light at dawn or the duration of time spent in the light. Expression of Pn CO, which is the closest described Pharbitis homolog to Arabidopsis CO and complements co mutations in Arabidopsis (Liu et al., 2001), was also tested under the same conditions used to analyze Pn CAB and Pn FT expression. The effects of dark-to-light and light-to-dark transitions on Pn CO mRNA expression was similar to that described for Pn CAB and clearly distinct from Pn FT (Figures 4D and 4F). Thus, the phases of Pn CAB and Pn CO expression are set by different mechanisms than the expression of Pn FT, suggesting that distinct circadian clocks control the oscillations of these genes.
Comparison of Pn FT Expression in Cultivars with Different Critical Night Lengths
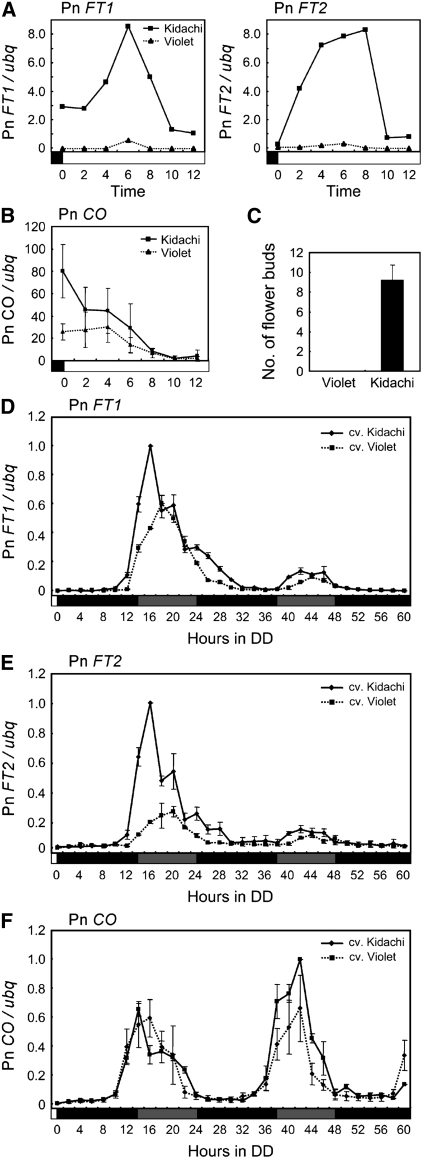
The Pharbitis cultivars Violet and Kidachi differ in the critical night length required to induce flowering. Kidachi requires 8- to 9-h nights at 25°C to promote flowering, whereas Violet requires longer nights at the same temperature (Imamura et al., 1966). To determine whether this effect could be explained by differential regulation of Pn FT expression in Violet and Kidachi, seedlings of both cultivars were grown under a regime of 15 h light/9 h dark (Figures 5A to 5C ; see Supplemental Figure 4B online), which induced flowering in Kidachi but not Violet (Figure 5C). Expression of the Pn FT1 and Pn FT2 mRNAs was clearly detected in Kidachi, whereas almost no Pn FT expression was observed in Violet (Figure 5A). To test whether this difference in expression of Pn FT between Kidachi and Violet was due to a difference in the phase of a circadian rhythm regulating Pn FT expression, both cultivars were grown under 10 h light/14 h dark and then shifted to constant darkness. Pn FT1 and Pn FT2 mRNA levels in Kidachi started to rise after 12 h in darkness and exhibited a peak at 16 h in darkness, whereas levels in Violet first rose after 14 h and exhibited a peak at 18 h in darkness (Figures 5D and 5E; see Supplemental Figure 4C online). Therefore the regulation of Pn FT differs between Kidachi and Violet, being expressed at an earlier phase in Kidachi. The Pn FT1 and Pn FT2 genes are also expressed at higher amplitude in Kidachi than Violet. These differences in Pn FT expression correlate with flowering of Kidachi occurring under shorter nights. The levels of Pn CO mRNA expression in 15 h light/9 h dark (Figure 5B; see Supplemental Figure 4B online) and the timing of its expression in continuous darkness were similar in Kidachi and Violet (Figure 5F; see Supplemental Figure 4C online), suggesting that alterations in Pn CO mRNA expression are not responsible for the differences in Pn FT expression.
