Abstract
CRY2 is a blue light receptor regulating light inhibition of hypocotyl elongation and photoperiodic flowering in Arabidopsis thaliana. The CRY2 protein is found primarily in the nucleus, and it is known to undergo blue light–dependent phosphorylation and degradation. However, the subcellular location where CRY2 exerts its function or undergoes blue light–dependent phosphorylation and degradation remains unclear. In this study, we analyzed the function and regulation of conditionally nuclear-localized CRY2. Our results show that CRY2 mediates blue light inhibition of hypocotyl elongation and photoperiodic promotion of floral initiation in the nucleus. Consistent with this result and a hypothesis that blue light–dependent phosphorylation is associated with CRY2 function, we demonstrate that CRY2 undergoes blue light–dependent phosphorylation in the nucleus. CRY2 phosphorylation is required for blue light–dependent CRY2 degradation, but only a limited quantity of CRY2 is phosphorylated at any given moment in seedlings exposed to blue light, which explains why continuous blue light illumination is required for CRY2 degradation. Finally, we showed that CRY2 is ubiquitinated in response to blue light and that ubiquitinated CRY2 is degraded by the 26S proteasome in the nucleus. These findings demonstrate that a photoreceptor can complete its posttranslational life cycle (from protein modification, to function, to degradation) inside the nucleus.
INTRODUCTION
Cryptochromes are photolyase-like blue light receptors found in plants and animals that regulate the circadian clock and developmental processes (Cashmore, 2003; Lin and Shalitin, 2003; Sancar, 2003). The Arabidopsis thaliana genome encodes at least two cryptochromes, CRY1 and CRY2, which primarily regulate deetiolation and photoperiodic flowering, respectively (Ahmad and Cashmore, 1993; Guo et al., 1998). Arabidopsis CRY3 is a chloroplast/mitochondria protein that acts as a single-strand DNA photolyase (Kleine et al., 2003; Selby and Sancar, 2006). Although the major function of CRY2 is to regulate photoperiodic promotion of floral initiation (Guo et al., 1998; El-Assal et al., 2001), it also mediates blue light inhibition of hypocotyl elongation (Lin et al., 1998; Mockler et al., 1999). Overexpression of the green fluorescent protein–CRY2 fusion protein can complement both long-hypocotyl and late-flowering phenotypes of the cry1 cry2 double mutant (Yu et al., 2007). CRY2 is ubiquitously expressed at different developmental stages (see Supplemental Figure 1 online) and in different tissues (data not shown), although a tissue-specific transgenic expression study showed that CRY2 regulates floral initiation in vascular cells (Endo et al., 2007). CRY1 and CRY2 are both found in the nucleus, CRY1 was reported to undergo nucleus/cytoplasm shuttling in response to light, but no such subcellular trafficking has been reported for CRY2 (Cashmore et al., 1999; Guo et al., 1999; Kleiner et al., 1999; Yang et al., 2001). Importantly, whether CRY1 and CRY2 exert their physiological functions inside the nucleus remains unclear. Given that the apparent subcellular localization of a protein is not necessarily where the protein functions in the cell and that CRY2 was reported to be involved in the blue light regulation of anion channels in the plasma membrane (Folta and Spalding, 2001), where CRY2 acts inside the cell needs to be determined experimentally.
Arabidopsis cryptochromes undergo blue light–dependent phosphorylation in vivo (Shalitin et al., 2002, 2003; Bouly et al., 2003; Moller et al., 2003). The blue light–induced phosphorylation of CRY2 is required for the photoactivation and the physiological functions of the photoreceptor (Shalitin et al., 2002; Yu et al., 2007). However, it is not clear where in the cell cryptochrome phosphorylation takes place. CRY2 is also known to be degraded in response to blue light (Ahmad et al., 1998; Lin et al., 1998), but it is not known where in the cell CRY2 is degraded, nor is it clear whether ubiquitination and the 26S proteasome are involved in CRY2 degradation. In this report, we show that CRY2 acts in the nucleus and that both CRY2 phosphorylation and degradation processes take place in the nucleus. Moreover, we also demonstrate that CRY2 is ubiquitinated in response to blue light and that CRY2 is degraded in a phosphorylation- and 26S proteasome–dependent manner in the nucleus.
RESULTS
CRY2 Mediates Blue Light Inhibition of Hypocotyl Elongation and Photoperiodic Regulation of Floral Initiation in the Nucleus
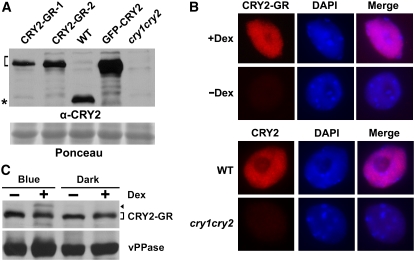
To investigate the exact subcellular compartment where CRY2 action and regulation take place, we used a conditional nuclear localization approach. We prepared transgenic Arabidopsis plants expressing the CRY2-GR (rat glucocorticoid receptor) fusion protein in the cry1 cry2 mutant background (referred to as CRY2-GR/cry1 cry2). The cry1 cry2 mutant develops a longer hypocotyl when grown in blue light and shows delayed flowering when grown in long-day photoperiods, whereas transgenic expression of active CRY2 can rescue both phenotypes (Yu et al., 2007). The GR fusion protein approach has been successfully used to study the nucleus-dependent function of many Arabidopsis nuclear proteins (Lloyd et al., 1994; Samach et al., 2000; Huq et al., 2003). Although originally discovered in mammals, transgenically expressed rat GR and GR fusion proteins also reside in the cytosol of Arabidopsis cells, and they are translocated into the nucleus in the presence of the synthetic corticosteroid, Dexamethasone (Dex) (Lloyd et al., 1994). We selected CRY2-GR/cry1 cry2 lines expressing CRY2-GR at a level not exceeding that of endogenous CRY2 in the wild-type plants (Figure 1A). Independent transgenic lines of CRY2-GR/cry1 cry2 were used to confirm that the phenotypic changes reported here were not due to T-DNA insertion mutagenesis (data not shown).
Figure 1.
Expression and Dex-Dependent Nuclear Localization of CRY2-GR.
(A) Immunoblot showing relative levels of expression of CRY2-GR in two independent lines (CRY2-GR-1 and CRY2-GR-2), the control GFP-CRY2 in the respective transgenic lines, and endogenous CRY2 in wild-type plants. Samples were prepared from etiolated seedlings of indicated genotypes, fractioned by a 10% SDS-PAGE gel, blotted, stained with Ponceau S (Ponceau), and probed with the anti-CRY2 antibody (α-CRY2). The bracket indicates CRY2 fusion proteins, and the asterisk indicates the endogenous CRY2.
(B) Immunostaining (red) showing accumulation of CRY2-GR (line CRY2-GR-1) in the nucleus (4′,6-diamidino-2-phenylindole stain, blue) in the presence of Dex. Top panel: nuclei were isolated from CRY2-GR/cry1 cry2 seedlings grown in Murashige and Skoog (MS) medium containing 30 μM Dex (+Dex) or mock solution (−Dex) in the dark. Bottom panel: CRY2 immunostaining of the nuclei isolated from wild-type and cry1 cry2 mutant plants. The immunostaining was probed with anti-CRY2 (α-CRY2) antibody and then the second antibody conjugated with the fluorescent dye rhodamine red-x.
