Abstract
RAD51, the eukaryotic homolog of the bacterial RecA recombinase, plays a central role in homologous recombination (HR) in yeast and animals. Loss of RAD51 function causes lethality in vertebrates but not in other animals or in the flowering plant Arabidopsis thaliana, suggesting that RAD51 is vital for highly developed organisms but not for others. Here, we found that loss of RAD51 function in the moss Physcomitrella patens, a plant of less complexity, caused a significant vegetative phenotype, indicating an important function for RAD51 in this organism. Moreover, loss of RAD51 caused marked hypersensitivity to the double-strand break-inducing agent bleomycin in P. patens but not in Arabidopsis. Therefore, HR is used for somatic DNA damage repair in P. patens but not in Arabidopsis. These data imply fundamental differences in the use of recombination pathways between plants. Moreover, these data demonstrate that the importance of RAD51 for viability is independent of taxonomic position or complexity of an organism. The involvement of HR in DNA damage repair in the slowly evolving species P. patens but not in fast-evolving Arabidopsis suggests that the choice of the recombination pathway is related to the speed of evolution in plants.
INTRODUCTION
Homologous recombination (HR), generally considered to be a precise DNA damage repair process, plays an important role in the maintenance of genome stability. The eukaryotic RecA homolog RAD51, one of the best-studied eukaryotic recombination proteins, has a central position in this pathway (Baumann and West, 1998; Paques and Haber, 1999; Shinohara and Ogawa, 1999; Sung et al., 2000, 2003; Symington, 2002). RAD51 was originally identified in yeast (Saccharomyces cerevisiae) as one of the genes leading to sensitivity to ionizing radiation when defective. Besides hypersensitivity to DNA damaging agents and meiotic defects, yeast rad51 mutants are viable and have no developmental abnormalities. However, RAD51 deficiency causes lethality in chicken DT40 and mouse cells (Thacker, 1999), and this phenotype has been considered typical for multicellular organisms for some time. More recently, rad51 mutants in Caenorhabditis elegans (Rinaldo et al., 2002; Alpi et al., 2003), Drosophila melanogaster (Staeva-Vieira et al., 2003), and the flowering plant Arabidopsis thaliana (Li et al., 2004) were found to be fully viable, suggesting that RAD51 is an essential gene in vertebrates only, but not in other organisms.
The moss Physcomitrella patens is a nonflowering plant and taxonomically quite distant from Arabidopsis (Reski, 1998; Cove, 2005; Cove et al., 2006). The P. patens genome contains two duplicated, functional RAD51 genes, Pp RAD51A and Pp RAD51B (Markmann-Mulisch et al., 2002), also described as PpaRad51.1 and PpaRad51.2, both of which encode genuine RAD51 proteins with all attributes of RAD51 (Ayora et al., 2002). To analyze the role of RAD51 in development and DNA damage repair in this plant, we used gene targeting to knock out both RAD51 genes. Surprisingly, complete loss of RAD51 function caused a marked vegetative phenotype, suggesting an important role of RAD51 in P. patens development.
RAD51 is involved in DNA damage repair, especially in the repair of double strand breaks (DSBs). These lesions can be repaired either by HR or nonhomologous end joining (NHEJ). HR is thought to prevail in yeast, while NHEJ is generally believed to be the predominant repair pathway in vertebrates and plants (Reiss, 2003; Dudas and Chovanec, 2004; Schuermann et al., 2005; Burma et al., 2006; Sonoda et al., 2006). Bleomycin is a potent inducer of DSBs with a mode of action comparable to ionizing radiation (Povirk, 1996; Ramotar and Wang, 2003). rad51 mutants in yeast (Game and Mortimer, 1974), C. elegans (Rinaldo et al., 2002), and D. melanogaster (Staeva-Vieira et al., 2003) are hypersensitive to ionizing radiation, indicating an involvement of RAD51 in somatic DNA damage repair in these organisms. To analyze its role in DNA damage repair in P. patens, we analyzed resistance to bleomycin. The rad51 mutant in Arabidopsis, At rad51-1 (Li et al., 2004), which was not yet analyzed in respect to DNA damage repair, was included in this analysis. While loss of RAD51 function substantially affected resistance to bleomycin in P. patens, it was hardly affected in Arabidopsis, suggesting that Arabidopsis and P. patens differ in the use of somatic DNA damage repair pathways. However, At rad51-1 is highly sensitive to the interstrand cross-linking (ICL) agent mitomycin C (Dronkert and Kanaar, 2001), indicating an essential role of RAD51 in ICL repair. Since mutants in RAD51 paralogs (Baumann and West, 1998; Shinohara and Ogawa, 1999; Thacker, 1999; West, 2003), another group of genes acting in the HR pathway, are also unaffected in their resistance to bleomycin but highly sensitive to mitomycin C in Arabidopsis (Bleuyard and White, 2004; Bleuyard et al., 2005), the major group of HR genes seems to be involved in ICL repair but not in DSB repair in Arabidopsis.
RESULTS
Inactivation of Pp RAD51A and Pp RAD51B by Gene Targeting
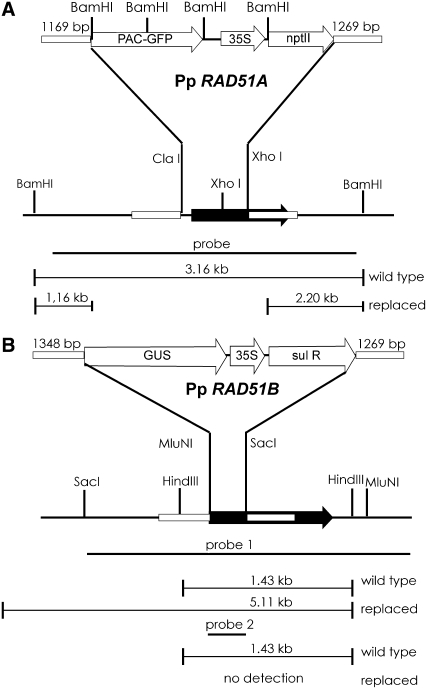
A gene targeting strategy likely to eliminate RAD51 function was devised that deletes a portion of the RAD51 coding region essential for protein–protein interaction and for binding to single-stranded DNA (Ayora et al., 2002). The deleted portion was replaced by a green fluorescent protein (GFP) reporter and a kanamycin resistance gene in the vector to target Pp RAD51A, pKOrad51A (Figure 1A). In the vector to target Pp RAD51B, pKOrad51B (Figure 1B), this region is replaced with a β-glucuronidase (GUS) reporter and a sulfonamide resistance gene.
