Abstract
The latent membrane protein 1 (LMP1) of the Epstein–Barr virus has transforming properties in rodent fibroblasts and is expressed in most of the cancers associated with Epstein–Barr virus (EBV) infection including posttransplant lymphomas, Hodgkin’s disease, nasopharyngeal carcinoma, and AIDS-related lymphomas. In this study, three lineages of LMP1 transgenic mice were established with LMP1 expressed under the control of the Ig heavy chain promoter and enhancer. Lymphoma developed in all three lineages, and the incidence of lymphoma increased significantly with age with lymphomas developing in 42% of transgenic mice over 18 months. The expression of LMP1 was detected at high levels in the lymphoma tissues but only at trace levels in normal lymphoid tissues. Gene rearrangement of the Ig heavy chain indicated monoclonality or oligoclonality in all lymphomas, some of the lymphoid hyperplastic spleens, and some histologically normal spleens. These data reveal that LMP1, without the expression of other EBV genes, is oncogenic in vivo and indicate that LMP1 is a major contributing factor to the development of EBV-associated lymphomas.
Epstein–Barr virus (EBV) is linked to the development of several malignancies, primarily of B cell and epithelial-cell origin, including lymphoma in the immunocompromised host and nasopharyngeal carcinoma (1). A key biologic property of EBV that underlies its clear link to cancer is its ability to alter B cell growth regulation and induce permanent growth transformation. The B cells immortalized in vitro are infected latently and express at least nine proteins including six Epstein–Barr virus-encoded nuclear antigens (EBNA 1–6), and three latent membrane proteins (LMP1, LMP2A, and LMP2B) (2). Among these, only LMP1 can induce morphologic transformation and tumorigenicity in rodent fibroblasts (3). The expression of LMP1 also induces many changes associated with B cell activation, including cell clumping, increased numbers of villous projections, and the increased expression of CD23, CD39, CD40, CD44, and vimentin as well as the cell adhesion molecules, lymphocyte function-associated antigen-1, intercellular adhesion molecule-1, and lymphocyte function-associated antigen-3 (2). LMP1 also up-regulates bcl-2 and A20 expression, and this up-regulation, at least in part, contributes to its protective effect against apoptosis (4–6).
Recently, LMP1 has been shown to interact with the signaling molecules, tumor necrosis factor receptor-associated factors (TRAFs), that associate with the family of tumor necrosis factor receptors (TNFRs) (7). Two hybrid studies first identified TRAF3 with both LMP1 and CD40 (7, 8). Both the activation of CD40 and the expression of LMP1 activate NF-κB and induce the expression of many of the same cellular genes (5, 9–11). The TRAF interacting domain of LMP1 is essential for transformation as is the transmembrane region of LMP1, which is necessary for aggregation in the cellular membrane (12, 13). These features suggest that LMP1 functions as a constitutively activated member of the family of TNFRs (14, 15). Members of this family, which includes TNFR 1 and 2, CD40, and Fas(CD95), can either activate cellular proliferation through the activation of NF-κB (16, 17) or induce apoptosis through their association with death domain-containing proteins such as Fas-associating protein with death domain (FADD) and receptor interacting protein (RIP; ref. 18).
The aberrant expression, activation, or inactivation of critical cellular regulatory functions such as c-myc, bcl-2, ras, or p53 have been shown to be oncogenic in transgenic mice and also to be affected in human tumors (19, 20). Constitutively activated growth-factor receptors such as erbB also are transforming in vitro and oncogenic in vivo (21). Although the members of the TNFR family clearly can regulate cellular growth, constitutively activated forms have not been identified in tumors. As a novel, constitutively active member of this family, LMP1 provides a mechanism with which to evaluate the oncogenic potential of this receptor family.
In this study, we successfully established three lineages that express LMP1 in B lymphocytes. The transgenic mice have an increased incidence of lymphoma in mice 1 year or older. In normal spleens, the protein was detected at very low levels, whereas in tissues with lymphoma, LMP1 expression was detected at levels similar to the level in transformed lymphocyte lines. This study reveals a high correlation between expression of LMP1 and the development of lymphoma and suggests that LMP1 is a critical oncogenic factor in the development of EBV-associated cancers.
MATERIALS AND METHODS
Construction of Plasmids and Transgenic Mice.
The coding sequences of LMP1 were cloned with the Ig heavy chain (IgH) promoter and enhancer into the pGEM2 vector (Promega). An EcoRI-PvuI fragment representing the Ig construct that expresses LMP1 was injected into fertilized oocytes from (C57BL/6 × SJL)F1 mice. Founders were crossed initially with strain C57BL/6 and then crossed with BALB/c mice. Animals were screened for the presence of transgene with Southern Blot analysis of tail DNA and hybridization to the XhoI fragment within LMP1 (22). The transgene copy number was determined by comparing the intensity of the hybridizing bands on an autoradiograph with a serial dilution of DNA from the Raji cell line that has a known copy number of 50 per cell.
Reverse Transcriptase–PCR (RT-PCR) Analysis of LMP1.
RNA and DNA were extracted from frozen-tissue specimens, and LMP1 specific RNA was analyzed by using an RT-PCR assay as described (23, 24).
