Abstract
Centrosome reproduction by duplication is essential for the bipolarity of cell division, but the molecular basis of this process is still unknown. Mutations in Saccharomyces cerevisiae CDC31 gene prevent the duplication of the spindle pole body (SPB). The product of this gene belongs to the calmodulin super-family and is concentrated at the half bridge of the SPB. We present a functional analysis of HsCEN3, a human centrin gene closely related to the CDC31 gene. Tran- sient overexpression of wild-type or mutant forms of HsCen3p in human cells demonstrates that centriole localization depends on a functional fourth EF-hand, but does not produce mitotic phenotype. However, injection of recombinant HsCen3p or of RNA encoding HsCen3p in one blastomere of two-cell stage Xenopus laevis embryos resulted in undercleavage and inhibition of centrosome duplication. Furthermore, HsCEN3 does not complement mutations or deletion of CDC31 in S. cerevisiae, but specifically blocks SPB duplication, indicating that the human protein acts as a dominant negative mutant of CDC31. Several lines of evidence indicate that HsCen3p acts by titrating Cdc31p-binding protein(s).
Our results demonstrate that, in spite of the large differences in centrosome structure among widely divergent species, the centrosome pathway of reproduction is conserved.
Keywords: centrosome, duplication, Ca2+-binding protein, yeast, Xenopus laevis
Introduction
Although centrosome structure is strikingly different between yeasts and animal cells, proteins involved in the centrosome-associated microtubule nucleating activity are conserved (Martin et al. 1998; Murphy et al. 1998; Tassin et al. 1998). Given the important role of centrosome reproduction for cell division, one may conjecture that the corresponding molecular mechanisms also were conserved. The genes involved in animal centrosome duplication are unknown. However, several genes were identified in the spindle pole body (SPB) duplication pathway in budding yeast (Byers 1981; Rose and Fink 1987; Winey et al. 1991, Winey et al. 1993; Sun et al. 1992; Biggins and Rose 1994; Donaldson and Kilmartin 1996; McDonald and Byers 1997), and the precise SPB duplication cycle has been described recently (Adams and Kilmartin 1999).
The best example of a gene implicated in an early step of SPB duplication is the budding yeast CDC31, an essential gene in which mutations prevent the initiation of SPB duplication (Baum et al. 1986; Spang et al. 1993). The product of CDC31 shows homology to calmodulin, which is characterized by the presence of four potential Ca2+ binding sites, called EF-hands (Baum et al. 1986). It is localized to the half bridge of the SPB, on which the satellite forms when the SPB duplicates (Spang et al. 1993). Recently, it has been proposed that the half bridge undergoes a Cdc31p-dependent contraction to enable insertion of the newly synthesized SPB in the nuclear envelope (Adams and Kilmartin 1999). Cdc31p localization depends on Kar1p (Biggins and Rose 1994), which is also localized to the half bridge of the SPB (Spang et al. 1995) and is required for both SPB duplication and karyogamy (Conde and Fink 1976; Rose and Fink 1987). Kar1p contains a hydrophobic tail that probably anchors it in the nuclear envelope and which is necessary for its function (Vallen et al. 1992). A direct interaction between Kar1p and Cdc31p has been described (Biggins and Rose 1994). KAR1 also has been shown to be in genetic interaction with DSK2 (for dominant suppressor of KAR1; Vallen et al. 1994), a gene encoding a ubiquitin-like protein (Biggins et al. 1996). Dsk2p is also involved in SPB duplication since its overexpression or the expression of the allele dsk2-1 prevents SPB duplication (Vallen et al. 1994). However, DSK2 is not an essential gene, as are CDC31 and KAR1. Dsk2p is homologous to another ubiquitin-like protein, Rad23p, and the double deletion, Δdsk2 Δrad23, inhibits SPB duplication (Biggins et al. 1996). Neither Dsk2p nor Rad23p were localized to the SPB, and their role in centrosome duplication is unclear.
From these data, one could infer that mammalian members of the centrin family, which are structurally related to Cdc31p, are candidates for a function in centrosome duplication. However, the green algae, Chlamydomonas reinhardtii, centrin has been shown to be required for the proper segregation of the flagellar apparatus during cell division, rather than for the duplication of the basal bodies (Kuchka and Jarvik 1982; Wright et al. 1985; Taillon et al. 1992). In this organism, centrin is localized in the lumen of the basal bodies and forms contractile fibers connecting the basal bodies and the nucleus (Huang et al. 1988a,Huang et al. 1988b; Salisbury et al. 1988). The mutation, vfl2, in the centrin gene prevents the formation of the nucleus–basal body connection and the segregation of the basal bodies (Taillon et al. 1992). Moreover, centrin from C. reinhardtii is unable to complement cdc31 mutants in yeast. However, basal body-associated centrin is still detected in the vfl2 mutant making it likely that C. reinhardtii contains an additional centrin gene implicated in basal body duplication.
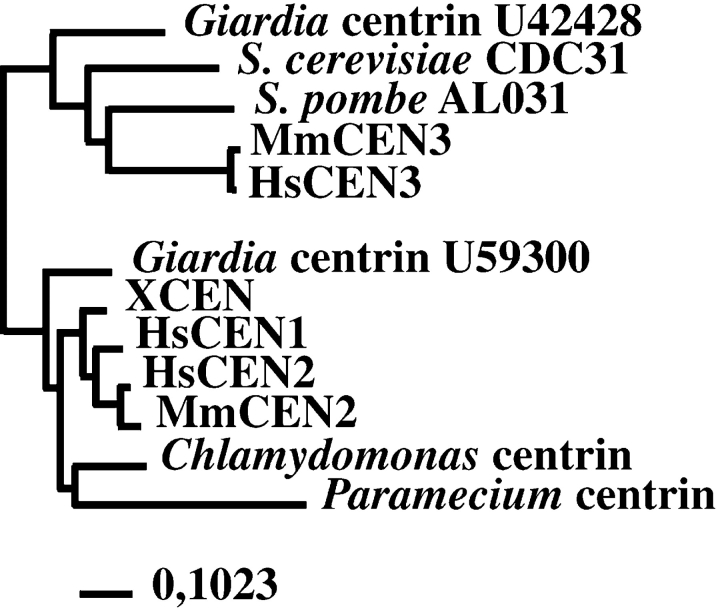
In human, three centrin genes have been described, named HsCEN1, HsCEN2, and HsCEN3 (Lee and Huang 1993; Errabolu et al. 1994; Middendorp et al. 1997; the symbols in the human genome database are CETN1, CETN2, and CETN3). The products of these genes are localized in the distal lumen of the centrioles and in the procentriole bud (Paoletti et al. 1996). Analysis of HsCEN2 revealed a possible function in cell cleavage since injection of recombinant HsCen2p in two-cell stage Xenopus laevis embryos induced undercleavage, leading to large blastomeres containing a variable number of microtubule asters (Paoletti et al. 1996). Sequence comparison revealed that HsCen3p shares more similarity with Cdc31p than the two other human centrin proteins, HsCen1p and HsCen2p (Middendorp et al. 1997), strongly suggesting the existence of two divergent subfamilies of centrin (see Fig. 1).
Figure 1.
CDC31 and centrin from Chlamydomonas reinhardtii define two divergent subfamilies. Note that human and murine CEN3 and CDC31 belong to the same subfamily, whereas HsCEN1, HsCEN2, and centrin from Chlamydomonas belong to the other subfamily. Accession numbers are indicated for the S. pombe centrin gene sequence, which has been found in the genome sequencing project and for the two divergent Giardia genes. Bar, mutation frequency.
