Abstract
How plant organs grow to reach their final size is an important but largely unanswered question. Here, we describe an Arabidopsis thaliana mutant, brassinosteroid-insensitive4 (bin4), in which the growth of various organs is dramatically reduced. Small organ size in bin4 is primarily caused by reduced cell expansion associated with defects in increasing ploidy by endoreduplication. Raising nuclear DNA content in bin4 by colchicine-induced polyploidization partially rescues the cell and organ size phenotype, indicating that BIN4 is directly and specifically required for endoreduplication rather than for subsequent cell expansion. BIN4 encodes a plant-specific, DNA binding protein that acts as a component of the plant DNA topoisomerase VI complex. Loss of BIN4 triggers an ATM- and ATR-dependent DNA damage response in postmitotic cells, and this response coincides with the upregulation of the cyclin B1;1 gene in the same cell types, suggesting a functional link between DNA damage response and endocycle control.
INTRODUCTION
The control of organ growth is a fundamental question in plant development, but the intrinsic mechanisms that mediate this process remain largely unknown. The final size of a plant organ is defined by genetic programs, but environmental factors such as nutrition, temperature, and light can also impinge on those programs to develop an organ size appropriate to the surrounding growth conditions. Several plant growth regulators, such as auxin, brassinosteroids (BRs), ethylene, and gibberellins, are also known to modulate organ growth. Given that the overall size of an organ depends on both the number and size of its cells, all of these upstream signals must be transduced into mechanisms that regulate cell proliferation and cell expansion. Recent genetic studies have identified a number of genes that influence these processes and have begun to uncover the molecular and cellular bases of organ growth in plants. The primary driver of organ growth is the addition of new cells into a developing organ; thus, a key control exists in determining the duration of cell proliferation during organogenesis. This process is mediated by several independent genetic pathways that either promote or terminate the maintenance of cellular meristematic competence (Anastasiou and Lenhard, 2007). In addition, many plant cell types undergo massive postmitotic cell expansion, and the resulting increase in mass also contributes to organ growth. Cell expansion is driven by water uptake into vacuoles and cell wall biogenesis; consistent with this, many mutations that result in defects in organ growth occur in genes involved in one or the other of these processes (Rojo et al., 2001; Cosgrove, 2005). In addition, another set of mutations that alters the final size of plant cells points to the control of cell expansion by endoreduplication, the amplification of chromosomal DNA without corresponding mitosis (Sugimoto-Shirasu and Roberts, 2003). The correlation between ploidy, nuclear DNA content, and cell size has long been reported in Arabidopsis thaliana and in many other plant species (Nagl, 1978; Galbraith et al., 1991; Melaragno et al., 1993), although the molecular details of how cells endoreduplicate and how an increase in ploidy might lead to an increase in cell size still remain elusive. Recent genetic studies have revealed a number of key cell cycle genes involved in the switch from the mitotic cell cycle to the endocycle (Inze and De Veylder, 2006) and other positive and negative regulators that function during the progression of successive endocycles (Hulskamp, 2004) or at their termination (Imai et al., 2006). It is well established that various checkpoint mechanisms ensure progression through the mitotic cell cycle. It is reasonable to predict that similar mechanisms operate during endocycles, but the existence of such controls has been poorly investigated.
To gain further insights into the molecular and cellular mechanisms that underlie plant organ growth, we have characterized a previously identified mutant in Arabidopsis, brassinosteroid-insensitive4 (bin4), that displays a severe dwarf phenotype (Yin et al., 2002). We show that the primary defect in bin4 is in cell expansion rather than in cell proliferation and that this is associated with a failure in the progression of endocycles in appropriate cell types. Our study clearly demonstrates a close link between cell size and ploidy and provides genetic evidence that cell expansion requires, and is coupled to, an increase in ploidy through endoreduplication. BIN4 encodes a plant-specific, DNA binding protein, and our genetic and in vivo evidence suggests that BIN4 is a new component of the plant DNA topoisomerase VI (topo VI) complex. We also show that the loss of functional BIN4, or other plant topo VI components, activates the expression of several DNA damage response genes. This response is mediated through the two upstream phosphatidylinositol 3-kinase (PI3K)–like protein kinases, ataxia telangiectasia–mutated (ATM) and ATM and Rad3–related (ATR), that are known to initiate various DNA damage repair processes during the mitotic cell cycle. We propose that the structural integrity of the genome is closely monitored during plant endocycles and that the cellular response to damaged DNA leads to an early arrest of endocycles.
RESULTS
The BR-Insensitive Mutant bin4 Has Cell and Organ Size Defects
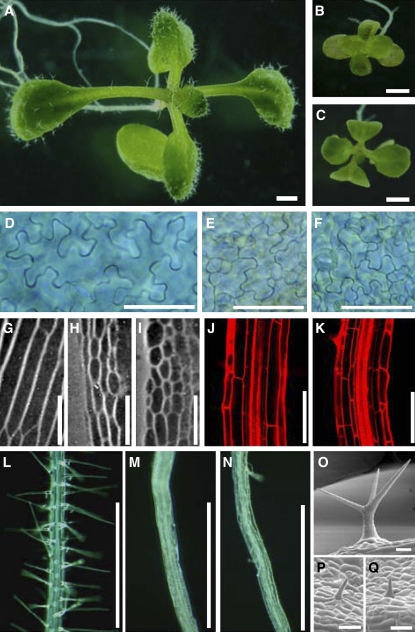
An extreme dwarf mutant, bin4, was originally identified in a screen for mutants that have reduced sensitivity to BR (Yin et al., 2002). Like other BR-insensitive mutants, organ growth is severely compromised in bin4. Twelve-day-old, light-grown bin4-1 seedlings have small cotyledons and true leaves with short petioles (Figures 1A to 1C; see Supplemental Table 1 online). The size of other organs, such as hypocotyls and roots, is also reduced to 15 to 35% of the wild-type level (see Supplemental Table 1 online). The application of exogenous BR or other plant growth regulators, including auxin, gibberellin, and ethylene, does not rescue the small size phenotype in bin4-1 (data not shown). To examine the cellular basis of these reduced organ growth phenotypes in bin4, we cleared cotyledons from 8-d-old, light-grown seedlings and measured cell number and cell size on the abaxial surface. As shown in Figure 1D, wild-type cotyledons develop many large, heavily interdigitated epidermal cells. We found that the size of this cell type is dramatically reduced in bin4-1 (Figure 1E), from an average of 5032 ± 2699 μm2 in the wild type to 790 ± 314 μm2 in bin4-1 (n = 50), whereas total estimated cell number per cotyledon surface is comparable between the wild type (1543 cells) and the mutant (1480 cells) (see Supplemental Table 1 online). The small cell size phenotype was also observed in hypocotyls (Figures 1G to 1I) and roots (Figures 1J and 1 K), and the quantification of cell size in these organs revealed that their cell sizes are reduced to ∼25% of wild-type levels (see Supplemental Table 1 online). This defect in cell size is not limited to the epidermal cell layer but is also present in cortex and endodermis (Figures 1G to 1K; see Supplemental Table 1 online). Both root hairs and leaf trichomes are composed of a single cell in Arabidopsis, and the reduced cell size in bin4-1 resulted in more pronounced developmental defects in these cell types (i.e., almost complete lack of root hair formation [Figures 1L to 1N] and unbranched, miniature trichomes [Figures 1O to 1Q]).
Figure 1.
Dwarf and Reduced Cell Size Phenotypes of bin4.
