Abstract
Many proteins of the photosynthesis complexes are encoded by the genome of the chloroplast and synthesized by bacterium-like ribosomes within this organelle. To determine where proteins are synthesized for the de novo assembly and repair of photosystem II (PSII) in the chloroplast of Chlamydomonas reinhardtii, we used fluorescence in situ hybridization, immunofluorescence staining, and confocal microscopy. These locations were defined as having colocalized chloroplast mRNAs encoding PSII subunits and proteins of the chloroplast translation machinery specifically under conditions of PSII subunit synthesis. The results revealed that the synthesis of the D1 subunit for the repair of photodamaged PSII complexes occurs in regions of the chloroplast with thylakoids, consistent with the current model. However, for de novo PSII assembly, PSII subunit synthesis was detected in discrete regions near the pyrenoid, termed T zones (for translation zones). In two PSII assembly mutants, unassembled D1 subunits and incompletely assembled PSII complexes localized around the pyrenoid, where we propose that they mark an intermediate compartment of PSII assembly. These results reveal a novel chloroplast compartment that houses de novo PSII biogenesis and the regulated transport of newly assembled PSII complexes to thylakoid membranes throughout the chloroplast.
INTRODUCTION
Photosynthesis involves several pathways that are highly compartmentalized within chloroplasts. For example, the Calvin cycle occurs in the stroma, which is analogous to the cytoplasm of a cell (Suss et al., 1993). The light-dependent reactions occur within the membranes of an elaborate network of flattened vesicles, the thylakoids. These reactions are performed by three multisubunit complexes: photosystem I (PSI), photosystem II (PSII), and the cytochrome b6/f complex (Staehelin, 2003). Thylakoids themselves are compartmentalized. PSII is localized in domains of thylakoid membranes that are appressed within the stacks of thylakoid vesicles (called grana), and PSI is localized in the nonappressed stroma thylakoid membranes that face the chloroplast stroma (Andersson and Anderson, 1980).
The biogenesis of chloroplasts also involves many well-characterized pathways (Zerges, 2000; Eckhardt et al., 2004; Benning et al., 2006), but much less is known about their spatial organization. In situ data are sparse regarding the locations of the biosynthesis and assembly of components of chloroplast compartments and the various steps in the expression of the chloroplast genome. Of the few reported in situ studies, two showed that proteins that function in chlorophyll synthesis and lipid transport to thylakoids are localized in punctate regions near the periphery of the chloroplast (Watanabe et al., 2001; Xu et al., 2005). These two processes thus appear to be more compartmentalized than was previously realized.
The biogenesis of PSII has been studied extensively. This complex uses energy from light to carry out the first charge separation reaction of oxygenic photosynthesis (Minagawa and Takahashi, 2004). Many of the 25 PSII subunits are encoded by nuclear genes, synthesized by cytoplasmic 80S ribosomes, imported into the chloroplast, and finally targeted to thylakoid membranes (Keegstra and Cline, 1999; Kessler and Schnell, 2006). However, the major integral membrane subunits of PSII are encoded by chloroplast genes, whose mRNAs are translated by bacterium-like 70S ribosomes within the chloroplast (Beligni et al., 2004). These include D1, D2, CP43, and CP47, which are encoded by psbA, psbD, psbC, and psbD, respectively. While all subunits are synthesized for de novo assembly of the PSII complex, the D1 subunit is also synthesized to replace D1 subunits damaged by photochemical reactions inherent to PSII activity. This D1 repair synthesis is elevated under high-light stress conditions (Aro et al., 1993).
A widely accepted model proposes that chloroplast-encoded PSII subunits are synthesized directly into thylakoid membranes. This model is supported by two lines of evidence. First, chloroplast polysomes synthesizing subunits of PSII and other thylakoid membrane proteins cofractionated with thylakoid membranes on sucrose density gradients (Chua et al., 1973, 1976; Margulies and Michaels, 1974, 1975; Margulies et al., 1975, 1987; Yamamoto et al., 1981; Margulies, 1983; Herrin and Michaels, 1985; Breidenbach et al., 1988; Klein et al., 1988; Zhang et al., 1999). These membranes were proposed to be stroma thylakoids based on results of thylakoid fractionation experiments (Yamamoto et al., 1981; Leto et al., 1985; Mattoo and Edelman, 1987; Adir et al., 1990; van Wijk et al., 1996). Second, in situ evidence for this model was provided by electron microscopy (EM) images of a few cells and isolated thylakoid membranes, which revealed chloroplast ribosomes or incompletely assembled PSII subcomplexes primarily at nonappressed stroma thylakoid membranes (Bourque et al., 1971; Margulies and Michaels, 1974; Chua et al., 1976; Yamamoto et al., 1981; de Vitry et al., 1989). While it has been assumed that thylakoid membrane protein synthesis and assembly occur in stroma thylakoids throughout the chloroplast, to our knowledge, the possibility that these processes could occur in specialized domains of stroma thylakoids has not been explored.
In an alternative model, the inner membrane of the chloroplast envelope has been proposed to house the synthesis and assembly of the chloroplast-encoded thylakoid membrane proteins. Chloroplast membranes that resemble the inner envelope membrane in buoyant density and pigment composition were found to be associated with RNA binding proteins (RBPs) and a homolog of poly(A) binding protein (Zerges and Rochaix, 1998; Zerges et al., 2002). Similar nonthylakoid membranes cofractionated with 50% of chloroplast ribosomes (Margulies and Weistrop, 1980). The credibility of this alternative model was increased by the discovery that thylakoid biogenesis requires vesicular transport from the inner envelope membrane (Hoober et al., 1991; White and Hoober, 1994; Kroll et al., 2001; Vothknecht and Soll, 2005). Although these data would seem to contradict the aforementioned evidence for thylakoids as the location of PSII subunit synthesis, they do not necessarily. For example, most of the EM images cited above were taken of isolated thylakoids, recognized by their stacked regions. However, other membranes with bound ribosomes in the same fractions would have been disregarded as contaminating rough endoplasmic reticulum (Chua et al., 1976). Moreover, the thylakoid membranes that were concluded to have bound chloroplast polysomes (cited above) were in sucrose density gradient fractions that are heavily contaminated by membranes that resemble the membranes of the chloroplast envelope (Joyard et al., 1980; Zerges and Rochaix, 1998). The latter membranes have been hypothesized to be undifferentiated thylakoid membranes of an unknown location in the chloroplast of Chlamydomonas reinhardtii (Zerges and Rochaix, 1998).
The location of the de novo assembly of the photosynthesis complexes has also been addressed in Synechocystis, the closest known bacterial relative of the chloroplasts of the green plants (which include Chlamydomonas) (Devereux et al., 1990; Gray, 1999). Results of cellular subfractionation experiments provided strong evidence for the assembly of PSI and PSII occurring in the plasma membrane (Zak et al., 2001), which is considered to be homologous with the inner membrane of the chloroplast envelope (Joyard et al., 1991). Yet, the three-dimensional structure of this bacterium obtained by electron tomography revealed that most ribosomes within 6 nm of a membrane (i.e., sufficiently close to be inserting a nascent protein) were on or near thylakoid membranes (van de Meene et al., 2006). The possibility that an unknown membrane type gives rise to thylakoid membranes was suggested by the description of unit membrane–like sheets that had dense clusters of ribosomes and were connected to thylakoid membranes. Similarly, in Chlamydomonas and pea (Pisum sativum), many of the chloroplast ribosomes seen by EM were in clusters located near isolated thylakoid membranes but not directly on them (Chua et al., 1976; Yamamoto et al., 1981).
Considering the uncertainty regarding the location of thylakoid membrane biogenesis, we have begun to address this problem by characterizing the locations of PSII subunit synthesis in the chloroplast of Chlamydomonas with fluorescence in situ hybridization (FISH), immunofluorescence (IF) staining, and confocal microscopy. This eukaryotic green alga is widely used as a model system for studies of chloroplast biogenesis (Rochaix et al., 1998). While exploring the aforementioned problems, we found that Chlamydomonas is ideally suited for characterization of the distribution of specific chloroplast mRNAs and proteins by these techniques because its single chloroplast has a definite anatomy that is easily recognizable in every cell examined.
RESULTS
The Chlamydomonas Chloroplast as Revealed by Fluorescence Confocal Microscopy
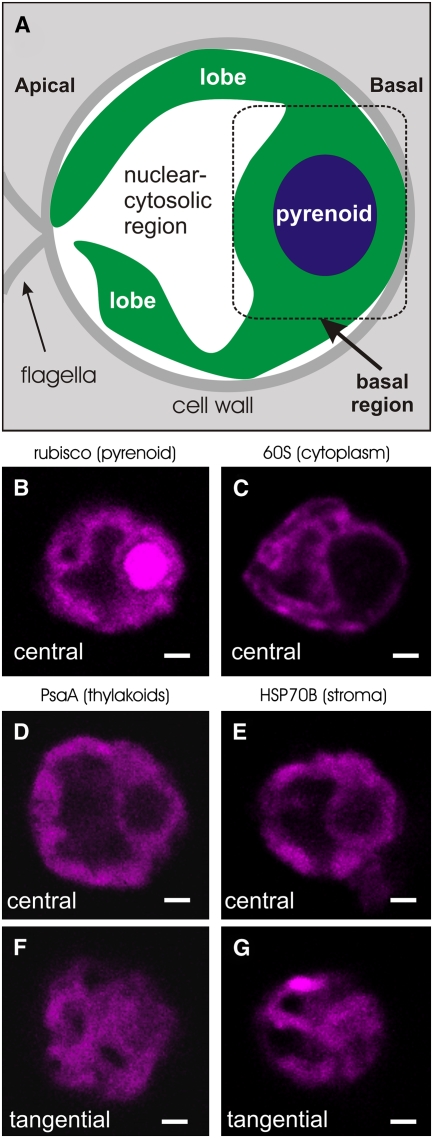
In order to validate our identification of relevant cellular compartments by fluorescence confocal microscopy, Figure 1 shows an illustration of a typical cell (A) and the IF staining patterns of four marker proteins (B to G). Each cell is shown with its apical-basal axis oriented from left to right (Figure 1A). The globular basal region of the chloroplast contains the pyrenoid, which was identified by its IF staining for ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) (Figure 1B) (Lacoste-Royal and Gibbs, 1987; Suss et al., 1995; Borkhsenious et al., 1998). In the images throughout this report, the pyrenoid is seen as a spherical region that is largely devoid of fluorescence signals from chloroplast mRNAs or marker proteins. The IF signal from a marker protein for the cytoplasm, an r-protein of the 60S subunit of the cytoplasmic ribosome, revealed the outer perimeter of the chloroplast (Figure 1C). The chloroplast extends apically from the basal region to skirt the cytosol, thereby enclosing a central region with the nucleus, cytoplasm, and other organelles. Thylakoids and stroma are interspersed throughout the chloroplast, as seen from the IF staining patterns of marker proteins for these compartments: PsaA (Figures 1D and 1F) and HSP70B (Figures 1E and 1G), respectively (Redding et al., 1999; Schroda et al., 2001).
