Abstract
The Arabidopsis thaliana MALE STERILITY1 (MS1) gene encodes a nuclear protein with Leu zipper–like and PHD-finger motifs and is important for postmeiotic pollen development. Here, we examined MS1 function using both cell biological and molecular biological approaches. We introduced a fusion construct of MS1 and a transcriptional repression domain (MS1-SRDX) into wild-type Arabidopsis, and the transgenic plants showed a semisterile phenotype similar to that of ms1. Since the repression domain can convert various kinds of transcriptional activators to dominant repressors, this suggested that MS1 functioned as a transcriptional activator. The Leu zipper–like region and the PHD motif were required for the MS1 function. Phenotypic analysis of the ms1 mutant and the MS1-SRDX transgenic Arabidopsis indicated that MS1 was involved in formation of pollen exine and pollen cytosolic components as well as tapetum development. Next, we searched for MS1 downstream genes by analyzing publicly available microarray data and identified 95 genes affected by MS1. Using a transgenic ms1 plant showing dexamethasone-inducible recovery of fertility, we further examined whether these genes were immediately downstream of MS1. From these results, we discuss a role of MS1 in pollen and tapetum development and the conservation of MS1 function in flowering plants.
INTRODUCTION
Pollen development is a postmeiotic process that produces mature pollen grains from microspores. In addition to activities within the microspore itself, sporophytic anther tissues play important roles in this process not only by providing physical support but also by supplying signals and materials necessary for pollen development. Especially, the tapetum, which is the innermost somatic cell layer of the anther locule, plays an essential role in pollen development (Goldberg et al., 1993). In Arabidopsis thaliana, the MALE STERILITY1 (MS1), ABORTED MICROSPORES (AMS), and At MYB103 genes are expressed in the tapetum after meiosis. The ms1 and ams mutants and transgenic lines with reduced At MYB103 expression are defective in microspore development (Wilson et al., 2001; Ito and Shinozaki, 2002; Higginson et al., 2003; Sorensen et al., 2003). The fact that these genes encode putative transcription factors suggests that they may control genes necessary for early stages of pollen development.
A crucial aspect of pollen development is the proper formation of pollen exine and pollen coat (for a review, see Piffanelli et al., 1998). Pollen exine is a highly sculptured extracellular matrix that helps to maintain the pollen structure and protects the pollen from biotic and abiotic stresses. A major constituent of exine is sporopollenin, which is made up of a series of related polymers derived from very-long-chain fatty acids and also contains modest amounts of oxygenated aromatic rings and phenylpropanoids. Several Arabidopsis genes involved in exine formation have been identified from analyses of sporophytic mutants defective in pollen development. The MS2 gene encodes a putative fatty acyl reductase that possibly reduces fatty acids to fatty alcohols and is expressed specifically in the tapetum at a stage shortly after microspores are released from the tetrad (Aarts et al., 1997). The DEFECTIVE IN EXINE FORMATION1 gene encodes a novel protein that is predicted to be membrane associated and contains potential calcium binding domains (Paxson-Sowders et al., 2001). The FACELESS POLLEN1 (FLP1) gene encodes a putative lipid transfer protein that is involved in pollen coat and exine formation (Ariizumi et al., 2003). The NO EXINE FORMATION1 gene encodes a novel protein that is predicted to be a plastid integral membrane protein (Ariizumi et al., 2004). MS2 and FLP1 are candidate genes involved in sporopollenin biosynthesis.
Exine formation is complete by the first pollen mitosis; afterwards, the interstices of the exine are covered by the pollen coat, or tryphine. The pollen coat is a lipidic matrix and contains proteins mediating species specificity of pollination and components necessary for pollination efficiency. Although exine and pollen coat formation is temporally separated, their biosyntheses include some common components, such as fatty acids (Piffanelli et al., 1998). In addition, major proteins in the Arabidopsis pollen coat have been isolated (Mayfield et al., 2001). The proteins include two extracellular lipases (EXL4 and 6) and five oleosins (GRP14, 16, 17, 18, and 19). The GRP17 protein was demonstrated to play a role in initiating pollination (Mayfield and Preuss, 2000). The lipases and oleosins make up >90% of the detectable pollen coat proteins, and the corresponding genes reside in two genomic clusters. Individual oleosins exhibited extensive divergence between Arabidopsis ecotypes, but the entire cluster remains intact. Analysis of the syntenic region in Brassica oleracea revealed even greater divergence but a similar clustering of the genes. These observations suggest that GRP proteins are important for speciation in plants (Mayfield et al., 2001).