(A) to (C) Twelve-day-old, light-grown seedlings.
(D) to (F) Differential interference contrast micrographs of 12-d-old cotyledon epidermis after clearing with chloral hydrate.
(G) to (I) Differential interference contrast micrographs of agar imprints of 3-d-old, dark-grown hypocotyls.
(J) and (K) Optical sections of 8-d-old, light-grown roots stained with propidium iodide (1 μg/mL).
(L) to (N) Twelve-day-old roots.
(O) to (Q) Scanning electron micrographs of mature trichomes.
(A), (D), (G), (J), (L), and (O) show wild-type Columbia ecotype; (B), (E), (H), (K), (M), and (P) show bin4-1; and (C), (F), (I), (N), and (Q) show bin4-2. Bars = 1 mm in (A) to (C), 50 μm in (D) to (I), 100 μm in (J) to (N), and 50 μm in (O) to (Q).
bin4 Is Defective in Increasing Ploidy by Endoreduplication
Various cell types in Arabidopsis support at least a few rounds of endocycle, and the resulting higher ploidy often positively correlates with larger cell size (Galbraith et al., 1991; Melaragno et al., 1993). Given that reduced cell size is the major contributor to the organ growth defect in bin4, we examined whether cells in bin4 are impaired in endoreduplication. Our flow cytometric analysis, using 15,000 nuclei from 12-d-old leaves, indicates that the ploidy of wild-type leaves ranges from 2C to 32C, having a highest peak at 4C (see Supplemental Figures 1A and 1P online). By sharp contrast with this, we found that the ploidy in 12-d-old leaf cells of bin4-1 reaches only 8C (see Supplemental Figures 1B and 1P online). Although the overall peak pattern is similar between the wild type and bin4-1, all of the peaks in bin4-1 are smaller and broader than those in the wild type and the interpeak signals tend to be higher in bin4-1, further suggesting mechanistic faults in the endocycle progression in bin4 (see Supplemental Figure 1B online). Moreover, our fluorescence microscopy revealed that nuclear size is significantly smaller in many cell types of bin4-1, including leaf trichomes (see Supplemental Figures 1E and 1K online), root hairs (see Supplemental Figures 1F and 1L online), leaf marginal epidermis (see Supplemental Figures 1G and 1M online), root cortex (see top panels of Supplemental Figures 1H and 1N online), and root metaxylem cells (see top panels of Supplemental Figures 1I and 1O online). It is generally thought that nuclear size and ploidy are tightly linked; accordingly, our immunocytochemistry of 5-bromo-2-deoxyuridine (BrdU)–incorporated nuclei demonstrates that the large nuclei in wild-type cells do arise from the replication of DNA (see lower panels of Supplemental Figures 1H and 1I online) and that this process ceases prematurely in bin4-1 (see lower panels of Supplemental Figures 1N and 1O online). The double mutant analyses with the overendoreduplicated, overbranched trichome mutants triptychon (try), rastafari (rfi), and kaktus2 (kak2) and the ectopic root hair mutant glabra2 (gl2) showed that none of these mutations modify the bin4 phenotype (see Supplemental Figure 2 online). These results suggest that BIN4 functions genetically in a pathway that is either downstream or independent of these negative regulators in trichome and root hair formation.
Colchicine Treatment Induces Polyploidization and Partly Rescues the Cell and Organ Size Phenotypes in bin4
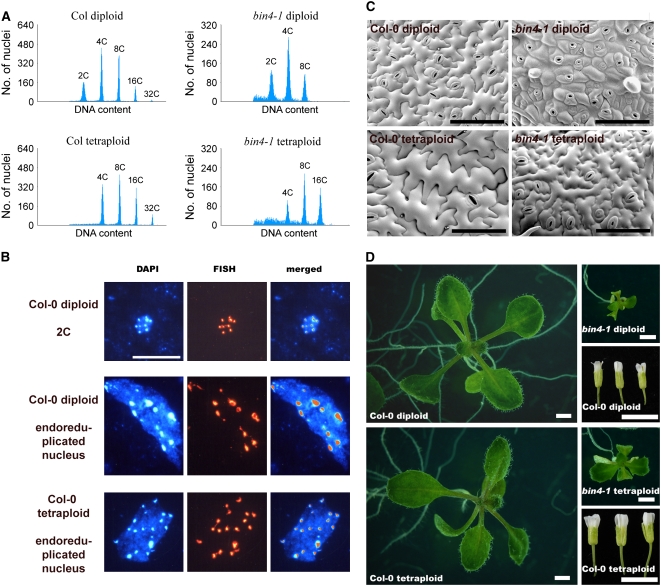
Increasing nuclear DNA content by endoreduplication is thought to provide a license for an increase in cell size (Sugimoto-Shirasu and Roberts, 2003), and the correlated cell size and ploidy phenotypes in bin4-1 further support this functional link. We generated tetraploid plants in Arabidopsis to determine whether an increase in nuclear DNA content is sufficient to promote cell expansion. We treated young shoot apices of wild-type and bin4-1 heterozygous plants with 0.05% colchicine and identified their putative tetraploid progeny from the self-fertilized plants. Our flow cytometric analysis confirmed that the ploidy peaks shifted to higher C values, starting from 4C in both wild-type and bin4-1 tetraploid plants (Figure 2A). Consistent with this, our fluorescence in situ hybridization (FISH) analysis using a probe specific to centromeric repetitive DNA revealed that nuclei in their tetraploid cells have twice the diploid number of centromeres (Figure 2B), suggesting that the number of chromosomes is doubled in tetraploid plants and that this contributes to the doubling of their total nuclear DNA content.
Figure 2.
Colchicine-Induced Polyploidization Partly Rescues the Cell and Organ Size Phenotype in bin4.
(A) Flow cytometric analysis of 12-d-old, light-grown seedlings.
(B) FISH analysis of endoreduplicated polytene nuclei in wild-type diploid plants and polyploid nuclei in colchicine-induced tetraploid plants.
(C) Scanning electron micrographs of 14-d-old leaf epidermis.
(D) Twelve-day-old, light-grown seedlings and mature flowers. Organ size is more than doubled in bin4 tetraploid plants. Polyploidization appears to have much less impact on the wild-type organ size, but its flower size is significantly increased.
Col and Col-0, Columbia ecotype; DAPI, 4′,6-diamidino-2-phenylindole. Bars = 10 μm in (B), 200 μm in (C), 1 mm in (D) (seedlings), and 5 mm in (D) (flowers).
We found that the size of leaf epidermal cells from 10-d-old wild-type tetraploid plants increased dramatically compared with diploid plants at the same age (Figure 2C), clearly demonstrating that increasing the nuclear DNA content promotes cell expansion. The cell size is also improved in bin4-1 tetraploid plants, suggesting that BIN4 is not directly involved in the cellular processes that underlie cell expansion. The frequency of trichomes with four branches was increased in wild-type tetraploid leaves, and overall trichome development was also improved in bin4-1 tetraploid leaves (data not shown). These increases in cell size in tetraploid plants contribute to the partial recovery of organ size in bin4-1 (Figure 2D).
