Abstract
Classical cadherins form parallel cis-dimers that emanate from a single cell surface. It is thought that the cis-dimeric form is active in cell–cell adhesion, whereas cadherin monomers are likely to be inactive. Currently, cis-dimers have been shown to exist only between cadherins of the same type. Here, we show the specific formation of cis-heterodimers between N- and R-cadherins. E-cadherin cannot participate in these complexes. Cells coexpressing N- and R-cadherins show homophilic adhesion in which these proteins coassociate at cell–cell interfaces. We performed site- directed mutagenesis studies, the results of which support the strand dimer model for cis-dimerization. Furthermore, we show that when N- and R-cadherins are coexpressed in neurons in vitro, the two cadherins colocalize at certain neural synapses, implying biological relevance for these complexes. The present study provides a novel paradigm for cadherin interaction whereby selective cis-heterodimer formation may generate new functional units to mediate cell–cell adhesion.
Keywords: cadherins, cell adhesion, heterophilic binding, synaptic adhesion, dimerization
Introduction
The cadherins are single-pass transmembrane proteins that mediate calcium-dependent cell–cell adhesion (Takeichi 1991; Gumbiner 1996). This superfamily consists of several subfamilies comprising the classical, desmosomal, and the CNR/protocadherins (Sano et al. 1993; Suzuki 1996; Suzuki et al. 1997; Takeichi et al. 1997; Kohmura et al. 1998; Wu and Maniatis 1999). Classical cadherins (for example, N-, E-, and R-) share a common primary structure with five tandemly repeated extracellular domains of ∼110 amino acids each, a transmembrane segment, and a short cytoplasmic domain (Hatta et al. 1988; Matsunami et al. 1993; Shapiro and Colman 1998). The cytoplasmic domain of the classical cadherins interacts with a complex that includes α- and β-catenins, and plakoglobin, all of which function in part to mediate connections between the cadherins and the cytoskeleton (Ozawa et al. 1989, Ozawa et al. 1990; Reynolds et al. 1992; Kemler 1993; Shibamoto et al. 1995; Yap et al. 1998).
The first extracellular domain (EC1) governs the binding specificity of the cadherins. The best evidence for this comes from experiments which show that specificities of cell adhesion between P- and E-cadherins could be switched when their NH2-terminal regions were exchanged (Nose et al. 1990). Furthermore, site-directed mutations in the EC1 domain can significantly alter or abolish adhesion (Tamura et al. 1998).
Crystal structures of the EC1 domain from N-cadherin suggested new ideas about the adhesive interface between cadherins. Two separate interfaces, the strand dimer (which mediates parallel or cis-dimerization) and the adhesion dimer, are thought to contribute to N-cadherin adhesive function (Shapiro et al. 1995). The strand dimer is formed by lateral association between the side chain of tryptophan 2 (Trp-2) and a pocket in the hydrophobic core of the partner molecule. Mutagenesis studies have shown that Trp-2, and also its acceptor pocket, are crucial for adhesion in both N- and E-cadherins (Tamura et al. 1998). The necessity for lateral cis-dimers is supported by experiments with a recombinant C-cadherin extracellular segment (Brieher et al. 1996), showing that only the dimeric form mediates strong adhesion. Furthermore, artificially induced multimerization of a chimeric tailless C-cadherin could strengthen adhesion in cells expressing this molecule (Yap et al. 1997). Thus, it appears likely that cis-dimers are a fundamental unit for cadherin function, at least for the classical cadherins. More recent crystallographic evidence has been presented for both N- and E-cadherins, however, which either shows Trp-2 disordered (Nagar et al. 1996; Tamura et al. 1998) or inserted into a pocket in its own hydrophobic core (Pertz et al. 1999), rather than that of a dimer partner. This evidence does not necessarily dispute the strand dimer model. By analogy, enzymes can have active forms and inactive forms, and it is well known that much of the functional diversity of proteins accrues due to their ability to adopt alternate conformations. Therefore, it seems possible that the strand dimer and other conformations of Trp-2 and other nearby regions could reflect different natural states of cadherins.
In nervous tissue, classical cadherins are critical for formation of brain nuclei (Beesley et al. 1995; Redies and Takeichi 1996; Uchida et al. 1996; Colman 1997; Redies 1997; Arndt et al. 1998; Uemura 1998) and establishment of neuronal connectivity, including synaptic target recognition (Fannon and Colman 1996; Uchida et al. 1996). It is known that dozens of cadherins are expressed in brain (Suzuki et al. 1991, Suzuki et al. 1997; Sano et al. 1993; Suzuki 1996), and at least some are coexpressed in the same neuron (Kohmura et al. 1998; Wohrn et al. 1999). Although N- and R-cadherins are related to each other, the expression patterns of N- and R-cadherins in the central nervous system differ substantially from one another (Redies et al. 1993; Ganzler and Redies 1995; Matsunami and Takeichi 1995; Redies 1997; Wohrn et al. 1998). Partial overlap in expression of N- and R-cadherins in the same cell groups has been demonstrated (Inuzuka et al. 1991; Redies et al. 1993; Redies 1997; Wohrn et al. 1999). Therefore, various interactions between N- and R-cadherins, and perhaps other cadherins, may exist to maintain the integrity of nervous and other tissues.
To investigate the possibility of heterodimerization between different cadherins, we expressed N-, E-, and R-cadherins in L cells and established stable single and double cadherin expressors. We chose to study N- and R-cadherins because regions of N-cadherin expression in the nervous system often abut regions of R-cadherin expression (Inuzuka et al. 1991; Redies et al. 1993; Redies 1995; Wohrn et al. 1998, Wohrn et al. 1999). In contrast, the expression pattern of E-cadherin shows no such correlation (Matsunami and Takeichi 1995; Fannon and Colman 1996). We show here that N- and R-cadherins can form stable cis-heterodimers, whereas E-cadherin cannot dimerize with either of these molecules. Site-directed mutagenesis experiments suggest that N/R cis-dimers are likely to adopt the strand dimer configuration. These heterodimers can interact with appropriate cadherins from opposing cells to mediate functional cell–cell adhesion. The correlated expression, both in time and space, observed for N- and R-cadherins suggests that the functional cis-heterodimers between N- and R-cadherins reported here may reflect their natural interaction in vivo. It is of interest that the formation of parallel dimers allows for cis-heterodimer formation, while maintaining homophilic adhesive interactions across the opposed bilayers.
Materials and Methods
Antibodies
Rat mAbs against the extracellular domain of mouse N-cadherin (MNCD2) and the extracellular domain of mouse R-cadherin (MRCD5) were kindly provided by Prof. M. Takeichi (Kyoto University, Japan; Matsunami and Takeichi 1995). Polyclonal antibodies against the EC1 domain of N-cadherin (anti-NEC1) and the EC5 domain of E-cadherin (anti-EEC5) were generated in rabbit and guinea pig, respectively (Fannon and Colman 1996). Rabbit polyclonal antibody against GFP was purchased from Clontech Laboratories. Mouse mAbs against myc tag and rabbit polyclonal antibodies against α- and β-catenin were purchased from Sigma Chemical Co.
cDNA Constructs
cDNAs encoding full-length N- and E-cadherin were isolated by screening a mouse brain cDNA library. The coding regions were sequenced and were identical to the published N- and E-cadherin sequences (Nagafuchi et al. 1987; Miyatani et al. 1989). R-cadherin cDNA in Bluescript was kindly provided by Prof. M. Takeichi (Matsunami et al. 1993). The N-, E-, and R-cadherin coding regions were each inserted into the pCXN2 expression vector for protein expression in L cells.