Figure 1.
Schematic Representation of P. patens RAD51 Genes and the Gene Targeting Strategy.
Schematic representations of Pp RAD51A (A) and Pp RAD51B (B). The gene targeting vectors used to inactivate the genes are shown above (not to scale). Restriction sites used for cloning or DNA gel blot analysis are shown. The probes used for DNA gel blotting and fragment sizes of authentic and replaced genes are shown below the sketch. Black arrows indicate the RAD51 target gene coding regions. Regions of homology (given in base pairs) between target loci and replacement vectors are shown as small boxes. The probes used for DNA gel blotting are shown as black bars. Open arrows denote genes and promoters. PAC-GFP, fusion protein of chloroplast-targeted pale cress and GFP; 35S, cauliflower mosaic virus 35S promoter; nptII, kanamycin resistance gene; sul R, sulfonamide resistance gene.
After transformation of wild-type P. patens protoplasts with pKOrad51A, 14 stable lines were obtained. The DNA gel blot analysis (Figure 2A) showed that RAD51A was targeted in five of them. However, only in one transformant, later named Pp rad51A, the genomic RAD51A gene was precisely replaced with the in vitro–modified copy, and no further insertions of targeting vector DNA including the cloning vector were detected. The others contained either a number of additional copies of the targeting vector integrated in the target locus, as is frequently observed in P. patens gene targeting (Reiss, 2003; Kamisugi et al., 2006), or an unmodified Pp RAD51A gene resided in the genome, as is often observed in gene targeting in plants (Reiss, 2003). The efficiency of gene targeting obtained with RAD51A was thus 36%, but the expected precise replacement of the endogenous gene with the modified copy of pKOrad51A occurred in only 7% of the transformants.
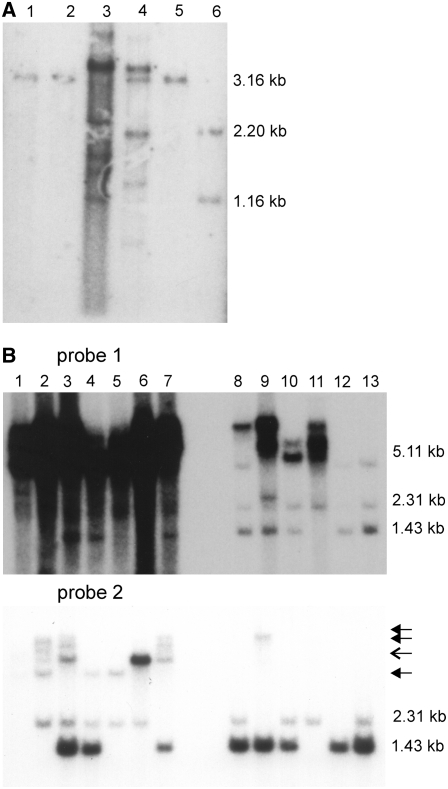
Figure 2.
DNA Gel Blot Analysis of P. patens RAD51 Transformants.
(A) Genomic DNA of transformants obtained by transformation of the wild type with pKOrad51A digested with BamHI, separated by agarose gel electrophoresis, blotted, and the blot hybridized with the probe shown in Figure 1A. Pp RAD51A is located on a 3.16-kb BamHI fragment that is split in fragments of 1.16 and 2.2 kb after gene replacement. Lane 1, transformant 2/1-4; lane 2, transformant 2/1-5; lane 3, transformant 2/1-6; lane 4, transformant 2/1-7, lane 5; the wild type; lane 6, transformant 2/1-9 (Pp rad51A).
(B) Genomic DNA of transformants obtained with pKOrad51B in the wild type (transformant names starting with 1) and the Pp rad51A knockout line (transformant names starting with 7) was digested with HindIII, separated by agarose gel electrophoresis, blotted, and the blot hybridized with two different probes. Probe 1 covers the entire gene but mainly detects a 1.43-kb fragment of Pp rad51A in the wild type that shifts to 5.11 bp after replacement. Probe 2 corresponds to the region of Pp RAD51B that is deleted from the genome after gene replacement. Both probes cross-hybridize with Pp RAD51A and detect a 2.31-kb fragment in the wild type that is lost in Pp rad51A knockout lines. Lane 1, transformant 7/2 (Pp rad51AB); lane 2, 1/3 (Pp rad51B); lane 3, 1/7; lane 4, 1/5; lane 5, 1/1 (Pp rad51B-2); lane 6, 1/2; lane 7, 7/1; lane 8, 1/4; lane 9, 7/4; lane 10, 1/6; lane 11, 1/8; lane 12, Pp rad51A; lane 13, the wild type. Closed arrows indicate bands obtained by cross-hybridization with vector sequences. The open arrow indicates a vector cross-hybridizing band, except for lanes 3 and 6 in which the signal is specific for Pp RAD51B sequences.
The eight stable lines obtained after transformation of wild-type protoplasts with pKOrad51B were characterized with PCR using a combination of 5′ and 3′ locus-specific and sulfadiazine resistance gene-specific primers. This analysis indicated that Pp RAD51B had correctly recombined with the gene targeting vector in six of the eight transformants, while the vector had integrated at random in the other two lines, suggesting a gene targeting efficiency of 75%. However, the DNA gel blot analysis (Figure 2B) showed that the portion foreseen to be deleted from the P. patens genome was actually absent in only three of the transformants; the others contained this portion, either in authentic size, or in rearranged form. Therefore, RAD51B was deleted in only 37%. In addition, additional copies of the targeting vector, including the cloning vector, were present in all transformants, indicating that the predicted precise gene replacement was not obtained in a single case. Moreover, several transformants turned out to be polyploid, reducing the number of useful RAD51B knockout lines to Pp rad51B and Pp rad51B-2. The data suggest that the mechanisms of gene targeting operating in P. patens produce unpredicted and unusual recombination products that cannot be sufficiently discriminated from the desired gene replacement by conventional PCR analysis. In addition, although the gene targeting efficiencies were as high (36% for Pp RAD51A and 75% for Pp RAD51B) as predicted (Kamisugi et al., 2005), such complex, unclear gene targeting events and polyploidization (Schween et al., 2005) had reduced the number of knockout lines useful for phenotypic analysis considerably.