Figure 5.
Expression Pattern of Pn FT1 and Pn FT2 mRNAs in Two Pharbitis Cultivars Reflects the Differences in Their Critical Night Length.
(A) Quantification of Pn FT1 and Pn FT2 transcript levels in cv Violet and Kidachi under 15 h light/9 h dark at 25°C. Cotyledons were harvested every 2 h for the first 12 h on day 5. Solid line, Kidachi; dotted line, Violet.
(B) Quantification of Pn CO transcript levels in cv Violet and Kidachi. Samples were harvested as in (A). Solid line, Kidachi; dotted line, Violet.
(C) Flowering times of Violet and Kidachi under 15 h light/9 h dark. Flower buds present at the first 15 nodes of the main axis were counted 4 weeks after the start of the experiment. Twenty-two plants of each cultivar were used.
(D) and (E) Pn FT1 and Pn FT2 expression in Violet and Kidachi after the shift to continuous darkness (DD). Plants were grown under 10 h light/14 h dark for 6 d and then transferred to DD. Cotyledons were harvested every 2 h for 60 h. RT-PCR was performed to detect Pn FT1, Pn FT2, and ubq. Solid line, Kidachi; dotted line, Violet.
(F) Pn CO expression in Violet and Kidachi after the shift to continuous darkness (DD). Analysis performed as described for (D) and (E). Solid line, Kidachi; dotted line, Violet. Error bars show se of three PCR experiments. A biological replicate provided a similar result.
DISCUSSION
We studied the mechanisms of daylength measurement controlling flowering of Pharbitis (I. nil). Putative orthologs of the Arabidopsis flowering-time gene FT were isolated (Pn FT1 and Pn FT2). Pn FT1 promotes flowering when overexpressed in transgenic plants, and expression of both genes is closely associated with floral induction by different photoperiodic treatments. Taken together, these experiments indicate that Pn FT1 and Pn FT2 are likely to promote flowering of Pharbitis. DNA gel blotting indicated that there are additional Pn FT genes that may also be involved in flowering control. In photoperiodic systems, a circadian clock provides the timing mechanism that allows measurement of daylength (Thomas and Vince-Prue, 1997). Circadian clocks are entrained (or synchronized) to the daily cycle of night and day through their responsiveness to light and dark as well as to high and low temperatures (Salomé and McClung, 2005). We demonstrated that the clock that regulates Pn FT expression is entrained specifically by the light-to-dark transition at dusk. Genetic variation that alters regulation of Pn FT expression by this clock changes photoperiod response. By contrast, distinct circadian clocks with different entrainment characteristics control the rhythms of Pn FT and Pn CAB. Similarly, phase control of circadian leaf movements in Pharbitis is not correlated with daylength-regulated flowering (Bollig, 1975). We conclude that Pharbitis contains a light-sensitive clock that measures night length and regulates Pn FT expression.
Comparison of the Photoperiodic Mechanisms of Pharbitis, Rice, and Arabidopsis
Both LD and SD response types are present in monocot and dicot families, suggesting that these differences evolved several times. Nevertheless, there are striking similarities in the broad mechanisms of their photoperiodic responses that suggest that the basic system was present in the progenitor of all angiosperms.
In Arabidopsis, rice, and Pharbitis, expression of FT (Hd3a in rice or Pn FT in Pharbitis) occurs only in daylengths that induce flowering and shows a diurnal pattern of expression suggestive of circadian clock control. Thus, in each species, response to photoperiod is mediated by transcriptional regulation of FT or FT-like genes through an intersection between clock regulation and daylength. However, the mechanisms that regulate FT differ between species. In Arabidopsis, exposure to light plays a major role in FT regulation (Figure 6). FT transcription is activated by CO under LDs but not under SDs because under LDs, CO mRNA expression coincides with exposure of plants to light leading to stabilization of CO protein (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002; Imaizumi et al., 2003; Valverde et al., 2004). Under SDs, CO mRNA only accumulates in darkness, and CO protein is rapidly degraded through a ubiquitination mechanism involving the SPA1 protein (Laubinger et al., 2006). Therefore, in Arabidopsis, regulation of the stability of CO protein in a light-dependent manner is an important factor in daylength discrimination.