For immunoprecipitation, R-cadherin was fused with GFP (green fluorescence protein) at its COOH terminus and N-cadherin with 6 copies of the myc epitope. An AgeI or XhoI site was created by PCR to substitute for the natural R- or N-cadherin stop codon by PCR. The AgeI site was used for ligation of the R-cadherin cDNA with the unique AgeI site of the GFP sequence in the plasmid pEGFP-N1 (Clontech Laboratories); the XhoI site was used for ligation of N-cadherin to the 6× myc tag. Finally, a full-length R-cadherin–GFP or N-cadherin–myc was inserted into the pCXN2 vector. The site-directed mutation in EC1 domain of N-cadherin, Trp-2→Ala (W2A), which causes dissociation of N-cadherin dimer and loss of adhesive activity, was generated as described before (Tamura et al. 1998). A deletion mutation of the N-cadherin EC1 domain was generated by removing the coding region from 710–1,256 by EcoNI and ligated. These mutations did not change the reading frame. To generate a chimeric construct ENEC1, the E-cadherin EC1 domain was substituted with the N-cadherin EC1 domain. For this procedure, a 1.5-Kb fragment of E-cadherin containing signal peptide and mature peptide EC1–EC3 region was cut out and subcloned into XbaI and XhoI site of Bluescript vector. A PCR fragment containing a new Tth111I restriction site in E-cadherin was engineered to match at the appropriate positions of N-cadherin, and ligated to a fragment from the N-cadherin EC1 domain, which was excised by Tth111I, to yield the replacement of N-cadherin EC1 domain in E-cadherin. Finally, the whole chimeric cadherin cDNAs were cloned into the pCXN2 vector. All PCR products and ligation sites were sequenced using the ABI computerized automated DNA sequencing.
Cell Culture and Transfection
L cells were cultured in a humidified atmosphere of 10% CO2 and in 90% DME containing 10% FBS. Neurons were prepared from hippocampi of embryonic day 18 Sprague Dawley rats as described previously (Benson and Tanaka 1998). In brief, neurons were dissociated by treatment with 0.25% trypsin for 15 min at 37°C, followed by triturating through a Pasteur pipette. Neurons were plated at a density of 3,600 cell/cm2 on poly-l-lysine–coated coverslips in MEM containing 10% horse serum. After 4 h, when neurons had attached, coverslips were transferred to dishes containing a monolayer of cortical astroglia, where they were maintained for up to 2 wk in MEM containing N2 supplement, sodium pyruvate (1 mM), and ovalbumin (0.1%). To obtain single or double transfectants, the cells were transfected by using Superfect (Qiagen Inc.), and then cultured in selective medium containing 800 μg/ml of G418 (GIBCO BRL). Colonies were isolated and examined for N-, R-, and E-cadherin expression by immunofluorescence and positive cells were expanded and used for further studies.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde, delipidated in 100% methanol, permeabilized with 0.1% Triton X-100, and blocked with 5% normal goat serum in PBS. After incubation for 1 h at 37°C with primary antibodies, cells were then incubated with fluorescent-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) at room temperature for 30 min. Coverslips were then mounted and examined by confocal laser microscopy.
Aggregation Assays
Monolayer cultures were treated with 0.01% trypsin in HCMF (Hepes-buffered Ca2+- and Mg2+-free Hanks' Solution) supplemented with 1 mM CaCl2 for 30 min at 37°C. The trypsinized cells were washed gently in HCMF containing calcium and 1% BSA at 4°C. This procedure can dissociate cell layers into single cells, leaving cadherins intact on the cell surface. After the cells were thoroughly dissociated, 5 × 105 cells per well were transferred to 24-well dishes for a final volume of 0.5 ml HCMF containing 1% BSA with or without 1 mM Ca2+. The plates were rotated at 80 rpm at 37°C for 45 min.
For mixed aggregation analysis, cells expressing different cadherin constructs were labeled with different lipophilic dyes before mixing. We used DiI (1,1-dioctadecyl-3,3,3,3-tetramethylindocarbocyanine) and DiO (3,3-dioctadecyloxacarbocyanine perchlorate), each purchased from Molecular Probes. Stock solutions of DiI were made by dissolving 2.5 mg DiI in 1 ml of 100% ethanol, and stocks of DiO were made by dissolving 2.5 mg DiO in 1 ml of 90% ethanol, 10% dimethylsulfoxide. These stock solutions were sonicated and filtered before use. To label cells with these dyes, they were incubated for 8 h at 37°C in serum-containing DME at final concentrations of 15 μg/ml and 10 μg/ml for DiI and DiO, respectively. The cells were washed extensively with HCMF containing calcium to prevent cross contamination of the dyes. After single cell suspensions were obtained as described above, 2.5 × 105 cells per well of each of two types were transferred to a 24-well dish. After rotating at 80 rpm at 37°C for 45 min, 50 μl of the fixed aggregates were removed, placed on a slide, and covered with a coverslip and examined by confocal microscopy.
Immunoblotting and Immunoprecipitation
For immunoblotting and immunoprecipitation experiments, 5 × 106 cells were cultured in 100-mm tissue culture dishes at 37°C for 3 d. In coculture experiments, 5 × 106 cells expressing N-cadherin wild-type and R-cadherin–GFP mixed in a ratio 1:1 were cultured in a 100-mm dish for 24 h. The confluent monolayer was then washed with PBS, scraped off the dish, pelleted for 5 min at 1,000 rpm and extracted in 1.0 ml of lysis buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1% NP-40, 1% Triton X-100, Ca2+ free leupeptin 0.5 μg/ml, pefabloc 0.1 mM, aprotinin 1 μg/ml) for 30 min. After cell lysis, the samples were centrifuged for 20 min at 15,000 rpm and the supernatants were separated. Protein concentration was determined with the BCA protein assay (Pierce Chemical Co.) and samples were run on 6% SDS-PAGE, transferred to nitrocellulose, membrane-blocked with 5% milk protein, and incubated overnight with primary antibodies. After secondary antibody incubation and routine washing, blots were developed with the chemiluminescence system (New England Nuclear Life Science Products). For immunoprecipitation, the supernatants were incubated with specific antibody (3 μl/sample) for 1 h at 4°C. The antigen–IgG complexes were precipitated by sequential incubation with protein-A–Sepharose 4B. The immunoprecipitates were washed extensively, and boiled with SDS-sample buffer, and the proteins were detected by immunoblotting with specific antibodies.
For N-cadherin or GFP immunodepletion experiments, 1 ml cell lysates were incubated with 20 μl of N-cadherin or 10 μl GFP antibodies. The antibodies with complexes were then removed by anti-rabbit IgG-agarose (Sigma Chemical Co.). 30 μl of each protein sample from the homogenates was subjected to SDS-PAGE and the separated proteins were electrophoretically transferred to a nitrocellulose membrane sheet. The membrane was processed to detect protein with specific antibodies.
Results
R- and N-Cadherins Form Heterophilic Cell–cell Interfaces
To evaluate and compare the adhesive properties of N-, R-, and E-cadherins, we generated stable L cell lines expressing these cadherins (LR, LN, and LE). Aggregation assays were performed to verify the adhesive properties of these cell lines. Stable transfectants of each cadherin showed rapid calcium-dependent formation of large aggregates. To test the binding specificity, we performed mixed aggregation assays with DiI (red) or DiO (green)-labeled cell lines. When LR cells were mixed with LN cells, heterotypic aggregates were produced, although the individual cell types still remained segregated (Fig. 1 a). In contrast, mixed assays of LE cells with either LN or LR cells showed that LE cell aggregates remained entirely segregated from LR or LN cell aggregates (Fig. 1b and Fig. c). These results were consistent with previous reports (Matsunami et al. 1993) and suggest that the heterophilic interaction of R- and N-cadherins may be weaker than their respective homophilic interactions. Similarly, this interaction also can be shown in LR and LN cell cocultures by immunostaining using N-cadherin antibodies and R-cadherin–GFP (Fig. 1, d–f). Both types of cadherins were present at the junctions between the two subpopulations, even when the two cell types were mixed randomly before plating on the dish. Complexes of R- and N-cadherins, however, could not be detected by coimmunoprecipitation using GFP antibody (see Fig. 3 A). These results indicate that N- and R-cadherins can form heteroadhesive clusters between adjacent cells, but these trans-complexes are too weak to maintain under condition used for routine immunoprecipitation.
