Abstract
Keratin polypeptides 8 and 18 (K8/18) are intermediate filament (IF) proteins that are expressed in glandular epithelia. Although the mechanism of keratin turnover is poorly understood, caspase-mediated degradation of type I keratins occurs during apoptosis and the proteasome pathway has been indirectly implicated in keratin turnover based on colocalization of keratin-ubiquitin antibody staining. Here we show that K8 and K18 are ubiquitinated based on cotransfection of His-tagged ubiquitin and human K8 and/or K18 cDNAs, followed by purification of ubiquitinated proteins and immunoblotting with keratin antibodies. Transfection of K8 or K18 alone yields higher levels of keratin ubiquitination as compared with cotransfection of K8/18, likely due to stabilization of the keratin heteropolymer. Most of the ubiquitinated species partition with the noncytosolic keratin fraction. Proteasome inhibition stabilizes K8 and K18 turnover, and is associated with accumulation of phosphorylated keratins, which indicates that although keratins are stable they still turnover. Analysis of K8 and K18 ubiquitination and degradation showed that K8 phosphorylation contributes to its stabilization. Our results provide direct evidence for K8 and K18 ubiquitination, in a phosphorylation modulated fashion, as a mechanism for regulating their turnover and suggest that other IF proteins could undergo similar regulation. These and other data offer a model that links keratin ubiquitination and hyperphosphorylation that, in turn, are associated with Mallory body deposits in a variety of liver diseases.
Keywords: ubiquitination, keratins, phosphorylation, intermediate filaments, Mallory bodies
Introduction
Keratins are epithelial-specific intermediate filament (IF) proteins that comprise more than 20 members (K1-K20; Fuchs and Weber 1994). They are subdivided into type I (K9-K20) and type II (K1-K8) keratins (Moll et al. 1982; Fuchs and Weber 1994). Various keratin combinations are expressed in an epithelial cell type–preferential manner, and form noncovalent heteropolymers of a 1:1 type I to type II ratio. For example, keratin 8 and 18 (K8/18) are found mainly in simple-type epithelia as in the liver, pancreas and intestine, while K5/14 and K1/10 are found in basal and suprabasal keratinocytes, respectively. All IF proteins possess a central α-helical rod domain and non–α-helical NH2-terminal (head) and COOH-terminal (tail) domains. The head and tail domains manifest significant structural heterogeneity among IF proteins as compared with the rod domains (Fuchs and Weber 1994), and contain the sites for several posttranslational modifications including phosphorylation and glycosylation (Omary et al. 1998).
Keratins are highly dynamic and reorganize during various cellular events such as mitosis and apoptosis. Although keratin functions remain poorly understood, except for their important role in helping cells cope with stress (Fuchs and Cleveland 1998; Ku et al. 1999), it is hypothesized that filament reorganization plays an integral role in keratin function. Potential mechanisms that are likely involved in filament reorganization include phosphorylation, proteolysis, and/or interaction with non-keratin proteins. In the case of K8/18, K8 serines (S) 23/73/431 and K18 S33/52 are known in vivo phosphorylation sites (Omary et al. 1998). Proteolysis of keratins has been described during virus infection (Zhang and Schneider 1994) and apoptosis (Caulin et al. 1997; Ku et al. 1997) and is mediated by viral proteases or caspases, respectively. Degradation, possibly by caspases, and abnormal keratin filaments have also been described in newborn rats that are exposed to ethanol in utero (Montes et al. 1996).
Keratin degradation via ubiquitination has also been implicated although direct evidence for this is lacking. For example, K8/18 and ubiquitin colocalize in hepatocyte cytoplasmic inclusions termed Mallory bodies (MB), as determined by immunofluorescence staining (Lowe et al. 1988; Ohta et al. 1988). MB inclusions are found in a variety of human liver diseases and can be induced in mice after chronic drug intoxication (Jensen and Gluud 1994). In addition, ubiquitin-conjugated K8 fragments, but not intact K8, accumulate in colorectal carcinomas (Nishibori et al. 1996). The significance of keratin ubiquitination in MB inclusions is unclear but it may be associated with keratin stabilization or, alternatively, with keratin degradation as occurs in a number of well-studied ubiquitinated proteins (Hershko and Ciechanover 1998; Laney and Hochstrasser 1999). In general, keratin turnover appears to be a very slow process as demonstrated in cultured hepatocytes (Denk et al. 1987) and colonocytes (Chou et al. 1992). However, the abundance of keratins (e.g., K8/18 make up ∼5% of total cultured cell protein; Chou et al. 1993) makes it difficult to appreciate subtle changes in keratin turnover. In this report, we provide direct evidence for K8/18 ubiquitination and subsequent degradation in vivo, and show that keratin phosphorylation modulates this process.