(C) Immunoblot comparing the level of CRY2-GR (line CRY2-GR-1) in 7-d-old seedlings grown in MS medium containing 30 μM Dex (+Dex) or mock solution (−Dex) in the dark or continuous blue light. The blot was probed with anti-CRY2, stripped, and reprobed with anti-vPPase (vacuolar pyrophosphatase) to show the relative loading. The arrowhead indicates phosphorylated CRY2-GR, and the bracket indicates unphosphorylated CRY2-GR.
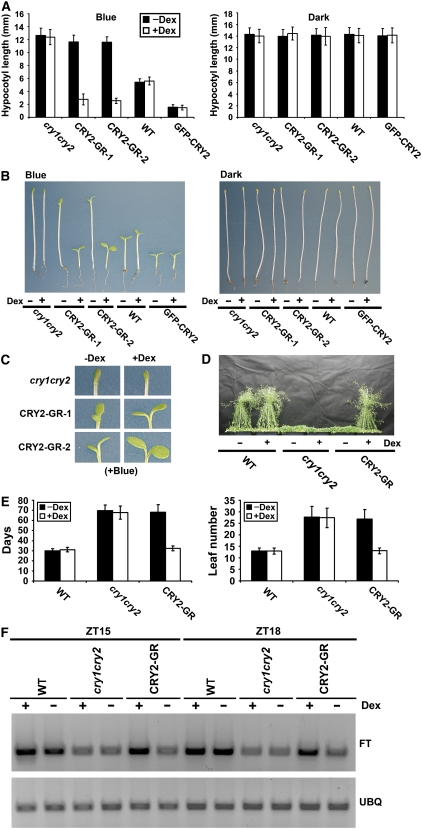
Dex-dependent nuclear localization of CRY2-GR was confirmed by nuclear immunostaining. As shown in Figure 1B, CRY2 was not detected in the nucleus of CRY2-GR/cry1 cry2 plants in the absence of Dex but abundantly present in the nucleus when CRY2-GR/cry1 cry2 plants were treated with Dex. The dramatically increased immunostain of CRY2-GR in the nucleus in response to the Dex treatment must result from nuclear translocation of CRY2-GR because seedlings treated with Dex showed no increase in the overall level of the CRY2-GR protein (Figure 1C). We next examined whether CRY2-GR located in the cytosol (−Dex) or nucleus (+Dex) may rescue the long-hypocotyl phenotype of the cry1 cry2 mutant grown in blue light. Figure 2A shows that, when grown in continuous blue light in the absence of Dex, CRY2-GR/cry1 cry2 seedlings developed long hypocotyls similar to those of the cry1 cry2 mutant. By contrast, CRY2-GR/cry1 cry2 seedlings developed short hypocotyls when grown in blue light in the presence of Dex, demonstrating that nuclear CRY2-GR rescued the long-hypocotyl defect of the parent in blue light (Figures 2A and 2B). We noticed that CRY2-GR/cry1 cry2 seedlings grown under blue light in the presence of Dex exhibited hypocotyls shorter than those of the wild type (Figures 2A and 2B), which, taken together with the fact that the level of CRY2-GR in neither transgenic line exceeds the level of the endogenous CRY2 in the wild type, suggests that CRY2-GR may be physiologically more active than the endogenous CRY2. The Dex treatment showed no effect on the light inhibition of hypocotyl elongation of the CRY2-GR/cry1 cry2 seedlings grown in red or far-red light (data not shown) or of the wild type and the cry1 cry2 mutant parent grown in blue light (Figures 2A and 2B). We concluded that nuclear-localized CRY2 is capable of mediating blue light inhibition of hypocotyl elongation. However, in contrast with the cry1 cry2 mutant parent, CRY2-GR/cry1 cry2 seedlings grown under blue light in the absence of Dex showed cotyledon opening (Figure 2C). There are at least two explanations for this Dex-independent activity of CRY2-GR. It is possible that cytosolic CRY2 may mediate blue light stimulation of cotyledon opening/expansion (Lin et al., 1998). Alternatively, a small fraction of CRY2-GR might leak into the nucleus in the absence of Dex. Indeed, transgenic plants overexpressing CRY2-GR to a level higher than that of the wild-type plants showed not only expanded cotyledons but also shortened hypocotyls in the absence of Dex (see Methods).
Figure 2.
Only Nuclear-Localized CRY2-GR Can Rescue the Long-Hypocotyl and Late-Flowering Phenotypes of the cry1 cry2 Mutant.
(A) Hypocotyl lengths of 5-d-old seedlings with indicated genotypes grown on soil watered with 30 μM Dex (+Dex) or mock solution (−Dex) in continuous blue light (16 μmol m−2 s−1) or in the dark. The means of hypocotyl lengths (n ≥ 20) and standard deviations of two independent lines (CRY2-GR-1 and CRY2-GR-2) are shown.
(B) Representative seedlings from samples shown in (A).
(C) A close-up cotyledon image of the seedling shown in the left panel of (B), with the indicated genotype and treatment shown.
(D) Representative 50-d-old plants of indicated genotypes (CRY2-GR/cry1 cry2 is line CRY2-GR-1). Plants were grown in long-day photoperiods (16 h light/8 h dark) and sprayed daily (from days 10 to 30) with 30 μM Dex (+Dex) or mock solution (−Dex).
(E) The flowering times of plants, for which the representatives are shown in (D). The flowering times are measured as days to flower (days) or number of rosette leaves (leaf number) at flowering, and the means and standard deviations (n ≥ 20) are shown.
(F) FT mRNA expression in the wild type, cry1 cry2, and CRY2-GR/cry1 cry2 (line CRY2-GR-1). Plants were grown on MS medium containing 30 μM Dex (+Dex) or mock solution (−Dex) in long-day photoperiods (16 h light/8 h dark). Samples were harvested at 15 (ZT15) and 18 (ZT18) h after light on, and the RT-PCR results are shown.
We then examined where in the cell CRY2 could regulate photoperiodic control of floral initiation by testing whether CRY2-GR might rescue the late-flowering phenotype of the cry1 cry2 mutant in a Dex-dependent manner. In this experiment, CRY2-GR/cry1 cry2 and control plants were grown in long-day photoperiods, plants were sprayed (or watered) with Dex or mock solution, and the flowering time was measured (Figures 2D and 2E). As shown in Figure 2D, the CRY2-GR/cry1 cry2 plants treated with the mock solution flowered as late as the cry1 cry2 mutant when grown in long-day photoperiods. Dex treatment apparently rescued this defect so that the CRY2-GR/cry1 cry2 plants treated with Dex flowered at about the same time as the wild type (Figures 2D and 2E). The Dex treatment had no effect on the flowering time of the wild type or the cry1 cry2 mutant parent (Figures 2D and 2E). Moreover, the Dex-dependent nuclear translocation of CRY2-GR rescued the impaired mRNA expression of the flowering-time gene FT in the cry1 cry2 mutant. The CRY2-GR/cry1 cry2 transgenic lines treated with Dex exhibited elevated levels of FT mRNA expression in comparison to the same lines treated with the mock solution (Figure 2F). Therefore, Dex-dependent nuclear translocation of CRY2-GR seems to promote floral initiation by a similar mechanism as that of the endogenous CRY2 (Yanovsky and Kay, 2002; Valverde et al., 2004). We concluded that CRY2 regulates photoperiodic floral initiation in the nucleus.