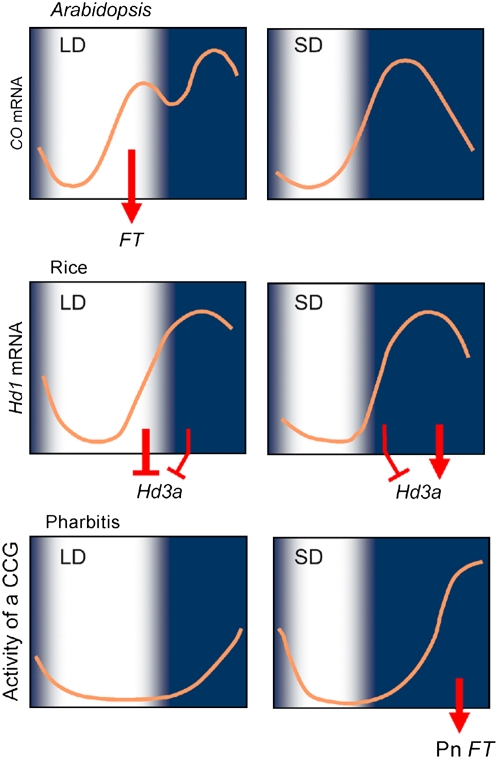
Figure 6.
A Model for the Timekeeping Mechanism in Pharbitis Compared with Those for Arabidopsis and Rice.
A model for control of flowering under LD (left) and SD (right) in Arabidopsis (top), rice (middle), and Pharbitis (bottom). In Pharbitis, the activity of a clock-controlled gene (CCG) that regulates the transcription of Pn FT is plotted. The activity of this gene is regulated by a specific circadian clock whose phase is reset by light off. Under LDs, Pn FT mRNA does not accumulate because the rhythm in the activity of the CCG is terminated by light at dawn before it reaches its peak phase. By contrast, under SDs, Pn FT mRNA does accumulate because the rhythm in expression of the CCG reaches a peak phase during the longer night. In Arabidopsis, expression of CO mRNA while the plants are exposed to light under LDs stabilizes CO protein, enabling activation of FT transcription under these conditions. Under SDs, FT expression is not activated because CO mRNA is only expressed in the dark and the protein does not accumulate under these conditions. In rice, under LDs, expression of Hd1 mRNA while the plants are exposed to light causes Hd1 protein to act as a repressor of Hd3a transcription and this inhibits flowering. Under these conditions, the repressor form is also proposed to repress Hd3a transcription during the short night. By contrast, under SDs, the Hd1 repressor form is made early during the long night, but later the activator form is made and this activates Hd3a transcription.
In both rice and Pharbitis, expression of FT homologs (Hd3a and Pn FT, respectively) rise in darkness at the end of the night, which is consistent with flowering of SD plants requiring exposure to long nights. Nevertheless, our data imply that despite the phenotypic similarities between rice and Pharbitis, these plants distinguish SDs and LDs by different mechanisms (Figure 6). In rice, Hd1, a CO homolog, was proposed to act both as an activator and a repressor of Hd3a expression (Izawa et al., 2002; Hayama et al., 2003). The repressor form is produced in response to the activation of phytochrome by light and is formed under LDs when Hd1 mRNA expression coincides with exposure of plants to light. Under SDs, Hd1 mRNA is not expressed in the light period, preventing formation of the repressor form and allowing Hd3a activation toward the end of the night. Activation of Hd3a also requires Hd1 because mutations in Hd1 reduce Hd3a expression (Izawa et al., 2002). The activator form of Hd1 is present under SDs because the active form of phytochrome (Pfr), which is required to produce the Hd1 repressor, decays during the long nights characteristic of SD conditions (Izawa et al., 2002). By contrast, the Pfr form of phytochrome must be stable for long enough in the dark to ensure that Hd1 made during the night under LDs remains in the repressor form (Figure 6) (Izawa et al., 2002). Thus, although the mRNA of Hd1 is expressed in a similar temporal pattern under LDs and SDs with a peak in the night (Izawa et al., 2002; Kojima et al., 2002; Hayama et al., 2003), posttranscriptional regulation of Hd1 under long nights ensures that Hd3a activation occurs only under SD conditions. However, the mechanism regulating Hd3a expression under SDs is likely to be more complex, and in some rice varieties, the Early heading date1 gene, which encodes a response regulator, also plays an important role in Hd3a transcriptional activation (Doi et al., 2004). Therefore, although mechanisms similar to the one we have described in Pharbitis have not been demonstrated or proposed in rice, their presence cannot be definitively excluded.