BIN4 Encodes a Nuclear DNA Binding Protein
By positional cloning, we found that the BIN4 locus corresponds to the gene At5g24630 (see Supplemental Figure 3A online). Sequencing of genomic DNA from bin4-1 identified a 2-bp (GC) deletion in the predicted BIN4 coding sequence, which should produce a truncated protein lacking the last 92 amino acids of the wild-type protein, with its final 5 amino acids substituted from QLIPQ to HDTTI (see Supplemental Figure 3B online). We identified a T-DNA insertion allele of this gene, named bin4-2 (SALK_110750), from the T-DNA mutant collection of the Salk Institute (Alonso et al., 2003) (see Supplemental Figure 3A online) and found that it has severe cell size and ploidy phenotypes indistinguishable from those of bin4-1 (Figure 1; see Supplemental Figure 1 online). In addition, F1 progeny produced by genetic crosses between bin4-1 and bin4-2 show similar dwarf phenotypes (data not shown), formally confirming the identity of the BIN4 gene. The BIN4 locus consists of 13 exons and 12 introns (see Supplemental Figure 3A online). As predicted, the bin4-1 mutation does not interrupt the transcription of full-length mRNA, but the T-DNA insertion within the third exon of bin4-2 completely abolishes its transcription (see Supplemental Figure 3C online). Our immunoblot analysis using a BIN4-specific antiserum, however, revealed that neither bin4-1 nor bin4-2 produces any detectable BIN4 protein (see Supplemental Figure 3D online), suggesting that functionally they are probably both null alleles.
A PHI-BLAST sequence search revealed that BIN4 is a single-copy gene in Arabidopsis and appears to be plant-specific. The predicted BIN4 protein does not show strong similarity to any protein in public databases, except that its C terminus contains short sequences that are similar to the DNA binding domain of a High-Mobility Group protein (see Supplemental Figure 4A online). Importantly, the corresponding sequences in BIN4 possess an RGR motif, also called an AT hook, that is found in most High-Mobility Group proteins and shows strong binding to AT-rich DNA sequences (see Supplemental Figure 4D online). Consistent with this, surface plasmon resonance experiments demonstrated that the prokaryotically overexpressed His:S:BIN4 protein strongly binds DNA in a concentration- and salt-dependent manner (see Supplemental Figures 4B and 4C online). Other motifs that are present in the BIN4 protein include a putative nuclear localization signal (KRGRPSKEKQPPAKKAR) located in the C-terminal part of the protein, suggesting that BIN4 might function within the nucleus. Indeed, we found that the BIN4:GFP (for green fluorescent protein) fusion protein, expressed under the control of a cauliflower mosaic virus 35S promoter in Arabidopsis leaf epidermal cells, is targeted to the nucleus (see Supplemental Figures 4F and 4G online). Our sequence analysis also identified two highly conserved repeat peptide domains in the middle part of BIN4 (see Supplemental Figures 4A and 4E online) that may have a role in BIN4 function.
BIN4 Is a Component of the Plant DNA Topoisomerase VI Complex
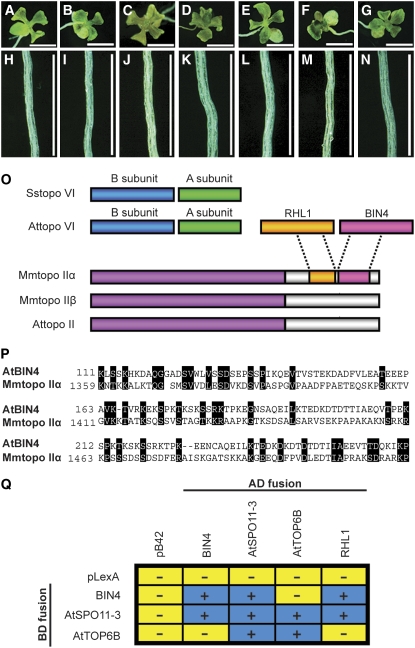
The cell size and ploidy phenotypes found in bin4 are very similar to those of a group of other dwarf mutants that lack components of the plant topo VI complex (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002, 2005; Yin et al., 2002) (Figures 3A to 3D and 3H to 3K). Therefore, we tested the genetic relationship between bin4 and the topo VI mutants hypocotyl7 (hyp7), rhl2, and hyp6. As shown in Figures 3E to 3G and 3L to 3N, the leaf and root phenotypes of bin4-1 hyp7, bin4-1 rhl2, and bin4-1 hyp6 double mutants are indistinguishable from those of their respective parents, suggesting that BIN4 acts in the same pathway or complex as RHL1/HYP7, At SPO11-3/RHL2/BIN5, and At TOP6B/RHL3/HYP6/BIN3. In addition, PHI-BLAST revealed that the N-terminal part of BIN4 has weak sequence homology with the C-terminal 104 amino acids of animal topo IIα, with a total of 28% identical and 12% similar amino acids (Figures 3O and 3P). We previously reported that the C-terminal region of animal topo IIα is weakly related to RHL1/HYP7 (Sugimoto-Shirasu et al., 2005). Strikingly, we found that this domain lies just proximal to the domain that has similarity to BIN4 (Figure 3O), strongly suggesting that BIN4 functions as a part of the plant topo VI complex. We performed a yeast two-hybrid assay to determine whether BIN4 can physically interact with components of the plant topo VI complex. As shown in Figure 3Q, BIN4 can interact with both At SPO11-3/RHL2/BIN5 (i.e., topo VI subunit A) and RHL1/HYP7 but not directly with At TOP6B/RHL3/HYP6/BIN3 (i.e., topo VI subunit B). BIN4 can also self-interact in vivo (Figure 3Q), suggesting that BIN4 may participate as a dimer, or dimers, in topo VI.
Figure 3.
Genetic and in Vivo Interactions between BIN4, RHL1/HYP7, At SPO11-3/RHL2/BIN5, and At TOP6B/RHL3/HYP6/BIN3.
(A) to (N) Ten-day-old, light-grown seedlings ([A] to [G]) and roots ([H] to [N]) of bin4-1 ([A] and [H]), hyp7 ([B] and [I]), hyp6 ([C] and [J]), rhl2 ([D] and [K]), bin4-1 hyp7 ([E] and [L]), bin4-1 hyp6 ([F] and [M]), and bin4-1 rhl2 ([G] and [N]). Bars = 2 mm in (A) to (G) and 0.5 mm in (H) to (N).
(O) Schematic representation of type II DNA topoisomerases from archaea, animals, and plants. Sstopo VI, topo VI from Sulfolobus shibatae; Attopo VI, topo VI from Arabidopsis; RHL1, RHL1 from Arabidopsis; BIN4, BIN4 from Arabidopsis; Mmtopo II, topo II from Mus musculus; Attopo II, topo II from Arabidopsis. RHL1/HYP7 and BIN4 exhibit partial sequence homology with the C-terminal domain of Mm topo IIα.
(P) Sequence alignment of the N-terminal domain of At BIN4 and the C-terminal domain of Mm topo IIα (GenBank accession number NP035753). Numbers refer to the amino acid length of each deduced protein, and black boxes represent similar or identical amino acids.
(Q) Yeast two-hybrid analysis of BIN4, RHL1/HYP7, At SPO11-3/RHL2/BIN5, and At TOP6B/RHL3/HYP6/BIN3 protein interactions. The BIN4, RHL1/HYP7, At SPO11-3/RHL2/BIN5, and At TOP6B/RHL3/HYP6/BIN3 genes were cloned into pLexA (BD, binding domain fusion) and pB42AD (AD, activator domain fusion) vectors, and their protein interactions were detected by induction (+, blue boxes) or no induction (−, yellow boxes) of the lacZ reporter gene.