Blue Light–Induced CRY2 Degradation Is a High-Irradiance Response That Requires Continuous Blue Light Illumination
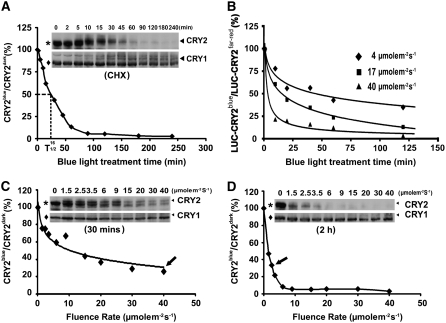
CRY2 is rapidly degraded in etiolated seedlings exposed to blue light, providing an explanation for why CRY2 plays only a minor role in deetiolation in comparison to CRY1 (Lin et al., 1998). To better understand this important blue light response of CRY2, we first examined the kinetics and fluence-dependence of blue light–induced CRY2 degradation. CRY2 degradation was studied only under the in vivo conditions because this reaction is extremely sensitive to in vitro conditions. CRY2 failed to show blue light–induced degradation in all the cell-free systems tested, including those used to study light-regulated degradation of other proteins (see Supplemental Figure 2 online; data not shown) (Osterlund et al., 2000; Seo et al., 2003; Vierstra, 2003; Valverde et al., 2004). In etiolated seedlings, CRY2 is a stable protein with a half-life of longer than 24 h (Figure 3; data not shown). By contrast, in etiolated seedlings exposed to blue light (16 μmol m−2 s−1), the half-life (t1/2) of CRY2 was reduced to ∼25 min (Figure 3A). In contrast with blue light–induced decline in the level of the CRY2 protein (Figure 3A), CRY2 mRNA levels increased slightly in response to blue light (see Supplemental Figure 3 online). The blue light–dependent CRY2 degradation occurs in vivo over a relatively wide temperature range from 10 to 45°C (see Supplemental Figure 4 online). Given that the level of CRY2 mRNA is regulated by blue light (see Supplemental Figure 3 online) and the circadian clock (Toth et al., 2001), we tested how a fusion protein of luciferase and CRY2 (LUC-CRY2) under control of the constitutive 35S promoter may respond to blue light. We found that luminescence of the transgenic seedlings expressing LUC-CRY2 declined in a blue light–dependent manner (Figure 3B), confirming that the blue light–dependent decrease of CRY2 is independent from its transcription. Similar to the endogenous CRY2, the LUC-CRY2 fusion protein also showed increased degradation in response to increased fluence rates of blue light (Figure 3B). Under similar fluence rates of blue light (16 to 17 μmol m−2 s−1), the LUC-CRY2 fusion protein showed a half-life and kinetics of degradation comparable to those of the endogenous CRY2 (Figures 3A and 3B).
Figure 3.
Kinetics of Blue Light–Dependent Degradation of Endogenous CRY2 in Wild-Type Seedlings or LUC-CRY2 in Transgenic Seedlings.
(A) Immunoblot showing endogenous CRY2 in etiolated wild-type seedlings exposed to blue light (16 μmol m−2 s−1) for the indicated time. Five-day-old etiolated seedlings were treated with cycloheximide (CHX) for 4 h before transferring to blue light for the indicated duration. Protein samples were extracted, fractioned by 10% SDS-PAGE, and blotted. The immunoblot was probed with anti-CRY2, stripped, and reprobed with anti-CRY1 antibodies. The relative level of CRY2 (including both phosphorylated upper bands and unphopsphorylated lower band) is calculated as described (see Methods) and presented as CRY2blue (level of CRY2 after blue light treatment)/CRY2dark (level of CRY2 before blue light treatment).
(B) Change in luminescence of LUC-CRY2 seedlings in response to blue light. LUC-CRY2 seedlings were grown on MS medium (∼150 seedlings per Petri dish) in far-red light for 5 d and exposed to blue light of the fluence rates and durations indicated. The luminescence of untreated and blue light–treated seedlings was measured using a CCD camera, and the relative luminescence per seedling is presented as the percentage of LUC-CRY2blue (luminescence after blue light treatment)/LUC-CRY2far-red (luminescence before blue light treatment).
(C) and (D) Immunoblot showing endogenous CRY2 in etiolated wild-type seedlings exposed to blue light with the fluence rates indicated for 30 min (C) or 2 h (D). The relative level of CRY2 is calculated as in (A). Arrows indicate the fluence rates by which ∼70% of CRY2 was degraded.
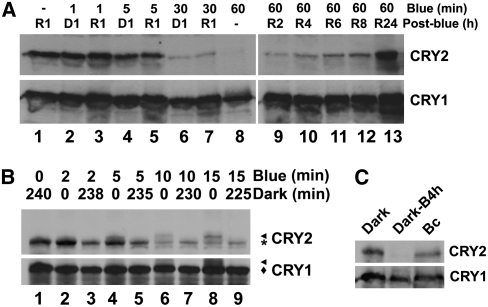
Depending on the fluence rate of blue light and illumination time, the same amount of photons caused different extents of CRY2 degradation in vivo. For example, ∼70% CRY2 was degraded in etiolated seedlings exposed to blue light of 40 μmol m−2 s−1 for 30 min, which equals a total fluence of 72,000 μmol m−2 s−1 (Figure 3C, arrow). By contrast, only approximately one-fourth of the total fluence, or ∼18,000 μmol m−2, was needed to result in a similar extent of CRY2 degradation in etiolated seedlings exposed to blue light of a lower fluence rate (2.5 μmol m−2 s−1) but for a longer time (2 h) (Figure 3D, arrow). Therefore, blue light–dependent CRY2 degradation is a high irradiance response that does not obey the law of reciprocity. This result predicts that CRY2 degradation requires continuous blue light illumination. To test this prediction, we treated etiolated seedlings with blue light (20 μmol m−2 s−1) for different durations up to 1 h, then transferred the seedlings to dark (or red light) for 1 h, and examined the level of CRY2 (Figure 4A). Figure 4A shows that, compared with etiolated seedlings treated with 1 h of blue light (Figure 4A, lane 8), those treated with blue light for 1 to 5 min followed by 1 h of posttreatment in the dark or red light showed markedly reduced CRY2 degradation (Figure 4A, lanes 2 to 5). Upon an almost complete depletion under blue light (Figure 4A, lane 9), CRY2 slowly (at least 24 h) reaccumulated in seedlings transferred from blue light to red light (Figure 4A, lanes 10 to 13). These results demonstrate that continuous blue light is required for CRY2 degradation.
Figure 4.
Continuous Blue Light and CRY2 Phosphorylation Is Required for CRY2 Degradation.
