Abstract
The sperm cell of flowering plants cannot migrate unaided and must be transported by the pollen tube cell of the male gametophyte to achieve successful fertilization. Long-distance pollen tube guidance is controlled by the seven-celled female gametophyte, the embryo sac. Previous reports showed that the synergid cell of the embryo sac is essential for pollen tube guidance. Here, we report the identification of a central cell guidance (ccg) mutant, which is defective in micropylar pollen tube guidance. CCG encodes a nuclear protein with an N-terminal conserved zinc β-ribbon domain that is functionally interchangeable with that of TFIIB in yeast. This suggests that CCG might act as a transcription regulator for pollen tube guidance. CCG is expressed in the central cell of the female gametophyte. Expression of CCG in the central cell alone is sufficient to restore the normal pollen tube guidance phenotype, demonstrating that the central cell plays a critical role in pollen tube guidance.
INTRODUCTION
Long-distance guided polar cell growth is common in development as seen during axon guidance in animals and pollen tube guidance in higher plants. The two share many similar features, although they occur in different kingdoms and serve different purposes (Palanivelu and Preuss, 2000). A typical feature of guided growth is that it requires the distant communication and interaction of two different cell or tissue types. In higher plants, pollen tube guidance requires the signaling between the male gametophyte and the female gametophyte to ensure the precise targeting of the pollen tube to the female gametophyte over a long distance (Hülskamp et al., 1995a, 1995b; Cheung and Wu, 2000; Johnson and Preuss, 2002; Higashiyama et al., 2003). The entire process of pollen tube guidance can be divided into two subsequent phases: the sporophytic phase and the gametophytic phase. The sporophytic phase refers to pollen tube growth within the transmitting tract of the pistil, whereas the latter starts when the pollen tube emerges at the surface of the placenta and grows along the funiculus and enters the micropylar opening of the ovule. The pollen tube growth and guidance process are very complex and need multiple signals to guide the pollen tube to its target (Pruitt, 1999; Johnson and Preuss, 2002; Higashiyama et al., 2003; Palanivelu and Preuss, 2006).
Signaling molecules involved in the sporophytic phase of pollen tube guidance have been identified recently. Long-chain lipids from the pollen coat (Fiebig et al., 2000) and hydrophobic stigma (Wolters-Arts et al., 1998) have been implicated in pollen hydration and germination, together with the abundant pollen coat protein GRP17 (Mayfield and Preuss, 2000) and the water channel protein aquaporin (Ikeda et al., 1997). Polar pollen tube growth is supported by arabinogalactans (Wang et al., 1993; Cheung et al., 1995, 2000; Wu et al., 1995, 2000; Sommer-Knudsen et al., 1996, 1998), pectin (Mollet et al., 2000), the lipid transfer protein SCA (Jauh et al., 1997; Park et al., 2000), chemocyanin (Kim et al., 2003; Dong et al., 2005), and γ-aminobutyric acid (Palanivelu et al., 2003), providing nutrition and guidance within the transmitting tract of the gynoecium. These sporophytic cues are not sufficient, however, to ensure successful guidance and fertilization. The female gametophyte plays a major role in guiding the pollen tube to the mycropylar opening of the ovule and the subsequent double fertilization event. Although the female gametophyte (embryo sac) is the source of these final pollen tube guidance cues (Hülskamp et al., 1995a; Ray et al., 1997; Shimizu and Okada, 2000), little is known about the signal molecules or how these molecules are perceived. The precise origin of some signals was shown by elegant cell ablation experiments in Torenia fournieri to be associated with the synergid cells of the embryo sac (Higashiyama et al., 2001, 2003). Further supporting evidence showed that loss of a synergid-expressed MYB98 gene abolished the ovule's ability to attract pollen tubes (Kasahara et al., 2005). These data suggested that the synergid cells are essential for pollen tube guidance, although it is not clear whether the synergids are the source of the attracting signal or just the physical interaction interface. In maize (Zea mays), RNA interference knockdown of the Zm EA1 gene, initially isolated from egg cells, abolished micropylar pollen tube guidance (Márton et al., 2005). Zm EA1 is expressed in both egg and synergid cells and encodes a small diffusible protein of 94 amino acids, suggesting that Zm EA1 is a potential signaling molecule for micropylar pollen tube guidance in maize. The expression of Zm EA1 in egg cells suggested that in addition to the synergids, other cells of the female gametophyte may play a role in micropylar pollen tube guidance.
Here, we present data demonstrating that other cells do indeed play a role in micropylar pollen tube guidance. In particular, we report a critical role played by the central cell, providing comprehensive phenotypic and molecular characterization of the Arabidopsis thaliana female gametophytic mutant central cell guidance (ccg), which abolishes micropylar pollen tube guidance. CCG encodes a nuclear protein with a conserved N-terminal C2C2-type zinc finger motif typical for the general transcription factor TFIIB family and functionally interchangeable in the yeast two-hybrid assay. We show that the expression of CCG in the central cell completely rescues pollen tube guidance in the ccg mutant and that the CCG gene is normally expressed in the central cell of the female gametophyte. These results suggest that the central cell plays a critical role in micropylar pollen tube guidance in Arabidopsis.
RESULTS
Isolation of Mutants Defective in Pollen Tube Guidance
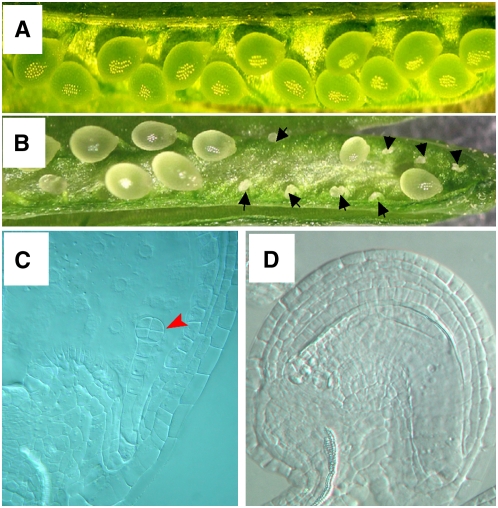
Failure of pollen tube guidance abolishes fertilization and subsequent seed development. Previous studies have shown that the gametophytic phase of pollen tube guidance is controlled by the female gametophyte. Therefore, we first screened for female gametophytic mutants via distorted Mendelian segregation (Howden et al., 1998) of the kanamycin resistance gene (Kanr) on the Ds element in Arabidopsis gene/enhancer trap lines (Sundaresan et al., 1995). This was followed by detailed analysis to determine if mutants affected pollen tube guidance. One such mutant that affected pollen tube guidance was designated ccg. This mutant plant line contained a single Ds insertion as revealed by DNA gel blot analysis using a 755-bp fragment at the 5′ end of the Ds element as a probe (data not shown). The hemizygous mutant line (Ds/-) showed a 1.10:1.00 ratio of Kanr:Kans in the progeny of selfed F2 plants (n > 1000). This suggested that the distorted segregation is most likely caused by a gametophytic mutation. We checked the seed set of both wild-type and mutant plants. Full seed set was observed in the wild-type plant (Figure 1A), while an obvious reduction in seed set was observed in the mutant (Figure 1B). Approximately 56% (1172:2092) of the ovules in the mutant developed into mature seeds, and 44% (920:2092) of the ovules were small and dehydrated. Whole-mount clearing of ovules in mutant siliques (Ds/-) 3 d after pollination showed that 56% of the seeds contained embryos at the octant stage (Figure 1C), and ∼44% (n > 1000) of the ovules did not contain embryos, suggesting that the ovules were unfertilized (Figure 1D). No morphological abnormalities in other floral and vegetative tissues were observed.
Figure 1.
The Seed Setting in Wild-Type and Mutant Plants.
(A) Wild-type siliques show full seed set.
(B) In mutant siliques (ccg/CCG), only approximately half of the ovules developed into seed; the others were shrunken and dried.
(C) A micrograph showing a wild-type octant embryo in ccg/CCG pistils at 60 h after pollination.
(D) A mutant ovule in the same pistil as in (C), showing no embryo structure.
Although the distorted segregation ratio observed in the mutant plants was close to the expected distortion for a sex-specific gametophytic mutation, it does not exclude the possibility that both sexes are partially affected. To investigate whether the mutation affects male and/or female gametophytic function, reciprocal crosses were performed. Reciprocal crosses with wild-type plants showed a 93.5% transmission efficiency of the Kanr gene through the male (n = 416) and a 14.2% transmission efficiency (n = 464) through the female, indicating a strong reduction in transmission through the female gametophyte. Tetrad analysis using the qrt2 mutant (Preuss et al., 1994; Copenhaver et al., 2000) showed that the majority of the pollen grains were morphologically indistinguishable from wild-type pollen. Only ∼4% of the pollen grains (n > 2000) aborted during early developmental stages, strongly suggesting that the mutation did not affect pollen development but disrupted the function of the female gametophyte. Despite a significant transmission of the Kanr gene through both gametophytes, homozygous plants were not recovered, implying that the mutation causes zygotic lethality.