The C-terminal domain of animal topo IIα may be required for the translocation of topo II into the nucleus (Adachi et al., 1997). We explored whether RHL1/HYP7 and BIN4 play similar functions in the plant topo VI complex by overexpressing previously reported At SPO11-3/RHL2/BIN5:GFP proteins (Sugimoto-Shirasu et al., 2002) in the rhl1-2 and bin4-2 background. These proteins are normally transported into the nucleus (Sugimoto-Shirasu et al., 2002), and as shown in Supplemental Figures 4H and 4I online, neither the rhl1 nor the bin4 mutation interferes with its nuclear translocation. Thus, RHL1/HYP7 and BIN4 appear to play some function other than bringing the topo VI complex into the nucleus.
BIN4 Is Expressed in Both Proliferating Cells and Endoreduplicating Cells
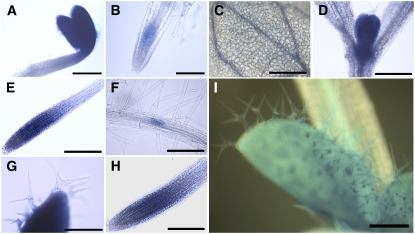
We examined the developmental pattern of the BIN4 promoter activity by fusing the 2-kb genomic region upstream of the BIN4 initiation codon to the β-glucuronidase (GUS) reporter gene. The histochemical analysis revealed that the BIN4 promoter is active in endoreduplicating cells, including those in young, rapidly expanding cotyledons (Figure 4A), vascular cells (Figure 4C), elongating root cells (Figures 4E and 4H), and developing leaf trichomes (Figures 4G and 4I). The GUS signal is also detectable in various tissues that contain proliferating cells, such as root and shoot apical meristems (Figures 4B, 4D, 4E, and 4H) and lateral root primordia (Figure 4F). The publicly available microarray data, using synchronized Arabidopsis cell cultures (Menges et al., 2005), revealed that BIN4 is ubiquitously expressed at low levels during the mitotic cell cycle (see Supplemental Figure 5 online). This pattern of expression is similar to that of other components of the plant topo VI complex (i.e., RHL1/HYP7, At SPO11-3/RHL2/BIN5, and At TOP6B/RHL3/HYP6/BIN3) but is markedly different from that of At TOPII (see Supplemental Figure 5 online), supporting our previous finding that plant topo II and topo VI have some distinct function in vivo (Sugimoto-Shirasu et al., 2005).
Figure 4.
BIN4 Is Expressed in Both Proliferating Cells and Endoreduplicating Cells.
pBIN4:GUS expression is detected in cotyledons (A) and root tips (B) of 2-d-old seedlings, in cotyledons (C) and primary leaves (D) of 4-d-old seedlings, in the root tip (E), at the lateral root initiation site (F), and in an emerging leaf (G) of 6-d-old seedlings, and in the root tip (H) and developing leaves (I) of 8-d-old seedlings. GUS expression was studied in more than five independent transformants generated with this construct, and they all showed the same expression pattern. Bars = 500 μm in (A), (B), and (I), 200 μm in (C) to (F) and (H), and 100 μm in (G).
The bin4 Mutation Triggers the Ectopic Expression of DNA Damage Response Genes in Nondividing Cells
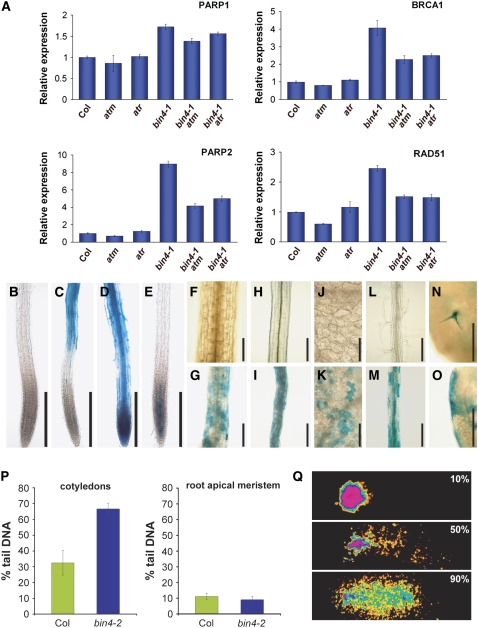
The predicted in vivo function of the plant topo VI complex is to untangle chromosomes during DNA replication, and any failure in this process leads to an accumulation of double-strand breaks (DSBs) in topo VI mutants (Hartung et al., 2002). To further explore these observations, we examined the level of DSBs by comet assays using 8-d-old cotyledons and root tips (Figure 5P). The majority of nuclei from cotyledons of this age undergo some level of endoreduplication, whereas nuclei in the root apical meristem remain in the mitotic cell cycle (data not shown). Arabidopsis plants accumulate more DSBs as they mature (Boyko et al., 2006); accordingly, nuclei isolated from wild-type cotyledons contain more DSBs (∼30% of total DNA) than those from root apical meristems (∼10% of total DNA) (Figure 5P). In addition, we found that the nuclei from bin4-2 cotyledons accumulate more than twice as many DSBs (∼65% of total DNA) as those from wild-type cotyledons (Figure 5P). By contrast, we detected no significant difference in DSB levels in nuclei prepared from wild-type and bin4-2 root meristems (Figure 5P). In addition, our real-time RT-PCR analysis revealed that several DSB-inducible genes, including poly (ADP-ribose) polymerase1 (At PARP1) (Doucet-Chabeaud et al., 2001), At PARP2 (Babiychuk et al., 1998), At RAD51 (Doutriaux et al., 1998), and breast cancer susceptibility1 (At BRCA1) (Lafarge and Montane, 2003), were significantly upregulated in bin4 (Figure 5A).
Figure 5.
The bin4 Mutation Triggers an ATM/ATR-Dependent DNA Damage Response in Postmitotic Cells.
(A) Quantitative real-time expression analysis of the DSB-inducible genes PARP1, PARP2, BRCA1, and RAD51 in 14-d-old wild-type, bin4-1, atm, atr, bin4-1 atm, and bin4-1 atr seedlings. The Actin2 expression level was used as a reference. The values represent averages of four independent replicates ± sd. Col, Columbia ecotype.
(B) to (E) pPARP2:GUS expression in wild-type primary roots (B), bin4-1 primary roots (C), wild-type primary roots exposed to 10 μg/mL bleomycin for 21 h (D), and wild-type primary roots exposed to 100 μM etoposide for 21 h (E).
(F) to (O) pPARP2:GUS expression in 7-d-old, light-grown hypocotyls ([F] and [G]), 4-d-old, dark-grown hypocotyls ([H] and [I]), mature cotyledons ([J] and [K]), mature roots ([L] and [M]), and mature leaves ([N] and [O]). (F), (H), (J), and (L) show wild-type organs, and (G), (I), (K), and (M) to (O) show bin4-1 organs.
(P) Statistical analysis of a comet assay comparing relative DNA damage in nuclei of 8-d-old wild-type and bin4-2 cotyledons (left panel) and root apical meristems (right panel). The means of 70 comets from five individual gels were assessed per sample. Error bars represent sd.
(Q) Examples of comets exhibiting 10% (top), 50% (middle), and 90% (bottom) tail DNA in comets.
Bars = 200 μm in (B) to (E) and (H) to (K), 100 μm in (F) and (G), and 250 μm in (L) to (O).