Materials and Methods
Cells and Reagents
Cell lines (HT29 [human colon], BHK [hamster kidney]) were obtained from the American Type Culture Collection. Antibodies (Abs) used were: L2A1 (anti-K8/18; Chou et al. 1992), M20, and TS1 (anti-K8), DC10 (anti-K18; Neomarkers), LJ4 (anti-K8 phospho-(p)S73; Liao et al., 1997), IB4 and 8250 (anti-K18 pS33; Ku et al. 1998a), 5B3 (anti-K8 pS431; Ku and Omary 1997), anti-ubiquitin (Ub) Ab (Dako Corp.), 8592 (anti-K8/18; Ku and Omary 1997), 3055 (anti-K18 pS52; Liao et al. 1995), and Ab 4668, which was raised against the nonphosphorylated K18 form of the peptide used to generate Abs 8250 and IB4 (Ku et al. 1998a). Other reagents used were: anti-His tag Ab, NiNTA™ Spin kit (Qiagen), and the proteasome inhibitor N-acetyl-Leu-Leu-norleucinal (ALLN; Calbiochem).
Cell Culture and Transfection
BHK cells, cotransfected with one or more of several keratin constructs and His-tagged wild-type (WT) ubiquitin (kindly provided by Dr. Ron Kopito, Stanford University), or HT29 cells were treated overnight with DMSO (0.1%) or with ALLN dissolved in DMSO (270 μM). Cells were then solubilized with SDS (2%)-containing sample buffer or used for immunoprecipitation or isolation of the ubiquitinated proteins. Transient transfection was carried out using LipofectAMINE™ (GIBCO BRL). The constructs used for transfection were: WT K8 and/or K18; K8 S431, S73 or S23→alanine (A; Ku et al. 1997; Liao et al. 1997); and K18 S52 or S33→A (Ku and Omary 1994; Ku et al. 1998a). Other K8 constructs (L71→P, S73→D, P74→L) were generated with a Transformer™ mutagenesis kit (CLONTECH Laboratories, Inc.). Metabolic labeling with 35S-met/cys (100 μCi/ml, 10 min) was done in RPMI 1640 medium lacking met (note K8 and K18 do not have any cys, not shown) and containing 10% dialyzed fetal calf serum. Cells were cultured in the labeling medium for 30 min before adding 35S-met/cys, then processed immediately or cultured in normal medium for various chase periods.
Analysis of Keratins
Immunoprecipitates of K8, K18, or K8/18 were obtained from HT29 cells or from transfected (35S-met labeled or unlabeled) BHK cells after solubilization. Fractionation into cytosolic and residual pellet fractions was done by three cycles of freeze-thawing of transfected cells in PBS containing 5 mM EDTA followed by centrifugation. Alternatively, ubiquitinated proteins were isolated using the NiNTA™ kit as recommended by the supplier (the presence of NP-40 interfered with binding to the nickel beads, not shown) followed by separation by SDS-PAGE, Coomassie staining and fluorography when relevant. Total cell lysates, cytosolic or remaining pellet fractions, NiNTA-isolated proteins, or immunoprecipitates were also analyzed by immunoblotting then visualization of the blotted proteins using enhanced chemiluminescence. Densitometry to quantify 35S-keratins was done with a Bio-Rad Fluor-S MultiImager system.