Figure 1.
IF Staining Patterns of Marker Proteins Reveal Chloroplast Anatomy and Morphology.
(A) An illustration of a typical longitudinal cell section shows the chloroplast (green) as composed of apical lobes and a basal region, which contains the pyrenoid (blue).
(B) to (G) Images show 0.2-μm central optical sections of cells that were single IF-stained for a marker protein for the pyrenoid and thylakoids (Rubisco) (B), the cytoplasm, an r-protein of the 60S subunit of the cytoplasmic ribosome (C), thylakoids (PsaA) (D), and stroma (HSP70B) (E). In tangential optical sections taken between the center and the periphery, (F) and (G) show the organization of lobes in the same cells shown in (D) and (E), respectively. Cells are shown with their apical–basal axis oriented from left to right. Bars = 1.0 μm.
Although the chloroplast is widely believed to resemble a cup, with the basal region as its base and a continuous rim with its brim near the apical cell pole (Ohad et al., 1967b), our images revealed more complex and variable chloroplast morphologies. Similar features were described previously based on three-dimensional reconstructions of a few Chlamydomonas cells from serial EM images. In one report, the chloroplast is described as having, instead of an apical rim, a few elongated finger-like lobes (Schotz et al., 1972). We observed many chloroplasts with such lobes, as seen outlined by the IF signal from the cytoplasmic r-protein (Figure 1C). The authors of another report likened the chloroplast to the white of a boiled egg with the narrowest (apical) end cut off to form an opening and having ∼11 large perforations in the resulting rim (Gaffal et al., 1995). We saw these features in other cells, in which chloroplast lobes appeared to connect at their apical ends, as seen in the tangential optical sections, which were taken within the forward-most lobes to the viewer (Figures 1F and 1G).
The psbA and psbC mRNAs Colocalize to Discrete Regions in the Chloroplast Basal Region during the Induction of PSII Subunit Synthesis and Assembly
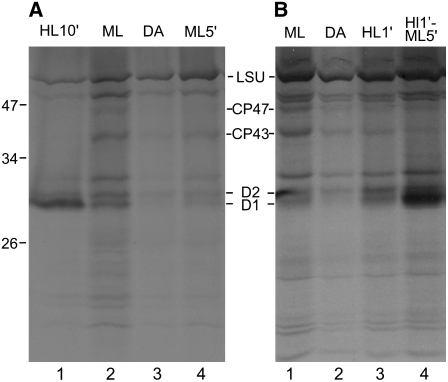
To determine where chloroplast mRNAs and translation components colocalize specifically for de novo PSII assembly, we first needed to establish conditions that rapidly induce translation for this process but do not significantly induce psbA translation for PSII repair. Moderate-intensity light (ML; 50 to 200 μE·m−2·s−1) activates the translation of multiple chloroplast mRNAs for the biogenesis of the photosynthesis complexes (Chua et al., 1976; Herrin et al., 1986; Lee and Herrin, 2002), while preferential psbA translation for D1 repair synthesis is evident only under high-intensity light (HL; >700 μE·m−2·s−1) (van Wijk et al., 1994; Kettunen et al., 1997). These two distinct modes of translational regulation were revealed by the results of in vivo 35S pulse-labeling experiments (Figure 2). Cells were treated with cycloheximide, which specifically inhibits 80S ribosomes, so that only chloroplast-encoded proteins would be radiolabeled. Cells cultured photoautotrophically under ML (∼150 μE·m−2·s−1) synthesized several proteins at higher rates than did cells from the same culture at the end of a 2-h incubation in the dark (Figures 2A, lanes 2 and 3, and 2B, lanes 1 and 2). Hereafter, these are called ML cells and dark-adapted (DA) cells, respectively. ML cells synthesized PSII subunits primarily for de novo complex assembly, because the bands corresponding to D1 and D2 were of similar intensities, consistent with their equal stoichiometry in the PSII complex. The synthesis of CP43 and CP47 also appeared to be positively regulated by light (see Figure 2 legend). Subunit synthesis for de novo PSII assembly was rapidly induced when DA cells were exposed to ML for 5 min (ML5′ cells; Figure 2A, lane 4). The similar synthesis levels of D1 and D2 also demonstrated that D1 synthesis was not induced significantly for PSII repair by this 5-min ML exposure. Preferential D1 synthesis for PSII repair was observed when DA cells were exposed to HL for 10 min, as seen by the predominant 35S labeling of D1 in these HL10′ cells (Figure 2A, lane 1). The synthesis of most other proteins was downregulated in HL10′ cells, as reported previously for the large subunit of Rubisco under less severe light stress conditions (Shapira et al., 1997). The results in Figure 2B, lanes 3 and 4, are described later in this report.
Figure 2.
Protein Synthesis for PSII Assembly and Repair Is Induced by Light.
Synthesis of the PSII subunits D1, D2, CP43, and CP47 was monitored in vivo by 35S pulse-labeling for 5 min. 80S cytoplasmic ribosomes were inhibited by cycloheximide. Of the proteins synthesized by chloroplast ribosomes, D1, D2, CP43, CP47, and the large subunit of Rubisco (LSU) were recognized (Ohnishi and Takahashi, 2001). Cells exposed to high-intensity light for 10 min (HL10′ cells) preferentially synthesized D1 for PSII repair ([A], lane 1). Cells cultured under moderate light (ML cells) synthesized D1, D2, CP43, and CP47 for de novo PSII assembly ([A], lane 2; [B], lane 1). Lower levels of synthesis were detected in 2-h dark-adapted cells (DA cells; [A], lane 3; [B], lane 2). The induction of the synthesis of subunits for de novo PSII assembly was detected during a 5-min exposure to moderate light (ML5′ cells; [A], lane 4) or following exposure to high light for 1 min (HL1′ cells; [B], lane 3). Light-induced synthesis was more evident for D1 and D2 than for CP43 and CP47, possibly due to lower levels of these larger proteins that undergo a complete round of synthesis during the 5-min labeling period. HL1′ cells exposed to moderate light for 5 min (HL1′ML5′ cells) underwent both PSII subunit synthesis for de novo assembly and preferential D1 synthesis for PSII repair ([B], lane 4).
The green FISH signal seen throughout this report is specific to the psbA mRNA because it was not detected in a psbA deletion mutant (see Supplemental Figure 1A online). Although a psbC deletion mutant is not available for this control, the psbC FISH signal probably also is specific, because the FISH probes used are not complementary to any other sequence in the chloroplast or nuclear genomes (Table 1). The FISH signals from the chloroplast rbcL and psaA mRNAs (see below) also were absent in deletion mutants for these genes (see Supplemental Figure 1 online).
Table 1.
Oligonucleotide FISH Probe Sequences
| Name | Probe Sequence (5′→3′) |
|---|---|
| psbA-1 | CTGAAACTGGTTCACGGATACCATCGATGTCTACTGGC |
| psbA-2 | ACGGAAAGATAATTCCCACTCACGACCCATGTAGCAGTATACACTAGA |
| psbA-3 | GAATACGATCATGAAGTTGAAAGTACCAGAGATACCTAAAGGCATACCGT |
| psbA-4 | GGAAGATTAGACGACCAAAGTAACCATGAGCAGCTACAATGTTGTAAGTT |
| psbC-1 | TACCTGACCACCAAGCAAAACCTGTTGTTTCTTGGTCACGGCCACCTACT |
| psbC-2 | ATAATTTCACCACCTGGACCTACACCGTAACCTAAAGTTGCGATGTGTGG |
| psbC-3 | GGGTTAGTAATAACACGAACGTCACCACCACCTGGAGCCCAAGTATCGTA |
| psbC-4 | AGTCTAAACCGTTTGGACCACGTAGAGGTTCTAACCATGGACCACGGAAG |
| psaA-1 | TTGGAACCATTCTAGTTTTGGAGCAGCTTTGTGGTAGTGGAACCAACCAG |
| psaA-2 | CTACGTGGTGGTGAGCAGTATCACTTAACCAAAGACCACCAGTAACAGGG |
| psaA-3 | AAGCTGTAACCGTACCCCAAACATCAGATTGCATCTTCCAGCTGAAGTGG |
| psaA-4 | AGTAGGAGGTGCGCCTTCCAGCGAATACATGATTTTTCCCCCTTCCAGTT |
| rbcL-1 | CCACATTCTTCAGGTGGAACACCTAGTTGTGGAGTCATACGGAATGCAGC |
| rbcL-2 | GCAGGTGGAATACGAAGGTCTTCAAGACGTAGAGCACGTAAAGCTTTGAA |
| rbcL-3 | GTGCATAGCACGGTGGATGTGTAGAAGAAGACCGTTGTCACGACAGTAGA |
| rbcL-4 | CACCTGGCATTGAACACCAGTCTTGAGTGAAGTAAATACCACGGCTACGG |
Boldface Ts indicate modified C6-dT residues that were labeled with either Alexa Fluor 488 or Alexa Fluor 555 (Molecular Probes).
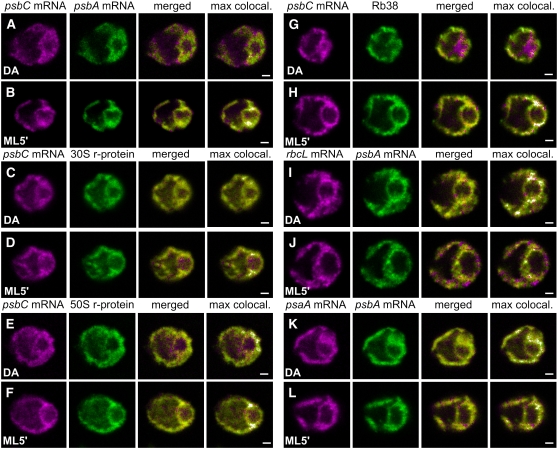
As a first approach to identify the location(s) of PSII assembly, FISH was used to determine where mRNAs encoding two PSII subunits colocalize specifically under conditions of their translation (i.e., in ML5′ cells but not in DA cells). In DA cells, the FISH signals from the psbA and psbC mRNAs were detected throughout the chloroplast and their colocalization was seen in a few scattered regions (Figure 3A). By contrast, in all ML5′ cells examined, a punctate colocalization pattern was seen at 0.5 to 1.0 μm from the pyrenoid, amid weaker psbA and psbC FISH signals throughout the chloroplast (Figure 3B). This punctate pattern was typically located lateral to the pyrenoid (i.e., above and below the pyrenoid in our images).
Figure 3.
Chloroplast mRNAs Encoding PSII Subunits and Chloroplast Translation Proteins Colocalize in Specific Regions under Conditions of PSII Assembly.
