Abstract
Methylcinnamate, which is widely distributed throughout the plant kingdom, is a significant component of many floral scents and an important signaling molecule between plants and insects. Comparison of an EST database obtained from the glandular trichomes of a basil (Ocimum basilicum) variety that produces high levels of methylcinnamate (line MC) with other varieties producing little or no methylcinnamate identified several very closely related genes belonging to the SABATH family of carboxyl methyltransferases that are highly and almost exclusively expressed in line MC. Biochemical characterization of the corresponding recombinant proteins showed that cinnamate and p-coumarate are their best substrates for methylation, thus designating these enzymes as cinnamate/p-coumarate carboxyl methyltransferases (CCMTs). Gene expression, enzyme activity, protein profiling, and metabolite content analyses demonstrated that CCMTs are responsible for the formation of methylcinnamate in sweet basil. A phylogenetic analysis of the entire SABATH family placed these CCMTs into a clade that includes indole-3-acetic acid carboxyl methyltransferases and a large number of uncharacterized carboxyl methyltransferase–like proteins from monocots and lower plants. Structural modeling and ligand docking suggested active site residues that appear to contribute to the substrate preference of CCMTs relative to other members of the SABATH family. Site-directed mutagenesis of specific residues confirmed these findings.
INTRODUCTION
Methylcinnamate and methyl-p-coumarate play important roles in plant–insect interactions and are widely distributed throughout the plant kingdom. They have been identified in both aerial and underground tissues from monocots and dicots as well as from ferns (Konda and Kawazu, 1979; Schaefers and Herrmann, 1982; Williams and Whitten, 1983; Hooper et al., 1984; Bandara et al., 1988; Cambie et al., 1990; Garcia et al., 1990; Seifert and Unger, 1994; Hiraga et al., 1996; Daayf et al., 1997a; Odell et al., 1999; Wu et al., 1999; Dias et al., 2003; Bruni et al., 2004; Azah et al., 2005; bin Jantan et al., 2005). Methylcinnamate, a known component of the floral scent from various orchid species, acts as a preferred attractant for euglossine bees and is electrophysiologically active toward excised bee antennae (Dodson et al., 1969; Ackerman, 1989; Schiestl and Roubik, 2003; Eltz and Lunau, 2005). Methylcinnamate production, however, is not strictly associated with flowers and has been found to accumulate to high levels in other tissues, including the leaves of sweet basil (Ocimum basilicum). In contrast with methylcinnamate, its p-hydroxylated derivative methyl-p-coumarate has been shown to have insecticidal or insect-deterrent (Hattori et al., 1992; Seifert and Unger, 1994) as well as antifungal (Bandara et al., 1988; Seifert and Unger, 1994) properties. Moreover, in cucumber (Cucumis sativus), methyl-p-coumarate can act as both an anticipin and an elicitor-inducible phytoalexin (Daayf et al., 1997a, 1997b, 2000).
The rapid growth, the presence of secretory glandular trichomes (glands) that produce a diversity of compounds from several different major biosynthetic pathways, and the availability of lines producing copious amounts of volatile compounds make sweet basil an excellent system to investigate the production of methylcinnamate and methyl-p-coumarate in planta. In sweet basil, the glands represent highly active metabolic factories that produce large amounts of terpenoids and phenylpropanoids as well as fatty acid–derived metabolites. Several different basil lines have been developed that produce different compounds from these various pathways as their major volatile and aroma components. Sweet Dani lemon basil (line SD) produces large amounts of citral, a mixture of the monoterpenoid compounds geranial and neral, which gives this variety its strong lemon scent. By contrast, basil lines SW and EMX-1 contain the phenylpropanoid pathway–derived phenylpropenes eugenol and methylchavicol, respectively, as well as mixtures of monoterpenoids and sesquiterpenoids. A fourth basil line, cinnamon basil (line MC), produces large amounts of methylcinnamate. The availability of these basil lines with distinct metabolite profiles and the development of methods for isolating the cells responsible for natural product biosynthesis have enabled us to utilize a comparative systems biology approach to identify genes involved in the production of the various volatile components unique to the different basil lines (Gang et al., 2001, 2002a, 2002b; Iijima et al., 2004a, 2004b).
The development of a large glandular trichome EST database made from these four distinct basil lines (J. Kapteyn, C. Soderlund, and D. Gang, unpublished data) allowed us to identify the genes responsible for the ultimate step in methylcinnamate biosynthesis (Figure 1). As described in this report, methylcinnamate is produced in sweet basil by the activity of a newly identified member of the SABATH family of carboxyl methyltransferases (D'Auria et al., 2003). This enzyme, designated p-coumaric acid/cinnamic acid carboxyl methyltransferase (CCMT), has a unique substrate preference for cinnamate as well as p-coumarate and represents a new addition to this family of S-adenosylmethionine (AdoMet)–dependent carboxyl methyltransferases. It is more closely related to indole-3-acetic acid carboxyl methyltransferase (IAMT) than to salicylic acid carboxyl methyltransferase (SAMT) and other characterized members utilizing structurally similar substrates. We have modeled the active site of CCMT based on the previously solved crystal structure of SAMT from Clarkia breweri (Zubieta et al., 2003). Altering selected residues in the predicted active sites successfully changed the substrate specificity of CCMT, permitting the utilization of salicylic acid (SA), a preferred substrate of distantly related SABATH family members. We also identified strictly conserved residues among all characterized SABATH enzymes and propose additional structural elements essential to the basic functionality of this family. In addition, we have identified new, nonplant members of this gene family in several microbial taxa, including both prokaryotes and eukaryotes, suggesting that this gene family may have originated in ancestral organisms and subsequently acquired diversified functions in plants.
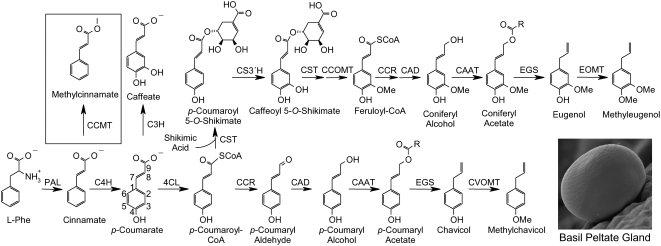
Figure 1.
Relationship of Methylcinnamate Biosynthesis to the Production of Other Phenylpropanoid Pathway–Derived Volatiles in Basil Glandular Trichomes.
Enzyme abbreviations are as follows: PAL, Phe ammonia lyase; CCMT, p-coumarate/cinnamate carboxyl methyltransferase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; C3H, p-coumarate 3-hydroxylase; CST, p-coumaroyl shikimate transferase; CS3′H, p-coumaroyl 5-O-shikimate 3′-hydroxylase; CCOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; CAAT, coniferyl alcohol acetyl transferase; EGS, eugenol (and chavicol) synthase; EOMT, eugenol O-methyltransferase; CVOMT, chavicol O-methyltransferase.
RESULTS
Gas Chromatography–Mass Spectrometry Analysis of Methylcinnamate in Basil Tissues
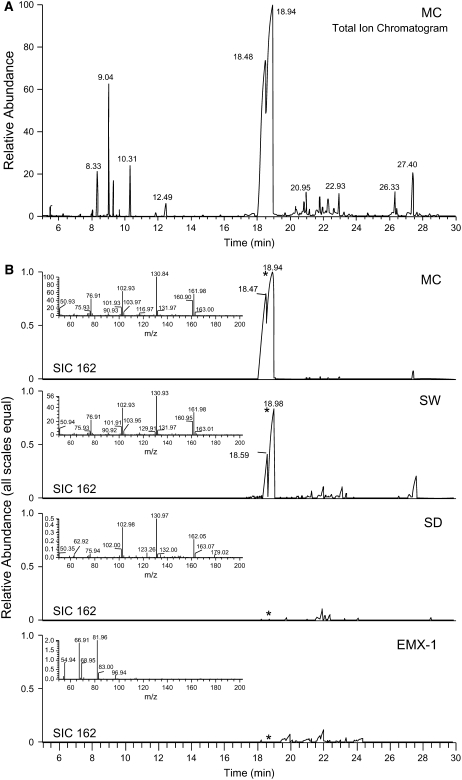
Methylcinnamate is the key volatile component of several basil varieties and chemotypes, one of which, line MC, was selectively bred to produce high levels of this compound. To determine the biosynthetic commitment of line MC toward methylcinnamate, we used gas chromatography–mass spectrometry (GC-MS) analysis to evaluate the quantity of methylcinnamate relative to other volatile components produced in the leaves across the four referenced basil lines. Methylcinnamate constitutes 63% (based on total ion chromatogram peak area) of the total extracted volatiles from line MC leaves (Figure 2A). Line SW contains much smaller levels of methylcinnamate (<10% of that produced by line MC, based on relative peak area); instead, it accumulates large amounts of eugenol. Line SD produces large amounts of citral but only trace amounts of methylcinnamate (Figure 2B); in line EMX-1, no methylcinnamate could be detected in ethyl acetate extracts from leaves, whereas large amounts of the phenylpropene methylchavicol were present instead. These differences in methylcinnamate production between the basil lines were used in a functional genomics approach to identify the enzyme(s) responsible for the formation of this methyl ester in basil.
Figure 2.
Metabolite Profiling Revealed High Levels of Methylcinnamate in Basil Line MC.
(A) Total ion chromatogram from GC-MS analysis of ethyl acetate extracts of line MC young leaves. The peaks at 18.5 and 19 min are Z- and E-methylcinnamate, respectively.
(B) Selected ion chromatograms for m/z 162 (molecular ion of methylcinnamate) for all four basil lines, with scales normalized based on internal standard peak area to allow for direct relative quantitative comparisons. Line MC produces much higher levels of methylcinnamate than other basil lines. Line EMX-1 does not produce any detectable methylcinnamate. Z- and E-methylcinnamate isomerize readily in the presence of light, with the E-isomer more thermodynamically favorable, so it is not clear whether the Z-isomer is also produced biosynthetically by basil or is an artifact of sample handling in the light. Major compounds by retention time (min) are as follows: 8.33, internal standard (trichlorobenzene); 9.04, 1,8-cineole; 9.25, E-β-ocimene; 9.7, cis-δ-terpineol; 10.31, linalool; 11.9, borneol; 12.49, α-terpineol; 18.48, Z-methylcinnamate; 18.94, E-methylcinnamate; 20.35, α-bergamotene; 20.8, α-humulene; 20.95, E-β-farnesene; 21.1, muurola-4,5-diene; 21.8, germacrene D; 21.95, β-selinene; 22.05, α-selinene; 22.6, α-bulnesene; 22.93, γ-cadinene; 24.7, Z-nerolidol; 26.33, cubenol; 27.4, α-cadinol.
