Abstract
Two ancient and highly divergent actin-based cytoskeletal systems have evolved in angiosperms. Plant genomes encode complex actin and actin binding protein (ABP) gene families, most of which are phylogenetically grouped into gene classes with distinct vegetative or constitutive and reproductive expression patterns. In Arabidopsis thaliana, ectopic expression of high levels of a reproductive class actin, ACT1, in vegetative tissues causes severe dwarfing of plants with aberrant organization of most plant organs and cell types due to a severely altered actin cytoskeletal architecture. Overexpression of the vegetative class actin ACT2 to similar levels, however, produces insignificant phenotypic changes. We proposed that the misexpression of the pollen-specific ACT1 in vegetative cell types affects the dynamics of actin due to its inappropriate interaction with endogenous vegetative ABPs. To examine the functionally distinct interactions among the major classes of actins and ABPs, we ectopically coexpressed reproductive profilin (PRF4) or actin-depolymerizing factor (ADF) isovariants (e.g., ADF7) with ACT1. Our results demonstrated that the coexpression of these reproductive, but not vegetative, ABP isovariants suppressed the ectopic ACT1 expression phenotypes and restored wild-type stature and normal actin cytoskeletal architecture to the double transgenic plants. Thus, the actins and ABPs appear to have evolved class-specific, protein–protein interactions that are essential to the normal regulation of plant growth and development.
INTRODUCTION
In plant cells, the actin cytoskeleton undergoes dramatic reorganization in response to external and internal cues and thus regulates several vital cellular processes, including cytoplasmic streaming, organelle movement and repositioning, establishment of cell polarity, tip growth, cell division, expansion, and differentiation, and reactions to pathogen attack, wounding, and hormones (reviewed in Staiger, 2000; Wasteneys and Galway, 2003; Hussey et al., 2006; Staiger and Blanchoin, 2006). The dynamic reorganization of actin is modulated by the specific activity of a plethora of actin binding proteins (ABPs) that either promote or inhibit actin polymerization. Actins and most ABPs in plants are encoded by multigene families (Meagher and Fechheimer, 2003). For example, the Arabidopsis thaliana genome contains eight actin genes that are grouped based on their phylogeny and expression pattern into two major classes: vegetative and reproductive (Figure 1A). The vegetative class actin genes are expressed constitutively in all vegetative organs and cell types, whereas the reproductive class actin genes are expressed predominantly in the mature pollen, growing pollen tubes, and in some cases in ovules (e.g., ACT11; Meagher et al., 1999b). The vegetative actins differ from the reproductive actins by 4 to 7% at the amino acid sequence level, a difference similar to or even greater than that among cytoplasmic and muscle actins of vertebrates. Moreover, these two ancient and differentially expressed plant actin classes have not shared a common ancestral gene for at least 300 million years (McDowell et al., 1996; Meagher et al., 2000). Profilins and actin-depolymerizing factors (ADFs), the two most highly expressed ABPs in plants, also contain ancient subclasses of genes that are differentially expressed in patterns clearly defined as vegetative or constitutive and reproductive (Hussey et al., 2002; Kandasamy et al., 2002b).
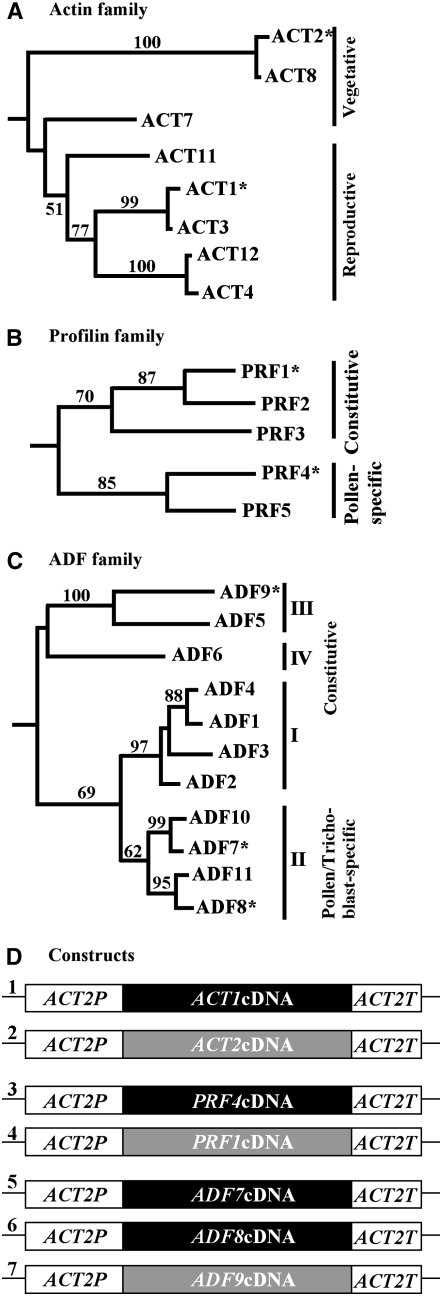
Figure 1.
Arabidopsis Actin, Profilin, and ADF Families.
(A) Actin tree showing two major classes of protein isovariants: vegetative and reproductive, which are encoded by three and five actin genes, respectively.
(B) Profilin tree showing two ancient classes of protein isovariants, constitutive and pollen specific, which are encoded by three and two profilin genes, respectively.
(C) ADF tree containing 11 proteins that are grouped into two major classes, constitutive and pollen/trichoblast specific, and encoded by four subclasses of genes (I to IV). The asterisks represent various protein isovariants that were overexpressed or misexpressed in this study. Branches with bootstrap values >50% are indicated in these neighbor-joining trees (see Methods).
(D) Various actin (1, A2P:A1; 2, A2P:A2), profilin (3, A2P:P4; 4, A2P:P1), and ADF (5, A2P:ADF7; 6, A2P:ADF8; 7, A2P:ADF9) constructs used in the ectopic expression and suppression studies. The ACT2 expression cassette is shown in two white boxes [left box: promoter, 5′ untranslated region, leader exon, to AUG codon of second exon; right box: 3′ untranslated region and poly(A) sequences]. The black boxes represent cDNAs from reproductive class genes, and the gray boxes indicate vegetative or constitutive cDNAs.
Significantly, the two classes of actin-based cytoskeletal systems appear to be functionally distinct in most angiosperms. This supposition is supported by the presence in maize (Zea mays) of two distinct classes of profilins that are different in their biochemical properties. The vegetative (class II) profilins of maize have higher affinity for poly-l-Pro, and they sequester more monomeric actin and disrupt the actin cytoplasmic architecture in live cells more rapidly than pollen-specific (class I) profilins (Kovar et al., 2000). Moreover, our studies of ectopic expression of a reproductive actin in vegetative tissues also suggest distinct roles for vegetative and reproductive classes of actins in Arabidopsis (Kandasamy et al., 2002a). We find that the misexpression of the pollen-specific reproductive actin ACT1, but not the overexpression of the vegetative actin ACT2, in vegetative tissues is extremely toxic and results in the formation of severely dwarfed plants with a highly altered organization of the actin cytoskeleton. We hypothesized that the high level ectopic expression of pollen actin in vegetative tissues affected actin dynamics, perhaps due to the weak interactions among the reproductive actin and the endogenous vegetative ABPs, thus leading to massive polymerization of actin into star- or sheet-like structures. This hypothesis predicts the existence of essential class-specific interactions among the actins and ABPs to properly streamline the formation of the actin microfilament arrays necessary for normal development of different plant organs and cell types. It appears that disruption in the balance between actin and the coexpressed accessory proteins leads to deleterious consequences for cell morphology and proliferation, resulting in aberrant plant development.
In yeast, which has a single actin gene, overproduction of actin is lethal. But when profilin is overexpressed together with actin, the deleterious effects of high levels of actin are suppressed (Magdolen et al., 1993). Profilin is a small (14 kD) actin monomer binding protein that forms 1:1 complexes with monomeric G-actin and buffers actin filament formation by sequestering G-actin from the soluble pool and participating in the polymerization of F-actin (Carlsson et al., 1977; Pollard and Cooper, 1986). To examine whether profilin can counteract the deleterious effects of overproduction of pollen-specific ACT1 in the vegetative organs and to illustrate the existence of class-specific interactions among actins and ABPs in vivo, we ectopically coexpressed reproductive actin and different profilin isovariants in the same transgenic plants. Here, we demonstrated that simultaneous overproduction of reproductive actin and reproductive profilin, but not reproductive actin and vegetative profilin, almost fully suppressed the morphological and cytoskeletal phenotypes of dwarf plants. Moreover, we have shown that the ADFs, which bind both monomeric and filamentous actin and thus regulate actin organization, also suppressed the ACT1 ectopic expression phenotypes in a class-specific manner. Only the pollen/trichoblast-specific class of ADF genes, but not the constitutive class of ADFs, significantly suppressed the ACT1-induced morphological and cellular phenotypes. Because the ADF proteins regulate actin dynamics by depolymerizing filaments from their pointed ends and by severing actin filaments and increasing the concentration of barbed ends for promotion of actin polymerization (Carlier, 1998; Bamburg, 1999; Chen et al., 2000), we assume that ectopically overproducing reproductive actin and the appropriate class of ADFs might have led to balanced, normal remodeling of actin filaments. Our results confirm the existence of functional specificity among the vegetative and reproductive classes of actins and ABPs and suggest that class-specific interactions between these two classes of cytoskeletal proteins in vivo are essential for proper control of actin assembly and normal development of plants.