As mentioned above, the Arabidopsis MS1 gene is important for early stages of pollen development. The ms1 mutant is male sterile and produces pollen that fails to properly form the exine layer at the uninucleate stage with subsequent abnormal vacuolation in both microspores and the tapetum (Dawson et al., 1993; Wilson et al., 2001; Ito and Shinozaki, 2002). Analyses using transmission electron microscopy (TEM) showed that ms1 pollen had abnormal exine at the uninucleate and bicellular stages, and the ms1 tapetum did not exhibit signs of programmed cell death (Ariizumi et al., 2005; Vizcay-Barrena and Wilson, 2006). The MS1 gene encodes a protein containing a Leu zipper–like sequence, a nuclear localization signal, and a putative PHD-finger motif (Wilson et al., 2001; Ito and Shinozaki, 2002). A fusion of the MS1 nuclear localization signal to green fluorescent protein was found to localize to the nucleus (Ito and Shinozaki, 2002). Using in situ mRNA hybridization analysis, MS1 was shown (Ito and Shinozaki, 2002) to be expressed specifically in the tapetal tissue and transiently near the time of tetrad formation at anther stage 7, as defined by Sanders et al. (1999). These results suggest that the MS1 gene expression in the sporophytic tapetum regulates the development of the gametophytic pollen, particularly the formation of the extracellular exine, as well as tapetum development itself.
The PHD-finger motif has been found in transcription and/or chromatin remodeling factors. For example, PHD domains of the NURF and the ING2 proteins bind to trimethylated Lys-4 of the histone H3 and mediate chromatin remodeling (Li et al., 2006; Pena et al., 2006; Shi et al., 2006; Wysocka et al., 2006). Although the presence of a putative PHD finger and Leu zipper in MS1 suggests a function in transcriptional regulation, the function of the MS1 PHD domain has not been tested.
Here, we show that a fusion construct between MS1 and a plant transcriptional repression domain causes otherwise wild-type transgenic Arabidopsis plants to exhibit ms1-like phenotypes, suggesting that MS1 functions as a transcriptional regulator. We examined in detail by electron microscopy the dominant-negative ms1-like transgenic plant and the ms1 mutant and showed that these plants are defective in the development of pollen exine and cytosol as well as tapetum. In addition, we show that the PHD and the Leu zipper in MS1 protein are required for its function. To further understand the MS1 function at the molecular level, we isolated putative target genes of MS1. From these results, we discuss pleiotropic functions of MS1. Finally, we discuss whether the MS1-dependent regulatory system is conserved in flowering plants.
RESULTS
Repression Domain Experiment and ms1 Mutant Analysis
We and others showed previously that the MS1 gene encodes a PHD-type protein containing a nuclear localization signal (Wilson et al., 2001; Ito and Shinozaki, 2002). Therefore, we hypothesized that MS1 is a transcriptional regulator. To test this hypothesis, we examined MS1 function using a plant transcriptional repression domain. We reasoned that if MS1 is a DNA binding protein, a fusion between MS1 and a repression domain can behave as a dominant-negative mutation by interfering with the normal MS1 protein. A plant transcriptional repression domain (EAR) was identified in the class II ethylene-responsive element binding factor (ERF) and TFIIIA-type zinc finger repressors, such as SUPERMAN (SUP) (Ohta et al. 2001; Hiratsu et al. 2002). When the EAR motif was fused to transcriptional activators, such as ETHYLENE-INSENSITIVE3 (EIN3) and CUP-SHAPED COTYLEDON1 (CUC1), the chimeric proteins acted as dominant repressors (Hiratsu et al., 2003). For example, a transgenic plant expressing EIN3-EAR exhibited ethylene insensitivity. This indicates that the EAR motif is active when fused to transcription factors.