The Mutant Is Defective in Micropylar Pollen Tube Guidance
To investigate whether pollen tube guidance was impaired, we examined pollen tube guidance in manually pollinated wild-type and mutant pistils by aniline blue staining and scanning electron microscopy. In wild-type pistils 24 h after pollination (HAP), pollen tubes were observed to grow on the surface of the funiculus after leaving the transmitting tissue and finally entered the micropyle (Figure 2A). Similar results have been observed in Arabidopsis by other studies (Wilhelmi and Preuss, 1997, 1999; Lush, 1999). With the aniline blue staining method, we found that ∼92% of the ovules (n = 603) had pollen tube entry in wild-type pistils (Figure 2A), while only 59% of ovules (n = 1371) (Ds/-) had pollen tube entry in mutant pistils (Figure 2B); the remaining 41% of ovules had no pollen tube entry (Figure 2C). To further investigate whether some pollen tubes could enter the mutant ovule and fertilize the female gametophyte, we pollinated the mutant and wild-type pistils with pollen grains containing the pollen tube–expressed marker gene ProAKV-GUS (Rotman et al., 2005). If pollen tube entry occurred, β-glucuronidase (GUS) activity would be detected at the micropylar portion of female gametophytes. In wild-type pistils, 92% of the ovules were GUS positive (n = 613) (Figure 2D), and 8% were GUS negative; while in mutant pistils, only 58.8% of the ovules (n = 779) showed a GUS signal, the remaining 41.2% ovules were GUS negative (Figure 2E), indicating that there was no pollen tube entry in these ovules. As a control, the ovules of unpollinated mutant pistils were shown to be GUS negative (data not shown). Taken together, the data suggest that the mutant ovules are defective in pollen tubes attraction.
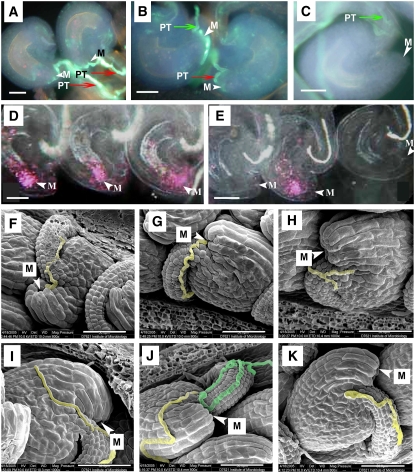
Figure 2.
Pollen Tube Guidance in Wild-Type and ccg/CCG Plants.
(A) to (C) Ovules stained with aniline blue to visualize the pollen tube (PT). M, micropyle.
(D) and (E) Whole-mount stained ovules pollinated with pollen grains carrying the ProAKV:GUS reporter.
(F) to (K) Scanning electron micrographs showing pollen tube guidance; the pollen tubes were false-colored with blue or yellow.
(A) An example of pollen tube (arrow) entry into the micropyle (arrowhead) in wild-type pistils.
(B) A micrograph showing two ovules in mutant pistils (ccg/CCG). Note pollen tube entry into the ovule at the right as in (A) but into the ovule at the left.
(C) A micrograph showing a mutant ovule without pollen tube in the same pistil as in (B). Note that the pollen tube grows along the funiculus and turns back at the micropyle region.
(D) A micrograph showing that all ovules in wild-type pistils had GUS activity carried by the pollen tube at the micropylar part of the embryo sacs (arrowheads), indicating successful pollen tube entry.
(E) A micrograph showing that in mutant pistils (ccg/CCG), approximately half of the ovules had no GUS activity, indicating no pollen tube entry.
(F) and (G) Scanning electron micrograph of wild-type ovules showing that pollen tubes grow along the funiculus and then turn abruptly to enter the micropyle (arrowheads).
(H) to (K) Aberrant pollen tube guidance in ccg ovules.
(H) A pollen tube stops growing near the micropyle (arrowhead).
(I) A pollen tube bypassing the micrpopyle and growing on the surface of the integument.
(J) Two pollen tubes grow along the funiculus but fail to enter the micropyle (arrowhead). One pollen tube bypasses the micropyle (yellow), and another pollen tube (blue) turns away from the micropyle.
(K) Another example showing that the pollen tube turns away from the micropyle.
Red arrow indicates pollen tube entry into the micropyle; green arrow indicates no pollen tube entry. Bar = 20 μm for (A) to (E) and 50 μm for (F) to (K).
To further clarify whether the mutation abolished funicular or micropylar pollen tube guidance, scanning electron microscopy analysis was performed. In the wild type, pollen tubes appeared tightly adhered to the funiculus and always grew toward the micropyle, precisely entering the micropylar opening of the ovule (Figure 2F). In mutant pistils (Ds/-), pollen tubes were found to grow along the funiculus and enter the micropyle of about half of the ovules (Figure 2G), while in the other half, the pollen tubes failed to find the micropylar opening and grew without direction (Figures 2H to 2K), with some ceasing to grow near the micropyle (Figure 2H), others turning away (Figure 2K) or bypassing the micropyle and growing on the ovule surface toward the chalazal end (Figure 2I). Occasionally, multiple pollen tubes arrived at the same ovule (Figure 2J). These observations demonstrate that mutant ovules have lost their ability to guide pollen tube growth to the micropyle, suggesting a defect in micropylar pollen tube guidance.
The Mutant Forms Morphologically Normal Female Gametophytes That Remain Unfertilized
To investigate whether the pollen tube guidance defect was caused by impaired gametophyte development as implicated by earlier works (Hülskamp et al., 1995a; Ray et al., 1997; Shimizu and Okada, 2000; Kasahara et al., 2005), confocal laser scanning microscopy (CLSM) was performed according to Christensen et al. (1998). Pistils at different developmental stages were collected from mutant (Ds/-) and wild-type plants and processed for CLSM analysis. Ovules within a pistil at the same floral stage from both wild-type and mutant plants were compared using CLSM. Results showed that ovule development in the mutant plant is the same as that in the wild-type plants as reported earlier (Christensen et al., 1998; Shi et al., 2005). The mutant pistils, which are heterozygous for the gametophytic ccg mutation, always contain 50% wild-type ovule (no mutation) and 50% mutated ovules. Therefore, we compared the development of the wild-type and the mutant ovules within the same pistil. No obvious morphological changes or developmental arrest could be observed in ovules of the same mutant pistil. For example, ovules (∼50) in a mutant pistil at floral stage 13c (Smyth et al., 1990) all reached the final developmental stage (FG7) (Figure 3A), indicating that the 50% mutated ovules developed as that of the wild-type ovules within the same pistil. In other words, the mutant ovules are indistinguishable by morphology from the wild type before pollen tube entry. Consistently, the synchronous development of the female gametophyte was also normal, as shown in Table 1 for the mutant pistils. Data for wild-type embryo sac development was not presented since this has been reported extensively elsewhere (Christensen et al., 1997; Shi et al., 2005). When 12 pistils from the same mutant inflorescence were analyzed, ovules within the each pistil were mostly at two adjacent developmental stages (Table 1), the same as that in the wild type (data not shown; Christensen et al., 1997; Shi et al., 2005). For example, in pistil 12, 7 h after emasculation at floral stage 12c, 29 ovules out of 32 were at stage FG7, indicating that both mutated and normal ovules reached the final developmental stage. These data showed that the mutation did not affect female gametophyte development.
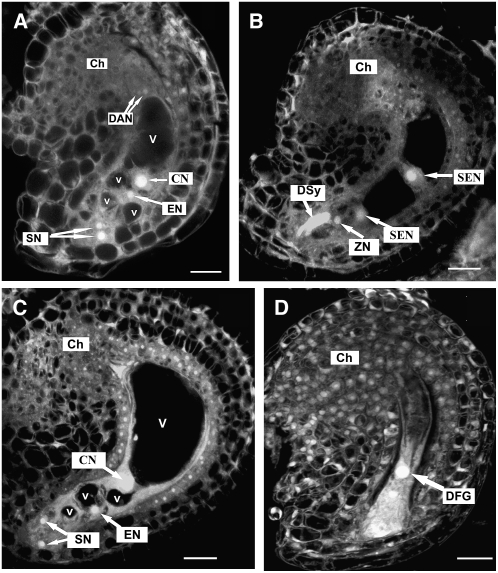
Figure 3.
Ovule Development in Mutant Plants as Revealed by CLSM Observation.
(A) A mutant ovule with mature four-celled embryo sac at early stage FG7. Note the polar nuclei fused to form a diploid central nucleus and degenerating antipodal nuclei.
(B) A wild-type ovule from a ccg/CCG pistil showing an embryo sac at 48 HAP, with one degenerated synergid cell, an intact synergid cell, a zygotic cell, and two secondary endosperm nuclei.
(C) An unfertilized mutant ovule from the same pistil as in (B) showing the four-celled embryo sac. The ovule noticeably increased in size.
(D) The unfertilized mutant embryo sac undergoes degeneration at 72 HAP. All images were projected from multiple 1-μm optical sections.
Ch, chalazal end; CN, central nucleus; DAN, degenerating antipodal cell; DFG, degenerated female gametophyte; DSy, degenerated synergid cell; EN, egg cell nucleus; SEN, secondary endosperm nucleus; SN, synergid nucleus; V, large vacuole; v, small vacuole; ZN, zygote nucleus. The developmental stages of embryo sac are defined according to Christensen et al. (1998). Bars = 10 μm.
Table 1.