Identification of CCMT Activity in Basil Glandular Trichomes
Possible routes to methylcinnamate biosynthesis could include either a methyltransferase that uses AdoMet as a methyl donor and the carboxyl group of cinnamate as the acceptor or an acyltransferase that couples the cinnamoyl moiety from cinnamoyl-CoA to methanol, similar to what was recently shown for the production of the flavor compound methyl anthranilate in Concord grape (Vitis vinifera) (Wang and Luca, 2005). In order to distinguish which enzyme type is in fact responsible for the formation of methylcinnamate in sweet basil, cell-free protein extracts were produced from both young leaf glandular trichomes and whole young leaves of all four basil lines and analyzed for potential enzyme activities.
To evaluate whether methylcinnamate in MC basil trichomes might be formed by an acyltransferase activity, we incubated MC gland protein extracts with cinnamoyl-CoA in the presence of D3-methanol (CD3OH) and analyzed the products by GC-MS. No labeled methylcinnamate was detected in these assays, suggesting that methylcinnamate production in sweet basil is not due to the activity of an acyltransferase.
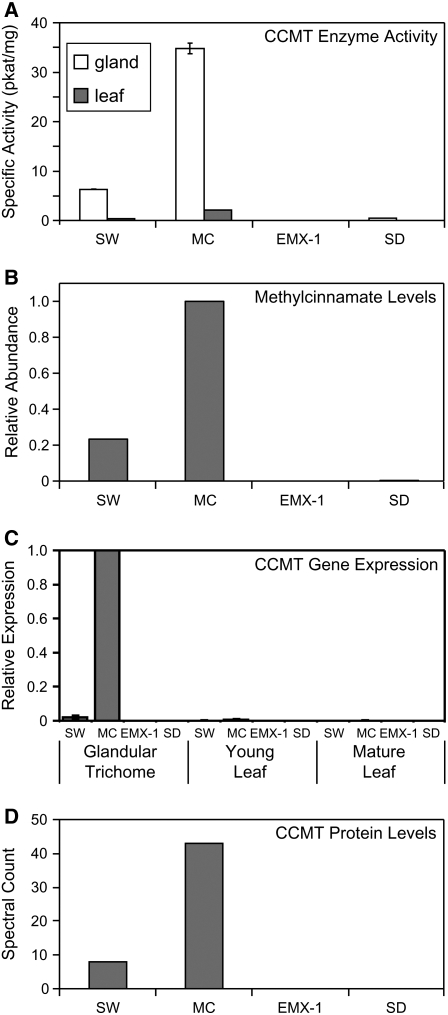
By contrast, the formation of methylcinnamate from cinnamic acid and AdoMet was readily detectable in several basil lines. The highest level of this methyltransferase activity was found in protein extracts from glandular trichomes of line MC (35 pkat/mg; Figure 3A), whereas protein extracts from young leaves of this line possessed only 6% of this activity. This dilution of activity in whole leaves relative to glandular trichomes suggested that CCMT is active specifically in the latter, a pattern that we observed previously for other characterized enzymes of specialized metabolism in sweet basil (Gang et al., 2001, 2002b; Iijima et al., 2004b). CCMT activities in the glandular trichomes from lines SW and SD were 18 and 1.4% of that for line MC, respectively, while no CCMT activity was detected in line EMX-1, a result that correlated positively with methylcinnamate levels in these lines (Figures 2 and 3B). Similar to line MC, a dilution of CCMT activity was observed in leaves versus glandular trichomes of lines SW and SD, again confirming that CCMT expression is also localized specifically to glandular trichomes in these two basil lines (Figure 3A).
Figure 3.
CCMT Enzyme Specific Activity and Transcript Profiling in Tissues of Four Basil Lines: SW, MC, EMX-1, and SD.
(A) CCMT specific activity in protein samples from glandular trichome cells (white bars) and whole young leaf tissues (gray bars).
(B) Relative levels of methylcinnamate in young leaves.
(C) Relative mRNA levels, as determined by quantitative RT-PCR (qRT-PCR), of CCMT in glandular trichomes, whole young leaves, and mature leaves.
(D) Relative levels of CCMT proteins, as determined by a label-free proteomics method.
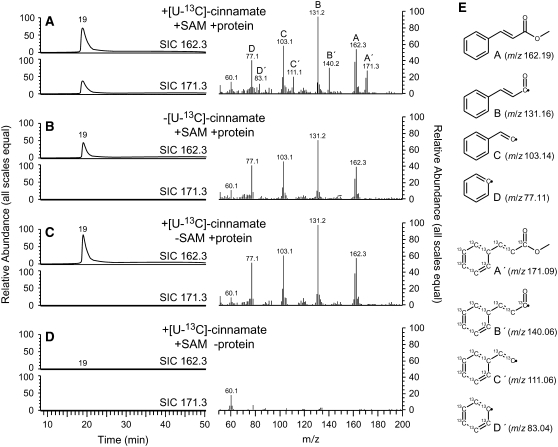
To unequivocally identify the product formed in initial radiolabel-based activity assays, we incubated [U-13C]cinnamate with unlabeled AdoMet and total protein isolated from MC glands. The use of [U-13C]cinnamate permitted us to discriminate between the endogenous and de novo–synthesized methylcinnamate in the GC-MS analysis of the products formed in these assays due to a shift of +9 m/z units for the molecular ion and diagnostic fragment ions in the mass spectrum of the product formed. Results of these assays clearly show de novo methylcinnamate formation (Figure 4).
Figure 4.
Formation of Methylcinnamate in Nondesalted Crude Protein Extracts of MC Glandular Trichomes.
Assay products were analyzed by GC-MS. [U-13C]Cinnamate was provided as substrate in all assays; other assay components are as indicated in the graphs. [U-13C]Methylcinnamate was formed only when AdoMet (SAM) and plant protein extracts were added (A). Endogenous methylcinnamate from the crude protein extracts is present in all samples ([A] to [C]) except for the no-protein control (D). In (E), fragment ions observed in the MS spectra are readily explained and support the identity of the product as [U-13C]methylcinnamate (compared with ions from natural abundance methylcinnamate).
We observed that CCMT activity from protein extracts of MC glands exhibited substrate inhibition, as AdoMet concentrations were increased from 10 μM to 1 mM, with significant inhibition observed at concentrations >200 μM (see Supplemental Figure 1 online), which matches a classical substrate inhibition profile (Cornish-Bowden, 1979). Assays with both nondesalted and desalted crude protein extracts from MC whole leaves or isolated trichomes displayed the same substrate inhibition effect, ruling out the possibility of a small endogenous metabolite present in the protein extracts as the cause of the inhibition. In addition, we used LC-MS to analyze the purity of the AdoMet used in these assays and found no detectible S-adenosylhomocysteine (AdoHcy), although this compound is readily detected by this analytical method (data not shown). This suggests that potential product inhibition by AdoHcy contamination in the AdoMet substrate was not the cause. Interestingly, this substrate inhibition effect was not observed in similar assays with recombinant CCMT enzymes (see below), even at high (2 mM) AdoMet concentrations. At present, the reason for the susceptibility of native CCMT activity but not recombinant CCMT enzymes to inhibition by AdoMet is unknown. This inhibition is likely to be caused by some form of posttranslational modification of the polypeptide chain, such as by proteolytic cleavage, phosphorylation of specific amino acid residues, etc. Future research will seek to answer this interesting question.
Identification of Basil Genes Encoding CCMT
In order to identify the gene(s) encoding CCMT from basil, we generated ESTs (7314 sequences) using a cDNA library constructed from MC glandular trichomes and added these to a larger EST database from the other basil lines described above (23,232 ESTs total). A total of 482 MC ESTs (6.6% of MC ESTs) in this database represent 241 cDNA clones that assemble into 22 contigs (two singletons) and encode proteins that are similar to SAMT-like proteins in the UniProt database. Three additional ESTs from line SD were assembled into the MC contigs, while no similar ESTs were identified from basil lines SW or EMX-1. The largest of these contigs consists of 132 ESTs and represents the fourth largest single contig found in the entire basil glandular trichome EST database (the largest composed of ESTs entirely from a single basil line).
Twelve of the 22 contigs contained full-length open reading frames, whereas 6 contigs encoded partial open reading frames of 200 amino acids or more. Alignment of the translated amino acids sequences and subsequent cluster analysis revealed that the 22 contigs represent allelic versions of three distinct although very similar (96 to 97% amino acids sequence identity) proteins identified as new members of the SABATH family. These three isoforms were designated CCMT1, CCMT2, and CCMT3 (see below for activity determination) in order of decreasing number of total cDNA clones (allelic variants) for each isoform. The basil CCMT proteins are 373 amino acids in length and 57% identical (75% similar) to Arabidopsis thaliana IAMT1, the closest biochemically characterized relative to basil CCMTs.
To estimate the number of CCMTs present in basil lines MC, SW, and SD and to determine why line EMX-1 lacked CCMT transcripts, we performed DNA gel blot analysis using genomic DNA from all four basil lines (see Supplemental Figure 2 online). Line MC appeared to possess several CCMTs, while lines SW, SD, and EMX-1 all appeared to possess fewer. Another interesting finding was that the banding pattern was nearly identical for lines SD and EMX-1, with the pattern for SW showing some similarities to these two, while the pattern for MC showed little resemblance to the other lines. The increased number of copies and the distinct pattern suggest that gene duplication(s) may have occurred in line MC relative to the other lines. It is possible that changes in genome structure and the distribution of CCMT copies in line MC are linked to the increased pattern of CCMT expression in this line. This approach shows the relative differences between basil lines, while sequencing of the genomes of these basil lines will provide in the future the exact copy number of CCMT genes.
qRT-PCR Analysis of CCMT Expression
To confirm the relative expression of CCMTs between basil lines predicted by EST abundance and to determine whether their expression is indeed specific to trichomes, RNA was isolated from glandular trichomes, young and mature leaves, young and mature stem sections, and roots of all four basil lines grown under standardized conditions. Isolated RNA was used for relative qRT-PCR (Figure 3C). CCMT transcripts were detected in lines SW, MC, and SD but not line EMX-1, despite high expression of the CVOMT gene in this line, used as a positive control. The glandular trichomes of line MC showed the highest CCMT expression, while expression in glands of line SW was only 2% of that for MC, and expression in line SD was barely detectable. CCMT expression in other tissues of line MC, such as leaves, was <1% of that for glands. For the three lines with detectable CCMT expression, the level of expression for leaf tissues decreased with increased leaf size/age (Figure 3C), consistent with gland-specific expression and a dilution effect of glandular trichome cells and transcripts by other leaf tissue cell types and their transcripts as leaves expand.