Our data and previous work suggest that photoperiodic flowering of Pharbitis is not regulated by the mechanism described above for rice. In Pharbitis, the circadian clock regulates Pn FT so that the phase of Pn FT expression always occurs at a constant time after dusk and is suppressed by exposure to light. This result is consistent with previous physiological experiments that suggested the existence of a dusk set circadian rhythm in Pharbitis (Lumsden and Furuya, 1986; Lumsden, 1998). The mechanism through which Pn FT expression is suppressed by light is not clear. Physiological experiments have shown that a night break that does not alter the phase of the photoperiodic response rhythm nevertheless suppresses flowering of Pharbitis. Therefore, suppression of Pn FT expression by light may not only be triggered by a change in the state of the circadian oscillator but also by a change in the activity of a circadian output rhythm by light (Lumsden and Furuya, 1986). Such a mechanism would be related to the external coincidence models proposed to regulate CO and Hd1 in Arabidopsis and rice, respectively (Suarez-Lopez et al., 2001; Izawa et al., 2002; Yanovsky and Kay, 2002).
The peak of Pn CO and Pn FT mRNA expression coincides during SDs, suggesting that Pn CO might activate Pn FT. However, as plants are exposed to increasing duration of light before their transfer to darkness, the peaks no longer coincide (Figure 4F), suggesting no direct relationship between Pn CO and Pn FT expression. Whether Pn CO activates Pn FT could be tested more directly by analyzing Pn FT expression in transgenic plants overexpressing Pn CO. Interestingly, in tomato, which is a member of the Solanaceae and therefore in the same order (Solanales) as Pharbitis, no relationship between the CO-like gene family and flowering could be established, although an FT homolog (SINGLE FLOWER TRUSS) plays a similar role in promoting flowering (Ben-Naim et al., 2006; Lifschitz et al., 2006). Therefore, CO-like genes might not be responsible for the regulation of FT-like genes in this group of plants. However, it cannot be excluded that other CO-like genes are present in Pharbitis and that the protein encoded by one of these activates Pn FT. Nevertheless, of the two characterized CO-like genes, Pn CO encodes a protein strikingly similar to Arabidopsis CO and complements the co-1 mutation of Arabidopsis (Liu et al., 2001).
Studies of photoperiodic flowering and the regulation of circadian rhythms in Arabidopsis suggest mechanisms by which the distinct clock systems described in Pharbitis could arise. All clock-regulated genes studied in detail in Arabidopsis are entrained by dawn and dusk, although dawn provides a stronger entrainment signal (Salomé and McClung, 2005). However, in the Arabidopsis elf3 mutant, rhythms are entrained by dusk and are arrested in constant light (McWatters et al., 2000; Covington et al., 2001). We show that the clock of Pharbitis that regulates Pn FT expression exhibits similar characteristics to that of Arabidopsis elf3 mutants, whereas a second clock similar to that of wild-type Arabidopsis regulates Pn CAB and Pn CO expression.
Regulation of circadian rhythms was shown to be cell autonomous in Arabidopsis (Thain et al., 2000, 2002), so that the coexistence of two clock systems in a single Pharbitis plant may be explained if they occur in different cell types. Pn FT and Pn CAB could be expressed in different cell types that use distinct clock systems. In Arabidopsis, FT is expressed specifically in the vascular tissue (Takada and Goto, 2003) and is proposed to encode a mobile protein that moves to the meristem (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007). Therefore, an attractive possibility is that in Pharbitis, a clock system entrained specifically by dusk acts in the phloem to regulate Pn FT in a dark-dependent manner, whereas a second system acts in the leaf mesophyll to regulate Pn CAB expression.