Synchrony of Female Gametophyte Development in the Mutant Plant
| No. of Female Gametophytes at Developmental Stages
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Pistil No. | FG0 | FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 |
| 1 | 33 | |||||||
| 2 | 30 | 2 | ||||||
| 3 | 5 | 17 | ||||||
| 4 | 10 | 16 | 4 | |||||
| 5 | 4 | 15 | 1 | |||||
| 6 | 3 | 19 | 2 | |||||
| 7 | 5 | 28 | 8 | |||||
| 8 | 9 | 11 | ||||||
| 9 | 2 | 6 | 11 | |||||
| 10 | 12 | 18 | ||||||
| 11 | 6 | 17 | ||||||
| 12 (Em7 h) | 3 | 29 | ||||||
Ovules from 12 pistils of the same inflorescence were analyzed by confocal microscopy, and their developmental stages (FG0 to FG7) were determined according to Christensen et al. (1998). Ovules within each pistil are mostly at two adjacent developmental stages, the same as that in wild-type plants described by Christensen et al. (1998) and Shi et al. (2005), indicating that the mutation does not affect developmental synchrony. Em7 h, 7 h after emasculation.
When analyzing pistils from wild-type and mutant (Ds/-) plants, 48 HAP with wild-type pollen, 98.5% of ovules (n = 80) in wild-type and 55% of ovules (n = 208) in mutant pistils had developed endosperm, as shown in Figure 3B. The remaining embryo sacs in the mutant (approximately half) were unfertilized. In these ovules, the egg cell and central cell nucleus and two synergid nuclei were still detectable (Figure 3C). Noticeably, the unfertilized ovules increased in size. This increase in size is mainly attributed to the continued growth of the integuments as revealed by the obvious enlargement of the endothelium of the inner integument (Figure 3C). This also occurred in wild-type ovules when unpollinated. At 72 HAP, the unfertilized mutant embryo sacs began to degenerate (Figure 3D). These results suggest that mutant ovules completely lost their ability to attract pollen tubes even if more time for pollination was provided.
To check whether the cellularization of the embryo sacs was defective, we introduced a plasma membrane–targeted fluorescent marker by crossing the mutant with the ProAKV-GFP-ROP6C transgenic line (Escobar-Restrepo et al., 2007). CLSM showed that the female gametophytes in wild-type plants underwent cellularization to form a seven-celled embryo sac at floral stage 12 (Smyth et al., 1990), as did all ovules (n > 300) from plants heterozygous for the mutation and homozygous for ProAKV-GFP-ROP6C. The green fluorescent protein (GFP) signal was detected in the cell membrane of the female gametophyte and revealed the same pattern of cellularization in all ovules, whether from wild-type or mutant pistils at floral stage 12c. An example is shown for an ovule at stage FG7 in Figure 4A. These findings indicate that the cellularization process in mutant ovules is normal and that the development of the mutant ovules is not affected by the mutation.
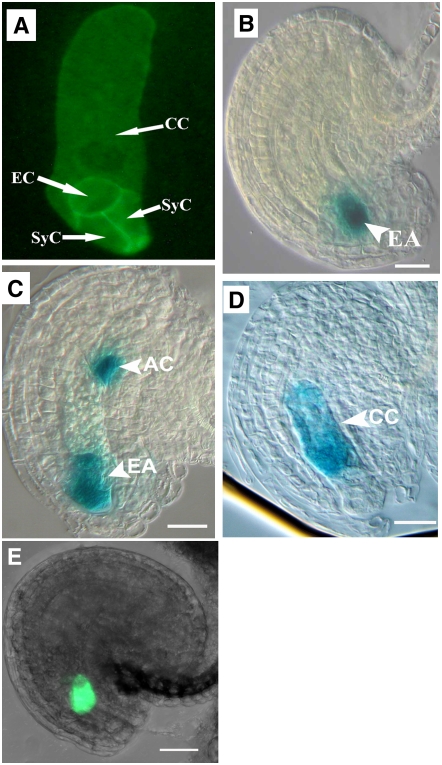
Figure 4.
Cell Fate Specification and Cellularization of Female Gametophytes in the Mutant.
Marker lines were crossed to the mutant line, and ovules were checked for marker expression pattern and statistic analysis. For ProMYB98-GFP, the reporter was introduced into the mutant by Agrobacterium tumefaciens–mediated transformation. For all five markers, the same pattern is observed in both wild-type and mutant ovules. AC, antipodal cell; CC, central cell; EA, egg apparatus; EC, egg cell; SyC, synergid cell. Bars = 20 μm.
(A) A CLSM projection showing the cellularization pattern of mature mutant embryo sacs expressing the ProAKV-GFP-ROP6C cell membrane GFP marker gene showing cellularization of the embryo sac.
(B) A mutant ovule showing egg apparatus–specific GUS marker expression (ET2632).
(C) A mutant ovule showing the expression of egg apparatus and antipodal cell marker (ET1811).
(D) A mutant ovule showing central cell–specific GUS marker expression (ET956).
(E) A CLSM image showing synergid-specific ProMYB98-GFP pattern in mutant ovules.
To further investigate whether the mutation affects cellular structures, such as the filiform apparatus, we performed transmission electron microscopy on >20 ovules from the mutant, and no abnormality in the ultrastructure of synergid cell, egg cell, central cell, and the filiform apparatus was observed (data not shown). This suggests that the mutation had no effect on the development of these gametophytic cells and the filiform apparatus.
The Gametophytic Cell Fates Were Specified Correctly in Mutant Ovules
The above phenotypic analysis indicated that the mutation did not affect female gametophyte morphology per se. These observations, however, cannot exclude the possibility that alterations in cell fate specification, not easily observed visually, could cause the pollen tube guidance defects observed. Therefore, we asked whether the gametophytic cells in mutant ovules were specified correctly, as suggested by their normal morphology. Several gametophytic cell-specific enhancer detector marker lines, including ET2632 for the egg apparatus (including the egg cell and two synergid cells), ET1811 for the egg apparatus and the antipodal cells, and ET956 as a specific central cell marker (U. Grossniklaus, unpublished data), were crossed with the mutant, and their segregation patterns in the F1 and F2 generations were studied. In addition, ProMYB98-GFP, which served to identify the synergids (Kasahara et al., 2005), was also included.
Ovules from F1 plants that were either heterozygous for both the mutation and the marker or only heterozygous for the marker were analyzed for GUS activity. Typical marker staining patterns in the mutant ovules are shown in Figure 4. As expected, approximately half of the ovules from wild-type plants heterozygous for the marker showed a GUS signal in gametophytic cells for all markers (Table 2). Similar results were obtained for ovules of plants heterozygous for both the mutation and the marker, where approximately half of the ovules showed a GUS signal localized specifically to gametophytic cells (Table 2). The GUS expression patterns and expression levels of the cell-specific markers were identical between mutant and wild-type ovules. One would expect a significant change in the number of GUS staining ovules in mutant pistils if the mutation were to change the fate of certain gametophytic cells. In transgenic plant lines heterozygous for the ccg mutation, homozygous for the ProMYB98-GFP transgene, mature ovules (n = 150) from three pistils investigated all displayed the same GFP pattern and fluorescent intensity (Figure 4E), as reported in wild-type ovules by Kasahara et al. (2005), and no ovules with changes in GFP signal were found, indicating that the synergids in mutant ovules express MYB98 as that in the wild type. From these data, we conclude that cell fate specification in mutant ovules is normal, suggesting that the mutation specifically affects pollen tube guidance and has no effect on ovule development and gametophytic cell fate specification.
Table 2.
Statistics of Ovules with GUS Signal in F1 Plants of Crosses between the Wild Type or ccg Mutant and the Marker Lines ET1811 (Egg Apparatus and Antipodal Cells), ET2632 (Egg Apparatus), and ET956 (Central Cell)
| Plant | No. of Ovules with GUS | No. of Ovules without GUS | Percentage of GUS Ovules | χ2 (1) |
|---|---|---|---|---|
| ET1811/+WT | 281 | 329 | 46.1% | 0.0083 (P > 0.05) |
| ET1811/+ccg/CCG | 249 | 295 | 45.8% | |
| ET2632/+WT | 206 | 244 | 45.8% | 0.059 (P > 0.05) |
| ET2632/+ccg/CCG | 117 | 144 | 44.8% | |
| ET956/+WT | 330 | 356 | 48.1% | 0.032 (P > 0.05) |
| ET956/+ccg/CCG | 236 | 260 | 47.6% |
Molecular Cloning of the CCG Gene
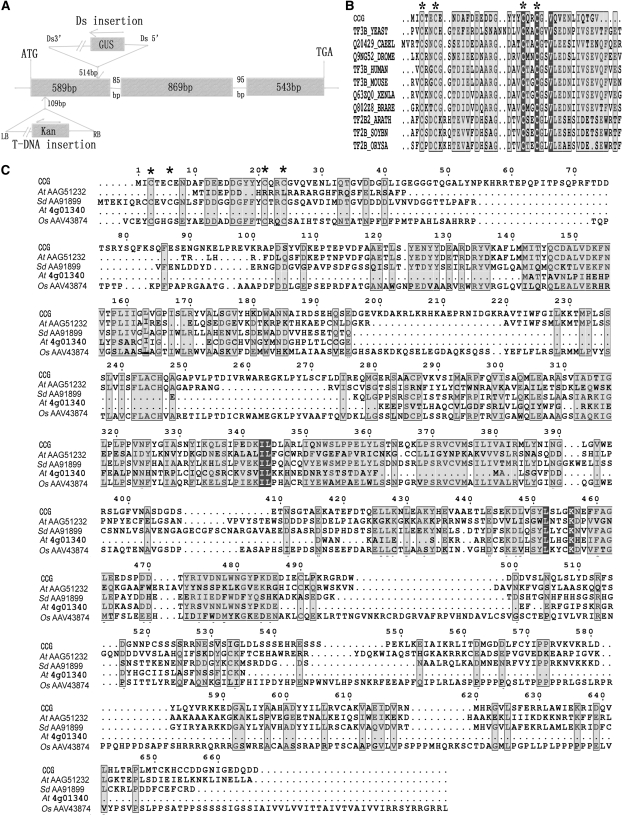
To identify the CCG gene, we used thermal asymmetric interlaced PCR (TAIL-PCR) to obtain the genomic sequences flanking the Ds element in the mutant. Sequence analysis of the TAIL-PCR products revealed that the Ds element had inserted into the first exon, 514 bp downstream of ATG of a predicted gene At2g02955 on chromosome II (Figure 5A) in the genome.