Results
K8 and K18 Are Ubiquitinated In Vivo
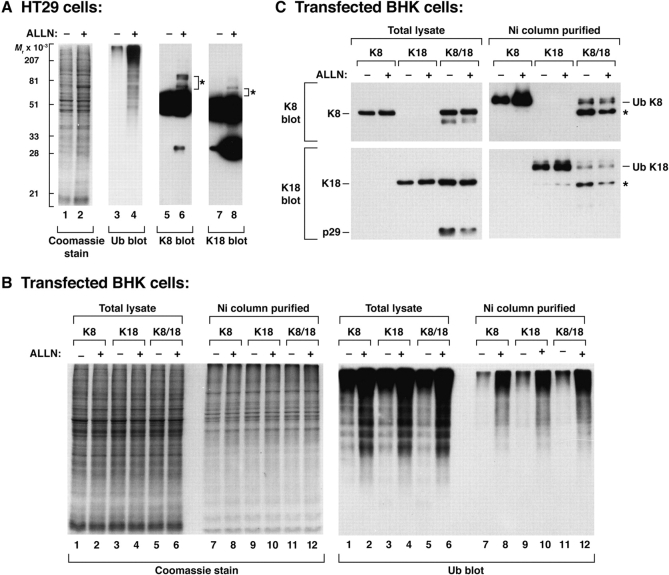
We tested the effect of proteasome inhibition on keratins in cultured HT29 cells (Fig. 1 A). ALLN increased the level of detectable ubiquitinated proteins (lane 4), and generated K8 (lane 6) and K18 (lane 8) ladders. However, the extent of ladder formation was minimal and required significant over-exposure for visualization, and several anti-Ub Abs did not bind K8/18 immunoprecipitates likely due to inability to precipitate the ubiquitinated keratins (not shown). This led us to test keratin ubiquitination in a cell transfection system that included cotransfection of His-tagged Ub with K8 and/or K18 to allow isolation of the ubiquitinated proteins. Proteasome inhibition of keratin-transfected BHK cells markedly increased the ability to detect ubiquitinated proteins as determined by blotting of total cell lysates or cell fractions that are collected with a Ni column in order to isolate His-tagged ubiquitinated proteins (Fig. 1 B). Confirmation of K8/18 ubiquitination was obtained by immunoblotting the Ni column–purified samples with K8- or K18-specific Abs (Fig. 1 C). Exposure of transfected cells to ALLN increased the singly ubiquitinated K8 and K18 species, while the level of ubiquitinated K8 and K18 was significantly less in cotransfected K8/18 cells as compared with single keratin-transfected cells. The K8 and K18 antibodies did not recognize any multiply-ubiquitinated keratins which may be due to the Ni column binding efficiency to single ubiquitinated species. Overall, these data indicate that K8 and K18 can be ubiquitinated in vivo, with enhanced levels of ubiquitinated species upon proteasome inhibition or upon expression of single keratins rather coexpression of type I and type II keratins.
Figure 1.
In vivo ubiquitination of K8 and K18. BHK cells, cotransfected with the indicated keratin(s) and His-Ub, or HT29 cells were treated overnight with DMSO (0.1%) or with an equal volume of ALLN (in DMSO). Total lysates were prepared by solubilizing the cell pellets in sample buffer, and His-ubiquitinated proteins from keratin-transfected BHK cells were purified using a Ni column after cell solubilization in 8-M urea. Total lysates and Ni column–purified samples were separated by SDS-PAGE then stained with Coomassie blue, or were immunoblotted with antibodies to Ub (His), K8, or K18. Asterisks in A highlight bands that likely correspond to ubiquitinated K8 or K18 in ALLN-treated HT29 cells. Asterisks in C represent species that likely correspond to ubiquitinated K8/18 fragments and/or unmodified K8/18 that is part of the keratin tetramer.
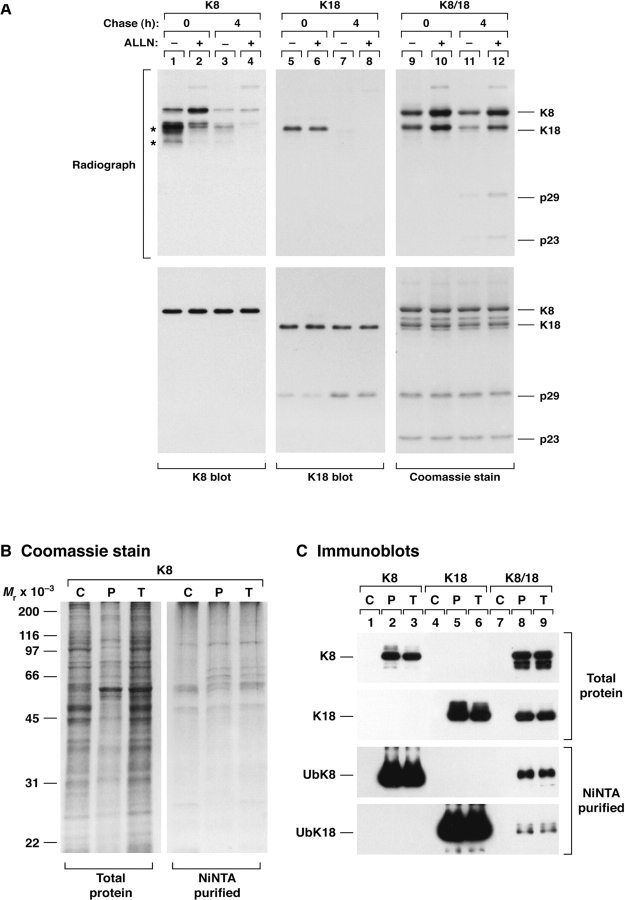
To assess if keratin ubiquitination plays a role in keratin degradation, we compared the turnover of 35S-methionine pulse-labeled K8, K18, and K8/18 in transfected BHK cells in the presence or absence of ALLN (Fig. 2 A). After a 10-min pulse, ALLN increased the level of detectable K8 (lane 1 vs 2) and K8/18 (lane 9 vs 10) but had no effect on singly expressed K18 (lane 5 vs 6). The effect of ALLN was not appreciable after 4 h of chase for singly transfected K8 or K18 due to rapid degradation but was clearly evident for cotransfected K8/18 due to the increased stability of the cotransfected keratins (Fig. 2 A, lanes 9–12). K18 fragments (p29 and p23) also form upon K18 transfection of BHK cells, as detected by fluorography after a 4-h chase (lane 12), due to caspase-mediated cleavage (Ku et al. 1997) as newly synthesized proteins accumulate. Hence, proteasome inhibition stabilizes newly synthesized K8 or K8/18, but not K18 alone, which supports keratin ubiquitination being an intermediate step for keratin degradation.