Distinct Temporal Patterns of Pn FT Expression Occur in Cultivars Exhibiting Different Critical Night Lengths
We found that the difference in critical night length between Kidachi and Violet is due to a difference in the phase of the circadian rhythm regulating Pn FT and/or in the amplitude of Pn FT expression. The Arabidopsis early-flowering mutant timing of cab1-1 (toc1-1) was previously shown to flower early under SDs by changing the phase of expression of genes controlling photoperiodic flowering, a mechanism related to that proposed here for Kidachi (Yanovsky and Kay, 2002). In the toc1-1 mutant, the expression of CO occurs at an earlier phase under SDs, and this causes coincidence between CO expression and exposure to light leading to activation of FT expression even under SDs (Yanovsky and Kay, 2002). In Pharbitis, alteration in the timing of expression of Pn CO does not explain the difference in critical night length shown by Kidachi and Violet because Pn CO expression was very similar in both accessions despite strong differences in Pn FT expression. Nevertheless, we propose that the shorter night length sufficient to induce flowering in Kidachi is caused by the phase of another clock-controlled gene that regulates Pn FT being shifted to an earlier time so that it is expressed earlier in the night. In addition, the clock output gene inducing Pn FT expression may be expressed at a higher amplitude in Kidachi. The difference in phase of Pn FT expression could be caused by a shorter period length of the clock in Kidachi compared with Violet, but this seems unlikely because under continuous darkness, the period length of both cultivars seemed similar; therefore, the difference seems to be due to changes in phase regulation rather than in period length. Altering the phase of regulators of expression of FT-like genes may represent a more general mechanism in which photoperiod regulation varies within species. Growth arrest in poplar is regulated by photoperiod, and Populus nigra FT expression causes termination of growth. Recently, P. nigra FT was shown to be expressed only under those daylengths in which growth arrest occurs, and FT expression in different poplar accessions correlated with altered timing of expression of the upstream regulator CO (Bohlenius et al., 2006). These differences correlated with the latitude at which the poplar accessions grow in nature, suggesting that they are important for the adaptation of accessions to these locations. This system is therefore analogous to the mechanism proposed here to regulate Pn FT expression and flowering in different Pharbitis accessions in that in both systems, variation in daylength response is caused by changing the phase of expression of regulatory genes, but in Pharbitis, a correlation between Pn CO and Pn FT expression was not observed.
METHODS
Plant Materials and Growth Conditions
Pharbitis nil seed of Ipomoea nil Choisy cv Violet and cv Kidachi were purchased from Marutane. Seeds were germinated as described by Liu et al. (2001). Germinated seedlings were grown on vermiculite in growth chambers at 25 to 28°C, except for the night break experiment where plants were grown at 22°C. LDs consisted of 16 h light and 8 h dark, and SDs were 10 h light and 14 h dark. Arabidopsis thaliana seeds were plated on soil, stratified at 4°C for 3 d, and grown in controlled environment chambers under SDs (8 h light/16 h dark).
Nucleic Acid Isolation and cDNA Synthesis
Total RNA was isolated using the RNeasy mini kit (Qiagen). The DNA-free kit (Ambion) was used for removal of DNA from the RNA samples. cDNA synthesis was performed using oligo(dT) and SuperScript II reverse transciptase (Invitrogen). DNA was isolated using the Phytopure kit (Amersham). All protocols were performed according to the manufacturer's instructions.
Isolation of FT Homologs from Pharbitis and DNA Gel Blotting
Partial clones of Pn FT1 were isolated by RT-PCR with primers designed in the Arabidopsis FT sequence. The primers used were 5′-CTCCATTGGTTGGTGACTGA-3′ and 5′-CTCGCGAGTGTTGAAGTT-3′. For Pn FT2, 5′-CAGAGTTGTTGGAGACGTTC-3′ and a primer based on the partial Pn FT1 clone, 5′-GCCGAAGCTTGCTCCTGTAGTTCC-3′, were used. Full-length clones were obtained with 5′-RACE using a system from Gibco BRL and with 3′-RACE.