Figure 5.
The Genomic Organization of CCG and Alignment of CCG Protein with Its Homologs from Other Organisms.
(A) The CCG gene has three exons, two small introns, and a full-length cDNA of 2001 bp. Ds inserted into the first exon at 514 bp downstream of ATG in ccg-1, with a small 90-bp deletion, and the T-DNA was inserted in the first exon at 109 bp downstream of ATG in ccg-2.
(B) Alignment of the CCG N-terminal zinc β-ribbon domain with its closest homologs.
(C) Alignment of the CCG protein with its homologs from other organisms.
CCG, Arabidopsis CCG protein; TF3B_YEAST, Saccharomyces cerevisiae TFIIIB; Q20429_CAEEL, Caenorhabditis elegance transcription factor homolog protein 1; Q9NG52_DROME: Drosophila RNA polymerase III transcription factor; TF3B_HUMAN, Homo sapiens TFIIIB 90KD subunit; TF3B_MOUSE, mouse BRF1 protein; Q63ZQ0_XENLA, Xenopus laevis locus 494758 protein; Q802Z8_BRARE, zebra fish BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIb; TF2B2_ARATH, TF2B from Arabidopsis; TF2B_SOYBN, TF2B from Glycine max; TF2B_ORYSA, TF2B from O. sativa. Other proteins are named by their GenBank accession number preceded by the species name: At, Arabidopsis thaliana; Sd, Solanum demissum; Os, Oryza sativa. The asterisk indicates the conserved four Cys residues in the Zn-finger domain.
To confirm that At2g02955 indeed codes for CCG, a 5.5-kb genomic DNA fragment of At2g02955, including the predicted promoter, transcribed region, and 3′-end untranscribed region, was amplified by high-fidelity PCR and subcloned into the binary vector pCAMBIA1300. The construct was then verified by sequencing and introduced into ccg/CCG mutant plants by Agrobacterium tumefaciens–mediated transformation. Eight independent transformants were obtained after double selection with kanamycin for the Ds insertion and hygromycin for the presence of the complementation transgene. Eight T1 transgenic lines were obtained, and siliques of these lines were checked for normal seed set. Furthermore, seeds from these T1 plants were plated on kanamycin plates to analyze the Ds segregation ratio. Among these T1 plants, five lines showed ∼22% increase in seed set (∼78%, n > 500) when compared with the seed set of the mutant (56%, n = 2092) (see Supplemental Table 1 online), and the remaining three transformed plant lines did not show significant increase in seed set. This indicated that the transgene had complemented the mutation restoring the pollen tube guidance function to the mutant female gametophytes. The T2 seedlings from these families showed a Kanr:Kans ratio of ∼2:1 (n > 500), a significant increase compared with the 1.1:1.0 ratio in selfed ccg/CCG progeny. According to Mendelian law, selfed progeny of the ccg/CCG, P-At2g02955/+ genotype should display a Kanr (ccg/ccg, P-At2g02955/P-At2g02955; ccg/ccg, P-At2g02955/+; ccg/CCG, P-At2g02955/P-At2g02955; ccg/CCG, P-At2g02955/+; and ccg/CCG, +/+) to Kans (CCG/CCG, P-At2g02955/P-At2g02955; CCG/CCG, P-At2g02955/+; and CCG/CCG,+/+) ratio of 2:1, assuming that the female gametophytes of the ccg,+ genotype would not be fertilized. The observed 2:1 ratio of Kanr:Kans in the transgenic lines suggests a complete complementation of the mutation by the transgene. Therefore, we concluded that the 5.5-kb DNA fragment of chromosome II complemented the semisterile phenotype of ccg/CCG plants.
We have searched databases for additional insertion lines in the At2g02955 gene, and a T-DNA insertion line, salk_077907, was identified. Molecular analysis revealed that the T-DNA element was inserted into the first exon of the At2g02955 gene, 109 bp downstream of the ATG (Figure 5A). Both genetic and phenotypic analysis showed that this mutant has a similar Kanr:Kans segregation ratio (1.1:1.0, n = 314) as shown by PCR analysis, seed abortion rate (42%, n = 2928), pollen tube entry rate (58.7%, n = 592), and identical pollen tube guidance phenotype to the ccg/CCG mutant (Figure 2). Further outcross tests showed that the transmission efficiency was 79% (n = 226) through male and 14% (n = 228) through female. Female gametophyte development in the 12 pistils analyzed was shown again to be normal, but micropylar pollen tube guidance was impaired. Both the complementation result and the analysis of the additional mutant allele demonstrate that our CCG gene is synonymous with At2g02955.
We obtained cDNA sequence by RT-PCR using primers based on The Arabidopsis Information Resource annotation of 2001 nucleotides in length (GenBank accession number AY735574). To confirm the function of the predicted open reading frame, we complemented the mutant with this full-length cDNA under the transcriptional control of the CCG promoter. The transgene restored the pollen guidance to mutant plants. These results show that the 2001-nucleotide mRNA sequence is sufficient to encode the functional CCG protein (data not shown).
CCG Encodes a Nuclear Protein with an N-Terminal Zinc β-Ribbon Domain That Is Functionally Interchangeable with the N-Terminal Domain of TFIIB
CCG encodes a 666–amino acid protein, which consists of a zinc β-ribbon domain (www.sanger.ac.uk//cgi-bin/Pfam/PF08271) at the N terminus at amino acid residues 1 to 36 (Figure 5B), and a C-terminal Ser-rich domain (residues 508 to 547). No obvious nuclear localization signal is present in the CCG protein. In addition, several Cys residues are present toward the C-terminal portion of the protein. A BLASTP search using the predicted CCG amino acid sequence showed that there were four homologous sequences in GenBank from Arabidopsis, Solanum demissum, and Oryza sativa. They share high similarity throughout the protein (Figure 5C), and the biochemical function of all these genes was unknown.
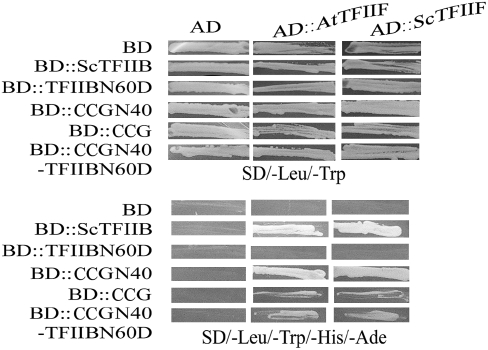
The zinc β-ribbon domain of ∼33 amino acids is present near the N terminus of the general transcription factor TFIIB and TFIIIB family proteins from many species (Figure 5B). This domain adopts a zinc ribbon-like structure that is involved in the interaction with RNA polymerase II and TFIIF and plays a crucial role in selecting the transcription initiation site (Buratowski et al., 1989; Ha et al., 1993; Zhu et al., 1996; Chen and Hahn, 2003; Bushnell et al., 2004). In yeast, the CCG N-terminal domain can interact with yeast and Arabidopsis TFIIF, resembling the TFIIB zinc β-ribbon domain (Figure 6). Yeast two-hybrid assays showed that CCGN40, containing the N-terminal zinc β-ribbon domain, strongly interacted with both yeast and Arabidopsis TFIIF. Yeast Sc TFIIB and the CCGN40-TFIIBN60D chimeric protein with the zinc β-ribbon domain of TFIIB substituted by the CCG N-terminal domain interacted with the yeast and Arabidopsis TFIIF proteins, respectively. Consistently, the CCG full-length protein can also interact with the yeast and Arabidopsis TFIIF, but the interaction was weaker. On the contrary, either TFIIBN60D (Figure 6) or CCGN40D (data not shown) without the N-terminal zinc β-ribbon domain lost their ability to interact with TFIIF. These findings suggest that the N-terminal zinc β-ribbon domain in CCG can interact with TFIIF and is functionally interchangeable with that of the basic transcription factor Sc TFIIB in yeast cells.
Figure 6.
CCG Interacts with TFIIF through the N Terminal Zinc β-Ribbon Domain in Yeast.
Yeast cells cotransformed with plasmids as indicated at the left side. On top, they were grown on SD/-Leu/-Trp (top panel), SD/-Leu/-Trp/-His (middle panel), or SD/-Leu/-Trp/-His/-Ade (bottom panel). Cell growth on SD/-Leu/-Trp/-His/-Ade indicates the interaction between the two cotransformed gene products. Note CCG N40 interacts strongly with At TFIIF and Sc TFIIF, and there is weaker interaction for CCG and CCGN40-TFIIBN60D chimeric protein with At TFIIF and Sc TFIIF. At TFIIF, Arabidopsis TFIIF; CCGN40, CCG N-terminal 40 amino acids; CCGN40-ScTFIIBN60D, chimeric protein between CCG N-terminal 40 amino acids and S. cerevisiae TFIIB with N-terminal 60–amino acid deletion; Sc TFIIB, S. cerevisiae TFIIB; Sc TFIIBN60D, Sc TFIIB with N-terminal 60–amino acid deletion; Sc TFIIF, S. cerevisiae TFIIF.