(A) Immunoblots showing the level of CRY2 in etiolated seedlings exposed to blue light for different durations and then transferred to red light or dark (post-blue) for 1 h (lanes 1 to 8) or the level of CRY2 in etiolated seedlings exposed to blue light for 60 min (blue) and then transferred to red light for different durations (lanes 9 to 13).
(B) Immunoblot showing changes in the levels of phosphorylated CRY2 and unphosphorylated CRY2 in etiolated seedlings exposed to blue light followed by a posttreatment in the dark. Five-day-old etiolated wild-type seedlings were exposed to blue light (15 μmol m−2 s−1) for the durations indicated (blue). Samples were either analyzed immediately after blue light exposure (lanes 2, 4, 6, and 8) or analyzed after a dark treatment for the durations indicated (lanes 3, 5, 7, and 9). Arrowhead and asterisk (or diamond) indicate phosphorylated and unphosphorylated CRY2 (or CRY1), respectively.
(C) Immunoblot showing CRY2 in 5-d-old etiolated seedlings (dark) exposed to blue light for 4 h (20 μmol m−2 s−1; Dark-B4 h) or 5-d-old seedlings germinated and grown in continuous blue light (20 μmol m−2 s−1; Bc). Similar amounts of total protein from different samples were loaded as shown by the similar level of CRY1 (bottom).
Why Is Continuous Blue Light Required for CRY2 Degradation?
Given that photoreceptors absorb photons almost instantaneously, that CRY2 appears to act as the photoreceptor for its own degradation (see Supplemental Figure 5 online), and that CRY2 undergoes blue light–dependent phosphorylation (Shalitin et al., 2002), we hypothesized that only phosphorylated CRY2 may be degradable but photon-excited CRY2 is phosphorylated only gradually in blue light. According to this hypothesis, CRY2 phosphorylation would be a rate-limiting step, and phosphorylated CRY2 represents only a small fraction of the CRY2 protein pool in the cell at any moment. Indeed, both phosphorylated and unphosphorylated CRY2 were detected in light-grown seedlings (Figure 3C) (Shalitin et al., 2002). To further test the relationship between CRY2 phosphorylation and degradation, we compared the relative levels of phosphorylated CRY2 and unphosphorylated CRY2 in etiolated seedlings exposed to blue light before and after a posttreatment in the dark. It has been shown previously that phosphorylated CRY2 can be detected as an upper-shifted band in immunoblots (Shalitin et al., 2002, 2003), so we used this method to distinguish phosphorylated CRY2 (Figure 4B, CRY2-Pi) from unphosphorylated CRY2 (Figure 4B, CRY2). Figure 4B shows that phosphorylated CRY2 was detected in etiolated seedlings exposed to blue light (15 μmol m−2 s−1) for 2, 5, 10, or 15 min (Figure 4B, CRY2-Pi in lanes 2, 4, 6, and 8), but the phosphorylated CRY2 disappeared after the seedlings were transferred to dark for ∼4 h (Figure 4B, note the absence of the CRY2-Pi bands in lanes 3, 5, 7, and 9). The level of unphosphorylated CRY2 did not increase concurrently to account for the disappearance of phosphorylated CRY2 during dark treatment (Figure 4B, note the similar signal intensity of the unphosphorylated CRY2 bands between lanes 2 and 3, 4 and 5, 6 and 7, or 8 and 9), suggesting that phosphorylated CRY2 was more likely degraded than dephosphorylated in the absence of light. Based on these results, we propose that the reason CRY2 degradation needs continuous blue light is because only part of the photoexcited CRY2 pool is phosphorylated at any given moment and only phosphorylated CRY2 is degradable. In other words, the biochemical reaction of degradation of phosphorylated CRY2 is independent of light. This dark reaction of CRY2 degradation may serve as a desensitization mechanism to remove photoexcited photoreceptor molecules. The results shown previously (Shalitin et al., 2002; Mockler et al., 2003) and in this study (Figure 4B) suggest that there must exist an equilibrium between unphosphorylated CRY2 and phosphorylated CRY2 in light-grown plants. This proposition predicts that plants grown under different light conditions should arrive at different equilibria between phosphorylated and unphosphorylated CRY2, resulting in a different steady state level of the CRY2 protein. Interestingly, etiolated seedlings and seedlings germinated and grown under continuous blue light accumulated similar amounts of CRY2 (Figures 1C and 4C), although etiolated seedlings exposed to blue light for a few hours contain almost no detectable CRY2 (Figure 4C). Therefore, etiolated seedlings exposed to blue light and seedlings germinated and grown in continuous blue light must arrive at different CRY2 equilibria, presumably resulting from combined light effects on synthesis (see Supplemental Figure 3 online), photoconversion (Lin et al., 1995b; Banerjee et al., 2007; Bouly et al., 2007), phosphorylation/dephosphorylation (Shalitin et al., 2002), and degradation of CRY2 (Figure 3). Among those possible points of control, CRY2 phosphorylation is likely the primary rate-limiting reaction in the establishment of CRY2 equilibrium (Figure 4B). Provided that photoreceptor molecules are most likely photoexcited instantaneously when plants are exposed to light, it is intriguing why the photoexcited CRY2 molecules cannot all be phosphorylated instantaneously. It is conceivable that the photoexcited CRY2 needs to be imported into the nucleus to be phosphorylated by a nuclear protein kinase, suggesting that the nuclear importation process or the concentration of the speculative nuclear kinase(s) may be the limiting factors. These hypotheses, however, remain to be tested.
CRY2 Is Ubiquitinated and Degraded via the 26S Proteasome Pathway in Response to Blue Light
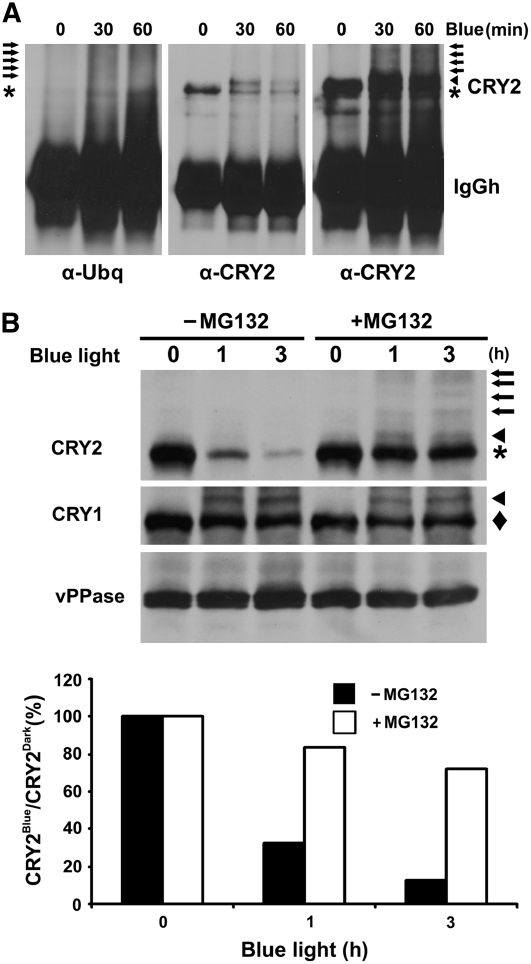
We next examined whether CRY2 may be ubiquitinated in vivo in response to blue light. In this experiment, CRY2 was purified by immunoprecipitation (IP) from etiolated seedlings with or without blue light treatment. The resulting CRY2-IP products were analyzed by immunoblots. The immunoblot was probed first with the anti-ubiquitin antibody, and the blot was then stripped and reprobed with the anti-CRY2 antibody (Figure 5A). As expected, little ubq-conjugate signal was detected from the CRY2-IP product prepared from etiolated seedlings (Figure 5A, left panel, α-Ubq, 0 min Blue), which is consistent with the fact that CRY2 is degraded only in response to blue light. By contrast, abundant ubq-conjugates with apparent molecular masses larger than that of CRY2 were detected in the CRY2-IP product prepared from seedlings exposed to blue light for 30 min (Figure 5A, left panel, α-Ubq, 30 min Blue, arrows). In addition, increased ubiquitin signals with molecular masses smaller than that of CRY2 were detected in seedlings exposed to blue light for 60 min (Figure 5A, left panel, α-Ubq, 60 min Blue), which may represent partially degraded ubq-CRY2 conjugates. The blue light–induced ubq-conjugates were also recognized by the anti-CRY2 antibody (Figure 5A, right panel). A possibility that those ubq-conjugates might result from higher level of background signals due to unequal loading of the CRY2-IP product is ruled out by examining the lighter exposure of the same immunoblot probed by the anti-CRY2 antibody (Figure 5A, middle panel). In contrast with the blue light–induced increase of ubq-conjugates (Figure 5A, left panel), a significantly reduced amount of CRY2 was immunoprecipitated from seedlings exposed to blue light (Figure 5A, middle panel). We concluded that CRY2 is polyubiquitinated in response to blue light.
