Abstract
Plasmodesmata establish a pathway for the intercellular trafficking of viral movement proteins and endogenous non-cell-autonomous proteins, such as the two closely related meristem-maintaining KNOTTED1-like homeobox (KNOX) proteins Zea mays KNOTTED1 (KN1) and Arabidopsis thaliana SHOOTMERISTEMLESS (STM). KNOX family members are DNA binding proteins that regulate the transcriptional activity of target genes in conjunction with BEL1-like homeodomain proteins. It has been shown previously, using in vivo transport assays, that the C-terminal domain of KN1, including the homeodomain, is necessary and sufficient for cell-to-cell transport through plasmodesmata. Here, using interaction and coexpression assays, we demonstrate that the microtubule-associated and viral movement protein binding protein MPB2C from Nicotiana tabacum, and its homolog in Arabidopsis, At MPB2C, are KN1/STM binding factors. Interaction between the MPB2C proteins and KN1/STM was mapped to the KN1 homeodomain, a region not essential for heterodimerization with BEL1. Expression of MPB2C in single cells prevented KN1 cell-to-cell movement. Furthermore, in vivo trichome rescue studies established that MPB2C negatively regulates KN1 association to plasmodesmata and, consequently, cell-to-cell transport. These findings are discussed in terms of the role played by MPB2C proteins in regulating the cell-to-cell trafficking of homeodomain proteins in plants.
INTRODUCTION
In plants, cell fate is generally determined through positional information. Exchange of non-cell-autonomous signals may provide the means by which cells assume their fate in relation to surrounding tissues. To this end, information molecules may cross the plasma membranes via secretion/internalization or follow a symplasmic route via plasmodesmata (PD). Cell-to-cell trafficking of macromolecules, such as RNA-protein complexes, is thought to occur through PD. Numerous studies on viral movement proteins (MPs) as well as endogenous plant proteins have provided direct evidence for the involvement of a PD-based non-cell-autonomous protein (NCAP) signaling pathway (Haywood et al., 2002; Lucas and Lee, 2004; Ruiz-Medrano et al., 2004).
An important class of NCAPs is represented by the KNOTTED1-like homeobox (KNOX) proteins that play key roles in meristems, including KNOTTED1 (KN1) in maize (Zea mays) (Lucas et al., 1995) and SHOOTMERISTEMLESS (STM) and KNAT1/BREVIPEDICELLUS (BP) in Arabidopsis thaliana (Kim et al., 2003, 2005b). These KN1-like transcription factors have a highly conserved structure, insofar as they contain KNOX, ELK, and homeodomain (HD) motifs, are expressed in shoot meristems, and are required to maintain cells in an undifferentiated state (Vollbrecht et al., 1991; Lincoln et al., 1994; Long et al., 1996; Venglat et al., 2002). Ectopic expression of STM, BP, or KN1 in Arabidopsis results in the formation of lobed leaves, irregular vein patterns, and ectopic meristem-like structures (Lincoln et al., 1994; Chuck et al., 1996; Byrne et al., 2000; Hibara et al., 2003).
The cell-to-cell movement capacity of a subclass of KNOX proteins was confirmed, in vivo, in Arabidopsis leaves and shoot apices (Kim et al., 2002, 2003, 2005b). An ingenious trichome rescue assay, developed by Kim et al. (2005b), provided independent proof that KN1 functions as an NCAP. In that study, subepidermal expression of a GLABROUS1-KN1 fusion protein in a trichome-deficient glabra1 (gl1) background line restored trichome development in the epidermal layer. This trichome rescue system confirmed the capacity of KN1 to mediate cell-to-cell trafficking of its mRNA and showed that the HD motif is necessary for NCAP function.
KNOX proteins have been found to interact with a number of other proteins. The KNOX motif is necessary for the formation of heterodimeric complexes with BEL1-like HD (BLH) proteins (Bellaoui et al., 2001; Muller et al., 2001), and, depending on their composition, KNOX-BLH heterodimers recognize specific DNA motifs (Bellaoui et al., 2001; Muller et al., 2001; Nagasaki et al., 2001; Smith et al., 2002; Chen et al., 2003, 2004; Bhatt et al., 2004). In general, as with the animal homologs of KNOX and BLH proteins, it is thought that the HD motif mediates binding to promoter sequences of genes that affect cell fate (Smith et al., 2002). Binding of BLH family members to STM occurs independently of the BLH HD motif and appears to trigger the nuclear import of STM (Cole et al., 2006).
Other KNOX-interacting partners are found among members of the Arabidopsis OVATE family proteins (At OFPs). In transient expression assays, At OFP1–green fluorescent protein (GFP) colocalizes with BP or BLH1 to form punctate structures along microtubules as well as at the cell periphery (Hackbusch et al., 2005). Thus, At OFP members may be involved in localizing HD proteins to the cytoskeleton. Interestingly, the punctate structures formed by OFP-GFP at the microtubules are very similar to those detected with the structurally unrelated microtubule-associated protein MPB2C (Kragler et al., 2003).
Microinjection experiments have established that maize KN1 has the capacity to interact with PD to increase the size exclusion limit (SEL) and mediate its cell-to-cell trafficking as well as that of its own mRNA (Lucas et al., 1995; Kragler et al., 2000). Most viruses encode specialized MPs that have the capacity to interact with host proteins to regulate their intercellular transport through PD, and such microinjection studies provided direct evidence in support of the hypothesis that KN1 and tobacco mosaic virus (TMV) MP utilize common components of the NCAP pathway (Kragler et al., 1998b, 2000).
There is evidence that the regulation of movement of the TMV MP-RNA complex into neighboring cells depends on the interactions of TMV MP with endogenous proteins such as the NON-CELL-AUTONOMOUS PATHWAY PROTEIN1 (NCAPP1) (Lee et al., 2003), PD-located casein kinases (Lee et al., 2005), cell wall–modifying enzymes such as pectin methyl esterase (Chen et al., 2000), or the Ca2+-sequestering protein calreticulin (Chen et al., 2005). Furthermore, within plant cells, the TMV MP association with microfilaments and microtubules appears to regulate the access of the TMV MP-RNA complex to the PD pathway (Heinlein et al., 1995; McLean et al., 1995; Boyko et al., 2000, 2007; Gillespie et al., 2002; Nelson and Citovsky, 2005; Wright et al., 2007).
Insight into the mechanism by which microtubules may negatively regulate TMV MP movement has been gained through the identification of MPB2C, an endogenous protein in Nicotiana tabacum that interacts with the TMV MP. In situ labeling experiments demonstrated that MPB2C localizes to microtubules, and transient expression assays revealed that it inhibits the cell-to-cell movement of TMV MP (Kragler et al., 2003). In transiently MPB2C-silenced Nicotiana benthamiana plants, the TMV MP association with microtubules was reduced; however, the protein was still transported from cell to cell (Curin et al., 2007). In addition, plants appear to regulate the levels of TMV MP by targeting it to the ubiquitin-dependent 26S proteasome degradation pathway (Reichel and Beachy, 2000).
In this study, we further explored the events underlying KN1 cell-to-cell trafficking by testing the capacity of KN1 to interact with MPB2C. Our studies revealed that MPB2C can interact with KN1 and STM but not with BEL1. MPB2C binding was mapped to the HD motif shown to be essential for the cell-to-cell movement of KN1. Movement assays provided evidence that MPB2C functions in vivo as a negative regulator of KN1 cell-to-cell movement activity. Based on these findings, we advance the hypothesis that plants use a common pathway involving MPB2C to regulate the entry of viral MPs and HD proteins into the NCAP pathway.
RESULTS
Identification of MPB2C Binding Partners
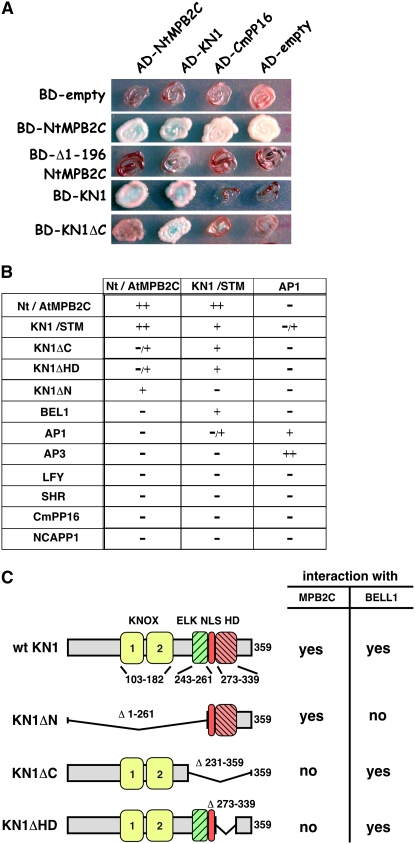
To identify interaction partner(s) common to TMV MP and plant NCAPs, we probed N. tabacum MPB2C (Nt MPB2C) for its capacity to bind to a range of structurally distinct NCAPs, including maize KN1 and Arabidopsis STM, LEAFY (LFY) (Sessions et al., 2000; Wu et al., 2003), and SHORT ROOT (SHR) (Nakajima et al., 2001; Gallagher et al., 2004). In yeast two-hybrid assays, we found that full-length Nt MPB2C, but not an N-terminally truncated Nt MPB2C (Δ1-196 Nt MPB2C), interacted with both KN1 and STM (Figures 1A and 1B).
Figure 1.
Protein Interaction Assays Showing Specific Interaction of KN1 with MPB2C.
(A) Yeast two-hybrid interaction assay using Nt MPB2C, Δ1-196 Nt MPB2C, KN1, and KN1ΔC fused to the GAL4 DNA binding domain (BD) and Nt MPB2C, KN1, and Cm PP16 fused to the GAL4 activation domain (AD). BD-empty and AD-empty strains express only the binding and activation domains, respectively. Note that BD-Nt MPB2C alone gives high background on the selective growth medium but shows no α-galactose activity (blue stain).
(B) Summary of the observed yeast two-hybrid interactions. The interaction according to the measured relative β-galactose activity and growth rate was characterized as high (++), with the maximum measured in the system, middle (+) with lower activity of the maximum, very low (−/+), with ∼10-fold less than the maximum activity of the particular construct, or not above background (−). For an example with BD-KN1ΔC, see Supplemental Table 1 online.
(C) KN1 domain structure and deletion constructs used in yeast two-hybrid assays and their capacity to interact with MPB2C or BEL1.