Immunoblot Analyses of LMP1, A20, Bcl-2, and c-Myc.
Frozen tissues were pulverized and resuspended in RIPA buffer (25 mM Tris, pH 7.5/150 mM NaCl/2 mM EDTA/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/10 mM sodium orthovanadate/50 mM NaF/50 mM β-glycerophosphate/2 μg/ml aprotinin/4 μg/ml leupeptin/3 μg/ml pepstatin/100 μg/ml phenylmethylsulfonyl fluoride) at 4°C. Samples were centrifuged at 7,500 × g for 10 min at 4°C, and the supernatants were collected for immunoblot analysis. Protein concentrations were determined with a Bradford assay (Bio-Rad). Protein samples (200 μg) were separated by SDS/PAGE (8% for A20, 8.5% for LMP1 and c-Myc, and 12% for Bcl-2) and transferred electrophoretically onto 0.45 μm Optitran membranes (Schleicher & Schuell). The equal loading of samples was evaluated by staining the filter with India ink. LMP1 was identified with the antibodies S12 or CS1–4 (Dako). Mouse monoclonal anti-A20 (at 1:1000 dilution), rabbit polyclonal anti-Bcl-2 (PC68-C; at 1:500 dilution; Calbiochem), and rabbit polyclonal anti-c-Myc (at 1:500 dilution; Santa Cruz Biotechnology) were used for A20, Bcl-2, and c-Myc immunoblots, respectively. For Bcl-2 immunoblots, a peptide competition assay was performed by incubating anti-Bcl-2 with the original peptide antigen, PP52 (Calbiochem), used to make the antibody, at 1:10 (antibody:peptide ratio) overnight at 4°C. The filters were washed in Tris-buffered saline with 0.5% Tween-20 three times for 15, 5, and 5 min, respectively. For LMP1 and A20 blots, the filters were incubated with 1:1,000 of horseradish peroxidase-conjugated goat anti-mouse κ light chain (Southern Biotechnology Associates) in blocking solution for 1 hr and washed again with five changes of Tris-buffered saline with Triton X-100 for 30 min. Horseradish peroxidase-conjugated anti-rabbit IgG (Amersham) was used for Bcl-2 and c-Myc blots, at 1:2,000. The filters were then incubated with SuperSignal Chemiluminescent Substrate for immunoblotting (Pierce) for 3 min and exposed for autoradiography.
IgH Gene Rearrangement.
Southern blots, prepared with DNA (8 μg) extracted from tissues and digested with EcoRI, were hybridized with 32P-labeled single-stranded RNA probe, PJ11, representing the JH region of the IgH gene (25).
Histopathologic Examination and Immunohistochemical Staining for LMP1.
Tissues were fixed in either 4% paraformaldehyde or 10% neutral-buffered formalin, for 12–24 h, embedded in paraffin, and sectioned 4 μm thick. Tissues were stained with hematoxylin and eosin for routine histologic examination. Sections selected for immunostaining were prepared on poly-l-lysine-coated slides, deparaffinized, rehydrated, and treated with proteinase K (Dako). The sections were incubated with a rabbit polyclonal anti-LMP1 (26) that was preabsorbed three times with crude mouse splenic extracts for 90 min at room temperature. Sections were incubated with peroxidase-conjugated goat anti-rabbit antibody (Amersham). Staining was visualized with diaminobenzidine (Sigma), and sections were lightly counterstained with methyl green. For immunofluorescence staining for LMP1, 5 μm frozen sections were stained with a fluorescein-conjugated goat anti-rabbit antibody. Fluorescein-conjugated anti-mouse CD3, phycoerythrin-conjugated anti-mouse B220 (PharMingen), and tetramethylrhodamine B isothiocyanate-conjugated anti-mouse IgG (Southern Biotechnology Associates) were used as staining for T and B lymphocyte markers, respectively.
RESULTS
Production of IgH LMP1 Transgenic Mice.
To target LMP1 expression to B cells, a construct was prepared in which the genomic coding sequence for LMP1 was cloned under the control of the mouse IgH enhancer and a VH promoter from a mouse hybridoma. We established four founders of IgH LMP1 transgenic mice and designated them 3, 6, 8, and 9. Founder 8 succumbed to lymphoma; therefore, a lineage was not established. Lineages were established successfully from founders 3, 6, and 9 with approximately 10 copies of the transgene in lineage 3, 2 copies in lineage 6, and 5 copies in lineage 9 (Table 1). The lineages have been maintained as heterozygous lines with all of the transgene insertions at autosomal sites. Interestingly, in lineage 3 apparently the transgene has integrated into the Ig locus. The digestion of DNA from lineage 3 by restriction enzymes produces two genomic BamHI fragments of 23 kb and 6.6 kb that hybridize to the heavy chain Ig joining region and represent the two Ig alleles. Hybridization with the LMP1 coding sequences also identified the larger, 23-kb fragment, indicative of insertion into the Ig locus.
Table 1.