To obtain lines in which the RAD51A and RAD51B genes were inactivated simultaneously, a double knockout mutant was produced by retransforming the Pp rad51A knockout line with the RAD51B targeting vector and the transformants characterized as described before (Figure 2B). The single line obtained in this approach, Pp rad51AB, displayed a developmental phenotype. To confirm that this phenotype is caused by a loss of RAD51 function, an independent double knockout line was produced by crossing Pp rad51A and Pp rad51B, each individually not showing such a phenotype. This line, Pp rad51AB-2, exhibited the same growth phenotype as Pp rad51AB, confirming that this defect was caused by a loss of RAD51 function.
To verify that the RAD51 genes were functionally inactivated in Pp rad51A and Pp rad51B, RNA gel blotting experiments were performed. These showed that the authentic RAD51A and RAD51B transcripts were lost in the knockout lines, as predicted (see Supplemental Figure 1A online). The complete loss of the corresponding transcripts in the Pp rad51A, Pp rad51B, and Pp rad51AB mutants was confirmed by RT-PCR (see Supplemental Figure 1B online).
Loss of RAD51 in P. patens Affects Growth
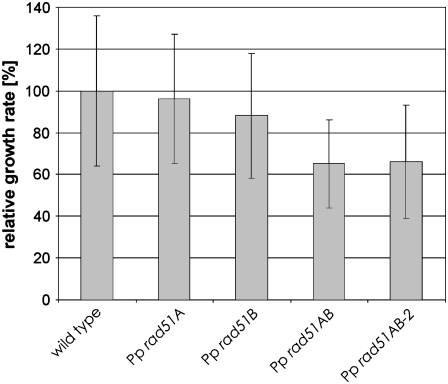
The growth rate of young protonema colonies was determined to analyze whether the mutants were affected in growth (Figure 3). While protonema growth of both single knockout lines was comparable to the wild type, both double knockout lines grew markedly slower, indicating that loss of RAD51 affects growth in the vegetative phase of the life cycle of P. patens. Since both lines were nearly identical in this assay, the further analysis of the developmental phenotype was restricted to Pp rad51AB.
Figure 3.
P. patens rad51 Double Knockout Mutants Are Affected in Growth.
The average increase in size of young Pp rad51A, Pp rad51B, Pp rad51AB, and Pp rad51B-2 P. patens colonies is shown relative to the wild type.
Loss of RAD51 in P. patens Affects Vegetative Development
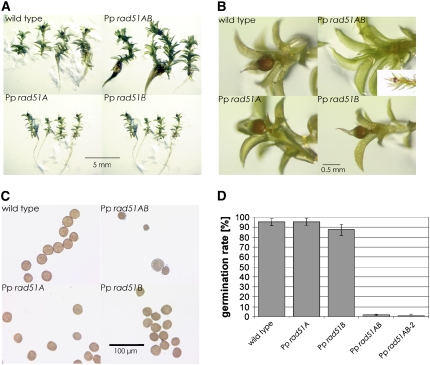
To analyze the vegetative developmental phenotypes, single young colonies were grown until maturity. The differences in colony morphology became apparent (Figure 4A) when the wild type had developed typical colonies with a dense center, intensively branched protonemata growing out from the colony, and the first young gametophores, the leafy shoots of moss, had developed. Both single knockout lines formed similar colonies, but these appeared slightly less compact, had a less pronounced dense center, and protonema filament outgrowth was slightly enhanced. By contrast, the colonies formed by the double knockout line Pp rad51AB were markedly reduced in size, consisted entirely of densely growing tissue, and gametophore formation was entirely inhibited. At later stages, when colonies matured, the differences between the wild type and the Pp rad51A and Pp rad51B single knockout mutants leveled off, possibly because colony morphology is dominated by gametophores by then, but the striking difference to the double knockout persisted (data not shown).
Figure 4.
Vegetative Developmental Phenotypes of P. patens rad51 Mutants.
(A) Morphology of wild-type, Pp rad51A, Pp rad51B, and Pp rad51AB P. patens colonies. The picture was taken after 2 weeks of growth.
(B) Close-up view of colonies shown in (A) to show the morphology of protonema filaments.
The macroscopically visible differences in colony morphology are likely to be caused by aberrations in protonema growth (Figure 4B). The filaments, including the apical cells of wild-type colonies, consisted predominantly of chloronema, cells densely packed with chloroplasts that form intensely green-colored filaments (Reski, 1998; Cove, 2005; Cove et al., 2006). By contrast, in Pp rad51A and Pp rad51B colonies, the apical cells were caulonema, a nearly colorless filament type that forms from chloronema and consists of relatively fast-growing cells with few chloroplasts. Another difference was observed in the formation of side branches. The primary filament in the wild type is highly branched shortly after the apical cell and the side branch initials originate in chloronema cells. In both the Pp rad51A and Pp rad51B single knockout lines, however, the first side branch initials are initiated on caulonema-like cells and only later, when the primary filament has formed secondary chloronema, side branches form on chloronema. This difference to the wild type may be the cause for the fact that highly branched structures formed only relatively late in the single knockout lines and thus branching seems delayed. In contrast with these relatively minor differences, the Pp rad51AB double knockout mutant did not form caulonema at all; rather, all filaments were chloronemata. Moreover, branching was strongly suppressed and the filaments formed few and irregular side branches rather than the regular pattern observed with the wild type. In summary, chloronema-to-caulonema transition and side branch initiation were inhibited nearly completely by the loss of RAD51 function.
These alterations in vegetative development could be the indirect consequence of a cell proliferation defect. A partial depletion of RAD51 function as expected to occur in either of the single knockouts could reduce growth slightly. Poor growth conditions induce caulonema formation in P. patens. Slow growth may be perceived as a signal indicating poor growth conditions and thus trigger caulonema development. However, RAD51 function could be highly important for the fast-proliferating caulonema cells. Thus, the complete loss of RAD51 function in the double knockout would inhibit the chloronema-to-caulonema transition, explaining the opposing phenotypes observed in the single and double knockout lines. Side branch initiation could have a similar requirement for RAD51, explaining the absence of these structures in the double knockout. However, side branches are formed in the single knockouts, but the initials originate mainly from caulonema instead of chloronema, explaining the alterations in the branching pattern observed in the single knockouts.