Proteomics Analysis Supports the Presence of All CCMT Isoforms in MC Trichomes
We used a shotgun proteomics approach to address the issue of whether all three major CCMT isoform proteins identified from the MC EST library were in fact present in the trichome proteome of line MC. In these experiments, >400 proteins were identified from 809 peptide sequences. We performed parallel experiments with the other three basil lines (Z. Xie, J. Kapteyn, and D. Gang, unpublished data), since several studies suggest that the sampling frequency (referred to as spectral count or label-free quantification) of proteins in a shotgun proteomics study corresponds to the abundance of those proteins in the biological sample (Pang et al., 2002; Gao et al., 2003; Liu et al., 2004). We identified 43 spectra corresponding to peptides from CCMTs present in line MC (5.3% of total spectra for this line, similar to 6.6% of ESTs) and 8 spectra in line SW, while no spectra were observed in lines SD and EMX. These results (Figure 3D) correlate well with the pattern observed for CCMT gene expression and activity in these lines (Figures 3A and 3C), suggesting that the levels of production of methylcinnamate in these lines may be determined by the activity of the CCMT enzyme(s) and regulated primarily at the level of gene transcription. Moreover, 11 unique peptide sequences were identified for the CCMTs (see Supplemental Table 1 online), one of which was unique to the proteome of line SW. Each of the three CCMT isoforms identified based on ESTs yielded a distinguishing tryptic peptide spanning amino acids 197 to 209 from the glandular trichome proteome of line MC (see Supplemental Table 1 online), clearly demonstrating that these three CCMT isoforms are in fact present in translated form in the glandular trichomes of line MC and providing a biological link between the genes identified using in silico methods and observed enzyme activity.
Biochemical Characterization of CCMT Isoforms
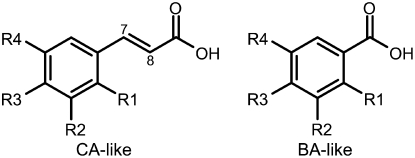
To determine the biochemical function of the three isolated CCMT isoforms (CCMT1, CCMT2, and CCMT3), their coding regions were subcloned into the expression vector pET-28a, which contains an N-terminal polyhistidine (6×His) tag, and the proteins were overexpressed in Escherichia coli. When cinnamate (5 mM) was added to the growth medium during CCMT overexpression, only cultures actively expressing a CCMT isoform produced methylcinnamate detectable by GC-MS (Figure 5). To verify whether cinnamate is the best substrate for these enzymes, affinity-purified recombinant CCMT proteins were assayed with a variety of potential substrates. Compounds tested included cinnamate as well as structurally related molecules with various substitutions on the aromatic ring and/or different side chain lengths and configurations. Of 20 tested compounds, the same 5 compounds served as substrates for all three isoforms: E-cinnamate (CA), p-coumarate, hydro-p-coumarate (4-hydroxyhydrocinnamate), hydrocinnamate, and benzoate (BA). The highest activity was found with CA, identifying it as a primary substrate for all three isoforms. CCMT2 had the highest specific activity with CA (6.6 nkat/mg purified protein), followed by CCMT1 and CCMT3 (59 and 32% of CCMT2 activity, respectively). While there was no difference in relative activities between the three isoforms for hydro-p-coumarate and BA (∼22 and ∼8% of activity with CA, respectively) (Table 1, Figure 6), significant differences were found for hydrocinnamate and p-coumarate. CCMT3 exhibited ∼2.3-fold higher relative activity with p-coumarate compared with CCMT1 and CCMT2, whose activities were nearly identical (Table 1). CCMT3 also displayed the highest activity with hydrocinnamate relative to CA, followed by CCMT1 and CCMT2 (39, 21, and 6% of each enzyme's maximum activity with CA, respectively). Interestingly, only CCMT1 showed a very low level of activity with m-coumarate (Table 1). The activity of all three CCMT enzymes with p-coumarate and the lack thereof with other cinnamate derivatives having substitutions elsewhere on the phenyl ring indicate that the substrate binding site may be rather restricted in these enzymes.
Figure 5.
GC-MS Analysis.
(A) Extracts of culture medium from E. coli cells expressing CCMT1 in the presence of cinnamate.
(B) Extracts of culture medium from E. coli cells expressing only the empty vector as a control in the presence of cinnamate.
(C) The product produced by the recombinant purified CCMT1.
Insets show mass spectra at time 19 min, corresponding in (A) and (C) to methylcinnamate formed by E. coli cells expressing CCMT1 and by purified recombinant CCMT1 protein. Cultures not expressing CCMT enzymes (B) were not able to produce methylcinnamate.
Table 1.
Relative Activities of Functionally Expressed Basil CCMTs and CCMT1-H160M with Different Substrates
| Substitution Pattern
|
Relative Specific Activity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrate | Structure | R1 | R2 | R3 | R4 | CCMT1 | CCMT2 | CCMT3 | CCMT1-H160M |
| CA-like | |||||||||
| trans-Cinnamic acid (CA) | H | H | H | H | 100 (1.7) | 100 (7.1) | 100 (15.1) | 100 (7.5) | |
| Hydrocinnamic acida | H | H | H | H | 21.4 (1.0) | 6.8 (3.4) | 39.3 (5.5) | 21.8 (1.9) | |
| p-Coumaric acid (pCA) | H | H | OH | H | 29.6 (3.9) | 24.5 (5.7) | 63.5 (8.0) | 80.2 (1.6) | |
| 4-Hydroxyhydrocinnamic acida | H | H | OH | H | 22.4 (4.8) | 19.1 (6.5) | 23.5 (2.0) | 0.9 (0.7) | |
| m-Coumaric acid | H | OH | H | H | 6.8 (1.5) | 0 | 0 | 0 | |
| o-Coumaric acid | OH | H | H | H | 0 | 0 | 0 | 0 | |
| Caffeic acid | H | OH | OH | H | 0 | 0 | 0 | 0 | |
| Hydrocaffeic acida | H | OH | OH | H | 0 | 0 | 0 | 0 | |
| Ferulic acid | H | OMe | OH | H | 0 | 0 | 0 | 0 | |
| Hydroferulic acida | H | OMe | OH | H | 0 | 0 | 0 | 0 | |
| 3,4-Dimethoxycinnamic acid | H | OMe | OMe | H | 0 | 0 | 0 | 0 | |
| Sinapic acid | H | OMe | OH | OMe | 0 | 0 | 0 | 0 | |
| BA-like | |||||||||
| Benzoic acid (BA) | H | H | H | H | 7.8 (0.8) | 10.1 (4.8) | 6.2 (1.3) | 161.9 (2.2) | |
| Salicylic acid (SA) | OH | H | H | H | 0 | 0 | 0 | 61.1 (0.4) | |
| Anthranilic acid | NH2 | H | H | H | 0 | 0 | 0 | 20.5 (0.7) | |
| Shikimic acid | H | OH | OH | OH | 0 | 0 | 0 | 0 | |
| Other | |||||||||
| Phenylacetic acid | 0 | 0 | 0 | 0 | |||||
| Phenylpyruvic acid | 0 | 0 | 0 | 0 | |||||
| Indole-3-acetic acid | 0 | 0 | 0 | 0 | |||||
| Jasmonic acid | 0 | 0 | 0 | 0 | |||||
These substrates have a single carbon bond between C7 and C8 and not the double bond shown in the generalized structures in the table.
Figure 6.
General Structures for CA-Like and BA-Like Compounds.
The molecular masses of the three recombinant CCMTs determined by gel filtration chromatography were ∼88 kD, whereas on SDS-PAGE the denatured proteins exhibited a single band corresponding to a molecular mass of ∼44 kD, suggesting that the active enzymes exist as homodimers in solution. CCMT proteins were active from pH 6.0 to 9.0, with maximum activity at pH 7.5, 90% of maximum at pH 7.0 and 8.0, and <50% of maximum at pH 6.0 and 9.0. CCMTs did not require cations for their activities; however, strong inhibition (>65%) was observed when Cu2+, Ni2+, or Zn2+ was added at 5 mM to the reaction. Fe2+, Mg2+, Mn2+, and NH4+ led to mild inhibition (<35%), and the addition of Ca2+, K+, or Na+ had no effect on enzyme activities. Since recombinant proteins were purified using Ni2+ agarose columns, the potential inhibitory effect of Ni2+ on enzyme activity was evaluated at six different concentrations: 0.01, 0.05, 0.1, 0.5, 1.0, and 5.0 mM. These experiments showed that Ni2+ concentrations of <1 mM had no effect on enzyme activity.
Steady state kinetics of each CCMT enzyme revealed that the recombinant proteins had a high affinity toward AdoMet, with apparent Km values ranging from 1 to 13 μM depending on the second substrate and CCMT isoform tested (Table 2). All three enzymes displayed similar apparent Km values for CA, ranging from 124 μM (CCMT1) to 196 μM (CCMT3). CCMT1 and CCMT2 had lower apparent Km values for p-coumarate and BA (from 1.8- to 2.7-fold lower) compared with that for CA, in contrast with CCMT3, which had an 11-fold lower apparent Km value for BA and an almost 2-fold higher apparent Km value toward p-coumarate. CCMT1 and CCMT2 had nearly identical catalytic efficiencies (kcat/Km ratio) with CA, which were almost fivefold higher than that for CCMT3, suggesting that the contribution of CCMT3 to the formation of methylcinnamate in the plant could be small. Interestingly, CCMT2 can use p-coumarate and CA with nearly equal efficiency, while for CCMT1 the difference between catalytic efficiencies with these two substrates was 1.3-fold, favoring CA. The similar catalytic efficiencies of CCMT1 and CCMT2 toward p-coumarate and CA in addition to the higher Vmax with CA suggest that these enzymes can use both substrates efficiently in vivo. Thus, we designated the identified enzymes CCMT, for S-adenosyl-l-Met:cinnamate/p-coumarate carboxyl methyltransferases. These parameters were similar to apparent kinetic parameters determined for CCMT activity in crude protein extracts from line MC glandular trichomes (e.g., apparent Km of ∼224 ± 34 and ∼245 ± 34 μM for CA and p-coumarate, respectively).
Table 2.