Increased incidence of lymphoma in transgenic mice
| Lineage | Copy no. | Age range, months | n | %Lymphoma (n)* |
|---|---|---|---|---|
| 3 | 10 | <12 | 13 | 0 |
| 12–18 | 12 | 41.7 (5) | ||
| >18 | 41 | 51.2 (21) | ||
| 6 | 2 | <12 | 8 | 0 |
| 12–18 | 11 | 18.2 (2) | ||
| >18 | 26 | 23.1 (6) | ||
| 9 | 5 | <12 | 3 | 33.3 (1) |
| 12–18 | 8 | 37.5 (3) | ||
| >18 | 12 | 50.0 (6) | ||
| Total | – | <12 | 24 | 4.2 (1) |
| 12–18 | 31 | 32.3 (10) | ||
| >18 | 79 | 41.8 (33) | ||
| Control | 0 | >18 | 43 | 11.6 (5) |
Fisher’s exact test indicates P values of 0.0001, 0.31, and 0.008 for lineages 3, 6, and 9, respectively, compared to negative controls.
Incidence of Lymphoma.
In comparison with negative littermates, lymphoma developed with increased incidence in all lineages. The development of lymphoma was rare in mice under 1 year of age but increased to 32% in mice over 1 year-old and to 42% in mice over 18 months (Table 1). The incidence of lymphoma was higher in lineages 3 and 9 with 42% and 38% of animals over 12 months and 50% of animals over 18 months developing lymphoma (Table 1). The incidence of lymphoma in elderly mice in lineages 3 and 9 was statistically significantly greater than strain- and age-matched negative transgene controls, housed under the same conditions, and given the same diet, with P values of 0.0001 and 0.008, respectively.
Most mice with lymphoma had massive enlargement of the spleen. In ≈60% of animals with lymphoma in all three lineages, the lymphoma was disseminated and frequently involved the liver, lungs, or the mesenteric, hilar, or cervical lymph nodes. Lymphoma was found infrequently in the bone marrow and never detected in the central nervous system. The lymphomas were predominantly follicular center-cell lymphoma with either predominantly large cells or mixed small and large cells (Fig. 1A). These neoplasms were composed of intermediate to large, cohesive lymphoid cells with irregularly shaped oval nuclei and moderate to abundant lightly eosinophilic cytoplasm. The chromatin was often condensed and marginated on the nuclear membrane with the nucleoli also often juxtaposed with the nuclear membrane. Mitotic figures were observed frequently. In a few animals without frank lymphoma, aggressive lymphoid infiltrates that invaded the adjacent normal tissue were detected (Fig. 1B). These infiltrates were monoclonal or oligoclonal by Ig gene rearrangement; however, the lymphocytes lacked the enlarged vesicular nuclei and prominent nucleoli characteristic of tumor cells.
Figure 1.

Histopathology and immunohistochemistry for LMP1 in lymphoma and lymphoid infiltrate. (A) Follicular center cell lymphoma (lineage 3), large cell type. The normal architecture of the spleen is effaced by sheets of large neoplastic cells with abundant cytoplasm. The nuclei are irregularly shaped, with clumped or marginated chromatin and sometimes prominent nucleoli. Mitotic figures are frequently present. (Hematoxylin and eosin; ×600.) (B) Lymphoid infiltrate in the lung. An aggressive lymphoid-cell infiltrate (lineage 6) has invaded the adjacent alveolar tissue. Analysis of DNA extracted from this tissue demonstrated monoclonal heavy chain rearrangement on Southern blot. A high level of LMP1 was detected by immunoblot analysis of a protein lysate prepared from this tissue. (Hematoxylin and eosin; ×200.) (C) Expression of LMP1 in the lymphoid infiltrate shown in B. The tissue was stained with rabbit polyclonal anti-LMP1 and detected with an immunoperoxidase-tagged goat anti-rabbit serum. LMP1 is detected as dark brown staining in the cell membrane, with some cells showing the characteristic capping. (×1,000.) (D and E) Expression of LMP1 in lymphoma tissue and negative-staining control (lineage 3). The splenic lymphoma tissue was stained with either rabbit polyclonal anti-LMP1 (D) or normal rabbit serum at same dilution (E). The staining was detected with fluorescein-conjugated goat anti-rabbit antibody. LMP1 is detected as green fluorescence in the plasma membrane, with some cells showing the characteristic capping. (×1,000.) (F) Presence of cytoplasmic Ig in the lymphoma. The same lymphoma tissue depicted in D and E was stained with tetramethylrhodamine B isothiocyanate -conjugated goat anti-mouse IgG. IgG is detected as red fluorescence in the cytoplasm of most cells. This lymphoma also expresses a high level of LMP1 and was shown to be monoclonal by IgH gene rearrangement (see also Fig. 4 A and B). (×1,000.)
Expression of LMP1.
LMP1 expression was analyzed by RT-PCR amplification of spliced mRNAs, immunoblot analyses, and immunohistochemistry. In almost all tissue samples from all transgenic animals, including liver, ovary, and intestine, LMP1 expression was detected by RT-PCR (data not shown). The detection of mRNA did not correlate with LMP1 expression detected on immunoblots nor with evidence of pathology. As it is unlikely that the IgH promoter and enhancer is active in all tissues, the detection of LMP1 transcription in tissues likely reflects transcription in infiltrating lymphocytes.