As controls for these experiments, we tested the interaction of MPB2C with three cell-autonomous proteins: the BLH protein BEL1 and the two MADS box proteins APETALA1 (AP1) (Sessions et al., 2000) and APETALA3 (AP3) (Jack et al., 1992). As shown in Figure 1B, no positive interactions were obtained with any of these cell-autonomous proteins. The specificity of the interaction between MPB2C and KN1/STM was supported by the failure of MPB2C to interact with a pumpkin (Cucurbita maxima) phloem NCAP, Cm PP16 (Xoconostle-Cázares et al., 1999) (Figures 1A and 1B). A similar negative result was obtained with Nt NCAPP1, a component of the NCAP pathway that mediates the delivery of Cm PP16 to PD (Lee et al., 2003).
A single Nt MPB2C–like gene was identified in the Arabidopsis genomic database (At MPB2C; At5g08120) with a similarity at the amino acid level of 62%. Yeast two-hybrid interaction assays demonstrated that At MPB2C exhibited properties equivalent to those of the N. tabacum homolog (Figures 1A and 1B). Both can form homodimers and specifically interact with KN1 and STM.
A Functional KN1 Cell-to-Cell Movement Motif Is Essential for MPB2C–KN1 Interaction
The inability of MPB2C to dimerize with the HD protein BEL1 implies that MPB2C-KN1/STM heterodimer formation depends on motif(s) distinct from those found in BEL1, allowing a BEL1-like protein to bind to KN1/STM. To identify such motifs, we used three KN1 deletion constructs: KN1ΔN, lacking both the KNOX and ELK motifs; KN1ΔC, devoid of the ELK, NLS, and HD motifs; and KN1ΔHD, lacking the HD motif (Figure 1C). In yeast two-hybrid assays, both KN1ΔC and KN1ΔHD interacted with STM and BEL1 but did not interact significantly with At MPB2C or Nt MPB2C (Figure 1B). These results indicated that the HD motif was necessary for MPB2C binding. Thus, the KN1–MPB2C interaction requires the presence of the HD motif that is also responsible for DNA binding (Smith et al., 2002; Viola and Gonzalez, 2006) and cell-to-cell movement (Kim et al., 2005b). In agreement with the reported function of the KNOX motif to mediate an interaction with BEL1-like proteins (Bellaoui et al., 2001; Muller et al., 2001), no KN1ΔN heterodimer formation was observed with BEL1.
Protein overlay assays were next performed to further test for the interaction between KN1 and MPB2C. Here, TMV MP was included as a positive control, since we earlier demonstrated binding to MPB2C under the conditions used in these overlay assays (Kragler et al., 2003). To probe MPB2C binding to the KN1 region essential for cell-to-cell transport, we used a mutant KN1 protein, KN1M6, which is defective in cell-to-cell movement capacity (Lucas et al., 1995). KN1M6 harbors three Lys-to-Ala substitutions (from Lys-265 to -267 to Ala-265 to -267) in a basic amino acid stretch adjacent to the HD motif.
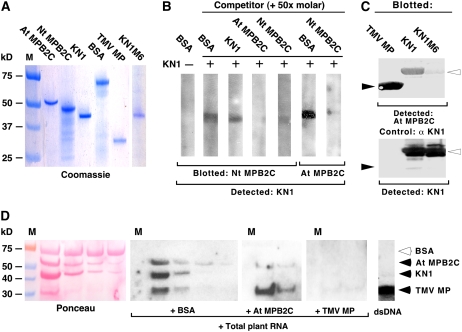
First, At MPB2C, Nt MPB2C, KN1, KN1M6, and TMV MP were expressed in Escherichia coli, isolated to near homogeneity, and quantified following SDS-PAGE (Figure 2A). Next, Nt MPB2C or At MPB2C was blotted onto a nitrocellulose membrane, which was then cut into strips and used for overlay assays. Incubating immobilized Nt MPB2C with BSA followed by KN1 immunodetection confirmed the specificity of the detection system; note the absence of signal (Figure 2B). Incubation of the immobilized Nt MPB2C with KN1 plus a 50-fold molar excess of BSA, or KN1, followed by KN1 immunodetection resulted in a clearly discernible signal.
Figure 2.
In Vitro Interactions of MPB2C, KN1, TMV MP, and Movement-Defective KN1M6 Mutant Protein.
(A) Recombinant protein samples (1 μg) of His-At MPB2C, His-Nt MPB2C, KN1 TMV MP, and KN1M6 after purification were separated by SDS-PAGE and stained with GelCode Blue reagent.
(B) Protein overlay assays with Nt MPB2C and At MPB2C. Purified Nt MPB2C or At MPB2C was electrotransferred after SDS-PAGE onto nitrocellulose membranes, cut into 5-mm strips (corresponding to ∼0.5 μg of blotted protein), and then incubated with KN1 (1 μg). The presence of KN1 bound to Nt MPB2C (0.5 μg) or At MPB2C (0.5 μg) was detected using KN1-specific antibodies. In the control with BSA (lane 1), no signal could be detected. Coincubation of KN1 with a 50-fold molar excess of BSA (lanes 2, 6) or KN1 (lane 3) produced KN1 signals at the expected sizes. In the presence of a 50-fold molar excess of At MPB2C or Nt MPB2C, no KN1 signal could be detected (lanes 4, 5, 7).
(C) Protein overlay interaction assay with TMV MP, KN1, and KN1M6 blotted onto nitrocellulose. Purified proteins (5 μg) were submitted to SDS-PAGE, electrotransferred on membranes, and incubated with 20 μg of At MPB2C. The presence of bound At MPB2C was monitored with specific antibodies directed against recombinant At MPB2C (top panel; closed arrowheads indicate binding to TMV MP, and open arrowheads indicate binding to KN1 and KN1M6). As a loading control (bottom panel), the same blot was incubated with specific antibodies directed against recombinant KN1. Note that the KN1M6 mutant protein produced a faint At MPB2C signal compared with KN1 and the loading control.
(D) RNA overlay interaction assay with TMV MP, KN1, and At MPB2C. All three recombinant proteins were mixed with BSA (1 μg) with decreasing concentrations (∼1, 0.5, 0.25, and 0.12 μg), size-separated via SDS-PAGE, and electrotransferred on membranes. The blotted proteins were incubated with [32P-γ]ATP-labeled total Arabidopsis RNA in the presence of a 10-fold molar excess of BSA, At MPB2C, or TMV MP. Note that at the positions of At MPB2C, KN1, and TMV MP, signals were detected (black arrowheads). An excess of At MPB2C selectively inhibited KN1 RNA binding, whereas surplus of TMV MP competed with RNA binding of all blotted proteins.
To further confirm the specificity of KN1 binding to Nt MPB2C, we next performed competition assays with a 50-fold molar excess of At MPB2C or Nt MPB2C. As shown in Figure 2B, excess levels of either Nt MPB2C or At MPB2C greatly diminished the KN1 signal. Equivalent results were obtained when we performed reciprocal overlay experiments in which At MPB2C was used as the immobilized protein (Figure 2B).
To test whether a movement-impaired KN1 protein is still capable of MPB2C interaction, we reversed the overlay system and blotted equal molar amounts of TMV MP, KN1, and the movement-deficient mutant KN1M6 onto the membrane and then incubated it with recombinant At MPB2C. Immunodetection of bound MPB2C revealed strong signals at the positions of the TMV MP and KN1, whereas only a very faint signal was present at the position of KN1M6. Equal amounts of KN1 and KN1M6 were loaded, as indicated by equivalent signals detected with the KN1-specific antibody (Figure 2C, bottom panel). These results are consistent with our yeast two-hybrid findings and support the notion that a functional KN1 cell-to-cell movement motif is essential for a strong interaction with MPB2C.
MPB2C Binds to RNA and Alters KN1 RNA Binding Capacity
Previous microinjection and trichome rescue experiments established that KN1 binds and traffics its own mRNA to neighboring cells (Lucas et al., 1995; Kragler et al., 1998b; Kim et al., 2005b). To test whether MPB2C displays RNA interaction activity and could alter KN1 RNA recognition, we submitted the recombinant proteins to RNA binding assays. Immobilized and renatured BSA, At MPB2C, KN1, and TMV MP were incubated with labeled total Arabidopsis RNA harvested from 2-week-old seedlings. An RNA signal was detected at the positions of At MPB2C, KN1, and TMV MP (Figure 2D). Showing the specificity of the RNA binding assay, no RNA was detected associated with the negative control protein BSA.
Next, by adding excess quantities of At MPB2C or TMV MP to the assay, we tested the potential effect of At MPB2C on the KN1 RNA binding activity. High amounts (10-fold excess) of At MPB2C reduced specifically detectable RNA signals at KN1 and MPB2C, whereas excess TMV MP diminished all RNA signals (Figure 2D). Neither KN1 nor At MPB2C bound to double-stranded DNA, whereas TMV MP could bind, in vitro, double-stranded DNA (Figure 2D, right panel).
KN1 Cell-to-Cell Trafficking Is Restricted by MPB2C
In previous studies, a negative effect of transiently expressed MPB2C was reported on the cell-to-cell movement capacity of coexpressed TMV MP (Kragler et al., 2003). Consequently, we next examined whether MPB2C has a similar effect on the ability of KN1 to move through PD. Here, microinjection methods were used to test KN1 movement in the presence of MPB2C. KN1 and MPB2C were first expressed in and purified from E. coli, fluorescently labeled, and then introduced into mesophyll cells of N. benthamiana leaves. Movement capacity was tested by coinjection of 12-kD F-dextran or 11-kD rhodamine-dextran: an increase in PD SEL occurs during NCAP trafficking, and this allows for the codiffusion of the reporter dextrans.
In control experiments, the small membrane-impermeant fluorescent tracer, lucifer yellow CH, was first injected into target mesophyll cells to confirm their symplasmic connectivity (Table 1). A second test probe, acridine orange, which is membrane-impermeable in its protonated form and binds to DNA, callose, and cell wall material (Karabetsos et al., 1987; Alche and Rodriguez-Garcia, 1997; He et al., 2007), also moved readily into neighboring cells. Green fluorescent nuclei became visible within seconds in the injected cells and after 30 s in neighboring cells (Table 1, Figure 3A).
Table 1.