We have undertaken a functional analysis of HsCen3p to test a potential role in centrosome duplication. In human cultured cells, we have demonstrated that centriolar targeting of HsCen3p requires a functional fourth EF-hand. Injection of recombinant wild-type HsCen3p or of RNA coding for a mutant form of HsCen3p in two-cell stage Xenopus embryos induces undercleavage, with blastomeres containing only one or two microtubule asters. Finally, HsCen3p is able to block cell growth by impairing SPB duplication in Saccharomyces cerevisiae. CDC31 can overcome this block in a dose-dependent manner. We have shown that HsCen3p binds the Cdc31p-binding protein Kar1p, but this interaction is not sufficient to explain the effect of HsCen3p.
Materials and Methods
Cloning of cDNA of Human Centrins in pCB6
To allow detection and localization of the overexpressed protein, cDNA coding for HsCen1p or HsCen3p was cloned in the mammalian expression vector pCB6 (Brewer and Roth 1991), in fusion with either a VSVG epitope in the NH2 terminus region or a six histidines tag in the COOH terminus region. HsCen1p or HsCen3p were amplified by PCR to introduce EcoRI and XbaI restriction sites, respectively, at the 5′ and 3′ ends of the cDNA. The PCR products were double digested by EcoRI and XbaI and ligated in the EcoRI/XbaI-digested pBS-KS vector containing the cDNA coding for the 15 amino acids of the VSVG protein recognized by the mAb, P5D4 (Soldati and Perriard 1991). The cDNA encoding the fusion between VSVG and HsCen1p or HsCen3p was excised by a KpnI/BamHI double digestion and inserted in the mammalian expression vector pCB6 under the control of CMV promoter. A histidine tag was introduced by a PCR also inserting restriction sites to enable cloning in pCB6. The mutants HsCen3p-D147,149,151A, where the three aspartates in position 147, 149, and 151 were replaced by alanines and the mutant HsCen3p-P99A, where the proline in position 99 was replaced by an alanine, were generated by PCR mutagenesis and cloned in pCB6 in fusion, respectively, with a VSVG or a six histidines tag.
Transfection of HeLa Cells
Exponential growing HeLa cells were transfected by electroporation. 5 × 106 HeLa cells were detached by trypsin, washed, and resuspended in 200 μl of medium containing 10% FCS and 15 mM Hepes, pH 7.5. 40 μg of plasmid and 20 μg of carrier DNA (salmon sperm DNA) were diluted in 50 μl of 210 mM NaCl solution and mixed to the cell suspension in a 4-mm electroporation cuvette. Cells were submitted to an electric pulse of 290 V, 960 μF, and an unlimited resistance in an electroporator (BioRad). Cells were then washed in 5 ml of medium containing 10% FCS and 15 mM Hepes, pH 7.5, and seeded either on collagen-coated coverslips for immunofluorescence analysis or on a petri dish for Western blot analysis. Immunofluorescence of HeLa cells was performed as described by Paoletti et al. 1996.
Western Blot Analysis
Samples were separated on 12% SDS-PAGE as described by Laemmli 1970 and electrophoretically transferred onto nitrocellulose and processed for immunoblotting as described by Paoletti et al. 1996. After saturation, membranes were incubated with 1/2,000 anti-HsCen1p, 1/250 anti-HsCen3p, 1/100 anti-Cdc31p rabbit sera, or 1/50 anti-Kar1p goat sera. Immune detection was carried out with anti-IgGs coupled to alkaline phosphatase (HsCen1p) or to HRP (HsCen3p), followed by enhanced chemiluminescence (ECL) detection (Nycomed Amersham, Inc.) according to the company's instructions. For detection of anti-Cdc31p and anti-Kar1p antibodies, biotin-conjugated secondary antibodies and alkaline phosphatase-conjugated streptavidin were used (Nycomed Amersham, Inc.).
Microinjection and Immunofluorescence Experiments in Xenopus Embryos
6 histidine-tagged HsCEN3 cDNA, generated by PCR inserting NdeI and BamHI restriction sites, was cloned in the bacterial expression vector, pET3b. Recombinant His-HsCen3p was induced in the BL21DE3 Escherichia coli strain and purified on NiNTA agarose (Qiagen Inc.) according to manufacturer's instructions. High molecular weight contaminants were eliminated by chromatography on superdex 75 (Pharmacia Biotech, Inc.). Microinjection and immunofluorescence experiments were performed as described by Paoletti et al. 1996. cDNA was cloned in pβGFP/RN3P (Zernicka-Goetz et al. 1996), replacing the GFP insert, and RNAs were transcribed in vitro as described by Jarmolowski et al. 1994, and diluted in 10 mM Hepes before injection.
Yeast Strain Genotypes
YPH500, MATa ade2 his3 leu2 ura3 trp1. YMK229, MATa ade2 his3 leu2 ura3 trp1 lys2 Δkar1::HIS3 pRS315-cdc31-16-LEU2.
Cloning of Centrin Genes in Saccharomyces cerevisiae Expression Vectors
cDNAs encoding HsCen1p, HsCen2p, HsCen3p, and Cdc31p were amplified by PCR, and KpnI and XbaI restriction sites were introduced, respectively, before the initiation codon and after the stop codon. The PCR products were cloned at the KpnI and XbaI sites of the 2μ multicopy plasmid, pYES-URA3, or in the centromeric plasmid, pRS315 (for HsCEN3), under the control of the Gal4 inducible promoter. To generate fusions between either amino acids 1–23 of HsCen3p and amino acids 18–161 of Cdc31p, or amino acids 1–17 of Cdc31p and amino acids 24–167 of HsCen3p, the corresponding fragments of cDNA were amplified by PCR, inserting restriction sites to enable ligation of the products in pYES-URA3. For the fragments corresponding to amino acids 1–17 of Cdc31p and amino acids 1–23 HsCen3p, KpnI and XhoI restriction sites were introduced, respectively, before the start codon and by mutagenesis of the last three codons of the cDNA. For the fragments corresponding to amino acids 18–161 of Cdc31p and amino acids 24–167 HsCen3p, XhoI and XbaI restriction sites were introduced, respectively, by mutagenesis of the first three codons of the cDNA and after the stop codon. The constructions were transformed in yeast strain YPH500 or YMK229 as described by Wimmer et al. 1992.
Growth of Yeast Strains
Yeast strains were grown on synthetic medium lacking specific amino acids. For growth on solid medium, two dilutions of each strain were spotted on glucose plates (repressing conditions) or on raffinose + galactose plates (inducing conditions), both lacking uracil or uracil and leucine, and then grown at 30°C. Raffinose was added to the galactose medium for the induction experiments because YPH500 and YMK229 yeast strains grow poorly on medium containing galactose as the only carbon source. For the cotransformation of HsCEN3 and CDC31, the URA3 gene in the plasmid harboring CDC31 was replaced by the LEU2 marker. Cells transformed with the two plasmids were grown on plates lacking uracil and leucine.
For growth in glucose or raffinose media lacking uracil, cells were diluted in fresh medium at the beginning of the experiments and grown at 30°C. To de-repress the GAL4 promotor before growth in galactose + raffinose-URA, cells were first washed twice and diluted in fresh raffinose-URA medium and grown for 2 h at 30°C. At the begining of the experiment, galactose was added to the cultures.
Immunofluorescence Experiments
Cells were fixed as described by Belgareh and Doye 1997 and incubated with anti-Cen3p antibody diluted at 1:200 in PBS containing 1% BSA and antitubulin antibody (Sigma Chemical Co.) diluted at 1:1,000, followed by Fluorescein-conjugated goat anti–rabbit and rhodamine-conjugated goat anti–mouse antibodies (Jackson ImmunoResearch Laboratories, Inc.) at a dilution of 1:700. For DNA staining, 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) was used. Western blot experiments showed that anti-Cen3p antibody was not able to detect the endogenous level of Cdc31p expressed in yeast. Alternatively, cells were fixed as described by Rout and Kilmartin 1990 and incubated with 45D10, an mAb directed against Spc110p, and with anti-Cen3p antibody.
Electron Microscopy
Cells were fixed and prepared for EM as described by Doye et al. 1994.