RESULTS
Ectopic Expression of Reproductive ACT1 in Vegetative Tissues Is Toxic to Plant Growth
Arabidopsis contains a modestly large actin gene family with eight expressed genes: three vegetative (ACT2, ACT7, and ACT8) and five reproductive (ACT1, ACT3, ACT4, ACT11, and ACT12; Figure 1A). Among the five reproductive class genes, ACT11 is expressed in both male and female reproductive tissues, whereas the rest of the four genes are predominantly expressed in mature pollen and growing pollen tubes. By contrast, the three vegetative class actin genes are expressed constitutively and strongly in all vegetative organs and tissues. Moreover, ACT7 is induced to higher levels in young and meristematic tissues and in response to hormones (Meagher et al., 1999b; Zimmermann et al., 2004). In addition to their differential expression patterns, the two classes of actin genes are functionally distinct. Ectopic expression of pollen-specific ACT1 under the control of vegetative ACT2 regulatory sequences (A2P:A1) results in the production of dwarf transgenic plants (Figures 2A, 2D, and 3) with altered organ morphology and aberrant actin cytoskeletal organization (Kandasamy et al., 2002a). In the dwarf plants, A2P:A1 expression increased the total actin levels three- to fourfold above the levels of endogenous vegetative actin. By contrast, transgenic plants with low levels of ACT1 expression (less than onefold above endogenous vegetative actin) have plant morphology very much similar to the wild type (data not shown). Also, none of the normal-looking transgenic plants contained higher levels of ACT1, and there was always a direct correlation between the level of ACT1 expression and the extent of defect in the morphology of the plant (Kandasamy et al., 2002a). On the other hand, overexpression of ACT2 under its own regulatory sequences (A2P:A2) increased the level of total actin three- to fourfold (Figure 2H) but did not result in any extreme dwarf phenotypes in the wild type (Figures 2B and 2E) or the act2-1 mutant background (data not shown). A small percentage of plants (<2%) overexpressing high levels of ACT2 was 50% or more smaller than the wild type, whereas a higher percentage (20%) of plants misexpressing similar levels of ACT1 was highly dwarf and abnormal (Figure 3). This suggests that perhaps even higher levels ACT2 (severalfold overproduction) may be necessary to reveal any aberrant phenotype.
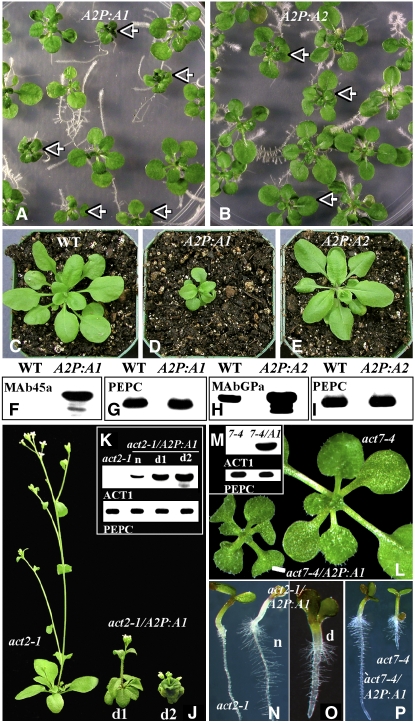
Figure 2.
Effect of Ectopic Expression of ACT1 Protein on the Morphology of Plants.
(A) Seventeen-day-old wild-type seedlings misexpressing pollen-specific ACT1. Dwarf plants expressing high levels of ACT1 are marked with arrowheads.
(B) Seventeen-day-old wild-type seedlings overexpressing vegetative ACT2. Plants expressing high levels of ACT2 are marked with arrowheads.
(C) Twenty-four-day-old wild-type control plant.
(D) Twenty-four-day-old transgenic plant misexpressing ACT1.
(E) Twenty-four-day-old transgenic plant overexpressing ACT2.
(F) A protein gel blot showing a 45-kD ACT1 band in the transgenic (A2P:A1) but not wild-type sample. Probed with the reproductive anti-actin antibody MAb45a (see Methods).
(G) A strip of the same blot in (F) probed with anti-PEP carboxylase (PEPC) antibody to show equal loading of total protein.
(H) A protein gel blot showing a fourfold overexpression of ACT2 in the A2P:A2 transgenic plant compared with the wild type. Probed with the general anti-actin antibody MAbGPa (see Methods).
(I) A strip of the same blot in (H) probed with anti-PEPC antibody to show equal loading of total protein.
(J) Thirty-day-old act2-1 plant and mutant plants (d1 and d2) misexpressing ACT1.
(K) Protein gel blot analysis of transgenic act2-1 mutant plants expressing different levels of ACT1. Note the absence of ACT1 in the act2-1 mutant. The bottom blot shows equal amount of PEPC in all samples. n, normal plant; d1 and d2, dwarf plants.
(L) Sixteen-day-old act7-4 mutant plant and a dwarf mutant plant expressing ACT1.
(M) Protein gel blot analysis of a transgenic act7-4 mutant plant expressing ACT1. Note the absence of ACT1 in act7-4 mutant. The bottom blot shows equal amount of PEPC in both samples.
(N) Four-day-old act2-1 mutant and normal act2-1 transgenic seedlings expressing low levels of ACT1. Note the complementation of stunted root hair phenotype in the transgenic seedling (n).
(O) Six-day-old dwarf act2-1 mutant seedling expressing high levels of ACT1.
(P) A 4-d-old act7-4 mutant and a normal act7-4 transgenic seedling expressing ACT1. Note the complementation of retarded root growth phenotype in the transgenic seedling (act7-4/A2P:A1).
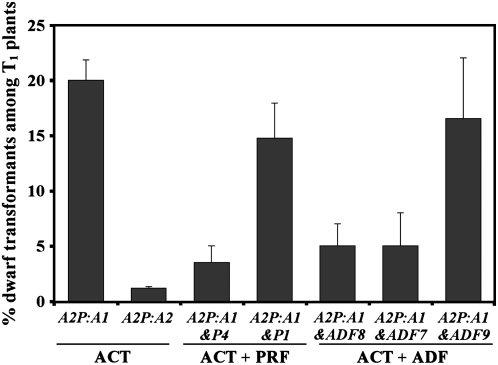
Figure 3.
Profilin and ADF-Mediated Suppression of the ACT1-Induced Dwarf Phenotype.
Note that the coexpression of reproductive and vegetative profilin or ADF isovariants with ACT1 differentially reduce the percentage of dwarf plants. The results represent at least two different vacuum infiltration experiments. More than 100 independent transgenic lines were observed for each ACT and ACT + PRF constructs, and 75 lines were examined for each ACT + ADF construct. The bars represent sd.
To further demonstrate that the misexpression, but not overproduction, of actin caused aberrant architecture of plants, we introduced ACT1 into mutant plants expressing significantly reduced amounts of total actin. For example, transformation of A2P:A1 (ACT1) into act2-1 mutant plants, which contain ∼40% reduced levels of total actin (data not shown), also produced extremely small and highly aberrant transformants (Figure 2J). Furthermore, the misexpression of ACT1 in the act2-1 mutants affected the morphology of the plants in an ACT1 protein concentration-dependant manner (Figure 2K). Transgenic mutant plants expressing high levels of ACT1 were severely dwarfed with highly curved, tiny leaves compared with the untransformed act2-1 plants (Figures 2J and 2K). Also, they were fully sterile. Even manual self-pollination or cross-pollination of stigmas of these severely dwarf plants with wild-type pollen resulted in the production of no siliques or seeds. The plants with slightly lower levels of ACT1 than these extremely dwarf plants were small, but they had mildly curved leaves (see d1 in Figures 2J and 2K), and they were partially fertile. By contrast, plants with very low levels of ACT1 (see n in Figure 2K) were normal like the wild type and fully fertile (data not shown). However, both normal (Figure 2N, right; Gilliland et al., 2002) and all dwarf transgenic plants (Figure 2O) revealed suppression of the stunted act2-1 mutant root hair phenotype (Figure 2N, left). Moreover, the introduction of the A2P:A1 transgene into act7-4 mutant plants that showed ∼30% reduction in the total protein levels (data not shown) also induced a dwarf plant phenotype (Figures 2L and 2M). However, the ectopic expression of ACT1 still suppressed the retarded root growth phenotype of the act7-4 mutant plants (Figure 2P). Thus, the ACT1 misexpression phenotype in wild-type plants is not due to too much actin alone because ACT1 expression induced the dwarf phenotype even in actin mutants that showed significant reduction in the endogenous vegetative actin levels. The manifestation of severely dwarf plant architecture is consequently due to the overexpression of the inappropriate class of actin. Also, the complementation of the vegetative actin phenotypes revealed in a subset of cell types in actin mutants suggests that ACT1 (also control ACT2; data not shown) overproduced in the vegetative cells is functional and most likely folded correctly.
Heterologous Expression of Arabidopsis Reproductive and Vegetative Actins in Yeast Affects Cell Growth and Cytoskeletal Architecture
Because the formation of actin filaments is delicately balanced by the interaction of actin (monomeric and filamentous) with a complex system of accessory ABPs in all eukaryotic cells, we presumed that the dwarf phenotype and aberrant cytoskeleton described above are caused by the poor interaction of the reproductive ACT1 with the endogenous vegetative ABPs expressed in the vegetative organs and cell types (Kandasamy et al., 2002a). To examine the behavior of plant actins in a heterologous system, we coexpressed a reproductive actin isovariant, ACT12, or a vegetative isovariant, ACT8, with yeast actin in yeast cells. For ease of cloning, the reproductive ACT12 and vegetative ACT8 genes were chosen for our studies with yeast cells, instead of the homologous ACT1 and ACT2, respectively. Our results showed that high levels of both classes of plant actin isovariants were toxic to yeast cells, as evidenced by their slow growth and altered cell morphology (large cell size) and cytoskeletal architecture. While trace amounts of ACT12 and ACT8 proteins were incorporated into F-actin patches and filaments, the majority of plant actins were found in unusual rod-like structures, thick cables and spots that are mostly found in the mother cells and stained only with plant actin-specific antibody (see Supplemental Figures 1B and 1C online) and not with phalloidin (data not shown). However, the F-actin binding reagent phalloidin stained actin patches heavily in the bud and actin filaments mainly in the mother cell of actively growing wild-type yeast cells (see Supplemental Figure 1A online). These results suggest that plant actins were incorrectly organized into rods or wrongly folded in yeast, perhaps because the fungal accessory proteins did not effectively recognize plant actin sequences.