The induction of subunit synthesis for PSII assembly that occurred while dark-adapted (DA) cells were exposed to moderate light for 5 min (ML5′ cells) correlated with the colocalization near the pyrenoid of the psbC mRNA with the psbA mRNA ([A] and [B]) and proteins of the chloroplast translation machinery ([C] to [H]). The percentage of cells with the localization patterns seen in each image set and the number of cells examined (n) are given for each condition. Image sets have the psbC mRNA FISH signal and the psbA mRNA FISH signal in a DA cell (90%; n = 20) and a ML5′ cell (100%; n = 20) ([A] and [B]); IF from the r-protein of the 30S chloroplast ribosomal subunit in a DA cell (83%; n = 40) and a ML5′ cell (100%; n = 20) ([C] and [D]); IF from the r-protein of the 50S subunit in a DA cell (83%; n = 40) and a ML5′ cell (100%; n = 20) ([E] and [F]); and IF from the RNA binding protein RB38 in a DA cell (88%; n = 40) and a ML5′ cell (100%; n = 20) ([G] and [H]). Noncolocalization in T zones can be seen for the psbA mRNA FISH signal with the rbcL mRNA FISH signal in a DA cell (100%; n = 8) (I) and a ML5′ cell (100%; n = 10) (J) and with the psaA mRNA FISH signal in a DA cell (100%; n = 10) (K) and a ML5′ cell (100%; n = 9) (L). The fourth column shows merged images in which the pixels with the strongest colocalized signals are highlighted in white (max colocal.). Each image shows a 0.2-μm optical section. Bars = 1.0 μm.
The visualization of the punctate colocalization pattern in ML5′ cells was hampered by the nonlocalized FISH signals, which probably were from nontranslated psbA and psbC mRNAs. This was expected because several Chlamydomonas chloroplast mRNAs accumulate in excess for their translation and for photoautotrophic growth (Eberhard et al., 2002; Lee and Herrin, 2003) and the majority of psbA mRNAs is not associated with polysomes (Yohn et al., 1996). To highlight this colocalization pattern, we used the Colocalization Finder plugin of ImageJ (http://www.rsb.info.nih.gov/ij; Abramoff et al., 2004) to display the pixels in which the signal was strongest in both channels in white, as seen in the fourth column of Figure 3 (max colocal). This analysis revealed in all ML5′ cells that the strongest colocalized FISH signals were in the discrete regions lateral to the pyrenoid (Figure 3B). This pattern is specific to conditions of PSII assembly (i.e., ML5′ cells), because in DA cells the pixels with the strongest colocalized FISH signals were fewer and dispersed within the basal region and some were in lobes (Figure 3A). This pattern is specific to the colocalized psbA and psbC mRNAs, because the pixels with only one strong FISH signal were not localized in any consistent pattern (data not shown).
These results raise the possibility that psbA and psbC translation for de novo PSII assembly occurs in the chloroplast basal region around the pyrenoid and in discrete regions lateral to it, hereafter called T zones (for translation zones). However, additional evidence was required to demonstrate that mRNAs are localized in T zones for their translation rather than for other steps of their expression or their degradation.
Chloroplast Ribosomal Proteins Colocalized with the psbC and psbA mRNA in T Zones Specifically during PSII Assembly
If psbA and psbC mRNAs colocalized in T zones for their translation, chloroplast ribosomes should colocalize with them specifically under conditions of de novo PSII assembly (i.e., in ML5′ cells but not in DA cells). To test this prediction, we analyzed the in situ distributions of r-proteins of both subunits of the chloroplast ribosome, each in pair-wise combinations with the psbC and psbA mRNA FISH signals and in both DA and ML5′ cells. In all ML5′ cells examined, T zones had the strongest colocalized r-protein IF signal and FISH signal from either mRNA (Figures 3D and 3F). By contrast, in most DA cells examined, maximal colocalization was more dispersed (Figures 3C and 3E). Thus, these results indicate that both subunits of the chloroplast ribosome colocalized with the psbA and psbC mRNAs in the T zones specifically in cells that were inducing the translation of these mRNAs for PSII assembly (ML5′ cells), consistent with protein synthesis for PSII assembly occurring in these regions.
Localization of the RNA Binding Protein RB38
We performed a similar analysis with RB38, an RBP that has been proposed to regulate the translation of the psbA mRNA (Barnes et al., 2004). An article in this issue identifies an RBP called RB40 as RB38 and demonstrates that this protein is specifically required for translation initiation on the chloroplast psbD mRNA for PSII biogenesis (Schwarz et al., 2007). In both DA and ML5′ cells, the RB38 IF signal was distributed throughout the chloroplast (Figures 3G and 3H). However, in most ML5′ cells, but not in DA cells, the strongest colocalized signals from RB38 and the psbC mRNA were primarily in T zones. Thus, the colocalization of this RBP with the psbC mRNA in T zones specifically in ML5′ cells further supports our hypothesis that the synthesis of PSII subunits occurs in these regions.
Statistical Analyses Confirmed the Colocalization Patterns
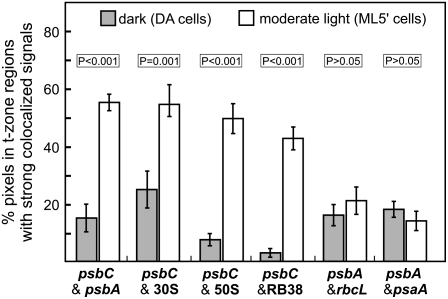
The colocalization patterns shown in Figure 3 were quantified and statistically tabulated across the DA and ML5′ conditions. We used Colocalization Finder (ImageJ) to determine the percentage of pixels that had strong signal from both molecules under comparison specifically in sampled regions (100 pixels) where T zones were observed in ML5′ cells. The mean percentage of pixels with strong colocalized signals in these regions of ML5′ cells, relative to that in DA cells, was higher by 3.6-fold (P < 0.001) for the psbA and psbC mRNAs, by 2.2-fold (P = 0.001) and 6.4-fold (P < 0.001) for the psbC mRNA and the r-proteins of the 30S and 50S ribosomal subunits, respectively, and by 14.3-fold (P < 0.001) for the psbA mRNA and RB38 (Figure 4).
Figure 4.
Statistical Analysis of Colocalization in Regions Where T Zones Are Located across the DA and ML5′ Conditions.
Within multiple images of DA cells and ML5′ cells, regions (100 pixels) located midway between the pyrenoid and the lateral cell periphery were subjected to analysis by Colocalization Finder (ImageJ) to determine the percentage of pixels with strong colocalized signals in the combinations indicated at bottom. Bars indicate mean values for DA cells (gray bars) and ML5′ cells (white bars). Error bars indicate 2 se. In ML5′ cells relative to DA cells, statistically significantly higher levels of percentage colocalization were observed for the psbA and psbC mRNAs and the psbC mRNA with respect to each of the chloroplast ribosomal subunits (30S and 50S) and RB38. In ML5′ cells relative to DA cells, no significant increase was observed in the mean percentage colocalization of the psbA mRNA with the rbcL mRNA or the psaA mRNA in the regions of T zones.
The mRNAs of rbcL and psaA Are Not Recruited to T Zones in ML5′ Cells
To determine whether chloroplast mRNAs encoding subunits of other photosynthesis complexes localize to T zones for their translation, we performed the analyses described above for the colocalization of the FISH signals of the mRNAs of rbcL and psaA relative to the psbA mRNA. rbcL and psaA encode the large subunit of Rubisco and the PsaA subunit of PSI, respectively (Malnoe et al., 1979; Kuck et al., 1987). Neither of these mRNAs showed any particular pattern of colocalization with the psbA mRNA; the pixels with the strongest colocalized signals were seen at apparently random locations throughout the chloroplast in both DA cells and ML5′ cells (Figures 3I to 3L). In regions where T zones are located, the mean percentage of pixels with the strongest colocalized FISH signals were not significantly different between DA and ML5′ cells (Figure 4) (P > 0.05). For the rbcL mRNA, this was not due to a constant level of its translation, because large subunit synthesis was higher in ML5′ cells than in DA cells (Figure 2A, lanes 3 and 4). Moreover, positive light regulation of rbcL translation was demonstrated in a more comprehensive study that controlled for the level of the rbcL mRNA (Herrin et al., 1986). Therefore, the rbcL mRNA is not localized specifically to T zones during the induction of its translation in ML5′ cells. However, it is not known whether psaA translation is activated by light. Therefore, it remains possible that localized psaA translation in T zones is not enhanced in ML5′ cells and, therefore, is not detectable by this approach. Nevertheless, this result supports our conclusion that the psbA and psbC mRNAs colocalized in T zones for their translation for de novo PSII assembly. Additional research is required to determine whether T zones house subunit synthesis for the assembly of thylakoid membrane complexes other than PSII.
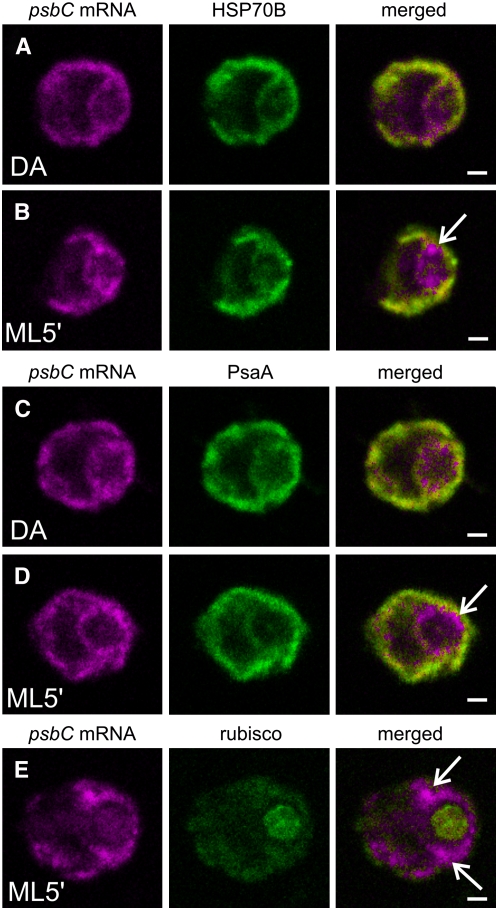
T Zones Are within Stroma and Overlap Thylakoids
To address the location of the T zones, we localized them with respect to the marker proteins for stroma and thylakoids, HSP70B and PsaA, respectively (Redding et al., 1999; Schroda et al., 2001). Although it was not possible to precisely determine where the psbA and psbC mRNAs localized due to the limited resolution of confocal microscopy and the interspersion of these compartments, a few important observations were made. The FISH signals from each of these mRNAs in T zones were within regions of HSP70B or PsaA IF, which were of variable intensities and extended throughout most of the chloroplast (Figures 5B and 5D). Clearly, T zones do not correspond to stroma and thylakoids throughout the chloroplast. Thus, our hypothesis that translation for PSII assembly occurs in T zones contradicts the current model that this process occurs generally at nonappressed thylakoid membranes (see Introduction). Our data are consistent with the initiation steps of psbA and psbC translation occurring in the stroma around the pyrenoid and the subsequent synthesis of D1 and CP43 into the neighboring thylakoid membranes (see Discussion).