Immunoprecipitation Experiments
Cell extracts were prepared by resuspending a frozen pellet corresponding to 50 ml of a 10-h culture (0.7 OD600) in 250 μl of lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 1% NP-40) containing protease inhibitors (Boehringer Mannheim Corp.). 0.4 g of glass beads were added and the samples were incubated with vigorous vortexing at 4°C for 30 min and then centrifuged 15 min at 20,000 g at 4°C. Protein G–Sepharose beads, equilibrated in PBS, were incubated with affinity-purified antibodies for 2 h at 4°C under mild agitation. Immunoglobulin-bound protein G–Sepharose beads were sedimented, washed three times in lysis buffer, and then incubated with cell extracts for 2 h at 4°C under mild agitation. Sedimented protein G–Sepharose beads were washed six times in lysis buffer and once in distilled water. The immunoprecipitates were solubilized from the Sepharose beads by incubation with the SDS-PAGE sample buffer (100°C, 5 min) and centrifugation.
Results
HsCen3p Targeting to Centrioles in Human Cells
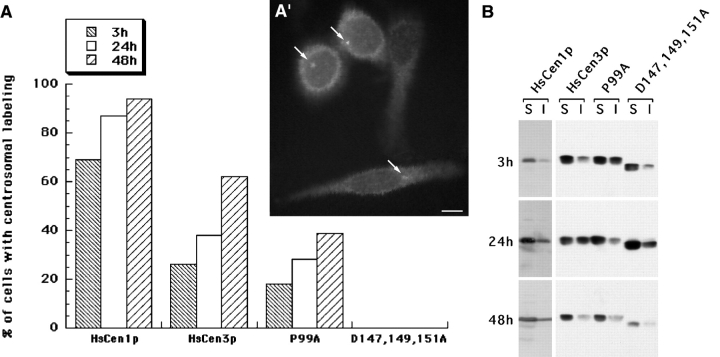
We first attempted to test HsCen3p function in human cultured cells. We undertook transient overexpression experiments of wild-type HsCEN3 or mutants of HsCEN3 in HeLa cells or diploid skin fibroblasts. We chose two mutants that were expected to be defective for HsCen3p function: the first, HsCen3p-P99A, affected a proline conserved among the Cdc31p-related centrins and located between the second and the third EF-hand; the second, HsCen3p-D147,149,151A, inactivated the fourth and most conserved EF-hand (Middendorp et al. 1997). The overexpressed proteins were fused either with 16 amino acids of the VSVG protein cloned at the NH2 terminus and recognized by the mAb P5D4, or with a 6 histidines epitope cloned at the COOH terminus. Transient overexpression of wild-type HsCen3p did not produce any obvious phenotype, such as centrosome duplication defect or abnormal mitosis. We observed that the protein was able to exchange with the centriolar endogenous HsCen3p (Fig. 2 A′), suggesting that these tags did not impair the protein function. However, when compared with overexpression of HsCen1p, we observed that localization of HsCen3p to the centrosome was slower than the localization of HsCen1p (Fig. 2 A), suggesting either that the half-life of HsCen3p is longer than the one of HsCen1p or that the centriolar targeting of the two proteins is differently regulated. The localization of the HsCen3p-P99A mutant is less efficient than that of the wild-type protein. The HsCen3p-D147,149,151A mutant was totally unable to exchange with the centriole-associated endogenous protein (Fig. 2 A). This suggested that centriolar localization depends upon the presence of a functional fourth EF-hand. We checked by Western blot experiments that the mutant protein was expressed (Fig. 2 B) and observed that, despite the lack of centriolar localization, part of this mutant protein was associated with a Triton X-100 insoluble fraction. However, overexpression of either of the two mutant proteins did not arrest cell growth, possibly due to the presence of a sufficient amount of wild-type protein.
Figure 2.
Mutation of the fourth EF-hand impairs centriole localization. A, HeLa cells were transfected either with VSV-tagged HsCEN1 or HsCEN3, or P99A mutant form of HsCEN3, or with His-tagged D147,149,151A mutant form of HsCEN3, fixed, and analyzed by double immunofluorescence with antitag and anticentriole antibodies. The percentage of cells with centrosomal localization of the tagged protein was estimated 3, 24, or 48 h after transfection. A′, HeLa cells transfected with HsCEN3 were processed for antitag immunofluorescence 48 h after transfection. Three cells in this field present centrosome labeling (arrows). Bar, 2 μm. B, Soluble (S) and insoluble (I) Triton X-100 proteins from 105 cells were migrated on 12% SDS-PAGE, transferred to nitrocellulose, and revealed either with anti-HsCen1p antibodies or with anti-HsCen3p antibodies. Note that overexpression of HsCen1p (detection in alkaline phosphatase) is more efficient than overexpression of wild-type or mutant HsCen3p (detection in ECL). In all cases, the major part of the overexpressed protein is extracted by Triton X-100 treatment. Note also the existence of an insoluble pool of D147,149,151A mutant protein, despite the lack of centriole localization.
As noted above, the amount of wild-type homologous protein was not critical in transient transfection experiments suggesting that, just like in yeast (Geier et al. 1996; see also Fig. 4), the amount of homologous protein is not critical. However, we consistently failed in establishing cell lines stably overexpressing HsCen3p, whereas we could easily do so with HsCEN1 and HsCEN2. This observation suggested that increase in the dosage of HsCen3p might have a long-term effect, deleterious only after one or two centrosome duplication cycles. This possibility has been addressed in the early developing Xenopus embryo, in which several rapid rounds of division take place.
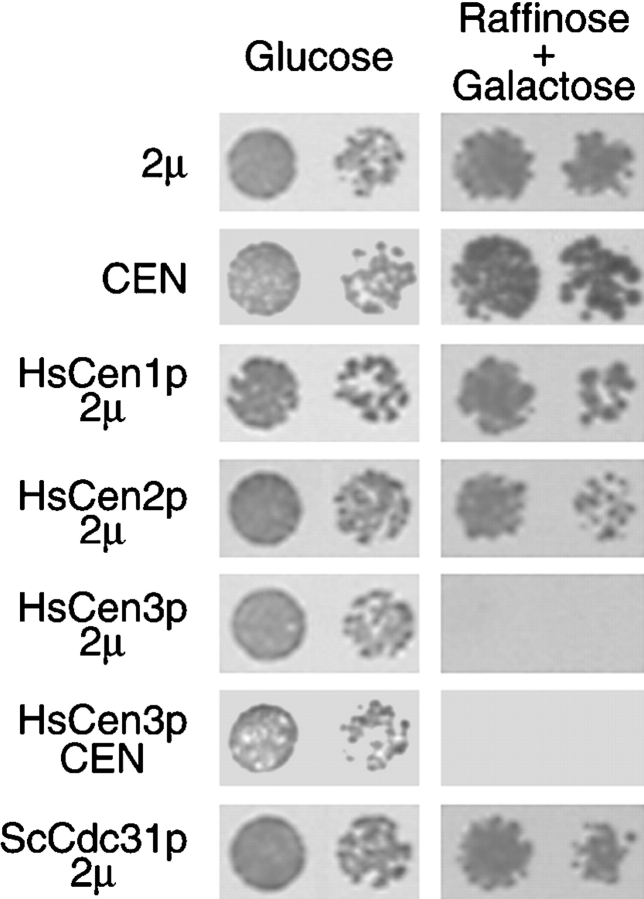
Figure 4.