Ectopic Expression of Reproductive Profilin Does Not Produce an Aberrant Phenotype
Arabidopsis encodes five highly divergent profilin isovariants that are grouped based on their phylogeny and expression patterns into two ancient classes: constitutive (PRF1-3) and pollen specific (PRF4 and 5) (Kandasamy et al., 2002b; Figure 1B). To examine whether misexpression of a pollen-specific profilin in vegetative tissues, like reproductive ACT1, has any toxic effect on plant growth, we ectopically expressed PRF4 under the control of the ACT2 regulatory sequences (A2P:P4). Protein gel blot analysis with a PRF4-specific monoclonal antibody revealed no endogenous expression of PRF4 in the leaf tissue of wild-type plants but showed various levels of PRF4 expression in different (>50) A2P:P4 transgenic plants examined (Figure 4C). Even plants containing very high levels of PRF4 in vegetative tissues looked morphologically indistinguishable from wild-type plants (Figures 4A and 4B). Immunolocalization confirmed the absence of any PRF4 protein in the leaf cells of wild-type seedlings and a high level of PRF4 expression in the transgenic plants, where it is seen dispersed uniformly throughout the cytoplasm (Figures 4D and 4E). Moreover, we constitutively overexpressed the vegetative PRF1 under the regulation of the ACT2 regulatory sequences (A2P:P1). On protein gel blots, a PRF1-specific monoclonal antibody detected a 14-kD faint band in the wild-type sample and a strong PRF1 band in the A2P:P1 transgenic plants (Figure 4H). Immunolabeling also revealed a relatively higher intensity of PRF1 staining in the transgenic leaf cells compared with the wild type (Figures 4I and 4J). Quantification of PRF1 bands on the protein gel blots of wild-type and dozens of transgenic plant leaf samples suggested that even a three- to sixfold overproduction of PRF1 in the transgenic plants did not produce any obvious morphological abnormality (Figures 4F to 4H). Thus, both the misexpression of PRF4 and overexpression of PRF1, even at high levels, were not harmful to the growth of Arabidopsis plants. These results were surprising because earlier studies of microinjection of purified recombinant birch pollen profilin into Tradescantia stamen hair cells showed drastic changes in cellular organization and cessation of cytoplasmic streaming (Staiger et al., 1994; Valster et al. 1997). These discrepancies may very well be due to the significantly higher concentrations of profilin injected into the stamen hair cells (i.e., ∼5- to 15-fold increase in cytoplasmic profilin concentration in interphase cells) compared with only a two- to fourfold overproduction of total profilin (endogenous profilin + transgenic PRF1 or PRF4) achieved in the transgenic Arabidopsis tissue.
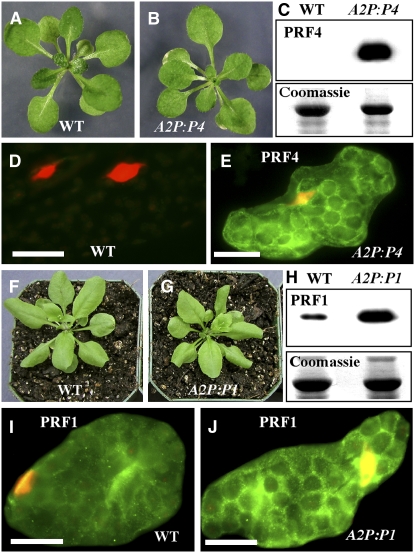
Figure 4.
Effect of Profilin Ectopic Expression and Overexpression on Plant Growth.
(A) Three-week-old wild-type seedling.
(B) Three-week-old transgenic seedling misexpressing PRF4.
(C) Protein gel blot (top panel) showing expression of PRF4 in transgenic (A2P:P4) but not wild-type leaf sample. Top blot reacted with MAbPRF45. The bottom panel shows a Coomassie blue–stained duplicate gel.
(D) Wild-type leaf cell labeled with MAbPRF45 showing no staining. Nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI) are shown in red.
(E) Transgenic (A2P:P4) leaf cell showing strong staining for PRF4. Orange color indicates nucleus.
(F) Twenty-five-day-old wild-type plant.
(G) Twenty-five-day-old transgenic plant overexpressing PRF1 (A2P:P1).
(H) Protein gel blot (top panel) showing strong expression of PRF1 in transgenic (A2P:P1) plant. Wild-type leaf sample shows a 14-kD faint band. Top blot reacted with PRF1-specific MAbPRF1. The bottom panel shows a Coomassie blue–stained duplicate gel.
(I) Wild-type leaf cell labeled with MAbPRF1 showing a weak staining. Nucleus stained with DAPI is shown in red.
(J) Transgenic (A2P:P1) leaf cell showing a strong staining for PRF1. Orange color indicates nucleus.
Bars = 20 μm.
High-Level Expression of Reproductive, but Not Vegetative, Profilin Suppresses the ACT1 Misexpression Phenotype
In yeast, coexpression of the actin monomer binding protein profilin had been shown to suppress the deleterious effects of the overexpression of actin by reducing actin assembly (Magdolen et al., 1993). So, to examine whether overproduction of profilin can suppress the dwarf phenotype and the aberrant cytoskeletal architecture of ACT1 misexpression in Arabidopsis, we coexpressed reproductive actin and profilin isovariants together in the same vegetative organs using the ACT2 regulatory sequences. First, we identified two A2P:P4 single insertion plant lines from the various PRF4-misexpressing transgenic lines by scoring the segregation ratios of antibiotic (hygromycin [Hyg]) resistant to sensitive seedlings (3:1). These lines were also tested for high levels of stable PRF4 protein expression in the vegetative tissue by protein gel blot analysis. ACT1 (A2P:A1) was then introduced separately into both of these PRF4-misexpressing transgenic lines by Agrobacterium tumefaciens–mediated transformation. More than 100 independent double transgenic lines (∼50 for each PRF4 line) were isolated based on antibiotic resistance (Hygr for PRF4 and kanamycinr [Kan] for ACT1). Examination of seedlings and adult plants revealed that <3% of these double transformants expressing both ACT1 and PRF4 were smaller than the wild type (50% or less the size), whereas 20% or more of the plants misexpressing ACT1 alone were dwarf (Figure 3). Protein gel blot analysis showed that several (∼15%) of the normal-looking A2P:A1 and P4 double transgenic plants contained high levels of ACT1, similar to the dwarf A2P:A1 single transformants (e.g., Figures 5A and 5C). The rest of the plants (∼85%) had only low-level expression of ACT1 and thus were expected to be morphologically normal. However, probing of duplicate blots using PRF4-specific monoclonal antibodies suggested that all these double transgenic plants contained high levels of PRF4 protein, whereas no PRF4 expression appeared in the wild-type or A2P:A1 dwarf plants (Figure 5D). These results indicate that coexpression of high levels of the reproductive PRF4 in vegetative tissues suppressed the ACT1-induced dwarf phenotype. In other words, although they contain high levels of ACT1, this group of 15% or more plants looked similar in size (>90% height) to the wild-type plants with large rosette leaves and thick inflorescence stems (Figures 5A and 5B), and they were fully fertile (data not shown). Next, we examined the actin cytoskeletal architecture of these suppressed plants by immunofluorescence microscopy analysis of leaf cells. Our qualitative observations showed that the A2P:A1 and P4 plant cells (Figures 6E and 6F) had more intense actin staining compared with wild-type cells (Figures 6A and 6B), but almost all of these cells lacked the aberrant star- or sheet-like aggregates of actin typically observed in the A2P:A1 plants (Figures 6C and 6D). Thus, the coexpression of reproductive PRF4 significantly reduced the abnormal actin structures induced by the misexpression of reproductive ACT1.
Figure 5.
Suppression of ACT1-Induced Dwarf Phenotype by Coexpression of Pollen-Specific PRF4.
(A) Approximately 7-week-old plants. A2P:A1, dwarf single transformant misexpressing ACT1; A2P:A1&P4, a double transformant misexpressing both ACT1 and PRF4 simultaneously. Note the suppression of dwarf phenotype in the double transgenic plant.
(B) Rosette leaves of just-bolted wild-type and single (A2P:A1) and double (A2P:A1&P4) transgenic plants.
(C) Protein gel blot analysis of ACT1 expression. Top panel shows a blot probed with MAb45a for ACT1. Both the dwarf single transformant misexpressing ACT1 and the normal double transgenic plant coexpressing ACT1 and PRF4 have same levels of ACT1 protein. The wild type has no detectable ACT1. Bottom panel shows a Coomassie blue–stained duplicate gel.
(D) Protein gel blot analysis of PRF4 expression. Top panel shows a blot probed with MAbPRF45 for PRF4 protein. Only the double transgenic plant shows strong PRF4 band. Bottom panel is a Coomassie blue–stained duplicate gel to show equal loading of total protein.
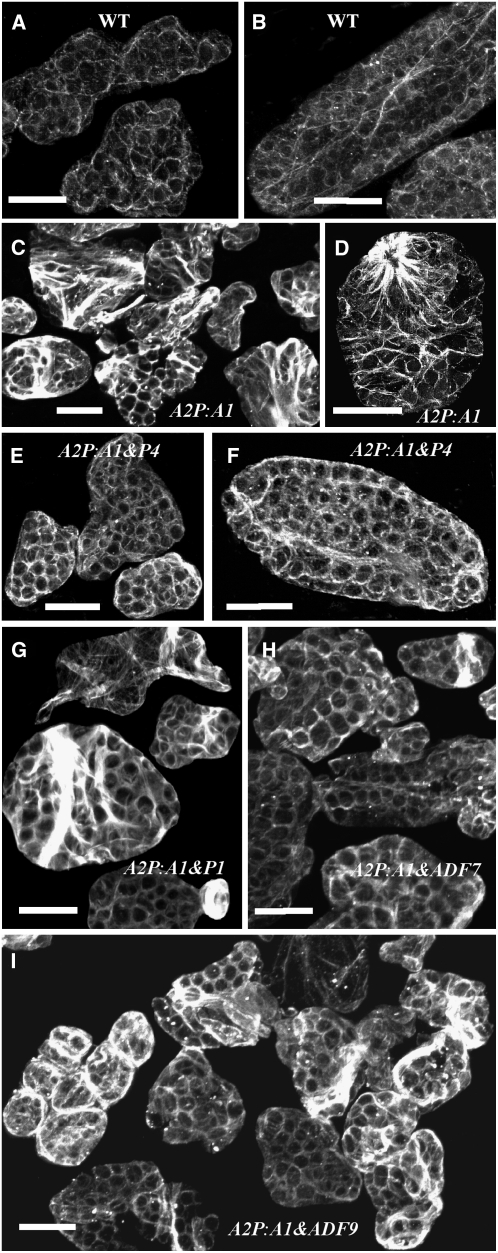
Figure 6.
Immunofluorescence Labeling of Actin in Leaf Cells.