Figure 5.
CRY2 Is Ubiquitinated in Response to Blue Light and Degraded by the 26S Proteasome.
(A) Immunoblots showing ubiquitin conjugates of the CRY2-IP products. Five-day-old etiolated wild-type seedlings were exposed to blue light (15 μmol m−2 s−1) for 30 or 60 min. CRY2 was immunoprecipitated with the anti-CRY2 antibody, fractionated by 10% SDS-PAGE, and blotted. Immunoblots were probed by the anti-ubiquitin antibody (left panel) or anti-CRY2 antibody (middle and right panels). The middle and right panels are different exposures of the same blot. Arrows, arrowhead, and asterisk indicate ubiquitinated CRY2, phosphorylated CRY2, and unphosphorylated CRY2, respectively.
(B) Inhibition of CRY2 degradation by the proteasome inhibitor MG132. Five-day-old wild-type seedlings grown in the dark were excised and incubated with MG132 or mock solution and exposed to blue light (12 μmol m−2 s−1) for 1 or 3 h. Top: the immunoblot was probed with anti-CRY2 antibody (CRY2; arrows, arrowhead, and asterisk indicate ubiquitinated CRY2, phosphorylated CRY2, and unphosphorylated CRY2, respectively), stripped, and reprobed with the anti-CRY1 antibody (CRY1; arrowhead and diamond indicate phosphorylated and unphosphorylated CRY1, respectively) and then with the antivacuolar pyrophosphatase antibody (vPPase). Bottom: the relative level of CRY2 was calculated as described (see Methods) and presented as the percentage of CRY2blue (level of CRY2 after blue light treatment)/CRY2Dark (level of CRY2 before blue light treatment).
Polyubiquitinated proteins are usually degraded by the 26S proteasome (Vierstra, 2003). To test this, we examined the effects of the 26S proteasome–specific inhibitor MG132 on blue light–dependent CRY2 degradation. In this experiment, etiolated seedlings were incubated in MG132 before blue light treatment. As shown in Figure 5B, in seedlings not treated with MG132, >80% of CRY2 was degraded within 3 h of blue light treatment (Figure 5B). By contrast, in seedlings preincubated with MG132, <30% of CRY2 was degraded after 3 h of blue light treatment (Figure 5B). Figure 5B also shows a modest increase of phosphorylated CRY2 (Figure 5B, arrowhead) and ubiquitinated CRY2 (Figure 5B, arrows) in samples treated with blue light and MG132 compared with etiolated samples or samples treated with blue light but not MG132. These observations are consistent with the hypothesis that phosphorylated CRY2 is ubiquitinated and degraded because both phosphorylated CRY2 and ubiquitinated CRY2 would be expected to accumulate in the absence of degradation. The fact that suppression of CRY2 degradation by MG132 resulted in only a modest increase of phosphorylated CRY2 (Figure 5B, arrowhead) may be explained by phosphorylated CRY2 being largely converted to Ubq-CRY2 conjugates (Figure 5B, arrows). We concluded that CRY2 is phosphorylated and ubiquitinated in response to blue light and that ubiquitinated CRY2 is degraded by the 26S proteasome.
Blue Light–Dependent CRY2 Phosphorylation and Degradation Occur in the Nucleus
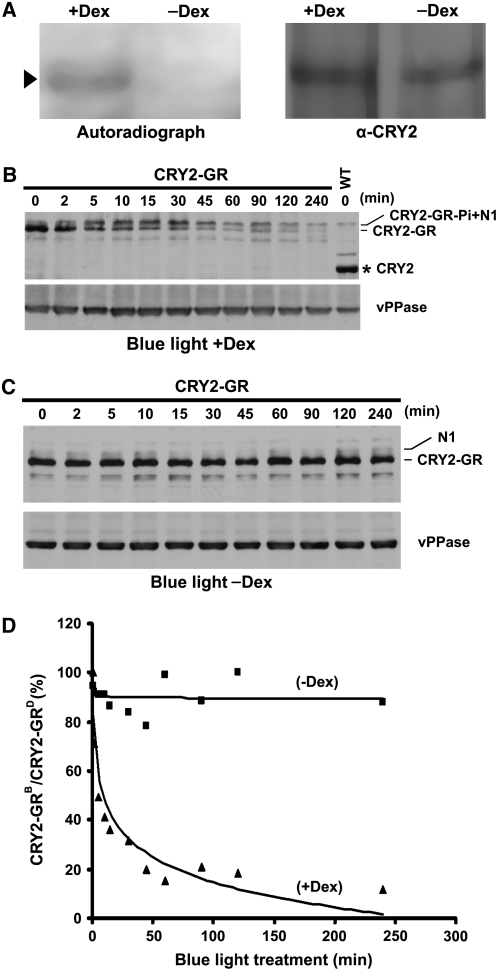
Upon translation in the cytosol, CRY2 may be phosphorylated before or after entering the nucleus. Similarly, nuclear CRY2-ubq conjugates may be exported to the cytosol for degradation or they may be degraded inside the nucleus. To distinguish these possibilities, we examined the location in the cell where CRY2 is phosphorylated or degraded using the conditional CRY2 nuclear localization approach. To test the subcellular localization of CRY2 phosphorylation, etiolated CRY2-GR/cry1 cry2 seedlings were grown on MS medium with Dex (or mock solution), labeled with radioactive 32P, and exposed to blue light (18 μmol m−2 s−1) for 15 min. CRY2-GR was then immunoprecipitated and analyzed for the 32P labeling. Figure 6A shows that CRY2-GR isolated from Dex-treated seedlings exposed to blue light was radioactively labeled with 32P, whereas CRY2-GR isolated from mock-treated seedlings exposed to blue light was not radioactively labeled. Therefore, CRY2 is phosphorylated primarily in the nucleus.