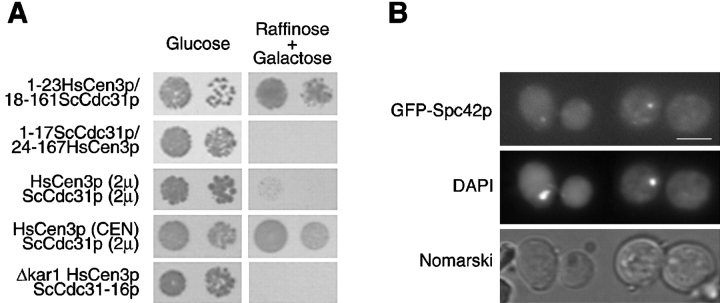
HsCen3p specifically blocks cell growth. Yeast strains transformed with a 2μ (pYES) or a CEN (centromeric) empty vector, or a 2μ vector encoding HsCen1p, HsCen2p, HsCen3p, yeast Cdc31p, or a CEN vector encoding HsCen3p were grown on glucose plates (repressive condition) or on raffinose + galactose plates (inducing condition) for 3 d at 30°C. For each condition, two dilutions were spotted. Cells expressing HsCen3p on a 2μ or on a CEN plasmid did not grow in inducing condition.
Dose-dependent HsCen3p Inhibition of Centrosome Duplication in the Xenopus Early Embryo
We investigated HsCen3p function in a heterologous animal system undergoing a simple cell division cycle, the Xenopus embryo, which has already been used to study HsCen2p function (Paoletti et al. 1996). Recombinant histidine-tagged HsCen3p (His-HsCen3p) was microinjected in one blastomere of two-cell stage embryos. Embryos were observed before mid-blastula transition (7 h after fertilization at 22°C) and after gastrulation (22 h after fertilization). 72% of the embryos injected with 12 mg/ml His-HsCen3p presented undercleavage, whereas only 2% of PBS-injected embryos presented a cleavage delay (Table ; Fig. 3 A). 22 h after fertilization, 25% of the His-HsCen3p-injected embryos had exogastrulated and 69% had lysed, compared with 23% and 2% for the control embryos, respectively. These effects were concentration-dependent as shown in Table . As a control, we injected heat- or protease-treated His-HsCen3p. As other centrins (Paoletti et al. 1996), His-HsCen3p is resistant to heat treatment (95°C for 1 h). Injection of 12 mg/ml heat-treated His-HsCen3p resulted in under-segmentation (Table ; Fig. 3 A, 3). The effect was, however, weaker than with the nonheated protein, as judged by embryo lysis at 22 h, suggesting partial heat denaturation. Protease-treated His-HsCen3p had no significant effect (Table ; Fig. 3 A, 4).
Table 1.
Short- and Long-term Effects of Heterologous Centrin 3 in Two-Cell Stage Injected Xenopus Embryos
| Pre-MBTdevelopment | Post-MBTdevelopment | ||||||
|---|---|---|---|---|---|---|---|
| Injected solution | N | U | L | N | E | L | Eggs |
| % | % | % | % | % | % | n | |
| Protein injection | |||||||
| HsCen3p (1.5 mg/ml) | 80 | 20 | 0 | 49 | 48 | 3 | 90 |
| HsCen3p (3 mg/ml) | 25 | 75 | 0 | 25 | 75 | 0 | 20 |
| HsCen3p (6 mg/ml) | 30 | 70 | 0 | 10 | 48 | 42 | 91 |
| HsCen3p (12 mg/ml) | 28 | 72 | 0 | 6 | 25 | 69 | 77 |
| Heat-treated HsCen3p | 31 | 69 | 0 | 10 | 77 | 13 | 97 |
| Protease-treated HsCen3p | 95 | 5 | 0 | 58 | 28 | 14 | 56 |
| PBS | 98 | 2 | 0 | 75 | 23 | 2 | 133 |
| RNA injection | |||||||
| WT HsCen3 | 63 | 32 | 5 | 30 | 46 | 24 | 106 |
| Mutant HsCen3 | 38 | 58 | 4 | 6 | 29 | 65 | 103 |
| GFP | 89 | 11 | 0 | 64 | 19 | 17 | 37 |
| Hepes | 94 | 5 | 1 | 64 | 25 | 11 | 79 |
25 ml of native or treated recombinant HsCen3p, or 25 ml of wild-type HsCEN3 (WT), mutant HsCEN3, or GFP RNA at 0.6 μg/ml or control solutions (PBS or Hepes) were injected in one blastomere of two-cell stage embryos. Short-term effects were observed 7 h (protein injection) or 11 h (RNA injection) after fertilization. The effect on blastomere cleavage was scored as normal (N), under-segmented (U), or lysis (L) of the injected half. Long-term effects were observed 22 h after fertilization. Development of the injected side of the embryo was scored as normal (N), participating in exogastrulation of the whole embryo (E), or lysed (L). Note that in the case of RNA injections, pre-MBT development was analyzed 11 h after fertilization to allow RNA translation.
Figure 3.
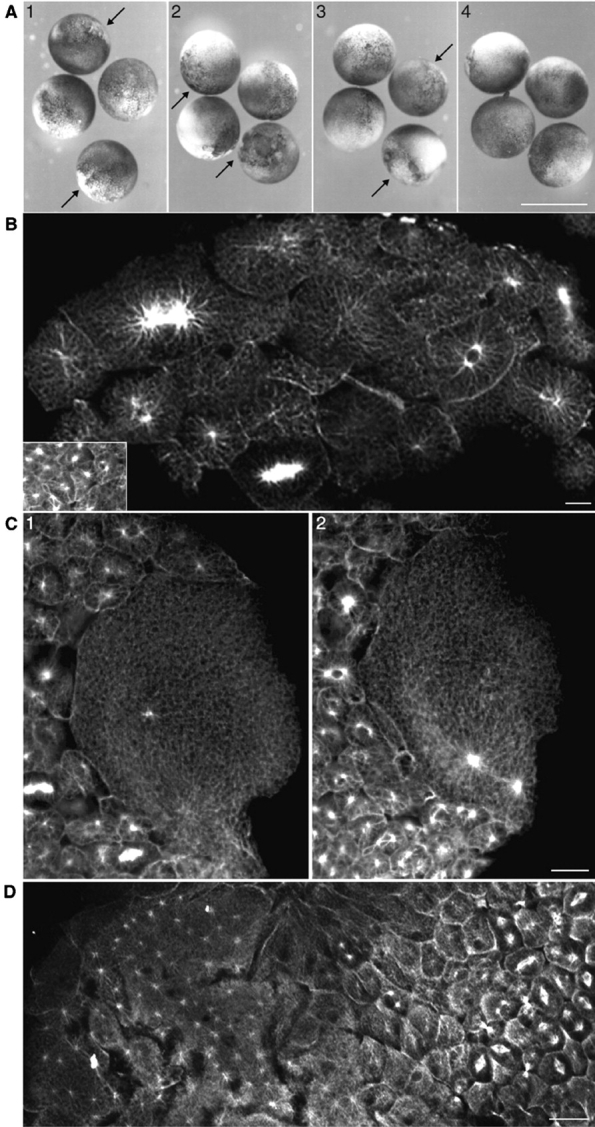
6His-HsCen3p induces blastomeres undercleavage in Xenopus laevis embryos. A, Two-cell stage embryos microinjected either with 6 mg/ml His-HsCen3p (1), 12 mg/ml His-HsCen3p (2), 12 mg/ml heat-treated His-HsCen3p (3), or 12 mg/ml protease-treated His-HsCen3p (4) were observed 7 h after fertilization. Undercleaved blastomeres are present in the first three conditions (arrows). Bar, 1 mm. B, An embryo, injected with His-HsCen3p, methanol-fixed 8 h after microinjection, and labeled with anti–α-tubulin antibodies, contains large blastomeres with one or two microtubule asters in the injected half, whereas the uninjected cells gave rise to small blastomeres with one or two asters (inset). Bar, 50 μm. C, Large blastomere of a His-HsCen3p–injected embryo exhibiting one aster in a confocal section (1) and two additional asters in another section (2), whereas the uninjected cells gave rise to small blastomeres with one or two asters (bottom left). Bar, 50 μm. D, An embryo, injected with 12 mg/ml HsCen2p, methanol-fixed 8 h after microinjection, and labeled with anti–α-tubulin antibodies, contains large blastomeres with numerous asters (left) in the injected half, whereas the uninjected cells gave rise to small blastomeres with one or two asters (right). Bar, 50 μm.