Microinjection Studies on KN1 Movement in the Presence of MPB2C
| Injected Probes | Injections | Movement (%) | Comment |
|---|---|---|---|
| Lucifer yellow CH | 20 | 20 (100) | Extensive; cells symplastically connected |
| Acridine orangea | 20 | 20 (100) | Nuclei, extensive; cells symplastically connected |
| 12-kD F-dextran + 11-kD rhodamine-dextran | 20 | 1 (5) | Both dyes remain in injected cells |
| Rhodamine-KN1 | 10 | 9 (90) | Extensive |
| Rhodamine-KN1 + 12-kD F-dextran | 12 | 9 (75) | Both extensive |
| FITC-Nt MPB2C | 10 | 0 (0) | Appears in punctae and cytosol |
| FITC-At MPB2C | 10 | 0 (0) | Appears in punctae and cytosol |
| Nt MPB2C + 12-kD F-dextran | 10 | 0 (0) | No increase of PD SEL; dye remains in injected cells |
| At MPB2C + rhodamine-KN1 + 12-kD F-dextran | 10 | 0 (0) | No increase of PD SEL; dye remains in injected cells |
| FITC-At MPB2C + rhodamine-KN1 | 12 | 0 (0) | Appears in punctae and cytosol |
| At MPB2C + TMV MP + 12-kD F-dextran | 24 | 0 (0) | No increase of PD SEL; dye remains in injected cells |
| At MPB2C + CMV MP + 12-kD F-dextran | 18 | 15 (84) | Extensive; increase of PD SEL |
Fluorescent probes were microinjected into N. benthamiana mesophyll cells and distribution was evaluated within 5 min. Equimolar amounts were used in probes in which two proteins were combined and adjusted to a final concentration of 2 μg/μL.
Acridine orange (molecular weight = 302).
Figure 3.
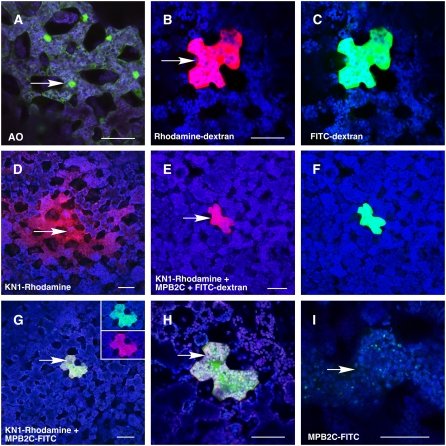
Microinjection Assays Establish the Inhibition of KN1 Cell-to-Cell Trafficking in the Presence of MPB2C.
Confocal microscopy images of mesophyll cells of 3- to 5-week-old N. benthamiana source leaves at 3 min after injection of fluorescent probes.
(A) Microinjection of the small dye acridine orange (molecular weight = 302) fluorescently staining cell walls, RNA, and DNA. A green fluorescence signal appeared in nuclei and cytoplasm of the injected (arrow) and neighboring cells, consistent with the dye having moved symplasmically between cells via PD. The confocal image was recorded avoiding the red channel.
(B) and (C) An equimolar mixture of 11-kD rhodamine-dextran (red) (B) and 12-kD F-dextran (green) (C) remains in the injected cell, confirming that PD are not dilated by the trafficking of endogenous NCAPs.
(D) Cell-to-cell movement of recombinant rhodamine–labeled KN1 (red).
(E) and (F) An equimolar mixture (2 μg/μL) of At MPB2C (no fluorescent tag) and rhodamine-KN1 (red) (E) with the fluorescent tracer 12-kD F-dextran (green) (F) remains in the injected cell.
(G) An equimolar mixture (2 μg/μL) of FITC-labeled At MPB2C (green) and rhodamine-KN1 (red) was used as a probe. No fluorescent signal of proteins could be detected in adjacent cells. The insets show the injected cell avoiding the red and the green channel, confirming the presence of both fluorescently tagged proteins.
(H) Higher magnification of the injected cell in (G).
(I) A single cell is shown after injection with FITC-labeled At MPB2C (green). The image was taken at low laser energy settings to visualize the appearance of fluorescently tagged At MPB2C at small green punctae in the cytosol.
Note that in merged images, coinciding signals of green and red channels appear yellow/orange. Fluorescent detection filter settings where as follows: blue, chloroplast 684 to 777 nm; green, FITC-tagged 505 to 537 nm; red, rhodamine-tagged 620 to 650 nm. Arrows indicate sites of injection. Bars = 80 μm.
A mixture of 11-kD rhodamine-dextran and 12-kD F-dextran was next injected to test for the presence of endogenous NCAP trafficking. Retention of both probes in the injected cells (Figures 3B and 3C) established that these mesophyll cells were not actively engaged in NCAP movement. To confirm the capacity of this tissue to traffic NCAPs, we next injected fluorescently labeled KN1. As expected, rhodamine-KN1 was observed to move from cell to cell in 90% of such injections (Figure 3D, Table 1). Rhodamine-KN1, similar to TRITC-labeled KN1 (Kragler et al., 1998b), induced an increase of the PD SEL, allowing 12-kD F-dextran to move into neighboring mesophyll cells (Table 1).
The potential for cell-to-cell movement by At MPB2C and Nt MPB2C, per se, was next tested by coinjection with 12-kD F-dextran. As shown in Table 1, both MPB2C proteins remained in the injected cells together with the 12-kD F-dextran. Thus, based on microinjection assays, MPB2C does not have the capacity to increase the PD SEL or to move from cell to cell. Subsequently, we determined whether MPB2C inhibits KN1 transport activity by injecting a mixture of rhodamine-KN1, At MPB2C, and 12-kD F-dextran into target mesophyll cells. In the presence of At MPB2C, rhodamine-KN1 and 12-kD F-dextran remained in the injected cells (Figures 3E and 3F). Equimolar mixtures of rhodamine-tagged KN1 and fluorescein isothiocyanate (FITC)–tagged MPB2C also persistently stayed in the injected cells (Table 1, Figures 3G and 3H). Microinjected At MPB2C often appeared in fluorescent punctae within the cytosol (Figure 3I); a similar phenomenon was earlier observed with the Nt MPB2C-DsRED fusion protein (Kragler et al., 2003). Consistent with a previously reported specific inhibitory effect of Nt MPB2C on TMV MP movement capacity, no increase in PD SEL was detected when TMV MP was microinjected in the presence of At MPB2C (Table 1). Finally, coinjection of MPB2C along with cucumber mosaic virus MP (CMV MP) did not block the MP-induced increase in SEL (Table 1). Together, these microinjection experiments indicated that At MPB2C has the capacity to interact with KN1 and TMV MP in the mesophyll cytoplasm to block their ability both to increase the PD SEL and to move cell to cell.
Transient Expression of MPB2C Inhibits KN1 Cell-to-Cell Movement in Arabidopsis
In Arabidopsis, transiently expressed KN1 was earlier shown to move between epidermal cells as well as from mesophyll cells into epidermal cells in KN1 transgenic plants (Kim et al., 2002, 2003, 2005b). Thus, the biolistic bombardment system was next employed to further investigate the inhibitory effects of MPB2C on KN1 movement capacity in epidermal tissues. We transiently expressed GFP-KN1 and MPB2C-DsRED in Arabidopsis epidermal cells. Two to 3 d after bombardment, we inspected this tissue for cell-to-cell transport of the fluorescent fusion proteins.
Control experiments were first performed to determine whether DsRED, GFP-KN1, At MPB2C-GFP, or Nt MPB2C-DsRED could move through epidermal PD. Consistent with the findings of Kim et al. (2002), GFP-KN1 was transported from bombarded cells to neighboring cells, albeit at a low rate of ∼10% (Figure 4, Table 2). Interestingly, in the presence of KN1, when the PD SEL should be increased, no movement of DsRED was detected (Figure 4B, Table 2). Also, Nt MPB2C-DsRED and At MPB2C-GFP remained in the bombarded cells (Table 2). Furthermore, both proteins were observed to be colocalized with GFP-KN1 in cytosolic punctae, and this association abolished KN1 cell-to-cell spread (Figures 4E to 4G, Table 2). Colocalization of At MPB2C fused to red fluorescent protein (RFP) was also observed with KN1 harboring GFP at the C terminus (KN1-GFP; see Supplemental Figure 1 online). These results were equivalent to those obtained in microinjection studies and offered additional support for the hypothesis that MPB2C interacts with KN1. In microinjection and transient expression assays, MPB2C interaction with KN1 prevents KN1 from moving cell to cell.
Figure 4.
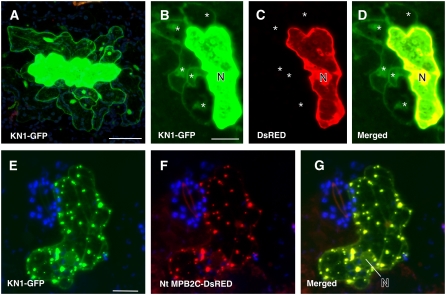
Cell-to-Cell Movement of Transiently Expressed KN1 Is Impaired by MPB2C in Arabidopsis Leaf Epidermal Tissue.
Two-week-old Arabidopsis C24 plants were bombarded with gold particles coated with plasmids expressing fluorescent proteins driven by the CaMV 35S promoter. Confocal images of fluorescent fusion proteins coexpressed in single epidermal cells were taken at 1 to 2 d after bombardment.
(A) KN1-GFP–emitted fluorescent signal (green) is present in cells adjacent to the bombarded cell at 2 d after bombardment.
(B) to (D) Coexpression of KN1-GFP and DsRED. KN1-GFP is found in neighboring cells (B), whereas DsRED remains in the expressing cell (C). (D) shows a merged image.
(E) to (G) Cell coexpressing KN1-GFP and Nt MPB2C-DsRED. KN1-GFP remains in the bombarded cell and appears in stationary punctae (E). Red channel image shows the presence of Nt MPB2C-DsRED in the same cell (F). In the merged image (G), colocalization of KN1 and Nt MPB2C fluorescent fusion proteins is visible (yellow).
N, nucleus. Stars indicate cells in which fluorescent proteins moved. Bars = 80 μm.
Table 2.