Comparison of Km, Kcat, and Kcat/Km Values for Basil CCMT Isoforms and CCMT1-H160M
| Substrate
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Property | Enzyme | CA | pCA | BA | SA | AdoMet (CA) | AdoMet (pCA) | AdoMet (BA) | AdoMet (SA) |
| Km (μM) | CCMT1 | 124 (18) | 70.4 (3.0) | 45.5 (2.3) | 5.42 (0.78) | 1.14 (0.10) | 2.03 (0.09) | ||
| CCMT2 | 189 (33) | 77.0 (9.3) | 76.2 (6.2) | 13.3 (1.5) | 2.59 (0.19) | 2.60 (0.15) | |||
| CCMT3 | 196 (23) | 388 (42) | 17.3 (1.0) | 8.14 (0.43) | 3.96 (0.20) | 1.17 (0.10) | |||
| CCMT1-H160M | 577 (23) | 1200 (80) | 2430 (80) | 422 (35) | 5.70 (0.55) | 4.22 (0.43) | 11.1 (1.2) | 8.2 (0.9) | |
| Kcat | CCMT1 | 0.188 (0.007) | 0.084 (0.004) | 0.015 (0.001) | 0.198 (0.005) | 0.081 (0.003) | 0.018 (0) | ||
| CCMT2 | 0.251 (0.016) | 0.134 (0.003) | 0.020 (0) | 0.317 (0.015) | 0.106 (0.004) | 0.024 (0.001) | |||
| CCMT3 | 0.063 (0.001) | 0.050 (0.002) | 0.003 (0) | 0.090 (0.003) | 0.054 (0.003) | 0.004 (0) | |||
| CCMT1-H160M | 0.019 (0.001) | 0.009 (0.001) | 0.035 (0.002) | 0.003 (0) | 0.013 (0.001) | 0.008 (0.001) | 0.028 (0.002) | 0.006 (0) | |
| Kcat/Km | CCMT1 | 1,560 (165) | 1,190 (9) | 323 (6) | 37,700 (4500) | 72,100 (2900) | 8,720 (400) | ||
| CCMT2 | 1,390 (180) | 1,790 (210) | 266 (19) | 24,100 (1400) | 41,500 (1800) | 9,440 (700) | |||
| CCMT3 | 327 (33) | 132 (11) | 192 (8) | 11,100 (400) | 14,700 (500) | 3,590 (200) | |||
| CCMT1-H160M | 33.4 (1.5) | 7.21 (0.12) | 14.4 (0.6) | 7.19 (0.59) | 2,180 (230) | 2,020 (210) | 2,490 (110) | 690 (60) | |
Structural Modeling of CCMT Proteins
Alignment of the basil CCMT genes with IAMT, SAMT, and several other biochemically characterized members of the SABATH family (Figure 7) revealed that all of the residues implicated in AdoMet binding, as well as other active site residues previously identified in the crystal structure of SAMT from C. breweri (Cb SAMT) (Zubieta et al., 2003), are conserved in basil CCMTs. More specifically, this includes residues equivalent to Trp-151 and Gln-25 of Cb SAMT, which have been implicated (Zubieta et al., 2003) as hydrogen bond donors for salicylate to ensure proper orientation and proximity to the donated methyl group of AdoMet. The most striking difference between the CCMT proteins and their closest relative, IAMT, is a gap of 11 amino acids in CCMT spanning residues 126 to 136 of IAMT1. These residues were not predicted to be in the active site based on the initial model of IAMT (Zubieta et al., 2003), and the impact of the gap on CCMT function could not be determined based on the alignment alone.
Figure 7.
Alignment of the CCMT1 Protein Sequence with Selected Characterized Members of the SABATH Family.
The important active site and substrate binding residues determined for C. breweri SAMT by Zubieta et al. (2003) are highlighted with stars for AdoMet/AdoHcy binding residues and with circles for salicylate binding residues. Boxshade was used to highlight residues, with white text on black background indicating an identical residue for more than half of the sequences.
To further investigate the role of specific residues in substrate binding and CCMT catalysis, we used the solved crystal structure of Cb SAMT to generate structural models of the three basil CCMTs and snapdragon (Antirrhinum majus) Am SAMT. We then compared these models against the structure of Cb SAMT as well as a modeled structure of Arabidopsis IAMT. The first models generated were based only on alignments between Cb SAMT and Ob CCMT amino acid sequences. These initial models suggested that the residues corresponding to Cb SAMT Met-150 and Met-308 are altered in CCMTs to His-160 and a null residue (deleted), respectively. These two Met residues have been proposed to provide a molecular clamp in Cb SAMT for the aromatic ring of SA (Zubieta et al., 2003). Based on these initial results, we replaced the His-160 codon in CCMT with a Met codon (CCMT1-H160M mutant) and generated the reciprocal mutation in Am SAMT (SAMT-M150H mutant) to determine whether these changes might alter the substrate preference of the enzymes as the models predicted. In an effort to generate the molecular clamp mentioned above, we also created a mutant CCMT1 protein with an introduced Met codon at amino acid position 323 (CCMT1-M323 insertional mutant) as well as a mutant CCMT1 protein containing both modifications (CCMT1-H160M/M323 double mutant).
Insertion of the Met residue at position 323 of CCMT1 completely abolished the activity of the enzyme in both the single mutant and the double mutant. The lack of any catalytic activity in the insertional mutant suggested that the initial alignment was likely incorrect at this position and that insertion of the Met residue led to a shift in structure affecting the active site or some other critical structural component of the enzyme, causing the protein to be nonfunctional. Later multiple sequence alignments with additional SABATH family member sequences, some newly available, led us to modify our initial alignments and structural models in which we had created a gap opposite Cb SAMT Met-308 and instead align CCMT Cys-322 at this position. Future research will pursue a C322M substitution to determine the effects of this mutation on the activity of the enzyme. CCMT1 with the single H160M change, however, retained activity with CA while demonstrating a dramatically increased activity toward BA as a substrate (Tables 1 and 2). This CCMT1-H160M mutant protein also acquired the ability to accommodate SA as a substrate, although with lower activity than that toward CA. Although Arabidopsis IAMT1 was the closest relative to CCMTs, none of the CCMT1 mutants were able to utilize indole-3-acetic acid as a substrate, suggesting that some other feature of the protein or active site conferred the selectivity of CCMT against indole-3-acetic acid, such as the substitution of a larger, more polar Ser-243 in CCMT versus the comparably positioned Gly-256 in IAMT. Once the crystal structure of IAMT is solved, this question may be able to be answered. New CCMT and Am SAMT models based on the revised alignments (Figure 7) predicted several nonconserved residues to be in the active site cavity and suggested that they may contribute to or be responsible for the differential substrate preference of these enzymes.
Docking Experiments Explain the Differences in Substrate Preferences of CCMTs and SAMTs
To relate the modeled protein structures to the experimentally determined substrate specificity of recombinant native proteins (CCMT1, CCMT2, CCMT3, and Am SAMT) and mutant proteins (CCMT1-H160M and Am SAMT M163H), we performed docking experiments using GOLD version 3.1.1. Figure 8 shows representative structures for CCMT1 and CCMT1-H160M with several potential substrates docked and for Am SAMT with SA docked.
Figure 8.
Structural Models of Ob CCMT1, Ob CCMT1-H160M, and Am SAMT Proteins with Potential Substrates Docked into the Active Sites.
Models are for Ob CCMT1 ([A], [C], [E], and [G]), Ob CCMT1-H160M ([B], [F], and [H]), and Am SAMT (D). Only those protein residues most relevant to substrate binding (possessing at least one amino acid residue atom within 5 Å of a substrate atom) are shown. (A) and (B) show docking with cinnamate; (C) shows docking with p-coumarate; (D) to (F) show docking with salicylate; and (G) and (H) show docking with BA.
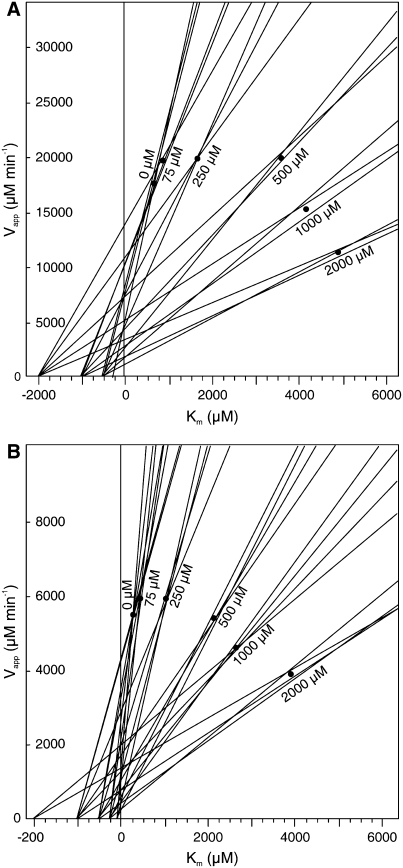
One of the most interesting of the docking experiment results was that of SA to the CCMT isoforms, which suggested that SA could be a good inhibitor of CCMT enzymes because it appeared to bind well in the active site in an orientation that prevented methyl group transfer (Figure 8E). In vitro inhibition studies validated this prediction: SA was found to be a good inhibitor of CA methylation by both CCMT1 and CCMT3 enzymes (CCMT2 was not tested due to its high similarity to CCMT1). However, the inhibition of CCMT1 and CCMT3 by SA did not fit classical competitive, noncompetitive, uncompetitive, or mixed linear inhibition models (Figure 9; see Supplemental Figure 4 online). Instead, it appeared to fit parabolic inhibition models most closely, with the inhibition approximating competitive inhibition at low (<250 μM) SA concentrations, leading to apparent Ki values of ∼85 and 90 μM for SA with CCMT3 and CCMT1, respectively. However, at higher concentrations (500 to 2000 μM), SA appeared to function more like a noncompetitive inhibitor. These results have significant implications regarding the catalytic mechanism and function of the CCMT enzymes. First, they suggest that SA binds with differing affinities to at least two sites on the functional CCMT proteins. Since these proteins exist as homodimers in solution, it is possible that these sites are the active sites of each of the monomers. These results also suggest that nonproductive binding of substrate to one site affects the affinity of the other (active) site for substrate. Such reaction kinetics have not yet been reported for members of the SABATH family of carboxyl methyltransferases. More detailed kinetics analysis will be required to determine the binding order of substrates and to identify where SA binds to CCMT enzymes.
Figure 9.
Direct Linear Plots of Velocity Versus Km for CCMT1 and CCMT3 with Varying SA and CA Concentrations.