Full-length open reading frames of Pn FT1 and Pn FT2 were amplified by PCR and purified using a QIAquick PCR purification kit (Qiagen). The resulting fragments were used as templates to synthesize probes by random labeling (as described in gene expression analysis). The Pn FT2 probe template contained an additional 111 bp of sequence upstream of the first exon. The primers used for amplification were PnFT1f (5′-CTAGCTAGGATGCGAAGGGGAA-3′), PnFT1r2 (5′-ATCGGCTGCCCAGGGCTCTGC-3′), PnFT2f (5′-ACATGCATGTAAAGATCCCA-3′), and PnFT2r5 (5′-TCGTCTCCGGCCTCCGGTGC-3′).
Gene Expression Analysis Using RT-PCR
Single-stranded cDNA was used to amplify genes. The products after 26 cycles for Pn FT1 and Pn FT2 and 21 for Ubiquitin were separated on an agarose gel and blotted onto a nylon membrane (Amersham). Numbers of cycles used to amplify genes for Supplemental Figure 4B online and Figures 5A and 5B were 25 for Pn CO, 28 for Pn FT1 and Pn FT2, and 19 for Ubiquitin (Ubq). These genes were probed with respective probes labeled with Redivue 32P-dCTP (Amersham). Probes were synthesized using the Rediprime II Random Prime labeling system (Amersham). Images were visualized using a PhosphorImager (Molecular Dynamics). The bands were quantitated using Image Quant software (Molecular Dynamics). Each RNA analysis was performed at least twice, and a representative blot is shown.
Quantitations using real-time PCR were performed either with iQ SYBR Green 2X mix (Bio-Rad) or a modified 10× buffer (Karsai et al., 2002) using an iCycler iQ real-time PCR detection system (Bio-Rad). Biological replicates were used, and each PCR was performed in triplicate. Each figure illustrates the mean for a single biological sample, but in each case, a biological replicate provided a similar result. Products were quantified against a standard curve using pooled cDNA from time points where expression was high. The primers used for amplification were Pn FT1 (5′-ACCCTGAGGGAATACCTCCACT-3′ and 5′-GGAAGGAGCAGGGTAATTAATCGG-3′), Pn FT2 (5′-TGAGGGAGTACCTACACTGGTTG-3′ and 5′-AGGGTGCGTCATTACGCATT-3′), Pn Ubq (5′-GGAGTCGACTCTTCACTTGG-3′ and 5′-TGGGACATTAGGGGATTCAG-3′), and Pn CAB (5′-GCCATTCTTGAGCTCCTTGACC-3′ and 5′-TTTTCCCGGAGCTGTTGTCC-3′). For Pn CO, primers (5′-AACGAGATGTCATGCGCAGTAG-3′ and 5′-GGGAGATTGAGGTATCACTCAAGG-3′) were used to amplify across an intron, and the resulting products were separated on a gel, transferred to a filter, and probed with a Pn CO probe. This protocol was necessary because Pn CO mRNA exists in spliced and unspliced forms, as described previously (Liu et al., 2001).
Transformation of Arabidopsis and Pharbitis with Pn FT1
Full-length Pn FT1 cDNA was amplified using primers with Gateway extensions (Invitrogen) to generate an entry clone. An LR reaction to a destination vector was performed to create a Pn FT1 clone expressed under the cauliflower mosaic virus 35S or a maize (Zea mays) Ubiquitin promoter. Arabidopsis co-2 plants were transformed using the floral dip method (Clough and Bent, 1998), and transformants selected on germination medium plates with 50 μg/mL of kanamycin. T2 plants from lines segregating in a 3:1 ratio were screened for the early flowering phenotype.
Pharbitis transformations were performed as previously described (Ono et al., 2000). Seeds were harvested from the regenerated hygromycin- or kanamycin-resistant plants (T0 plants). Ten to fifteen seeds from each independent transformant were grown to obtain T1 plants. These T1 plants were used to analyze the segregation of the transgene and obtain flowering time data. In cases where the T0 plants produced only two to three seeds, the segregation analysis was performed in the T2 generation.