(A) and (B) The wild type.
(C) and (D) Dwarf plant cells misexpressing ACT1.
(E) and (F) Cells from a suppressed, normal double transgenic plant coexpressing ACT1 and PRF4.
(G) Cells from a dwarf double transgenic plant coexpressing ACT1 and PRF1.
(H) Cells from a normal double transgenic plant coexpressing ACT1 and ADF7.
(I) Cells from a dwarf double transgenic plant coexpressing ACT1 and ADF9.
All samples were labeled with MAbGPa. Bars = 20 μm.
In addition, we co-overproduced reproductive ACT1 and vegetative PRF1, which is expressed constitutively in all organs, including the pistil (Kandasamy et al., 2002b), in the vegetative cell types using the ACT2 regulatory sequences. As described above for the generation of A2P:A1 and P4 plants, we first isolated two high-level PRF1 protein–overexpressing A2P:P1 lines and then transformed them with A2P:A1. We produced >100 double transgenic plants and evaluated them for morphological, molecular, and cellular phenotypes by observing the size and fertility of the plants and by performing protein gel blot and immunocytochemical analyses. Unlike the A2P:A1 and P4 double transgenic plants, almost 15% of A2P:A1 and P1 plants still revealed the dwarf phenotype (Figure 3). They contained smaller leaves and weaker inflorescence stems (Figures 7A and 7B) and showed highly reduced fertility. Protein gel blot analysis revealed that none of the large, normal-looking plants contained high levels of ACT1. Furthermore, all the dwarf double (A2P:A1 and P1) transgenic plants had approximately the same amount of ACT1 as the dwarf, high-level ACT1 (A2P:A1) expressing plants (Figure 7C). Probing of duplicate blots with a PRF1-specific monoclonal antibody revealed that all the small plants contained 4 to 5 times more PRF1 protein compared with its levels in wild-type or A2P:A1 dwarf single transformants (Figure 7D). Thus, even with greatly increased levels of coexpression of PRF1, high levels of ACT1 expression in shoots resulted in a dwarf phenotype. Our successful complementation studies of prf1 mutants with the A2P:P1 transgene suggested that the overexpressed PRF1 protein is functional in transgenic plants (data not shown). Immunofluorescence microscopy analysis revealed that ∼50% of cells of the dwarf double A2P:A1 and P1 transformants still contained aggregates of actin in the form of sheets (Figure 6G), although the extent of aberration is slightly less severe compared with the severely dwarfed A2P:A1 plants (Figures 6C and 6D). These observations suggest that co-overproduction of vegetative profilin and reproductive actin did not fully suppress the ACT1-induced cellular and morphological phenotypes. There may be a marginal effect due to the coexpression of PRF1 protein because we observed a small drop in the percentage of dwarf A2P:A1 and P1 plants (15%) compared with A2P:A1 plants (20%; Figure 3) and less severe cellular and morphological phenotypes at least in some dwarf double transformants.
Figure 7.
Lack of Suppression of the Dwarf Phenotype by Coexpression of Constitutive PRF1 with Reproductive ACT1.
(A) Approximately 7-week-old plants. A2P:A1, dwarf single transformant misexpressing ACT1; A2P:A1&P1, a double transformant misexpressing ACT1 and overexpressing PRF1 simultaneously. Note the double transgenic plant still exhibiting dwarf phenotype.
(B) Rosette leaves (the largest) of wild-type and single (A2P:A1) and double (A2P:A1&P1) transformants.
(C) Protein gel blot analysis of ACT1 expression. Top panel shows a blot probed with MAb45a for ACT1. Bottom panel shows a strip of the same blot probed for PEPC.
(D) Protein gel blot analysis of PRF1 expression. Top panel shows a blot probed with MAbPRF1 for PRF1 protein. Bottom panel shows a strip of the same blot probed for PEPC.
Class-Specific Suppression of ACT1-Induced Dwarf Phenotype by ADF Isovariants
ADFs are another class of small (∼17 kD) ABPs that regulate actin dynamics by binding both monomeric and filamentous actin in eukaryotic cells. In Arabidopsis, there are 11 diverse ADF genes that are broadly grouped, based on their expression pattern, into two major classes: constitutive (ADF1 to 6 and 9) and pollen/trichoblast specific (ADF7, 8, 10, and 11). Based on the phylogeny, they are further subdivided into four subclasses (Figure 1C; Ruzicka et al., 2007). To examine whether co-overexpression of ADF protein isovariants with ACT1 protein could suppress the dwarf phenotype manifested by ACT1 misexpression, we ectopically overproduced ACT1 with ADF7, ADF8, or ADF9 in the vegetative tissue. ADF7 and 8 are closely related in sequence (78% identical at the amino acid sequence level) but show completely different expression patterns. The ADF7 gene is active predominantly in the mature pollen, as revealed by ADF7 promoter-β-glucuronidase fusion gene expression (Figure 8A), whereas ADF8 is strongly expressed in the root trichoblast cells and developed root hairs (Figure 8B). On the other hand, ADF9, which is ∼50% identical to ADF7 and ADF8 at the amino acid sequence level, is constitutively expressed in all organs and cell types, except pollen (Figure 8C). After examining several dozen independent transgenic plants, we found that the expression of any of these three ADFs alone under the control of ACT2 regulatory sequences had no harmful effect on the growth of plants. Thus, the single transgenic plants either misexpressing ADF7 (Figure 8D) or ADF8 (Figure 8F) or overexpressing ADF9 (Figure 9A) were indistinguishable from wild-type plants at the seedling or adult stage of development.
Figure 8.
Suppression of ACT1-Induced Dwarf Phenotype by Coexpression of Pollen/Trichoblast-Specific ADFs.
(A) to (C) ADF promoter-β-glucuronidase fusion gene expression.
(A) Flower showing pollen-specific expression of ADF7.
(B) Two-day-old germinating seedling showing root hair–specific expression of ADF8.
(C) Two-week-old seedling showing constitutive expression of ADF9.
(D) Twenty-four-day-old wild-type plant, single transformants misexpressing ADF7 (A2P:ADF7) and ACT1 (A2P:A1), and a double transformant (A2P:A1&ADF7) coexpressing ACT1 and ADF7.
(E) Protein gel blot analysis of ACT1 and ADF7 expression in leaves. Top blot probed with reproductive actin-specific antibody MAb45a to show ACT1 protein. The weak bottom bands represent breakdown products. Middle blot from a duplicate gel is probed with MAbADF8, which recognizes ADF7. Bottom blot is probed with anti-PEPC antibody to show equal loading of proteins. Note the wild-type sample has no ACT1 and ADF7 proteins, the ADF7-misexpressing normal plant has no ACT1 protein, and the ACT1-misexpressing dwarf plant has no ADF7 protein.
(F) Twenty-four-day-old wild-type plant, single transformants misexpressing ADF8 (A2P:ADF8) and ACT1 (A2P:A1), and a double transformant (A2P:A1&ADF8) coexpressing ACT1 and ADF8.
(G) Protein gel blot analysis of ACT1 and ADF8 expression in leaf samples. Top blot probed with MAb45a to show ACT1 protein. The breakdown products were not detectable in this blot. Middle blot from a duplicate gel is probed with MAbADF8 to show ADF8 protein. Bottom panel probed with anti-PEPC antibody.
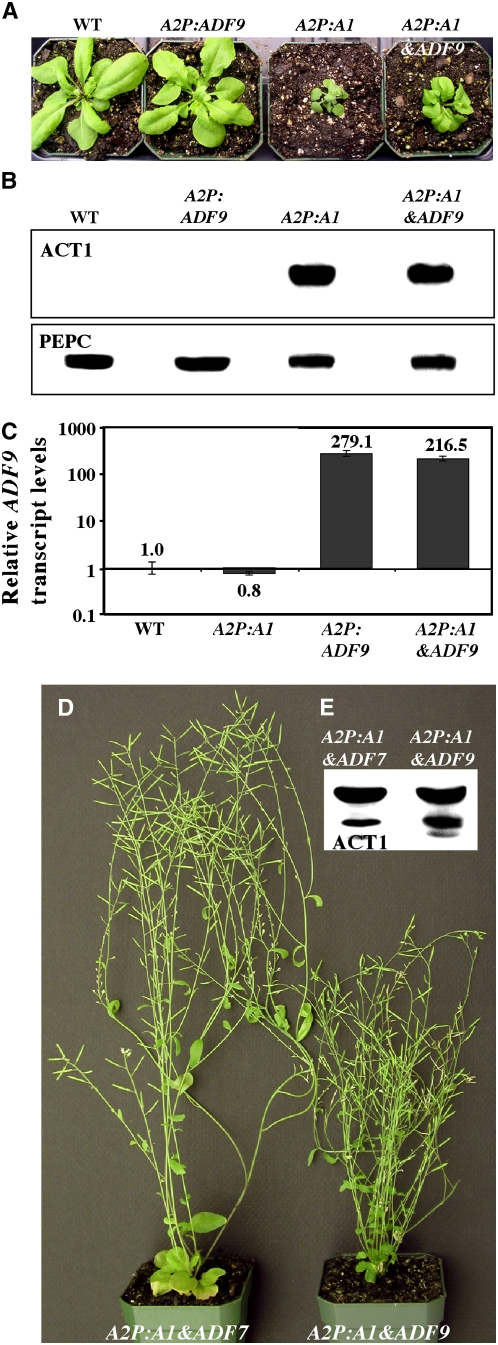
Figure 9.
Effect of Coexpression of Reproductive ACT1 and Constitutive ADF9 on Plant Development.
(A) Twenty-five-day-old wild-type plant, single transformants overexpressing ADF9 (A2P:ADF9) and misexpressing ACT1 (A2P:A1), and a double transformant (A2P:A1&ADF9) coexpressing ACT1 and ADF9. Note the dwarf stature of the double transformant.
(B) Protein gel blot analysis of ACT1 in leaf samples. Top blot probed with MAb45a to show ACT1 protein. Bottom panel probed with anti-PEPC antibody. Wild-type and the normal single transformant overexpressing ADF9 contain no ACT1 protein.
(C) RT-PCR analysis of ADF9 expression. The single A2P:ADF9 transformants and the double A2P:A1&ADF9 transgenic plants have >100 times the level of ADF9 transcripts than the wild type and plants misexpressing ACT1. Because of lack of ADF9-specific antibody, the levels of ADF9 expression are monitored here by RT-PCR analysis.