Lines were calculated using the equation Vapp = v + v/s × Km, according to Eisenthal and Cornish-Bowden (1974), where v (the y intercept) is the observed velocity of the reaction at a given substrate concentration (s) and the term v/s is the slope of the line. SA acts like a competitive inhibitor at low concentrations and like a mixed inhibitor at high concentrations for both enzymes CCMT1 (A) and CCMT3 (B).
Phylogenetic Relationships of CCMTs within the SABATH Family
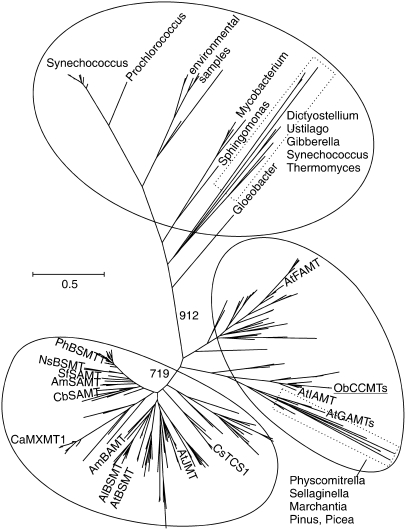
We conducted an exhaustive search of currently available protein and genomic sequence databases to identify all sequences available (from all organisms) having homology with the SABATH gene family. In total, >400 sequences were identified from dozens of plant taxa (including Cb SAMT, all known and characterized SABATH members, and our basil CCMT sequences) as well as many sequences representing nonplant genes and sequences derived from marine environmental sampling. A maximum likelihood tree produced from SABATH family sequences representing as much taxonomic diversity as sequence and alignment quality would permit (Figure 10) suggests that the SABATH family is split into three primary clusters. The first cluster comprises microbial sequences that are distinct from terrestrial plant sequences. The second cluster consists of two subclades that are not distinct based on bootstrap support and includes the basil CCMT genes, At IAMT, and Arabidopsis farnesoic and gibberellic acid carboxyl methyltransferases (At FAMT and At GAMT), along with several groups of genes of unknown function from Arabidopsis, Brassica napus, poplar (Populus sp), rice (Oryza sativa), and other monocots as well as all genes identified from conifers and lower plants. The third major cluster comprises sequences from higher plants and includes the characterized SAMTs, BSMTs, BAMT, JMT, and the N-methyltransferases as well as several distinct clusters of uncharacterized monocot and eudicot SAMT-like sequences. It was surprising that basil CCMT does not cluster with benzenoid carboxyl methyltransferases (SAMTs, BSMTs, BAMT, etc.), whereas jasmonic acid carboxyl methyltransferase and the N-methyltransferases do.
Figure 10.
Phylogenetic Tree of the SABATH Family of Carboxyl Methyltransferases Produced Using Maximum Likelihood.
The biochemically characterized plant proteins shown in Figure 7 are indicated, as are two gibberellic acid carboxyl methyltransferases from Arabidopsis (gi:62319595 [GAMT1] and gi:30696725 [GAMT2]). The locations of microbial and lower plant taxa are also shown.
DISCUSSION
Conservation of Primary Structure in SABATH Family Proteins
The newly available basil CCMT protein sequences along with those from recently cloned and characterized SABATH genes allowed us to obtain an updated perspective on important primary sequence features of SABATH family proteins and to identify regions of conservation among the protein sequences of the expanded gene family. While the residues surrounding the presumed active site cavities can be quite variable, those that were previously indicated by the solved crystal structure of C. breweri SAMT to be involved in AdoMet/AdoHcy binding (Zubieta et al., 2003) are well conserved in basil CCMTs, as they are in most plant-derived members of this gene family that have been characterized to date (indicated by stars in Figure 7).
The strict conservation of many residues across the SABATH proteins characterized to date suggests an essential role in this enzyme family, even if this role is not yet known. There are 24 amino acid residues distributed throughout the primary amino acid sequences that are absolutely conserved in the 14 biochemically characterized SABATH family members from plants but that are not identified as active site residues or implicated as having direct interaction with the substrates or products based on solved crystal structures and structural models. Thus, individual solved crystal structures or models, while essential to help us understand the function of a protein and powerful for predictive purposes, may not by themselves explain the contribution of each building block of an enzyme structure to its function.
Substrate Preference and Active Site Structure
The characteristic feature of the three basil CCMT proteins is their strong preference for p-coumarate and CA (as well as saturated side chain analogs) and their complete lack of activity with SA and most of the other substrates preferred by other characterized members of the SABATH family, with the exception of a low level of activity with BA (Table 1). Previously, the benzenoid-specific enzymes of the SABATH family were subdivided based on their methyl acceptor preferences in two categories, SAMT and BAMT types (Effmert et al., 2005). The SAMT types share the conserved Met-150/Met-308 clamp for the aromatic ring of SA and demonstrate a strong preference for SA over BA as a substrate (Zubieta et al., 2003). All other SABATH family members, including basil CCMTs, possess a His residue at the site equivalent to Cb SAMT Met-150, while residues aligned with Cb SAMT Met-308 are somewhat more divergent. However, as Chen et al. (2003) pointed out, this active site feature is not absolutely essential for the utilization of SA as a substrate, as both At BSMT1 and Al BSMT1 can methylate SA (while possessing a His at this position), although they do strongly prefer BA as substrate. Our docking results (Figure 8) also explain why most members of the SABATH family of carboxyl methyltransferases that are able to efficiently use SA as a substrate have replaced the otherwise highly conserved His residue with Met. Met is not able to form the same hydrogen bond as His, which allows SA to retain its intramolecular hydrogen bond between the 2-hydroxyl group and the carboxylate carbonyl, thereby allowing the carboxylate group to bind in a position and orientation suitable for attack on the reactive methyl of AdoMet. Evaluation of the CCMT1-H160M mutant enzyme activity further supports these conclusions regarding general binding cavity structure and substrate binding, in which SA and BA became much better substrates for this mutated version of the CCMT1 enzyme compared with the nonmutated recombinant form (Tables 1 and 2, Figure 8).
Evolution of Methylcinnamate Production in Plants
Identification of the basil CCMT genes answers a long-standing question regarding the biogenesis of methylcinnamate and methyl-p-coumarate in plants and demonstrates that members of the SABATH family of carboxyl methyltransferases are responsible for the production of these compounds in sweet basil and presumably in many other plants as well. The strong substrate preference and high catalytic efficiencies, as well as the low apparent Km values of CCMTs for p-coumarate and CA, further substantiate the biological role of these enzymes in the production of the methylated forms of these compounds. An alternative means of production, involving an acyltransferase responsible for the transfer of cinnamate/p-coumarate from the corresponding CoA esters to methanol, was eliminated as a potential biosynthetic route in these plants. A third alternative route via a second type of acyltransferase has also been proposed. A 1-O-trans-cinnamoyl-β-d-glucopyranose:alcohol cinnamoyltransferase was partially purified from fruits of Physalis peruviana (cape gooseberry) and was shown to be capable of transferring cinnamate from cinnamoyl-β-d-glucopyranose to methanol, forming methylcinnamate (Latza and Berger, 1997). This avenue of synthesis of methylcinnamate is supported by the recent identification of a UDP-glucose:cinnamate glucosyltransferase from strawberry (Fragaria sp) (Lunkenbein et al., 2006), which also makes methyl and ethyl esters of cinnamic acid, although activity for the subsequent acyltransferase has not yet been reported. Thus, it may be possible that the production of methylcinnamate could have evolved convergently via different biochemical mechanisms in different plant lineages, although this has yet to be proven conclusively.
The distribution of methylcinnamate production throughout several plant families, presumably via biosynthesis by the CCMT homologs identified in this phylogenetic analysis, suggests the possibility that this function may have arisen early in plant evolution, possibly prior to the divergence of monocots and dicots. Several sequence homologs of basil CCMT and Arabidopsis IAMT were found for rice, but it remains to be determined whether these genes encode true CCMTs or IAMTs. The biochemical functions of the related genes from lower plants and of the microbial enzymes are also unknown. Future functional characterization of these genes will be essential to identify whether CCMT activity represents an ancient biochemical process that has been conserved throughout the history of plant evolution or whether it arose via convergent (repeated) evolution in multiple instances throughout evolution of the plant kingdom (Pichersky and Gang, 2000; Gang, 2005). In either case, the evolutionary selective pressures responsible for driving the development of this biochemical function remain to be determined, and future expanded functional genomics data, coupled with detailed metabolic profiling of the respective species, will provide keys to solving this puzzle.
Origin and Evolution of the SABATH Family
SABATH family members were identified in diverse photosynthetic organisms, including three taxa of cyanobacteria, the phytoplankton Thalassiosira, and taxa representing the major lineages of terrestrial plants, including the bryophytes Physcomitrella (moss) and Marchantia (liverwort), the lycophyte Selaginella, several gymnosperm taxa, and numerous monocot and eudicot flowering plants, again suggesting the origin of this gene family in taxa ancestral to all land plant lineages. Among eukaryotes, SABATH homologs were distributed throughout several major groups in addition to the terrestrial plants, including fungi, amoebozoa, and stramenophiles. No SABATH sequences were identified in any taxa from the Archaebacteria. The possibility of lateral transfer events between divergent taxa cannot be eliminated, and homologs were observed in both maize (Zea mays) and its fungal pathogen Ustilago maydis, suggesting potential opportunities for horizontal gene transfer. The identification of SABATH family members in taxa spanning many of the major branches of the tree of life suggests the possibility of an early evolutionary origin of this gene family in an ancient common ancestor and an evolutionarily conserved functionality across diverse taxa.
Phylogenetic analysis based on the maximum likelihood tree (Figure 10) suggests that the progenitor of the SABATH gene family evolved long before the divergence of vascular plants, since several taxa of eubacteria representing at least three different major groups, the cyanobacteria Synechococcus, Prochlorococcus, and Gloeobacter, all possess SABATH family members. The presence of these genes among a group of taxa thought to be involved in endosymbiosis with early eukaryotes suggests that this family of genes may have been inherited from prokaryotes. BLAST searches, however, did not result in the identification of SABATH homologs among any sequence data available from green or red algae, including Chlamydomonas reinhardtii and Cyanidioschyzon merolae, for which the entire or extensive genome sequences are available, indicating potential gene loss in these taxa. The identification of a homolog in the genome sequence of the diatom Thalassiosira, however, suggests that the loss of this gene in red algae may have occurred after the secondary endosymbiotic event that gave rise to the modern brown algae. This raises interesting questions about the biological role of these SABATH homologs in the taxa in which they have been maintained versus those for which the sequence has been lost. Nevertheless, the tree topology suggests that plant SABATH genes most likely originated from an ancient ancestor shared with the microbial taxa and that the ancestral form of this gene may have undergone a duplication early in plant evolutionary history prior to the divergence of monocots and eudicots, with subsequent duplication events and additional diversification taking place later to generate the contemporary groups and diverse functions within the gene family seen here.