Coexpression after Particle Delivery and Cell-to-Cell Movement of KN1-GFP in the Presence of RFP Fusion Proteins in Arabidopsis
| GFP Fusion
|
RFP Fusion
|
|||||
|---|---|---|---|---|---|---|
| GFP Construct | RFP Construct | No. of Cells | Neighboring Cells (%) | Comment | Neighboring Cells (%) | Comment |
| None | DsRED | 193 | n/a | n/a | 0 (0) | Nucleus, cytosol |
| GFP | DsRED | 84 | 70 (83) | Nucleus, cytosol | 0 (0) | Nucleus, cytosol |
| GFP | Nt MPB2C-DsRED | 137 | 110 (80) | Nucleus, cytosol | 0 (0) | Punctae, microtubules, aggregates |
| GFP-KN1 | DsRED | 124 | 16 (13) | Nucleus, cytosol | 0 (0) | Nucleus, cytosol |
| GFP-KN1 | Nt MPB2C-DsRED | 209 | 0 (0) | Punctae, microtubules | 0 (0) | Punctae, microtubules, aggregates |
| At MPB2C-GFP | Nt MPB2C-DsRED | 128 | 0 (0) | Punctae, microtubules, aggregates | 0 (0) | Punctae, microtubules, aggregates |
| None | KN1-mRFP1 | 94 | n/a | n/a | 9 (10) | Cytosol, nucleus |
DNA encoding fluorescent fusion protein constructs was coated on gold particles and delivered into Arabidopsis epidermal cells. Two- to 4-week-old soil-grown plants were used, and fluorescent fusion protein distribution was analyzed at 2 to 3 d after bombardment using a confocal microscope. When two constructs were combined, an approximately equimolar mixture was used to coat the gold particles. Exclusively, cells were evaluated showing high levels of detectable red and/or green fluorescence. n/a, not applicable.
MPB2C Reduces the Trichome Rescue Efficiency of Non-Cell-Autonomous GL1-KN1HD Fusions in Transgenic Plants
The potential of At MPB2C to limit KN1 trafficking was further tested using the trichome rescue system developed by Kim et al. (2005b). For these experiments, we transformed transgenic gl1/ProRbcS:GFP-GL1-KN1HD trichome rescue lines and Arabidopsis wild-type control lines with a construct expressing a tap-tagged At MPB2C driven by the cauliflower mosaic virus (CaMV) 35S promoter. As an assay for cell-to-cell movement, the number of trichomes present on the adaxial surface of the third and fourth leaves was counted (Table 3). In Pro35S:AtMPB2C-TAG transgenic control lines, the number and distribution of trichomes did not change significantly compared with those in wild-type plants (Figure 5A). The trichome rescue line gl1/ProRbcS:GFP-GL1-KN1HD (Figure 5B) showed ∼66% of the trichomes observed with the control lines (Table 3). The rescue line developed on average 21 ± 3.4 trichomes per leaf, while the trichome rescue lines harboring the Pro35S:AtMPB2C developed on average 14 ± 7 trichomes per leaf (Table 3). A statistical Student's t test analysis revealed a P value of 1.0E-4, suggesting that the difference of trichome numbers observed on the trichome rescue lines was statistically significant. Thus, trichome rescue plants ectopically expressing At MPB2C (Figures 5C and 5D) developed significantly fewer trichomes than the parent rescue plant. Two independent gl1 rescue lines highly expressing At MPB2C relative to the GL1-KN1HD construct (see Supplemental Figure 2 online) over three generations consistently developed fewer than five trichomes on the third or fourth leaf. Also, these lines maintained a limited number of trichomes on leaves appearing at later stages (Figure 5C; see Supplemental Figure 2 online). Furthermore, as exemplified by the plant shown in Figure 5D, two T1 lines had uneven distributions of trichomes over the leaf surface. The negative effect of At MPB2C overexpression on ProRbcS:GFP-GL1-KN1HD–mediated trichome rescue activity is consistent with the hypothesis that MPB2C limits the cell-to-cell movement of GL1-KN1HD.
Table 3.
At MPB2C Overexpression Alters Trichome Recovery Mediated by GFP-GL1-KN1HD in Transgenic gl1 Trichome Rescue Plants
| Genotype (Number of T1 Lines)a | Total No. of Trichomes (No. of Leaves Evaluated)b | Average No. per Leaf | Relative Changec | Comments |
|---|---|---|---|---|
| Wild-type Col-0 (10) | 638 (20) | 32 ± 3.9 | 100% | All plants with normal trichome distribution |
| Wild-type Col-0/Pro35S:AtPB2C (7) | 442 (14) | 32 ± 2.7 | 100% | All plants with normal trichome distribution |
| gl1/ProRbcS:GFP-GL-KN1HD (45) | 1475 (90) | 21 ± 3.4 | 66% | All plants with >15 trichomes per leaf |
| gl1/ProRbcS:GFP-GL-KN1HD/Pro35S:AtMPB2C (21) | 8439 (596) | 14 ± 7 | 44% | Fifty-two plants (T2) show fewer than three trichomes per leaf; two plants (T1) show irregular trichome distribution across one leaf |
Number indicates the number of T1 lines representing independently transformed plants except for gl1/ProRbcS:GFP-GL-KN1HD, which was a stable T6 line.
Leaf trichomes appearing on the adaxial side on the third and fourth leaves of T2 plants (≥10) were counted.
Percentage of trichomes calculated against Col-0 plants transformed with Pro35S:AtMPB2C.
Figure 5.
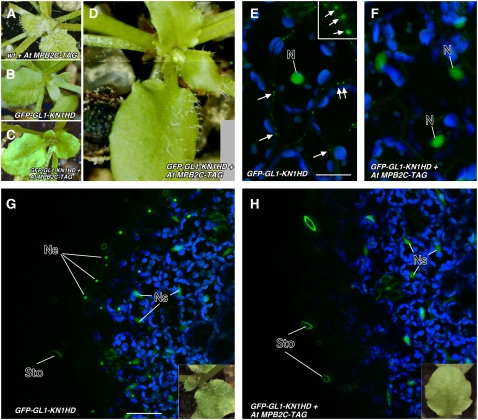
At MPB2C Overexpression Alters the Number of Trichomes Developing on GFP-GL1-KN1HD Trichome Rescue Lines.
(A) Wild-type Arabidopsis Columbia (Col-0) plant transformed with Pro35S:AtMPB2C-TAG showing normal trichome development.
(B) Trichome rescue in gl1 mutant lines transformed with ProRbcS:GFP-GL1-KN1HD that form trichomes on the leaf surface due to GL1 transport mediated via the KN1 HD (Kim et al., 2005b).
(C) and (D) Transgenic gl1 plants harboring ProRbcS:GFP-GL1-KN1HD and Pro35S:At MPB2C-TAG develop trichomes at lower numbers (C) or in T1 plants appear irregularly distributed at the adaxial surfaces of the leaves (D).
(E) to (H) Confocal images of transgenic trichome rescue lines and cellular distribution of GFP-GL1-KN1HD in abaxial leaf cells.
(E) In control lines, GFP-GL1-KN1HD (green) accumulates in the nucleus and at PD-like structures at the cell borders (arrows). The box shows high magnification of GFP-labeled PD-like structures at the cell wall. Bar = 40 μm.
(F) In trichome rescue lines overexpressing At MPB2C-TAG, the GFP-GL1-KN1HD–mediated fluorescent signals appear exclusively in the nuclei. No PD-like structures are detected at cell walls.
(G) and (H) Confocal images of epidermal and subepidermal cells of transgenic trichome rescue lines harboring GFP-GL1-KN1HD. Insets show a leaf from the transgenic line used for confocal imaging. Bar = 80 μm.
(G) Trichome rescue line expressing GFP-GL1-KN1HD and forming trichomes. The GFP fusion protein is detected in nuclei of epidermal cells and subepidermal cells. Note that symplasmically isolated guard cells show no green fluorescent nuclei.
(H) Transgenic trichome rescue line expressing At MPB2C-TAG and developing no trichomes. The green fluorescent signal emitted by GFP-GL1-KN1HD is detected exclusively in the nuclei of subepidermal cells.
Ne, nuclei of epidermal cells; Ns, nuclei of subepidermal cells; Sto, stomata.
MPB2C Expression Abolishes GFP-GL1-KN1HD Presence at PD
The subepidermal expression of GL1-KN1HD as a GFP fusion protein allowed us to ask whether the cellular distribution was altered due to the coexpression of At MPB2C. In ProRbcS:GFP-GL1-KN1HD lines, fluorescent signal was detected in spots in the region of the cell walls thought to resemble accumulation at PD (Figure 5E; see Supplemental Figure 3 online) (Kim et al., 2002) as well as in nuclei of epidermal and subepidermal cells (Figure 5G; see Supplemental Figure 3 online). To confirm the inhibition of GFP-GL1-KN1HD transport through PD by MPB2C, we examined the distribution of GFP-GL1-KN1HD in trichome rescue lines harboring Pro35S: At MPB2C-TAG. In plants with few or no trichomes, GFP-GL1-KN1HD fluorescent signal was detected neither in punctae at cell walls nor in nuclei of epidermal cells (Figure 5F; see Supplemental Figure 3 online). These lines showed fluorescent fusion protein exclusively in the nuclei of subepidermal cells, indicating stable expression of the GFP-GL1-KN1HD construct (Figure 5H).
KN1 Shows Altered Subcellular Distribution in the Presence of MPB2C
To further confirm the potential of KN1 to interact with MPB2C, we investigated the subcellular distribution of KN1 and MPB2C fluorescent fusion proteins expressed after agroinfiltration (Figure 6). First, we established the subcellular distribution of KN1-GFP alone. At all stages examined, KN1-GFP appeared in the cytosol and in the nuclei of ∼80% of cells (Figures 6A and 6B). In weakly expressing cells up to 36 h after infiltration, KN1-GFP was detected in the cytosol and nucleus. In highly expressing cells, KN1-GFP accumulated at the nuclear envelope and at endoplasmic reticulum (ER)–like structures, which collapsed after heat treatment (Figure 6A). At later time points, KN1-GFP appeared mainly as an evenly distributed protein in the cytosol and formed, in addition, fluorescent aggregates of varying sizes (Figure 6B).
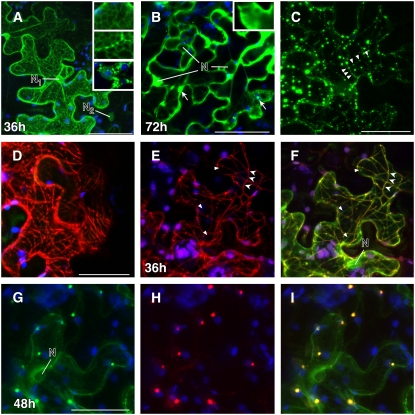
Figure 6.