(D) Adult double transgenic plants coexpressing ACT1 and ADF7 (left) and ACT1 and ADF9 (right).
(E) Protein gel blot showing the levels of ACT1 protein in the double transgenic plants shown in (D). Probed with reproductive actin-specific MAb45a.
When we coexpressed pollen-specific ADF7 and ACT1 by transforming single insertion lines carrying A2P:ADF7 with the A2P:A1 transgene, <5% of the double transgenic plants revealed the dwarf phenotype in contrast with 20% or more dwarf transgenic plants carrying ACT1 alone (Figure 3). The rest of the plants were morphologically identical to the wild-type plants or plants carrying only ADF7 (Figure 8D). Protein gel blot analysis on those normal-looking plants with ACT1-specific monoclonal antibodies revealed that several (∼15%) of them contained high levels of the misexpressed ACT1 protein, similar to the single A2P:A1 dwarf plants (Figures 8D and 8E). Moreover, those double transformants with high levels of ACT1 all contained high levels of ADF7 protein (Figure 8E, middle panel). Thus, the ectopic expression of pollen-specific ADF7 protein suppressed the toxic effect of ACT1 in plants containing high levels of this reproductive actin protein. Interestingly, the closely related trichoblast/root hair–specific ADF8 also showed an effect similar to ADF7 when coexpressed along with ACT1 (Figures 3, 8F, and 8G). As a result, plants containing high levels of ACT1 in vegetative tissue have large rosette leaves and looked like normal wild-type plants in the presence of ADF8 (Figure 8F). On the other hand, when the vegetative ADF9 protein was coexpressed along with the reproductive ACT1, this ADF isovariant had only marginal effects on the suppression of dwarf phenotype. Nearly 17% of the double transformants containing A2P:A1 and A2P:ADF9 transgenes were dwarf compared with 5% or less dwarf plants in those with A2P:A1 and A2P:ADF7 or A2P:ADF8 transgenes (Figure 3). Because even high-level expression of ADF9, as determined by quantitative RT-PCR analysis of the transcripts in leaves (Figure 9C), could not suppress the toxic effect of ACT1, those plants containing high levels of ACT1 in the vegetative tissue were still very small compared with wild-type plants (Figures 9A and 9B). The ADF isovariant-specific suppression of the ACT1-induced dwarf phenotype is clearly evident in Figure 9D. Although they had about the same level of ACT1 protein (Figure 9E), the adult plant expressing ADF7 (Figure 9D, left) was almost twice the size of the plant expressing ADF9 (Figure 9D, right). Immunocytochemical analysis suggested that comisexpression of ADF7 (Figure 6H) or ADF8 (data not shown) with ACT1 greatly reduced the aberrant actin cytoskeletal structures in most (∼95% normal) leaf cells, whereas co-overexpression ADF9 had only a little effect because a lot of cells (40 to 50%) still contained aggregates of actin bundles (Figure 6I). Even in ACT1 expressing dwarf single transformants only 50 to 60% of leaf cells showed obvious aberrant actin structures. Thus, the coexpression of pollen/root hair-specific ADF7 or ADF8, but not the constitutive ADF9, with the reproductive ACT1 considerably suppressed the ACT1-induced aberrant plant and cellular phenotypes.
DISCUSSION
The Extent of Functional Redundancy in Plant Actin Cytoskeleton
Complex multicellular organisms like plants contain gene families encoding actin and various ABPs that together modulate the dynamics of the actin-based cytoskeleton (Meagher and Fechheimer, 2003). For example, the Arabidopsis genome encodes eight actin, five profiling, and 11 ADF genes, which are broadly grouped into constitutive or vegetative and reproductive (pollen-specific) classes (Figure 1). There are at least 14 additional families of ABPs encoded by multiple genes, but they are less well characterized in plants (Cvrckova, 2000; Meagher and Fechheimer, 2003). The significance of our actin misexpression studies must therefore be considered in light of the complex actin cytoskeletal system. Molecular genetic analyses of null mutations in many of the differentially expressed plant cytoskeletal genes reveal only mild phenotypes that are restricted to a few cell types. Even mutation in the strongly and constitutively expressed ACT2 gene shows only a stunted root hair phenotype, and the lack of this actin isovariant has minimal effects on the survival and growth of individual plants (Gilliland et al., 2002; Ringli et al., 2002; Nishimura et al., 2003). Moreover, the root hair development phenotype of the act2-1 mutation can be complemented with most other actin isovariants (M.K. Kandasamy, E.C. McKinney, and R.B. Meagher, unpublished data), including the highly divergent reproductive class actin ACT1 (Gilliland et al., 2002). Similarly, mutations in the other vegetative actin gene, ACT7, which is active in all young developing tissues and responds to phytohormones, reveal retarded root growth and reduced ability to form callus on hormone-containing medium as the only phenotypes. Otherwise the ACT7-deficient plant morphology is quite comparable to the wild type (Kandasamy et al., 2001; Gilliland et al., 2003). However, mutations in both these actin genes together (act2-1/act7-4) are very deleterious and result in extremely dwarf plants whose viability could be rescued only by growing them initially on medium containing sucrose as carbon source (M.K. Kandasamy and R.B. Meagher, unpublished data). Knockout mutations in individual reproductive class late-pollen actin genes also reveal no detrimental effect on fertility and seed set (Pawloski et al., 2006). Similarly, knockout or knockdown mutations in several of the ABP genes, including profilins (McKinney et al., 2001) and ADFs (D.R. Ruzicka and R.B. Meagher, unpublished data), also showed only moderate to no effect on the growth and overall architecture of plants. The weak phenotypes of single mutants suggest that there is some degree of functional redundancy among these cytoskeletal genes in plants. However, in simple unicellular organisms such as yeast, where there is only a single copy for most of these cytoskeletal genes, null mutations in actin and some ABPs (e.g., cofilin and profilin) are lethal or show severe deleterious phenotypes approaching lethality (Shortle et al., 1982; Haarer et al., 1993; Moon et al., 1993).
Phylogenetic analysis suggests that the vegetative and reproductive class plant actin genes have not shared a common ancestor for 300 to 400 million years, and even the actin genes encoding highly similar proteins within each class (e.g., ACT2 and 8; ACT1 and 3 with single amino acid differences) have existed as duplications in plant genomes for approximately the past 50 million years (McDowell et al., 1996; Meagher et al., 2000). If vegetative and reproductive actin-based cytoskeletal systems have been functioning independently in different organs and tissues for this length of time, it might not be surprising to find both novel and dysfunctional interactions among proteins from these two different systems (Meagher et al., 1999a). Moreover, the various plant actin proteins have an unusually large number of nonconservative amino acid substitutions (6 to 10%), which map to the surface of the molecule, in comparison to far fewer total changes (3 to 7%) and only a few surface changes among the 500–million-year-old animal muscle and cytoplasmic actin subclasses (Hightower and Meagher, 1986; McDowell et al., 1996). All these amino acid substitutions in plant actins should have significant effect on protein–protein interactions because even the few changes in animal actin isovariants lead to different physical properties in vitro (Garrels and Gibson, 1976). For example, despite their relative similarity, the vertebrate nonmuscle and muscle actins display differential binding capacity for the actin monomer binding proteins profilin and thymosin (Larsson and Lindberg, 1988; Oshima et al., 1989; Weber et al., 1992). Moreover, dominant-negative amino acid changes (e.g., V163L, V163M, and R183G) resulting from mutations in human α-skeletal muscle actin cause nemaline myopathy and distinct pathological phenotypes, such as formation of cytoplasmic nemaline bodies and intranuclear rods (Ilkovski et al., 2004). A single amino acid change (Ile-76 to Val-76) also rendered Drosophila flight muscle actin inactive (Fyrberg et al., 1998). Furthermore, the different classes of plant profilin and ADF protein isovariants exhibit varying biochemical properties (Kovar et al., 2000; Allwood et al., 2002). These observations combined with differential gene regulation strongly favor the view that the different classes and subclasses of cytoskeletal protein isovariants have functional relevance in plants, as in animals.
To understand the extent of functional redundancy and/or specificity among the cytoskeletal proteins in plants, we have ectopically expressed pollen-specific actin (ACT1) in the vegetative tissues of wild-type plants (Kandasamy et al., 2002a) and in act2-1 and act7-4 mutants containing markedly reduced amounts of total actin (this study). Misexpression of high levels of ACT1 in vegetative tissues retarded the growth of both wild-type and mutant plants and severely altered the architecture of most plant organs. The overproduction of ACT1 in vegetative cells, where there is no expression of reproductive accessory proteins, resulted in the formation of sheet- or star-like aberrant actin structures and thick transverse actin cables instead of the longitudinal arrays of thin actin filaments seen in the wild type (Kandasamy et al., 2002a). However, overexpression of ACT2 in vegetative tissues had little effect on the morphology of the plant or assembly of actin filaments. This clearly shows that the degree of redundancy among more divergent actin isovariants in the same plant is not high and that there are definitely functional differences between the two different classes of actin isovariants. Because of this functional specificity, the misexpressed pollen actin might have interacted poorly with various ABPs present in the vegetative cells, and this might have caused an imbalance between these two groups of proteins and resulted somehow in abnormal polymerization and arrangement of actin filaments. Surprisingly, however, the misexpression of diverse isovariants of two ABPs alone, profilins and ADFs, did not reveal any harmful effects. We propose that in these plants the misexpressed reproductive ADFs and profilins might interact weakly with the native vegetative actin and cause no toxicity to the assembly of actin cytoskeleton and thus have little effect on the morphology of the plant. The activity of native vegetative actin is normally buffered by the endogenous profilins and ADFs and myriad other ABPs. However, profilin microinjection studies have shown drastic effects on the actin cytoskeleton and thereby the streaming of cytoplasm in Tradescantia stamen hair cells (Staiger et al., 1994; Valster et al., 1997). However, in transgenic plants, we are not overexpressing PRF4 or PRF1 to the levels even one half that of the microinjected recombinant profilin, and this may be one of the reasons for not seeing any phenotype with profilin overexpression. Also, an increase in cellular concentration of the small molecular weight foreign (ectopically expressed) ABPs (e.g., PRF4 and ADF7) may not be as toxic as the foreign reproductive actin (ACT1) proteins that tend to aggregate or polymerize abnormally in the absence of proper interacting partner APBs, as further revealed by aberrant cytoskeleton resulting from the heterologus expression of plant actins in yeast cells.
