Abstract
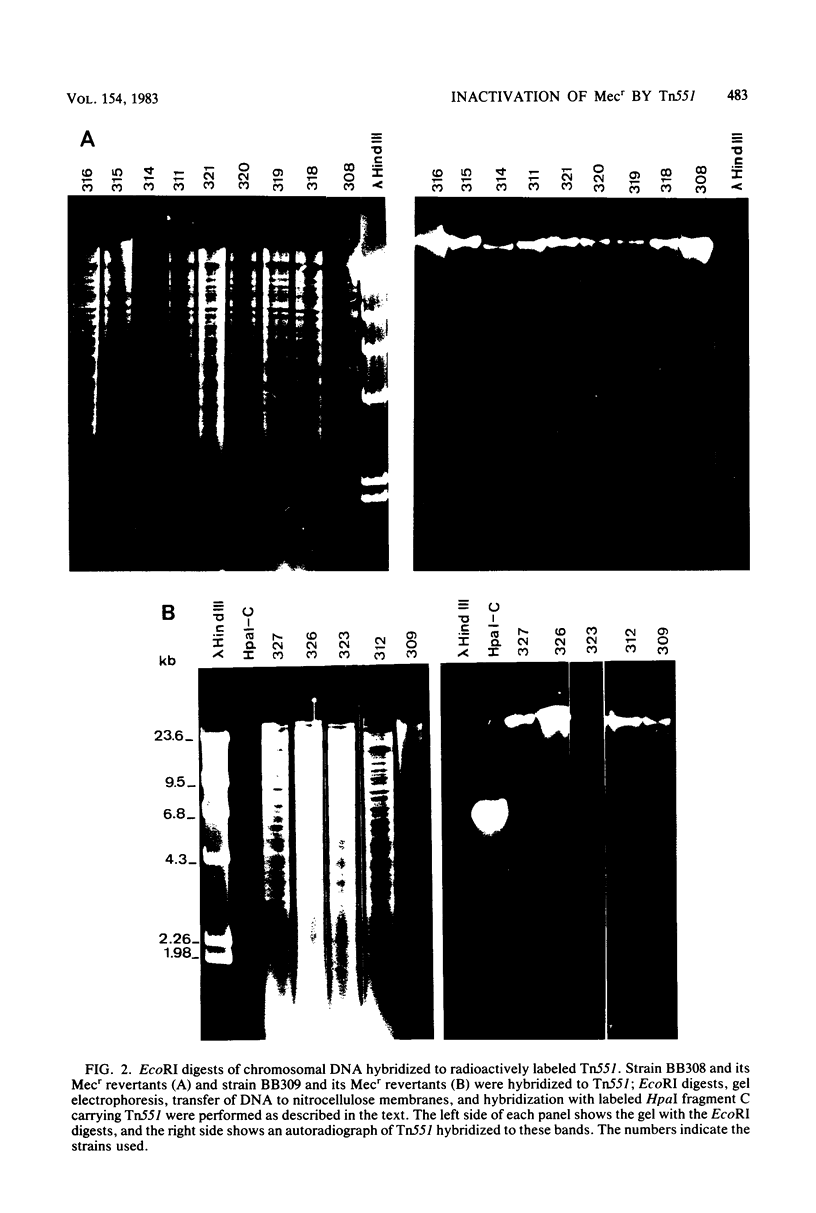
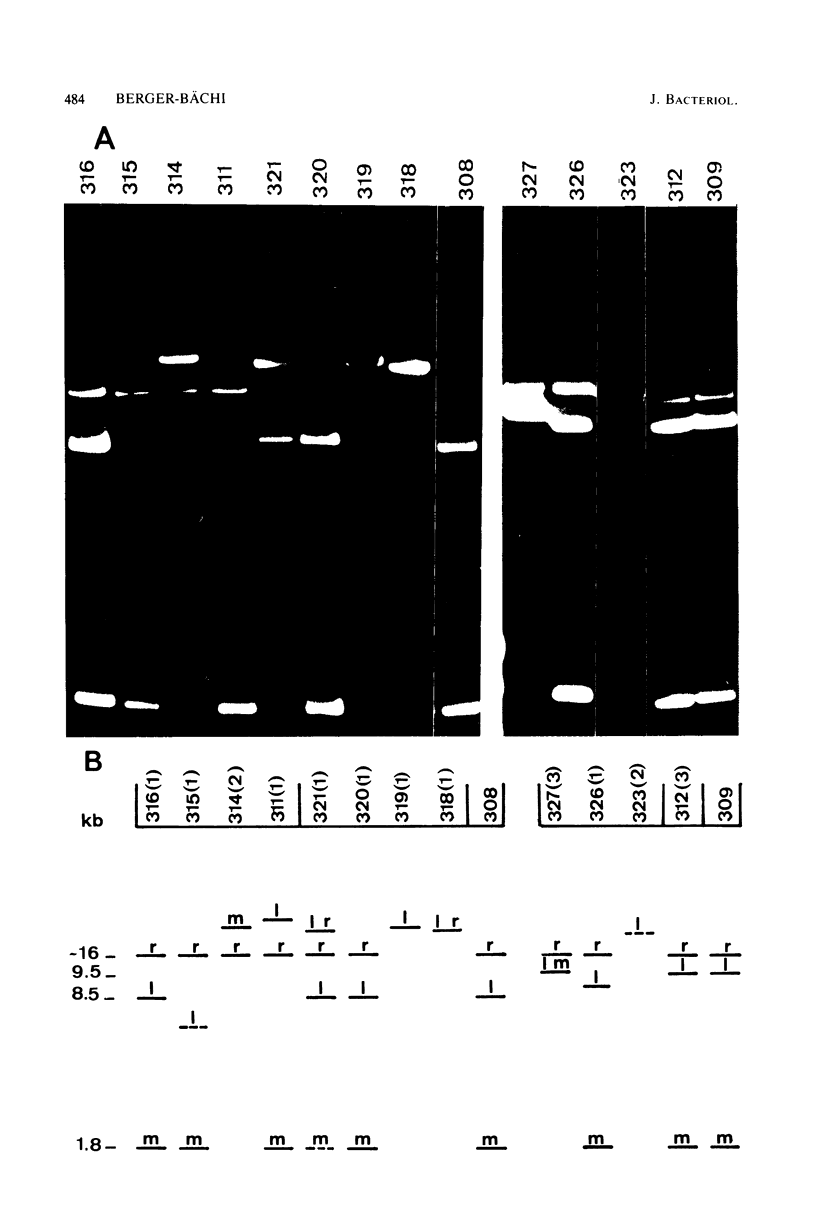
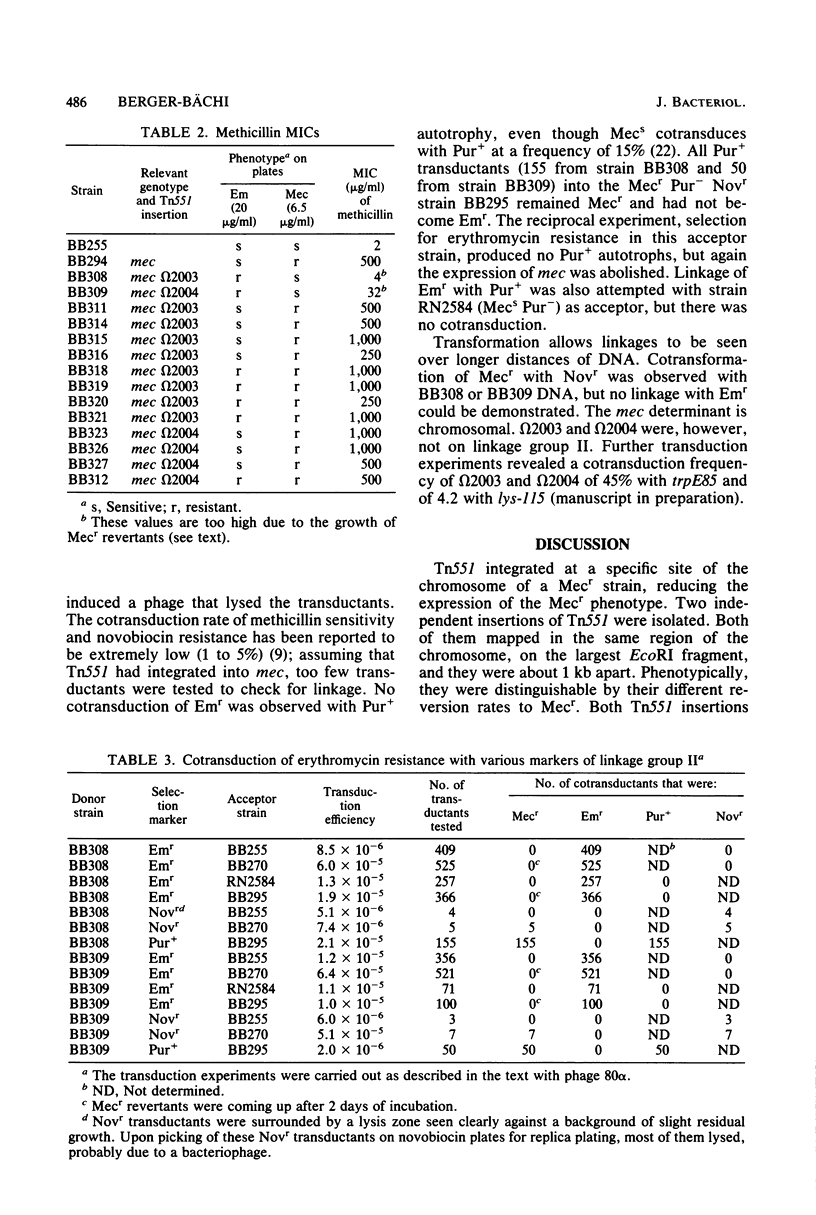
Transposon Tn551 was translocated into the chromosome of a methicillin-resistant (mec) strain of Staphylococcus aureus by heat inactivation of a thermo-sensitive plasmid carrying Tn551 and selection for erythromycin-resistant (Emr) survivors. Two independent chromosomal insertions of Tn551 were obtained which reduced the level of the methicillin resistance by a factor of 50 to 100, making the strains phenotypically methicillin sensitive (Mecs). Each of the Tn551 insertions was on the largest fragment produced by EcoRI digestion of the chromosomal DNA of these strains. The integration sites lie about 1 kilobase apart. These Mecs strains reverted to Mecr at frequencies of 2.4 X 10(-8) and 3.6 X 10(-5), respectively. The majority of Mecr revertants still were Emr; only a few lost the Emr phenotype concomitantly with reversion to the Mecr phenotype. Hybridization data with labeled Tn551 showed complex rearrangements and deletions in the region of the insertion. These two Tn551 insertions do not lie on the same linkage group, II, as the mec determinant. The phenotypic expression of methicillin resistance, therefore, is also dependent upon a chromosomal genetic marker not physically linked to the mec determinant.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bächi B. Physical mapping of the BglI, BglII, PstI and EcoRI restriction fragments of staphylococcal phage phi 11 DNA. Mol Gen Genet. 1980;180(2):391–398. doi: 10.1007/BF00425853. [DOI] [PubMed] [Google Scholar]