Colocalization and Intracellular Distribution of KN1 and MPB2C Fluorescent Fusion Proteins.
Confocal images of N. benthamiana source leaves expressing fluorescent fusion proteins at 1 to 3 d after agroinfiltration.
(A) After 36 h, KN1-GFP is detected in the nuclei and at the nuclear envelope (N1). Also, a cellular distribution similar to that in ER-associated proteins (ER-GFP) is observed in high-expressing cells. In low-expressing cells, no association with the nuclear envelope is detected (N2). After heat treatment (2 min of high fluorescent light), the KN1-GFP–tagged ER-like structures collapse and appear as large bodies outside the nucleus (insets).
(B) High levels of KN1-GFP appear as soluble protein in the cytosol and as aggregates (arrows) at 72 h after infiltration.
(C) and (D) Images exemplifying the cellular distribution of At MPB2C-GFP (C) and At MPB2C-RFP (D). Green and red fluorescent punctae and filaments are detected aligned with arrays previously shown to resemble microtubules (Kragler et al., 2003).
(E) to (I) Tissue coexpressing KN1-GFP and Nt MPB2C-DsRED after 36 h.
(E) Nt MPB2C-DsRED appears at thin filaments (arrowheads) and in small punctae.
(F) The same cell shows the expression of Nt MPB2C-DsRED colocalizing with KN1-GFP in the merged image. Triangles indicate KN1-GFP not colocalizing with Nt MPB2C-DsRED. Note that the majority of detected KN1-GFP and Nt MPB2C is present at filamentous structures resembling microtubules (arrowheads).
(G) Cells coexpressing KN1-GFP after 48 h.
(H) Cells coexpressing At MPB2C-mRFP after 48 h.
(I) Merged image of (G) and (H). Green signal emitted by GFP fused to KN1 is detected with At MPB2C-mRFP in punctae.
Blue indicates chloroplast autofluorescence. N, nucleus. Bars = 40 μm.
In contrast with KN1-GFP, At MPB2C-GFP and At MPB2C-RFP were not observed in nuclei and appeared transiently at static filaments and later in punctate arrays, suggesting an association with the cytoskeleton (Figures 6C and 6D). At MPB2C-RFP punctae did not colocalize with ER-GFP or the F-actin marker Talin-GFP (see Supplemental Figure 1 online). In the later stages of expression, or in highly expressing cells, we observed that At MPB2C-GFP generally formed large aggregates. In coexpression experiments after 1 d, At MPB2C-GFP punctae colocalized with its ortholog Nt MPB2C-DsRED (see Supplemental Figure 1 online).
Next, we asked whether MPB2C induces changes in KN1 cellular distribution. Within 2 d after agroinfiltration, KN1-GFP and Nt MPB2C-DsRED2 became visible in small punctae and at microtubule-like structures (Figures 6E and 6F). This colocalization pattern was transient, and at later stages, KN-GFP and Nt MPB2C-DsRED2 appeared together in small punctae (Figures 6G to 6I). Comparable results were also obtained with KN1-GFP coexpressed with At MPB2C-mRFP1 in Arabidopsis epidermal cells after particle delivery (see Supplemental Figure 1 online). Combined with the reported altered cellular distribution of TMV MP due to overexpressed MPB2C (Kragler et al., 2003), these findings suggest that MPB2C has the potential to induce KN1 accumulation at microtubules in vivo.
The STM KNOX HD Mediates MPB2C Interaction
To test whether STM, similar to KN1, colocalizes and interacts with At MPB2C, we employed the bimolecular fluorescence complementation system (Walter et al., 2004). This split yellow fluorescent protein (YFP) system tests for in vivo interaction of fusion proteins consisting of N- and C-terminal truncated YFP and interaction partners. Once the fusion partners interact, high levels of YFP fluorescence can be detected in the cells.
First, we expressed in N. benthamiana epidermal cells split YFP fusion proteins (NYFP-STMΔHD and CYFP-STMΔHD) harboring a mutant form of STM lacking the C-terminal ELK and HD motifs (amino acids 252 to 382). High fluorescence levels were detected in these cells, confirming that the STMΔHD constructs were functional and that the remaining KNOX motif was sufficient to mediate STMΔHD homodimerization (Figure 7A). As with KN1-GFP (Figures 6A and 6B), the STMΔHD constructs appeared at ER-like structures and in the cytosol. By contrast, coexpressed NYFP-STMΔHD and CYFP-At MPB2C emitted weaker YFP signals exclusively in the cytosol that was excluded from nuclei (Figure 7B). Similar to At MPB2C (Figures 6C and 6J), coexpression of CYFP-At MPB2C together with full-length STM fused to NYFP (NYFP-STM) appeared in small mobile and highly fluorescent punctae (Figure 7C), which formed larger aggregates at 2 to 3 d after infiltration (Figure 7D).
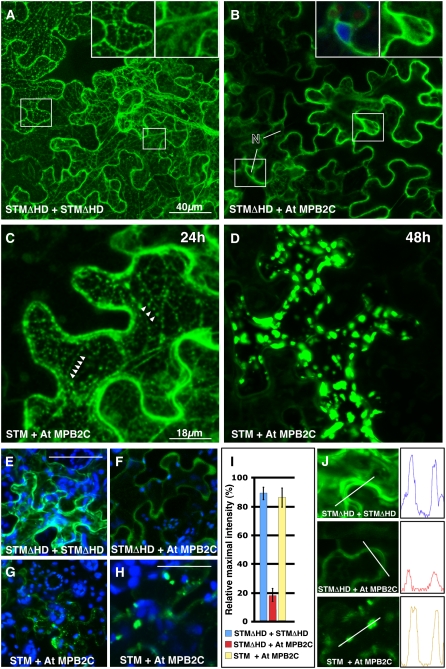
Figure 7.
At MPB2C Interaction with STM Depends on the Presence of the HD in Split YFP Interaction Assays.
Confocal images of N. benthamiana source leaves expressing split YFP constructs after agroinfiltration.
(A) Cells expressing NYFP-STMΔHD and CYFP-STMΔHD. Two days after agroinfiltration, high YFP fluorescence signals were present in the cytosol and in association with ER-like structures. Insets show magnified regions with fluorescent signals in ER-like structures and the cytosol.
(B) Cells expressing NYFP-STMΔHD and CYFP-At MPB2C at 2 d after agroinfiltration. Relatively low YFP signals were detected exclusively in the cytosol of epidermal cells. Insets show magnified regions indicated by rectangles with nucleus (4′,6-diamidino-2-phenylindole [DAPI]–stained) and cytosol. Note that no yellow fluorescent signal was detected in nuclei (N). Blue indicates DAPI staining, and green indicates YFP fluorescence.
(C) and (D) Cells expressing NYFP-STM and CYFP-At MPB2C.
(C) One day (24 h) after agroinfiltration, high YFP fluorescence signals were present in mobile punctate structures (arrowheads) and the cytosol.
(D) After 2 d (48 h), large fluorescent punctae appeared and no fluorescent signals were detected. Note that the YFP signal in the cytosol disappeared and no YFP signals were detected in nuclei.
(E) to (J) Quantification of YFP fluorescence in epidermal cells expressing split YFP constructs. Note that to facilitate measurements, the confocal detection system was set to low sensitivity to avoid signal saturation and the tissues were scanned with the same confocal laser intensity, pinhole, gain, number of stack settings, and z-axis distance from the cell surface.
(E) and (F) Cells expressing NYFP-STMΔHD and CYFP-STMΔHD emit a high fluorescence signal (E) compared with the very low signals in the cytosol detected in cells expressing NYFP-STMΔHD and CYFP-At MPB2C (F). Bar = 50 μm.
(G) Cells expressing NYFP-STM and CYFP-At MPB2C emitted high YFP fluorescence signals in punctate structures.
(H) Higher magnification of a different cell and tissue with similar punctate fluorescence signals. Bar = 20 μm. Blue indicates chloroplast autofluorescence, and green indicates YFP fluorescence.
(I) Column chart of relative fluorescence intensity measurements in cells expressing the split YFP constructs. Confocal images of epidermal cells were analyzed for their maximal YFP fluorescence signal levels. The maximal signals of nine cells in four different infiltrated leaves were measured and the average was calculated (error bars = average deviation).
(J) Examples of the analyzed cell regions and the measured maximal emission in these cells.
In the split YFP system, the intensity of the YFP signal reflects the capacity of two proteins to interact. Thus, we scanned the tissues with fixed confocal settings and measured the relative fluorescent signal intensities in cells expressing the YFP constructs (Figures 7E to 7J). Compared with cells expressing the two STMΔHD constructs (Figures 7A and 7E), very low YFP signals could be detected in cells coexpressing NYFP-STMΔHD and CYFP-At MPB2C (Figure 7F). High fluorescence levels were detected again in cells expressing full-length STM and At MPB2C split YFP fusion constructs (Figures 7G and 7H).
The fluorescence signals emitted by the split YFP fusion proteins were measured on a linear scale and compared with each other (Figures 7I and 7J). Coexpressed STMΔHD fusion proteins yielded similar high levels of fluorescent signals as did NYFP-STM expressed together with CYFP-At MPB2C. Supporting the notion that the HD mediates interaction, NYFP-STMΔHD coexpressed with CYFP-At MPB2C showed an approximately fivefold weaker signal (Figures 7I and 7J).
MPB2C Promoter Is Active in Young Tissues
The MPB2C-mediated downregulation of KN1 trafficking through PD implies that, in wild-type Arabidopsis plants, At MPB2C should be coexpressed or expressed in neighboring tissues where KN1-related proteins, such as STM and KNAT1, are produced. To test this prediction, we examined At MPB2C promoter activity in Col-0 plants transformed with ProAtMPB2C:GUS (for β-glucuronidase) constructs. Two to 3 d after germination, ProAtMPB2C:GUS–driven GUS production was detected in the vascular tissues of cotyledons, leaf primordia, and the area of the shoot apical meristem (Figures 8A and 8B). In later developmental stages, these plants showed GUS staining in carpels and ovules (Figures 8C and 8D). Here, GUS activity was detected in the funiculus and integuments (Figure 8D). In roots, ProAtMPB2C:GUS plants showed weak GUS activity in a limited number of epidermal cells in proximity to the root apical meristem (Figure 8E).