Figure 2.
Stabilization of transfected keratins by proteasome inhibition and partitioning of ubiquinated keratins with the insoluble pool. (A) BHK cells were transfected with K8 and/or K18. After 36 h, transfected cells were incubated overnight with DMSO or ALLN as in Fig. 1, then labeled with 35S-met (10 min) followed by immediate harvesting or chasing for 4 h. K8, K18 or K8/18 were precipitated with Abs TS1, DC10, and L2A1, respectively. Immunoprecipitates were analyzed by SDS-PAGE then Coomassie staining and fluorography, or were blotted with antibodies to K8 or K18. Due to significant K8 and K18 degradation and their low overall levels when transfected alone, their immunoprecipitates were visualized by blotting rather than Coomassie staining. Asterisks (lane 1) correspond to induced K18 (transfection of K8 likely induces endogenous hamster K18 expression, see also Fig. 4 C) and to K8 and/or K18 degradation products. (B and C) BHK cells were transfected with K8 and/or K18 and with His-Ub followed by isolation of a cytosolic (C), remaining pellet (P) and total cell lysates (T). Ubiquitinated proteins (NiNTA-purified) were also isolated from each of these fractions followed blotting with anti-K8 and anti-K18 Abs.
We also examined the solubility of ubiquitinated species in transfected BHK cells. The detergent-free cytosolic keratin pool under these conditions is undetectable (Fig. 2 C, lanes 1, 4, and 7), and all the ubiquitinated keratin species are present in the remaining insoluble pellet (Fig. 2 C, compare lanes P with T). However, the cytosolic pool did contain ubiquitinated proteins as exemplified in lane C of the NiNTA-purified proteins isolated from cells transfected with K8 (Fig. 2 B), K8/18 or K18 (not shown but very similar profile to that shown in Fig. 2 B). Hence, the ubiquitinated keratin species are preferentially noncytosolic.
Phosphorylated Keratins Accumulate upon Proteasome Inhibition and Are Less Ubiquitinated
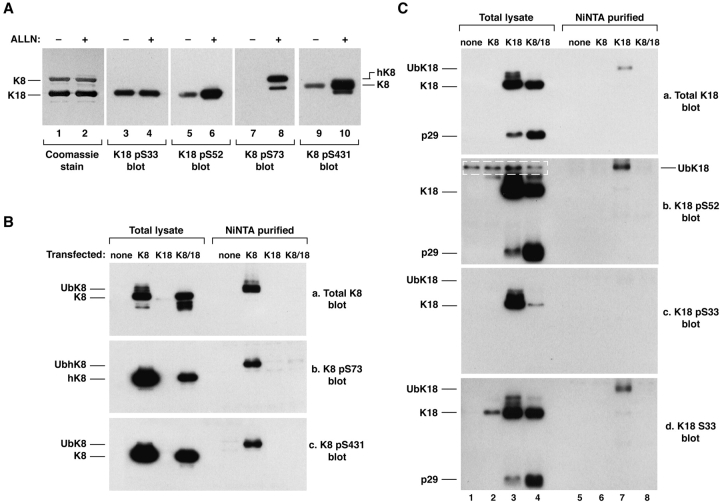
Given that several proteins undergo Ub-mediated degradation in a phosphorylation-dependent fashion, we examined the effect of proteasome inhibition on keratin phosphorylation in HT29 cells and in transfected BHK cells. HT29 cells, cultured with ALLN, accumulated phosphorylated keratins including K8 pS73, K8 pS431, and K18 pS52, but not K18 pS33 (Fig. 3 A). This suggested that keratin phosphorylation may be a protective mechanism against keratin degradation or, alternatively, that it may promote keratin degradation. To distinguish between these two possibilities, we compared the phospho- and total-keratin ubiquitinated species in BHK cells cotransfected with His-Ub and K8, K18, or K8/18 using epitope-specific anti-phosphokeratin antibodies. Ubiquitinated fractions of K8 pS73/pS431 and K18 pS52 were easily detected, while ubiquitinated K18 pS33 was barely detected (Fig. 3B and Fig. C). However, the relative stoichiometries of ubiquitinated K8 pS73/431 were significantly less than that of total ubiquitinated K8 (Fig. 3 B, i.e., the relative intensities of [Ub-phospho-K8]/[total K8] from b and c are significantly less than [total Ub-K8]/[total K8] from a). This suggests that K8 S73/431 phosphorylation may protect against ubiquitination. The apoptotic p29 K18 fragment was detected more efficiently upon transfection of K8/18 as compared with transfection of K18 alone (C, blot a, lane 3 vs 4). This may be due to more overall caspase cleavage and less overall K18 ubiquitination when K18 is stabilized by K8, as compared with K18 expression alone. The minimal K18 S33 phosphorylation in the ubiquitinated K18 fraction suggests that K18 S33 phosphorylation may serve a protective role in modulating K18 ubiquitination.