Anti–α-tubulin staining demonstrated that most of the under-segmented blastomeres (91%) contained two large microtubule asters (Fig. 3B and Fig. C; Table ). The size of the center of these asters was comparable to that of control blastomeres, making it unlikely that they contain numerous duplicated, but unseparated, centrosomes. A few blastomeres contained a single aster (5%) and 4% presented three or four asters. These observations, in marked contrast with the effect of HsCen2p injection (Fig. 3 D), strongly suggest that His-HsCen3p inhibits cleavage by impairing centrosome duplication or inhibits both cleavage and centrosome duplication independently.
Table 2.
Number of Asters per Blastomere in the Injected Half of Embryos
| Number of asters/blastomere | ||||||
|---|---|---|---|---|---|---|
| Injection | 1 | 2 | 3 | 4 | 5 or more | Blastomeres |
| % | % | % | % | % | n | |
| HsCen3p | 5 | 91 | 1 | 3 | 0 | 103 |
| WT HsCEN3 RNA | 10 | 78 | 3 | 3.5 | 5.5 | 111 |
| Mutant HsCEN3 RNA | 18 | 68.5 | 3 | 2.5 | 8 | 126 |
7 h (protein injection) or 11 h (RNA injection) after fertilization, embryos were fixed and labeled with monoclonal anti-α–tubulin. For each condition, the number of asters per large blastomere in the injected half of the embryo was scored in two embryos from two independent experiments. One or two asters were observed in blastomeres cleaved at normal rate.
We also tested whether the fourth EF-hand mutant, which failed to localize to the centrosome (see above), could also disturb centrosome duplication. As it is not possible to perform anticentrosome immunofluorescence in whole Xenopus embryos, we microinjected the nucleus of Xenopus somatic cells with the plasmid coding for this mutant form. We checked that the fourth EF-hand mutant was unable to localize to the centrosome in Xenopus, whereas the wild-type form did (data not shown). As we were not able to concentrate the mutant protein at 12 mg/ml due to its insolubility, we injected the in vitro transcribed RNA. As a control, we injected RNA encoding wild-type HsCen3p or GFP, all at 0.6 μg/ml. Injection of wild-type HsCEN3 RNA confirmed results obtained with HsCen3p (Table ). The RNA encoding the mutated HsCen3p induced under-segmentation, demonstrating a stronger effect than wild-type HsCEN3 RNA. Injection of GFP RNA, or of Hepes buffer, had a minimal effect (Table ). Anti–α-tubulin staining showed that most of the RNA-injected blastomeres contained two asters when observed 11 h after fertilization (Table ). Compared with protein-injected embryos, blastomeres with either one aster, or three or more asters were more numerous. This variation is probably dose-dependent. It is noteworthy that, in the case of RNA injection, the local amount of protein is difficult to assess as it depends on both diffusion and translation of RNA.
HsCen3p Inhibits SPB Duplication in Budding Yeast
Finally, another approach to support a possible involvement of HsCEN3 in centrosome duplication was to test whether HsCEN3 could complement S. cerevisiae strains bearing temperature-sensitive mutations in CDC31 (cdc31-100 D144V, cdc31-101 F39I, cdc31-102 K123E) or a complete deletion of CDC31. Expression of HsCen3p did not rescue the temperature-sensitive phenotype of the mutants nor did it complement the deletion of ScCDC31 (data not shown). However, overexpression of HsCen3p in a wild-type background was lethal: cells transformed with a galactose inducible 2μ or CEN (centromeric) plasmid encoding HsCen3p did not grow when expression was induced (Fig. 4). This effect was specific because overexpression of HsCen1p, HsCen2p, or Cdc31p (Fig. 4; see also Geier et al. 1996) did not inhibit growth of wild-type cells. We checked by Western blotting that HsCen1p, HsCen2p, HsCen3p, and Cdc31p were expressed to comparable levels in the strains transformed with the 2μ plasmid (data not shown).
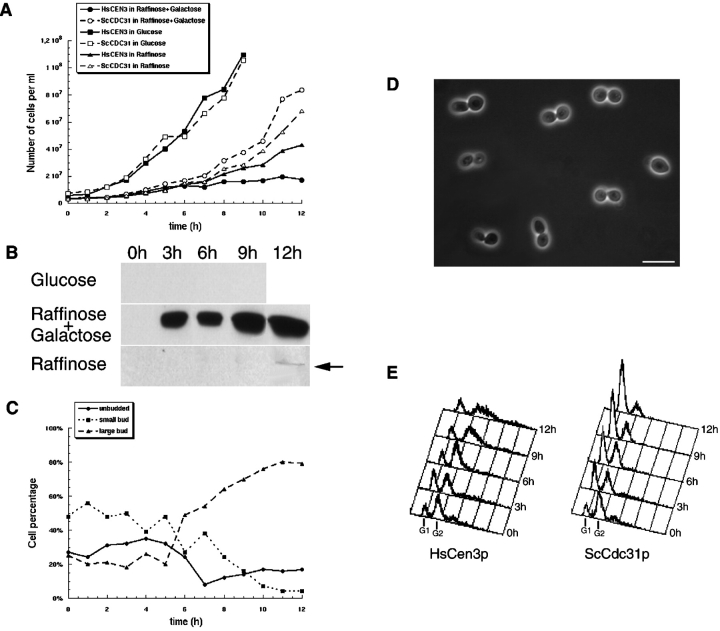
To further investigate the effect of HsCen3p, yeast strains bearing HsCEN3 or CDC31 on a galactose inducible plasmid were grown in liquid medium complemented with glucose (repression), raffinose (low level of expression), or raffinose + galactose (induction). Whereas cell growth was identical in glucose medium for both strains, cells expressing HsCen3p stopped growing 6 h after induction (Fig. 5 A). Moreover, cell growth of this strain in raffinose, where a very low expression of HsCen3p was detected (Fig. 5 B), decreased after a 10-h culture. This indicates that even a very low level of HsCen3p is able to impair cell proliferation. This was further confirmed by the effect of HsCEN3 cloned on a centromeric plasmid, which blocked cell proliferation (see Fig. 4). The effect of HsCen3p overexpression was not reversible: cells grown in raffinose + galactose medium for 12 h were not rescued from the block when seeded onto glucose plates.
Figure 5.
Yeast cells expressing HsCen3p present a large bud. A, Yeast strain expressing HsCen3p or Cdc31p were grown in inducing condition (raffinose + galactose), repressing condition (glucose), or in raffinose medium (the Gal promotor is neither repressed or induced). The number of cells were determined at hourly intervals. Cells expressing HsCen3p stopped growing in raffinose + galactose and in raffinose medium at 6 and 10 h, respectively. B, Proteins from yeast strain expressing HsCen3p growing in glucose, raffinose, or raffinose + galactose medium were prepared and analyzed by Western blot with anti-HsCen3p antibodies. HsCen3p is rapidly and efficiently induced in raffinose + galactose and a very low signal is detected in raffinose after a 12-h culture. C, Yeast strain expressing HsCen3p was grown in inducing condition (raffinose + galactose). The number of unbudded, small-budded, or large-budded cells were determined at hourly intervals. Cells presenting a large bud start accumulating at 6 h. D, Phase-contrast microscopy of yeast strain expressing HsCen3p grown in inducing conditions. Most of the cells present a large bud. Bar, 10 μm. E, FACS analysis of yeast strain expressing HsCen3p or Cdc31p grown in inducing conditions (raffinose + galactose). At 12 h, cells expressing HsCen3p have mainly a G2 DNA content, whereas control cells have a G1 DNA content.