Figure 1.
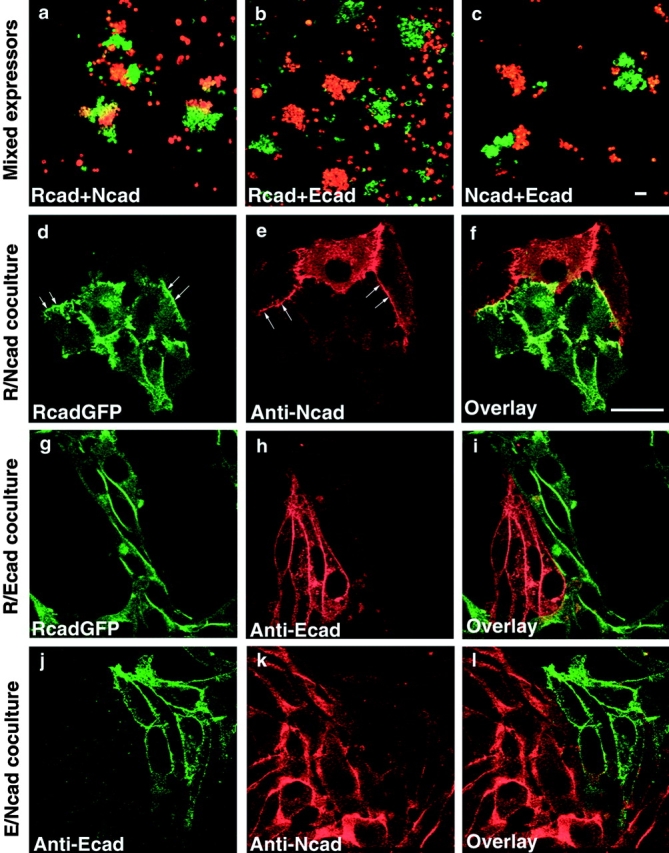
Cell aggregation assays and immunofluorescence microscopy of LR, LN, and LE cells in mixed cultures. LR cells were labeled with DiO (green in a and b); LN cells with DiI (red in a) or DiO (c); and LE cells with DiI (red in b and c). Mixed aggregation assays showed weak coaggregation between LR cells and LN cells, but the cells still preferentially sorted out and clustered in homotypic groups (a). LE cells did not coaggregate with LR (b) or LN cells (c). N- and R-cadherin proteins formed heteroadhesive clusters located at the same areas of cell–cell contacts (arrows) in LR and LN coculture (d–f), but heteroadhesive clusters did not establish at areas of cell–cell contacts between either LR and LE (g–i) or LE and LN cocultures (k and l). Bars, 25 μm.
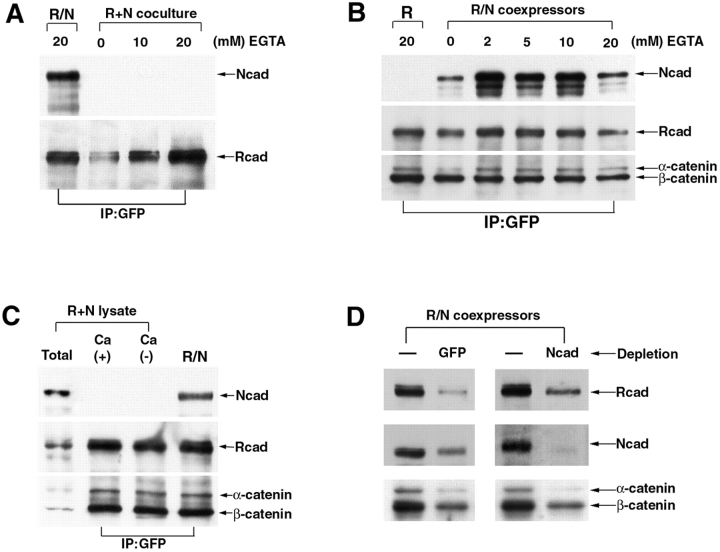
Figure 3.
The strand heterodimer of R-/N-cadherin is resistant to EGTA treatment and forms directly in coexpressing cells. A, LR/N cells and cocultured cells of LN and LR were treated by EGTA (∼20 mM) for 10 min before lysis, immunoprecipitated with GFP antibody (IP: GFP), and immunoblotted. In this coculture system, the heterophilic adhesive complexes were undetectable. B, In contrast, complexes of lateral heterodimers from LR/N cells were detected in any concentration of EGTA treatment. C, Lysates from LN and LR cells were mixed together, incubated in IP buffer with or without calcium for 1 h, immunoprecipitated with GFP antibody, and then detected by immunoblotting. The LR and LN cell lysates were loaded (Total lane) before immunoprecipitation. D, The proteins from LR/N cells were depleted with GFP or NEC1 antibodies, and immunoprecipitated by anti-rabbit IgG-agarose. The amount of R- and N-cadherin in the lysate before (−) or after depletion was detected by R- and N-cadherin antibodies.
Cocultures of E- and R-cadherin expressors, and E- and N-cadherin expressors, were also examined (Fig. 1, g–l). Immunolocalization of the respective cadherins showed that N- and R-cadherins each formed cell junctions that were clearly separate from E-cadherin–mediated junctions. This is in contrast to N/R cocultures which clearly show regions of heterotypic adhesion between N- and R-cadherins. This leads us to suggest that the differential adhesive properties of these molecules are important factors in controlling their localization at cell junctions.
R- and N-Cadherins Form Stable Heterodimers in Coexpressing Cell Lines
We investigated whether different cadherins could form cis-heterodimers by using cell lines coexpressing R- and N-cadherins or R- and E-cadherins (LR/N, LR/E; Fig. 2 A). We used an R-cadherin fused with GFP to enable detection in coimmunoprecipitation experiments. Total cell lysates from LN, LR, and LR/N were immunoprecipitated with GFP antibody and detected by Western blotting with N-, R-, or E-cadherin antibodies. The R-cadherin–GFP fusion protein was able to coprecipitate with N-cadherin from the LR/N cell lysate (Fig. 2 B). In contrast, E-cadherin was not detected in immunoprecipitates from LR/E cell lines (Fig. 2 C). These two proteins, however, accumulate at cell membranes (Fig. 2 A). These data indicate the absence of interaction between R- and E-cadherins. In both LR/N and LR/E cell lines, cadherins were associated with α- and β-catenin and these proteins could be precipitated together with GFP antibody, suggesting that catenins do not control the heterointeractions between these cadherins (Fig. 2B and Fig. C).
Figure 2.