- Cohen S., Sweeney H. M., Basu S. K. Mutations in prophage phi11 that impair the transducibility of their Staphylococcus aureus lysogens for methicillin resistance. J Bacteriol. 1977 Jan;129(1):237–245. doi: 10.1128/jb.129.1.237-245.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B., Tomasz A. Altered penicillin-binding proteins in methicillin-resistant strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1981 May;19(5):726–735. doi: 10.1128/aac.19.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Kayser F. H., Benner E. J., Troy R., Hoeprich P. D. Experimental and clinical aspects of resistance determinants. Mode of resistance against beta-lactam antibiotics in staphylococci. Ann N Y Acad Sci. 1971 Jun 11;182:106–117. doi: 10.1111/j.1749-6632.1971.tb30649.x. [DOI] [PubMed] [Google Scholar]
- Kayser F. H., Wüst J., Corrodi P. Transduction and elimination of resistance determinants in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1972 Sep;2(3):217–223. doi: 10.1128/aac.2.3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser F. H., Wüst J., Santanam P. Genetic and molecular characterisation of resistance determinants in methicillin-resistant Staphylococcus-aureus. J Med Microbiol. 1976 May;9(2):137–148. doi: 10.1099/00222615-9-2-137. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Terminal nucleotide sequences of Tn551, a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid. 1980 Sep;4(2):148–154. doi: 10.1016/0147-619x(80)90004-9. [DOI] [PubMed] [Google Scholar]