Loss of RAD51 in P. patens Affects Generative Development
Gametophore formation proceeded normally in both single knockout mutants and was nearly normal in the double knockout mutant. However, fewer gametophores formed in Pp rad51AB, and those that formed were larger and grew in a more extended form (Figure 5A). Sporophytes and spore capsules developed like the wild type in the single knockout mutants also, but development was delayed in the double knockout mutant (Figure 5B), although maturation was normal (Figure 5B, inset). Spores in the Pp rad51A and Pp rad51B single knockout mutants were produced in the same number and with the same size and appearance as the wild type (Figure 5C). Pp rad51AB double knockout mutant spore capsules contained fewer spores, and most of these were considerably smaller than the wild type and irregular in shape.
Figure 5.
Phenotypes of P. patens rad51 Mutants at Later Developmental Stages.
(A) Wild-type, Pp rad51A, Pp rad51B, and Pp rad51AB gametophores at the stage of spore capsule maturation.
(B) Close-up view of the gametophores in (A), showing normal spore capsule maturation in Pp rad51A and Pp rad51B and a delay in development in Pp rad51AB. The inset in the Pp rad51AB panel shows a gametophore at a later stage that carries a normally matured spore capsule.
(C) Spores obtained from mature wild-type, Pp rad51A, Pp rad51B, and Pp rad51AB spore capsules.
(D) Germination efficiencies of spores obtained from Pp rad51A, Pp rad51B, and Pp rad51AB spore capsules.
To analyze this phenotype further, spore germination was tested. The spore germination rate of Pp rad51A was identical to the wild type, and that of Pp rad51B was only slightly reduced. However, Pp rad51AB and Pp rad51AB-2 spores barely germinated (Figure 5D). Moreover, nearly all wild-type and single knockout mutant spores established a viable protonema, while those of the double knockout mutants rarely grew, reducing the survival rate further to 0.18% for Pp rad51AB and 0.33% for Pp rad51AB-2.
Spores are the direct product of meiosis. Therefore, meiosis is likely to be defective, indicating an important function of RAD51 in this process. However, the defect appears less severe than in Arabidopsis where seed set was virtually abolished (Li et al., 2004). Thus, RAD51 may play a less important role in meiosis in P. patens, the mechanisms of meiotic recombination may differ between P. patens and Arabidopsis, or spore development is a more robust process than the analogous processes are in Arabidopsis.
RAD51 Plays an Essential Role in DSB Repair in P. patens
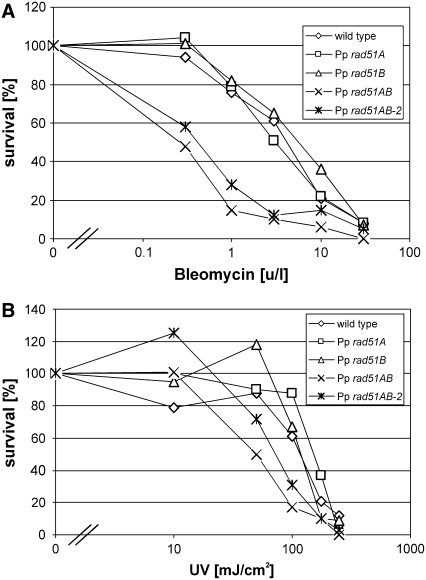
The radiomimetic bleomycin induces DSBs, lesions that cause lethality. To analyze whether RAD51 plays a role in DSB repair in P. patens, protonema was treated with increasing concentrations of bleomycin and the survival rates determined (Figure 6A). While survival of both single knockout lines was comparable to the wild type, the double knockout lines were significantly affected in resistance to bleomycin. The shoulder in the survival curves for the wild type and single RAD51 knockouts indicates that DNA damage in the low dosage range has successfully been repaired in these cells. This shoulder is absent in the curves of both Pp rad51AB and Pp rad51AB-2 double knockout lines, indicating that in abolishing RAD51 function, an important part of the DNA damage repair process has been eliminated.
Figure 6.
Resistance of P. patens rad51 Mutants to DNA Damage.
(A) Resistance to bleomycin. Survival curves of P. patens wild-type, Pp rad51A, Pp rad51B, Pp rad51AB, and Pp rad51B-2 protonema fragments obtained after treatment with 0, 0.3, 1, 3, 10, and 30 units/liter of bleomycin.
(B) Resistance to UV light. Survival curves obtained with 5-d-old protonema colonies of P. patens wild type, Pp rad51A, Pp rad51B, Pp rad51AB, and Pp rad51AB-2 after treatment with 0, 10, 50, 100, 175, and 250 mJ/cm2 UV.
To confirm this result with another type of DNA damage, resistance to UV light was analyzed (Figure 6B). Both single knockout lines, Pp rad51A and Pp rad51B, were as resistant to UV light as the wild type. However, survival of the double knockout lines was significantly more affected at high doses of UV light, indicating that RAD51 has an important function in the repair of such DNA damage also.
RAD51 Is Essential for ICL but Not for DSB Repair in Arabidopsis
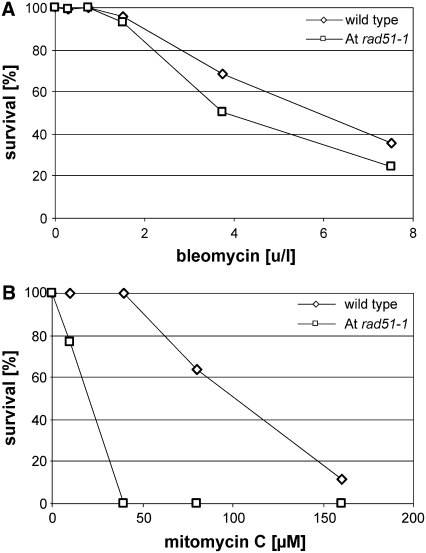
To analyze the role of RAD51 in DNA damage repair in Arabidopsis and to compare these data to those previously obtained with the RAD51 paralogs, resistance of the At rad51-1 mutant to bleomycin (Figure 7A) and mitomycin C (Figure 7B) was analyzed. The survival curves obtained with bleomycin for wild-type and homozygous At rad51-1 mutant seedlings were very similar, and only at high concentrations did At rad51-1 individuals survive slightly less. However, the survival curves for At rad51-1 and the wild type have an identical shoulder and decline with nearly identical slope, indicating that resistance to bleomycin was not significantly affected by loss of RAD51 function in Arabidopsis. By contrast, the survival curves obtained with mitomycin C were strikingly different for At rad51-1 and the wild type. While the growth of the wild type was not affected by concentrations up to 40 μM mitomycin C, mutant plants had completely lost viability at this concentration already. Therefore, loss of RAD51 function caused marked hypersensitivity to mitomycin C in Arabidopsis, indicating an important function of this gene in ICL repair in this organism.