Figure 3.
Keratin phosphorylation in HT29 and transfected BHK cells. (A) HT29 cells were incubated with DMSO or ALLN as in Fig. 1 followed by immunoprecipitation of K8/18 using Ab L2A1. Precipitates were blotted with site-specific phosphokeratin antibodies: K18 pS33 (Ab 8250), K18 pS52 (Ab 3055), K8 pS73 (Ab LJ4), K8 pS431 (Ab 5B3). Note that ALLN treatment generates the previously described hyperphosphorylated form of K8 (due to pS73), termed hK8 (Liao et al. 1997), that is seen as a faint band just above K8 (lane 2) and is recognized by Ab LJ4 (lane 8). (B and C) BHK cells were cotransfected with His-Ub and vector alone (labeled as none), K8 and/or K18. After 2 d, cells were solubilized directly in sample buffer or used to purify Ub-conjugated proteins with a Ni column. The total lysate and the Ni column–purified proteins were blotted with anti-K8 (B) or anti-K18 (C) antibodies. The bands enclosed within the dotted lines (C, blot b) are nonspecific since they are also observed in vector alone–transfected cells. The antibodies used were: M20 (total K8 pool), LJ4 (K8 pS73), 5B3 (K8 pS431), DC10 (total K18 pool), 3055 (K18 pS52), IB4 (K18 pS33), and 4668 (K18 S33, i.e., the nonphosphorylated epitope). Immunoblotting with Ab 8250, which recognizes K18 pS33 also gave a similar result to that in Fig. 3 C, c (not shown).
Mutation of Keratin Phosphorylation Sites Increases Keratin Ubiquitination and Turnover
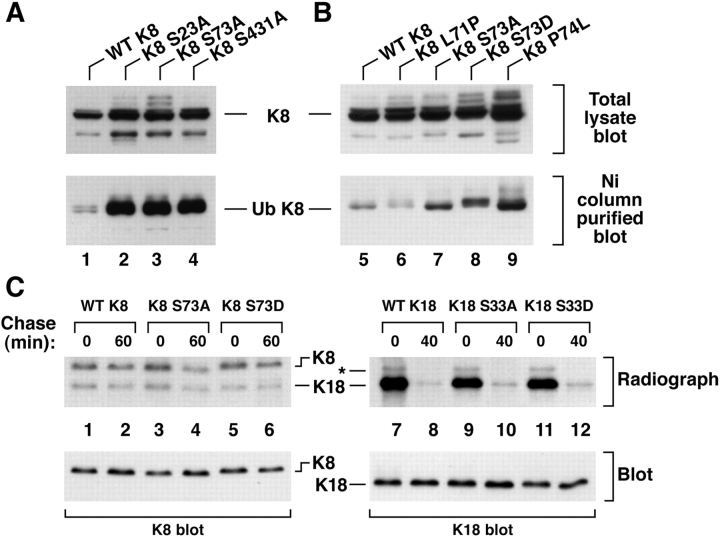
Given the results of Fig. 3, which suggested that keratin phosphorylation may play a role in modulating keratin turnover, we tested the effect of mutating K8/18 phosphorylation sites on the yield of ubiquitinated keratins in transfected cells. Mutation of any of the three K8 phosphorylation sites (S23/73/431) increased the yield of ubiquitinated K8 (Fig. 4 A) which is consistent with the results of Fig. 3 B. The association of K8 ubiquitination with its turnover was then assessed by comparing the turnover of WT versus phosphorylation-mutant K8. As shown in Fig. 4 C, 14% of 35S-met labeled WT K8 degrades after a 1-h chase (lane 1 vs 2) while 52% of K8 S73→A degrades after the same chase period (lane 3 vs 4). Substitution of an aspartate, which can mimic a phosphate, provided partial protection from degradation in that 26% of K8 was degraded.