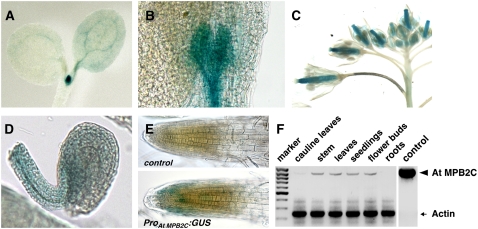
Figure 8.
Promoter Activity of At MPB2C in Arabidopsis.
(A) to (E) Seedling and flower tissues of ProAtMPB2C:GUS plants stained for GUS activity.
(A) to (C) The promoter drives GUS production in the vasculature of cotyledons (A), tissue associated with leaf primordia, and apical meristem (B) and carpels (C).
(D) Weak GUS activity in ovules of ProAtMPB2C:GUS plants.
(E) Root tips of wild-type (control) and ProAtMPB2C:GUS plants.
(F) RT-PCR of At MPB2C and Actin as a control. Note the very low levels of At MPB2C transcripts relative to Actin in wild-type Arabidopsis Col-0 plants.
Consistent with the promoter–GUS activity observed in Arabidopsis wild-type lines, RT-PCR assays (Figure 8F) indicated that At MPB2C mRNA is either not produced or only weakly produced in cauline leaves and root tissues. Thus, the presence of MPB2C transcripts correlates with the staining patterns observed with the corresponding promoter–GUS constructs. Collectively, the observed At MPB2C expression pattern suggests that the gene is active in tissues neighboring or overlapping those with STM and KNAT1/BP function (Lincoln et al., 1994; Long et al., 1996). At MPB2C mRNA is produced in distinct plant parts, such as mersistems, carpels, and developing ovules, in which STM-related interaction partners like the BEL1-like proteins function (Modrusan et al., 1994; Reiser et al., 1995; Cole et al., 2006).
DISCUSSION
Intercellular trafficking of HD transcription factors, such as KN1, STM, and KNAT1, is likely to act as an important developmental signal in plants. HD protein trafficking is thought to occur via PD and should be under strict control to ensure the proper separation of differentiation domains. Developmental modifications in PD are known to affect free GFP diffusion and to regulate selective KN1 transport (Itaya et al., 1998; Oparka et al., 1999; Kim et al., 2002, 2003, 2005a, 2005b). However, almost nothing is known about the potential PD transport regulators suggested to exist for KN1 or STM. Here, we show that MPB2C is an RNA and HD motif binding protein that functions as a component in the regulation of KN1 trafficking.
KN1 Subcellular Distribution
Our observation that ectopically expressed KN1 is not located exclusively to the nucleus by default is not surprising. STM, the Arabidopsis homolog of KN1, had also been reported to be cytoplasmic upon ectopic overexpression in epidermal leaf cells (Cole et al., 2006). In other studies, however, coexpressed STM and KNAT1 fluorescent fusion proteins had been reported to be located to nuclei (Hackbusch et al., 2005). Cole et al. (2006) suggested that this inconsistency might be an artifact. However, in our experiments, despite an often observed accumulation of KN1-GFP at the nuclear surface, single confocal sections revealed true nuclear localization, with visible exclusion from the nucleoli, of the construct in most cells of agroinfiltrated and particle-bombarded tissues. Kuijt et al. (2004) examined the subcellular localization of three rice (Oryza sativa) KNOX class I family members. They reported complex and heterogenous patterns of intracellular localization varying between tissues and different KNOX class I members. Intriguingly, highly motile punctate structures were also detected with Os KN1, which is the closest relative to maize KN1 in rice (Kuijt et al., 2004). The described localization patterns coincided, except for the transient reticulate structure, with the observed patterns displayed by KN1-GFP. These various intracellular localization patterns detected with KNOX proteins might be caused by a tight regulatory mechanism imposed by KNOX binding partners such as the OVATE proteins, BLH proteins, and MPB2C.
MPB2C as a Molecular Regulator of HD Protein Transport
The microtubule-associated Nt MPB2C was originally described as a specific TMV MP–interacting partner inhibiting the cell-to-cell trafficking of TMV MP but not that of the CMV MP (Kragler et al., 2003). As it seems unlikely that plants evolved a conserved MPB2C protein to limit the spread of one distinct class of viruses, we probed the MPB2C interaction with other plant endogenous NCAPs. Interaction assays indicated that both MPB2C orthologs from N. tabacum and Arabidopsis specifically bind to KN1 and STM. Intriguingly, MPB2C does not bind to other NCAPs, such as SHR, LFY, or Cm PP16. MPB2C seems to recognize specifically non-cell-autonomous class I KNOX proteins, represented by KN1 and STM, and not other HD proteins such as BEL1 that are suggested to be cell-autonomous (Kim et al., 2005b). Thus, MPB2C may regulate the intercellular distribution of an NCAP subclass and seems to be part of a pathway controlling KN1/STM availability for translocation across cell borders. Indeed, abundant MPB2C interferes with the cell-to-cell transport activity of KN1 in microinjection assays, in transient expression assays, and in trichome rescue assays. These observations are in agreement with the previously suggested function of MPB2C as a microtubule-associated TMV MP binding protein with the capacity to interrupt cell-to-cell transport by enhancing the microtubular association of TMV MP (Curin et al., 2007).
At present, we cannot exclude the possibility that the observed inhibition of cell-to-cell transport is a secondary effect of the high expression levels of MPB2C. However, this scenario seems unlikely, as in our experiments, KN1 and STM interact directly with MPB2C via their HD. Also, the endogenous promoter activity pattern of MPB2C in Arabidopsis suggests that MPB2C functions within or in the proximity of meristematic tissues, where STM and BP/KNAT1 are expressed and moving from cell to cell.
KN1 and STM colocalize in the cytosol with MPB2C in a punctate manner previously shown to coincide with microtubules (Kragler et al., 2003). A similar pattern was observed with subsets of OVATE family proteins interacting with distinct BLH and KNOX proteins (Hackbusch et al., 2005). However, a function of OVATE regarding cell-to-cell transport was not reported. MPB2C, although of similar size, harbors no motifs related to OVATE family members and seems to have a distinct function directed specifically toward regulating the trafficking of KNOX proteins. For other KNOX motif binding proteins, such as BLH proteins, a negative role on KN1 cell-to-cell transport seems unlikely. This notion finds support based on the following: (1) BLH proteins bind to the KNOX motif of KN1 dispensable for cell-to-cell transport, and (2) BLH proteins facilitate the nuclear import of KNOX proteins (Cole et al., 2006).
Considering that HD proteins function to define cell identity, it is logical to assume that mechanisms exist to control both HD protein levels in the appropriate (expressing) tissues and their cell-to-cell transport to neighboring tissues. Support for this notion is derived from the failure of MPB2C to interact with mutant KN1 and STM proteins lacking a functional HD transport motif, such as KN1M6, KN1ΔHD (Lucas et al., 1995; Kim et al., 2005b), and STMΔHD. Also, in transgenic trichome rescue plants, MPB2C inhibits the KN1HD-mediated cell-to-cell transport of GL1 toward PD and, consequently, to the epidermis (Figure 5; see Supplemental Figure 3 online). Therefore, we conclude that MPB2C negatively controls KN1 trafficking activity by binding to the HD motif.
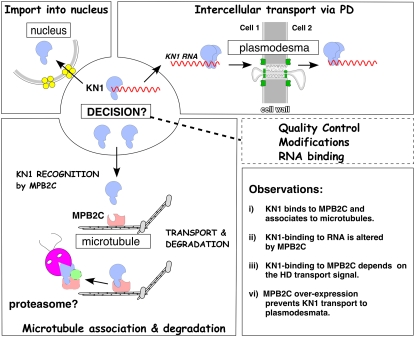
Sequence analysis of MPB2C cDNAs and predicted amino acid sequences indicates that no significantly related genes are found in other kingdoms than plants and that the gene is present only once per plant genome. As discussed for OVATE proteins (Hackbusch et al., 2005), MPB2C seems to provide a link for HD proteins to the microtubules. Here, as suggested for the TMV MP pathway (Curin et al., 2007), MPB2C could constitute the entry point for an HD protein sequestration pathway operating with the help of proteasomes to degrade excess cytosolic KN1 (Figure 9).
Figure 9.
Model of MPB2C Regulating KN1 Transport.
A hypothetical model depicting MPB2C as a KN1 transport-controlling factor. KN1 is produced in the cytosol; there, a decision is made between nuclear import (right panel), cell-to-cell transport via PD (left panel), and MPB2C-mediated association to microtubules (bottom panel). MPB2C binding to KN1 depends on a functional HD motif, which also constitutes the KN1 cell-to-cell transport signal. Once a KN1-mRNA complex enters the PD translocation pathway, the KN1 HD transport motif is modified or occupied. By this means, a KN1-mRNA complex escapes nuclear import and gains access to adjacent cells. However, aberrant or unbound KN1 molecules, not immediately entering the PD pathway, could be recognized by MPB2C. In such a scenario, KN1 could associate with MPB2C at the cytoskeleton. Here, KN1, as suggested for TMV MP (Kragler et al., 2003, Curin et al., 2007), might rapidly enter the proteasome-dependent degradation pathway. By this means, MPB2C could regulate two aspects: (1) the quality and amount of KN1 present in the shoot apical meristem, and (2) the pace of KN1-mRNA cell-to-cell transport to neighboring tissues.
In tobacco and Arabidopsis, endogenous MPB2C levels are very low (Kragler et al., 2003) (Figure 8) and at the time of trichome initiation may not be sufficient to block KN HD transport from cell to cell, thus allowing trichome rescue in the ProRbcS:GFP-GL1-KN1HD line. On the other hand, in transgenic trichome rescue plants, high levels of MPB2C blocked KN1HD-mediated transport. However, in wild-type plants, class I HD proteins are expressed in a limited number of cells and endogenous MPB2C might contribute sufficiently to modulate non-cell-autonomous KNOX protein trafficking. MPB2C could restrict the number of KN1/STM macromolecules available to enter the PD pathway in the shoot apical meristem. By this means, MPB2C may ensure that transport-competent HD transcription factors are not distributed extensively across multiple cell layers. As depicted in the model presented in Figure 9, MPB2C may directly regulate the HD protein levels available for intercellular transport.