Class-Specific Interaction among Actin and ABPs in Vivo
Our suppression data further support functional specificity and class-specific interaction in vivo among the two major classes of actin and two classes of ABPs. When we ectopically co-overproduced reproductive actin ACT1 and reproductive profilin PRF4 simultaneously in the same vegetative tissue, there was almost full suppression of the toxic effect of misexpressed ACT1. Thus, plants overproducing both ACT1 and PRF4 looked quite similar to wild-type plants, and immunocytochemical studies revealed that the suppression worked by reducing abnormal actin assembly (aggregation) because their cells contain a typical, but intensely staining, actin cytoskeleton. However, when we overproduced reproductive ACT1 and vegetative profilin PRF1, there was only a marginal effect on the suppression of dwarf morphological phenotype, and the dwarf plant cells still exhibited the aggregated actin cellular phenotype. This clearly showed that there is preferential, class-specific interaction between the actin and profilin isovariants in plant cells, but it was still rather surprising to see that overexpression of reproductive class profilin alone was sufficient to compensate for the overproduction of ACT1. The actin assembly and disassembly in eukaryotic cells generally involves profilin and a host of other associated accessory proteins (Pollard et al., 2000; Paavilainen et al., 2004; Staiger and Blanchoin, 2006). Because the manifestation of the dwarf phenotype was ACT1 concentration dependent, even sequestering moderate amounts of overproduced ACT1 monomers by PRF4 might be enough to bring the actin concentration below the toxic threshold level and prevent actin aggregation into sheet- or star-like structures.
Moreover, we also examined the effect of coexpressing three different isovariants of ADF (ADF7, ADF8, and ADF9) on the toxic effects of ACT1. The ADF protein is also a critical player in the remodeling of the plant actin cytoskeleton (Maciver and Hussey, 2002). Similar to the results with profilin, only the pollen-specific ADF7, but not the constitutive ADF9, was able to suppress the dwarf phenotype of ACT1-misexpressing plants. Interestingly, the trichoblast/root hair–specific ADF8 was also able to interact in vivo with ACT1 and suppress the deleterious effects of its misexpression. ADF8 is very closely related in amino acid sequence to the pollen-specific ADF7, compared with the constitutive ADF9 (Maciver and Hussey, 2002; Feng et al., 2006). Also, both pollen tubes and root hairs are fast, tip-growing cells requiring highly dynamic, polarized actin cytoskeleton and tip-directed vesicle trafficking modulated by actin (Hepler et al., 2001; Cole and Fowler, 2006; Samaj et al., 2006), and these similar cell types may have some common components involved in actin assembly. Therefore, the pollen-specific ADF7 and root hair–specific ADF8 were able to interact with ACT1 in vivo and suppress the dwarf phenotype.
The differential expression of ABPs may also explain why ACT1 was able to complement the stunted root hair phenotype of act2-1 mutants, when ectopically expressed using the ACT2 promoter (Gilliland et al., 2002; this study). In the ACT1-misexpressing act2-1 mutant plants, the root hair growth is restored, but the organization of all other vegetative organs and cell types are severely altered. Because root hairs endogenously express ADF8, which was shown here to suppress the dwarf plant phenotype when coexpressed along with ACT1, it is possible that the endogenous root hair–specific ADFs (ADF8 and ADF11) might interact properly with ACT1 and therefore might complement the act2-1 root hair phenotype. Moreover, ACT1 complements the retarded root growth phenotype of the act7-4 mutant. From both these complementation studies, it is clear that ACT1 is able to suppress a subset of vegetative actin mutant phenotypes while adversely affecting the development and organization of other organs. This may be due to the differential expression of some of the ABPs that interact properly with ACT1 (e.g., root trichoblast–specific ADF8 and 11) or may be due to the expression of constitutive ABPs in roots (e.g., ADF6, which is expressed both in pollen, root, and other vegetative tissue) that may moderately interact with the reproductive actins. In the aerial vegetative organs, there is no expression of ADF8 or ADF11 or enough expression of other ABPs with similar properties; hence, the pollen-specific ACT1 may not be able to interact normally with any of the endogenous APBs, resulting in aberrant cytoskeletal organization and development.
Actin Dynamics in ACT1-Misexpressed Dwarf and Suppressed Normal Plants
Actin is generally organized in plant cells into arrays of thin cables and fine filaments and baskets surrounding the nucleus and chloroplasts (Kost et al., 1998; Kandasamy and Meagher, 1999; Sheahan et al., 2004). The proper assembly and organization of actin depends upon the expression of an appropriate mixture of ABPs and signals mediated by various cellular signaling pathways involving molecules like the Rho family of GTPases (Staiger, 2000; Valster et al., 2000; Vantard and Blanchoin, 2002). When the delicate balance between the various cytoskeletal components is impaired, there are unusual consequences for actin assembly and cellular architecture, as observed in the ACT1-misexpressed dwarf plants. Although we assume that weak and/or inappropriate interactions of reproductive actin with the endogenous vegetative ABPs in the cells might be the cause for formation of abnormal star- or sheet-like actin structures, the understanding of the exact biochemical mechanism behind this unusual process requires further analysis. However, the genetic suppression of the ACT1-induced dwarf phenotype and the aberrant actin organization by the coexpression of reproductive profilin or ADF isovariants supports the need for class-specific interaction in vivo among actin and various ABP isovariants for proper assembly of the actin cytoskeleton. In plant cells, as reported for maize and Papaver pollen and tobacco suspension cells where profilin is massively abundant, a remarkably small percentage (5 to 10% and 1 to 2%, respectively) of total actin is present in the filamentous form (Gibbon et al., 1999; Snowman et al., 2002; Wang et al., 2005), and the large monomer pool is predicted to be in a complex with profilin (Staiger and Blanchoin, 2006) and other ABPs. The massive overproduction of pollen actin in the vegetative cells of dwarf plants might result in lack of or insufficient levels of appropriate actin monomer sequestering proteins like profilin. Moreover, poor interaction of endogenous vegetative profilin and other ABPs with the pollen actin isovariant might have left the cells with excess free actin monomers. As observed in yeast mutants deficient in profilin or other ABPs, where actin is deposited into bar-like structures (Magdolen et al., 1993), the excess reproductive actin monomers in the dwarf (A2P:A1) plant cells are arranged into star- or thick cable-like structures (see the model in Supplemental Figure 2B online). Similarly, the plant actins expressed in yeast cells were incorporated into rods, thick cables, and unusual patches, probably because of their poor affinity for the yeast accessory proteins. However, when we coexpressed reproductive actin and reproductive profilin or ADF in the same plant cells (A2P:A1 and A2P:ADF7 or PRF4), there appeared to be proper interaction between the two cytoskeletal components. The reproductive ABPs balance out the excess of reproductive actin monomers in the cell; hence, there was normal F-actin polymerization, albeit with more actin filaments (see Supplemental Figure 2C online). We suggest that because of poor affinity, the endogenous or coexpressed vegetative ABPs could not balance the concentration of excessive reproductive actin monomers (in A2P:A1 and A2P:ADF9 or PRF1 double transformants) and resulted in plants that were still dwarfed with abnormal actin structures (see Supplemental Figure 2D online). In the control plants overexpressing vegetative actin alone (A2P:A2), the levels of endogenous vegetative ABPs may be sufficient to buffer the excess actin and regulate polymerization to form more actin filaments but avoid formation of abnormal actin structures (see Supplemental Figure 2E online).
Biochemical evidence for the differential binding of reproductive and vegetative ABPs with the two major classes of plant actin would provide further support to our genetic suppression studies and our model. In this regard, it is worth mentioning that Chris Staiger's group (Purdue University) has recently found marked differences in the preference of the Arabidopsis vegetative PRF2 and reproductive PRF4 for plant and vertebrate actins. Specifically, they discovered that PRF4 has ∼3.5-fold higher affinity for monomeric actin from plants than it does for vertebrate actin, whereas PRF2 shows equal binding to both types of actin monomers (F. Chaudhry, S. Huang, S. Kasina, and C.J. Staiger, unpublished data). The vegetative PRF1 used in this study is the closest homolog to PRF2. The findings from Staiger's lab obviously lend support to our assumption that the reproductive and the vegetative ABPs differentially interact with ACT1. Moreover, future cross-linking and immunoprecipitation studies with isovariant-specific antibodies will elucidate the differential binding properties of various vegetative and reproductive cytoskeletal proteins in plants that are wild-type or have different transgenic backgrounds. More efficient binding of ACT1 to PRF4 or ADF7 than ACT1 to PRF1 or ADF9 would explain why the vegetative and reproductive ABPs differentially suppress the ectopic expression phenotypes of pollen-specific ACT1 protein in vegetative cell types. In summary, our data on the ectopic expression of ACT1 and ABP isovariant-specific suppression of the dwarf morphological and actin cellular phenotypes provide strong evidence for the existence of functional differences among the two classes of actin and ABP isovariants in vivo.
METHODS
Plant Material and Generation of Transgenic Plants
Wild-type (Columbia), mutant (act2-1 and act7-4), and transgenic Arabidopsis thaliana plants were cultivated in growth chambers at 22°C with 16 h of light and 8 h of dark periods. As described earlier (Gilliland et al., 2002), act2-1 is a null mutant allele with a T-DNA insertion just five nucleotides upstream of ATG in the vegetative class ACT2 gene. The act7-4 null mutant allele has a T-DNA insertion in the leader intron of the vegetative ACT7 gene (Gilliland et al., 2003). Seven different constructs were made for this study of suppression of ACT1 ectopic expression phenotypes (Figure1D): (1) A2P:A1, an actin misexpression construct that contains a 1.1-kb full-length ACT1 cDNA inserted between a 1.3-kb promoter and the terminator region of ACT2; (2) A2P:A2, a control actin overexpression construct in which the ACT1 cDNA was replaced with a 1.1-kb full-length ACT2 cDNA. Similarly, different profilin and ADF full-length cDNAs replaced ACT1 cDNA in the rest of the constructs: (3) A2P:P4, a profilin misexpression construct with a 405-bp PRF4 cDNA; (4) A2P:P1, a profilin overexpression construct containing a 396-bp PRF1 cDNA; (5) A2P:ADF7, an ADF misexpression construct with a 414-bp ADF7 cDNA; (6) A2P:ADF8, another ADF misexpression construct with a 423-bp ADF8 cDNA; (7) A2P:ADF9, an ADF overexpression construct with a 426-bp ADF9 cDNA. The various steps involved in cloning A2P:A1 and A2P:A2 constructs were described previously (Kandasamy et al., 2002a).