Figure 6.
Blue Light–Dependent Phosphorylation and Degradation of CRY2-GR Occur in the Nucleus.
(A) Autoradiograph showing CRY2-GR was phosphorylated only in the presence of Dex. CRY2-GR/cry1 cry2 seedlings were grown on MS medium with (+Dex) or without (−Dex) Dex, preincubated with 32P for 2 h, and exposed to blue light (18 μmol m−2 s−1) for 15 min. CRY2 was isolated by IP, fractionated, and blotted. After autoradiography (left), the blot was probed with anti-CRY2 antibody (right) to show the level of CRY2. Arrowhead indicates phosphorylated CRY2-GR.
(B) and (C) Immunoblots showing that CRY2-GR underwent blue light–dependent degradation only in the presence of Dex. Five-day-old etiolated CRY2-GR/cry1 cry2 seedlings were treated with Dex (B) or mock solution (C) for 2 h and then exposed to blue light (15 μmol m−2 s−1) for the durations indicated. Protein extracts were fractioned by 10% SDS-PAGE gels, and the immunoblots were probed using anti-CRY2 and then anti-vPPase antibodies. CRY2-GR and CRY2-GR-Pi indicate unphosphorylated or phosphorylated CRY2-GR, respectively, N1 indicates the position of a band nonspecifically recognized by the anti-CRY2 antibody, and the asterisk indicates endogenous CRY2.
(D) The CRY2-GR signals were digitized and calculated as described (see Methods) and presented as the percentage of CRY2-GRB (level of CRY2-GR after blue light treatment)/CRY2-GRD (level of CRY2-GR before blue light treatment).
To test where CRY2 was degraded, CRY2-GR/cry1 cry2 seedlings were preincubated with Dex or mock solution and exposed to blue light (15 μmol m−2 s−1) for up to 4 h. As shown in Figures 6B to 6D, CRY2 degradation was clearly detected in Dex-treated seedlings but not in mock-treated seedlings. After 4 h of blue light treatment, nucleus-localized CRY2-GR (+Dex) was almost completely degraded, whereas cytosol-localized CRY2 (−Dex) showed no clear sign of degradation (Figures 6B to 6D). Under similar fluence rates of blue light (15 to 16 μmol m−2 s−1), the kinetics of degradation of the nuclear-localized CRY2-GR (Figure 6D) is comparable to that of the endogenous CRY2 (Figure 3A). Consistent with the in vivo 32P-labeling experiment (Figure 6A), phosphorylated CRY2-GR that migrates slower was detected only in seedlings treated with blue light and Dex (Figure 1C, arrowhead; Figures 6B and 6C, compare CRY2-GR-Pi+N1 in [B] and N1 in [C]). We conclude that CRY2 phosphorylation and degradation both take place in the nucleus.
DISCUSSION
Most photoreceptors are known to shuttle between different subcellular compartments in response to light. For example, rhodopsins undergo arrestin-dependent endocytosis after photoactivation (Kiselev et al., 2000); phototropins are released from the plasma membrane to the cytoplasm in response to blue light (Sakamoto and Briggs, 2002); phytochromes are imported into the nucleus from the cytosol in a light-dependent manner (Sakamoto and Nagatani, 1996; Nagy et al., 2001; Nagatani, 2004); and cryptochromes also shuttle between the cytoplasm and nucleus (Cashmore et al., 1999; Panda et al., 2002). In this report, we show that Arabidopsis CRY2 completes its posttranslational life cycle in the nucleus: CRY2 mediates blue light inhibition of hypocotyl elongation and photoperiodic regulation of floral initiation in the nucleus, and both blue light–dependent CRY2 phosphorylation and degradation take place in the nucleus. This finding is consistent with the notion that regulation of nuclear gene expression is the major molecular mechanism underlying the function of CRY2 and the discovery that cryptochromes are the major blue light receptors regulating genome-wide gene expression changes in response to blue light (Lin, 2000; Ma et al., 2001; Ohgishi et al., 2004). However, we should point out that the fact that nuclear CRY2 mediates blue light inhibition of hypocotyl elongation and photoperiodic control of floral initiation does not necessarily exclude a possibility that cytosolic CRY2 may possess some additional physiological functions. Similarly, our findings that CRY2 is phosphorylated, ubiquitinated, and degraded in the nucleus do not necessarily rule out additional biochemical or cell biological processes associated with CRY2 located in other cellular compartments.
Our observation that CRY2 undergoes blue light–dependent phosphorylation in the nucleus raises an interesting question concerning cryptochrome autophosphorylation. Arabidopsis CRY1 is known to bind ATP and to have autophosphorylation activity (Bouly et al., 2003; Shalitin et al., 2003; Brautigam et al., 2004; Ozgur and Sancar, 2006). However, under the same experimental condition used to show CRY1 autophosphorylation, autophosphorylation activity of Arabidopsis CRY2 was not detected in vitro (Ozgur and Sancar, 2006; X. Yu and C. Lin, unpublished data). The finding that CRY2 was not phosphorylated in the cytosol in response to blue light (Figure 6A) would be consistent with the possible lack of CRY2 autophosphorylation and existence of a nuclear protein kinase that phosphorylates photoexcited CRY2. However, a seemingly improbable scenario that CRY2 might turn on its autophosphorylation activity only in the nucleus cannot be ruled out at present.
Blue light–dependent degradation of CRY2 has been proposed to play an important role in regulating CRY2 function (Lin et al., 1998; Mockler et al., 2003). We found that CRY2 was ubiquitinated in response to blue light and that the ubiquitinated CRY2 was degraded by the 26S proteasome in the nucleus. We also showed that CRY2 degradation is a high-irradiance response that requires continuous blue light, that only phosphorylated CRY2 is degraded, and that plants grown under different blue light conditions have different steady state levels of CRY2 degradation. In etiolated seedlings exposed to continuous blue light, a continuous phosphorylation and degradation of CRY2 is favored until most CRY2 in the cell is degraded (Figures 3 and 4C). In seedlings germinated and grown under continuous blue light, a different equilibrium is established to allow accumulation of significant amounts of CRY2 (Figure 4C). These results indicate that plants exposed to different light conditions establish different equilibria of CRY2 phosphorylation, which in turn determines the relative amount of bioactive CRY2 in the nucleus. The blue light control of the equilibrium of phosphorylated and unphosphorylated CRY2 may serve an adaptive value enabling plants to respond to an ever-changing light environment in nature.