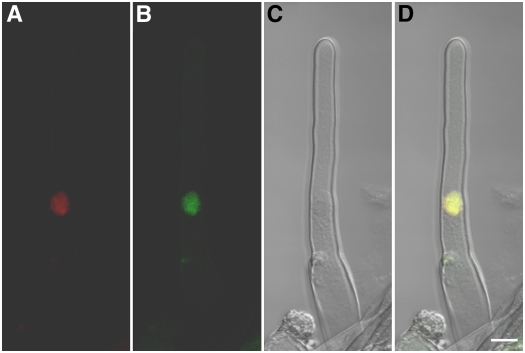
To investigate the subcellular localization, a CCG-GFP fusion driven by the CCG promoter was made and introduced into the wild-type plant. We failed to detect any GFP fluorescence in either the root cell or central cell. This indicated that the CCG promoter is a weak promoter. Therefore, a CCG-GFP fusion driven by the constitutive cauliflower mosaic virus 35S promoter was constructed and introduced into wild-type Arabidopsis plants. Three transgenic lines were obtained that complemented the ccg mutation (see Supplemental Table 1 online). In the two transgenic lines checked, the GFP signal was detected in the nucleus of root cells, as shown in a root hair in Figure 7. This indicated that CCG is indeed a nuclear protein.
Figure 7.
Nuclear Localization of CCG Protein in Arabidopsis Transformed with the 35S∷CCG-GFP Gene.
(A) DNA 4′,6-diamidino-2-phenylindole (DAPI) staining of a root hair cell.
(B) GFP signal also detected in the same root hair cell.
(C) Bright-field image of the same cell shown in (A) and (B).
(D) A merged image of (A) to (C) showing colocalization of CCG-GFP and DAPI staining. DAPI fluorescence is coded red, and GFP signal is green. CCG-GFP localized in the nucleus. Bar = 10 μm.
CCG Is Expressed in Seedlings and Reproductive Organs
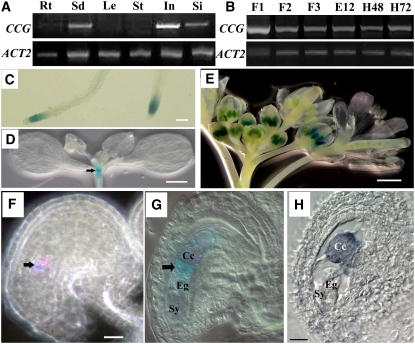
To study the expression pattern of the CCG gene, RT-PCR, GUS reporter assays, and RNA in situ hybridization experiments were performed. RT-PCR analysis showed that CCG is expressed at high levels in seedlings, inflorescences, and young siliques and at lower levels in roots, but it was not detected in leaves and stems (Figure 8A). During reproductive development, CCG transcripts were also expressed in flowers at different developmental stages and in siliques shortly after fertilization (i.e., 48 and 72 HAP) (Figure 8B).
Figure 8.
Expression Pattern of the CCG Gene in Wild-Type Plants.
CCG expression pattern revealed by RT-PCR in different organs (A) and floral stages (B) as indicated and ProCCG-GUS-3T transgene ([C] to [G]) and RNA in situ hybridization (H). Bar = 1mm in (C) and (E), 50 μm in (D), and 20 μm in (F) and (H).
(A) RT-PCR results showing CCG expression in seedlings (Sd), inflorescences (In), and siliques (Si) with the ACT2 gene as an internal reference. Rt, root; Le, leaf; St, stem.
(B) RT-PCR results showing CCG expression in flowers at stages FG2-3 (F1), FG4-5 (F2), and FG6-7 (F3) (Christensen et al., 1998). CCG is also expressed in pistils 12 h after emasculation (E12), 48 HAP (H48), and 72 HAP, indicating CCG expression in mature ovules and fertilized ovules.
(C) A micrograph of the transgenic root showing CCG expression in root tip.
(D) A micrograph showing CCG expression in the shoot apex (arrow).
(E) A micrograph showing CCG expression in anthers.
(F) A Nomarski micrograph of an ovule at FG6 showing CCG expression in the central cell (arrow) of the ovule.
(G) A Nomarski micrograph of an ovule at FG7 showing CCG expression in the central cell (arrow; Cc). Note no expression in egg cell (Eg) and synergids (Sy).
(H) An oblique section of an ovule at FG7 hybridized with CCG antisense RNA probe showing CCG expression in the central cell.
To gain insight into the cellular pattern of CCG expression, a ProCCG-GUS-3T reporter construct was made and transformed into Arabidopsis. In T2 transgenic lines, GUS activity was detected in root tips (Figure 8C) and shoot apical meristems (Figure 8D). In male reproductive organs, strong GUS signals were detected in anthers at various developmental stages (Figure 8E), primarily in microspores with weaker expression in mature pollen grains (Figure 8E). In female reproductive organs, the GUS signal was detected in the central cell of the mature female gametophyte (Figures 8F and 8G; see Supplemental Figure 1 online). The expression of GUS in central cell of the mature female gametophyte was pollination independent (E12; Figure 8B). No GUS staining was observed in other cells such as synergid, egg cell, or antipodal of the ovules. After fertilization, the GUS signal was detected in endosperm cells and the fertilized egg cells; thereafter, the GUS signal became weaker in octant embryo and endosperm, and no GUS signal was detectable in ovules after the early globular stage (data not shown).
To confirm the surprising central cell expression of the CCG gene as revealed by the promoter GUS reporter assay, RNA in situ hybridization experiments were also performed using digoxigenin-labeled antisense and sense RNA probes. CCG transcripts were detected in microspores and pollen grains (data not shown), and again a strong signal was observed in the central cell (Figure 8H; see Supplemental Figure 2 online) of the female gametophyte. No hybridization signals were found in other gametophytic cells, such as synergid, egg cell, or antipodal cells, of the ovules (see Supplemental Figure 2 online). This confirmed the central cell expression of CCG as revealed by the promoter GUS-reporter assay, suggesting that the central cell plays a role in pollen tube guidance.
Specific Expression of CCG in the Central Cell Rescues the Mutant Phenotype
All earlier studies suggested that the synergid cell is responsible for pollen tube guidance (Higashiyama et al., 2001; Kasahara et al., 2005; Márton et al., 2005). Our results suggested that the central cell also plays a role in this process. To exclude that undetectable CCG expression levels in synergid cells may be responsible for the role in pollen tube guidance, we used a central cell–specific promoter to restrict the expression of CCG to the central cell. This approach should provide evidence for the role of CCG expression in the central cell in micropylar pollen tube guidance. The FIS2 promoter exhibits central cell–specific activity before fertilization, as shown in ProFIS2-GUS-3T transgenic plants (Chaudhury et al., 1997; Luo et al., 2000). We adapted this FIS2 promoter construct, replacing the GUS reporter gene with the CCG cDNA to produce the ProFIS2-CCG-3T construct. We introduced this transgene into the ccg/CCG mutant plant line. Three independent transgenic plants were obtained that showed significant increase in seed set (73 to 77%) and Kanr:Kans of 1.7:1.9 (see Supplemental Table 1 online). For example, in line 2, the seed set increased from 58 to 77% (n = 691) after complementation by the transgene, and the Kanr:Kans ratio of T2 transgenic plants increased from 1.1:1.0 to 1.9:1 (n = 331). All T2 transgenic mutant plants, homozygous for the ProFIS2-CCG-3T transgene (ccg/CCG; ProFIS2-CCG-3T/ProFIS2-CCG-3T) showed restored fertility (705 ovules from 15 siliques were analyzed). Among them, 536 developed into mature embryos and 169 were arrested at zygotic stages. These findings indicate that ProFIS2-CCG-3T completely rescued the defective pollen tube guidance phenotype. Taken together, these data demonstrate that the central cell of the female gametophyte plays a role in micropylar pollen tube guidance in Arabidopsis.
DISCUSSION
The Central Cell Plays a Critical Role in Micropylar Pollen Tube Guidance
In Arabidopsis, the intact and mature female gametophyte is indispensable for pollen tube guidance (Hülskamp et al., 1995b; Ray et al., 1997; Shimizu and Okada, 2000). Recently, laser ablation experiments and genetic analyses indicated that the synergid cells of the female gametophyte are instrumental to micropylar pollen tube guidance. Ablation of the synergid cell, but not other gametophytic cells, in T. fourieri abolished the pollen tube guidance process (Higashiyama et al., 2001). Loss of function of the synergid-expressed genes At MYB98 in Arabidopsis and Zm EA1 in maize also abolished micropylar pollen tube guidance (Kasahara et al., 2005; Márton et al., 2005). These data led to the conclusion that the synergids, and no other gametophytic cells, play an essential role in micropylar pollen tube guidance (Higashiyama et al., 2003).