To test whether cells were arrested at a specific stage of the cell cycle upon HsCen3p overexpression, we monitored the budding index throughout the induction. As shown in Fig. 5C and Fig. D, cells with a large bud started to accumulate 6 h after induction, reaching a maximum value of 80% at 12 h. At this time, cells overexpressing Cdc31p were mainly unbudded, only 10% of them having a large bud (data not shown). We demonstrated by flow cytometry analysis that the large budded HsCen3p-expressing cells had a G2 DNA content (Fig. 5 E) and an increased size, compared with control cells in G2-M.
Immunofluorescence analysis of HsCen3p-induced large budded cells with antitubulin antibody did not show the spindle characteristic of the G2-M phase (Fig. 6 A). mAb 45D10, which recognizes the SPB component Spc110p (Rout and Kilmartin 1990), revealed only one dot (Fig. 6 B). A single dot corresponding to SPB was also observed in a yeast strain expressing HsCen3p and GFP-Spc42p (Fig. 6C and Fig. C′). Cytoplasmic background, but no staining of the SPB, could be detected with anti-HsCen3p antibodies, whatever the fixation method used, indicating that HsCen3p was not able to localize to the SPB. These results, together with the DAPI staining showing the nucleus most often trapped into the neck (Fig. 6A and Fig. B), strongly suggested that cells failed to complete mitosis due to unduplicated SPB or unseparated SPBs.
Figure 6.
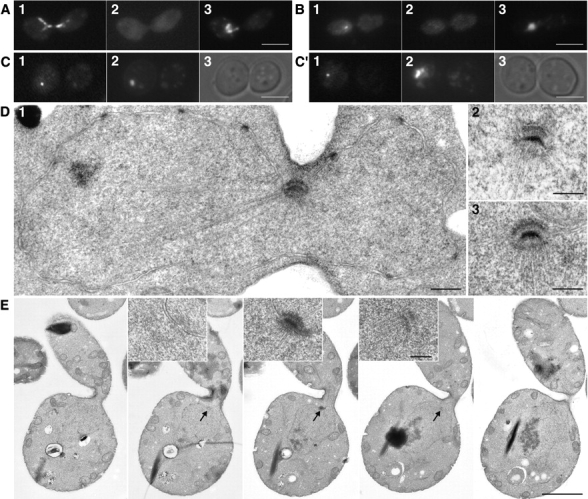
Yeast cells expressing HsCen3p contain an unduplicated SPB. A and B, 12 h after induction, cells expressing HsCen3p were fixed and double labeled with an anti–α-tubulin (A1) or an anti-Cen3p (A2) antibody, or an anti-Spc110p (B1) or an anti-Cen3p (B2) antibody. DNA was labeled with DAPI (A3 and B3). Large-budded cells did not contain any spindle, but diverging microtubules radiating from the nucleus trapped in the bud neck (A1). Anti-Spc110p antibody detected only one dot on the nucleus trapped in the bud neck (B1). No specific localization of HsCen3p was detected (A2 and B2). Bar, 5 μm. C and C′, 12 h after induction, GFP-Spc42p bearing strain overexpressing HsCen3p was observed for GFP in the fluorescein channel (1), for DNA staining (DAPI; 2), and in Nomarski optics (3). In both cases, a single dot per nucleus was detected. Bar, 5 μm. D 1, Electron micrograph of a cell 12 h after induction of HsCen3p expression. A single SPB is observed at the bud neck. The contour of the nucleus can be easily identified by the nuclear envelope bearing nuclear pores (black dots). Microtubules diverging from the SPB can be seen in the nucleus and in the cytoplasm. D 2 and 3, SPBs of two individual cells. Note the bent profile of the unduplicated SPB. Bars, 0.3 μm. E, Five serial sections of a cell expressing HsCen3p 12 h after induction. A single unduplicated SPB is observed in sections 2, 3, and 4 (arrows) and is enlarged tenfold in the corresponding insets. Bars, 2 μm and 0.2 μm, respectively.
To discriminate between these two possibilities, cells were fixed for EM. The observation of SPB in 35 sagittally sectioned cells blocked in G2-M showed neither an associated satellite nor additional SPB (Fig. 6 D). This was further established by serial sectioning (five successive nuclear sections of five individual cells). In all cases, a single unduplicated SPB was observed (Fig. 6 E). The second, third, and fourth sections always showed part of the single SPB and a few microtubules radiating from the SPB, whereas the first and the fifth sections did not contain any SPB. Examination of the unduplicated SPB at higher magnification showed that their size had not been affected. Altogether, this data shows that HsCEN3 behaves as a dominant negative mutant of CDC31, inhibiting the initiation of SPB duplication.
The most divergent region between HsCen3p and Cdc31p lies in the NH2 terminus region (Middendorp et al. 1997). To test whether this region was responsible for the dominant effect of HsCen3p, the 5′ region of the two cDNAs were swapped. The cDNA encoding amino acids 1–17 of Cdc31p in fusion with amino acids 24–167 of HsCen3p still blocked cell growth, whereas the reciprocal construction (1–23HsCen3p/18–161Cdc31p) had no effect (Fig. 7 A). This demonstrated that the effect of HsCen3p was not due to the divergent NH2 terminus end.
Figure 7.
Cdc31p can overcome HsCen3p-induced growth arrest in a dose-dependent manner. A, Cells expressing: a fusion between either amino acids 1–23 of HsCen3p and amino acids 18–161 of Cdc31p, or amino acids 1–17 of Cdc31p and amino acids 24–167 of HsCen3p; or overexpressing 2μ plasmids coding for HsCen3p and Cdc31p; or overexpressing both HsCen3p cloned on a centromeric (CEN) plasmid and Cdc31p cloned on a 2μ plasmid; or expressing HsCen3p and Cdc31-16p in a Δkar1 background were grown on glucose plates (repressive condition) or on raffinose + galactose plates (inducing condition) for 3 d at 30°C. For each condition, two dilutions were spotted. Cells expressing a fusion between amino acids 1–17 of Cdc31p and amino acids 24–167 of HsCen3p, both HsCen3p and Cdc31p, or expressing HsCen3p and Cdc31-16p in a Δkar1 background did not grow in inducing condition. B, 12 h after induction, GFP-Spc42p bearing strain overexpressing HsCen3p in a Δkar1 background was observed for GFP in the fluorescein channel, for DNA staining (DAPI), and in Nomarski optics. In both cases, a single dot per nucleus was detected. Bar, 5 μm.
It is possible that HsCen3p acts by titrating either Cdc31p or a protein that normally interacts with Cdc31p. If such a complex between HsCen3p and Cdc31p or a Cdc31p-binding protein exists, overexpressing Cdc31p might restore the native complex. To test this possibility, we performed cooverexpression of HsCen3p and Cdc31p. A yeast strain containing a multicopy plasmid expressing HsCen3p was transformed with a multicopy plasmid bearing CDC31 cDNA. As shown in Fig. 7 A, these cells did not grow under inducing conditions. However, when we attempted to find yeast genes able to suppress the effect of HsCen3p expression by transforming a yeast strain expressing HsCen3p cloned on a centromeric plasmid with a genomic library of S. cerevisiae cloned on a 2μ multicopy plasmid, we found only one gene able to rescue the phenotype, namely CDC31 (Fig. 7 A). In this condition, Western blot analysis showed that Cdc31p was expressed to a higher level than HsCen3p. This result suggested that HsCen3p might interact with either Cdc31p itself or with Cdc31p-binding protein(s) with a higher affinity than Cdc31p.