We further investigated the expression pattern of At PARP2 by introducing the pPARP2:GUS construct (Babiychuk et al., 1998) into the bin4-1 background. The histochemical analysis revealed that the At PARP2 gene is normally expressed at an undetectable level (Figures 5B, 5F, 5H, 5J, and 5L). To our surprise, however, we found that the bin4 mutation induces the expression of At PARP2 predominantly in postmeristematic cells (Figure 5C). This is in a sharp contrast with the bleomycin-treated wild-type roots, which exhibited a broad pattern of At PARP2 expression in both proliferating cells and postmeristematic cells (Figure 5D). Arabidopsis topo II (At TOPII) is implicated in playing a key role in the mitotic cell cycle (Sugimoto-Shirasu et al., 2005), and in agreement with this, wild-type roots, treated with etoposide, a topo II poison, display meristem-specific At PARP2 expression (Figure 5E). In addition, we detected similar ectopic expression of At PARP2 in other postproliferating tissues, including light- and dark-grown hypocotyls (Figures 5G and 5I, respectively), cotyledons (Figure 5K), mature part of roots (Figure 5M), leaf trichomes (Figure 5N), and leaf marginal cells (Figure 5O), in bin4. These results suggest that the induced transcriptional activation of DNA damage response genes is restricted to the nonproliferating cells that have undergone several endocycles.
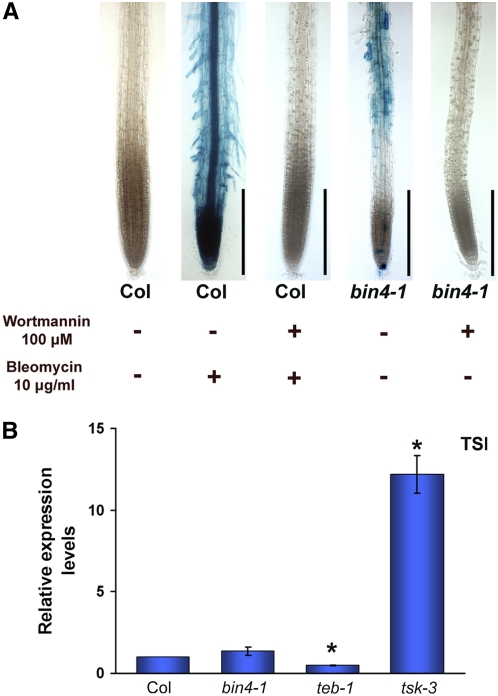
Several Arabidopsis mutants, including fasciata1 (fas1) and brushy1/tonsoku/mgoun3 (bru1/tsk/mgo3), exhibit constitutive upregulation of DNA damage genes, and this response is associated with the stochastic release of transcriptional gene silencing (TGS) (Takeda et al., 2004). This is not the case, however, for the tebichi (teb) mutant, in which elevated expression of DNA damage genes is not linked to the instability of epigenetic states (Inagaki et al., 2006). To test whether bin4 is defective in maintaining TGS, we examined the expression level of transcriptionally silent information (TSI) repeats that are known to be an endogenous target of TGS in Arabidopsis (Steimer et al., 2000). As shown in Figure 6B, our real-time RT-PCR analysis revealed that the expression of TSI repeats in bin4 is indistinguishable from that in the wild type and is strongly repressed compared with that of the tsk3 mutant, which accumulates abnormal levels of TSI transcripts.
Figure 6.
The PI3K-Like Kinase Inhibitor Wortmannin Can Block the DNA Damage Response in bin4, and Transcriptional Gene Silencing Is Not Released in bin4.
(A) The PI3K-like kinase inhibitor wortmannin strongly inhibits the bleomycin-induced pPARP2:GUS expression in wild-type roots and the ectopic expression of pPARP2:GUS in bin4 roots. Five-day-old wild-type and bin4 seedlings that carry the pPARP2:GUS construct were grown in the presence (+) or absence (−) of 100 μM wortmannin and 10 μg/mL bleomycin for 2 d. The activation of PARP2 promoter activity was assayed by histochemical GUS staining. Col, Columbia ecotype. Bars = 200 μm.
(B) Quantitative real-time PCR analysis of TSI expression in 2-week-old wild-type and bin4-1 seedlings. Transcriptional gene silencing is not released in bin4. The teb1 and tsk3 mutants were used as negative and positive controls for TSI expression. Values that are statistically different from those of the wild type (Student's t test; P < 0.05) are marked by asterisks.
The DNA Damage Response in bin4 Is Dependent on ATM and ATR
The DNA damage checkpoint senses various kinds of DNA damage during the mitotic cell cycle and coordinates its repair with cell cycle progression. The two upstream PI3K-like protein kinases ATM and ATR are key players in this response, and previous studies in plants showed that most of the transcriptional responses to DSBs are mediated by ATM (Garcia et al., 2003; Culligan et al., 2006; De Schutter et al., 2007), while the transcriptional responses to stalled replication forks are mediated by ATR (Culligan et al., 2004; De Schutter et al., 2007). To test whether the upregulation of DNA damage genes in bin4 is mediated by such checkpoint mechanisms, we generated atm bin4 and atr bin4 double mutants. Our real-time RT-PCR analysis revealed that the expression level of several DNA damage response genes, including PARP1, PARP2, BRCA1, and RAD51, is 1.7 to 9 times higher in bin4 compared with wild-type seedlings. In addition, we found that both atm and atr mutations partially suppress these transcriptional activations in bin4 (Figure 5A), indicating that the DNA damage response in bin4 is dependent on both ATM and ATR. A fungal PI3K-like kinase inhibitor, wortmannin, blocks the ATM kinase activity in animals (Burma et al., 2001), and we found that the application of 100 μM wortmannin to Arabidopsis wild-type roots strongly blocks bleomycin-induced pPARP2:GUS expression (Figure 6A), demonstrating that wortmannin inhibits plant ATM and/or other upstream PI3K-like kinases involved in the DNA damage response. We treated 5-d-old bin4 seedlings with 100 μM wortmannin and found that it strongly blocks pPARP2:GUS expression in bin4 (Figure 6A), further suggesting the involvement of ATM and/or other PI3K-like kinases in the bin4 damage response. The atm bin4 and atr bin4 double mutants are indistinguishable from bin4 single mutants (data not shown), indicating that the absence of at least one of these checkpoint mechanisms does not result in any growth alterations in bin4.
The bin4 Mutation Induces the Ectopic Expression of a Mitotic G2/M-Specific Gene in Nondividing Cells
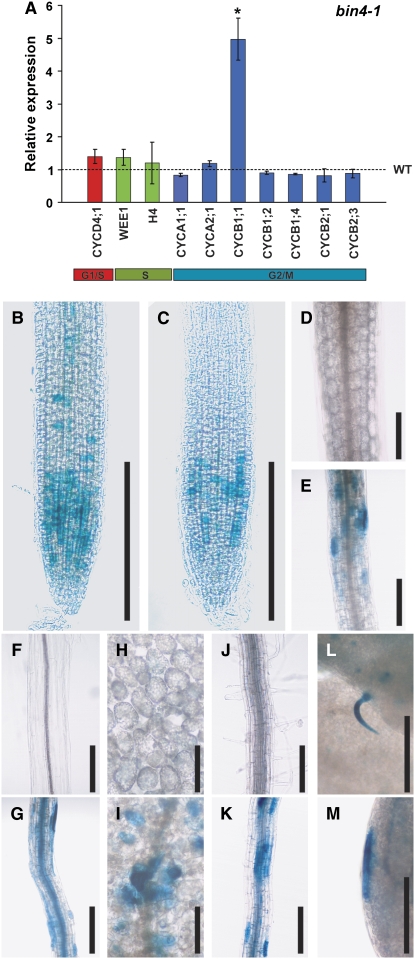
Recent studies show that the DNA damage response during the mitotic cell cycle leads to an accumulation of several cell cycle gene transcripts in plants (Culligan et al., 2006; De Schutter et al., 2007; Ramirez-Parra and Gutierrez, 2007). We examined whether the expression of several mitotic cell cycle genes, such as CYCA, CYCB, and CYCD, is altered in bin4. Most of these cell cycle–regulated genes are usually expressed only during the mitotic cell cycle, and their expression declines as cells switch from mitotic cell cycle to endocycle (Inze and De Veylder, 2006). As shown in Figure 7A, however, our real-time RT-PCR analyses showed that the transcript level of a G2/M-specific gene, cyclin B1;1 (CYCB1;1), was increased significantly in bin4-1. The expression of other G2/M-specific genes (CYCA1;1, CYCB1;2, CYC1;4, CYCB2;1, and CYCB2;3), a G1/S-specific gene (CYCD4;1), and S-specific genes (WEE1 and histone H4) does not appear to differ between the wild type and bin4 (Figure 7A).