MPB2C May Function as a Chaperone
Interestingly, the wide diversity of functions performed by the tubulin-based cytoskeleton and its associated factors, in conjunction with the high degree of structural conservation of interacting cytoskeletal factors observed across all kingdoms, suggest that a microtubule-associated protein, such as MPB2C, is present in nonplant organisms. However, an MPB2C-like protein could not be found in other kingdoms, which would facilitate a functional prediction for plants. In a hypothetical scenario, MPB2C constitutes a component of a larger microtubule-associated complex, whose function may be similar to those of chaperones that discriminate between correctly assembled and aberrant NCAPs. MPB2C would regulate two aspects of trafficking. First, the quality of NCAPs: only functional protein signals (e.g., in the form of correctly folded and/or modified KN1 protein bound to its mRNA) would bypass MPB2C and hence gain entry to adjacent cells. Second, the quantity of NCAPs: MPB2C would prevent abnormal amounts of non-cell-autonomous homedomain proteins produced in a single mutant meristematic cell from entering neighboring cells. Both functional aspects would constitute a fail-safe mechanism and ensure that tissues differentiate according to their developmental program (Figure 9).
We propose that non-cell-autonomy of KN1, based on transport via PD, is regulated by the microtubule-associated and TMV MP–interacting protein, MPB2C. In the cytosol, a decision is made regarding whether modified, or aberrant, KN1 proteins bind to MPB2C present at microtubules. Such an interaction determines whether KN1 enters the nucleus or moves as an RNA-protein complex through PD into neighboring cells. Once KN1 is recognized by MPB2C, the complex is delivered to an as yet unknown destination or to a degradation pathway. This would ensure that cell-to-cell transport of HD proteins is tightly regulated.
Our observations allow us to extend the proposed negative regulatory function of microtubules in the cell-to-cell transport of TMV to endogenous HD proteins. Our findings will enable us to further examine KN1/STM protein transport function in the complex intracellular and intercellular signaling networks existing in meristematic tissues.
METHODS
Plant Materials
Nicotiana sp and Arabidopsis thaliana plants were grown in controlled environmental chambers under light intensity in the range of 800 to 1000 μmol·m−2·s−1 and temperatures of 22°C for 12 h of light for Nicotiana species and 16 h of light for Arabidopsis.
Cloning, Expression, and Purification of Recombinant Proteins
Specific primers were designed based on the cDNA sequences of Nt MPB2C (GenBank accession number AAL95696) and the genomic sequences of At MPB2C (The Arabidopsis Information Resource accession number At5g08120) and used in PCR to clone the respective cDNAs or genomic fragments into TOPO or pENTR/D-TOPO vectors (Invitrogen) for further cloning.
EcoRI/BamHI cDNA RT-PCR cDNA fragments of At MPB2C were obtained from total RNA isolated from 2-week-old Arabidopsis Col-0 seedlings as template. EcoRI/BamHI cDNA PCR fragments of Zm KN1, Nt NCAPP1, Cm PP16, At LFY (a gift from D. Weigel, Max Planck Institute for Developmental Biology), At AP1 (a gift from E. Meyerowitz, California Institute of Technology), At AP3 (a gift from J. Bowman, University of California-Davis), and At SHR (a gift from P. Benfey, Duke University) were obtained from plasmid templates harboring the appropriate cDNAs and cloned via PCR into the yeast two-hybrid vectors pGAD424 or pGADT7 for activation domain fusions and pGBT9 or pGBKT7 for binding domain fusions (Matchmaker Systems; Clontech). KN1 deletion mutants were produced with the QuikChange site-directed mutagenesis kit (Stratagene) using the pGBT9-KN1 yeast shuttle vector as a template and verified by sequencing.
For His-tagged At MPB2C and Nt MPB2C protein expression, cDNA NcoI/BamHI fragments were cloned into the Escherichia coli expression vector pET30a. His-tagged MPB2C proteins were purified to near homogeneity using Ni-column chromatography (Amersham). For At MPB2C transient expression assays, NcoI/BamHI PCR cDNA fragments were cloned into the transient expression vector ala-dsRED (Kragler et al., 2003) or ala-mRFP1, obtained by replacing DsRED with an mRFP1 (Campbell et al., 2002) SalI/XbaI PCR fragment. The resulting fusion constructs contain a 10× Ala linker between the cDNAs and the RFP cDNAs. GFP expression vectors producing GFP, DsRED, or Nt MPB2C-DsRED were described elsewhere (Kragler et al., 2003).
Binary vectors expressing At MPB2C-GFP, At MPB2C-TAP, At MPB2C-mRFP1, NYFP-STM, NYFP-STMΔHD, CYFP-STMΔHD (lacking the C-terminal amino acids 252 to 382 with the ELK and HD motifs), and CYFP-At MPB2C for infiltration experiments were cloned using the Gateway system (Invitrogen) following the supplier's manual. In short, the appropriate cDNA PCR fragments of At MPB2C, STM, or STMΔHD lacking stop codons were transferred into pDONR ZEO, pENTR/D-TOPO, or pENTR4 (Invitrogen). After verification of the PCR-produced DNA sequences, the inserts were recombined into the 35S CaMV promoter harboring binary vectors pEarleyGate 103 for expression of GFP fusion proteins, pEarleyGate 205 (Earley et al., 2006) for expression of At MPB2C TAP-tag fusion proteins (At MPB2C TAG), pCR112 for N-YFP, or pCR113 (a kind gift from M. Huelskamp, Institute of Botany, University of Cologne) for C-YFP N-terminal fusion proteins. A pENTR4 plasmid harboring a cDNA NcoI/XbaI fragment of At MPB2C fused to ala-mRFP1 cDNA served as a source, and the binary Gateway vector pMDC32 (Curtis and Grossniklaus, 2003) served as a target to produce a binary vector carrying Pro35S:AtMPB2C-ala-mRFP1. The binary vector harboring Pro35S:Nt MPB2C-DsRED and Δ1-66 Nt MPB2C-DsRED was constructed by ligation of NcoI(partial)/XbaI-digested pGCN binary vector with appropriately digested PCR fragments obtained with pNt MPB2C-ala-DsRED and pΔ1-66 Nt MPB2C-ala-DsRED (Kragler et al., 2003) as templates.
Yeast Two-Hybrid Interaction Assays and in Vitro Binding
The MPB2C interaction assays were performed following an established protocol (Kragler et al., 1998a). In short, the interaction mapping was conducted with the Saccharomyces cerevisiae strains HF7c (MATa) or AH109 (MATa, GAL-MEL1) and the mating partner strains PCY3 (MATα) or Y187 (MATα, GAL-MEL1). To test for interaction of the activation domain and binding domain fusion proteins, the transformed strains were crossed and tested for His/Ade auxotrophy and β-galactosidase and/or α-galactosidase activity on selective media. All transformed and diploid strains were selected and grown at 30°C on the appropriate selective synthetic complete medium supplied with 2% glucose or galactose and, if high unspecific activity of the binding domain fusion occurred, supplied with 5 to 20 mM 3-amino-1,2,4-triazole to limit the background growth on His-lacking medium.
KN1, KN1M6, TMV MP, and CMV MP proteins were expressed in E. coli and purified as described (Lucas et al., 1995; Kragler et al., 1998b). His-tagged At/Nt MPB2C were purified to near homogeneity via Ni-column purification (Amersham) following the supplier's protocols. Conditions for the renatured blot overlay assay are based on previously described assays (Kragler et al., 2000, 2003). In short, proteins were transferred onto nitrocellulose membranes (Protean 2-μm membranes; Amersham), washed twice with water, washed three times with overlay buffer containing 2% BSA (Sigma-Aldrich) for 2 h, and incubated for 24 h at 10°C with overlay buffer (300 μL/cm2) containing 0.5 μg/mL purified KN1, MPB2C, or BSA (Sigma-Aldrich). For competition experiments, an ∼50-fold molar excess of purified protein was added to the overlay buffer. After incubation, the strips were washed five times for 2 min each time with overlay buffer avoiding BSA and treated with 2% (v/v) diglutaraldehyde (Sigma-Aldrich) for 1 min at room temperature. After washing five times with 1× PBS containing 5% BSA, bound recombinant proteins were detected following standard protein gel blot procedures using purified rabbit polyclonal antibodies (1:500) directed against KN1 or At MPB2C. Bound antibodies were detected with secondary anti-rabbit IgG conjugated with ALP or HR (1:10,000) followed by nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche) color-staining reaction or the Amersham ECL Plus Western blotting detection system (Amersham) by exposure to x-ray film (Fuji) following protocols supplied by the manufacturer. RNA overlay assays and 5′ labeling with [32P-γ]ATP of total RNA from 2-week-old Arabidopsis seedlings were done as described (Yoo et al., 2004).
Transient Expression and Microinjection Assays
Fluorescent labeling of MPB2C and KN1 with reactive rhodamine or FITC (Molecular Probes) and microinjection assays with recombinant proteins were performed on mesophyll cells of 3- to 4-week-old N. benthamiana source leaves as described previously (Fujiwara et al., 1993; Noueiry et al., 1994; Kragler et al., 2000). Infiltration assays on N. benthamiana leaves were performed with LBA4404 or AGL1 Agrobacterium tumefaciens strains harboring the appropriate binary vectors grown at 28°C in selective Luria-Bertani medium to an OD600 of 0.8, harvested, and resuspended in infiltration solution to obtain a final OD600 of 1.6, as described (Yoo et al., 2004). Coexpression of fluorescent proteins was achieved by coinfiltration of a 1:1 mixture (OD600 of 3.2) of AGL1 agrobacterial strains harboring the appropriate binary vectors. Transient expression of fluorescent fusion proteins in Arabidopsis was achieved by particle pressure delivery, using a Biolistic PDS-1000-He apparatus (Bio-Rad), of plasmid DNA coated onto 1-μm gold particles (Bio-Rad). Images of expressed fluorescent proteins or microinjected probes were obtained with a Leica SP1 or SP4 confocal microscope. GFP, FITC, acridine orange, and lucifer yellow CH as well as mRFP1, DsRED, and rhodamine fluorescent probes were excited at 476/488 nm and 568 nm, respectively. GFP, FITC, lucifer yellow CH, and acridine orange were detected at 500 to 520 nm (green channel), mRFP1, DsRED, and rhodamine at 610 to 630 nm (red channel), and chloroplast fluorescence at 680 to 710 nm (blue channel). All tissues/cells harboring green and red fluorescent probes were also scanned in sequential mode, switching between the 476/488-nm and 568-nm laser excitations and according to detection channels to ensure specific identification of red and green fluorescent probes in their appropriate channels. The images were assembled using ImageJ 1.32 freeware (http://rsb.info.nih.gov/ij/) and Canvas 8 (Deneba Systems).