In this report, we showed that the central cell is also critical for the female control of pollen tube guidance in Arabidopsis. First, our phenotypic analysis clearly showed that the mutation did not have an impact on female gametophyte development, and all cell types are correctly specified in the mutant, indicating that mutant embryo sacs reached maturity. Second, the two independent mutant alleles ccg-1 and ccg-2 showed the same pollen tube guidance defective phenotype, indicating that the loss of CCG function is solely responsible for the pollen tube guidance phenotype. Third, CCG is expressed in the central cell of the female gametophyte, although it is also expressed in other tissues and cell types. CCG expression has not been observed in the synergid or egg cell by RNA in situ hybridization analysis or by GUS reporter assays, nor is it among the genes regulated by MYB98 (Punwani et al., 2007). This indicates that CCG expression is central cell specific. It is likely that CCG protein (75 kD) is also restricted to the central cell, since only molecules <10 kD can freely diffuse between the central cell and the synergid as shown in Torenia (Han et al., 2000). Finally, ProFIS2-CCG-3T completely rescued the pollen tube guidance phenotype, indicating that central cell expression of CCG is sufficient to restore pollen tube guidance in mutant ovules.
Consistent with our findings, several lines of evidence imply that other gametophyic cells, in addition to the synergid cells, may play a role in pollen tube guidance. In maize, for example, the Zm EA1 gene is implicated in the attraction of pollen tubes to the embryo sac, and although this gene is expressed in the synergids (Márton et al., 2005), it is also expressed in the egg cell where it was initially isolated (Dresselhaus et al., 1994). Therefore, a role for this gene in pollen tube guidance resulting from its expression in the egg cell cannot be excluded. Stronger evidence is found in the Arabidopsis mutant, magatama, which is also defective in micropylar pollen tube guidance. This mutation affects the polar nuclei causing a fault that prevents their fusion, suggesting that a functional central cell or mature female gametophyte is essential for pollen tube guidance (Shimizu and Okada, 2000). These data cannot exclude the possibility that cells in addition to the synergids of the female gametophyte may play a critical role in micropylar pollen tube guidance in Arabidopsis.
CCG Might Play a Regulatory Role in Micropylar Pollen Tube Guidance
It is unclear how CCG functions in the central cell to control micropylar pollen tube guidance. One possibility is that CCG acts as a transcriptional regulator controlling downstream genes essential for the pollen tube guidance. CCG is a nuclear protein with a conserved N-terminal zinc β-ribbon domain typical for the general transcription factor TFIIB family (Zhu et al., 1996; Chen et al., 2000). It has been reported that the zinc β-ribbon domain is critical for the binding of TFIIB to RNA polymerase II and recruitment of polymerase II to the transcription preinitiation complex (Buratowski et al., 1989; Barberis et al., 1993; Buratowski and Zhou, 1993; Ha et al., 1993; Tubon et al., 2004) and selection of the polymerase II transcription start site (Pinto et al., 1994; Pardee et al., 1998). Site-specific mutagenesis suggested that the zinc β-ribbon domain interacts with a conserved surface of the polymerase II Dock domain (Chen and Hahn, 2003; Bushnell et al., 2004). Therefore, the zinc β-ribbon domain of TFIIB plays a pivotal role in transcription. We showed here that in yeast two-hybrid assays, the N-terminal zinc β-ribbon domain of CCG is interchangeable with that of TFIIB from both Arabidopsis and yeast, suggesting that the zinc β-ribbon domain of the CCG protein may also have a similar function in interacting with TFIIF and RNA polymerase II to regulate transcription. Further experiments are required to demonstrate such interactions in vivo. In addition to the zinc β-ribbon domain, there are several Cys residues and a Ser-rich domain at the C terminus whose role also remains to be investigated. Nevertheless, it is plausible to speculate that via its N-terminal domain, CCG may regulate a subset of genes that are essential for the central cell control of pollen tube guidance. Undoubtedly, the elucidation of the biochemical function of CCG and the identification of genes regulated by the CCG protein will shed light on the underlying molecular mechanism of pollen tube guidance. With the help of laser microdissection technology, now it is possible to identify genes regulated in the central cell by the CCG protein.
Another issue raised by this study is whether the central cell controls pollen tube guidance directly or via the synergid and/or the egg cell. Morphologically, the micropylar portion of the central cell surrounding the synergid and the egg cell is in close proximity to the micropyle, such that signals from the central cell could diffuse readily to the micropylar opening. An alternative scenario is that the central cell exerts its role by modulating synergid function. It has been observed that at the junction region of the egg cell, central cell, and synergid cell, the cell wall is absent or discontinuous, and the plasma membranes of these cells are in direct contact with one another. This eliminates the cell wall barrier that obstructs signal transduction (Huang and Russell, 1992; Christensen et al., 1997). Cell–cell communication between the egg apparatus and the central cell can be achieved preferentially via intercellular symplastic connections, such as plasmadesmata (Diboll and Larson, 1966; Han et al., 2000), that may help to establish and maintain their identity and position during embryo sac maturation (Dresselhaus, 2006). It is thus possible that signal produced in the central cell gets transported to the synergids or that CCG regulates signals that affect synergid function.
In conclusion, we showed here that the loss of CCG function specifically impairs the female control of micropylar pollen tube guidance. The expression of CCG in the central cell is sufficient to restore the pollen tube guidance defect in the mutant, demonstrating that the central cell plays a critical role in micropylar pollen tube guidance in Arabidopsis. Together with the established role of the synergid cell, we propose that pollen tube guidance requires multiple signals from the female gametophyte to ensure successful fertilization. The identification of the downstream genes regulated by CCG will be instrumental to understand the underlying mechanisms.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Landsberg erecta plants were grown in an air-conditioned room at 22°C under 16-h-light/8-h-dark cycles. Seed germination and selection were performed as described previously by Sundaresan et al. (1995). Briefly, surface-sterilized seeds were plated onto Murashige and Skoog (1962) agar medium supplemented with or without antibiotics. For kanamycin selection, 50 mg/L of kanamycin (Sigma-Aldrich) was supplemented. Similarly, 20 mg/L of hygromycin (Roche) was added for hygromycin selection. Salk_077907 seeds were obtained form the ABRC. T-DNA insertion was confirmed by PCR and DNA gel blot as described by Alonso et al. (2003). Plant transformation was performed according to Bechtold and Pelletier (1998).
Scanning Electron Microscopy
Pistils from both wild-type and mutant flowers were partially dissected and immediately fixed with 3% glutaraldehyde in 50 mM cacodylate buffer, pH 7.0. After an initial fixation for 2 h at room temperature under a gentle vacuum, pistils were kept in fresh fixative at 4°C overnight. Specimens were then washed three times in cacodylate buffer and post-fixed in 2% osmium tetroxide in cacodylate buffer overnight at 4°C. The fixed pistils were rinsed three times in cacodylate buffer, dehydrated in a graded ethanol series, and critical point dried in liquid carbon dioxide. The dry pistils were mounted on stubs and further dissected. After sputter coating with gold, pistils were analyzed with a JSM-5310LV scanning electron microscope (JEOL).
Fluorescence Staining of Pollen Tubes
To visualize pollen tubes, siliques were processed as described previously (Huck et al., 2003). Siliques were opened with a fine needle under a stereoscope and fixed immediately at 4°C overnight in Lavdowsky's FAA solution containing 1.5% formaldehyde, 2% acetic acid, and 30% ethanol. The fixed sample was hydrated by passing through an alcohol series (70, 50, 30, and 10%) with 10 min for each step. The sample was further softened with 10% chloral hydrate at 60°C for 10 min, subsequently rinsed twice with 100 mM sodium phosphate buffer, pH 7, and then in 5 M NaOH at 60°C for 5 min. Pollen tubes were stained with 0.1% aniline blue (Sigma-Aldrich) for 4 h and washed three times with the sodium phosphate buffer before observation. Stained samples were observed using a Zeiss Axioplan microscope (Carl Zeiss) equipped with an epifluorescence UV filter set (excitation filter at 365 nm, dichroic mirror at 395 nm, barrier filter long-pass at 420 nm).
GUS Assays
GUS staining was performed according to Vielle-Calzada et al. (2000). Pistils and siliques were opened and incubated in GUS staining solution (1 mg/mL X-Gluc [Biosynth], 2 mM Fe2+CN, 2 mM Fe3+CN, 10 mM EDTA, 0.1% Triton X-100, and 100 mg/mL chloramphenicol in 50 mM sodium phosphate buffer, pH 7.0) for 2 to 3 d at 37°C. Vegetative tissues were stained only for 1 d. The stained sample was either cleared in 20% lactic acid/20% glycerol solution or fixed with glataraldehyde and embedded in Historesin (Leica) for sectioning. Stained ovules and sections were observed on a Zeiss Axioplan microscope with Nomarski and dark-field optics.
Pollination Experiment
The stamen at floral stage 12c (Smyth et al., 1990) was emasculated by carefully removing the sepal and petal. After 12 h of emasculation, pollen grains from wild-type or mutant plants were dispersed onto the papillar cells of the recipient stigma. Pistils were allowed to set seeds or checked microscopically at different times after pollination for pollen tube entry.