Figure 7.
Transcriptional Expression Analyses Reveal the Ectopic Expression of the G2/M Phase Marker CYCB1;1 in Postmitotic Cells of bin4 Mutants.
(A) Quantitative real-time expression analysis of several core mitotic cell cycle genes. Actin2 expression level was used as a reference. Colored columns represent the relative expression of individual genes in bin4-1 compared with the wild-type expression levels, indicated as a dashed line. The values represent averages of four independent replicates ± sd, and the one that is statistically different from wild type (Student's t test; P < 0.05) is marked by an asterisk.
(B) to (M) Expression patterns of pCYCB1;1:GUS in 7-d-old primary root tips ([B] and [C]), 7-d-old, light-grown hypocotyls ([D] and [E]), 4-d-old, dark-grown hypocotyls ([F] and [G]), mature cotyledons ([H] and [I]), mature roots ([J] and [K]), and mature leaves ([L] and [M]). (B), (D), (F), (H), and (J) show wild-type organs, and (C), (E), (G), (I), and (K) to (M) show bin4-1 organs. Bars = 250 μm in (B) and (C), 200 μm in (D) to (G) and (J) to (M), and 100 μm in (H) and (I).
To further explore the spatial pattern of ectopic CYCB1;1 expression in bin4, we introduced the CYCB1;1:GUS construct (Colon-Carmona et al., 1999) into the bin4-1 background and examined the accumulation of CYCB1;1 transcripts. In the wild type, CYCB1;1 was predominantly expressed in proliferating tissues, and very little GUS signal was detected in mature tissues (Colon-Carmona et al., 1999) (Figures 7B, 7D, 7F, 7H, and 7J). We detected similar, or slightly reduced, levels of CYCB1;1 expression in the bin4 root meristems (Figure 7C), suggesting that bin4 is not greatly impaired in G2/M progression during the mitotic cell cycle. Unexpectedly, however, we found strong CYCB1;1 accumulation in nondividing cells, including those in light- and dark-grown hypocotyls (Figures 7E and 7G, respectively), cotyledons (Figure 7I), the mature part of roots (Figure 7K), leaf trichomes (Figure 7L), and leaf marginal cells (Figure 7M).
DISCUSSION
Role of BIN4 in Endoreduplication
In this study, we demonstrate that BIN4 encodes a novel plant-specific protein that is required for normal organ growth in Arabidopsis. Loss of BIN4 function results in a severe dwarf phenotype caused primarily by reduced cell expansion. Our study shows that BIN4 is expressed in both proliferating cells and expanding cells. However, the function of BIN4 does not appear to be essential during cell proliferation, because bin4 has the normal number of cells. Instead, our data clearly show that bin4 is defective in increasing ploidy in all cell types that normally endoreduplicate up to 16C or above. Furthermore, we demonstrate that doubling the chromosome number, and hence the ploidy level, by colchicine treatment can partially rescue the cell size defects in bin4, strongly suggesting that BIN4 is involved in the process of endoreduplication but not directly in increasing cell size. The endocycle is often viewed as a shortcut of the mitotic cell cycle that does not require its own molecular machinery. However, this study indicates that BIN4 is directly and specifically required for endoreduplication, suggesting that DNA replication during an endocycle requires some unique proteins. We also found that BIN4 is required only for polytene chromosome formation during endoreduplication but not for the colchicine-induced polyploidization, providing genetic evidence that these two processes are mechanistically distinct. We need to note that the colchicine-induced polyploidization of bin4 does not fully restore the wild-type ploidy level and thus its plant size. These data are consistent with the idea that BIN4 has some additional functional roles during plant development, for example, in maintaining the integrity of global chromatic structure or modifying the level of gene expression.
Our genetic and in vivo interaction studies revealed that BIN4, through its interaction with At SPO11-3/RHL2/BIN5 and RHL1/HYP7, functions as an integral and essential part of the plant topo VI complex. BIN4, like RHL1/HYP7, also has some vital function in the plant topo VI complex, since its loss of function leads to a phenotype as severe as that of topo VI mutants. Both RHL1/HYP7 and BIN4 possess sequence similarity to the C-terminal region of animal topo IIα, which is implicated in some regulatory function (Gadelle et al., 2003). Thus, it is likely that RHL1/HYP7 and BIN4 act together to enhance the decatenation activity of the plant topo VI. We have shown that both RHL1/HYP7 and BIN4 exhibit stable DNA binding in vitro, and we hypothesize that these DNA interactions help the plant topo VI to hold the substrate DNA during the decatenation reaction.
Ploidy, Cell Size, and Cell Differentiation
The occurrence of endoreduplication has been reported for various cell types in Arabidopsis, but very few studies have actually investigated its causal relationships. In this study, we first extended these observations and showed that vascular xylem cells also undergo several rounds of endocycle. The root metaxylem cells in bin4 are smaller than in the wild type, but their overall morphology appears to be normal. Thus, endoreduplication seems to be a prerequisite for cell expansion but not for differentiation. Furthermore, our study of cell size in isogenic diploid and tetraploid wild-type and bin4 plants provides strong evidence that increasing ploidy is coupled with, and contributes directly to, an increase in cell size. Whether cells have an active mechanism to sense an increase in the total nuclear DNA content and to control the extent of cell growth still remains unclear, but given that both polyteny and polyploidy have similar positive effects on cell size, this mechanism does not appear to differentiate between these two chromosome configurations.