All the ACT, PRF, and ADF cDNAs were PCR amplified from a mature flower library in the plasmid vector pCDNAII (Invitrogen), except for ADF8, which was amplified from a root cDNA library. The expression plasmids were mobilized into the Agrobacterium tumefaciens strain C58C1 and transformed into wild-type or mutant Arabidopsis plants by vacuum infiltration. Transformants were selected by plating the seeds on medium containing 35 mg/L Kan or 50 mg/L Hyg. For generating double transgenic plants, we first identified single insertion lines for different PRF and ADF transgenes by scoring the segregation ratio of antibiotic resistant to sensitive (3:1) plants in the T2 generation, and then we selected two plant lines expressing high levels of the respective protein isovariants by performing protein gel blot analyses. Finally, we transformed them with A2P:A1. Phenotypic assessment of wild-type and antibiotic-resistant single and double transgenic plants was made at different stages of development. To determine the plant size, we measured the diameter of the whole rosette at the time of bolting and the height of adult plants when they were 7 to 8 weeks old.
Heterologous Expression of Plant Actins in Yeast
The vegetative ACT8 (U42007; An et al., 1996) and the pollen-specific ACT12 (U27982; Huang et al., 1996) cDNAs were cloned into the yeast expression plasmid p413-HIS3 with the TEF (translation elongation factor) promoter. These constructs were transformed into the yeast strain DDY384 (Drubin et al., 1993). Transformed yeast cells were plated on minimal medium lacking Leu, uracil, and histitine to maintain the pDD41 and p413 plasmids, and glucose was used as a carbon source (Sachs et al., 1987). Because the strain DDY384 contains a deletion of the only essential yeast actin yACT1, it was complemented by the presence of the plasmid pDD41 containing wild-type yACT1.
Antibodies
Two monoclonal antibodies were used to detect actin either by protein gel blot analysis or by confocal immunofluorescence microscopy: (1) MAbGPa, a general plant actin–specific antibody that detects equally all eight expressed Arabidopsis actins (Kandasamy et al., 1999); and (2) MAb45a, a reproductive actin-specific antibody that reacts uniformly with actins ACT1, ACT3, ACT4, and ACT12 (Kandasamy et al., 1999). We used two profilin monoclonal antibodies to detect profilin at the protein gel blot or cellular level: (1) MAbPRF45 reacted with both pollen-specific profilin isovariants PRF4 and PRF5; and (2) MAbPRF1 was specific to the constitutive profilin isovariant PRF1 (Kandasamy et al., 2002b). ADF7 and ADF8 proteins were detected on protein gel blots with MAbADF8, which was raised against ADF8 recombinant protein, but recognized all four ADFs (ADF7, ADF10, ADF8, and ADF11) of the pollen/trichoblast-specific subclasses (see Figure 1C; Ruzicka et al., 2007). An anti-PEP carboxylase polyclonal antibody (Rockland) was used as control to monitor variation in protein loading and uniform transfer during electroblotting.
Protein Gel Blot Analysis
For detection of actin on protein gel blots, protein samples from frozen wild-type, mutant, and transgenic Arabidopsis leaves were prepared as described previously (Kandasamy et al., 1999) using an extraction buffer containing 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM EDTA, and complete protease inhibitor cocktail (one tablet/10 mL; Roche Diagnostics). The protein extracted in the extraction buffer was precipitated with trichloroacetic acid, and the pellet was washed with cold acetone, dried, and dissolved in solubilization buffer (11 mg Na2CO3 per mL 100 mM DDT). The protein suspension was mixed with equal volume of 2× sample buffer (Laemmli, 1970), boiled (5 min), and then loaded onto SDS-PAGE gels. However, for profilin analysis, the frozen leaf samples were extracted in extraction buffer, and after centrifugation the supernatant was directly mixed 1:1 with 2× sample buffer, boiled for 5 min, and then loaded onto the gels. On the other hand, ADF was assayed by directly extracting the frozen samples in 2× sample buffer. Different extraction procedures were followed to optimize for maximum content of each desired protein in a sample. Equal loading of proteins was monitored by Coomassie Brilliant Blue staining of duplicate gels, and uniform transfer of protein to the polyvinylidene fluoride membrane was determined by probing identical blots or strips of blots (>80 kD) with anti-PEP carboxylase antibody. The protein bands that were detected using the ECL kit (Amersham) were quantified using the NIH Sci Image program.
RT-PCR Analysis of ADF9 Expression
RNA was isolated from leaf tissues of wild-type and various transgenic plants using the RNeasy plant mini kit (Qiagen), and it was treated with RQ1 RNase-free DNase (Promega) before reverse transcription. Three micrograms of treated RNA were added to RT reactions using the Invitrogen SuperscriptIII first-strand synthesis kit with random hexamer primers to make cDNA. Real-time PCR was used to analyze cDNA populations using 18S primers (18S-RT2S, 5′-GGGGGCAATCGTATTTCATA-3′, and 18S-RT2A, 5′-TTCGCAGTTGTTCGTCTTTC-3′) as the endogenous control. The primers used for ADF9 detection are ADF9totalS (5′-TGGTGTTCACTACGAGCTTCA-3′) and ADF9totalA (5′-GATAAAATCCAGGACCGGG-3′). Reactions were performed on an Applied Biosystems 7500 real-time PCR system using SYBR Green detection chemistry (Applied Biosystems) as described previously (Deal et al., 2007). The 2−(ddCT) method (Livak and Schmittgen, 2001) of relative quantification was used in all experiments.
Immunofluorescence Labeling and Confocal Microscopy of Plant Cells
Cryofixation and freeze-substitution of leaves from young seedlings were done as described elsewhere (Kandasamy et al., 2002a). In brief, the samples were rapidly frozen in liquid propane (−180°C), freeze-substituted in acetone at −80°C for 72 h, and then gradually brought to room temperature over an 8-h period. After rehydration through a graded acetone series, the seedlings were washed in PME (50 mM PIPES, pH 7.0, 5 mM EGTA, 1 mM MgSO4, and 0.5% casein), permeabilized by treating with 1% Cellulysin (Calbiochem) and 0.1% Pectolyase (Sigma-Aldrich) in PME containing protease inhibitors (complete mini EDTA-free protease inhibitor tablets) for 20 min, washed again in PME (5 min) and PBS (2× 10 min), and squashed onto chrom-alum and gelatin-coated slides. Following air-drying, the leaf cells attached to the slides were further permeabilized in 0.5% Triton X-100 in PBS for 20 min and −20°C methanol for 10 min. After rinsing in PBS, the slides were blocked for 1 h in TBST-BSA-GS (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween 20, 2.5% BSA, and 10% goat serum) and then incubated in the primary antibody diluted (5 μg/mL) in TBST-BSA-GS. After overnight incubation, the slides were rinsed with PBS, and then labeled for 3 h with fluorescein isothiocyanate–conjugated anti-mouse secondary antibody (Sigma-Aldrich) at 1:100 dilution. The slides were rinsed in PBS (3× 10 min) and mounted with 80% glycerol in PBS containing 1 mg/mL p-phenylenediamine (Sigma-Aldrich). The actin microfilaments in the labeled cells were visualized with a Leica confocal laser scanning microscope (TCS-SP2). For profilin, after labeling with antibodies, the cells were stained with DAPI (Sigma-Aldrich) and observed with a Leica fluorescence microscope.
Localization of Plant and Native Actin in Yeast
Wild-type and transformed yeast cells were fixed in 4% paraformaldehyde freshly prepared in PME containing the protease inhibitor cocktail (Roche Diagnostics) for 1 h. After washing in PME containing 1.2 M sorbitol, the fixed cells were permeabilized by treating with Zymolyase (25 μg/mL; ICN) and Glusulase (55 μL/mL; NEN) for 30 min, washed again in PME as above, and immobilized onto chrom-alum and gelatin-coated slides. The cells were further permeabilized in cold methanol (6 min) and acetone (30 s), air-dried, and immunolabeled with plant actin-specific MAbGPa as described above for plant cells. For F-actin staining in wild-type cells, the blocked slides were incubated with Texas Red–conjugated phalloidin (0 to 25 μM; Molecular Probes) for 2 to 3 h, rinsed with PBS, mounted with 80% glycerol in PBS containing 1 mg/mL p-phenylenediamine, and observed with a Leica confocal microscope.
Sequence Comparisons and Phylograms
Sequences were aligned with ClustalW 1.82 (Higgins and Sharp, 1988) using default settings, and phylogenies were constructed with PAUP 4.0 (Rogers and Swofford, 1999) using the neighbor-joining tree building method. Bootstrap support values were based on 100 replicates using a full heuristic search. Actin, ADF, or profilin sequences of Saccharomyces cerevisiae and Chlamydomonas reinhardtii were used to root these trees. Specific sequence identifiers are given in Supplemental Data Sets 1 (Actin), 2 (profilin), and 3 (ADF) online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At ACT1, U39449 (At2g37620); At ACT2, U41998 (At3g18780); At PRF1, U43322 (At2g19760); At PRF4, U43324 (At4g29340); At ADF7, NM_118691 (At4g25590); At ADF8, NM_116293 (At4g00680); At ADF9, NM_119663 (At4g34970). The Arabidopsis Genome Initiative locus numbers are given in parenthesis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Actin Localization in Yeast Cells.
Supplemental Figure 2. Models for Class-Specific Interaction of PRF and ADF Isovariants with Actin and Regulation of Actin Assembly.
Supplemental Data Set 1. Amino Acid Sequences Used in Construction of the Actin Tree in Figure 1A.
Supplemental Data Set 2. Amino Acid Sequences Used in Construction of the Profilin Tree in Figure 1B.
Supplemental Data Set 3. Amino Acid Sequences Used in Construction of the ADF Tree in Figure 1C.