Figure 5.
Localization of the psbC mRNA Was Compared with the Distributions of Chloroplast Stroma and Thylakoids under Differential Conditions for de Novo PSII Assembly.
The distributions of the psbC mRNA and marker proteins for chloroplast stroma and thylakoids are compared before and after the induction of PSII subunit synthesis in DA and ML5′ cells, respectively. Image sets show the psbC mRNA FISH signal and IF from the marker protein for stroma (HSP70B) in a DA cell (100%; n = 20) and a ML5′ cell (100%; n = 20) ([A] and [B]), the marker protein for thylakoids (PsaA) in a DA cell (100%; n = 20) and a ML5′ cell (100%; n = 20) ([C] and [D]), and the marker protein complex for the pyrenoid (Rubisco; 100%; n = 20) (E). Arrows indicate where the psbC FISH signal is localized in T zones. The percentage of cells with the localization patterns seen in each image set (and described in the text) and the number of cells examined (n) are given above. Each image shows a 0.2-μm optical section. Bars = 1.0 μm.
The similar distributions of both HSP70B (Figures 5A and 5B) and PsaA (Figures 5C and 5D) in DA and ML5′ cells exclude the possibility that the localization patterns that formed in the T zones (Figures 3B, 3D, 3F, and 3H) resulted from the redistribution of stroma or thylakoids. For example, the T zones clearly are not pockets of stroma with free chloroplast mRNAs, chloroplast ribosome subunits, and RB38.
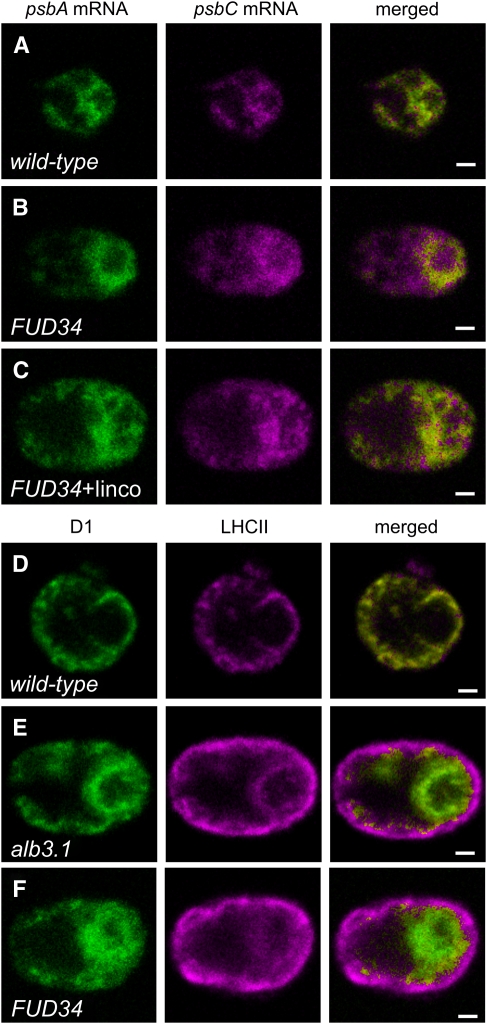
In the FUD34 Mutant, psbA mRNAs Are Localized Near the Pyrenoid by Their Translation
We took advantage of the repertoire of Chlamydomonas mutants that are affected in PSII assembly to address the location of translation for de novo PSII assembly. FUD34 is specifically defective for translation of the psbC mRNA due to a mutation in its 5′ untranslated region (Rochaix et al., 1989; Zerges et al., 1997). Our initial rationale was to examine FUD34 cells for the absence of a psbC mRNA localization pattern that is formed in wild-type cells to identify the location of CP43 synthesis for PSII assembly. However, this was not possible because detection of the relevant localization patterns in wild-type cells (Figure 3) required the induction of de novo PSII biogenesis with light, a process that requires functional PSII (Trebitsh and Danon, 2001). As FuD34 is deficient for PSII, this mutant and the wild-type control strain had to be cultured heterotrophically in the dark (see Methods).
Unexpectedly, much of the psbA FISH signal was localized within ∼1 μm of the pyrenoid in >90% of the FUD34 cells analyzed (Figure 6B). By contrast, 92% of wild-type cells did not show this pattern under the same conditions (Figure 6A). It was not unexpected that the psbA mRNA could be translated in the dark under these heterotrophic growth conditions, because the acetate provided as a reduced carbon source is known to induce protein synthesis in the Chlamydomonas chloroplast in the dark (Michaels and Herrin, 1990). This result seemed consistent with our hypothesis that protein synthesis for de novo PSII assembly occurs near the pyrenoid, considering that FUD34 translates the psbA mRNA at a wild-type level for the assembly of PSII complexes lacking CP43 (de Vitry et al., 1989) and the FUD34 cells did not translate the psbA mRNA for PSII repair (because they were cultured in the dark).
Figure 6.
Analyses of Two PSII Assembly-Impaired Mutants: FUD34 and an alb3.1 Mutant.
(A) to (C) Translation-dependent localization of the psbA mRNA near the pyrenoid was detected in the psbC translation mutant FUD34. The FISH signals from psbA and psbC mRNAs are shown in a wild-type cell (92%; n = 36) (A), an FUD34 mutant cell (90%; n = 20) (B), and an FUD34 mutant cell treated with lincomycin, an inhibitor of translation initiation by chloroplast ribosomes (100%; n = 20) (C).
(D) to (F) Unassembled D1 and the incompletely assembled PSII subcomplex were detected around the pyrenoid in the alb3.1 mutant and FUD34, respectively, where we propose they mark the location of PSII subunit synthesis and assembly. The IF signals from D1 and LHCII are seen in a wild-type cell (100%; n = 20) (D), an alb3.1 mutant cell (81%; n = 89) (E), and an FUD34 mutant cell (90%; n = 20) (F).
The percentage of cells with the localization patterns seen in each image set (and described in the text) and the number of cells examined (n) are given above. Each image shows a 0.2-μm optical section. Bars = 1.0 μm.
To determine whether these localized psbA mRNAs are translated and anchored by their nascent D1 chains to membranes around the pyrenoid, we asked whether the abolition of translation in the chloroplast delocalizes them. All FUD34 cells treated for 10 min with an inhibitor of translation initiation by chloroplast ribosomes, lincomycin (Schmidt et al., 1983), showed a dispersed distribution of the psbA FISH signal (Figure 6C). This result strongly suggests that the majority of the psbA mRNAs localized near the pyrenoid in this mutant are translated and that this localization requires their translation. Since, in FUD34, the psbA mRNA is translated for (incomplete) PSII assembly, these results provide further support of our hypothesis that de novo PSII subunit synthesis occurs near the pyrenoid in the wild type.
The psbA mRNA FISH signal that encircled the pyrenoid in FUD34 is distinct from the colocalization patterns in T zones, which are punctate and typically located lateral to the pyrenoid (Figure 3). However, in ML5′ cells, we also observed a similar ring of colocalized psbA and psbC mRNAs and of the psbC mRNA colocalized with the r-proteins or RB38, which was not observed in most DA cells (cf. the merged images of DA cells with ML5′ cells in Figure 3). This region weakly IF-stained for the marker proteins for stroma and thylakoids (Figures 5B and 5D, respectively).
In an ALBINO3.1 Mutant and FUD34, Intermediates of de Novo PSII Assembly Localize Near the Pyrenoid
Additional evidence for PSII assembly in this region around the pyrenoid was obtained from in situ localization studies of the incompletely assembled PSII subcomplex in FUD34 and the unassembled D1 in an ALBINO3.1 mutant. We predicted that these PSII assembly intermediates might mark the location of the synthesis and assembly of chloroplast-encoded PSII subunits (Leto et al., 1985). ALBINO3.1 functions in the incorporation of D1 during de novo PSII assembly; in the alb3.1 mutant, ac29, D1 is synthesized and integrated into the thylakoid membrane at a wild-type rate, but only 10% is assembled into functional PSII complexes (Bellafiore et al., 2002; Ossenbuhl et al., 2004). D1 and the light-harvesting complex II (LHCII) are components of the PSII-LHCII supercomplex in thylakoid membranes (reviewed in Nield and Barber, 2006). In alb3.1 mutant cells, LHCII associates only with these assembled PSII complexes (Ossenbuhl et al., 2004). Therefore, to characterize the localization of the unassembled D1 in this mutant, we determined where the IF signal of D1 is in excess over that of LHCII. In all wild-type cells, the IF signals from D1 and LHCII colocalized throughout the chloroplast (except within the pyrenoid; Figure 6D), consistent with previously reported results of immunogold EM (Vallon et al., 1985). By contrast, the majority of the alb3.1 cells had a dramatic localization of D1 in the chloroplast basal region, particularly within ∼1 μm of the pyrenoid (Figure 6E). This region only weakly IF-stained for LHCII, suggesting that the unassembled D1 in this mutant is localized there. This is in contrast with the lobes and the margin of the basal region, where LHCII IF was stronger than D1 IF, consistent with the proper localization of the fewer PSII-LHCII supercomplexes that assemble in this mutant. Similarly, most FUD34 cells showed localization of D1 in this pattern (Figure 6F). This D1 IF signal was mostly or entirely from the incompletely assembled PSII subcomplex, because a previous study found all detectable D1 with this subcomplex in FUD34 (de Vitry et al., 1989). In FUD34, LHCII probably accumulates in the absence of its association with PSII, as has been found in a mutant that is defective in the association between these complexes (Swiatek et al., 2001).
A general disorganization of thylakoid membranes does not localize unassembled D1 and the PSII subcomplex near the pyrenoid in the alb3.1 mutant and FUD34, respectively, because the LHCII distribution in both mutants appeared wild type. Thus, these results further support our hypothesis that protein synthesis occurs for de novo PSII assembly in the region surrounding the pyrenoid (see Discussion).