CLSM
To study the cytological structure of the female gametophyte, ovules were fixed and observed as described previously (Christensen et al., 1997; Shi et al., 2005). Inflorescences were fixed in 4% glutaraldehyde in 12.5 mM cacodylate buffer, pH 6.9, and dehydrated through a conventional ethanol series and subsequently cleared in 2:1 of benzyl benzoate:benzyl alcohol. Then, siliques were opened with a 30.5-gauge syringe along the replum, and ovules were mounted with immersion oil and sealed under No. 0 cover slips (ProSciTech) with fingernail polish. The developmental stages of ovules were according to Christensen et al. (1997). The sample was then viewed with a Zeiss LSM510 META laser scanning microscope with a 488-nm argon laser and a long-pass 530 filter. Serial optic sections were collected and projected with Zeiss LSM Image Browser software (Zeiss) and Photoshop version 7.0 software (Adobe).
In Situ Hybridization
Floral buds and pistils were fixed and processed according to Yang et al. (1999). Eight-micrometer-thick sections were hybridized with digoxigenin-labeled antisense and sense probe. For probe labeling, a 650-bp cDNA fragment amplified by PCR with primers 5′-GTGAAGTAAAAGATGCTAAGAG-3′ and 5′-TGTCTCTGAATCACCGTCACTAG-3′ was subcloned in pGEMT-easy vector (Promega). The vector used as template was then linearized with SalI and NcoI, respectively. Antisense and sense RNA were in vitro transcribed with SP6 and T7 RNA polymerases (Promega), respectively. Hybridized slides were rinsed twice with 2× SSC at room temperature for 30 min and then with 1× SSC twice at 60°C for 10 min each. Antibody staining and coloring were performed according to the manufacturer's recommendations (Roche). The slides were observed under a Zeiss Axioplan microscope (Zeiss) with Nomarski optics and photographed with a Nikon Coolpix 995 digital camera.
Molecular Cloning
To clone the CCG gene, genomic DNA was extracted by the CTAB method and used as template for the amplification of Ds flanking sequence and CCG gene. TAIL-PCR was performed as described previously (Liu et al., 1995; Grossniklaus et al., 1998; Yang et al., 1999) to obtain the Ds flanking sequences. The flanking sequence was further confirmed by PCR and sequence analysis. A 5.5-kb CCG genomic fragment, from 3000 bp upstream of ATG start codon to 600 bp downstream of the TGA stop codon, was amplified with primers 5′-AGAGGTACCTCATTGGCCAGTTGTTGTTGT-3′ and 5′-GAAGAGCTCATTCTGATCTAATGTCACAAG-3′ using LA DNA polymerase (Takara). The fragment was then cloned into pCAMBIA1300 (CAMBIA) at KpnI and SacI sites, giving rise to pCAMBIA1300-CCG construct and verified by subsequent sequencing.
To construct the ProCCG-GUS-3T reporter, the 3000-bp genomic fragment upstream the ATG start codon was amplified by PCR with a 5′ primer containing a XbaI site (5′-GGATCTAGATGGGTTCCAGTTTGAGTC-3′) and a 3′ primer with a BamHI site (5′-AGAGGATCCGTAATTGAAGAGACTAGAAAG-3′), respectively. Similarly, a 650-bp 3′ untranslated region downstream of the TGA stop codon was also obtained with primers with a SacI site (5′-GAAGAGTCTAAGTTCATAAGAAGCAGAAG-3′ and 5′-GAAGAGCTCATTCTGATCTAATGTCACAAG-3′). PCR fragments with expected size were sequenced and subcloned into pBI101.1 (Clonetech), giving rise to the ProCCG-GUS-3T reporter construct.
To construct the CCG-GFP protein fusion, the coding region of CCG was amplified using cDNA as template with primers 5′ AGAGGATCCATGATCTGCACGGAGTGCGA-3′ and 5′ GAGGGTACCCAATCATCTTGATCTTCTCC-3′ containing a BamHI and KpnI site, respectively. The PCR product was subcloned into pEGFP vector at BamHI and KpnI to give rise to pCCG-GFP. Then, the CCG-GFP was released from pCCG-GFP by BamHI and XbaI and subcloned behind the 35S promoter in pCAMBIA1300, yielding pCAMBIA1300-35S-CCG-GFP.
For constructing the ProFIS2-CCG-3T gene, the 3900-bp FIS2 genomic fragment contains 3189 bp of nucleotide sequence upstream of the predicted translational start site, exons 1, 2, and 3, the first 39 bp of exon 4 (Luo et al., 2000); and the first three introns were subcloned in fusion with CCG cDNA into pCAMBIA1300 vector.
ProMYB98-GFP construction was according to Kasahara et al. (2005). The 1674-bp promoter plus 87 bp coding sequences of MYB98 was amplified by PCR with primers 5′-CCCAAGCTTGCGGCGGAGATAGTGGCTGAGAGGTG-3′ and 5′-AAAACTGCAGATCTTCTTCTTTCATGTGTTCTTG-3′. The fragment was subsequently cloned in p1300-GFP at HindIII and PstI sites to yield ProMYB98-GFP.
RT-PCR Analysis
Total RNA was isolated from different tissues of wild-type plants with TRIzol reagent (Invitrogen). RNA quality and quantity were analyzed by electrophoresis and spectrophotometry. For cDNA synthesis, 3 μg of total RNA from different tissue was used for reverse transcription with AMV reverse transcriptase (Takara) according to the manufacturer's recommendations. One microliter of the reaction product was then used as template for PCR amplification with primers 5′-AGAGGATCCATGATCTGCACGGAGTGCGA-3′ and 5′-AGGTCTAGATCAATCATCTTGATCTTCTC-3′ containing BamHI and XbaI, respectively. ACT2 was also used as an internal control with primers 5′-GAAGATTAAGGTCGTTGCACCACCTG-3′ and 5′-ATTAACATTGCAAAGAGTTTCAAGGT-3′ to amplify a 477-bp DNA fragment with mRNA or the 563-bp one with genomic DNA. PCR products were checked by 1% agarose gel electrophoresis. Amplified CCG cDNA was gel purified and subcloned into pGEM-T vector for sequencing.
Yeast Two-Hybrid Assay
The coding regions of At TFIIF and Sc TFIIF (yeast TFIIF) were PCR amplified using primer At TFIIF-F (5′-GGAGAATTCATGGAAGATATTCATAATCTC-3′) and primer AtTFIIF-R (5′-GGACTCGAGTCACTGCCCACCTGTATCATC-3′); primer Sc TFIIF-F (5′-GGAGAATTCATGAGCAGTGGTTCAGCAG-3′) and primer Sc TFIIF-R (5′-GGACTCGAGCTAAACGACATCTTCCATTTC-3′), respectively, and subsequently cloned into the EcoRI and XhoI sites of pGADT7 vector (Clontech), resulting in plasmids pGADT7∷AtTFIIF and pGADT7∷ScTFIIF, respectively.
The coding region of CCG and the CCGN40 sequence for the N terminal 40 amino acids was PCR amplified using primer CCGY-F (5′-GAACCATGGTGATCTGCACGGAGTGCGA-3′) and primer CCGY-R (5′-AGAGGATCCTCAATCATCTTGATCTTCTC-3′); primer CCGN40-F (5′-GAACCATGGTGATCTGCACGGAGTGCGA-3′) and primer CCGN40-R (5′-GGAGAATTCATCGCCGTCGTCGACGCCGGT-3′), respectively, and subsequently cloned into the NcoI and BamHI sites of pGBKT7 vector (Clontech) for CCG and NcoI and EcoRI sites for At CCGN40, resulting in plasmids pGBKT7∷CCG and pGBKT7∷CCGN40.
The coding region of Sc TFIIB and TFIIBN60D (Sc TFIIB with an N-terminal 60–amino acid deletion) was PCR amplified using primer Sc TFIIB-F (5′-GGAGAATTCATGATGACTAGGGAGAGCATAG-3′) and Sc TFIIB-R (5′-GGACTGCAGTTCAACGCCCGGTAAGTTATC-3′); primer TFIIBN60D-F (5′-GGAGAATTCTCAAATGATGATCACAACGGTG-3′) and TFIIBN60D-R (5′-GGACTGCAGTTCAACGCCCGGTAAGTTATC-3′), respectively, and subsequently cloned into the EcoRI and PstI sites of pGBKT7 vector (Clontech), resulting in plasmids pGBKT7∷ScTFIIB and pGBKT7∷TFIIBN60D. The TFIIBN60D fragment was also cloned into the EcoRI and PstI sites of pGBKT7-CCGN40, giving rise the plasmid pGBKT7∷CCGN40-TFIIBN60D.
The derived plasmids were sequenced to confirm the absence of errors from PCR amplification and the correct in-frame fusion with the GAL4 DNA binding and activation domains. All the pGBKT7 vectors (either empty or encoding gene) were respectively cotransformed with pGADT7 vectors (either empty or encoding gene) into the yeast strain AH109 using the lithium acetate method.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession number At2g02955 (CCG).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GUS Reporter Expression in the Central Cell of the Female Gametophyte.
Supplemental Figure 2. CCG Expression in Ovules Revealed by RNA in Situ Hybridization.
Supplemental Table 1. Summary of Complementation Data.