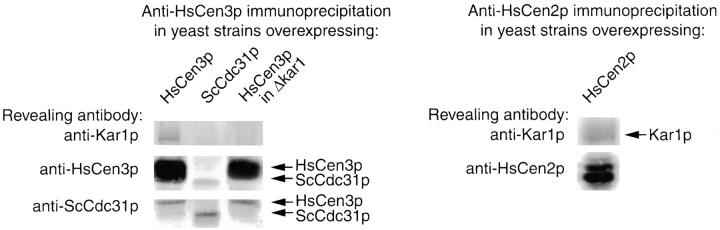
To discriminate between these two possibilities, we tested whether Cdc31p or Kar1p, a Cdc31p-binding protein involved in SPB duplication, were coimmunoprecipitated by anti-HsCen3p antibodies. As shown in Fig. 8, anti-HsCen3p antibodies did not immunoprecipitate Cdc31p in the yeast strain overexpressing HsCen3p, despite their capacity to weakly cross-react with Cdc31p, as observed in the strain overexpressing Cdc31p. By contrast, anti-HsCen3p antibodies immunoprecipitated Kar1p in the HsCen3p-overexpressing strain, whereas they did not in the Cdc31p-overexpressing strain. As expected, Kar1p was not immunoprecipitated in the Δkar1 YMKH229 strain overexpressing HsCen3p (see below). It is noteworthy that when we performed anti-HsCen2p immunoprecipitation in yeast strain overexpressing HsCEN2, we found that HsCen2p was able to interact in vivo with Kar1p, as previously shown in vitro (Geier et al. 1996). As HsCen2p does not impair SPB duplication, this result suggests that the specific blocking effect of HsCen3p in S. cerevisiae is dependent on interaction of HsCen3p with ScCdc31p-binding protein(s) other than Kar1p.
Figure 8.
HsCen3p interacts with Kar1p extracts of yeast cells overexpressing HsCen3p, Cdc31p. HsCen3p in a Δkar1/cdc31-16 background, or HsCen2p were immunoprecipitated with anti-HsCen3p affinity-purified antibodies or with anti-HsCen2p antibodies. Immunoprecipitated proteins were separated on SDS-PAGE, transferred onto nitrocellulose, and revealed with anti-HsCen3p, anti-Cdc31p, or anti-Kar1p antibodies, or with anti-HsCen2p and anti-Kar1p antibodies. In cells overexpressing HsCen3p, HsCen3p and Kar1p are coimmunoprecipitated. It is noteworthy that in the conditions we have used, Cdc31p and Kar1p are not coimmunoprecipitated, whereas HsCen2p and Kar1p are.
To investigate this possibility, we tested whether HsCen3p could inhibit growth of the YMK229 strain that is deleted for KAR1 and expresses cdc31-16, the product of which is able to localize to the SPB in absence of Kar1p (Vallen et al. 1994). To facilitate the phenotypic analysis, we used a YMK229 strain in which Spc42p is expressed in fusion with GFP. If HsCen3p acts by sequestering Kar1p only, expression of HsCen3p in a Δkar1 strain would be expected to be without effect. We observed the opposite result: expression of HsCen3p in this genetic background remained lethal (Fig. 7 A), 80% of the cells accumulating with a large bud after a 12-h induction (data not shown). Moreover, a single dot corresponding to GFP-Spc42p was observed in this strain, indicating that the SPB failed to duplicate (Fig. 7 B). This strongly suggests that HsCen3p interacts with at least another unidentified Cdc31p-binding protein involved in SPB duplication.
Discussion
To study the possible involvement of HsCen3p in centrosome duplication, we performed overexpression of wild-type and mutant forms of HsCen3p in human cultured cells, but did not succeed in disturbing the centrosome duplication cycle in transient transfection experiments. The overexpression of HsCen3p was found to be consistently less efficient than overexpression of HsCen2p, suggesting that either the level of HsCen3p is more tightly regulated than that of HsCen2p in human cells or that the half life of the two proteins are quite different. We observed that inactivation of the fourth EF-hand prevents centriole localization of the protein. However, part of this mutant protein can associate with Triton X-100 insoluble structures. A long-term effect of high dosage of HsCen3p was suggested by the fact that we could not establish stable cell lines expressing HsCen3p, whereas we could easily do it with the two other human centrin genes. We reasoned that a high dosage of HsCen3p could have a deleterious effect only after one or two centrosome duplication cycles, a possibility which was difficult to test in culture cells. Thus, we turned to the Xenopus embryo, which we previously used to study HsCen2p function (Paoletti et al. 1996). In this rapidly dividing system, HsCen2p and HsCen3p led to completely different effects, although undercleavage was observed in both cases: undercleaved blastomeres produced by injection of HsCen2p contained numerous asters, indicating that the centrosome duplication process itself was not prevented. Thus, HsCen2p could disturb either cytokinesis itself or any upstream event, for example, the timing of centrosome duplication or its coupling with cytokinesis. On the contrary, HsCen3p is shown in this work to impair centrosome duplication. As a consequence, cleavage is inhibited. Our results do not exclude, however, that HsCen3p also impairs events downstream of centrosome reproduction, including the cleavage process itself. This could explain that a few uncleaved blastomeres contained more than two microtubule asters. Injection of RNA encoding the mutant HsCen3p-D147,149,151A protein, which is unable to localize to the centrosome, also impairs centrosome duplication, suggesting that HsCen3p acts by interacting with a cytoplasmic XCen3p-binding protein.
We observed that HsCEN3 could not complement S. cerevisiae strains bearing temperature-sensitive mutations in CDC31 or a complete deletion of CDC31. On the contrary, HsCen3p inhibited cell growth and SPB duplication, whereas the two other human centrin proteins were without effect. We have shown that HsCen3p interacts with Kar1p, a protein which is also involved in SPB duplication (Biggins and Rose 1994), whereas the other known Cdc31p-binding protein, Kic1p, is involved in cell wall integrity (Sullivan et al. 1998). We also found that HsCen2p, which did not impair cell growth when overexpressed in yeast, is also able to interact with Kar1p as previously described in vitro (Geier et al. 1996). This suggests that HsCen3p disturbs SPB duplication by titrating other Cdc31p-binding proteins. This was further confirmed: HsCen3p still inhibits SPB duplication in a yeast strain expressing the cdc31-16 allele that can grow in the absence of Kar1p. It is noteworthy that, in the conditions we used to demonstrate the interaction between HsCen3p and Kar1p, we could not detect an interaction between Cdc31p and Kar1p. This result suggests that Kar1p has a higher affinity for HsCen3p and for HsCen2p than for Cdc31p or, more likely, that the functional Cdc31p/Kar1p complex, which is part of the half bridge, is not soluble in the conditions we used. No SPB localization has been detected for HsCen3p, making it possible that HsCen3p sequesters Cdc31p-binding proteins in the cytoplasm. The existence of complexes between HsCen3p and Cdc31p-binding protein(s) is in favor of a functional conservation between HsCen3p and Cdc31p.
Altogether, our data strongly argue in favor of the existence of two functionally distinct centrin families (see Fig. 1): a first one implicated in centrosome duplication, to which Cdc31p and HsCen3p belong; and a second family that participates in other cell division events, such as centrosome segregation or cytokinesis, and which includes centrin from C. reinhardtii and HsCen2p (Paoletti et al. 1996). The single centrin gene of the budding yeast might be able to fulfill both functions. It has been shown that, in addition to SPB duplication, Cdc31p regulates the activity of Kic1p, a kinase involved in cell integrity and in cell separation (Sullivan et al. 1998).
Recently, several studies have shown that cyclin-CDKs are required for driving centrosome duplication in animals. During embryonic cell cycles, cyclin E-CDK2 activity is required for centrosome reproduction (Hinchcliffe et al. 1999; Lacey et al. 1999), whereas cyclin A-CDK2 is also required in somatic cells (Matsumoto et al. 1999; Meraldi et al. 1999). Altogether, these data suggest that the cell cycle machinery indeed regulates centrosome reproduction, coupling it with the cell cycle. However, the centrosomal targets of cyclins-CDK2 are unknown. It is possible that these kinases transcriptionally regulate genes involved in centrosome duplication. Identification of genes coding for centrosomal proteins regulated by cyclin-CDKs will be a crucial step in understanding centrosome duplication regulation.