Immunohistochemical detection was not possible with the LMP1 mouse monoclonals; therefore, an LMP1-specific rabbit antiserum was used. This reagent could weakly detect LMP1 in 60% to 70% of EBV-infected lymphoid cell lines by using a fluorescein-conjugated goat anti-rabbit secondary antibody (26). In tissues with lymphoma, LMP1 was detected in a significant portion of cells with the characteristic capping of LMP1 in some cells (Fig. 1D), whereas normal rabbit serum did not stain the lymphoma tissue (Fig. 1E). These same samples also stained positive for B220 (data not shown) and cytoplasmic IgG (Fig. 1F), but not CD3, a T cell marker (data not shown), indicating that the LMP1 was expressed in B cells. Similarly, LMP1 was detected in membrane patches in a lymphoid infiltrate with a peroxidase-conjugated goat anti-rabbit secondary antibody (Fig. 1C). A high level of LMP1 was also detected by immunoblot analysis of a protein lysate prepared from this tissue, confirming immunohistochemical detection.
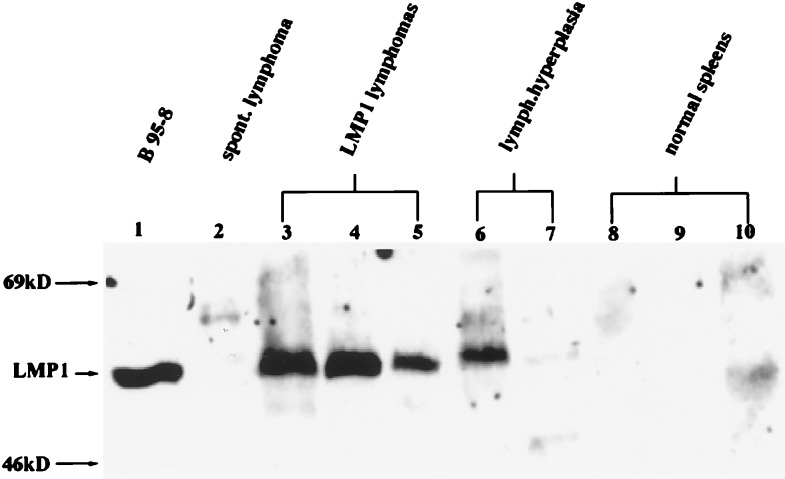
Immunoblot detection of LMP1 in tissues involved with lymphoma revealed high levels of expression in all but one sample examined (Fig. 2). In five occurrences of lymphoid hyperplasia, where the lymphoid tissue was greatly enlarged with increased numbers of blast cells and mitotic figures but the lymphocytes appeared to be nonneoplastic, LMP1 was also detected but at variable levels (Fig. 2, lanes 6 and 7). LMP1 was not detected frequently in normal spleen tissue but was detected at very low levels in some samples (Fig. 2, lane 10). The frequency of detection of LMP1 in normal spleens increased with age. In mice under 12 months, LMP1 was detected at low levels in 35% of normal spleens, whereas in mice older than 18 months, LMP1 was detected in 75% of normal spleens.
Figure 2.
Levels of the expression of LMP1 correlate with lymphoid pathologies. Immunoblots were prepared with total protein tissue lysate (200 μg) and 50 μg of protein lysate from the B95–8 EBV-positive lymphoid cell line (lane 1) and reacted with the CS1–4 mouse monoclonal antibody. A spontaneous lymphoma was obtained from a 2-year-old LMP1-negative littermate (lane 2). Lanes 3–5 are lymphoma tissues; lanes 6–7 are lymphoid hyperplasia; lanes 8–10 are normal lymphoid tissues. Lanes 3–10 are transgenic splenic tissues from lineage 3.
Analysis of Ig gene rearrangement indicated that all of the lymphoma samples were monoclonal or possibly oligoclonal (Fig. 3). Hybridization with the PJ11 probe to samples from lineage 3 revealed the standard 6.6-kb BamHI fragment representing the unrearranged heavy chain allele and an additional 23-kb fragment that also hybridized to the LMP1 probe (data not shown). Thus, all tissues from lineage 3 contained two genomic fragments with LMP1 integrated in close proximity to the IgH sequences. Ig gene rearrangement was detected in all tissue samples with lymphoma, and in some lymphoma samples, multiple rearranged bands could be detected. In a lymphoma from lineage 9 (Fig. 3, lane 4) two rearranged alleles were detected. This finding may represent a clonal lymphoma with a rearrangement of both Ig alleles or could represent a biclonal lymphoma. An example of lymphoma that developed in lineage 3 had three detectable rearranged alleles, which would also suggest the proliferation of more than one clone (Fig. 3, lane 2). Ig gene rearrangement was detected in all tissue samples with lymphoma and was also detected in the hyperplastic nodes, lymphoid infiltrates, and some normal spleens (Fig. 3). The detection of clonal JH rearrangement not only confirms the B cell origin of tumors but also indicates that there can be clonal expansion without histologic evidence of pathology.
Figure 3.