Supplementary Material
Acknowledgments
We thank Zuo-Shun Tang (Institute of Genetics and Developmental Biology) for assistance with the χ2 test. We also thank De Ye (China Agricultural University) and Venketesan Sundaresan at (University of California, Davis) for initial help with the mutant screen, Mark Curtis (Institute of Plant Biology, University of Zurich) for critical reading of the manuscript, and the ABRC for the T-DNA insertion lines. U.G. was supported by the University of Zurich and the Cold Spring Harbor President's Council. W.-C.Y. was supported by the Bai Ren Ji Hua Program of the Chinese Academy of Sciences and a grant (2007CB947600) from the Ministry of Science and Technology, China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wei-Cai Yang (wcyang@genetics.ac.cn).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–665. [DOI] [PubMed] [Google Scholar]
- Barberis, A., Muller, C.W., Harrison, S.C., and Ptashne, M. (1993). Delineation of two functional regions of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90 5628–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Buratowski, S., Hahn, S., Guarente, L., and Sharp, P.A. (1989). Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56 549–561. [DOI] [PubMed] [Google Scholar]
- Buratowski, S., and Zhou, H. (1993). Functional domains of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90 5633–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, D.A., Westover, K.D., Davis, R.E., and Kornberg, R.D. (2004). Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303 983–988. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Luo, M., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.-T., and Hahn, S. (2003). Binding of TFIIB to RNA polymerase II: Mapping the binding site for TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12 437–447. [DOI] [PubMed] [Google Scholar]
- Chen, H.T., Legault, P., Glushka, J., Omichinski, J.G., and Scott, R.A. (2000). Structure of a (Cys3His) zinc ribbon, a ubiquitous motif RNA polymerase in archaeal and eucaryal transcription. Protein Sci. 9 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., Wang, H., and Wu, H.-M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82 383–393. [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.-M. (2000). Pollen tube guidance – Right on target. Science 293 1441–1442. [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., Wu, H.-M., Di Stilto, V., Glaven, R., Chen, C., Wong, E., Gdahl, J., and Estavillo, A. (2000). Pollen–pistil interactions in Nicotiana tabacum. Ann. Bot. (Lond.) 85 29–37. [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202 136–151. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Keith, K.C., and Preuss, D. (2000). Tetrad analysis in higher plants. A budding technology. Plant Physiol. 124 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diboll, A.G., and Larson, D.A. (1966). An electron microscopic study of the mature megagametophyte in Zea mays. Am. J. Bot. 53 391–402. [PubMed] [Google Scholar]
- Dong, J., Kim, S.T., and Lord, E.M. (2005). Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 138 778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus, T. (2006). Cell–cell communication during double fertilization. Curr. Opin. Plant Biol. 9 41–47. [DOI] [PubMed] [Google Scholar]
- Dresselhaus, T., Lörz, H., and Kranz, E. (1994). Representative cDNA libraries from few plant cells. Plant J. 5 605–610. [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo, J., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W., and Grossniklaus, U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317 656–660. [DOI] [PubMed] [Google Scholar]
- Fiebig, A., Mayfield, J.A., Miley, N.L., Chau, S., Fischer, R.L., and Preuss, D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Ha, I., Roberts, S., Maldonado, E., Sun, X., Kim, L.U., Green, M., and Reinberg, D. (1993). Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 7 1021–1032. [DOI] [PubMed] [Google Scholar]
- Han, Y.Z., Huang, B.Q., Zee, S.Y., and Yuan, M. (2000). Symplastic communication between the central cell and the egg apparatus cells in the embryo sac of Torenia fournieri Lind. before and during fertilization. Planta 211 158–162. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003). Pollen tube guidance: Beacons from the female gametophyte. Curr. Opin. Plant Biol. 6 36–41. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293 1480–1483. [DOI] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.-Q., and Russell, S.D. (1992). Female germ unit: Organization, isolation, and function. Int. Rev. Cytol. 140 233–292. [Google Scholar]
- Huck, N., Moore, J.M., Federer, M., and Grossniklaus, U. (2003). The Arabidopsis mutant forenia disrupts the female gametophytic control of pollen tube reception. Development 130 2140–2150. [DOI] [PubMed] [Google Scholar]
- Hülskamp, M., Kopczak, S.D., Horejsi, T.F., Kihl, B.K., and Pruitt, R.E. (1995. a). Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 8 703–714. [DOI] [PubMed] [Google Scholar]
- Hülskamp, M., Schneitz, K., and Pruitt, R.E. (1995. b). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, S., Nasrallah, J.B., Dixit, R., Preiss, S., and Nasrallah, M.E. (1997). An aquaporin-like gene required for the Brassica self-incompatibility response. Science 276 1564–1566. [DOI] [PubMed] [Google Scholar]
- Jauh, G., Eckard, K., Nothnagel, E., and Lord, E. (1997). Adhesion of lily pollen tubes on an artificial matrix. Sex. Plant Reprod. 10 859–865. [Google Scholar]
- Johnson, M.A., and Preuss, D. (2002). Plotting a course: Multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. [DOI] [PubMed] [Google Scholar]
- Kasahara, R.D., Portereiko, M.F., Sandaklie-Nikolova, L., Rabiger, D.S., and Drews, G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Mollet, J.-C., Dong, J., Zhang, K., Park, S.Y., and Lord, E.M. (2003). Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 100 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–464. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush, W.M. (1999). Whither chemotropism and pollen tube guidance? Trends Plant Sci. 4 413–418. [DOI] [PubMed] [Google Scholar]
- Márton, M.L., Cordts, S., Broadhvest, J., and Dresselhaus, T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307 573–576. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., and Preuss, D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2 128–130. [DOI] [PubMed] [Google Scholar]
- Mollet, J.C., Park, S.Y., Nothnagel, E.A., and Lord, E.M. (2000). A lily stylar pectin is necessary for pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12 1737–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 15 473–497. [Google Scholar]
- Palanivelu, R., Brass, L., Edlund, A.F., and Preuss, D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114 47–59. [DOI] [PubMed] [Google Scholar]
- Palanivelu, R., and Preuss, D. (2000). Pollen tube targeting and axon guidance: Parallels in tip growth mechanisms. Trends Cell Biol. 10 517–524. [DOI] [PubMed] [Google Scholar]
- Palanivelu, R., and Preuss, D. (2006). Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol. 6 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, T.S., Bangur, C.S., and Ponticelli, A.S. (1998). The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem. 273 17859–17864. [DOI] [PubMed] [Google Scholar]
- Park, S.Y., Jauh, G.Y., Mollet, J.C., Eckard, K.J., Nothnagel, E.A., Walling, L.L., and Lord, E.M. (2000). A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, I., Wu, W.-H., Na, J.G., and Hampsey, M. (1994). Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 269 30569–30573. [PubMed] [Google Scholar]
- Preuss, D., Rhee, S.Y., and Davis, R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264 1458–1460. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.E. (1999). Complex sexual signals for the male gametophyte. Curr. Opin. Plant Biol. 2 419–422. [DOI] [PubMed] [Google Scholar]
- Punwani, J.A., Rabiger, D.S., and Drews, G.N. (2007). MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19 2557–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S., Park, S.-S., and Ray, A. (1997). Pollen tube guidance by the female gametophyte. Development 124 489–498. [DOI] [PubMed] [Google Scholar]
- Rotman, N., Durbarry, A., Wardle, A., Yang, W.C., Chaboud, A., Faure, J.E., Berger, F., and Twell, D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15 244–248. [DOI] [PubMed] [Google Scholar]
- Shi, D.Q., Liu, J., Xiang, Y.H., Ye, D., Sundaresan, V., and Yang, W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K.K., and Okada, K. (2000). Attractive and repulsive interactions between female gametophytes in Arabidopsis pollen tube guidance. Development 127 4511–4518. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer-Knudsen, J., Clarke, A.E., and Bacic, A. (1996). A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J. 9 71–83. [DOI] [PubMed] [Google Scholar]
- Sommer-Knudsen, J., Lush, W.M., Bacic, A., and Clarke, A.E. (1998). Re-evaluation of the role of a transmitting tract-specific glycoprotein on pollen tube growth. Plant J. 13 529–535. [Google Scholar]
- Sundaresan, V., Springer, P.S., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R.A. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Tubon, T.C., Tansey, W.P., and Herr, W. (2004). A nonconserved surface of the TFIIB zinc ribbon domain plays a direct role in RNA polymerase II recruitment. Mol. Cell. Biol. 24 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.-P., Basker, R., and Grossiniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404 91–94. [DOI] [PubMed] [Google Scholar]
- Wang, H., Wu, H.-M., and Cheung, A.Y. (1993). Development and pollination regulated accumulation and glycosylation of a stylar transmitting tissue-specific proline-rich protein. Plant Cell 5 1639–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi, L.K., and Preuss, D. (1997). Blazing new trails: Pollen tube guidance in flowering plant. Plant Physiol. 113 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi, L.K., and Preuss, D. (1999). The mating game: Pollination and fertilization in flowering plants. Curr. Opin. Plant Biol. 2 18–22. [DOI] [PubMed] [Google Scholar]
- Wolters-Arts, M., Lush, W.M., and Mariani, C. (1998). Lipids are required for directional pollen tube growth. Nature 392 818–821. [DOI] [PubMed] [Google Scholar]
- Wu, H.-M., Wang, H., and Cheung, A.Y. (1995). A floral transmitting tissue specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82 383–393. [DOI] [PubMed] [Google Scholar]
- Wu, H.M., Wong, E., Ogdahl, J., and Cheung, A.Y. (2000). A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J. 22 167–176. [DOI] [PubMed] [Google Scholar]
- Yang, W.-C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for sporogenesis and encodes a novel protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., Zeng, Q., Colangelo, C.M., Lewis, M., Summers, M.F., and Scott, R.A. (1996). The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat. Struct. Biol. 3 122–124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.