Measurement of the Flowering Response in Pharbitis
Flowering time for Pharbitis was measured as the average number of flower buds per plant. Visible buds in each leaf axil on the primary axis, the axillary flower buds of the bracts, and the terminal flower are included in the number of flower buds.
Phylogenetic Analysis
FT sequences were aligned using ClustalW (Higgins, 1994), as shown in Supplemental Figure 1 online. The phylogenetic tree was constructed using the Phylip 3.65 package with the neighbor-joining method (Felsenstein, 1989). One thousand replicates were used to generate the bootstrap values.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers bankit1020846 (Pn FT1) and bankit1020856 (Pn FT2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Sequences of FT-Like Proteins.
Supplemental Figure 2. Pharbitis Genomic DNA and BAC Clones of Pharbitis DNA Containing Pn FT1 or Pn FT2 Hybridized with Pn FT1 or Pn FT2 Probes.
Supplemental Figure 3. Effects of Different Light/Dark Regimes on Pn FT1 and Pn FT2 Expression and on Flowering.
Supplemental Figure 4. Gel Blot Data Used for Quantifications Shown in the Figures.
Supplemental Data Set 1. Alignments Used to Generate Figure 1A.
Supplementary Material
Acknowledgments
We thank Hans Henning Steinbiss for his advice on transformation protocols. We also thank Aidyn Mouradov for help in the early steps of the characterization of the Pn FT genes. R.H. was supported by a fellowship from the Japanese Society for the Promotion of Science. The laboratory of G.C. is supported by a core grant from the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: George Coupland (coupland@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Ben-Naim, O., Eshed, R., Parnis, A., Teper-Bamnolker, P., Shalit, A., Coupland, G., Samach, A., and Lifschitz, E. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46 462–476. [DOI] [PubMed] [Google Scholar]
- Bohlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A.M., Jansson, S., Strauss, S.H., and Nilsson, O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043. [DOI] [PubMed] [Google Scholar]
- Bollig, I. (1975). Photoperiodic time measurement and circadian leaf movement in Pharbitis nil controlled by the same clock? Z. Pflanzenphysiol. 77 54–69. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., and Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. [DOI] [PubMed] [Google Scholar]
- Covington, M.F., Panda, S., Liu, X.L., Strayer, C.A., Wagner, D.R., and Kay, S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., Yano, M., and Yoshimura, A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1l. Genes Dev. 18 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5 164–166. [Google Scholar]
- Hamner, K.C., and Bonner, J. (1938). Photoperiodism in relation to hormones as factors in floral initiation and development. Bot. Gaz. 100 388–431. [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 719–722. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G. (1994). CLUSTAL V: Multiple alignment of DNA and protein sequences. Methods Mol. Biol. 25 307–318. [DOI] [PubMed] [Google Scholar]
- Hsu, C.Y., Liu, Y., Luthe, D.S., and Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18 1846–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi, T., and Kay, S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11 550–558. Erratum. Trends Plant Sci. 11: 567. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Tran, H.G., Swartz, T.E., Briggs, W.R., and Kay, S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426 302–306. [DOI] [PubMed] [Google Scholar]
- Imamura, S. (1967). Physiology of Flowering in Pharbitis nil. (Tokyo: Japanese Society of Plant Physiologists).