The intriguing possibility that γ-tubulin, a protein involved in microtubule nucleation and stability, is required for basal body assembly in Paramecium suggests that controlling microtubule dynamics might also regulate centriole/basal body duplication and probably that the regulation of centriolar microtubule assembly shares common steps with cytoplasmic microtubule nucleation (Ruiz et al. 1999). Centrin 3 is the first centriole-associated protein, actually concentrated in the distal lumen of each centriole and in the early procentriole bud, to be shown to participate in the initiation of centrosome duplication in animals. Identification of proteins interacting with Cen3p in a Ca2+-dependent manner should be critical for further study of the regulation of the centrosome duplication. As HsCen3p blocks yeast SPB and frog centrosome duplication most likely by competing with Cdc31p and with Xenopus Cen3p for their physiological targets, the two experimental systems used in this study provide valuable tools to identify new proteins involved in SPB or centrosome duplication.
Acknowledgments
We would like to thank E. Schiebel for providing yeast strains and expression plasmids, J. Kilmartin for the gift of mAb 45D10, K. Ryan for the gift of the pβGFP/RN3P plasmid, C. Celati for the production and the purification of the anti-HsCen3p antibodies, L. Cabanié for the chromatography on superdex 75, T. Ruiz for valuable help in EM, and D. Meur for the art work. We thank B. Goud, G. Keryer, J. Moggs, A.M. Tassin, and F. Tournier for critical reading of the manuscript, A. Echard for sequence comparison, E. Bailly, N. Belgareh, and V. Doye for technical advice with the yeast experiments, and H. Philippe for his expertise in centrin phylogeny.
S. Middendorp received a fellowship from le Ministère de l'Enseignement Supérieur et de la Recherche and from l'Association pour la Recherche sur le Cancer. T. Küntziger received a fellowship from le Ministère de l'Enseignement Supérieur et de la Recherche and from the Grand-Duchy of Luxembourg Ministère de l'Éducation Nationale et de la Formation Professionnelle. This work was supported by Centre National de la Recherche Scientifique, Institut Curie, and by a European Economic Community grant (HCP CHRX CT 94-0642) to M. Bornens.
Footnotes
Sandrine Middendorp's present address is Laboratoire de Cytophysiologie et Toxicologie Cellulaire, Université Paris 7, 2 Place Jussieu, tour 53, 75251 Paris Cedex 05, France.
Abbreviations used in this paper: CEN, centromeric; HsCEN1, Homo sapiens centrin1 (CETN1 in the human genome database); HsCEN2, Homo sapiens centrin2 (CETN2 in the human genome database); HsCEN3, Homo sapiens centrin3 (CETN3 in the human genome database); SPB, spindle pole body.
References
- Adams I.R., Kilmartin J.V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae . J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Furlong C., Byers B. Yeast gene required for spindle pole body duplicationhomology of its product with Ca2+-binding protein. Proc. Natl. Acad. Sci. USA. 1986;83:5512–5536. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Rose M.D. Direct interaction between yeast spindle pole body componentsKar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 1994;125:843–852. doi: 10.1083/jcb.125.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Ivanovska I., Rose M.D. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer C.B., Roth M.G. A single amino acid change in the cytoplasmic domain alters the polarized delivery of influenza virus hemagglutinin. J. Cell Biol. 1991;114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Multiple roles of the spindle pole bodies in life cycle of Saccharomyces cerevisiae In Molecular Genetics in Yeast. D. von Wettstein, J. Friis, M. Kielland-Brandt, and A. Stenderup, editors. Munksgaard, Copenhague. 119–133.
- Conde J., Fink G.R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A.D., Kilmartin J.V. Spc42pa phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V., Wepf R., Hurt E.C. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errabolu R., Sanders M.A., Salisbury J.L. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- Geier B.M., Wiech H., Schiebel E. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J. Biol. Chem. 1996;271:28366–28374. doi: 10.1074/jbc.271.45.28366. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Li C., Thompson E.A., Maller J.L., Sluder G. Requirement of Cdk2-cyclin activity for repeated centrosome reproduction in Xenopus egg extracts. Nature. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Huang B., Mengersen A., Lee V.D. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding proteinhomology in its protein sequence with calmodulin and the yeast CDC31 gene product J. Cell Biol 107 1988. 133 140a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Watterson D.M., Lee V.D., Schibler M.J. Purification and characterization of a basal body-associated Ca2+-binding protein J. Cell Biol 107 1988. 121 131b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens W.C., Izaurralde E., Mattaj I.W. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka M.R., Jarvik J.W. Analysis of flagellar size control using a mutant of Chlamydomonas reinhardtii with a variable number of flagella. J. Cell Biol. 1982;92:170–175. doi: 10.1083/jcb.92.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey K.R., Jackson P.K., Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee V.D., Huang B. Molecular cloning and centrosomal localization of human caltractin. Proc. Natl. Acad. Sci. USA. 1993;90:11039–11043. doi: 10.1073/pnas.90.23.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin O.C., Gunawardane R.N., Iwamatsu A., Zheng Y.X. Xgrip109a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gamma TuRC) assembly and centrosome function. J. Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Hayashi K., Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- McDonald H.B., Byers B. A proteasome cap subunit is required for spindle pole body duplication in yeast. J. Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A.M., Bartek J., Nigg E.A. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2–Cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Middendorp S., Paoletti A., Schiebel E., Bornens M. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA. 1997;94:9141–9146. doi: 10.1073/pnas.94.17.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.M., Urbani L., Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J. Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J.L., Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Fink G.R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Kilmartin J.V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F., Beisson J., Rossier J., DupuisWilliams P. Basal body duplication in Paramecium requires gamma-tubulin. Curr. Biol. 1999;9:43–46. doi: 10.1016/s0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Salisbury J.L., Baron A.T., Sanders M.A. The centrin-based cytoskeleton of Chlamydomonas reinhardtiidistribution in interphase and mitotic cells. J. Cell Biol. 1988;107:635–641. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T., Perriard J.C. Intracompartmental sorting of essential myosin light chainsmolecular dissection and in vivo monitoring by epitope tagging. Cell. 1991;66:277–289. doi: 10.1016/0092-8674(91)90618-9. [DOI] [PubMed] [Google Scholar]
- Spang A., Courtney I., Fackler U., Matzner M., Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Courtney I., Grein K., Matzner M., Schiebel E. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 1995;128:863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D.S., Biggins S., Rose M.D. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 1998;143:751–765. doi: 10.1083/jcb.143.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G.H., Hirata A., Ohya Y., Anraku Y. Mutations in yeast calmodulin cause defect in spindle pole body functions and nuclear integrity. J. Cell Biol. 1992;119:1625–1639. doi: 10.1083/jcb.119.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon B.E., Adler S.A., Suhan J.P., Jarvik J.W. Mutational analysis of centrinan EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas . J. Cell Biol. 1992;119:1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A.M., Celati C., Moudjou M., Bornens M. Characterization of the human homologue of the yeast Spc98p and its association with gamma-tubulin. J. Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Hiller M.A., Scherson T.Y., Rose M.D. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J. Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Ho W., Winey M., Rose M.D. Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae . Genetics. 1994;137:407–422. doi: 10.1093/genetics/137.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer C., Doye V., Grandi P., Nehrbass U., Hurt E.C. A new subclass of nucleoporins that functionally interact with nuclear pore protein NSP1. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS2novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Hoyt M.A., Chan C., Goetsch L., Botstein D., Byers B. NDC1a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R.L., Salisbury J., Jarvik J.W. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 1985;101:1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Pines J., Ryan K., Siemering K.R., Haseloff J., Evans M.J., Gurdon J.B. An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development. 1996;122:3719–3724. doi: 10.1242/dev.122.12.3719. [DOI] [PubMed] [Google Scholar]