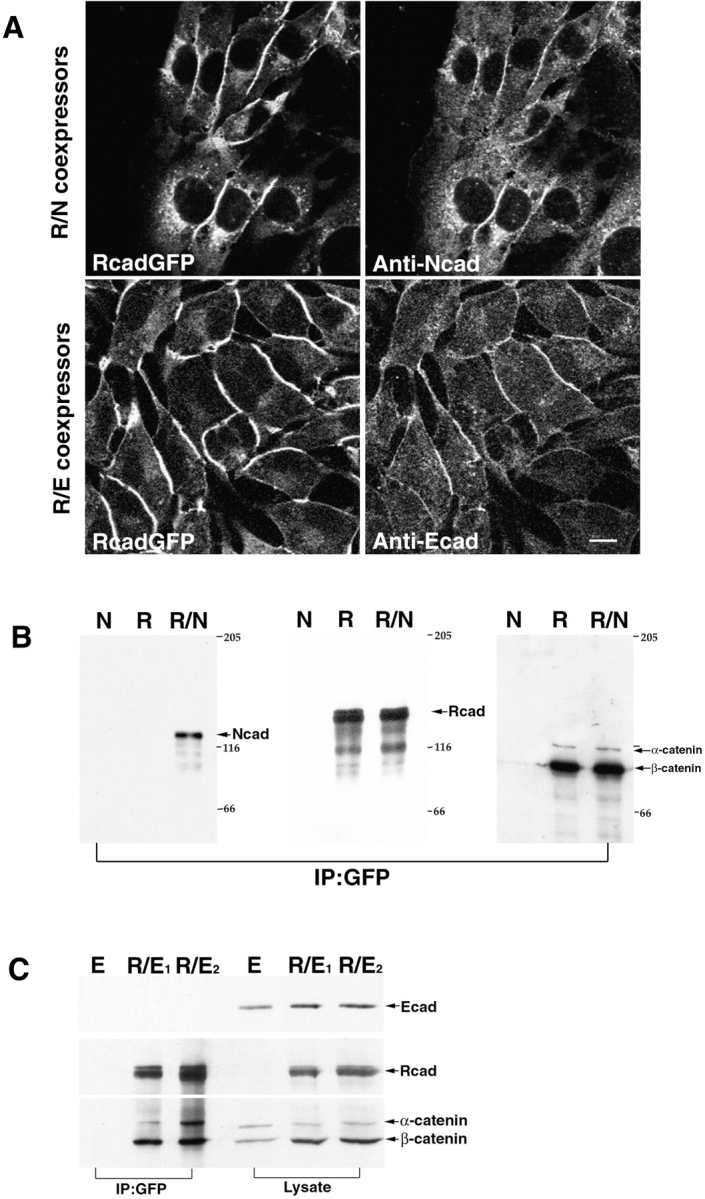
R-/N-cadherin, when coexpressed in L cells, forms heterocomplexes. A, Immunostaining of double transfectants showed that different cadherins were coexpressed in L cells. Bar, 10 μm. B, Lysates from LR, LN, and LR/N cells were immunoprecipitated with GFP antibody (IP: GFP) and detected by N- and R-cadherin and α- and β-catenin antibodies. N-cadherin was found to be coprecipitated with R-cadherin in LR/N cells. C, Lysates or immunoprecipitates from LE, LR/E1, and LR/E2 cell lines were detected by E- and R-cadherin and α- and β-catenin antibodies.
Heterophilic interaction in a calcium-dependent manner has been shown between LR and LN cells, both in our experiments and in those previously reported (Matsunami et al. 1993). Therefore, we used a calcium chelator (EGTA) to disrupt the adhesive interaction, so that we could determine whether the R-/N-cadherin complexes obtained by immunoprecipitation originated from the same or opposing cells. We made lysates from LN and LR cocultures treated with or without EGTA and performed immunoprecipitations with GFP antibody. R-cadherin–GFP did not coprecipitate N-cadherin in mixed monoexpressing LN and LR cocultures, either in the presence or absence of EGTA (Fig. 3 A). These results imply that the trans-heterophilic adhesive complex between R- and N-cadherin may be considerably weaker than the respective homophilic counterparts and undetectable in our assay system. In contrast, when immunoprecipitations were performed on lysates from the double-transfectant, LR/N, in the presence of EGTA (∼20 mM), R-cadherin–GFP coprecipitated N-cadherin equally as well as in the presence of Ca2+ (Fig. 3 B). Taken together, our data indicate that lateral dimerization accounts exclusively for the interaction between R- and N-cadherins observed in the coprecipitation experiments, and that the maintenance of this interaction is independent of Ca2+. Similar data have shown that lateral dimerization of E-cadherin is also independent of Ca2+ (Chitaev and Troyanovsky 1998).
We took great care to ensure that lateral dimerization between R- and N-cadherins could form only in cotransfected cells, and was not an artifact created in the lysate when the two proteins were initially mixed together. The lysates from LN and LR were pooled, incubated with or without Ca2+ for 1 h, and subjected to immunoprecipitation with GFP antibody. R-/N-cadherin complexes were detected only in cells in which these proteins were coexpressed and not when individual LR and LN lysates were mixed (Fig. 3 C). Furthermore, when the LR/N cell lysate was depleted with GFP and NEC1 antibodies, although the protein amount of R- and N-cadherin was reduced, both cadherins were still detectable (Fig. 3 D). Thus, cells stably transfected with R- and N-cadherin probably contain three association forms: R-/N-, N-/N-, and R-/R-cadherin dimers. The existence of monomeric forms also cannot be ruled out.
Tryptophan 2 of N-Cadherin Is Essential for Heterodimerization of R- and N-Cadherins
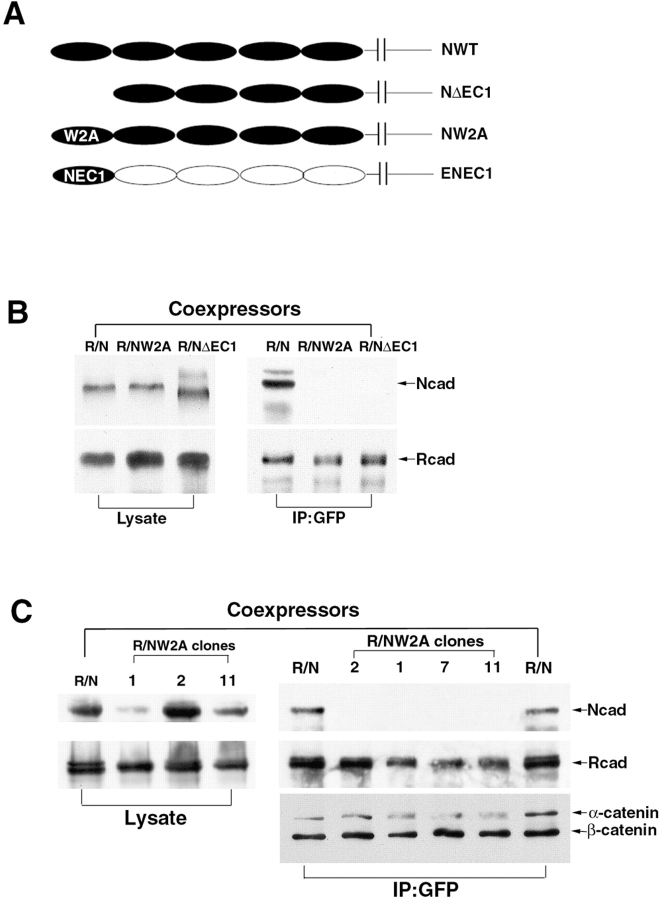
Previous studies demonstrated that Trp-2 in the N-cadherin EC1 domain is critical for strand dimer formation (Shapiro et al. 1995; Shapiro and Colman 1998; Tamura et al. 1998). To evaluate whether this model is relevant to R-/N-cadherin heterodimer formation, several constructs were produced. These included: N-cadherin lacking the EC1 domain (ΝΔEC1); Trp-2 in N-cadherin mutated to Ala (NW2A); and a chimeric molecule of E-cadherin whose EC1 domain was replaced by the N-cadherin EC1 domain (ENEC1; Fig. 4 A). R-cadherin–GFP and mutated N-cadherin were cotransfected into L cells to generate coexpressing cell lines, LR/NΔEC1 and LR/NW2A, and immunoprecipitations were performed using GFP antibody. Either deletion of the EC1 domain or the NW2A mutation completely abolished the coimmunoprecipitation of N-cadherin with R-cadherin (Fig. 4B and Fig. C). Therefore, the EC1 domain, and particularly Trp-2, is required for R-/N-cadherin heterodimer formation.