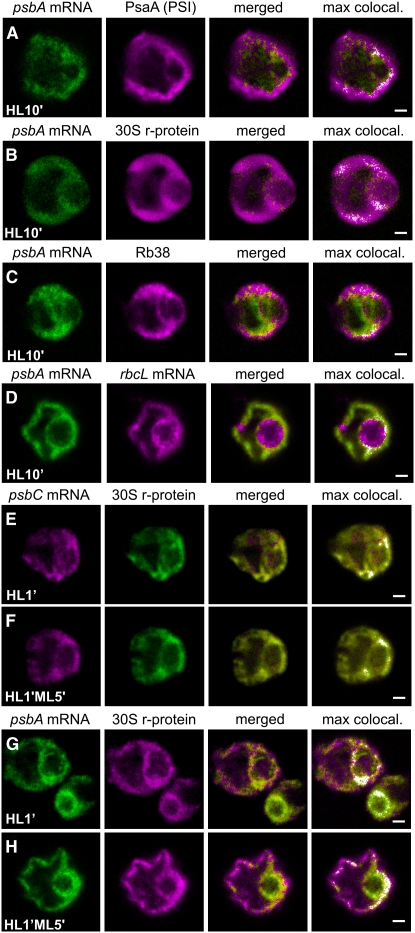
In Situ Evidence for D1 Repair Synthesis at Thylakoids throughout the Chloroplast
The incorporation of newly synthesized D1 subunits into photodamaged PSII complexes during high-light stress conditions is believed to occur at stroma thylakoid membranes, based on previously reported results of thylakoid subfractionation experiments (Adir et al., 1990). However, to our knowledge, there are no reported in situ data regarding the location of this process. Therefore, we used fluorescence confocal microscopy to address this problem. In HL10′ cells, the specific induction of D1 synthesis for PSII repair (Figure 2A, lane 1) correlated with a diffuse distribution of the psbA FISH signal throughout the chloroplast, where it colocalized with IF from the thylakoid marker protein PsaA, a chloroplast r-protein, and RB38 (Figures 7A to 7C). This pattern is not common to all chloroplast mRNAs during this high-light stress condition, because the rbcL mRNA was more concentrated around the pyrenoid in HL10′ cells (Figure 7D). This localization pattern will be described further in a subsequent report. These results are consistent with the current model that D1 repair synthesis occurs generally at nonappressed thylakoid membranes, because they did not reveal a specific location of psbA translation in cells carrying out this process.
Figure 7.
Under Conditions of D1 Repair Synthesis, the psbA mRNA Colocalized with Thylakoids and Translation Components throughout the Chloroplast.
(A) to (D) DA cells were exposed to HL for 10 min (HL10′ cells) to induce PSII photodamage and D1 repair synthesis and then FISH-probed for the psbA mRNA and concurrently IF-stained for the thylakoid marker protein PsaA (100%; n = 20) (A), the r-protein of the 30S subunit of the chloroplast ribosome (100%; n = 20) (B), the RNA binding protein RB38 (100%; n = 20) (C), or the rbcL mRNA (D).
(E) to (H) Cells were IF-stained for the r-protein of the 30S chloroplast ribosomal subunit and co-FISH-probed for the psbC mRNA ([E] and [F]) or the psbA mRNA ([G] and [H]). The psbA and psbC mRNAs colocalized with the 30S ribosomal subunit r-protein in T zones during the induction of PSII assembly in DA cells that were exposed to HL for 1 min ([E] and [G], respectively). Distinct localization of the psbA and psbC mRNAs during the D1 repair synthesis was induced during a subsequent 5-min incubation under ML ([E] and [G], respectively).
The percentage of cells with the localization patterns seen in each image set (and described in the text) and the number of cells examined (n) are given above. The fourth column shows merged images in which the pixels with the strongest colocalized signals are highlighted in white. Each image shows a 0.2-μm optical section. Bars = 1.0 μm.
Affirmative results in support of this conclusion were obtained when we looked for colocalization of psbA mRNA with the r-protein of the 30S ribosomal subunit that formed during the induction of PSII repair in cells that had localized psbA and psbC mRNAs in T zones. In this experiment, the initial localization of the psbA mRNA in T zones provided a reference for the identification of a repair-specific localization pattern. The psbC mRNA localized in T zones provided a negative control as an mRNA whose translation is not induced during PSII repair (Figure 2A, lane 1) (Minai et al., 2006). When DA cells were exposed to HL for only 1 min (HL1′ cells), D1 and D2 synthesis were induced for de novo PSII assembly (Figure 2B, lanes 2 and 3) and the strongest colocalized signals from the 30S subunit r-protein and either the psbA mRNA or the psbC mRNA were in T zones (Figures 7E and 7G, respectively), as was observed in ML5′ cells (Figure 3). This localization pattern was more distinct for the psbC mRNA than for the psbA mRNA, possibly because these cells were also inducing psbA translation for PSII repair, which became evident after a 5-min incubation under ML (Figure 2B, lane 4). This delayed induction of D1 repair synthesis occurred amid a sustained subunit synthesis for PSII assembly (Figure 2B, lane 4). During this 5-min incubation under ML, the strongest colocalized signals from the psbC mRNA and the 30S r-protein remained localized in T zones in most HL1′ML5′ cells (Figure 7F). However, in these HL1′ML5′ cells, the strongest colocalized signals from the psbA mRNA and the 30S subunit r-protein were broadly distributed throughout the basal region and within the lobes (Figure 7H). Therefore, the appearance of this dispersed distribution pattern correlated with the induction of D1 repair synthesis, and this was not seen for the psbC mRNA and the 30S subunit r-protein. These results further support the localization of the psbA translation throughout regions of the chloroplast with thylakoid membranes for the repair of photodamaged PSII complexes.
DISCUSSION
Our results provide in situ evidence for the current model that D1 synthesis for PSII repair occurs in stroma thylakoid membranes throughout the chloroplast (Figure 7) (Adir et al., 1990). They also contribute to the emergent realization that the processes underlying chloroplast biogenesis are highly compartmentalized by providing three lines of evidence showing that PSII subunit synthesis and assembly occur in a specific region of the chloroplast. First, during the rapid induction of PSII assembly, we observed in punctate regions lateral to the pyrenoid, termed T zones, the colocalization of multiple components of PSII subunit synthesis: the psbC and psbA mRNAs, chloroplast r-proteins, and RB38 (Figure 3). Weaker colocalized signals from these mRNAs and translation proteins were localized with stroma and thylakoids around the pyrenoid, specifically in ML5′ cells (Figures 3B, 3D, 3F, and 3H). The T zones were typically within this region. Second, in the psbC translation-deficient mutant FUD34, ongoing psbA translation for attempted PSII assembly correlated with the translation-dependent localization of the psbA mRNA around the pyrenoid (Figures 6B and 6C). Third, in two PSII assembly mutants, unassembled D1 and a partially assembled PSII subcomplex accumulate around the pyrenoid (Figures 6E and 6F), where we propose that they mark a compartment of PSII subunit assembly (see below).
On the Location of the PSII Assembly Compartment
Most T zones did not contact the pyrenoid (Figure 5E). T zones probably are within stroma, based on comparisons of the psbC mRNA FISH signal and IF from HSP70B (Figure 5B). Close inspection of ML5′ cells that were FISH-probed for either mRNA and co-IF-stained for the thylakoid marker protein PsaA revealed that most T zones overlapped with regions of the basal chloroplast that were densely populated by thylakoids (Figure 5D).
Previous EM studies revealed that the pyrenoid is surrounded by starch plates, then by a sphere of stroma, and that stroma and thylakoids are interspersed throughout the rest of the chloroplast (Ohad et al., 1967b). Chua et al. (1976) proposed that translation initiation of the Chlamydomonas chloroplast mRNAs encoding thylakoid membrane proteins occurs in the stroma and that these ribosomes subsequently localize at thylakoid membranes for the insertion of their polypeptide products. Our results are consistent with these steps occurring specifically in stroma and thylakoids within the T zones and generally around the pyrenoid.
The chloroplast envelope appears not to be a major location of PSII subunit synthesis and assembly (see Introduction): the chloroplast periphery was not enriched in chloroplast mRNAs, chloroplast r-proteins, or RB38 under any of the conditions examined. However, our data and most of the evidence for the synthesis and assembly of photosynthesis complexes at the inner envelope membrane can be reconciled if thylakoid biogenesis occurs in an internal chloroplast membrane compartment that has properties of the chloroplast envelope (see Introduction; reviewed in Zerges, 2000). It will be important to characterize T zones and the membranes around the pyrenoid to determine whether or not they contain known photosystem assembly factors, enzymes of chlorophyll and thylakoid membrane lipid biosynthesis, and the light-activated RBPs that have been proposed to localize chloroplast mRNAs for thylakoid biogenesis (Zerges and Rochaix, 1998; Zerges et al., 2002; Minagawa and Takahashi, 2004).
In published EM images of the chloroplast basal region, we noticed discrete structures with the same approximate size and location as T zones that appeared to be distinct from thylakoids and more electron-dense than stroma (Ohad et al., 1967b). Thylakoids connected with these structures, and a few thylakoid vesicles extended into them. Only one report mentioned these structures, in Chlamydomonas moewusii, and proposed that they contain chloroplast DNA because they were seen to contain fibers (Ris and Plaut, 1962).
Transcription could not have generated the colocalization patterns involving the mRNAs in T zones because their formation was not affected by rifampicin, an inhibitor of the chloroplast RNA polymerase in Chlamydomonas (see Supplemental Figure 2A online). Moreover, chloroplast nucleoids (the structures that contain multiple copies of the chloroplast genome) did not consistently colocalize with the mRNAs in T zones stained with 4′,6-diamidino-2-phenylindole (see Supplemental Figure 2B online). Also, the psbA FISH probes hybridize across exon junctions and, therefore, do not detect the psbA gene or unspliced mRNAs (see Methods). It is improbable that mature transcripts could be generated from a 6.6-kb gene with four introns within the 2 to 3 min that the psbA mRNAs localized to T zones in HL1′ cells (Figure 7G).
Light Regulates Translation for PSII Assembly and Repair
The results of 35S pulse-labeling experiments shown in Figure 2 indicate distinct modes of light regulation of protein synthesis in chloroplasts for PSII assembly and repair. While these modes of regulation have been described previously, the distinction between them has not been emphasized in most publications. The mechanisms underlying these modes of light regulation, at least in Chlamydomonas, probably are distinct, because the psbA 5′ untranslated region mediates the former (Franklin et al., 2002; Mayfield and Schultz, 2004) but not the latter (Minai et al., 2006). One study showed a clear distinction between the regulation of psbA translation by autofeedback repression for PSII assembly and by HL for D1 repair synthesis (Minai et al., 2006).
The Morphology of the Chlamydomonas Chloroplast Is Complex
Chloroplasts in our images did not have the generally accepted cup-like morphology with the apical chloroplast as a continuous rim (Figure 1). Rather, we observed the chloroplast morphology seen in three-dimensional reconstructions of a few cells that were made from serial EM images (Schotz et al., 1972; Gaffal et al., 1995). This morphology is retained by isolated chloroplasts (Mason et al., 2006), suggesting that it is maintained by a plastoskeleton (Reski, 2002). Clearly, the morphology of the Chlamydomonas chloroplast is complex and additional research is required to characterize it and the molecules and mechanisms underlying it.