METHODS
CCMT Activity Assays
CCMT activity was determined by measuring the formation of radiolabeled methylcinnamate by total protein extracted from isolated glandular trichomes and intact leaves of four basil (Ocimum basilicum) lines. Assays were performed in 50-μL reactions consisting of 50 mM potassium phosphate buffer, pH 8.0, 9.33 μM S-[methyl-14C]adenosyl-L-Met, 1 mM cinnamate, and 200 ng to 10 μg of protein. Reactions were incubated at 30°C for 30 to 60 min and stopped by the addition of 6 μL of 6 n HCl. Labeled methylcinnamate was extracted with ethyl acetate, and a portion of the separated organic phase was used for scintillation counting. Assays were performed in quadruplicate for each protein sample. Controls included assays with all reagents to which no protein was added and assays including protein with cinnamate omitted. Assays to detect the activity of an acyltransferase capable of producing methylcinnamate were performed using the same assay conditions (similar to those used previously for acyltransferase assays [Gang et al., 2002a]), with the exceptions that 50 μM cinnamoyl-CoA (synthesized according to a method reported previously [Beuerle and Pichersky, 2002] and purified using a Chromabond C18 column [Macherey-Nagel] and 25 μM D3-methanol [343854; Sigma-Aldrich]) was substituted for AdoMet and cinnamate, the reactions were incubated for 2 h, and detection was performed via GC-MS rather than radiometrically.
The standard assay mixture for the recombinant proteins consisted of 20 μL of 250 mM potassium phosphate buffer at pH 7.5 with 10 mM β-mercaptoethanol, 1 μL of 50 mM disodium EDTA, 2 μL of acid substrate dissolved in DMSO, 0.2 to 2 μL of enzyme solution (48 to 480 ng), 5 μL of radiolabeled AdoMet (389 μM; specific activity of 10.28 mCi/mmol), and water to a final volume of 100 μL. After incubation for 15 to 30 min, the reactions were stopped by the addition of 150 μL of organic solvent (hexane or ethyl acetate) and vortexing for 5 s After centrifugation (1 min at 16,600g) to separate the phases, 75 μL of the organic phase was used for liquid scintillation counting. Verification of the assay product was determined using nonradiolabeled AdoMet in a 100× scaled reaction and analyzing the concentrated organic phase by GC-MS.
GC-MS Analysis of Basil Tissues and CCMT Product
Volatiles were extracted as reported previously (Ma and Gang, 2005) by adding 2 mL of ethyl acetate containing internal standard (1,3,4-trichlorobenzene) to 0.5 g of fresh young basil leaves (<1.5 cm in length) and shaking overnight. A portion (100 μL) of the ethyl acetate extract was removed and used for analysis by GC-MS. Areas for all chromatogram peaks for a given sample were normalized to internal standard peak area to allow for the relative quantitation of methylcinnamate and other volatile compounds between samples. For the analysis of methylcinnamate production by Escherichia coli cells expressing recombinant CCMT, the culture medium (25 mL), grown as described below, was extracted with 5 mL of hexane and the hexane phase was subsequently concentrated to 200 μL and analyzed by GC-MS.
The identity of the product formed in the radiometric CCMT assays was confirmed by GC-MS analysis of ethyl acetate extracts from assays performed using stably labeled cinnamate and unlabeled AdoMet. Briefly, [U-13C]l-Phe (Cambridge Isotope Laboratories) was enzymatically converted into cinnamic acid using Phe ammonia-lyase from Rhodotorula glutinis (P1016; Sigma-Aldrich) and subsequently purified. Purified [U-13C]cinnamate was resuspended in ethanol, and the purity and concentration were checked using thin layer chromatography. CCMT enzyme assays were performed as described above with the exception of a higher AdoMet concentration and longer assay period (200 μM, 3 h). The product was extracted with ethyl acetate and analyzed by GC-MS. Controls were performed as described above, with the addition of assays from which AdoMet was omitted. Methylcinnamate was identified via library matching of mass spectra, and the mass spectrum and fragment pattern of labeled methylcinnamate were compared with those of an unlabeled, authentic methylcinnamate standard. Verification of the product formed by recombinant CCMT enzyme was performed by adding nonradiolabeled AdoMet to a 100× scaled reaction and analyzing the concentrated organic phase by GC-MS.
GC-MS analysis was performed using a Thermo Electron Trace GC Ultra apparatus linked to a DSQ mass spectrometer. Volatile compounds were separated on an Alltech ECONO-CAP-EC-5 capillary column (30 m × 0.25 i.d. × 0.25 mm film thickness) and identified using a previously reported method (Ma and Gang, 2005).
cDNA Library Construction for Line MC
A glandular trichome cDNA library for basil line MC was constructed as described previously (Gang et al., 2001). Sequencing of randomly selected cDNA clones was performed at the Arizona Genomics Institute. Sequence base calls, vector trimming, and low-quality sequence filtering were performed by the Arizona Genomics Computation Laboratory as described elsewhere (Udall et al., 2006). EST sequences were initially clustered using PaCE (Kalyanaraman et al., 2003) and subsequently assembled using CAP3 (Huang and Madan, 1999). Functional annotations for each EST and contig consensus sequence were generated by BLASTing (Altschul et al., 1990) against the UniProt database with a cutoff of 1e-10.
DNA Gel Blot Analysis
Genomic DNA was isolated from young leaves of each of the four basil lines using Qiagen DNeasy Plant Maxi kits. DNA (10 μg) was digested separately with EcoRI and XbaI. Blotting and hybridization were performed using standard procedures (Sambrook and Russel, 2001). A 484-bp CCMT probe was amplified from a coding region of the CCMT1 cDNA that was highly conserved (97.5% identity or higher) across the three identified basil CCMT isoforms. The following primers were used for probe amplification: forward CCMT1pf, 5′-CGGAATTCTCAGCCTTCTTCTGC-3′; reverse CCMT1pr, 5′-CTCGAAATTAAGCCCTCTTGC-3′.
qRT-PCR
Basil plants used to compare relative CCMT expression and activity were grown in a growth chamber at 34°C with a photoperiod of 16 h of light/8 h of dark and an illumination intensity of 260 μmol·m−2·s−1. Glandular trichomes were isolated from young leaves obtained from 36 plants at a growth stage at which the sixth or seventh leaf pair was emerging. RNA for other tissues was obtained from four to five plants. Total RNA was isolated from 100 mg of glandular trichomes or basil tissues using RNeasy Plant Mini kits (Qiagen). RNA was isolated and cDNA was synthesized for four independent biological samples for each tissue. For each sample, 2 μg of total RNA was treated with DNase I and subsequently reverse-transcribed using SuperScript III (Invitrogen) and SuperScript III First-Strand Synthesis Supermix (Invitrogen) for qRT-PCR, which includes both oligo(dT)20 and random hexamers. Quantitative PCR was performed using Platinum SYBR Green qPCR Supermix-UDG and ROX (Invitrogen) as a reference dye on an ABI PRISM 7000 apparatus. Two separate qPCRs were performed for each cDNA using either CCMT or 18S ribosomal gene primer pairs. Cycling parameters were a single step of 50°C for 2 min, a single step of 95°C for 10 min, and 40 cycles of 95°C for 15 s followed by 60°C for 60 s. CCMT transcript levels for each sample were normalized to 18S. Primer sequences for qPCR were as follows: for CCMT, forward CCMTq186F, 5′-TGCTTATAAATATTTGGAGGTGGATTT-3′, and reverse CCMTq186R, 5′-CAATCAAACCTAAACCGAACCA-3′; for 18S DNA, forward Ob18Sq150F, 5′-GCAAGCCTACGCTCTGGATA-3′, and reverse Ob18Sq150R, 5′-TCATAAATCCAAGAATTTCACCTCTGA-3′.
To further assess the relative gene expression levels for the three major CCMT isoforms identified from the EST analysis, we attempted to design gene-specific primers for qRT-PCR. However, the number of adjacent nucleotides that were polymorphic between the gene sequences and that could be confidently used for primer design was very low. Primer pairs were designed that were 3′ anchored at one to two polymorphic bases, but the amplification efficiency of these primers under PCR conditions sufficiently stringent to give discriminatory amplification was too low to permit confidence in any resulting relative quantification. Thus, the qRT-PCR results could not yield any further data regarding differential transcriptional regulation of the three putative CCMT isoforms identified in the basil EST database.
Proteomics Analysis
Total soluble protein was isolated from glandular trichomes of each basil line, which were obtained using the same method as for RNA isolation (Gang et al., 2001), with some modifications. RNase inhibitor was replaced by 1 mM phenylmethylsulfonyl fluoride (PMSF) during gland isolation. Settled glands were suspended in a 10-fold volume of 5 mM Tris-HCl, pH 8.0, with 1.0 mM PMSF and disrupted by sonication. The lysate was centrifuged for 15 min at 10,000g and 4°C to remove insoluble cell debris. The supernatant was subsequently centrifuged at 100,000g for 1 h at 4°C to separate soluble and membrane-associated protein fractions. The soluble proteins were precipitated with 0.25 volume of 100% trichloroacetic acid for 15 min on ice, followed by centrifugation for 15 min at 16,000g and 4°C. The resulting protein sample was applied to MudPIT (Washburn et al., 2001) and the SDS-PAGE gel slice-LC-MS/MS analysis methods as described previously (Breci et al., 2005). Proteins were identified by SEQUEST (Link et al., 1999), searching against a custom protein database translated from basil EST tentative consensus sequences and the plant protein database of The Arabidopsis Information Resource (Huala et al., 2001). Proteomics analyses were performed by the Proteomics Analysis Laboratory at the University of Arizona.
Recombinant Protein Expression and Purification
The coding regions of all CCMTs were amplified from full-length cDNA clones with the sense 27-mer oligonucleotide 5′-GCCATATGGCGAGAAAAGAGAACTATG-3′ that introduced an NdeI site at the initiating ATG codon and the antisense 36-mer oligonucleotide 5′-GCGGATCCTTAGTTCGATGCAATTTTAGACTAAACC-3′ that introduced a BamHI site downstream of the stop codon. The PCR-amplified 1.2-kb fragments were subcloned into the NdeI-BamHI site of the expression vector pET-28a (Novagen) and used to transform E. coli Rosetta cells. Site-directed mutagenesis was performed using PCR primers with mismatched nucleotides designed to encode the desired amino acid alteration in the resultant PCR products, which were then assembled into full-length mutagenized open reading frames via overlap extension PCR and subsequently cloned, expressed, and purified as described above.