Transgenic Arabidopsis Lines, Expression Constructs, and Assays
All transgenic Arabidopsis lines were produced using the flower-dipping/silwet agroinfiltration method (Clough and Bent, 1998) using binary vector harboring AGL1 agrobacteria grown in selective Luria-Bertani medium. Transgenic gl1 plants harboring ProRbcS:GFP-GL1-KN1HD (a gift from D. Jackson) were selected on germination plates containing hygromycin (50 μg/mL). Plants transformed with pEarleyGate 103 (Pro35S:AtMPB2C-GFP) or pEarleyGate 205 (Pro35S:AtMPB2C-TAP) vectors (Earley et al., 2006) were screened for BASTA resistance by spraying twice at 2 to 3 weeks after germination with 200 mg/L BASTA solution (Raiffeisen Warehouse). Genomic PCR fragments containing genomic 495-bp upstream sequences of At MPB2C (ProAtMPB2C) from the start codons were made using FK231 (5′-CACCCCTTTCTCGATGCAGTGATCAC-3′) and FK250 (5′-TTCAGTGTTCATCAAAACT-3′) primers and cloned into pENTRD-TOPO vector (Invitrogen). The binary vector used to produce the plant expression vector ProAtMPB2C:GUS via gateway recombination was pKGWFS7 (Karimi et al., 2002).
RT-PCR analysis was done as described (Kragler et al., 2003) using the primers FK227 (5′-CACCATGTATGAGCAGCAGCAAC-3′) and FK228 (5′-ATAATATGTAAAGGCTAGTGATTG-3′) for At MPB2C, FK296 (5′-GGAAGAAATGCTTCTCAAAC-3′) and FK398 (5′-GACTTCCCCGCGGAATTCGC-3′) for At MPB2C-TAP, FK259 (5′-GTATAGTTCATCCATGCCATGTG-3′) and FK296 (5′-GGAAGAAATGCTTCTCAAAC-3′) for At MPB2C-GFP, and FK346 (5′-ATGTACAGTGGCTATCTAAGCTCGCTC-3′) and FK347 (5′-CGCCGAGCCGGTACAGCCCGCC-3′) for KN1HD. To standardize the expression levels, we used FK424 (5′-GGAAGGATCTGTACGGTAAC-3′) and FK425 (5′-TGTGAACGATTCCTGGACCT-3′) primers directed against an Actin cDNA (The Arabidopsis Information Resource accession number At3g18780).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Nt MPB2C (AAL95696), At MPB2C (At5g08120), Zm KN1 (AAP76321), At BEL1 (At5g41410), At STM (At1g62360), At LFY (At5g61850), At SHR (At4g37650), At AP1 (At1g69120), At AP3 (At3g54340), Nt NCAPP1 (AF307094), and Cm PP16-1 (AF079170).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Subcellular Distribution of At MPB2C and Nt MPB2C, GFP-KN1, ER-GFP, and Talin-GFP Expressed in Epidermal Cells.
Supplemental Figure 2. Comparison of the Relative At MPB2C and GL1-KN1HD Expression Levels with Trichome Numbers in Pro35S:AtMPB2C Transgenic Trichome Rescue Lines.
Supplemental Figure 3. Confocal Images of GL1-GFP-KN1HD Signals Detected in Leaves of Arabidopsis Trichome Rescue Lines.
Supplemental Table 1. Example for the Yeast Two-Hybrid Interaction Categorization as Shown in Figure 1B.
Acknowledgments
We thank Bill Lucas for his generous financial support in the initial phase of this project and with material, access to the confocal microinjection unit, critical discussions, and comments on the manuscript; Dave Jackson for critical comments on the manuscript and providing transgenic GFP-GL1-KN1HD trichome rescue plants and the plasmid expressing KN1 tagged with GFP and BD-STM; and Andrea Mair, Andreas Rasner, Rainer Leitner, and Regina Pohn for technical assistance in the expression of recombinant proteins and antibody production. This work was supported by Austrian Science Fund Project P16928-Kragler.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Friedrich Kragler (friedrich.kragler@univie.ac.at).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alche, J.D., and Rodriguez-Garcia, M.I. (1997). Fluorochromes for detection of callose in meiocytes of olive (Olea europaea L.). Biotech. Histochem. 72 285–290. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Etchells, J.P., Canales, C., Lagodienko, A., and Dickinson, H. (2004). VAAMANA, a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111. [DOI] [PubMed] [Google Scholar]
- Boyko, V., Ferralli, J., Ashby, J., Schellenbaum, P., and Heinlein, M. (2000). Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2 826–832. [DOI] [PubMed] [Google Scholar]
- Boyko, V., Hu, Q., Seemanpillai, M., Ashby, J., and Heinlein, M. (2007). Validation of microtubule-associated tobacco mosaic virus RNA movement and involvement of microtubule-aligned particle trafficking. Plant J. 51 589–603. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Banerjee, A.K., and Hannapel, D.J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38 276–284. [DOI] [PubMed] [Google Scholar]
- Chen, H., Rosin, F.M., Prat, S., and Hannapel, D.J. (2003). Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 132 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H., Sheng, J., Hind, G., Handa, A.K., and Citovsky, V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H., Tian, G.W., Gafni, Y., and Citovsky, V. (2005). Effects of calreticulin on viral cell-to-cell movement. Plant Physiol. 138 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cole, M., Nolte, C., and Werr, W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curin, M., Ojangu, E.L., Trutnyeva, K., Ilau, B., Truve, E., and Waigmann, E. (2007). MPB2C, a microtubule-associated plant factor, is required for microtubular accumulation of tobacco mosaic virus movement protein in plants. Plant Physiol. 143 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T., Giesman-Cookmeyer, D., Ding, B., Lommel, S.A., and Lucas, W.J. (1993). Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the red clover necrotic mosaic virus movement protein. Plant Cell 5 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, K.L., Paquette, A.J., Nakajima, K., and Benfey, P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14 1847–1851. [DOI] [PubMed] [Google Scholar]
- Gillespie, T., Boevink, P., Haupt, S., Roberts, A.G., Toth, R., Valentine, T., Chapman, S., and Oparka, K.J. (2002). Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch, J., Richter, K., Muller, J., Salamini, F., and Uhrig, J.F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 102 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.): S303–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F., Huang, F., Wilson, K.A., and Tan-Wilson, A. (2007). Protein storage vacuole acidification as a control of storage protein mobilization in soybeans. J. Exp. Bot. 58 1059–1070. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., Epel, B.L., Padgett, H.S., and Beachy, R.N. (1995). Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270 1983–1985. [DOI] [PubMed] [Google Scholar]
- Hibara, K., Takada, S., and Tasaka, M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36 687–696. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Woo, Y.M., Masuta, C., Bao, Y., Nelson, R.S., and Ding, B. (1998). Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Karabetsos, J.H., Pappelis, A.J., and Russo, V.M. (1987). Visualization of halos in the epidermal cell wall of Allium cepa caused by Colletotrichum dematium f. circinans and Botrytis allii using fluorochromes. Mycopathologia 97 137–141. [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, I., Kobayashi, K., Cho, E., and Zambryski, P.C. (2005. a). Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 102 11945–11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y., Rim, Y., Wang, J., and Jackson, D. (2005. b). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y., Yuan, Z., Cilia, M., Khalfan-Jagani, Z., and Jackson, D. (2002). Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4103–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y., Yuan, Z., and Jackson, D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130 4351–4362. [DOI] [PubMed] [Google Scholar]
- Kragler, F., Curin, M., Trutnyeva, K., Gansch, A., and Waigmann, E. (2003). MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol. 132 1870–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler, F., Lametschwandtner, G., Christmann, J., Hartig, A., and Harada, J.J. (1998. a). Identification and analysis of the plant peroxisomal targeting signal 1 receptor NtPEX5. Proc. Natl. Acad. Sci. USA 95 13336–13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler, F., Monzer, J., Shash, K., Xoconostle-Cazares, B., and Lucas, W.J. (1998. b). Cell-to-cell transport of proteins: Requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J. 15 367–381. [Google Scholar]
- Kragler, F., Monzer, J., Xoconostle-Cazares, B., and Lucas, W.J. (2000). Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 19 2856–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt, S.J., Lamers, G.E., Rueb, S., Scarpella, E., Ouwerkerk, P.B., Spaink, H.P., and Meijer, A.H. (2004). Different subcellular localization and trafficking properties of KNOX class 1 homeodomain proteins from rice. Plant Mol. Biol. 55 781–796. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Taoka, K., Yoo, B.C., Ben-Nissan, G., Kim, D.J., and Lucas, W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., Rojas, M.R., Gomez-Ospina, N., Staehelin, L.A., and Lucas, W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299 392–396. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., and Lee, J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5 712–726. [DOI] [PubMed] [Google Scholar]
- McLean, B.G., Zupan, J., and Zambryski, P.C. (1995). Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27 13–23. [DOI] [PubMed] [Google Scholar]
- Nagasaki, H., Sakamoto, T., Sato, Y., and Matsuoka, M. (2001). Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13 2085–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 307–311. [DOI] [PubMed] [Google Scholar]
- Nelson, R.S., and Citovsky, V. (2005). Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiol. 138 1809–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry, A.O., Lucas, W.J., and Gilbertson, R.L. (1994). Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76 925–932. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I.M., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97 743–754. [DOI] [PubMed] [Google Scholar]
- Reichel, C., and Beachy, R.N. (2000). Degradation of tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83 735–742. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Kragler, F. (2004). The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 7 641–650. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Yanofsky, M.F., and Weigel, D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289 779–782. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola, I.L., and Gonzalez, D.H. (2006). Interaction of the BELL-like protein ATH1 with DNA: Role of homeodomain residue 54 in specifying the different binding properties of BELL and KNOX proteins. Biol. Chem. 387 31–40. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Veit, B., Sinha, N., and Hake, S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350 241–243. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wright, K.M., Wood, N.T., Roberts, A.G., Chapman, S., Boevink, P., Mackenzie, K.M., and Oparka, K.J. (2007). Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: Evidence from FRAP. Traffic 8 21–31. [DOI] [PubMed] [Google Scholar]
- Wu, X., Dinneny, J.R., Crawford, K.M., Rhee, Y., Citovsky, V., Zambryski, P.C., and Weigel, D. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130 3735–3745. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Yoo, B.C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]