METHODS
Preparation and Analyses of CRY2-GR Transgenic Lines
The Arabidopsis thaliana accession Columbia (Col), Wassilewskija (Ws), the phytochrome mutants, and the cry1 cry2 mutant (in Col background) used in this study are as described (Guo et al., 1998; Mockler et al., 1999; Franklin et al., 2003; Yu et al., 2007). The truncated rat GR containing the amino acid residues 508 to 795, which is sufficient for Dex-dependent nuclear translocation, was used (Sablowski and Meyerowitz, 1998). The CRY2-GR fusion protein expression construct was prepared by fusing the N terminus (residue 508) of the GR fragment (Sablowski and Meyerowitz, 1998) via a 5–amino acid linker (GPPPG), to the C terminus (residue 612) of the full-length CRY2, at the XhoI and SacI sites of the binary vector pKYLX (Lin et al., 1995a). The 35S:CRY2-GR was introduced into the cry1 cry2 mutant background by the floral dip transformation method. Transgenic plants expressing CRY2-GR under control of the 35S promoter were screened first to select those lines that showed flowering time and/or hypocotyl length similar to that of the parental cry1 cry2. These lines were subject to a secondary screen to select those that expressed CRY2-GR protein but at a level not significantly higher than the endogenous CRY2 in the wild type. Two of which, CRY2-GR-1 and CRY2-GR-2, were shown in this report. During the analyses, we found that higher levels of expression of the CRY2-GR fusion protein were able to rescue the cry1 cry2 mutant phenotype in the absence of Dex, presumably because a higher cellular concentration of CRY2-GR might overwhelm the plasma membrane/cytosol retention HSP90 chaperon machinery to allow leaks of CRY2-GR into the nucleus in the absence of Dex. Hypocotyl length and flowering time were measured as described previously (Yu et al., 2007).
For phenotypic analysis of CRY2-GR/cry1 cry2 seedlings in response to Dex, seedlings were grown in compound soil watered with Dex solution (30 μM Dex, 0.1% ethanol, and 0.01% Tween 20) or mock solution (0.1% ethanol and 0.01% Tween 20) under different light conditions for 4 d before measurement. Dex stock (30 mM) was prepared in 99% ethanol and stored at −20°C until use. Hypocotyls of at least 20 seedlings from different genotypes were measured as described previously (Yu et al., 2007). For flowering-time analysis, plants were grown in compound soil watered with Dex solution or the mock solution until flowering or sprayed daily with Dex or mock solution 10 d after germination until flowering or the time indicated.
LUC-CRY2 Transgenic Lines and Analyses
The LUC-CRY2 fusion protein expression construct was prepared by fusing CRY2 to the C terminus of firefly luciferase in the binary vector pKYLX. Expression of LUC-CRY2 was under control of the 35S promoter. The LUC-CRY2 transgene was introduced into the cry1 mutant. For the LUC-CRY2 degradation assay, seedlings were grown on MS medium in Petri dishes (∼150 seedlings per Petri dish) under continuous far-red light (5 μmol m−2 s−1) for 5 d, and dishes were then exposed to blue light of different fluence rates (4, 17, or 40 μmol m−2 s−1) for 0, 10, 20, 40, 60, or 120 min. After blue light treatment, seedlings were sprayed with luciferin solution (1 mM luciferin and 0.01% Triton X-100) and covered with foil. Luciferase activity was measured 5 min after luciferin application using a CCD camera (Princeton Instruments), digitized using the ImageJ software (http://rsb.info.nih.gov/ij/), and reported as luminescence/seedling after background subtraction and division by number of seedlings tested. The relative level of LUC-CRY2 was calculated by dividing the luminescence/seedling after blue light treatment with that before blue light treatment (grown in far-red light) and presented as LUC-CRY2blue/LUC-CRY2far-red (%).
Immunostaining, Immunoblot, and Phosphorylation Analysis
The enhanced chemiluminescence immunoblot method was as described (Lin et al., 1996). Briefly, proteins were extracted in 4× SDS sample buffer, fractionated in 10% SDS-PAGE mini gels, blotted to nitrocellulose membranes, stained with Ponceau S to verify equal loading, washed, probed with appropriate primary antibodies and the secondary antibody conjugated with horseradish peroxidase, and the signals developed using the enhanced chemiluminescence method (Whitehead et al., 1983). The SDS-PAGE running time, voltage, and temperature appear to determine the fine resolution associated with migration alterations resulting from protein phosphorylation. CRY2 signals were quantified by digitizing the band signal (ImageJ), subtracting the background signal, normalizing against the internal control (CRY1 or vPPase), and the relative level calculated against the value of the dark control. The relative level of CRY2 or CRY2-GR (Figures 3, 5, and 6) is calculated by the formula [(CRY2n − b2n)/(Cn − b1n)]/[(CRY20 − b20)/(C10 − b10)], in which CRY2n represents the signal intensity of CRY2 from a light-treated sample, b2n represents the background signal of the respective CRY2n lane in the immunoblot, C1n represents signal intensity of the loading control (CRY1 or vPPase) from a light-treated sample, b1n represents the background signal of the respective C1n lane in the immunoblot, and CRY20, b20, C10, and b10 represent respective CRY2, loading control, or background signals from the dark controls.
For nuclear immunostaining, seedlings were fixed in ice-cold fixation buffer (4% formaldehyde, 10 mM Tris-HCl, pH 7.5, 10 mM EDTA, and 100 mM NaCl), finely chopped using a razor blade, and suspended in sorting buffer (100 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM MgCl, 0.05% Tween 20, and 5% sucrose). Nuclei were sorted using a Falcon polystyrene cell strainer (Becton-Dickinson Labware). Nuclei were incubated with anti-CRY2 antibody (1/100 dilution), washed (10 mM sodium phosphate, pH 7.0, and 143 mM NaCl), incubated with Rhodamine Red-x conjugated goat anti-rabbit IgG (1/200 dilution) (Jackson ImmunoResearch Lab), and washed thoroughly. Images were captured at ×100 magnification using a Zeiss Axioskop 2 microscope with a Zeiss Axiocam HRC color digital camera or Zeiss AxioImager Z1 microscope with a Hamamatsu Orca-ER camera, analyzed using Zeiss Axio Vision software, and processed using Adobe Photoshop (Adobe Systems).
32P-radioactive labeling of CRY2 was as described previously (Shalitin et al., 2002, 2003; Yu et al., 2007). For the MG132 treatment, 5-d-old wild-type seedlings grown on MS medium in the dark were excised into 2- to 5-mm-long sections and incubated in 50 μM MG132 (EMD Chemicals) in 0.1% DMSO (prepared from a fresh stock of 50 mM in DMSO) or 0.1% DMSO mock control in the dark for 5 h. Explants were then exposed to blue light (12 μmol m−2 s−1) for the indicated durations. Proteins were extracted and boiled in 4× SDS-PAGE sample buffer, separated in 10% SDS-PAGE gels, blotted onto nitrocellulose, and probed with antibodies. The anti-Ubq antibody was purchased from Santa Cruz Biotechnology, and the anti-CRY2 and anti-vPPase antibodies are as previously described (Shalitin et al., 2002; Yu et al., 2007). To analyze CRY2 degradation in the absence of protein synthesis, sterilized seeds were sown on a filter paper placed on top of MS medium. MS medium was purchased as a premixed powder that contained sucrose (Sigma-Aldrich). Five-day-old seedlings were lifted together with the filter paper, incubated in 10 μM cycloheximide for 4 h, and then exposed to blue light.