Figure 4.
Schematic representation of N-cadherin wild-type and mutated constructs and coimmunoprecipitation experiments. A, The constructs of wild-type N-cadherin (NWT), EC1 domain deletion (NΔEC1), Trp-2 mutated to Ala in N-cadherin (NW2A), and a chimeric molecule of E-cadherin whose EC1 domain was replaced by the N-cadherin EC1 domain (ENEC1). B, Western blot analysis of lysates and immunoprecipitates from LR/N, LR/NΔEC1, and LR/NW2A cells. C, Four LR/NW2A clones expressing different levels of R- and N-cadherin (NW2A) were detected, and then immunoprecipitated by GFP antibody and analyzed by immunoblotting.
The Specificity of Heterodimerization Is Localized to the NH2-terminal Domain
Switching the cell-to-cell adhesive specificity of cadherins by EC1 domain exchange was demonstrated previously (Nose et al. 1990). To test whether the specificity of cis-dimer formation might also be encoded in the EC1 domain, we engineered a chimeric protein, ENEC1, formed by replacing the EC1 domain of E-cadherin by the EC1 domain of N-cadherin. We tested the adhesive and lateral interaction properties of this chimeric molecule in L cells (LENEC1). Immunocytochemical analyses showed that the chimeric cadherin protein was expressed on the cell surface and was concentrated at cell–cell boundaries (Fig. 5 A). LENEC1 cell lines exhibited strong self-aggregation. These cells also formed aggregates with LN cells, but showed far weaker aggregation with LE or LR cells (Fig. 5 B), indicating that ENEC1 was a functional artificial cadherin with adhesive specificity appropriate for the N-cadherin type. To determine whether ENEC1 interacted with R-cadherin, we established LR/ENEC1 coexpressors by cotransfecting R-cadherin–GFP and ENEC1 into L cells. The R-cadherin and ENEC1-chimeric proteins accumulate at sites of cell–cell contacts by immunostaining with E-cadherin antibody and R-cadherin–GFP fluorescence (Fig. 5 A). Using LR/N and LR/E cell lines as positive and negative controls, LR/ENEC1 cell lysates were treated with GFP antibody and the immunoprecipitates were probed with EEC5, NEC1, and RCD5 antibodies. These experiments (Fig. 5 C) showed that ENEC1 could be precipitated together with R-cadherin–GFP, suggesting that heterodimers were formed between R-cadherin and ENEC1. This suggests that the EC1 domain of classic cadherins plays an important role in determining the specificity of cis-heterodimer formation.
Figure 5.
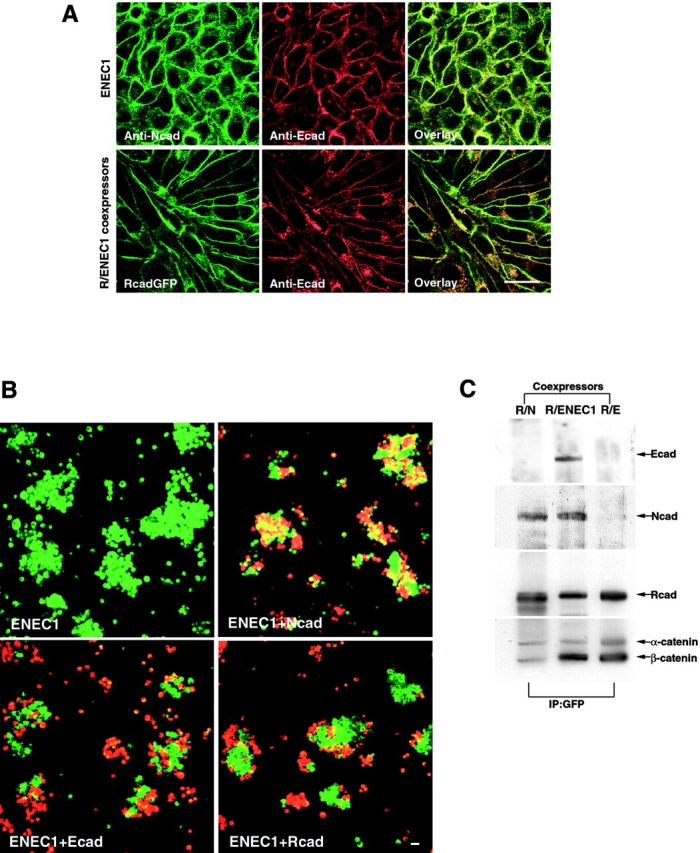
Binding specificity of ENEC1 and heterointeraction between R-cadherin and ENEC1. (A) LENEC1 or LR/ENEC1 was immunostained by NEC1 and EEC5 antibodies, showing that chimeric E-cadherin and R-cadherin colocalized at cell–cell contacts. B, LENEC1 cells coaggregated with LN cells, but aggregated to a far lesser extent with LE and LR cells. LENEC1 was labeled with DiO (green), LN, LE and LR cells with DiI (red). C, Immunoprecipitation of LR/N, LR/ENEC1, and LR/E cell lines with GFP antibody. Precipitates were detected with anti-EEC5, N-cadherin, R-cadherin, and α- and β-catenin, showing that ENEC1-cadherin coprecipitated with R-cadherin. Bars, 25 μm.
The R/N Heterodimer Constitutes a Functional Unit for Cell–Cell Adhesion
Can coexpressing LR/N cell lines mediate functional cell adhesion? And, if so, are R/N heterodimers involved in this process? To answer these questions, we first performed aggregation assays between different cadherin expressing cell lines. LR/N cells aggregated strongly with their own type and exhibited high affinity with both LN and LR cell lines, but segregated from LE cells (Fig. 6 A), suggesting that LR/N cell lines hold broad-range adhesive specificity. Second, we generated another cell line, LNmyc, which is a chimera of N-cadherin fused with a myc epitope tag, and cocultured these cells with LR/N, and LR/NW2A cells. The cell lysates were immunoprecipitated with either myc or GFP antibody. These experiments (Fig. 6 B) showed that R-/N-cadherin heterodimers could be coprecipitated with N-cadherin–myc in the LR/N and LNmyc cocultures, but not in the LR/NW2A and LNmyc coculture system, implying that the R-/N-cadherin heterodimer from one cell was functionally associated with N-cadherin from the opposing cell. This is notably different from the observation that R-cadherin–GFP cannot precipitate N-cadherin from an opposing cell, suggesting that the N/N association is stronger than the N/R association (see Fig. 8 C). We have performed experiments which showed that the W2A mutant of N-cadherin is a simple loss of function mutant, with no discernable dominant-negative effects (Fig. 6 A, e–h). This indicates that cells cotransfected with an adhesive cadherin and the NW2A mutant cadherin exhibit adhesive behavior identical to that of the adhesive active cadherin.
Figure 6.