Figure 7.
Resistance of the Arabidopsis rad51-1 Mutant to DNA Damage.
(A) Resistance to bleomycin. At rad51-1 and wild-type seedlings were germinated in the presence of 0, 0.3, 0.75, 1.5, 3.75, and 7.5 units/liter bleomycin, survival was scored, homozygous At rad51-1 individuals were detected by genotyping, and the data normalized and plotted.
(B) Resistance to mitomycin C. At rad51-1 and wild-type seedlings were germinated in the presence of 0, 10, 40, 80, and 160 μM mitomycin C, survival was scored, homozygous At rad51-1 individuals were detected by genotyping, and the data normalized and plotted.
The data were confirmed using a callus growth assay (Bleuyard and White, 2004). Proliferation of callus induced from roots of homozygous At rad51-1 plants was markedly more impaired in the presence of mitomycin C than of wild-type callus. However, growth was comparable to the wild type in the presence of bleomycin (data not shown). Therefore, the repair pathways of rapidly proliferating, undifferentiated cells are comparable to those of seedlings in respect to the requirement of RAD51 in ICL and DSB repair in Arabidopsis.
DISCUSSION
In contrast with chicken and mouse cells, loss of RAD51 function seems neither to affect growth nor vegetative development in nonvertebrates (Alpi et al., 2003; Staeva-Vieira et al., 2003), including the plant Arabidopsis (Li et al., 2004). These observations suggest that RAD51 has a vital function in vertebrates but not in other organisms. However, the RAD51 data obtained with P. patens described here seem to contradict this paradigm. Although loss of RAD51 has not caused lethality as in vertebrates, it affected growth and vegetative development in P. patens, indicating that RAD51 has an important function in this organism. Therefore, other criteria than taxonomic position or morphological complexity seem to determine the role of RAD51 in vegetative development.
The lethality of rad51 mutants in vertebrates is thought to be based on the tight interaction of RAD51 with tumor suppressor proteins, which establishes an indispensable role for this protein at the interface of DNA damage repair and cell cycle control (discussed in Li et al., 2004). However, the decisive difference could be that absence of RAD51 triggers apoptosis in vertebrates only, but not in other organisms. RAD51 in P. patens, but not in Arabidopsis, could play a role in DNA damage sensing and signal transduction to cell cycle control similar to vertebrates. However, instead of causing lethality, loss or RAD51 could impact cell cycle progression and cause a severe delay. Alternatively, however, the defects in DNA damage repair could have a similar effect. Genomes are constantly challenged by a number of intrinsic and extrinsic threats. External factors like UV, x-rays, or γ-rays and genotoxic chemicals permanently cause damage to DNA. The DNA molecule itself is unstable and metabolism generates oxidative and alkylating damage. The damage caused by DNA hydrolysis alone is estimated to be in the thousands per cell in organisms with a large genome (Lindahl, 1993). The Pp rad51AB double mutant is extremely sensitive to DNA damage. It is therefore possible that the repair of naturally occurring damage is considerably delayed in the absence of RAD51, thus causing indirectly a delay in cell proliferation. The marked developmental phenotype of the double mutant is likely to be the indirect consequence of this delay in cell proliferation.
Loss of RAD51 in P. patens caused a marked hypersensitivity to bleomycin. By contrast, resistance to bleomycin was hardly affected in the rad51 mutant in Arabidopsis. Bleomycin causes DSBs with two free ends as ionizing radiation (Povirk, 1996; Ramotar and Wang, 2003). Hence, the predominant pathway to repair two-sided DSBs is RAD51 dependent in P. patens but not in Arabidopsis. In this respect, P. patens is comparable to other organisms like yeast (Game and Mortimer, 1974), D. melanogaster (Rinaldo et al., 2002), and C. elegans (Staeva-Vieira et al., 2003), for which viable rad51 mutants exist that show a striking sensitivity to ionizing radiation. By contrast, bleomycin resistance is barely affected in At rad51-1; consequently, RAD51-independent processes must be used mostly in Arabidopsis. In this respect, Arabidopsis may be comparable to vertebrates since radiation resistance appears not to be dependent on RAD51 in these organisms (Lambert and Lopez, 2000). Therefore, plants appear as variable in their choice of recombination mechanisms as other organisms. In addition, the data suggest that the DNA damage repair pathways used by an organism are independent of its phylogenetic position or morphologic complexity.
The HR pathway consists of several branches, some of which may be RAD51 independent. One of them would be single-strand annealing. The major activities involved in this pathway in yeast are RAD52 and RAD59. However, sequence homologs of these genes do not exist in Arabidopsis. Another possibility would be a pathway depending on the RAD51 paralogs (Baumann and West, 1998; Shinohara and Ogawa, 1999; Thacker, 1999; Symington, 2002; West, 2003). However, those are also not involved in the repair of bleomycin-induced DNA damage in Arabidopsis (Bleuyard and White, 2004; Bleuyard et al., 2005). Since RAD51, the last not yet analyzed gene potentially involved, is not required, none of the known HR genes with strand-exchange or annealing activity appears to play a major role in somatic DSB repair in Arabidopsis. Two-sided DSBs can be repaired either by NHEJ or HR. Therefore, NHEJ must be the predominant repair pathway in somatic cells of higher plants, if not, other yet unidentified HR genes exist.