Transgenic lymphomas and lymphoid hyperplasia are clonal. Southern blots were prepared with DNA from samples of lymphoma, hyperplasia, or normal spleens and hybridized to a PJ11 probe representing the JH region of the heavy chain locus. “Normal spleen” is a nontransgenic control spleen with the genomic 6.6-kb EcoRI fragment. The upper genomic band (23 kb) in mice of lineage 3 is caused by the integration of the transgene into the heavy chain locus in this lineage (lanes 1, 2, 3, 6, 9, and 10). Lanes 7, 8, 11, and 13 are from lineage 6. Lanes 4, 5, and 12 are from lineage 9.
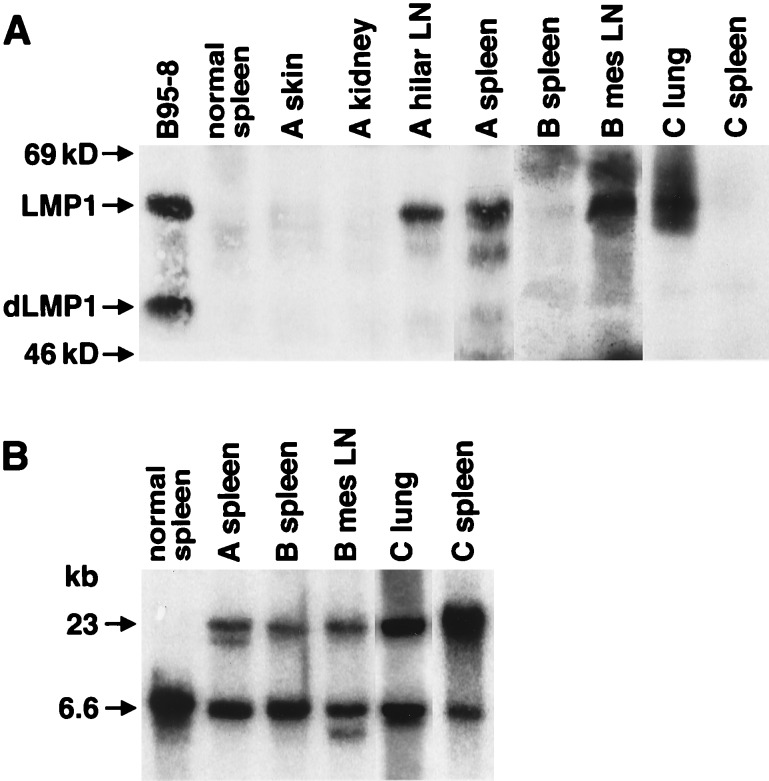
The expression of LMP1 correlated with the detection of clonality and lymphoma involvement. In the normal spleens that were LMP1 positive, Ig rearrangement was detected in 57% of the samples but was detected in only 15% of normal spleens that lacked the expression of LMP1. In animals with lymphoma, the expression of LMP1 was detected only in tissue samples with evidence of lymphoma and not in uninvolved organs (Fig. 4A). In the animal designated A, LMP1 was expressed in the hilar lymph node and spleen that had lymphoma but not in the skin or kidney. In animal B, LMP1 was detected in the mesenteric lymph node, which contained lymphoma, but not in the uninvolved, normal spleen. Similarly, in animal C, the lymphoma tissue in the lung sample contained LMP1, but LMP1 was not detected in the normal spleen of this animal. Ig gene rearrangement correlated with detection of LMP1 and identification of lymphoma (Fig. 4B).
Figure 4.
Expression of LMP1 correlates with IgH gene rearrangement and lymphoma detection. (A) Immunoblots were prepared with protein lysates from lymphoma samples and control tissues from three mice and the B95–8 cell line. Mouse A had lymphoma in hilar LN (lymph nodes) and the spleen, but not the skin and kidney. Mouse B had lymphoma in the mesenteric LN but not in the spleen. Mouse C had lymphoma in the lungs but not the spleen. “Normal spleen” was a spleen from a nontransgenic control mouse. (B) IgH gene rearrangement. Southern blots were hybridized to the PJ11 probe and showed the correlation of the lymphoma phenotype in the organs described in A with clonal rearrangement. In mouse A, gene rearrangement was analyzed and detected only in the spleen. All three mice were from lineage 3, which contained two genomic bands. The upper band (23 kb) resulted from the integration of the transgene into the heavy chain locus.
Oncogene Expression.
LMP1 has profound effects on cellular gene expression through the activation of NF-κB and the TRAF pathway (2, 6, 7, 11). LMP1 induces expression of B lymphocyte activation markers and antiapoptotic molecules such as Bcl-2, Mcl-1, and A20 (4–6, 27, 28). In vivo, bcl-2 and c-myc are expressed at high levels in EBV-infected tumors (29), suggesting that the activation of these genes is an important consequence of EBV infection. Many of these genes, including c-myc and A20, have been shown to be regulated by NF-κB transcription factors (5, 30).