Figure 4.
Accumulation and turnover of ubiquitinated K8 upon mutation of its phosphorylation sites. (A and B) The indicated K8 constructs and His-Ub were transfected into BHK cells. Cells were then divided into two equivalent fractions, one of which was used to prepare a total lysate and the second was used to isolate ubiquitinated proteins for immunoblotting with anti-K8 Ab. The multiple bands above K8 (lanes 3, 8, and 9) represent hyperphosphorylated and/or ubiquitinated K8. (C) BHK cells were transfected with the indicated K8 (lanes 1–6) or K18 (lanes 7–12) constructs, labeled with 35S-met for 10 min (after incubating with ALLN for 6 h), then chased for 60 min (K8) or 40 min (K18) followed by immunoprecipitation as described in Fig. 2. Equal amounts of the K8 and K18 precipitates were loaded as confirmed by blotting with anti-K8 or anti-K18 antibodies. K8 levels (see text) were quantified by densitometric scanning of the radiolabeled K8 bands after fluorography and exposure to x-ray film. Note that transfection with K8 stabilizes the endogenous low levels of BHK hamster K18 (lanes 1–6). The band indicated by an asterisk in lanes 7–12 likely corresponds to hamster K18.
We also examined K8 residues that are proximal to K8 S73, namely L71 and P74 of the sequence 69QSLLSPL, which are conserved in all type II keratins. Mutations of the corresponding K8 L71 and P74 in epidermal keratins (K1 L160→P and K5 P156→L) have been described in patients with keratin-related skin diseases (Chipev et al. 1992; Muller et al. 1998). K8 L71→P (69QSPLSPL) generates a K8 S70 potential phosphorylation site that indeed becomes phosphorylated, while K8 P74→L (69QSLLSLL) abolishes K8 S73 phosphorylation (not shown). Mutation of K8 S73→A/D or P74→L (i.e., conditions that decrease K8 phosphorylation in proximity to S73) increased K8 ubiquitination (Fig. 4 B) and, in contrast, mutation of K8 L71→P (which increases K8 phosphorylation, not shown) decreased K8 ubiquitination. Taken together, these data indicate that K8 phosphorylation protects against ubiquitination and subsequent degradation.
In contrast to the apparent involvement of K8 phosphorylation with its ubiquitination, mutation of K18 ser52/33 did not alter the levels of Ni column isolated K18 in His-Ub transfected BHK cells (not shown). In addition, the turnover of 35S-met labeled K18 S33→A/D is not significantly different as compared with 35S-met labeled WT K18 (Fig. 4 C).
Discussion
The overall findings of this study are: (a) K8 and K18 can be ubiquitinated in vivo, particularly when each keratin is expressed individually (Fig. 1), and the ubiquitinated species are associated with the noncytosolic fraction (Fig. 2B and Fig. C). (b) Keratin ubiquitination is associated with its degradation since proteasome inhibition also inhibits keratin degradation (Fig. 2). Although keratins are highly stable proteins, ALLN inhibition of keratin degradation despite coexpression of types I and II keratins (Fig. 2 A) suggests that keratin turnover by ubiquitination is a physiologic process that is likely difficult to appreciate due to its rapidity and to keratin abundance. (c) Proteasome inhibition results in accumulation of phosphorylated K8 and K18 with hyper- or neo-phosphorylation of most (i.e., K8 S431/73, K18 S52) but not all sites (e.g., K18 S33; see Fig. 3). In the case of K8, its phosphorylation protects from, but does not prevent, keratin ubiquitination (Fig. 4). This protection does not appear to be site specific, and involves K8 S23/73/431 which undergo phosphorylation during several biologic contexts (Omary et al. 1998). In the case of K18, an association of its phosphorylation with ubiquitination remains unclear. Although Ub-mediated degradation of many proteins is facilitated by phosphorylation (Hershko and Ciechanover 1998; Laney and Hochstrasser 1999), several proteins are protected by phosphorylation as noted for p53 (Chehab et al. 1999), c-fos (Okazaki and Sagata 1995), c-Jun (Musti et al. 1997), and K8 (shown herein). It is, however, possible that other keratin phosphorylation sites may turn out to serve a degradation-promoting role. Similar modes of ubiquitination and degradation, in phosphorylation-dependent or -independent fashions, are likely to involve other IF proteins.