DNA Damage Response during Endocycle
An unexpected finding in our study is that the bin4 mutation triggers the transcriptional activation of DNA damage response genes during endocycles, and the cell types exhibiting this response also ectopically express CYCB1;1, which is normally transcribed only at the G2- and M-phases of the mitotic cell cycle. Mutations in other components of the plant topo VI complex also upregulate CYCB1;1 transcription (data not shown), indicating that the observed phenotype is a general response to the loss of the topo VI complex. The DNA damage checkpoint during the mitotic cell cycle has been studied extensively, but very little is known as to whether similar checkpoint mechanisms operate in postmitotic cells. It is generally believed that DSBs are not a critical risk for cells that have entered the endocycle, since they do not undergo further chromosome segregation and cell division; indeed, one reported study suggests that mature tissues in Arabidopsis do not initiate a DNA damage response against the DSBs induced by ionizing radiation (Hefner et al., 2006). However, this study clearly demonstrates that various cell types that have completed several rounds of endocycle do respond to the DNA damage induced by a lack of functional topo VI or the application of DNA-damaging agents such as bleomycin. We show that ATM and ATR contribute almost equally and nonredundantly to the activation of DNA damage genes in bin4. We speculate that the ATR pathway is activated in response to stalled replication forks caused by torsional stress, while the ATM pathway is activated by DSBs that are generated by persistent replication problems (e.g., collapsed replication forks). We cannot rule out the possibility that the loss of BIN4 also leads to other defects, such as the global disruption of chromatin states that cause misexpression of genes. However, based on our observations that heterochromatin organization in interphase nuclei is not disrupted in bin4 (data not shown) and that TGS is not released in bin4, misexpression of DNA damage genes in bin4 is not likely to be a consequence of general chromosomal disruption. In agreement with this, Ramirez-Parra and Gutierrez (2007) recently reported that the transcriptional activation of several DNA damage genes in response to the fas1 mutation or zeocin, a DSB-inducing drug, is due to selective epigenetic modifications in their promoter sequences and not to global chromosome remodeling. Interestingly, the loss of checkpoints in atm bin4 or atr bin4 did not lead to the partial alleviation of the bin4 phenotype or the kind of cell death seen in aphidicolin-treated atr root tips (Culligan et al., 2004). It has been suggested that ATM and ATR play some overlapping functions in the DNA damage response (Culligan et al., 2006), and this possibility needs to be investigated in future studies using atm atr bin4 triple mutants.
The abnormal accumulation of CYCB1;1 transcripts in nondividing cells of bin4 is intriguing. Such abnormal accumulation of CYCB1;1 has been reported in several Arabidopsis mutants, including teb, mgo3/bru1/tsk, fas1, and fas2 (Suzuki et al., 2005; Endo et al., 2006; Inagaki et al., 2006; Schonrock et al., 2006), and in response to various genotoxic stresses (Chen et al., 2003; Culligan et al., 2006). Because these transcriptional activations are restricted to subsets of G2/M genes, they are thought to reflect an active cellular response to DNA damage rather than, or in addition to, G2 cell cycle arrest (Culligan et al., 2006; Schonrock et al., 2006). This study also supports the view that the transcriptional upregulation of CYCB1;1 in bin4 is not due to arrest at the G2/M phase of the cell cycle but is a specific transcriptional response induced by DNA damage. We need to note that the transcription of other G2/M genes (e.g., CYCB1;2) is downregulated in response to ionizing irradiation (Culligan et al., 2006), while the expression level of those transcripts remains relatively constant in bin4. These differences may arise from the fact that both ATM- and ATR-dependent responses are activated in bin4. The CYCB genes are normally expressed only in the mitotic cell cycle, and their downregulation during the endocycle is thought to be critical for its successive progression (Inze and De Veylder, 2006). It is plausible, therefore, that the ectopic transcription of CYCB1;1 and maybe several other cell cycle genes that are not covered in this study leads to an early cessation of endocycles in bin4.
METHODS
Plant Materials and Growth Conditions
The bin4-1 mutant of Arabidopsis thaliana (ecotype Columbia) was isolated as a BR-like dwarf from the Chory laboratory collection of T-DNA–transformed plant lines (Yin et al., 2002). The second allele of At BIN4, bin4-2 (SALK_110750; ecotype Columbia), was obtained from the T-DNA insertion mutant collection of the Salk Institute Genome Analysis Laboratory (Alonso et al., 2003). The pPARP2∷GUS and pCYCB1-1:GUS constructs were introduced into the bin4 mutant background by genetic crosses. Unless described otherwise, plants were grown on plates containing Murashige and Skoog salts, pH 5.8, 1% (w/v) sucrose, and 0.5% (w/v) phytagel under continuous light at 25°C.
Ploidy Measurements and Detection of DNA Replication
Ploidy levels were measured by flow cytometry and fluorescence microscopy as described previously (Sugimoto-Shirasu et al., 2005). Endoreduplicating nuclei were detected by the incorporation of 10 μM BrdU into replicating chromosomes and subsequent labeling using the BrdU immunocytochemistry kit (Roche), following the manufacturer's instructions.
Polyploidization by Colchicine Treatment
The shoot apices of 7-d-old wild-type and bin4-1 heterozygous plants were treated with 0.05% (w/v) colchicine dissolved in 0.1% (w/v) agarose and covered with plastic films overnight to keep under high humidity. Colchicine was rinsed off with distilled water the following day, and the plants were grown on rock wool under continuous light at 23°C. Plants that developed thicker leaves than normal diploid plants were selected as putative tetraploid lines and self-fertilized to collect seeds. Colchicine induces a chimera of diploid and tetraploid cells in shoot apices of the first generation, leading to the segregation of diploid and tetraploid plants in the next generation. Tetraploid plants were identified based on their overall morphology and size, and the increased ploidy was confirmed by flow cytometry.
FISH Analysis
Chromosome spreads were prepared from 14-d-old leaf cells according to Ross et al. (1996) with minor modifications. The spreads were then treated with 0.1 mg/mL RNase A at 37°C for 30 min, with 12.5 μg/mL pepsin at 37°C for 2 min, and with 4% formaldehyde at room temperature for 10 min before being dehydrated in an ethanol series. FISH probes were synthesized by PCR-amplifying the centromere-specific 180-bp repeat sequence (Heslop-Harrison et al., 1999). PCR products were fluorescently labeled using a nick translation kit (Amersham Biosciences) and an ARES Alexa Fluor 488 DNA labeling kit (Invitrogen) and Ready-To-Go DNA labeling beads (GE Healthcare). Prepared probes were dissolved in 50% formamide/2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and applied to the chromosome spreads. Chromosome spreads were incubated at 37°C for >16 h, washed with 50% formamide/2× SSC, 2× SSC, and 1× SSC at 37°C for 30 min, and counterstained with 1.5 μg/mL 4′,6-diamidino-2-phenylindole.
DNA Damage Assay
DNA damage was detected by comet assay using the CometAssay kit (Trevigen). Root tips and cotyledons of 8-d-old seedlings were chopped with a razor blade in 1 mL of Partec CyStain UV Precise P extraction buffer and filtered through CellTric 30-μm nylon filters (Partec). Further sample preparation was performed as described by Wang and Liu (2006). The percentage of DNA in each comet tail was evaluated with Comet Score software (http://www.autocomet.com). DNA damage for each tissue and genotype was calculated by averaging the values for percentage tail DNA from five individual slides, scoring 70 comets per slide. Sensitivity to methyl methane sulfonate was assayed as described previously (Stacey et al., 2006).
Quantitative Real-Time RT-PCR Analysis
Quantitative real-time RT-PCR was performed using SYBR Green Supermix (Toyobo). Actin mRNA concentrations were determined for each RNA sample to normalize total RNA levels. Primer sets used for quantitative real-time RT-PCR are listed in Supplemental Table 2 online.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under accession number At5g24630 (BIN4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Endoreduplication Stalls at 8C in bin4.
Supplemental Figure 2. The bin4 Mutation Is Epistatic to the Trichome Overbranched Mutants try, rfi, and kak2 and to the Ectopic Root Hair Mutant gl2.
Supplemental Figure 3. Positional Cloning and Transcript and Protein Expression Analyses of bin4-1 and bin4-2.
Supplemental Figure 4. BIN4 Is a Novel Nuclear Protein That Exhibits Stable DNA Binding Properties in Vitro.