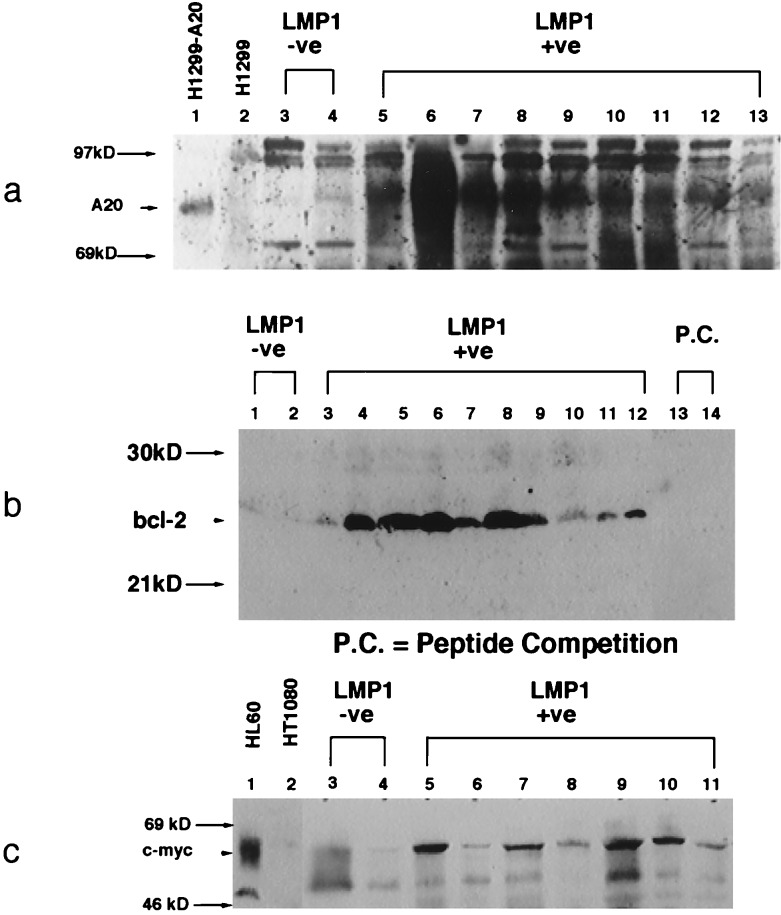
To determine whether these genes contributed to the development of lymphoma in LMP1-induced transgenic tumors, expression of A20, Bcl-2, and c-Myc was detected by immunoblot analysis. LMP1-positive lymphoma samples were compared with known positive controls and lymphoma samples that developed in LMP1-negative transgenic littermates. A20 is an unusual zinc finger containing protein that has been shown recently to interact with TRAF2 (31) and to inhibit specifically p53-mediated apoptosis (6). A cell line in which A20 is expressed at high levels under the control of the human cytomegalovirus immediate early promoter (H1299-A20) and the A20-negative parental line were used as controls for specificity of detection (6). A20 was found to be expressed at high levels in all of the LMP1-positive transgenic lymphomas but was not detected in the nontransgenic lymphoma samples (Fig. 5). Similarly, high levels of Bcl-2 were detected in all LMP1-positive lymphoma samples, but not in nontransgenic lymphoma tissues. The Bcl-2 peptide specific for the antisera effectively blocked this detection. Although the expression of c-Myc was more variable, this protein was also expressed at elevated levels in LMP1-induced lymphomas but at low levels in nontransgenic lymphomas (Fig. 5). c-Myc was detected at high levels in the known positive HL60 line but was not detected in the negative HT1080 line. The absence of expression of any of these genes in the lymphomas that developed spontaneously in the negative littermates suggests that other growth-stimulating pathways have been activated in these tumors. However, the consistent detection of elevated expression of A20, Bcl-2, and c-Myc in the LMP1 lymphomas suggests that induction of their expression is an important component of LMP1-induced lymphomagenesis.
Figure 5.
A20, Bcl-2, and c-Myc are expressed at elevated levels in transgenic lymphoma. Immunoblots were prepared with 200 μg total protein lysates from lymphomas that developed in nontransgenic littermates and transgenic lymphoma tissues and reacted with mouse monoclonal anti-A20, rabbit polyclonal anti-Bcl-2, and rabbit polyclonal anti-c-Myc.
DISCUSSION
Transgenic mice are a powerful tool for determining the effects of the expression of specific genes in specific tissues in a living animal (19). Many genes that have been identified as potential oncogenes as partners in chromosomal translocations or by retroviral acquisition have been proven to be oncogenic in transgenic models. Studies in transgenic mice of viral oncogenes such as simian virus 40 large T antigen have identified critical cellular pathways that are affected by the expression of the viral genes (32, 33). Similar studies for the human tumor virus EBV are more complex, because multiple viral genes are expressed in EBV-transformed lymphoid cell lines and in EBV-associated tumors, and several viral proteins contribute to the transformed and tumorigenic phenotype. Several studies have analyzed EBV genes in transgenic mice; however, in most cases expression was restricted to unusual tissues. The EBNA2 protein is essential for transformation and is thought to represent a constitutively activated member of the Notch signaling pathway (34, 35). The expression of EBNA2 under the control of the simian virus 40 early promoter and enhancer fused to the lymphoid-specific EBV promoter within BamHI W resulted in formation of renal adenocarcinomas (36). Although EBNA2 mRNA could be detected in most tissues, the protein was detected only at low levels in some tubule cells. This finding suggested that the unique pathology reflected expression that actually was restricted to kidney cells. In contrast, the expression of the EBV leader protein induced heart failure but did not induce tumors (37). A single study has succeeded in expressing an EBV protein in lymphocytes. Two lineages that express EBNA1 under the control of the polyoma early promoter and the heavy chain enhancer have an increased incidence of the development of lymphoma in elderly mice (38). Surprisingly, EBNA1 was expressed in all mice at all ages and was found at equivalent levels in normal and tumor tissues from these mice.