PSII Biogenesis May Occur in a Spatiotemporal Pathway
Our proposal that the synthesis and assembly of PSII subunits occur near the pyrenoid requires a process by which newly assembled complexes get to thylakoids throughout the chloroplast. Several of our results suggest the existence of a spatiotemporal pathway of PSII biogenesis. In this model, T zones would house the earliest stages of PSII subunit synthesis and assembly. Indeed, the colocalization patterns were detected in these regions within 1 min of light exposure (Figures 7E and 7G). An intermediate stage in this pathway could be located around the pyrenoid, where we observed the accumulation of unassembled D1 and the PSII subcomplex in steady state conditions in the alb3.1 mutant and FUD34, respectively (Figures 6E and 6F). The colocalized mRNAs and translation machinery components that we observed distinctly around the pyrenoid specifically in ML5′ cells, and the translation-dependent localization of the psbA mRNA in this region in FuD34 (Figure 6B), also could reflect protein synthesis for PSII biogenesis in this proposed intermediate compartment.
Such a spatiotemporal pathway of PSII biogenesis could involve a branch of the vesicular transport system that functions within chloroplasts for thylakoid biogenesis (Ohad et al., 1967b; Hoober et al., 1991; White and Hoober, 1994; Kroll et al., 2001; Vothknecht and Soll, 2005). Alternatively, newly assembled PSII complexes might laterally migrate within preexisting thylakoid membranes, which are largely contiguous between the basal region and the entire length of the lobes (Ohad et al., 1967b). As a third possibility, thylakoids could be built upon their termini in the basal region, where they are anchored at the pyrenoid and, consequently, being pushed in an apical direction either into the lobes or even to generate the lobes during chloroplast differentiation. The pyrenoid has a system of tubules into which thylakoid vesicles extend (Ohad et al., 1967a). The function(s) of these tubules, and why they contain thylakoid vesicles, are long-standing mysteries. In this third model, pyrenoid tubules could serve as the anchors.
In the context of this spatiotemporal model for PSII assembly, our observation that unassembled D1 and the PSII subcomplex localize around the pyrenoid in the alb3.1 mutant and FUD34, respectively (Figures 6E and 6F), suggests the existence of a quality-control checkpoint that ensures that only assembled PSII complexes are transported outward. This would be analogous to the system that retains nonnative and incompletely assembled complexes in the rough endoplasmic reticulum (Ellgaard and Helenius, 2003). Indeed, the half-lives of this unassembled D1 and the PSII subcomplex are >1 h (de Vitry et al., 1989; Ossenbuhl et al., 2004), suggesting that they should disperse by diffusion unless they are retained by some mechanism, such as a quality-control checkpoint in an active transport system.
Many problems remain to be addressed. The mechanisms that target chloroplast-encoded proteins are only beginning to be identified. It will be important to determine whether mRNAs are localized for PSII assembly and repair by RNA binding proteins, by interactions between the Chlamydomonas homolog of the conserved signal sequence binding protein cpSRP54 (AF238499), or by other mechanisms. We expect that the cytological organization of PSII assembly and repair described here will provide a context to frame future research into thylakoid membrane biogenesis.
METHODS
Culture Conditions
For the DA, ML, HL, and ML5′ conditions, cells were cultured photoautotrophically in high-salt minimal medium (Harris, 1989) until midlog phase (∼3 × 106 cells/mL) at 24°C and under a light intensity of 100 to 150 μE·m−2·s−1. Dark-adaptation was accomplished by a 2-h incubation in flasks wrapped with two layers of aluminum foil on an orbital shaker at 24°C. HL (2000 μE·m−2·s−1) was generated by a slide projector (Kodak) to which small (2 to 30 mL) cultures were exposed at a distance of 10 cm with manual shaking. The alb3.1 mutant (ac29; CC-245), FUD34 (CC-2518), and the control wild-type cells used in experiments with these mutants (Figure 6) were cultured on Tris-acetate-phosphate medium (Gorman and Levine, 1965) in the dark. When lincomycin was used, it was added to a final concentration of 200 μg/mL at 10 min prior to cell fixation. The chloroplast transcription inhibitor, rifampicin, was added to a final concentration of 350 μg/mL at 60 min prior to the end of the 2-h dark-adaptation period for the control experiment. The rifampicin was active because it was lethal at this concentration after 24 h (data not shown).
In Vivo 35S Pulse-Labeling Experiments
The level of 35S labeling of each subunit during a 5-min pulse-labeling period reflects its synthesis rate, because previous studies found that the levels of the mRNA encoding these subunits do not change dramatically in response to light exposure (Malnoe et al., 1988; Lee and Herrin, 2002) and degradation of these subunits was not detected within 30 min of their synthesis (de Vitry et al., 1989). Each pulse-labeling experiment was performed for 5 min with 1.2 × 107 cells in 0.3 mL of high salt minimal medium lacking NH4SO4. Cycloheximide was added to a final concentration of 10 μg/mL at 5 min prior to the addition of 80 μCi of [35S]SO4. Cells were not deprived of SO4 for more than the ∼15 min required for centrifugation and resuspension and treatment with cycloheximide prior to pulse-labeling, during which time 100 μM SO4 was available from the trace element mixture present in this medium. Pulse-labeling was performed on an orbital shaker for 5 min under the light conditions described. Cells were pelleted by centrifugation at 4000g for 2 min, resuspended and lysed in 100 μL of SDS-PAGE loading buffer (250 mM Tris, pH 6.8, 2% SDS, 20% glycerol, 50 mM DTT, and 2% 2-mercaptoethanol), and incubated at room temperature for 60 min, and then 50 μL of each sample was loaded on a 13% denaturing SDS-polyacrylamide gel (with 8M urea) on the basis of cell number (6 × 106 cells/lane). Following electrophoresis, the gels were dried and 35S-labeled proteins were revealed by autoradiography. Coomassie Brilliant Blue staining confirmed that comparable amounts of total protein were in the lanes (data not shown). The light stimulation of the 35S labeling of these proteins does not reflect effects on the rate of [35S]SO4 uptake, because a 200-fold excess of sulfate immediately prior to light exposure did not affect the pattern of labeled proteins (data not shown).
FISH
Cell fixation and permeabilization and FISH were performed as described previously (Colon-Ramos et al., 2003). Four FISH probes of 50 nucleotides were designed to hybridize to each of the mRNAs (Table 1) (Dron et al., 1982; Erickson et al., 1984; Kuck et al., 1987; Rochaix et al., 1989). The probes against the psbA mRNA hybridize across exon junctions. The psbA, rbcL, and psaA mRNA FISH signals were highly specific because they were not detected in deletion mutants for these genes (Bennoun et al., 1986; Redding et al., 1999; Satagopan and Spreitzer, 2004) (see Supplemental Figure 1 online). FISH probes were labeled with either Alexa Fluor 488 or Alexa Fluor 555 (Molecular Probes) according to the manufacturer's protocol on five modified C6-dT residues. Labeled probes were purified from a denaturing 13% polyacrylamide gel by excising the bands, manually crushing them, and overnight incubation in elution buffer (0.1% SDS, 10 mM magnesium acetate, and 0.5 M ammonium acetate) at 37°C. Triton X-100 at 1% (v/v) was used to permeabilize the fixed cells. The hybridization buffer contained 10 mM vanadyl ribonucleoside complex (VRC). The mounting medium was ProLong Gold Anti-fade (Molecular Probes).
IF Staining
Cells that had been subjected to FISH were incubated in blocking solution (PBS [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4], 0.1% BSA, and 2 mM VRC) for 30 min at room temperature, followed by a 75-min incubation with diluted primary antiserum in blocking buffer (with 10 mM VRC) at 37°C. After washing twice in PBS, cells were incubated in a 1:200 dilution of an anti-rabbit secondary antibody in blocking buffer (with 10 mM VRC) at room temperature for 45 min. The secondary antibody was anti-rabbit IgG–TRITC or IgG–fluorescein isothiocyanate (Sigma-Aldrich). ProLong Gold Anti-fade (Molecular Probes) was used for mounting.
The in situ IF staining patterns of D1, PsaA, and the r-proteins were highly specific, because each of the antibodies detected a protein of the expected molecular weight on immunoblots of total cellular protein (data not shown). The specificities of the IF signals from PsaA and D1 were also demonstrated by their absence in deletion mutants for psaA (Redding et al., 1999) and psbA (FuD7) (Bennoun et al., 1986), respectively (see Supplemental Figures 1D and 1E online). Staining with the secondary antisera alone did not generate a signal (data not shown). The r-protein of the 60S subunit of the cytoplasmic ribosome was detected by the antiserum described previously and called L4; it very weakly cross-reacts against two smaller r-proteins, L47 (60S) and S26 (30S) (data not shown) (Fleming et al., 1987a, 1987b). A chicken antiserum against D1 (AgriSera) was used in conjunction with a rabbit antiserum against LHCII (Figures 6D to 6F). It was critical to titer the primary antisera (especially those against thylakoid membrane proteins) and use them at the lowest concentration required to give a signal to avoid artifactual highly punctate IF staining patterns. The 50S subunit r-protein of the 50S subunit, L2, was detected by an antiserum described previously as L-1 (Schmidt et al., 1983, 1984; Randolph-Anderson et al., 1989). The antiserum against the r-protein of the 30S subunit of the Chlamydomonas reinhardtii chloroplast ribosome was called S-21 (Schmidt et al., 1983, 1984; Randolph-Anderson et al., 1989). Although the molecular weight of the r-protein detected with the S-21 antiserum is similar to that of the Chlamydomonas ortholog of Escherichia coli S21, they drastically differ in pI (Schmidt et al., 1983; Yamaguchi et al., 2002). The protein detected by this antiserum is most similar to the r-protein PSRP-4 in molecular mass, pI, and nuclear location of the gene(s) (Schmidt et al., 1983; Yamaguchi et al., 2002). Despite this uncertainty, these antisera (L-1 and S-21) detect bona fide r-proteins, because they were raised against proteins from highly purified chloroplast ribosome subunits and their levels were reduced severalfold in a mutant that accumulates ∼30% of the wild-type level of chloroplast ribosomes (immunoblot data not shown).
Microscopy
Fluorescence signals were visualized in serial 0.2-μm optical sections obtained with a Leica TCS SP2 confocal laser-scanning microscope and image acquisition software version 2.61 (Leica). Argon and green helium neon lasers were used to produce the 488- and 543-nm stimulation of fluorophores Alexa Fluor 488 and Alexa Fluor 555, respectively. Deconvolution was not used. Cells were observed under immersion oil with an HCX PL APO objective lens of 100× and numerical aperture of 1.4. Images were acquired in 512 × 512 format with digital zoom set at 4.8× with a pinhole size of 0.84 airy. Glow-Over (Leica) images were obtained after adjusting the maximal signal in each section to just below saturation. 4′,6-Diamidino-2-phenylindole was visualized with a Zeiss Axioplan fluorescence microscope with UV-G 365 filter cube, 100× oil objective with numerical aperture of 1.3, and SPOT Insight color camera model 3.2.0.