- Imamura, S., Muramatsu, M., Kitajo, S., and Takimoto, A. (1966). Varietal difference in photoperiodic behavior of Pharbitis nil. Bot. Mag. Tokyo 79 714–721. [Google Scholar]
- Ishikawa, R., Tamaki, S., Yokoi, S., Inagaki, N., Shinomura, T., Takano, M., and Shimamoto, K. (2005). Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Tokutomi, S., Okuno, K., and Shimamoto, K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22 391–399. [DOI] [PubMed] [Google Scholar]
- Jaeger, K.E., and Wigge, P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17 1050–1054. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Karsai, A., Muller, S., Platz, S., and Hauser, M.T. (2002). Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32 790–792, 794–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J., Moon, J., Lee, I., Maeng, J., and Kim, S.R. (2003). Molecular cloning and expression analysis of a CONSTANS homologue, PnCOL1, from Pharbitis nil. J. Exp. Bot. 54 1879–1887. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Laubinger, S., Marchal, V., Gentilhomme, J., Wenkel, S., Adrian, J., Jang, S., Kulajta, C., Braun, H., Coupland, G., and Hoecker, U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222. [DOI] [PubMed] [Google Scholar]
- Lifschitz, E., Eviatar, T., Rozman, A., Shalit, A., Goldshmidt, A., Amsellem, Z., Alvarez, J.P., and Eshed, Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.K., Belanger, H., Lee, Y.J., Varkonyi-Gasic, E., Taoka, K., Miura, E., Xoconostle-Cazares, B., Gendler, K., Jorgensen, R.A., Phinney, B., Lough, T.J., and Lucas, W.J. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19 1488–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Yu, J., McIntosh, L., Kende, H., and Zeevaart, J.A. (2001). Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 125 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden, P., Thomas, B., and Vince-Prue, D. (1982). Photoperiodic control of flowering in dark-grown seedlings of Pharbitis nil choisy: The effect of skeleton and continuous light photoperiods. Plant Physiol. 70 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden, P.J. (1998). Photoperiodic induction in short-day plants. In Biological Rhythms and Photoperiodism in Plants. P.J. Lumsden and A.J. Millar, eds (Oxford, UK: BIOS Scientific Publishers), pp. 167–181.
- Lumsden, P.J., and Furuya, M. (1986). Evidence for two actions of light in the photoperiodic induction of flowering in Pharbitis nil. Plant Cell Physiol. 27 1541–1551. [Google Scholar]
- Mathieu, J., Warthmann, N., Kuttner, F., and Schmid, M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17 1055–1060. [DOI] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408 716–720. [DOI] [PubMed] [Google Scholar]
- Ono, M., Sage-Ono, K., Kawakami, M., Hasebe, M., Ueda, K., Masuda, K., Inoue, M., and Kamada, H. (2000). Agrobacterium-mediated transformation and regeneration of Pharbitis nil. Plant Biotechnol. 17 211–216. [Google Scholar]
- Ray, P.M., and Alexander, W.E. (1966). Photoperiodic adaptation to latitude in Xanthium strumarium. Am. J. Bot. 53 806–816. [Google Scholar]
- Saji, H., Furuya, M., and Takimoto, A. (1984). Photoperiod preceding a flower-inductive dark period in dark-grown seedling of Pharbitis nil. Plant Cell Physiol. 25 715–720. [Google Scholar]
- Salomé, P.A., and McClung, C.R. (2005). What makes Arabidopsis tick: Light and temperature entrainment of the circadian clock. Plant Cell Environ. 28 21–38. [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Searle, I., and Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., Amasino, R.A., and Coupland, G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal, P., Gleissner, R., Corbesier, L., Apel, K., and Melzer, S. (2004). Modulation of flowering responses in different Nicotiana varieties. Plant Mol. Biol. 55 253–262. [DOI] [PubMed] [Google Scholar]
- Soltis, P.S., Soltis, D.E., and Chase, M.W. (1999). Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402 402–404. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto, A., and Hamner, K.C. (1965). Studies on red light interruption in relation to timing mechanisms involved in the photoperiodic response of Pharbitis nil. Plant Physiol. 40 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki, S., Matsuo, S., Wong, H.L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316 1033–1036. [DOI] [PubMed] [Google Scholar]
- Thain, S.C., Hall, A., and Millar, A.J. (2000). Functional independence of circadian clocks that regulate plant gene expression. Curr. Biol. 10 951–956. [DOI] [PubMed] [Google Scholar]
- Thain, S.C., Murtas, G., Lynn, J.R., McGrath, R.B., and Millar, A.J. (2002). The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 130 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B., and Vince-Prue, D. (1997). Photoperiodism in Plants. (San Diego, CA: Academic Press).
- Turner, A., Beales, J., Faure, S., Dunford, R.P., and Laurie, D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034. [DOI] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Vince-Prue, D., and Gressel, J. (1985). Pharbitis nil. In Handbook of Flowering, Vol. IV, A.H. Halevy, ed (Boca Raton, FL: CRC Press), pp. 47–81.
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.