The effect of the expression of LMP1 has been evaluated in transgenic mice where LMP1 was expressed under the control of the polyoma early promoter and enhancer (39). The expression of LMP1 induced epidermal hyperplasia with altered keratin gene expression. Replacement of the polyoma enhancer with IgH enhancer resulted in embryonic lethality with epidermal hyperplasia. In these mice, LMP1 could be detected only in the skin in both polyoma and Ig enhancer transgenes. Many studies have revealed toxicity associated with high levels of expression of LMP1, and the high level of lethality in transgenic mice likely reflects this toxicity (40).
In the IgH LMP1-transgenic lineages, the EBV gene is successfully targeted to the lymphoid compartment of transgenic mice. The increased detection of LMP1 in normal spleens in aging mice may reflect the promoter activity in activated B cells and suggests that other events contribute to the activation of the promoter or the survival of B cells that express LMP1. However, whatever events contribute to the expression of LMP1, it correlates well with pathology. In normal spleens in which LMP1 is detected, LMP1 is expressed at very low levels, perhaps reflecting occasional expression in a subset of cells. The detection of clonal proliferation in normal spleens may reflect the growth of these LMP1-expressing cells. In contrast, in lymphoma tissue, LMP1 is apparently expressed in most cells at levels equivalent to the level in transformed B cells. Although the follicular center-cell phenotype of the transgenic lymphomas is quite distinct from EBV-positive lymphomas, this distinct pathology likely reflects the mouse strain. Studies have shown that the same transgene results in distinctive lymphomas dependent on the mouse background (41).
Similar studies targeting the expression of cellular oncogenes to B cells have had striking effects on B cell maturation and lymphoma development. Ig-enhancer-regulated c-myc transgenic mice have greatly enlarged spleens, with polyclonal pre-B cell hyperplasia in young animals with 80% developing lymphoma by 14 weeks of age (42). In contrast, Ig-enhancer bcl-2 transgenic mice developed splenic hyperplasia, but only 11% of animals over 1 year old developed lymphoma (43). In dually transgenic mice, c-myc and bcl-2 are complementary, with mice developing lymphoma by 5–6 weeks of age (44). Although the LMP1 transgenic animals have elevated expression of both c-myc and bcl-2, the phenotype is less severe than that of deregulated c-myc but has greater malignant potential than deregulated bcl-2. These differences in malignant potential may reflect the fact that these genes are still regulated through their natural promoters and that LMP1 has activated their expression through pathways operative in normal B cells.
However, the significantly elevated incidence of lymphoma in these mice indicates that the constitutive activation of the TNFR family’s signaling pathways is oncogenic. The data presented in this study suggest that NF-κB is activated in the lymphomas, because two genes, A20 gene and c-myc, thought to be regulated by NF-κB (5, 30), are expressed. LMP1 has been shown to induce A20 expression by activating NF-κB, thereby blocking p53-mediated apoptosis (5, 6). Stimulation of a murine B lymphoma cell line with CD40L, a condition probably comparable to activation by LMP1, induces and sustains NF-κB binding to the sites known for transcriptional regulation of c-myc, thereby maintaining c-myc RNA levels and rescuing the cells from anti-surface-IgM-induced apoptosis (30). LMP1 has also been shown to up-regulate bcl-2 expression in B lymphocytes and thus protect the cells from apoptosis (4). The elevated levels of A20, Bcl-2, and c-Myc detected in the transgenic lymphomas suggest that the expression of their genes is a critical component in LMP1-mediated lymphomagenesis. However, the advanced age of the mice that developed tumors and the clonality of the tumors suggest that other genes or pathways that complement the pathway(s) activated by LMP1 must also become activated and contribute to the development of lymphoma. In EBV-transformed lymphocytes, similarly complementary pathways may be activated by other viral gene products.
Acknowledgments
The authors thank Dr. Hermann Eibel for the plasmid construct of Ig promoter and enhancer, Dr. Elliot Kieff for rabbit polyclonal anti-LMP1, Dr. David Thorley-Lawson for S12 monoclonal anti-LMP1, Dr. Vishva Dixit for monoclonal anti-A20, Katherine Fries for the H-1299 cell line, Dr. Jenny P.-Y. Ting for the HL60 cell line, Dr. Michael Schell for statistical analysis, Dr. Marty Mayo for helpful discussion, and the Lineberger Cancer Center Histopathology Core Facility for tissue section preparation. This work was supported by National Institutes of Health Grant CA 19014 (to N.R.-T.).