Supplementary Material
Acknowledgments
We thank Roger Deal and Gay Gragson for critical reading of the manuscript and the anonymous reviewers for their useful comments. Confocal microscopy was conducted at the Center for Advanced Ultrastructural Research at the University of Georgia. We thank Beth Richardson for her help with rapid freezing of samples for actin localization. This work was supported by the National Institutes of Health (GM-36397).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard B. Meagher (meagher@uga.edu).
Online version contains Web-only data.
References
- Allwood, E.G., Anthony, R.G., Smertenko, A.P., Reichelt, S., Drobak, B.K., Doonan, J.H., Weeds, A.G., and Hussey, P.J. (2002). Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 14 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R. (1999). Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15 185–230. [DOI] [PubMed] [Google Scholar]
- Carlier, M.F. (1998). Control of actin dynamics. Curr. Opin. Cell Biol. 10 45–51. [DOI] [PubMed] [Google Scholar]
- Carlsson, L., Nyström, L.-E., Sundkvist, I., Markey, F., and Lindberg, U. (1977). Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 115 465–483. [DOI] [PubMed] [Google Scholar]
- Chen, H., Bernstein, B.W., and Bamburg, J.R. (2000). Regulating actin-filament dynamics in vivo. Trends Biochem. Sci. 25 19–23. [DOI] [PubMed] [Google Scholar]
- Cole, R.A., and Fowler, J.E. (2006). Polarized growth: Maintaining focus on the tip. Curr. Opin. Plant Biol. 9 579–588. [DOI] [PubMed] [Google Scholar]
- Cvrckova, F. (2000). Are plant formins integral membrane proteins? Genome Biol. 1 Research001.1–001.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, R.B., Topp, C.N., McKinney, E.C., and Meagher, R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D.G., Jones, H.D., and Wertman, K.F. (1993). Actin structure and function: Roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol. Biol. Cell 4 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y., Liu, Q., and Xue, Q. (2006). Comparative study of rice and Arabidopsis actin-depolymerizing factors gene families. J. Plant Physiol. 163 69–79. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E.A., Fyrberg, C.C., Biggs, J.R., Saville, D., Beall, C.J., and Ketchum, A. (1998). Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 36 271–287. [DOI] [PubMed] [Google Scholar]
- Garrels, J.I., and Gibson, W. (1976). Identification and characterization of multiple forms of actin. Cell 9 793–805. [DOI] [PubMed] [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999). Latrunculin B had different effects on pollen germination and tube growth. Plant Cell 11 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland, L.U., Kandasamy, M.K., Pawloski, L.C., and Meagher, R.B. (2002). Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2–1 mutation. Plant Physiol. 130 2199–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland, L.U., Pawloski, L.C., Kandasamy, M.K., and Meagher, R.B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33 319–328. [DOI] [PubMed] [Google Scholar]
- Haarer, B.K., Petzold, A.S., and Brown, S.S. (1993). Mutational analysis of yeast profilin. Mol. Cell. Biol. 13 7864–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., and Sharp, P.M. (1988). CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73 237–244. [DOI] [PubMed] [Google Scholar]
- Hightower, R.C., and Meagher, R.B. (1986). The molecular evolution of actin. Genetics 114 315–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., An, Y.Q., McDowell, J.M., McKinney, E.C., and Meagher, R.B. (1996). The Arabidopsis thaliana ACT4/ACT12 actin gene subclass is strongly expressed throughout pollen development. Plant J. 10 189–202. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., Allwood, E.G., and Smertenko, A.P. (2002). Actin-binding proteins in the Arabidopsis genome database: properties of functionally distinct plant actin-depolymerizing factors/cofilins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, P.J., Ketelaar, T., and Deeks, M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57 109–125. [DOI] [PubMed] [Google Scholar]
- Ilkovski, B., Nowak, K.J., Domazetovska, A., Maxwell, A.L., Clement, S., Davies, K.E., Laing, N.G., North, K.N., and Cooper, S.T. (2004). Evidence for a dominant-negative effect in ACTA1 nemaline myopathy caused by abnormal folding, aggregation and altered polymerization of mutant actin isoforms. Hum. Mol. Genet. 13 1727–1743. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., Gilliland, L.U., McKinney, E.C., and Meagher, R.B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (1999). The late pollen-specific actins in angiosperms. Plant J. 18 681–691. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002. a). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002. b). Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil. Cytoskeleton 52 22–32. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., and Meagher, R.B. (1999). Actin-organelle interaction: Association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil. Cytoskeleton 44 110–118. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16 393–401. [DOI] [PubMed] [Google Scholar]
- Kovar, D.R., Drobak, B.K., and Staiger, C.J. (2000). Maize profilin isoforms are functionally distinct. Plant Cell 12 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Larsson, H., and Lindberg, U. (1988). The effect of divalent cations on the interaction between calf spleen profilin and different actins. Biochim. Biophys. Acta 953 95–105. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Maciver, S.K., and Hussey, P.J. (2002). The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 3 reviews3007.1–3007.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen, V., Drubin, D.G., Mages, G., and Bandlow, W. (1993). High levels of profilin suppress the lethality caused by overproduction of actin in yeast cells. FEBS Lett. 316 41–47. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Huang, S., McKinney, E.C., An, Y.Q., and Meagher, R.B. (1996). Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142 587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney, E.C., Kandasamy, M.K., and Meagher, R.B. (2001). Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development. Plant Cell 13 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., and Fechheimer, M. (2003). The Arabidopsis cytoskeletal genome. In The Arabidopsis Book, E.M. Meyerowitz and C.R. Somerville, eds (Rockville, MD: American Society of Plant Biologist), 10.1199/tab.0096, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Meagher, R.B., McKinney, E.C., and Kandasamy, M.K. (1999. a). Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell 11 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Kandasamy, M.K. (2000). The significance of diversity in the plant actin gene family: Studies in Arabidopsis. In Actin: A Dynamic Framework for Multiple Plant Cell Functions, C.J. Staiger, F. Baluska, D. Volkmann, and P. Barlow, eds (Dordrecht, The Netherlands: Klewer Academic Publishers), pp. 3–27.
- Meagher, R.B., McKinney, E.C., and Vitale, A.V. (1999. b). The evolution of new structures: Clues from plant cytoskeletal genes. Trends Genet. 15 278–284. [DOI] [PubMed] [Google Scholar]
- Moon, A.L., Janmey, P.A., Louie, K.A., and Drubin, D.G. (1993). Cofilin is an essential component of the yeast cortical cytoskeleton. J. Cell Biol. 120 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., Yokota, E., Wada, T., Shimmen, T., and Okada, K. (2003). An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polymerization, reveals its distinctive function in root development. Plant Cell Physiol. 44 1131–1140. [DOI] [PubMed] [Google Scholar]
- Oshima, S., Abe, H., and Obinata, T. (1989). Isolation of profilin from embryonic chicken skeletal muscle and evaluation of its interaction with different isoforms. J. Biol. Chem. 105 855–857. [DOI] [PubMed] [Google Scholar]
- Paavilainen, V.O., Bertling, E., Falck, S., and Lappalainen, P. (2004). Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 14 386–394. [DOI] [PubMed] [Google Scholar]
- Pawloski, L.C., Kandasamy, M.K., and Meagher, R.B. (2006). The late pollen actins are essential for normal male and female development in Arabidopsis. Plant Mol. Biol. 62 881–896. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., Blanchoin, L., and Mullins, R.D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29 545–576. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and Cooper, J.A. (1986). Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55 987–1035. [DOI] [PubMed] [Google Scholar]
- Ringli, C., Baumberger, N., Diet, A., Frey, B., and Keller, B. (2002). ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 129 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J.S., and Swofford, D.L. (1999). Multiple local maxima for likelihoods of phylogenetic trees: a simulation study. Mol. Biol. Evol. 16 1079–1085. [DOI] [PubMed] [Google Scholar]
- Ruzicka, D.R., Kandasamy, M.K., McKinney, E.C., Burgos-Rivera, B., and Meagher, R.B. (September 18, 2007). The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exibit novel and differential expression. Plant J. http://dx.doi.org/10.1111/j.1365-313x.2007.03257.x. [DOI] [PubMed]
- Sachs, A.B., Davis, R.W., and Kornberg, R.D. (1987). A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj, J., Muller, J., Beck, M., Bohm, N., and Menzel, D. (2006). Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 11 594–600. [DOI] [PubMed] [Google Scholar]
- Sheahan, M.B., Staiger, C.J., Rose, R.J., and McCurdy, D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136 3968–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle, D., Haber, J.E., and Botstein, D. (1982). Lethal disruption of the yeast actin gene by integrative DNA transformation. Science 217 371–373. [DOI] [PubMed] [Google Scholar]
- Snowman, B.N., Kovar, D.R., Shevchenko, G., Franklin-Tong, V.E., and Staiger, C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14 2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, C.J. (2000). Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 257–288. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., and Blanchoin, L. (2006). Actin dynamics: Old friends with new stories. Curr. Opin. Plant Biol. 9 554–562. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Yuan, M., Valenta, R., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr. Biol. 4 215–219. [DOI] [PubMed] [Google Scholar]
- Valster, A.H., Hepler, P.K., and Chernoff, J. (2000). Plant GTPases: The Rhos in bloom. Trends Cell Biol. 10 141–146. [DOI] [PubMed] [Google Scholar]
- Valster, A.H., Pierson, E.S., Valenta, R., Hepler, P.K., and Emons, A.M.C. (1997). Probing the plant actin cytoskeleton during cytokinesis and interphase by profilin injection. Plant Cell 9 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantard, M., and Blanchoin, L. (2002). Actin polymerization processes in plant cells. Curr. Opin. Plant Biol. 5 502–506. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Yu, Y., Chen, Z.L., and Xia, G.X. (2005). Functional characterization of Gossypium hirsutum profilin 1 gene (GhPFN1) in tobacco suspension cells. Characterization of in vivo functions of a cotton profilin gene. Planta 222 594–603. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., and Galway, M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54 691–722. [DOI] [PubMed] [Google Scholar]
- Weber, A., Nachmias, V.T., Pennise, C.R., Pring, M., and Safer, D. (1992). Interaction of thymosin b4 with muscle and platelet actin: Implications for actin sequestration in resting platelets. Biochemistry 31 6179–6185. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.