- Kuhl S. A., Pattee P. A., Baldwin J. N. Chromosomal map location of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1978 Aug;135(2):460–465. doi: 10.1128/jb.135.2.460-465.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg M., Sjöström J. E., Johansson T. Transformation of chromosomal and plasmid characters in Staphylococcus aureus. J Bacteriol. 1972 Feb;109(2):844–847. doi: 10.1128/jb.109.2.844-847.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Novick R. P. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979 Aug;175(1):19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Latta P. D., Swanson E. C., Pattee P. A. Translocatable elements in Staphylococcus aureus. Contrib Microbiol Immunol. 1979;6:41–55. [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Schwesinger M. D., Gruss A. D., Swanson E. C., Pattee P. A. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):400–404. doi: 10.1073/pnas.76.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Studies on plasmid replication. III. Isolation and characterization of replication-defective mutants. Mol Gen Genet. 1974;135(2):131–147. doi: 10.1007/BF00264781. [DOI] [PubMed] [Google Scholar]
- Pattee P. A. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J Bacteriol. 1981 Jan;145(1):479–488. doi: 10.1128/jb.145.1.479-488.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattee P. A., Neveln D. S. Transformation analysis of three linkage groups in Staphylococcus aureus. J Bacteriol. 1975 Oct;124(1):201–211. doi: 10.1128/jb.124.1.201-211.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Shafer W. M., Iandolo J. J. Transduction of staphylococcal enterotoxin B synthesis: establishment of the toxin gene in a recombination-deficient mutant. Infect Immun. 1980 Jan;27(1):280–282. doi: 10.1128/iai.27.1.280-282.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöström J. E., Löfdahl S., Philipson L. Transformation reveals a chromosomal locus of the gene(s) for methicillin resistance in Staphylococcus aureus. J Bacteriol. 1975 Sep;123(3):905–915. doi: 10.1128/jb.123.3.905-915.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. F., Wilkinson B. J. Differential methicillin susceptibilities of peptidoglycan syntheses in methicillin-resistant Staphylococcus aureus. J Bacteriol. 1981 Nov;148(2):610–617. doi: 10.1128/jb.148.2.610-617.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart G. C., Rosenblum E. D. Genetic behavior of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1980 Dec;144(3):1200–1202. doi: 10.1128/jb.144.3.1200-1202.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler P. W., Sweeney H. M., Cohen S. Absence of circular plasmid deoxyribonucleic acid attributable to a genetic determinant for methicillin resistance in Staphylococcus aureus. J Bacteriol. 1973 Nov;116(2):771–777. doi: 10.1128/jb.116.2.771-777.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]