Preparation of Crude Cell-Free Extracts from E. coli
Single isolated colonies from plates containing Luria-Bertani medium supplemented with 50 μg/mL kanamycin and 37 μg/mL chloramphenicol were used to inoculate 25-mL liquid cultures with equilavent antibiotics. These cultures were grown overnight at 37°C and used to inoculate 1-liter cultures. Once the larger cultures reached an OD600 of 0.5, they were induced by the addition of 0.4 mM isopropylthio-β-galactoside. After incubation for 24 h at 18°C, the cultures were harvested by centrifugation and the pellets were resuspended in 40 mL of 50 mM potassium phosphate, pH 7.5, 500 mM NaCl, 1 mM disodium EDTA, 10 mM β-mercaptoethanol, 10% glycerol (v/v), and 1 mM PMSF. Cells were lysed by the addition of lysozyme (at a final concentration of 50 μg/mL) and incubation on ice for 30 min followed by sonication on a pulsed probe-tip sonicator. The resulting slurry was clarified by centrifugation at 25,000g for 45 min at 4°C.
Partial Enzyme Purification
The CCMT proteins were purified using methods adapted from previous reports (Negre et al., 2003; Kaminaga et al., 2006). Crude extracts made from 4 liters of culture were applied to a column containing 0.5 mL of nickel-nitrilotriacetic acid agarose (Qiagen) using gravity flow. This was followed by washing with 50 mL of a solution containing 20 mM potassium phosphate, pH 7.5, 500 mM NaCl, and 20 mM imidazole. Washing proceeded until the OD280 of the eluate matched approximately that of the buffer prior to application on the column. Elution was performed with a solution of 20 mM potassium phosphate, pH 7.5, 500 mM NaCl, and 500 mM imidazole. Fractions were collected and assayed for CCMT activity, and those containing the highest CCMT activity were pooled and desalted on Econo-Pac 10DG columns (Bio-Rad Laboratories) into 50 mM potassium phosphate, pH 7.5, and 10% (v/v) glycerol. SDS-PAGE followed by staining with Coomassie Brilliant Blue and band densitometry was used to estimate purity. Protein concentrations were determined by the Bradford method (Bradford, 1976) using Bio-Rad protein reagent and BSA as a standard.
Determination of Native Molecular Mass
Molecular mass of the recombinant CCMT proteins was determined by gel filtration on a Superdex 200-HR (Pharmacia-Biotech) column (1 × 30 cm) calibrated with the following markers: β-amylase (200 kD), γ-globulin (160 kD), BSA dimer (132 kD), ovalbumin dimer (90 kD), BSA monomer (66 kD), carbonic anhydrase dimer (58 kD), ovalbumin monomer (45 kD), carbonic anhydrase monomer (29 kD), lactalbumin dimer (28.4 kD), chymotrypsin (25 kD), lactalbumin (14.2 kD), and cytochrome c (12.4 kD). Buffer containing 50 mM potassium phosphate, pH 7.5, and 10 mM β-mercaptoethanol was used for column equilibration and elution. Fractions of 1 mL were collected at a flow rate of 0.5 mL/min and analyzed for CCMT activity. Denaturing SDS-PAGE was performed on 12% gels to determine the subunit molecular mass. The gels were calibrated with molecular mass standards in the range 6.8 to 198 kD (Bio-Rad).
Enzyme Characterization
The pH values for optimum CCMT activities were determined using 50 mM Tris/Na phosphate/Na citrate buffer with pH ranging from 5.5 to 9.0. Enzyme assays were performed with one of the following cations present in the assay buffer at a final concentration of 5 mM: Na+, Mn2+, K+, Ca2+, NH4+, Cu2+, Zn2+, Fe2+, and Mg2+. (In the presence of divalent cations, EDTA was omitted from the reaction buffer.) Substrate specificity was tested with acids at 2.4 mM and AdoMet at 19.45 μM. Comparisons were made between hexane and ethyl acetate to determine extraction conditions giving maximum yield of product for each substrate tested. Those extracted with hexane included methylbenzoate, methylcinnamate, methylsalicylate, and methyl-4-hydroxyhydrocinnamate. All others were extracted using ethyl acetate.
Kinetic Properties
For kinetic analysis, an appropriate enzyme concentration was chosen so that the reaction velocity was proportional to the enzyme concentration and was linear with respect to incubation time for 15 to 30 min (depending on the substrate being tested). All assays were performed at 24°C. Kinetic data were evaluated by hyperbolic regression analysis (Hyper.exe version 1.01; J.S. Easterby). Triplicate assays were performed for all analyses.
Modeling of CCMT Structures
Amino acid sequence alignments for the three basil CCMTs, Arabidopsis thaliana IAMT, and snapdragon (Antirrhinum majus) SAMT proteins versus Clarkia breweri SAMT were used by Modeller version 8.2 (Sali and Blundell, 1993) to produce eight independent models for each of the proteins. All resulting models for each protein were very similar, and the model with the lowest calculated minimization energy was selected for further analysis. The modeled protein structures were visualized in O (Jones et al., 1991) or in Chimera (Pettersen et al., 2004). Images for display were produced by POV-Ray (Persistence of vision; http://www.povray.org) using parameters written in Molscript (Kraulis, 1991). Docking of ligands (potential substrates) to modeled protein active sites was performed using GOLD version 3.1.1 (Cambridge Crystallographic Data Centre).
Phylogenetic Analysis of the SABATH Family
Sequence database searches were performed using BLAST and psi-BLAST algorithms (Altschul et al., 1990, 1997) against the nr database of the National Center for Biotechnology Information (NCBI) as well as tBLASTn searches against dbEST and the gss, wgs, and htgs genomic sequence databases. Large numbers of EST hits for several taxa were assembled into consensus sequences at the species or genus level using GCG (Accelrys). In addition to the NCBI databases, we performed BLAST searches against sequence data available for Physcomitrella patens and Selaginella moellendorfii available at Physcobase (http://moss.nibb.ac.jp/). We also conducted BLAST searches against the Joint Genome Institute eukaryotic genome databases and the Integrated Microbial Genome database (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) as well as the Plant Genome Database (http://www.plantgdb.org/) to access additional sequence data not yet synched with NCBI. The resultant BLAST hits were filtered to eliminate redundancy as well as fragmentary sequences of <200 amino acids (nonplant sequences were filtered at a cutoff of 120 amino acids). Multiple sequence alignments were performed using ClustalX 1.83 (Thompson et al., 1997) using the BLOSUM protein weight matrix. A subset of difficult-to-align sequences were removed and aligned to the realigned primary group of sequences in profile mode. Alignments were manually refined by eye, and some difficult sequences were also manually aligned with the aid of RPS-BLAST (Marchler-Bauer et al., 2002) results generated from sequence searches against the Conserved Domain Database (Marchler-Bauer et al., 2005) and the resulting alignment to the PSSM (Pssm identifier 67,129) for the SABATH methyltransferase domain (Pfam accession number PF03492) (Bateman et al., 2004). Prior to performing maximum likelihood analysis, the alignment was further refined by removing highly variable and uninformative regions of the multiple sequence alignment as well as partial length sequences lacking sequence data for the included regions of the alignment. Phylogenetic analysis was performed for 132 residues and 231 sequences using Phyml (Guindon and Gascuel, 2003) with the JTT substitution model and 1000 bootstrap replicates, and tree files were viewed using TreeExplorer.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data library under accession numbers EU033968 (Ob CCMT1), EU033969 (Ob CCMT2), and EU033970 (Ob CCMT3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. AdoMet Inhibition of CCMT Activity in MC Glandular Trichomes.
Supplemental Figure 2. DNA Gel Blot Analysis of CCMT Copy Number and Organization in the Genomes of the Four Basil Lines: SW, MC, EMX-1, and SD.
Supplemental Figure 3. Sequence Alignment Used to Generate the Maximum Likelihood Tree Displayed in Figure 10.
Supplemental Figure 4. Additional Plots Demonstrating the Parabolic Nature of SA Inhibition of CCMT1 and CCMT3 Activities.
Supplemental Figure 5. Energy-Minimized Structures of Potential Substrates (in Free Acid and Carboxylate Forms) for CCMT Enzymes Used in Docking Experiments, Displayed from Two Sides.
Supplemental Table 1. Peptide Identification of CCMT Isoforms from MC and SW Glandular Trichome–Soluble Protein Samples Using a Proteomics Approach.
Supplemental Methods.
Supplemental Data Set 1. Text File of the Alignment of CCMT1 Protein Sequences Presented in Figure 7.
Supplemental Data Set 2. Text File of the Sequence Alignment Used to Generate the Maximum Likelihood Tree Displayed in Figure 10.
Supplementary Material
Acknowledgments
We thank Joseph Noel, Florence Pojer, and Jeannine Ross for valuable discussion and advice on the protein modeling and structural analysis; Joseph Bischoff for valuable discussion regarding the SABATH matrix and phylogenetic analysis; James Simon for valuable discussion regarding methylcinnamate formation in sweet basil; Brenda Jackson for assistance with GC-MS maintenance and operation; Paul Haynes for advice on the shotgun proteomics experimental design; and Linda Breci, George Tsaprailis, and Fatimah Hickman at the Arizona Proteomics Consortium for assistance with proteomics sample analysis and data mining. We thank the National Science Foundation (Grant MCB-0210170 to D.R.G. and Grant MCB-0615700 to N.D.) and the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (Grant 2005-35318-16207 to N.D.) for financial support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David R. Gang (gang@ag.arizona.edu).
Online version contains Web-only data.