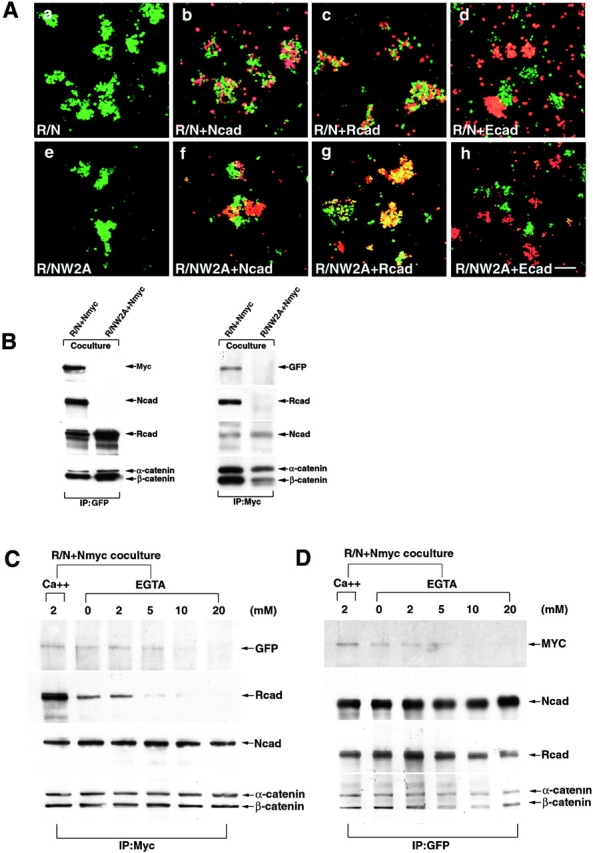
Analysis of LR/N cells adhesion function and adhesive complexes between LR/N and LNmyc cells. A, LR/N cells labeled with DiO (green) aggregated with their own type (a) and coaggregated with either LN (b) or LR (c) cells labeled with DiI (red), but segregated with LE (d) cells labeled with DiI; e–h show that LR/NW2A cells cotransfected with R-cadherin and NW2A mutant cadherin exhibited adhesive behavior identical to that of single R-cadherin transfectant. Bar, 100 μm. B, Lysates from coculture cells of LR/N and LNmyc or LR/NW2A and LNmyc were immunoprecipitated with either GFP or myc antibody, and detected with anti-myc, GFP, N- and R-cadherin, and α- and β-catenin sera. C and D, Cocultures of LR/N and LNmyc cells were treated with EGTA before lysis, the lysates were immunoprecipitated by myc or GFP antibodies, and detected with anti-myc, GFP, N- and R-cadherin, and α- and β-catenin sera.
Figure 8.
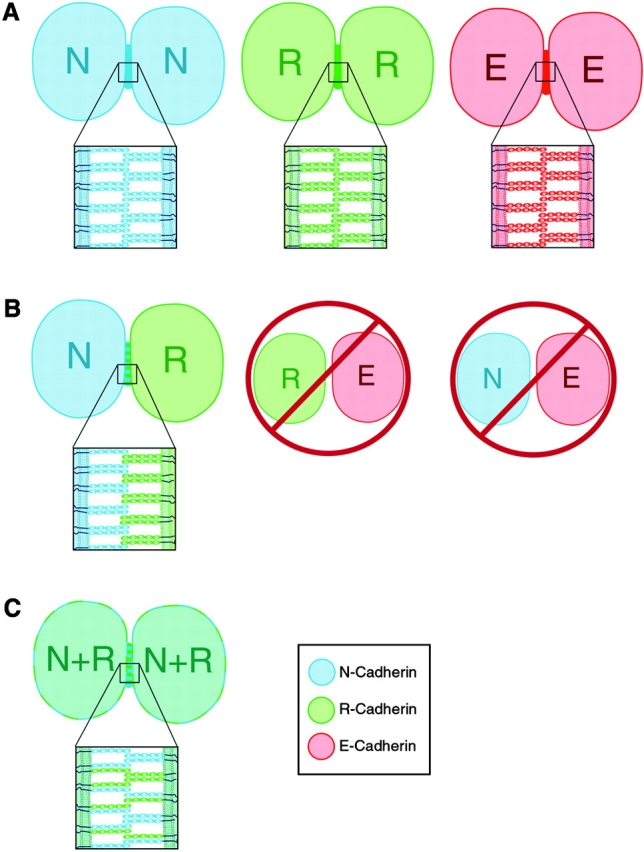
Schematic representation of the potential interactions between cadherin molecules at adjoining cell surfaces. A, Parallel dimers and homophilic adhesion exist between cadherins in cells expressing the same cadherin molecules. B, Parallel dimers and heterophilic binding between R- and N-cadherin molecules can form, but cannot form between R- and E-cadherin or N- and E-cadherin. C, Parallel heterodimers and homophilic interactions form between cadherins in R-/N-cadherin coexpressing cells.
To investigate whether the association between N-cadherin–myc and R-/N-cadherin heterodimers depend on cell–cell contacts, the cocultured cells were incubated for 10 min with different concentrations of EGTA before lysis. Here, data showed that N-cadherin–myc interacted with R-/N-cadherin heterodimers in a calcium-dependent manner. Dimers between cells dissociated from each other in high concentrations of EGTA (Fig. 6C and Fig. D), whereas R-/N-cadherin cis-heterodimers were stable to EGTA treatment (Fig. 6 D). These data reveal that sensitivity to EGTA treatment is different between cis-heterodimers and the cell-to-cell adhesion dimers.
Overexpressed R- and N-Cadherins Colocalize at Synaptic Junctions
Overlapping expression of R- and N-cadherin in the same neuron or fiber fascicle has been observed in the CNS (Redies 1997; Wohrn et al. 1999). Whether both cadherins can be present at the same synaptic junction is still unclear. We coexpressed R-cadherin–GFP and N-cadherin–myc in cultured hippocampal neurons. After a 72-h cotransfection, the neurons were fixed and stained using epitope tag antibodies. The results showed that R- and N-cadherins colocalize at the same synaptic junctions (Fig. 7, A–D, a–d). This colocalization suggests that the associations we observed between R- and N-cadherins in L cells may reflect similar interactions in neurons, where they may naturally be found together.
Figure 7.
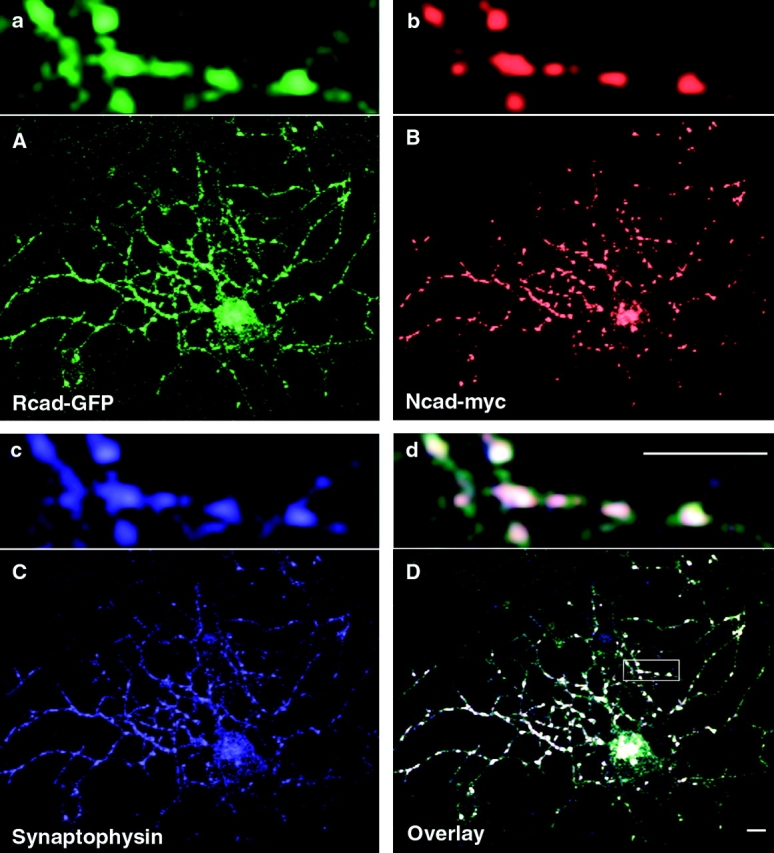
The distribution of R- and N-cadherins after expression in hippocampal neurons. Cell surface labeling of R-cadherin–GFP (A and a), N-cadherin–myc (B and b), and synaptophysin (C and c) was present at synaptic sites. Localization of R- and N-cadherins and synaptophysin is shown in D and d. Bar, 5 μm.