Loss of RAD51 in Arabidopsis had a dramatic effect on ICL repair. This requirement is the same as for the RAD51 paralogs in Arabidopsis (Bleuyard and White, 2004; Abe et al., 2005; Bleuyard et al., 2005), suggesting that this entire set of HR genes has an essential and nonredundant role in ICL repair in Arabidopsis. Mitomycin C was shown to induce replication-associated DSBs in mammalian cells (Niedernhofer et al., 2004; Cipak et al., 2006). Such lesions arise when a replication fork encounters an unrepaired single-strand break, giving rise to a collapsed replication fork. Supporting a DSB repair model for ICL repair, ICLs induce sister-chromatid exchanges, suggesting that these replication-associated DSBs are often resolved by HR (Niedernhofer et al., 2005; Thompson, 2005; Wilson and Thompson, 2007). In contrast with two-sided DSBs, replication-associated DSBs have only one free end to initiate repair and cannot be repaired by NHEJ (Helleday, 2003). Therefore, RAD51 and its paralogs in Arabidopsis may mainly function in the repair of replication fork-associated DSBs and may be vital in the restart of collapsed replication forks. This function is likely to be important in P. patens as well since double knockout mutants are hypersensitive to UV light. Although UV light, like ICLs, mainly induces modifications that are repaired by other mechanisms, unrepaired lesions interfere with replication and are likely to induce daughter-strand gaps and consequently DSBs. Supporting this model, UV treatment also induces sister-chromatid exchanges (Kadyk and Hartwell, 1993; Sonoda et al., 1999).
The differences with respect to somatic DSB repair between P. patens and Arabidopsis are striking. Arabidopsis is considered a fast-evolving species, and indeed a high degree of sequence divergence between ecotypes exists, suggesting that mutations still occur at high frequency (Jander et al., 2002; Koornneef et al., 2004; Mitchell-Olds and Schmitt, 2006), in support of NHEJ as an important DNA damage repair pathway. A high mutation rate causes genetic variability and thus may be the driving force for the rapid evolution of this species. By contrast, P. patens is a slowly evolving species (Reski, 1998), compatible with an error-free repair mechanism that preserves genome integrity. HR mostly prevents the fixation of mutations and thus counteracts genetic variability. Therefore, the choice of repair mechanism in DNA damage repair might be linked directly to the mechanisms of evolution in plants.
METHODS
Plant Material and Growth Conditions
The Gransden Wood wild-type strain of Physcomitrella patens was used in this work (Ashton and Cove, 1977). Long term cultures were cultured on minimal medium [0.8 g/L Ca(NO3)2 × 4H2O, 0.25 g/L MgSO2 × 7H2O, 1 mL KH2PO4/KOH (250 g/L KH2PO4 adjusted to pH 6.5 with KOH), 0.0125 g/L FeSO4 × 7H2O, 1 mL alternative TES (55 mg CoSO4 × 5H2O, 614 mg H3BO4, 55 mg CoCl2 × 6H2O, 25 mg Na2MoO4 × 2H2O, 55 mg ZnSO4 × 7H2O, 389 mg MnCl2 × 4H2O, 28 mg KI, adjusted to 1 liter with H2O), and 7 g/L agar, autoclaved for 20 min at 121°C]. For regular culture, this medium was supplemented with 0.5 g/L of diammonium tartrate and was overlaid with cellophane (standard medium). Culture was at 26°C in continuous light for 1 week, after which the material was subcultured. For this, four such cultures were collected in water, blended for 90 s with a Miccra-homogenizer D8 equipped with a P8 homogenizer tool (ART-Moderne Laborgeräte), diluted with an equal volume of water, and 1/20th plated. Spores were produced on minimal medium from protonema tissue plated at very low density and germinated on spore germination medium (0.25 g/L MgSO4 × 7H2O, 1.01 g/L KNO3, 0.0125 g/L FeSO4, 1 mL/L alternative TES, 1 mL KH2PO4/KOH, and 7 g/L agar) as described (Knight et al., 1988). To cross strains, equal amounts of freshly grown tissue were mixed, blended, and then grown under sporulation conditions. Spores were germinated on spore germination medium and transferred to selection medium after 7 d. For production of Pp rad51AB-2, protonema tissue was cultured for 2 weeks on 100 mg/L sulfadiazine and 50 mg/L G418 and transferred to 50 mg/L sulfadiazine for another 2 weeks.
The culture conditions for Arabidopsis thaliana and the At rad51-1 mutant were as described (Li et al., 2004).
Biological Assays
The growth rates in P. patens were determined by plating blended 5-d-old protonema tissue at low density on solid standard media. Pictures were taken with a digital camera connected to a digital imaging system (EDAS 260; Kodak) after 5 and 7 d of growth, and colony sizes were determined by pixel counting using imaging software (Kodak 1D Image analysis software). Growth rates were determined for individual colonies by calculating the differences in pixel counts obtained at days 5 and 7. Tissue blending generates a heterogeneous mixture of tissue fragments. Therefore, a broad range of different colony sizes are produced by this method. To reduce variability of the data, the size distribution in each sample was analyzed and colonies deviating from Gaussian distribution excluded from further analysis. Data were further processed using standard statistical procedures.
For phenotypic analysis of colony and filament morphology, freshly blended protonema were grown under standard conditions for 7 d, and small protonema colonies of 2 mm size were transferred on standard media without cellophane overlay as described (Thelander et al., 2004) and grown at 26°C in continuous light for several weeks. Pictures were taken at different time intervals. Close-ups were taken with a stereomicroscope (Leica MZFLIII) or microscope (Leica DMRB) connected to a DISCUS video camera and imaging system.
For the UV sensitivity assays, 5-d-old protonema tissue was plated and grown for 5 d on standard media. The plates were irradiated with UV 256 nm in a Stratalinker (Stratagene) with the doses indicated and directly returned back to light. For growth measurement, pictures were taken directly before irradiation and 48 h later.
For the bleomycin sensitivity assays, 5-d-old protonema tissue cultured under standard conditions was treated with the activities of bleomycin (Sigma-Aldrich; units as defined by the supplier) described in the figure legends in 10 mL of water for 1 h, washed with 50 mL water, blended, plated on solid standard medium in low density, and cultured in light at 26°C. The number of colonies established after 7 d of growth was determined and the data normalized and plotted.
The Arabidopsis resistance assays were performed essentially as described (Bleuyard and White, 2004). In brief, At rad51-1 seed obtained from a heterozygous plant and Columbia-0 seed were sown on agar media containing increasing concentrations of bleomycin or mitomycin C (Sigma-Aldrich) and grown until at least two true leaves had developed on plates that did not contain drug. At this time point, seedlings were scored resistant when plants had two or more true leaves and sensitive when development stalled at the cotyledon stage. At rad51-1 seedlings were genotyped by PCR as described (Li et al., 2004) after scoring survival to identify homozygous At rad51-1 individuals.