References
- Ackerman, J.D. (1989). Geographic and seasonal variation in fragrance choices and preferences of male euglossine bees. Biotropica 21 340–347. [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azah, M.A.N., Sam, Y.Y., Mailina, J., and Chua, L.S.L. (2005). (E)-Methyl cinnamate: The major component of essential oils of Alpinia malaccensis var. nobilis. J. Trop. For. Sci. 17 631–633. [Google Scholar]
- Bandara, B.M.R., Hewage, C.M., Karunaratne, V., and Adikaram, N.K.B. (1988). Methyl-ester of para-coumaric acid: Antifungal principle of the rhizome of Costus speciosus. Planta Med. 54 477–478. [DOI] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32 D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuerle, T., and Pichersky, E. (2002). Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal. Biochem. 302 305–312. [DOI] [PubMed] [Google Scholar]
- bin Jantan, I., Yalvema, M.F., Ayop, N., and Ahmad, A.S. (2005). Constituents of the essential oils of Cinnamomum sintoc Blume from a mountain forest of peninsular Malaysia. Flavour Frag. J. 20 601–604. [Google Scholar]
- Bradford, M.M. (1976). Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Breci, L., Hattrup, E., Keeler, M., Letarte, J., Johnson, R., and Haynes, P.A. (2005). Comprehensive proteomics in yeast using chromatographic fractionation, gas phase fractionation, protein gel electrophoresis, and isoelectric focusing. Proteomics 5 2018–2028. [DOI] [PubMed] [Google Scholar]
- Bruni, R., Medici, A., Andreotti, E., Fantin, C., Muzzoli, M., Dehesa, M., Romagnoli, C., and Sacchetti, G. (2004). Chemical composition and biological activities of Ishpingo essential oil, a traditional Ecuadorian spice from Ocotea quixos (Lam.) Kosterm. (Lauraceae) flower calices. Food Chem. 85 415–421. [Google Scholar]
- Cambie, R.C., Lal, A.R., Rickard, C.E.F., and Tanaka, N. (1990). Chemistry of Fijian plants. V. Constituents of Fagraea gracilipes A. Gray. Chem. Pharm. Bull. (Tokyo) 38 1857–1861. [Google Scholar]
- Chen, F., D'Auria, J.C., Tholl, D., Ross, J.R., Gershenzon, J., Noel, J.P., and Pichersky, E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36 577–588. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden, A. (1979). Fundamentals of Enzyme Kinetics. (London: Butterworth).
- Daayf, F., Bel-Rhlid, R., and Belanger, R.R. (1997. a). Methyl ester of p-coumaric acid: A phytoalexin-like compound from long English cucumber leaves. J. Chem. Ecol. 23 1517–1526. [Google Scholar]
- Daayf, F., Ongena, M., Boulanger, R., El Hadrami, I., and Belanger, R.R. (2000). Induction of phenolic compounds in two cultivars of cucumber by treatment of healthy and powdery mildew-infected plants with extracts of Reynoutria sachalinensis. J. Chem. Ecol. 26 1579–1593. [Google Scholar]
- Daayf, F., Schmitt, A., and Belanger, R.R. (1997. b). Evidence of phytoalexins in cucumber leaves infected with powdery mildew following treatment with leaf extracts of Reynoutria sachalinensis. Plant Physiol. 113 719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria, J.C., Chen, F., and Pichersky, E. (2003). The SABATH family of MTs in Arabidopsis thaliana and other plant species. Rec. Adv. Phytochem. 37 253–283. [Google Scholar]
- Dias, C., Dias, M., Borges, C., Almoster Ferreira, M.A., Paulo, A., and Nascimento, J. (2003). Structural elucidation of natural 2-hydroxy di- and tricarboxylic acids and esters, phenylpropanoid esters and a flavonoid from Autonoe madeirensis using gas chromatographic/electron ionization, electrospray ionization and tandem mass spectrometric techniques. J. Mass Spectrom. 38 1240–1244. [DOI] [PubMed] [Google Scholar]
- Dodson, C.H., Dressler, R.L., Hills, H.G., Adams, R.M., and Williams, N.H. (1969). Biologically active compounds in orchid fragrances. Science 164 1243–1249. [DOI] [PubMed] [Google Scholar]
- Effmert, U., Saschenbrecker, S., Ross, J., Negre, F., Fraser, C.M., Noel, J.P., Dudareva, N., and Piechulla, B. (2005). Floral benzenoid carboxyl methyltransferases: From in vitro to in planta function. Phytochemistry 66 1211–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenthal, R., and Cornish-Bowden, A. (1974). The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem. J. 139 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltz, T., and Lunau, K. (2005). Antennal response to fragrance compounds in male orchid bees. Chemoecology 15 135–138. [Google Scholar]
- Gang, D.R. (2005). Evolution of flavors and scents. Annu. Rev. Plant Biol. 56 301–325. [DOI] [PubMed] [Google Scholar]
- Gang, D.R., Beuerle, T., Ullmann, P., Werck-Reichhart, D., and Pichersky, E. (2002. a). Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol. 130 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang, D.R., Lavid, N., Zubieta, C., Chen, F., Beuerle, T., Lewinsohn, E., Noel, J.P., and Pichersky, E. (2002. b). Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang, D.R., Wang, J.H., Dudareva, N., Nam, K.H., Simon, J.E., Lewinsohn, E., and Pichersky, E. (2001). An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol. 125 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., Opiteck, G.J., Friedrichs, M.S., Dongre, A.R., and Hefta, S.A. (2003). Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2 643–649. [DOI] [PubMed] [Google Scholar]
- Garcia, R., Erazo, S., Canepa, A., Lemus, I., and Erazo, S. (1990). Secondary metabolites of Escallonia illinita Presl. An. Real Acad. Farm. 56 539–542. [Google Scholar]
- Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Hattori, M., Sakagami, Y., and Marumo, S. (1992). Oviposition deterrents for the limabean pod borer, Etiella zinckenella (Treitschke) (Lepidoptera: Pyralidae) from Populus nigra L. cv. Italica leaves. Appl. Entomol. Zool. (Jpn.) 27 195–204. [Google Scholar]
- Hiraga, Y., Chen, L., Kurokawa, M., Ohta, S., Suga, T., and Hirata, T. (1996). Structure-activity relationships of cinnamic acid derivatives as germination inhibitor of the fern Gleichenia japonica. Nat. Prod. Lett. 9 21–26. [Google Scholar]
- Hooper, S.N., Jurgens, T., Chandler, R.F., and Stevens, M.F.G. (1984). Methyl p-coumarate: A cytotoxic constituent from Comptonia peregrina. Phytochemistry 23 2096–2097. [Google Scholar]
- Huala, E., et al. (2001). The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.Q., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima, Y., Davidovich-Rikanati, R., Fridman, E., Gang, D.R., Bar, E., Lewinsohn, E., and Pichersky, E. (2004. a). The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol. 136 3724–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima, Y., Gang, D.R., Fridman, E., Lewinsohn, E., and Pichersky, E. (2004. b). Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 134 370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kalyanaraman, A., Aluru, S., Brendel, V., and Kothari, S. (2003). Space and time efficient parallel algorithms and software for EST clustering. IEEE Trans. Parallel Distrib. Syst. 14 1209–1221. [Google Scholar]
- Kaminaga, Y., et al. (2006). Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 281 23357–23366. [DOI] [PubMed] [Google Scholar]
- Konda, A., and Kawazu, K. (1979). Biologically active substances from the leaves of Platycarya strobilacea Sieb. et Zucc. III. Kagawa Daigaku Nogakubu Gakujutsu Hokoku 30 215–218. [Google Scholar]
- Kraulis, P.J. (1991). Molscript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24 946–950. [Google Scholar]
- Latza, S., and Berger, R.G. (1997). 1-O-Trans-cinnamoyl-beta-D-glucopyranose: Alcohol cinnamoyltransferase activity in fruits of cape gooseberry (Physalis peruviana L.). Z. Naturforsch. C. J. Biosci. 52 747–755. [Google Scholar]
- Link, A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates, J.R. (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17 676–682. [DOI] [PubMed] [Google Scholar]
- Liu, H.B., Sadygov, R.G., and Yates, J.R. (2004). A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76 4193–4201. [DOI] [PubMed] [Google Scholar]
- Lunkenbein, S., Bellido, M., Aharoni, A., Salentijn, E.M.J., Kaldenhoff, R., Coiner, H.A., Munoz-Blanco, J., and Schwab, W. (2006). Cinnamate metabolism in ripening fruit. Characterization of a UDP-glucose:cinnamate glucosyltransferase from strawberry. Plant Physiol. 140 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.-Q., and Gang, D.R. (2005). Metabolic profiling of in vitro micropropagated and conventionally greenhouse grown ginger (Zingiber officinale). Phytochemistry 67 2239–2255. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2005). CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 33 D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Panchenko, A.R., Shoemaker, B.A., Thiessen, P.A., Geer, L.Y., and Bryant, S.H. (2002). CDD: A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre, F., Kish, C.M., Boatright, J., Underwood, B., Shibuya, K., Wagner, C., Clark, D.G., and Dudareva, N. (2003). Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15 2992–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell, E., Raguso, R.A., and Jones, K.N. (1999). Bumblebee foraging responses to variation in floral scent and color in snapdragons (Antirrhinum: Scrophulariaceae). Am. Midl. Nat. 142 257–265. [Google Scholar]
- Pang, J.X., Ginanni, N., Dongre, A.R., Hefta, S.A., and Opiteck, G.J. (2002). Biomarker discovery in urine by proteomics. J. Proteome Res. 1 161–169. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., and Ferrin, T.E. (2004). UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pichersky, E., and Gang, D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 5 439–445. [DOI] [PubMed] [Google Scholar]
- Sali, A., and Blundell, T.L. (1993). Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schaefers, F.I., and Herrmann, K. (1982). Analysis of methyl and ethyl esters of hydroxybenzoic and hydroxycinnamic acids in plant material. J. Chromatogr. 240 387–396. [Google Scholar]
- Schiestl, F.P., and Roubik, D.W. (2003). Odor compound detection in male euglossine bees. J. Chem. Ecol. 29 253–257. [DOI] [PubMed] [Google Scholar]
- Seifert, K., and Unger, W. (1994). Insecticidal and fungicidal compounds from Isatis tinctoria. Z. Naturforsch. C. J. Biosci. 49 44–48. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J.A., et al. (2006). A global assembly of cotton ESTs. Genome Res. 16 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and Luca, V.D. (2005). The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J. 44 606–619. [DOI] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates, J.R. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19 242–247. [DOI] [PubMed] [Google Scholar]
- Williams, N.H., and Whitten, W.M. (1983). Orchid floral fragrances and male euglossine bees: Methods and advances in the last sesquidecade. Biol. Bull. 164 355–395. [Google Scholar]
- Wu, T.-S., Leu, Y.-L., and Chan, Y.-Y. (1999). Constituents of the fresh leaves of Aristolochia cucurbitifolia. Chem. Pharm. Bull. (Tokyo) 47 571–573. [DOI] [PubMed] [Google Scholar]
- Zubieta, C., Ross, J.R., Koscheski, P., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.