Supplemental Figure 5. Expression of At TOPII and Four Components of the Arabidopsis topo VI Complex during the Mitotic Cell Cycle.
Supplemental Table 1. Comparison of Various Cell and Organ Sizes in Arabidopsis Wild Type, bin4-1, and bin4-2.
Supplemental Table 2. Primer Sets Used for Real-Time RT-PCR to Analyze the Expression of Genes Involved in Cell Cycle Progression and DNA Repair.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Martin Hulskamp (University of Cologne, Germany) for providing try, rfi, and kak2 mutants; Liam Dolan (John Innes Centre) for providing the gl2 mutant; Peter Doerner (University of Edinburgh, United Kingdom) for providing the pCYCB1;1:GUS line; Elena Babiychuk and Mark Van Montagu (University of Gent, Belgium) for providing the pPARP2:GUS line; Kevin Culligan and Anne Britt (University of California, Davis) for providing atm and atr mutants; Soichi Inagaki (Nagoya University, Japan) and Atsushi Morikami (Meijo University, Japan) for providing teb1 and tsk3 mutants; and the Salk Institute Genomic Analysis Laboratory for providing T-DNA insertion mutants. We are grateful to Detlef Weigel (Max Planck Institute, Germany) for his help with AtGenExpress to positionally clone BIN4; to Kim Findlay (John Innes Centre) for her technical assistance with scanning electron microscopy; and to Takeshi Yoshizumi (RIKEN) for his advice on quantitative RT-PCR. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) to A.M., K.R., and K.S.-S.; by a grant from the U.S. Department of Agriculture to J.C.; by a grant from the Japan Society for the Promotion of Science (JSPS) to H.T.; and by BBSRC David Phillips Fellowships to K.S.-S. and C.E.W. K.S.-S. and C.B. were recipients of a JSPS Postdoctoral Fellowship and a European Union Marie Curie Early Stage Training studentship, respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Keiko Sugimoto-Shirasu (sugimoto@psc.riken.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adachi, N., Miyaike, M., Kato, S., Kanamaru, R., Koyama, H., and Kikuchi, A. (1997). Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nucleic Acids Res. 25 3135–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anastasiou, E., and Lenhard, M. (2007). Growing up to one's standard. Curr. Opin. Plant Biol. 10 63–69. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Cottrill, P.B., Storozhenko, S., Fuangthong, M., Chen, Y., O'Farrell, M.K., Van Montagu, M., Inze, D., and Kushnir, S. (1998). Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J. 15 635–645. [DOI] [PubMed] [Google Scholar]
- Boyko, A., Zemp, F., Filkowski, J., and Kovalchuk, I. (2006). Double-strand break repair in plants is developmentally regulated. Plant Physiol. 141 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma, S., Chen, B.P., Murphy, M., Kurimasa, A., and Chen, D.J. (2001). ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276 42462–42467. [DOI] [PubMed] [Google Scholar]
- Chen, I., Haehnel, U., Altschmied, L., Schubert, I., and Puchta, H. (2003). The transcriptional response of Arabidopsis to genotoxic stress—A high density colony array study (HDCA). Plant J. 35 771–786. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance. Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861. [DOI] [PubMed] [Google Scholar]
- Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K.M., Robertson, C.E., Foreman, J., Doerner, P., and Britt, A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48 947–961. [DOI] [PubMed] [Google Scholar]
- De Schutter, K., Joubes, J., Cools, T., Verkest, A., Corellou, F., Babiychuk, E., Van Der Schueren, E., Beeckman, T., Kushnir, S., Inze, D., and De Veylder, L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Chabeaud, G., Godon, C., Brutesco, C., de Murcia, G., and Kazmaier, M. (2001). Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genomics 265 954–963. [DOI] [PubMed] [Google Scholar]
- Doutriaux, M.P., Couteau, F., Bergounioux, C., and White, C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257 283–291. [DOI] [PubMed] [Google Scholar]
- Endo, M., Ishikawa, Y., Osakabe, K., Nakayama, S., Kaya, H., Araki, T., Shibahara, K., Abe, K., Ichikawa, H., Valentine, L., Hohn, B., and Toki, S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25 5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadelle, D., Filee, J., Buhler, C., and Forterre, P. (2003). Phylogenomics of type II DNA topoisomerases. Bioessays 25 232–242. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V., Bruchet, H., Camescasse, D., Granier, F., Bouchez, D., and Tissier, A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, F., Angelis, K.J., Meister, A., Schubert, I., Melzer, M., and Puchta, H. (2002). An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 12 1787–1791. [DOI] [PubMed] [Google Scholar]
- Hefner, E., Huefner, N., and Britt, A.B. (2006). Tissue-specific regulation of cell-cycle responses to DNA damage in Arabidopsis seedlings. DNA Repair (Amst.) 5 102–110. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S., Murata, M., Ogura, Y., Schwarzacher, T., and Motoyoshi, F. (1999). Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp, M. (2004). Plant trichomes: A model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5 471–480. [DOI] [PubMed] [Google Scholar]
- Imai, K.K., Ohashi, Y., Tsuge, T., Yoshizumi, T., Matsui, M., Oka, A., and Aoyama, T. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, S., Suzuki, T., Ohto, M.A., Urawa, H., Horiuchi, T., Nakamura, K., and Morikami, A. (2006). Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40 77–105. [DOI] [PubMed] [Google Scholar]
- Lafarge, S., and Montane, M.H. (2003). Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 31 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell-size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Nagl, W. (1978). Endopolyploidy and Polyteny in Differentiation and Evolution. (Amsterdam: Elsevier).
- Ramirez-Parra, E., and Gutierrez, C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Gillmor, C.S., Kovaleva, V., Somerville, C.R., and Raikhel, N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1 303–310. [DOI] [PubMed] [Google Scholar]
- Ross, K.J., Franz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4 507–516. [DOI] [PubMed] [Google Scholar]
- Schonrock, N., Exner, V., Probst, A., Gruissem, W., and Hennig, L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281 9560–9568. [DOI] [PubMed] [Google Scholar]
- Stacey, N.J., Kuromori, T., Azumi, Y., Roberts, G., Breuer, C., Wada, T., Maxwell, A., Roberts, K., and Sugimoto-Shirasu, K. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48 206–216. [DOI] [PubMed] [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Scheid, O.M., and Paszkowski, J. (2000). Endogenous targets of transcriptional silencing in Arabidopsis. Plant Cell 12 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Roberts, G.R., Stacey, N.J., McCann, M.C., Maxwell, A., and Roberts, K. (2005). RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl. Acad. Sci. USA 102 18736–18741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6 544–553. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Stacey, N.J., Corsar, J., Roberts, K., and McCann, M.C. (2002). DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12 1782–1786. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Nakajima, S., Inagaki, S., Hirano-Nakakita, M., Matsuoka, K., Demura, T., Fukuda, H., Morikami, A., and Nakamura, K. (2005). TONSOKU is expressed in S phase of the cell cycle and its defect delays cell cycle progression in Arabidopsis. Plant Cell Physiol. 46 736–742. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Tadele, Z., Hofmann, I., Probst, A.V., Angelis, K.J., Kaya, H., Araki, T., Mengiste, T., Scheid, O.M., Shibahara, K., Scheel, D., and Paszkowski, J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., and Liu, Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y.H., Cheong, H., Friedrichsen, D., Zhao, Y.D., Hu, J.P., Mora-Garcia, S., and Chory, J. (2002). A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc. Natl. Acad. Sci. USA 99 10191–10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.