Statistical Analysis of Colocalization
Two areas of 10 × 10 pixels were cropped per cell from the regions located midway between the pyrenoid and the cell periphery (i.e., where T zones were observed in ML5′ cells) from DA and ML5′ cells using Corel PHOTO-PAINT. A crop in one channel (red) was taken at the precise location of the crop in the other channel (green) using the xy coordinates. Colocalization was compared for both signals with the ImageJ plugin Colocalization Finder. To determine the percentage of intense colocalized pixels in each sampled region, only pixels with both channels above a fluorescence intensity of 175 were selected from the scatterplot generated by this plugin (an 80 × 80 square in the upper right corner of the plot).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Control Experiments Revealed High Specificities of FISH and IF Signals.
Supplemental Figure 2. psbA Transcription Does Not Generate the Localized psbA mRNAs in T Zones.
Supplementary Material
Acknowledgments
We thank Grant Brown, Daniel Colon-Ramos, Michel Goldschmidt-Clermont, Elizabeth Harris, Ursula Oberholzer, Jean-David Rochaix, Michael Schroda, Robert Spreitzer, Olivier Vallon, and Francis-Andre Wollman for helpful comments and technical advice; Marc Champagne and Julio Vasquez for invaluable advice regarding confocal microscopy and image analyses; Andrea Auchincloss, Marc Champagne, and Ursula Oberholzer for critical reading of the manuscript; Elizabeth Harris, Michael Hippler, Stephen Mayfield, Kevin Redding, and Michael Schroda for antisera; Kevin Redding for the psaA deletion mutant; and Genhai Zhu and Pioneer Hi-Bred International for the rbcL deletion mutant MX3312. This work used the confocal microscope of the Centre for Structural and Functional Genomics (Concordia University) and was funded by Operating Grant 217566-03 from the National Science and Engineering Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: William Zerges (zerges@alcor.concordia.ca).
Online version contains Web-only data.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics International 11 36–42. [Google Scholar]
- Adir, N., Shochat, S., and Ohad, I. (1990). Light-dependent D1 protein synthesis and translocation is regulated by reaction center II. Reaction center II serves as an acceptor for the D1 precursor. J. Biol. Chem. 265 12563–12568. [PubMed] [Google Scholar]
- Andersson, B., and Anderson, J.M. (1980). Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta 593 427–440. [DOI] [PubMed] [Google Scholar]
- Aro, E.M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143 113–134. [DOI] [PubMed] [Google Scholar]
- Barnes, D., Cohen, A., Bruick, R.K., Kantardjieff, K., Fowler, S., Efuet, E., and Mayfield, S.P. (2004). Identification and characterization of a novel RNA binding protein that associates with the 5′-untranslated region of the chloroplast psbA mRNA. Biochemistry 43 8541–8550. [DOI] [PubMed] [Google Scholar]
- Beligni, M.V., Yamaguchi, K., and Mayfield, S.P. (2004). The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth. Res. 82 315–325. [DOI] [PubMed] [Google Scholar]
- Bellafiore, S., Ferris, P., Naver, H., Gohre, V., and Rochaix, J.D. (2002). Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14 2303–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning, C., Xu, C., and Awai, K. (2006). Non-vesicular and vesicular lipid trafficking involving plastids. Curr. Opin. Plant Biol. 9 241–247. [DOI] [PubMed] [Google Scholar]
- Bennoun, P., Spierer-Herz, J.E., Girard-Bascou, J., Pierre, Y., Delosome, M., and Rochaix, J.-D. (1986). Characterization of photosystem II mutants of Chlamydomonas rienhardtii lacking the psbA gene. Plant Mol. Biol. 6 151–160. [DOI] [PubMed] [Google Scholar]
- Borkhsenious, O.N., Mason, C.B., and Moroney, J.V. (1998). The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiol. 116 1585–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, D.P., Boynton, J.E., and Gillham, N.W. (1971). Studies on the structure and cellular location of various ribosome and ribosomal RNA species in the green alga Chlamydomonas reinhardtii. J. Cell Sci. 8 153–183. [DOI] [PubMed] [Google Scholar]
- Breidenbach, E., Jenni, E., and Boschetti, A. (1988). Synthesis of two proteins in chloroplasts and mRNA distribution between thylakoids and stroma during the cell cycle of Chlamydomonas reinhardtii. Eur. J. Biochem. 177 225–232. [DOI] [PubMed] [Google Scholar]
- Chua, N.-H., Blobel, G., Siekevitz, P., and Palade, G.E. (1973). Attachment of chloroplast polysomes to thylakoid membranes in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 70 1554–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, N.H., Blobel, G., Siekevitz, P., and Palade, G.E. (1976). Periodic variations in the ratio of free to thylakoid-bound chloroplast ribosomes during the cell cycle of Chlamydomonas reinhardtii. J. Cell Biol. 71 497–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Ramos, D.A., Salisbury, J.L., Sanders, M.A., Shenoy, S.M., Singer, R.H., and Garcia-Blanco, M.A. (2003). Asymmetric distribution of nuclear pore complexes and the cytoplasmic localization of beta2-tubulin mRNA in Chlamydomonas reinhardtii. Dev. Cell 4 941–952. [DOI] [PubMed] [Google Scholar]
- Devereux, R., Loeblich, A.R., III, and Fox, G.E. (1990). Higher plant origins and the phylogeny of green algae. J. Mol. Evol. 31 18–24. [DOI] [PubMed] [Google Scholar]
- de Vitry, C., Olive, J., Drapier, D., Recouvreur, M., and Wollman, F.A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron, M., Rahire, M., and Rochaix, J.D. (1982). Sequence of the chloroplast DNA region of Chlamydomonas reinhardtii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J. Mol. Biol. 162 775–793. [DOI] [PubMed] [Google Scholar]
- Eberhard, S., Drapier, D., and Wollman, F.A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: Relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31 149–160. [DOI] [PubMed] [Google Scholar]
- Eckhardt, U., Grimm, B., and Hortensteiner, S. (2004). Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 56 1–14. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4 181–191. [DOI] [PubMed] [Google Scholar]
- Erickson, J.M., Rahire, M., and Rochaix, J.D. (1984). Chlamydomonas reinhardtii gene for the 32,000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 3 2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, G.H., Boynton, J.E., and Gillham, N.W. (1987. a). The cytoplasmic ribosomes of Chlamydomonas reinhardtii: Characterization of antibiotic sensitivity and cycloheximide-resistant mutants. Mol. Gen. Genet. 210 419–428. [DOI] [PubMed] [Google Scholar]
- Fleming, G.H., Boynton, J.E., and Gillham, N.W. (1987. b). Cytoplasmic ribosomal proteins from Chlamydomonas reinhardtii: Characterization and immunological comparisons. Mol. Gen. Genet. 206 226–237. [DOI] [PubMed] [Google Scholar]
- Franklin, S., Ngo, B., Efuet, E., and Mayfield, S.P. (2002). Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J. 30 733–744. [DOI] [PubMed] [Google Scholar]
- Gaffal, K.P., Arnold, C.-G., Friedriches, G.J., and Gemple, W. (1995). Morphodynamical changes of the chloroplast of Chlamydomonas reinhardtii during the 1st round of division. Arch. Protistenkd. 145 10–23. [Google Scholar]
- Gorman, D.S., and Levine, R.P. (1965). Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.W. (1999). Evolution of organellar genomes. Curr. Opin. Genet. Dev. 9 678–687. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Herrin, D., and Michaels, A. (1985). The chloroplast 32 kDa protein is synthesized on thylakoid-bound ribosomes in Chlamydomonas reinhardtii. FEBS Lett. 184 90–95. [Google Scholar]
- Herrin, D.L., Michaels, A.S., and Paul, A.L. (1986). Regulation of genes encoding the large subunit of ribulose-1,5- bisphosphate carboxylase and the photosystem II polypeptides D-1 and D-2 during the cell cycle of Chlamydomonas reinhardtii. J. Cell Biol. 103 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober, J.K., Boyd, C.O., and Paavola, L.G. (1991). Origin of the thylakoid membranes in Chlamydomonas reinhardtii y-1 in the light or dark. Plant Physiol. 96 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard, J., Block, M.A., and Douce, R. (1991). Molecular aspects of plastid envelope biochemistry. Eur. J. Biochem. 199 489–509. [DOI] [PubMed] [Google Scholar]
- Joyard, J., Douce, R., Siebertz, H.P., and Heinz, E. (1980). Distribution of radioactive lipids between envelopes and thylakoids from chloroplasts labelled in vivo. Eur. J. Biochem. 108 171–176. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, F., and Schnell, D.J. (2006). The function and diversity of plastid protein import pathways: A multilane GTPase highway into plastids. Traffic 7 248–257. [DOI] [PubMed] [Google Scholar]
- Kettunen, R., Pursiheimo, S., Rintamaki, E., Van Wijk, K.J., and Aro, E.M. (1997). Transcriptional and translational adjustments of psbA gene expression in mature chloroplasts during photoinhibition and subsequent repair of photosystem II. Eur. J. Biochem. 247 441–448. [DOI] [PubMed] [Google Scholar]
- Klein, R.R., Mason, H.S., and Mullet, J.E. (1988). Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J. Cell Biol. 106 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, D., Meierhoff, K., Bechtold, N., Kinoshita, M., Westphal, S., Vothknecht, U.C., Soll, J., and Westhoff, P. (2001). VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98 4238–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck, U., Choquet, Y., Schneider, M., Dron, M., and Bennoun, P. (1987). Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardtii: Evidence for in vivo trans-splicing. EMBO J. 6 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste-Royal, G., and Gibbs, S.P. (1987). Immunocytochemical localization of ribulose-1,5-bisphosphate carboxylase in the pyrenoid and thylakoid region of the chloroplast of Chlamydomonas reinhardtii. Plant Physiol. 83 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., and Herrin, D. (2002). Assessing the relative importance of light and the circadian clock in controlling chloroplast translation in Chlamydomonas reinhardtii. Photosynth. Res. 72 295–306. [DOI] [PubMed] [Google Scholar]
- Lee, J., and Herrin, D.L. (2003). Mutagenesis of a light-regulated psbA intron reveals the importance of efficient splicing for photosynthetic growth. Nucleic Acids Res. 31 4361–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leto, K.J., Bell, E., and McIntosh, L. (1985). Nuclear mutation leads to an accelerated turnover of chloroplast-encoded 48 kD and 34.5 kD polypeptides in thylakoids lacking photosystem II. EMBO J. 4 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoe, P., Mayfield, S.P., and Rochaix, J.D. (1988). Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. J. Cell Biol. 106 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoe, P., Rochaix, J.D., Chua, N.H., and Spahr, P.F. (1979). Characterization of the gene and messenger RNA of the large subunit of ribulose 1,5-diphosphate carboxylase in Chlamydomonas reinhardtii. J. Mol. Biol. 133 417–434. [DOI] [PubMed] [Google Scholar]