Discussion
The most important findings presented here are: 1, in cotransfected cell lines, R- and N-cadherins form stable cis-heterodimers; 2, Trp-2 in the EC1 domain of N-cadherin is required for heterodimer formation, suggesting association in a strand dimer conformation; 3, the heterodimer is a functional unit for mediating cell–cell adhesion; and 4, the cis-heterodimer and the adhesive dimer exhibit different requirements for Ca2+, implying that different interfaces mediate these interactions. These observations suggest that selective formation of strand heterodimers between different cadherins may play an important role in specific cell adhesion, and present the possibility that cis-heterodimers may be functionally relevant in vivo. While the data presented here does not prove that the oligomers identified by immunoprecipitation correspond to dimers, the circumstantial case is quite strong. The ability to abrogate these oligomers by specific mutation of Trp-2, the central structural residue in the crystallographically observed strand dimer, is strong supportive evidence.
In addition to the data presented here and the crystallographic data, dimeric interaction near the NH2-terminal domain of E-cadherin was also observed by EM (Tomschy et al. 1996). These studies showed that the adhesive interaction between NH2-terminal domains occurs by a two-step mechanism: antiparallel pairs from different molecules form a complex only after parallel pair formation. X-ray crystallographic studies of the NH2-terminal of the E-cadherin 1 and 2 domains suggested a different parallel dimer (Nagar et al. 1996). Experiments with a recombinant C-cadherin extracellular segment (CEC1-5) also revealed the existence of cis-dimeric interactions. Dimers were observed by chemical cross-linking, both in the presence and absence of calcium, and it was also shown that the CEC1-5 dimers had greater homophilic binding activity than monomers (Brieher et al. 1996). Consistent with this, a tailless E-cadherin can be cross-linked as a functional dimer on the cell surface (Ozawa and Kemler 1998). Chitaev and Troyanovsky 1998 showed that E-cadherin formed stable lateral dimers in epithelial junctions. Our present results provide the first evidence for parallel heterodimer formation between R- and N-cadherin, and suggest that heterodimer formation is specific to cadherin type. Furthermore, since R-cadherin makes heterodimers with a recombinant E-cadherin molecule whose EC1 domain was replaced by the N-cadherin EC1 domain, we conclude that the EC1 domain contributes to the recognition of parallel partner molecules, and that it controls the specificity of heterodimer formation.
In the current work, we show that mutating Trp-2 in N-cadherin can destroy heterodimer formation between R- and N-cadherin. For E-cadherin, the double mutation (Trp-2→Ala/Val-3→Gly) within the EC1 domain completely abolished dimerization, suggesting that dimer formation in E-cadherin was also driven by Trp-2 and hydrophobic core interaction (Chitaev and Troyanovsky 1998), as is the case for N-cadherin homodimers. This region may therefore be critical for both homo- and heterodimerization. It is difficult to predict the structural determinants of cis-dimer specificity based on the one existing crystal structure. While it appears that Trp-2, which is entirely conserved throughout the classical cadherins, is the central residue in the interface, other regions may function in determining specificity. We have not attempted to model these interactions due to the highly speculative nature of such studies. We do note, however, that the EC1 domain of N- and R-cadherins are far more similar (73% identity) than that of N- and E-cadherins (61%) or R- and E-cadherins (54%).
Our investigation raises the question of whether cadherin cis-heterodimers play a natural functional role in cell adhesion. We have shown that LR/N cells have broad-range adhesive specificity and high affinity with both LR or LN cells. The R-/N-cadherin heterodimer forms adhesive complexes with N-cadherin from opposing cells, revealing that the heterodimer is a functional cell–cell adhesive unit. Furthermore, N- and R-cadherins colocalize at neuronal synapses on the same cell surface, suggesting the likelihood that they naturally form heterodimers. Although the exact nature of cadherin adhesive contact is still unknown, strand dimer formation seems to be a critical step for subsequent adhesive dimer formation, and potentially for the formation of zipper-like adhesive structures proposed previously (Shapiro et al. 1995). Homophilic and heterophilic binding (Fig. 8, A–C) were thought to mediate strong and weak adhesion for different processes in central nervous system development, such as neurite outgrowth and synaptic junction formation (Colman 1997; Tanaka et al. 2000). Cis-heterodimeric interactions (Fig. 8 C) between different cadherins may be a biologically relevant mechanism in cell–cell adhesion, which could play an important role in controlling different states of adhesiveness between cells.
It is generally believed that the generation of adhesive specificities depends on amino acid differences in EC1 of a classical cadherin. The conservation of the Trp-2, as well as those residues forming the hydrophobic pocket that it inserts into, suggests the possibility that heterodimer formation might be feasible in those cells where more than one cadherin is expressed. Heterodimer formation between cadherins expressed by the same cell could open up interesting evolutionary possibilities for the testing of new adhesive specificities through gene duplication and natural selection, while at the same time preserving one member of the dimer pair in an unaltered functional homophilic adhesive bond. Furthermore, the possibility of titration of adhesive strength using different combinations of strong and weak cadherins would provide a way for cells to exploit a wide spectrum of adhesive responses, and even radically change their intercellular associations and affinities.
Acknowledgments
We thank Dr. M. Takeichi for providing R-cadherin cDNA and antibodies, and Jill Gregory for preparing the figures for this manuscript.
This work was supported by grants NS 20147 from the National Institute of Neurological Disorders and Stroke (National Institutes of Health) and from the National Multiple Sclerosis Society to D.R. Colman, and an Irma T. Hirschl Career Scientist Award to L. Shapiro.
Footnotes
Abbreviations used in this paper: DiI, 1,1-dioctadecyl-3,3,3,3-tetramethylindocarbocyanine; DiO, 3,3-dioctadecyloxacarbocyanine perchlorate; EC1, first extracellular domain; GFP, green fluorescence protein; Trp-2, tryptophan 2.