Targeted Gene Replacements
A fragment containing the Pp RAD51A genomic gene sequence was obtained from P. patens genomic DNA by PCR (Roche long template PCR kit) using primers pprad51a-1 (5′-CTGTGTGCGTAGTTATTCAACTCAAG-3′) and pprad51a-2 (5′-TCATTCCCTCCCTTCAAACTTC-3′) and then cloned in pCR2.1 (Invitrogen). The Pp RAD51A gene-specific gene replacement vector pKOrad51A was constructed by assembly of two subclones. The first one contained the 5′ end of the Pp RAD51A gene fused to a GFP reporter gene. To construct this plasmid, the Pp RAD51A genomic clone was digested with ClaI, the ends filled in with Klenow DNA polymerase fragment, and the fragment containing the 5′ end of Pp RAD51 excised by digestion with XbaI. A fragment encoding a PAC-GFP fusion protein was excised from pS139 (Meurer et al., 1998) with SmaI and HindIII and both fragments cloned together in XbaI- and HindIII-digested pUC19. The second subclone contained the 3′ end of Pp RAD51A fused to the kanamycin resistance (nptII) gene under control of the cauliflower mosaic virus 35S promoter, a derivative of the vector described by Wada et al. (1994), in which the HindIII site was converted to SalI by linker addition. This plasmid was obtained by digestion of the Pp RAD51A genomic clone with SacI and XhoI and ligation of the fragment containing the 3′ end of Pp RAD51A together with an EcoRI-SalI fragment containing the nptII gene in pBluescript SK digested with EcoRI and SacI. pKOrad51A was obtained by insertion of an XhoI and Ecl136II fragment from subclone 1 containing the Pp RAD51A 5′ end and GFP reporter gene between the XhoI and filled-in ClaI site of subclone 2. To release the targeting fragment, pKOrad51A was digested with SacI and XhoI prior to transformation.
A fragment containing the Pp RAD51B genomic sequence was obtained from P. patens genomic DNA by PCR (Roche long template PCR kit) using primers pprad51b-1 (5′-GGGGATCCAGATCTAAGTTATTGACGGTTGTGAG-3′) and pprad51b-2 (5′-GGGGATCCTGGCTATGCAGAAACTGACCATG-3′) and cloned in pGem-T (Stratagene). To obtain the Pp RAD51B gene replacement construct (pKOrad51B), a SpeI-MluNI fragment containing the 5′ portion and a SacI-SalI fragment containing the 3′ end of the Pp RAD51B gene was excised from the subclone and ligated together with a SmaI-EcoRI fragment (obtained from pBI101) containing the GUS reporter gene and an EcoRI-SacI fragment containing the sulfonamide resistance marker gene (obtained from the progenitor of pS001 described in Reiss et al., 1996) into SpeI- and SalI-digested pBluescript KS. To release the targeting fragment, pKOrad51B was digested with SpeI prior to transformation.
For transformation of P. patens, protonema were grown on standard medium supplemented with 0.5% glucose. Five-day-old tissue was harvested, protoplasts isolated, and 450,000 protoplasts transformed with 15 μg restriction enzyme–digested plasmid DNA as described previously (Schaefer and Zryd, 1997). Transformants were selected on 50 mg/L G418 (Duchefa) for pKOrad51A or 50 mg/L sulfadiazine (Sigma-Aldrich) for pKOrad51B transformants.
Stable transformants were selected by four cycles of growth on selective and nonselective media as described (Schaefer et al., 1991). Stable transformants were grown up and total DNA prepared and analyzed by DNA gel blotting and PCR.
Blotting Techniques
The procedures for preparation of P. patens genomic DNA and poly(A)+ RNA, preparation of radioactively labeled probes, DNA gel blotting, and RNA gel blotting were as described by Markmann-Mulisch et al. (2002).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: AJ344152 (Pp RAD51A), AJ344153 (Pp RAD51B), and At5g20850 (At RAD51).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. RAD51 Transcript Analysis of P. patens rad51 Mutants.
Supplementary Material
Acknowledgments
We thank Didier Schaefer, David Cove, Celia Knight, Andrew Cummings, Enzo Russo, and Jean-Pierre Zryd for their help with P. patens and the European Community (PREGENE, QLK3-CT-2000-00365; GENINTEG, FP6-503303) and the Deutsche Forschungsgemeinschaft (Graduiertenkolleg Molekulare Analyse von Entwicklungsprozessen) for financial support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Bernd Reiss (reiss@mpiz-koeln.mpg.de).
Online version contains Web-only data.
References
- Abe, K., Osakabe, K., Nakayama, S., Endo, M., Tagiri, A., Todoriki, S., Ichikawa, H., and Toki, S. (2005). Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 139 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi, A., Pasierbek, P., Gartner, A., and Loidl, J. (2003). Genetic and cytological characterization of the recombination protein RAD51 in Caenorhabditis elegans. Chromosoma 112 6–16. [DOI] [PubMed] [Google Scholar]
- Ashton, N.W., and Cove, D.J. (1977). The isolation and preliminary characterization of auxotrophic and analog resistant mutants of the moss Physcomitrella patens. Mol. Gen. Genet. 154 87–96. [Google Scholar]
- Ayora, S., Piruat, J.I., Luna, R., Reiss, B., Russo, V.E.A., Aguilera, A., and Alonso, J.C. (2002). Characterization of two highly similar Rad51 homologs of Physcomitrella patens. J. Mol. Biol. 316 35–49. [DOI] [PubMed] [Google Scholar]
- Baumann, P., and West, S.C. (1998). Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23 247–251. [DOI] [PubMed] [Google Scholar]
- Bleuyard, J.-Y., and White, C.I. (2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard, J.Y., Gallego, M.E., Savigny, F., and White, C.I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41 533–545. [DOI] [PubMed] [Google Scholar]
- Burma, S., Chen, B.P.C., and Chen, D.J. (2006). Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst.) 5 1042–1048. [DOI] [PubMed] [Google Scholar]
- Cipak, L., Watanabe, N., and Bessho, T. (2006). The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat. Struct. Mol. Biol. 13 729–733. [DOI] [PubMed] [Google Scholar]
- Cove, D. (2005). The moss Physcomitrella patens. Annu. Rev. Genet. 39 339–358. [DOI] [PubMed] [Google Scholar]
- Cove, D., Bezanilla, M., Harries, P., and Quatrano, R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57 497–520. [DOI] [PubMed] [Google Scholar]
- Dronkert, M.L.G., and Kanaar, R. (2001). Repair of DNA interstrand cross-links. Mutat. Res. 486 217–247. [DOI] [PubMed] [Google Scholar]
- Dudas, A., and Chovanec, M. (2004). DNA double-strand break repair by homologous recombination. Mutat. Res. 566 131–167. [DOI] [PubMed] [Google Scholar]
- Game, J.C., and Mortimer, R.K. (1974). A genetic study of X-ray sensitive mutants in yeast. Mutat. Res. 24 281–292. [DOI] [PubMed] [Google Scholar]
- Helleday, T. (2003). Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 532 103–115. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., Last, R.L., Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk, L.C., and Hartwell, L.H. (1993). Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics 133 469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi, Y., Cuming, A.C., and Cove, D.J. (2005). Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 33 E173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi, Y., Schlink, K., Rensing, S.A., Schween, G., von Stackelberg, M., Cuming, A.C., Reski, R., and Cove, D.J. (2006). The mechanism of gene targeting in Physcomitrella patens: Homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 34 6205–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, C.D., Cove, D.J., Boyd, P.J., and Ashton, N.W. (1988). The isolation of biochemical and developmental mutants in Physcomitrella patens. In Methods in Bryology, J.M. Glime, ed (Miyazaki, Japan: The Hattori Botanical Laboratory), pp. 47–58.