- Margulies, M.M. (1983). Synthesis of photosynthetic membrane proteins directed by RNA from rough thylakoids of Chlamydomonas reinhardtii. Eur. J. Biochem. 137 241–248. [DOI] [PubMed] [Google Scholar]
- Margulies, M.M., and Michaels, A. (1974). Ribosomes bound to chloroplast membranes in Chlamydomonas reinhardtii. J. Cell Biol. 60 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, M.M., and Michaels, A. (1975). Free and membrane-bound chloroplast polyribosomes Chlamydomonas reinhardtii. Biochim. Biophys. Acta 402 297–308. [DOI] [PubMed] [Google Scholar]
- Margulies, M.M., Tiffany, H.L., and Hattori, T. (1987). Photosystem I reaction center polypeptides of spinach are synthesized on thylakoid-bound ribosomes. Arch. Biochem. Biophys. 254 454–461. [DOI] [PubMed] [Google Scholar]
- Margulies, M.M., Tiffany, H.L., and Michaels, A. (1975). Vectorial discharge of nascent polypeptides attached to chloroplast thylakoid membranes. Biochem. Biophys. Res. Commun. 64 735–739. [DOI] [PubMed] [Google Scholar]
- Margulies, M.M., and Weistrop, J.S. (1980). Sub-thylakoid fractions containing ribosomes. Biochim. Biophys. Acta 606 20–33. [DOI] [PubMed] [Google Scholar]
- Mason, C.B., Bricker, T.M., and Moroney, J.V. (2006). A rapid method for chloroplast isolation from the green alga Chlamydomonas reinhardtii. Nat. Protocols 1 2227–2230. [DOI] [PubMed] [Google Scholar]
- Mattoo, A.K., and Edelman, M. (1987). Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32-kDa herbicide-binding protein. Proc. Natl. Acad. Sci. USA 84 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield, S.P., and Schultz, J. (2004). Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 37 449–458. [DOI] [PubMed] [Google Scholar]
- Michaels, A., and Herrin, D.L. (1990). Translational regulation of chloroplast gene expression during the light-dark cell cycle of Chlamydomonas: Evidence for control by ATP/energy supply. Biochem. Biophys. Res. Commun. 170 1082–1088. [DOI] [PubMed] [Google Scholar]
- Minagawa, J., and Takahashi, Y. (2004). Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth. Res. 82 241–263. [DOI] [PubMed] [Google Scholar]
- Minai, L., Wostrikoff, K., Wollman, F.A., and Choquet, Y. (2006). Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18 159–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nield, J., and Barber, J. (2006). Refinement of the structural model for the Photosystem II supercomplex of higher plants. Biochim. Biophys. Acta 1757 353–361. [DOI] [PubMed] [Google Scholar]
- Ohad, I., Siekevitz, P., and Palade, G.E. (1967. a). Biogenesis of chloroplast membranes. II. Plastid differentiation during greening of a dark-grown algal mutant (Chlamydomonas reinhardtii). J. Cell Biol. 35 553–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, I., Siekevitz, P., and Palade, G.E. (1967. b). Biogenesis of chloroplast membranes. I. Plastid dedifferentiation in a dark-grown algal mutant (Chlamydomonas reinhardtii). J. Cell Biol. 35 521–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi, N., and Takahashi, Y. (2001). PsbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. J. Biol. Chem. 276 33798–33804. [DOI] [PubMed] [Google Scholar]
- Ossenbuhl, F., Gohre, V., Meurer, J., Krieger-Liszkay, A., Rochaix, J.D., and Eichacker, L.A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph-Anderson, B.L., Gillham, N.W., and Boynton, J.E. (1989). Electrophoretic and immunological comparisons of chloroplast and prokaryotic ribosomal proteins reveal that certain families of large subunit proteins are evolutionarily conserved. J. Mol. Evol. 29 68–88. [DOI] [PubMed] [Google Scholar]
- Redding, K., Cournac, L., Vassiliev, I.R., Golbeck, J.H., Peltier, G., and Rochaix, J.D. (1999). Photosystem I is indispensable for photoautotrophic growth, CO2 fixation, and H2 photoproduction in Chlamydomonas reinhardtii. J. Biol. Chem. 274 10466–10473. [DOI] [PubMed] [Google Scholar]
- Reski, R. (2002). Rings and networks: The amazing complexity of FtsZ in chloroplasts. Trends Plant Sci. 7 103–105. [DOI] [PubMed] [Google Scholar]
- Ris, H., and Plaut, W. (1962). Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 13 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.D., Goldschmidt-Clermont, M., and Merchant, S. (1998). The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Rochaix, J.D., Kuchka, M., Mayfield, S., Schirmer-Rahire, M., Girard-Bascou, J., and Bennoun, P. (1989). Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 8 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satagopan, S., and Spreitzer, R.J. (2004). Substitutions at the Asp-473 latch residue of Chlamydomonas ribulosebisphosphate carboxylase/oxygenase cause decreases in carboxylation efficiency and CO(2)/O(2) specificity. J. Biol. Chem. 279 14240–14244. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Myers, A.M., Gillham, N.W., and Boynton, J.E. (1984). Immunological similarities between specific chloroplast ribosomal proteins from Chlamydomonas reinhardtii and ribosomal proteins from Escherichia coli. Mol. Biol. Evol. 1 317–334. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Richardson, C.B., Gillham, N.W., and Boynton, J.E. (1983). Sites of synthesis of chloroplast ribosomal proteins in Chlamydomonas. J. Cell Biol. 96 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotz, F., Bathelt, H., Arnold, C.-G., and Schimmer, O. (1972). Die Architektur und Organisation der Chlamydomonas-Zelle Ergebnisse der Elektronenmikroskopie von Serienschnitten und der daraus resultierenden dreidimensionalen Rekonstruktion. Protoplasma 75 229–254. [DOI] [PubMed] [Google Scholar]
- Schroda, M., Vallon, O., Whitelegge, J.P., Beck, C.F., and Wollman, F.-A. (2001). The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell 13 2823–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, C., Elles, I., Kortmann, J., Piotrowski, M., and Nickelsen, J. (2007). Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19 3627–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira, M., Lers, A., Heifetz, P.B., Irihimovitz, V., Osmond, C.B., Gillham, N.W., and Boynton, J.E. (1997). Differential regulation of chloroplast gene expression in Chlamydomonas reinhardtii during photoacclimation: Light stress transiently suppresses synthesis of the Rubisco LSU protein while enhancing synthesis of the PS II D1 protein. Plant Mol. Biol. 33 1001–1011. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A. (2003). Chloroplast structure: From chlorophyll granules to supra-molecular architecture of thylakoid membranes. Photosynth. Res. 76 185–196. [DOI] [PubMed] [Google Scholar]
- Suss, K.H., Arkona, C., Manteuffel, R., and Adler, K. (1993). Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc. Natl. Acad. Sci. USA 90 5514–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suss, K.H., Prokhorenko, I., and Adler, K. (1995). In situ association of Calvin cycle enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, ferredoxin-NADP+ reductase, and nitrite reductase with thylakoid and pyrenoid membranes of Chlamydomonas reinhardtii chloroplasts as revealed by immunoelectron microscopy. Plant Physiol. 107 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek, M., Kuras, R., Sokolenko, A., Higgs, D., Olive, J., Cinque, G., Muller, B., Eichacker, L.A., Stern, D.B., Bassi, R., Herrmann, R.G., and Wollman, F.-A. (2001). The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13 1347–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh, T., and Danon, A. (2001). Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc. Natl. Acad. Sci. USA 98 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon, O., Wollman, F.A., and Olive, J. (1985). Distribution of intrinsic and extrinsic subunits of the PS II protein complex between appressed and non-appressed regions of the thylakoid membrane: An immunocytochemical study. FEBS Lett. 183 245–250. [Google Scholar]
- van de Meene, A.M., Hohmann-Marriott, M.F., Vermaas, W.F., and Roberson, R.W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184 259–270. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J., Andersson, B., and Aro, E.M. (1996). Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 271 9627–9636. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J., Nilsson, L.O., and Styring, S. (1994). Synthesis of reaction center proteins and reactivation of redox components during repair of photosystem II after light-induced inactivation. J. Biol. Chem. 269 28382–28392. [PubMed] [Google Scholar]
- Vothknecht, U.C., and Soll, J. (2005). Chloroplast membrane transport: Interplay of prokaryotic and eukaryotic traits. Gene 354 99–109. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Che, F.-S., Iwano, M., Takayama, S., Yoshida, S., and Isogai, A. (2001). Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J. Biol. Chem. 276 20474–20481. [DOI] [PubMed] [Google Scholar]
- White, R.A., and Hoober, J.K. (1994). Biogenesis of thylakoid membranes in Chlamydomonas reinhardtii y1 (a kinetic study of initial greening). Plant Physiol. 106 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C., Fan, J., Froehlich, J.E., Awai, K., and Benning, C. (2005). Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17 3094–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., Prieto, S., Beligni, M.V., Haynes, P.A., McDonald, W.H., Yates, J.R., III, and Mayfield, S.P. (2002). Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: Identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14 2957–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Burke, J., Autz, G., and Jagendorf, A.T. (1981). Bound ribosomes of pea chloroplast thylakoid membranes: Location and release in vitro by high salt, puromycin, and RNase. Plant Physiol. 67 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn, C.B., Cohen, A., Danon, A., and Mayfield, S.P. (1996). Altered mRNA binding activity and decreased translational initiation in a nuclear mutant lacking translation of the chloroplast psbA mRNA. Mol. Cell. Biol. 16 3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak, E., Norling, B., Maitra, R., Huang, F., Andersson, B., and Pakrasi, H.B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 98 13443–13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation in chloroplasts. Biochimie 82 583–601. [DOI] [PubMed] [Google Scholar]
- Zerges, W., Girard-Bascou, J., and Rochaix, J.D. (1997). Translation of the chloroplast psbC mRNA is controlled by interactions between its 5′ leader and the nuclear loci TBC1 and TBC3 in Chlamydomonas reinhardtii. Mol. Cell. Biol. 17 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges, W., and Rochaix, J.D. (1998). Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 140 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges, W., Wang, S., and Rochaix, J.D. (2002). Light activates binding of membrane proteins to chloroplast RNAs in Chlamydomonas reinhardtii. Plant Mol. Biol. 50 573–585. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Paakkarinen, V., van Wijk, K.J., and Aro, E.M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274 16062–16067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.