ABBREVIATIONS
- EBNA
Epstein–Barr virus-encoded nuclear antigen
- EBV
Epstein–Barr virus
- IgH
Ig heavy chain
- LMP
latent membrane protein
- RT-PCR
reverse transcriptase–polymerase chain reaction
- TNFR
tumor necrosis factor receptor
- TRAF
tumor necrosis factor receptor-associated factor
References
- 1. Raab-Traub N. Semin Virol. 1996;7:315–323. [Google Scholar]
- 2.Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2343–2396. [Google Scholar]
- 3.Wang D, Liebowitz D, Kieff E. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 4.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 5.Laherty C D, Hu H M, Opipari A W, Wang F, Dixit V M. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 6.Fries K L, Miller W E, Raab-Traub N. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 8.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 9.Hammarskjöld M L, Simurda M C. J Virol. 1992;66:6496–6501. doi: 10.1128/jvi.66.11.6496-6501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paine E, Scheinman R I, Baldwin A S, Jr, Raab-Traub N. J Virol. 1995;69:4572–4576. doi: 10.1128/jvi.69.7.4572-4576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller W E, Mosialos G, Kieff E, Raab-Traub N. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaye K M, Izumi K M, Kieff E. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izumi K M, Kaye K M, Kieff E D. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatzivassiliou E, Miller W E, Raab-Traub N, Kieff E, Mosialos G. J Immunol. 1998;160:1116–1121. [PubMed] [Google Scholar]
- 16.Rothe M, Sarma V, Dixit V M, Goeddel D V. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 17.Kilger E, Kieser A, Baumann M, Hammerschmidt W. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu H, Shu H-B, Pan M-G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 19.Adams J M, Cory S. Science. 1991;254:1161–1167. doi: 10.1126/science.1957168. [DOI] [PubMed] [Google Scholar]
- 20.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 21.Frykberg L, Palmieri S, Beug H, Graf T, Hayman M J, Vennström B. Cell. 1983;32:227–238. doi: 10.1016/0092-8674(83)90513-5. [DOI] [PubMed] [Google Scholar]
- 22.Raab-Traub N, Flynn K. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 23.Raab-Traub N, Hood R, Yang C S, Henry B, Pagano J S. J Virol. 1983;48:580–590. doi: 10.1128/jvi.48.3.580-590.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C-L, Sadler R H, Walling D M, Su I-J, Hsieh H-C, Raab-Traub N. J Virol. 1993;67:6303–6308. doi: 10.1128/jvi.67.10.6303-6308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcu K B, Banerji J, Penncavage N A, Lang R, Arnheim N. Cell. 1980;22:187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- 26.Hennessy K, Fennewald S, Hummel M, Cole T, Kieff E. Proc Natl Acad Sci USA. 1984;81:7207–7211. doi: 10.1073/pnas.81.22.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowe M, Peng-Pilon M, Huen D S, Hardy R, Croom-Carter D, Lundgren E, Rickinson A B. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Rowe M, Lundgren E. Cancer Res. 1996;56:4610–4613. [PubMed] [Google Scholar]
- 29.Rickinson A B, Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2397–2446. [Google Scholar]
- 30.Schauer S L, Wang Z, Sonenshein G E, Rothstein T L. J Immunol. 1996;157:81–86. [PubMed] [Google Scholar]
- 31.Song H Y, Rothe M, Goeddel D V. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks J R, Lin J, Hinds P, Miller D, Levine A J. J Virol. 1989;63:790–797. doi: 10.1128/jvi.63.2.790-797.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy S A, Symonds H S, Van Dyke T. Proc Natl Acad Sci USA. 1994;91:3979–3983. doi: 10.1073/pnas.91.9.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh J J, Hayward S D. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 36.Törnell J, Farzad S, Espander-Jansson A, Matejka G, Isaksson O, Rymo L. Oncogene. 1996;12:1521–1528. [PubMed] [Google Scholar]
- 37.Huen D S, Fox A, Kumar P, Searle P F. J Gen Virol. 1993;74:1381–1391. doi: 10.1099/0022-1317-74-7-1381. [DOI] [PubMed] [Google Scholar]
- 38.Wilson J B, Bell J L, Levine A J. EMBO J. 1996;15:3117–3126. [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson J B, Weinberg W, Johnson R, Yuspa S, Levine A J. Cell. 1990;61:1315–1327. doi: 10.1016/0092-8674(90)90695-b. [DOI] [PubMed] [Google Scholar]
- 40.Hammerschmidt W, Sugden B, Baichwal V R. J Virol. 1989;63:2469–2475. doi: 10.1128/jvi.63.6.2469-2475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yukawa K, Kikutani H, Inomoto T, Uehira M, Bin S H, Akagi K, Yamamura K-I, Kishimoto T. J Exp Med. 1989;170:711–726. doi: 10.1084/jem.170.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams J, Harris A, Pinkert C, Corcoran L, Alexander W, Cory S, Palmiter R, Brinster R. Nature (London) 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 43.McDonnell T J, Korsmeyer S J. Nature (London) 1991;349:254–256. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- 44.Strasser A, Harris A W, Bath M L, Cory S. Nature (London) 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]