Ubiquitination and degradation may be coupled or uncoupled since degradation requires an active proteasome. Examples of uncoupling that apply to most if not all IF proteins are the codecoration of Ub and IF proteins as reported in several cytoplasmic deposits in association with a number of diseases (e.g., Lowe et al. 1988; Ohta et al. 1988; Akamatsu et al. 1995). One potential explanation is that the inclusion-associated diseases result in states whereby the proteasome capacity is exceeded (Johnston et al. 1998) or inhibited, with subsequent accumulation of ubiquitinated proteins within aggresomes. In the case of MB, which stain with antibodies to keratins and Ub, keratin hyperphosphorylation (Stumptner et al. 2000) and increased K8 and K18 synthesis (Cadrin et al. 1995; Hutter et al. 1993) occur as an early event during their formation. These data suggest a model whereby liver injury induces increased keratin synthesis and phosphorylation. Increased keratin synthesis, particularly if unbalanced between types I and II keratins, may then predispose to keratin ubiquitination. Keratin hyperphosphorylation in vivo protects against liver injury (Ku et al. 1998b) and as reported herein may also protect (in the case of K8) but not prevent keratin ubiquitination. Ubiquitinated and hyperphosphorylated keratins may then accumulate if the proteasome capacity is overloaded or suppressed.
Acknowledgments
We are grateful to Dr. Ron Kopito for supplying the His-tagged ubiquitin construct, and to Kris Morrow for preparing the figures.
This work was supported by Veterans Administration Merit and Career Development awards and National Institutes of Health grant DK52951.
Footnotes
Abbreviations used in this paper: Ab, antibody; ALLN, N-acetyl-Leu-Leu-norleucinal; IF, intermediate filament(s); K, keratin; MB, Mallory bodies; Ub, ubiquitin.
References
- Akamatsu M., Hori S., Tsutsumi Y., Osamura R.Y., Ohkido M. Ubiquitinated cytokeratin inclusions in lichen amyloidosusan immunohistochemical analysis. Pathol. Int. 1995;45:116–122. doi: 10.1111/j.1440-1827.1995.tb03431.x. [DOI] [PubMed] [Google Scholar]
- Cadrin M., Anderson N.M., Aasheim L.H., Kawahara H., Franks D.J., French S.W. Modifications in cytokeratin and actin in cultured liver cells derived from griseofulvin-fed mice. Lab. Invest. 1995;72:453–460. [PubMed] [Google Scholar]
- Caulin C., Salvesen G.S., Oshima R.G. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J. Cell Biol. 1997;138:1379–1394. doi: 10.1083/jcb.138.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab N.H., Malikzay A., Stavridi E.S., Halazonetis T.D. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA. 1999;961:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipev C.C., Korge B.P., Markova N., Bale S.J., DiGiovanna J.J., Compton J.G., Steinert P.M. A leucine→proline mutation in the H1 subdomain of keratin 1 causes epidermolytic hyperkeratosis. Cell. 1992;70:821–828. doi: 10.1016/0092-8674(92)90315-4. [DOI] [PubMed] [Google Scholar]
- Chou C.-F., Smith A.J., Omary M.B. Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J. Biol. Chem. 1992;267:3901–3906. [PubMed] [Google Scholar]
- Chou C.-F., Riopel C.L., Rott L.S., Omary M.B. A significant soluble keratin fraction in “simple” epithelial cellsLack of an apparent phosphorylation and glycosylation role in keratin solubility. J. Cell Sci. 1993;105:433–445. doi: 10.1242/jcs.105.2.433. [DOI] [PubMed] [Google Scholar]
- Denk H., Lackinger E., Zatloukal K., Franke W.W. Turnover of cytokeratin polypeptides in mouse hepatocytes. Exp. Cell Res. 1987;173:137–143. doi: 10.1016/0014-4827(87)90339-9. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Weber K. Intermediate filamentsstructure, dynamics, function and disease. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Cleveland D.W. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hutter H., Zatloukal K., Winter G., Stumptner C., Denk H. Disturbance of keratin homeostasis in griseofulvin-intoxicated mouse liver. Lab. Invest. 1993;69:576–582. [PubMed] [Google Scholar]