- Koornneef, M., Alonso-Blanco, C., Vreugdenhil, D., Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Lambert, S., and Lopez, B.S. (2000). Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J. 19 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.X., Chen, C.B., Markmann-Mulisch, U., Timofejeva, L., Schmelzer, E., Ma, H., and Reiss, B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, T. (1993). Instability and decay of the primary structure of DNA. Nature 362 709–715. [DOI] [PubMed] [Google Scholar]
- Markmann-Mulisch, U., Hadi, M.Z., Koepchen, K., Alonso, J.C., Russo, V.E.A., Schell, J., and Reiss, B. (2002). The organization of Physcomitrella patens RAD51 genes is unique among eukaryotic organisms. Proc. Natl. Acad. Sci. USA 99 2959–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Grevelding, C., Westhoff, P., and Reiss, B. (1998). The pac protein affects the maturation of specific chloroplast messenger RNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258 342–351. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds, T., and Schmitt, J. (2006). Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441 947–952. [DOI] [PubMed] [Google Scholar]
- Niedernhofer, L.J., Lalai, A.S., Hoeijmakers, J.H., Niedernhofer, L.J., Lalai, A.S., and Hoeijmakers, J.H.J. (2005). Fanconi anemia (cross)linked to DNA repair. Cell 123 1191–1198. [DOI] [PubMed] [Google Scholar]
- Niedernhofer, L.J., et al. (2004). The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24 5776–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk, L.F. (1996). DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat. Res. 355 71–89. [DOI] [PubMed] [Google Scholar]
- Ramotar, D., and Wang, H. (2003). Protective mechanisms against the antitumor agent bleomycin: Lessons from Saccharomyces cerevisiae. Curr. Genet. 43 213–224. [DOI] [PubMed] [Google Scholar]
- Reiss, B. (2003). Homologous recombination and gene targeting in plant cells. Int. Rev. Cytol. 228 85–139. [DOI] [PubMed] [Google Scholar]
- Reiss, B., Klemm, M., Kosak, H., and Schell, J. (1996). RecA protein stimulates homologous recombination in plants. Proc. Natl. Acad. Sci. USA 93 3094–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reski, R. (1998). Development, genetics and molecular biology of mosses. Bot. Acta 111 1–15. [Google Scholar]
- Rinaldo, C., Bazzicalupo, P., Ederle, S., Hilliard, M., and La Volpe, A. (2002). Roles for Caenorhabditis elegans RAD51 in meiosis and in resistance to ionizing radiation during development. Genetics 160 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, D., Zryd, J.P., Knight, C.D., and Cove, D.J. (1991). Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 226 418–424. [DOI] [PubMed] [Google Scholar]
- Schaefer, D.G., and Zryd, J.P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11 1195–1206. [DOI] [PubMed] [Google Scholar]
- Schuermann, D., Molinier, J., Fritsch, O., and Hohn, B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21 172–181. [DOI] [PubMed] [Google Scholar]
- Schween, G., et al. (2005). Large-scale analysis of 73,329 Physcomitrella plants transformed with different gene disruption libraries: Production parameters and mutant phenotypes. Plant Biol. 7 228–237. [DOI] [PubMed] [Google Scholar]
- Shinohara, A., and Ogawa, T. (1999). Rad51/RecA protein families and the associated proteins in eukaryotes. Mutat. Res. 435 13–21. [DOI] [PubMed] [Google Scholar]
- Sonoda, E., Hochegger, H., Saberi, A., Taniguchi, Y., and Takeda, S. (2006). Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst.) 5 1021–1029. [DOI] [PubMed] [Google Scholar]
- Sonoda, E., Sasaki, M.S., Morrison, C., Yamaguchi-Iwai, Y., Takata, M., and Takeda, S. (1999). Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeva-Vieira, E., Yoo, S., and Lehmann, R. (2003). An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P., Krejci, L., Van Komen, S., and Sehorn, M.G. (2003). Rad51 recombinase and recombination mediators. J. Biol. Chem. 278 42729–42732. [DOI] [PubMed] [Google Scholar]
- Sung, P., Trujillo, K., and Van Komen, S. (2000). Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451 257–275. [DOI] [PubMed] [Google Scholar]
- Symington, L.S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker, J. (1999). A surfeit of RAD51-like genes? Trends Genet. 15 166–168. [DOI] [PubMed] [Google Scholar]
- Thelander, M., Olsson, T., and Ronne, H. (2004). Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 23 1900–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.H. (2005). Unraveling the Fanconi anemia-DNA repair connection. Nat. Genet. 37 921–922. [DOI] [PubMed] [Google Scholar]
- Wada, M., Klein, C., Schell, J., and Reiss, B. (1994). A functional assay for Taq DNA-polymerase in PCR. Biotechniques 16 26–28, 30. [PubMed] [Google Scholar]
- West, S.C. (2003). Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4 435–445. [DOI] [PubMed] [Google Scholar]
- Wilson III, D.M., and Thompson, L.H. (2007). Molecular mechanisms of sister-chromatid exchange. Mutat. Res. 616 11–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.