RT-PCR
Seeds of the Arabidopsis wild type, cry1 cry2, and the CRY2-GR transgenic line were sterilized and sown on MS medium with or without 30 μM Dex. The plates were stratified at 4°C for 4 d, exposed to white light for 4 h, and placed in long-day photoperiodic conditions (16 h light/8 h dark). Seven-day-old seedlings were harvested at 15 (ZT15) and 18 (ZT18) h after the light was on. Total RNA was isolated using the RNeasy plant mini kit (Qiagen) and treated with RQ1 DNase I (2 units of DNase per 5.0 μg in a 20-μL reaction) according to the manufacturer's instructions (Promega). cDNA was prepared from 5.0 μg of RQ1 DNase I-treated RNA using the SuperScript first-strand cDNA synthesis system according to the manufacturer's instructions (Invitrogen). RT-PCR was performed as described previously (Mockler et al., 2004), for which the PCR conditions of the logarithmic phase were predetermined, and multiple biological samples were examined.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CRY2 (AT1G04400), CRY1 (AT4G08920), FT (AT1G65480), and UBQ (At4g05320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunoblot Showing CRY2 Protein Expression in Plants at Different Ages.
Supplemental Figure 2. CRY2 Failed to Be Degraded in the Cell-Free Extract in Response to Blue Light.
Supplemental Figure 3. RNA Gel Blot Showing CRY2 mRNA Expression Is Not Suppressed by Blue Light.
Supplemental Figure 4. Immunoblot Showing the Temperature Dependence of Blue Light–Dependent CRY2 Degradation.
Supplemental Figure 5. Immunoblots Showing That Blue Light–Induced CRY2 Degradation Does Not Require CRY1 or Phytochromes Tested.
Supplementary Material
Acknowledgments
We thank G. Whitelam for providing mutants impaired in multiple phytochrome genes, R.W. Sablowski and E.M. Meyerowitz for providing the glucocorticoid receptor vector, S. Kay for providing luciferase reporter vector, E. Tobin and S. Knowles for LUC analyses, and G Villa for assistance in experiments. This work is supported in part by the National Institutes of Health (GM56265 to C.L.), a Changjiang scholarship (to C.L.), and the 211 and 985 higher education enhancement funds to Hunan University. J.L. was partially supported by the UC MEXUS-CONACYT postdoctoral fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chentao Lin (clin@mcdb.ucla.edu).
Online version contains Web-only data.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., and Cashmore, A.R. (1998). Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, R., Schleicher, E., Meier, S., Viana, R.M., Pokorny, R., Ahmad, M., Bittl, R., and Batschauer, A. (2007). The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem. 282 14916–14922. [DOI] [PubMed] [Google Scholar]
- Bouly, J.P., Giovani, B., Djamei, A., Mueller, M., Zeugner, A., Dudkin, E.A., Batschauer, A., and Ahmad, M. (2003). Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem. 270 2921–2928. [DOI] [PubMed] [Google Scholar]
- Bouly, J.P., Schleicher, E., Dionisio-Sese, M., Vandenbussche, F., Van Der Straeten, D., Bakrim, N., Meier, S., Batschauer, A., Galland, P., Bittl, R., and Ahmad, M. (2007). Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282 9383–9391. [DOI] [PubMed] [Google Scholar]
- Brautigam, C.A., Smith, B.S., Ma, Z., Palnitkar, M., Tomchick, D.R., Machius, M., and Deisenhofer, J. (2004). Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101 12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R. (2003). Cryptochromes: Enabling plants and animals to determine circadian time. Cell 114 537–543. [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284 760–765. [DOI] [PubMed] [Google Scholar]
- El-Assal, S.E.-D., Alonso-Blanco, C., Peeters, A.J., Raz, V., and Koornneef, M. (2001). A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29 435–440. [DOI] [PubMed] [Google Scholar]
- Endo, M., Mochizuki, N., Suzuki, T., and Nagatani, A. (2007). CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta, K.M., and Spalding, E.P. (2001). Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26 471–478. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., Davis, S.J., Stoddart, W.M., Vierstra, R.D., and Whitelam, G.C. (2003). Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Duong, H., Ma, N., and Lin, C. (1999). The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 19 279–287. [DOI] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363. [DOI] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., and Quail, P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35 660–664. [DOI] [PubMed] [Google Scholar]
- Kiselev, A., Socolich, M., Vinos, J., Hardy, R.W., Zuker, C.S., and Ranganathan, R. (2000). A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron 28 139–152. [DOI] [PubMed] [Google Scholar]
- Kleine, T., Lockhart, P., and Batschauer, A. (2003). An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 35 93–103. [DOI] [PubMed] [Google Scholar]
- Kleiner, O., Kircher, S., Harter, K., and Batschauer, A. (1999). Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 19 289–296. [DOI] [PubMed] [Google Scholar]
- Lin, C. (2000). Photoreceptors and regulation of flowering time. Plant Physiol. 123 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., and Cashmore, A.R. (1996). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light- dependent regulation of plant growth and development. Plant J. 10 893–902. [DOI] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., Gordon, D., and Cashmore, A.R. (1995. a). Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc. Natl. Acad. Sci. USA 92 8423–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Jorns, M.S., Dutton, P.L., and Cashmore, A.R. (1995. b). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269 968–970. [DOI] [PubMed] [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A.M., Schena, M., Walbot, V., and Davis, R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266 436–439. [DOI] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y.C., Dolan, S., and Lin, C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082. [DOI] [PubMed] [Google Scholar]
- Mockler, T.C., Yu, X., Shalitin, D., Parikh, D., Michael, T.P., Liou, J., Huang, J., Smith, Z., Alonso, J.M., Ecker, J.R., Chory, J., and Lin, C. (2004). Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. USA 101 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, S.G., Kim, Y.S., Kunkel, T., and Chua, N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7 708–711. [DOI] [PubMed] [Google Scholar]
- Nagy, F., Kircher, S., and Schafer, E. (2001). Intracellular trafficking of photoreceptors during light-induced signal transduction in plants. J. Cell Sci. 114 475–480. [DOI] [PubMed] [Google Scholar]
- Ohgishi, M., Saji, K., Okada, K., and Sakai, T. (2004). Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Ozgur, S., and Sancar, A. (2006). Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry 45 13369–13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda, S., Hogenesch, J.B., and Kay, S.A. (2002). Circadian rhythms from flies to human. Nature 417 329–335. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92 93–103. [DOI] [PubMed] [Google Scholar]
- Sakamoto, K., and Briggs, W.R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell 14 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10 859–868. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sancar, A. (2003). Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103 2203–2237. [DOI] [PubMed] [Google Scholar]
- Selby, C.P., and Sancar, A. (2006). A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 103 17696–17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417 763–767. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yu, X., Maymon, M., Mockler, T., and Lin, C. (2003). Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, R., Kevei, E.E., Hall, A., Millar, A.J., Nagy, F., and Kozma-Bognar, L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- Whitehead, T.P., Thorpe, G.H.G., Carter, T.J.N., Groucutt, C., and Kricka, L.J. (1983). Enhanced luminescence procedure for sensitive determination of peroxidase-labelled conjugates in immunoassay. Nature 305 158–159. [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Yu, X., Shalitin, D., Liu, X., Maymon, M., Klejnot, J., Yang, H., Lopez, J., Zhao, X., Bendehakkalu, K.T., and Lin, C. (2007). Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. USA 104 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.