- Jensen K., Gluud C. The Mallory bodymorphological, clinical and experimental studies. Hepatology. 1994;20:1061–1077. doi: 10.1002/hep.1840200440. [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. Aggresomesa cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.-O., Omary M.B. Identification of the major physiologic phosphorylation site of human keratin 18potential kinases and a role in filament reorganization. J. Cell Biol. 1994;127:161–171. doi: 10.1083/jcb.127.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.-O., Omary M.B. Phosphorylation of human keratin 8 in vivo at conserved head domain serine 23 and at epidermal growth factor-stimulated tail domain serine 431. J. Biol. Chem. 1997;272:7556–7564. doi: 10.1074/jbc.272.11.7556. [DOI] [PubMed] [Google Scholar]
- Ku N.-O., Liao J., Omary M.B. Apoptosis generates stable fragments of human type I keratins. J. Biol. Chem. 1997;272:33197–33203. doi: 10.1074/jbc.272.52.33197. [DOI] [PubMed] [Google Scholar]
- Ku N.-O., Liao J., Omary M.B. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins EMBO (Eur. Mol. Biol. Organ.) J. 17 1998. 1892 1906a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.-O., Michie S.A., Soetikno R.M., Resurreccion E.Z., Broome R.L., Omary M.B. Mutation of a major keratin phosphorylation site predisposes to hepatotoxic injury in transgenic mice J. Cell Biol. 143 1998. 2023 2032b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.-O., Zhou X., Toivola D.M., Omary M.B. The cytoskeleton of digestive epithelia in health and disease. Am. J. Physiol. 1999;277:G1108–G1137. doi: 10.1152/ajpgi.1999.277.6.G1108. [DOI] [PubMed] [Google Scholar]
- Laney J.D., Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Liao J., Lowthert L.A., Ku N.-O., Fernandez R., Omary M.B. Dynamics of human keratin 18 phosphorylationPolarized distribution of phosphorylated keratins in simple epithelial tissues. J. Cell Biol. 1995;131:1291–1301. doi: 10.1083/jcb.131.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Ku N.-O., Omary M.B. Stress, apoptosis, and mitosis induce phosphorylation of human keratin 8 at ser73 in tissues and cultured cells. J. Biol. Chem. 1997;272:17565–17573. doi: 10.1074/jbc.272.28.17565. [DOI] [PubMed] [Google Scholar]
- Lowe J., Blanchard A., Morrell K., Lennox G., Reynolds L., Billett M., Landon M., Mayer R.J. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and Mallory bodies in alcoholic liver disease. J. Pathol. 1988;155:9–15. doi: 10.1002/path.1711550105. [DOI] [PubMed] [Google Scholar]
- Moll R., Franke W.W., Schiller D.L., Geiger B., Krepler R. The catalog of human cytokeratinspatterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Montes J.F., Estrada G., Lopez-Tejero M.D., Garcia-Valero J. Changes in the enterocyte cytoskeleton in newborn rats exposed to ethanol in utero. Gut. 1996;38:846–852. doi: 10.1136/gut.38.6.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F.B., Kuster W., Bruckner-Tuderman L., Korge B.P. Novel K5 and K14 mutations in German patients with the Weber-Cockayne variant of epidermolysis bullosa simplex. J. Invest. Dermatol. 1998;111:900–902. doi: 10.1046/j.1523-1747.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Musti A.M., Treier M., Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- Nishibori H., Matsuno Y., Iwaya M., Osada T., Kubomura N., Iwamatsu A., Kohno H., Sato S., Kitajima M., Hirohashi S. Human colorectal carcinomas specifically accumulate Mr 42,000 ubiquitin-conjugated cytokeratin 8 fragments. Cancer Res. 1996;56:2752–2757. [PubMed] [Google Scholar]
- Ohta M., Marceau N., Perry G., Manetto V., Gambetti P., Autilio-Gambetti L., Metuzals J., Kawahara H., Cadrin M., French S.W. Ubiquitin is present on the cytokeratin intermediate filaments and Mallory bodies of hepatocytes. Lab. Invest. 1988;59:848–856. [PubMed] [Google Scholar]
- Okazaki K., Sagata N. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:5048–5059. doi: 10.1002/j.1460-2075.1995.tb00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M.B., Ku N.-O., Liao J., Price D. Keratin modifications and soluble properties in epithelial cells and in vitro. Subcell. Biochem. 1998;31:105–139. [PubMed] [Google Scholar]
- Stumptner C., Omary M.B., Denk H., Zatloukal K. Hepatocyte cytokeratins are hyperphosphorylated at multiple sites in human alcoholic hepatitis and in a Mallory body mouse model. Am. J. Pathol. 2000;156:77–90. doi: 10.1016/S0002-9440(10)64708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Schneider R.J. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J. Virol. 1994;68:2544–2555. doi: 10.1128/jvi.68.4.2544-2555.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]