References
- Arndt K., Nakagawa S., Takeichi M., Redies C. Cadherin-defined segments and parasagittal cell ribbons in the developing chicken cerebellum. Mol. Cell. Neurosci. 1998;10:211–228. doi: 10.1006/mcne.1998.0665. [DOI] [PubMed] [Google Scholar]
- Beesley P.W., Mummery R., Tibaldi J. N-cadherin is a major glycoprotein component of isolated rat forebrain postsynaptic densities. J. Neurochem. 1995;64:2288–2294. doi: 10.1046/j.1471-4159.1995.64052288.x. [DOI] [PubMed] [Google Scholar]
- Benson D.L., Tanaka H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J. Neurosci. 1998;18:6892–6904. doi: 10.1523/JNEUROSCI.18-17-06892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher W.M., Yap A.S., Gumbiner B.M. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J. Cell Biol. 1996;135:487–496. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaev N.A., Troyanovsky S.M. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J. Cell Biol. 1998;142:837–846. doi: 10.1083/jcb.142.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman D.R. Neurites, synapses, and cadherins reconciled. Mol. Cell. Neurosci. 1997;10:1–6. doi: 10.1006/mcne.1997.0648. [DOI] [PubMed] [Google Scholar]
- Fannon A.M., Colman D.R. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- Ganzler S.I., Redies C. R-cadherin expression during nucleus formation in chicken forebrain neuromeres. J. Neurosci. 1995;15:4157–4172. doi: 10.1523/JNEUROSCI.15-06-04157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M. Cell adhesionthe molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hatta K., Nose A., Nagafuchi A., Takeichi M. Cloning and expression of cDNA encoding a neural calcium-dependent cell adhesion moleculeits identity in the cadherin gene family. J. Cell Biol. 1988;106:873–881. doi: 10.1083/jcb.106.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Miyatani S., Takeichi M. R-cadherina novel Ca(2+)-dependent cell–cell adhesion molecule expressed in the retina. Neuron. 1991;7:69–79. doi: 10.1016/0896-6273(91)90075-b. [DOI] [PubMed] [Google Scholar]
- Kemler R. From cadherins to cateninscytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- Kohmura N., Senzaki K., Hamada S., Kai N., Yasuda R., Watanabe M., Ishii H., Yasuda M., Mishina M., Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Matsunami H., Miyatani S., Inoue T., Copeland N.G., Gilbert D.J., Jenkins N.A., Takeichi M. Cell binding specificity of mouse R-cadherin and chromosomal mapping of the gene. J. Cell. Sci. 1993;106:401–409. doi: 10.1242/jcs.106.1.401. [DOI] [PubMed] [Google Scholar]
- Matsunami H., Takeichi M. Fetal brain subdivisions defined by R- and E-cadherin expressionsevidence for the role of cadherin activity in region-specific, cell–cell adhesion. Dev. Biol. 1995;172:466–478. doi: 10.1006/dbio.1995.8029. [DOI] [PubMed] [Google Scholar]
- Miyatani S., Shimamura K., Hatta M., Nagafuchi A., Nose A., Matsunaga M., Hatta K., Takeichi M. Neural cadherinrole in selective cell–cell adhesion. Science. 1989;245:631–635. doi: 10.1126/science.2762814. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A., Shirayoshi Y., Okazaki K., Yasuda K., Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature. 1987;329:341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- Nagar B., Overduin M., Ikura M., Rini J.M. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Nose A., Tsuji K., Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell. 1990;61:147–155. doi: 10.1016/0092-8674(90)90222-z. [DOI] [PubMed] [Google Scholar]
- Ozawa M., Kemler R. The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerization and negatively regulates adhesion activity. J. Cell Biol. 1998;142:1605–1613. doi: 10.1083/jcb.142.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Baribault H., Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO (Eur. Mol. Biol. Organ.) J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Ringwald M., Kemler R. Uvomorulin–catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc. Natl. Acad. Sci. USA. 1990;87:4246–4250. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O., Bozic D., Koch A.W., Fauser C., Brancaccio A., Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C. Cadherin expression in the developing vertebrate CNSfrom neuromeres to brain nuclei and neural circuits. Exp. Cell Res. 1995;220:243–256. doi: 10.1006/excr.1995.1313. [DOI] [PubMed] [Google Scholar]
- Redies C. Cadherins and the formation of neural circuitry in the vertebrate CNS. Cell Tissue Res. 1997;290:405–413. doi: 10.1007/s004410050947. [DOI] [PubMed] [Google Scholar]
- Redies C., Takeichi M. Cadherins in the developing central nervous systeman adhesive code for segmental and functional subdivisions. Dev. Biol. 1996;180:413–423. doi: 10.1006/dbio.1996.0315. [DOI] [PubMed] [Google Scholar]
- Redies C., Engelhart K., Takeichi M. Differential expression of N- and R-cadherin in functional neuronal systems and other structures of the developing chicken brain. J. Comp. Neurol. 1993;333:398–416. doi: 10.1002/cne.903330307. [DOI] [PubMed] [Google Scholar]
- Reynolds A.B., Herbert L., Cleveland J.L., Berg S.T., Gaut J.R. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 1992;7:2439–2445. [PubMed] [Google Scholar]
- Sano K., Tanihara H., Heimark R.L., Obata S., Davidson M., St T., John S., Taketani, Suzuki S. Protocadherinsa large family of cadherin-related molecules in central nervous system. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Colman D.R. Structural biology of cadherins in the nervous system. Curr. Opin. Neurobiol. 1998;8:593–599. doi: 10.1016/s0959-4388(98)80086-x. [DOI] [PubMed] [Google Scholar]
- Shapiro L., Fannon A.M., Kwong P.D., Thompson A., Lehmann M.S., Grubel G., Legrand J.F., Als-Nielsen J., Colman D.R., Hendrickson W.A. Structural basis of cell–cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Shibamoto S., Hayakawa M., Takeuchi K., Hori T., Miyazawa K., Kitamura N., Johnson K.R., Wheelock M.J., Matsuyoshi N., Takeichi M. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J. Cell Biol. 1995;128:949–957. doi: 10.1083/jcb.128.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Sano K., Tanihara H. Diversity of the cadherin familyevidence for eight new cadherins in nervous tissue. Cell Regul. 1991;2:261–270. doi: 10.1091/mbc.2.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S.C., Inoue T., Kimura Y., Tanaka T., Takeichi M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol. Cell. Neurosci. 1997;9:433–447. doi: 10.1006/mcne.1997.0626. [DOI] [PubMed] [Google Scholar]
- Suzuki S.T. Protocadherins and diversity of the cadherin superfamily. J. Cell. Sci. 1996;109:2609–2611. doi: 10.1242/jcs.109.11.2609. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Takeichi M., Uemura T., Iwai Y., Uchida N., Inoue T., Tanaka T., Suzuki S.C. Cadherins in brain patterning and neural network formation. Cold Spring Harbor Symp. Quant. Biol. 1997;62:505–510. [PubMed] [Google Scholar]
- Tamura K., Shan W.S., Hendrickson W.A., Colman D.R., Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Shan W.S., Phillips G.R., Arndt K., Bozdagi O., Shapiro L., Huntley G.W., Benson D.L., Colman D.R. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;In press doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- Tomschy A., Fauser C., Landwehr R., Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Honjo Y., Johnson K.R., Wheelock M.J., Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T. The cadherin superfamily at the synapsemore members, more missions. Cell. 1998;93:1095–1098. doi: 10.1016/s0092-8674(00)81452-x. [DOI] [PubMed] [Google Scholar]
- Wohrn J.C., Puelles L., Nakagawa S., Takeichi M., Redies C. Cadherin expression in the retina and retinofugal pathways of the chicken embryo. J. Comp. Neurol. 1998;396:20–38. [PubMed] [Google Scholar]
- Wohrn J.C., Nakagawa S., Ast M., Takeichi M., Redies C. Combinatorial expression of cadherins in the tectum and the sorting of neurites in the tectofugal pathways of the chicken embryo. Neuroscience. 1999;90:985–1000. doi: 10.1016/s0306-4522(98)00526-0. [DOI] [PubMed] [Google Scholar]
- Wu Q., Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Yap A.S., Brieher W.M., Pruschy M., Gumbiner B.M. Lateral clustering of the adhesive ectodomaina fundamental determinant of cadherin function. Curr. Biol. 1997;7:308–315. doi: 10.1016/s0960-9822(06)00154-0. [DOI] [PubMed] [Google Scholar]
- Yap A.S., Niessen C.M., Gumbiner B.M. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]