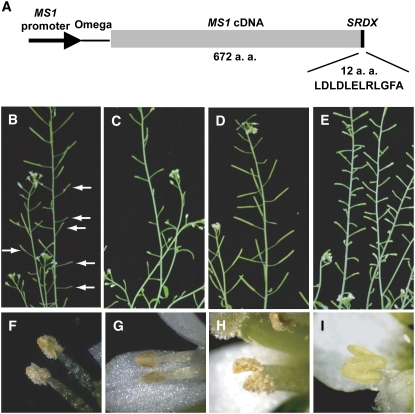
We fused the MS1 cDNA to a DNA sequence for a 12–amino acid EAR motif, SRDX (LDLDLELRLGFA) (Figure 1A). Since the cauliflower mosaic virus (CaMV) 35S promoter was not able to drive proper MS1 expression (see below), the MS1-SRDX chimeric gene was fused with the MS1 promoter, and this construct (ProMS1:MS1-SRDX) was introduced into wild-type Arabidopsis plants. The transformants were reduced in fertility to various extents. We show two representative transgenic plants exhibiting intermediate and severe loss of fertility (Figures 1B and 1C, respectively). We recognized light-yellow pollen grains on dehisced anthers of the intermediate sterile plant (Figure 1F) and a few brownish pollen grains in anther locules of severely sterile plants (Figure 1G). These phenotypes are intermediate between the wild type (Figures 1D and 1H) and the mutant (Figures 1E and 1I). RT-PCR analysis showed that these plants expressed both endogenous MS1 and the introduced MS1-SRDX genes (data not shown).
Figure 1.
A Construct and Representative Transgenic Plants Expressing MS1-SRDX.
(A) ProMS1:MS1-SRDX construct. MS1-SRDX encoding a fusion protein was driven by the MS1 promoter. Omega is a tobacco mosaic virus (TMV) omega sequence. This construct was transformed to Columbia (Col-0) plants. a.a., amino acids.
(B) to (E) Inflorescence stems.
(F) to (I) Anthers.
(B) and (F) The ProMS1:MS1-SRDX transgenic T1 plant showing intermediate sterility. Arrows indicate very small siliques.
(C) and (G) The ProMS1:MS1-SRDX transgenic T1 plant showing severe phenotype. All of the siliques are small.
(D) and (H) A Col-0 plant.
(E) and (I) ms1-1 mutant plant (Ler background). All of the siliques are small.
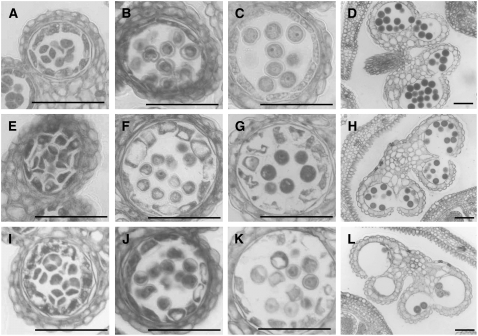
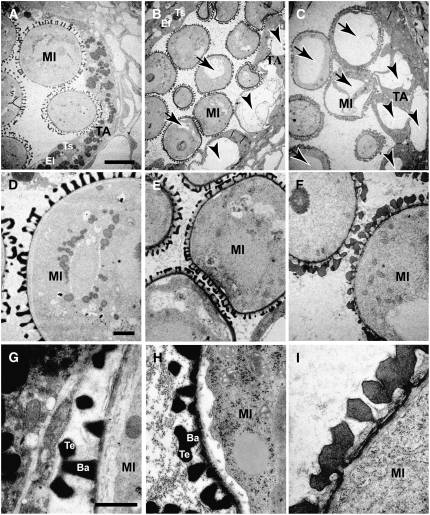
We further examined the phenotype of these anthers using light microscopy. At the tetrad stage, we could not observe phenotypic difference between wild-type and the transgenic plants (Figures 2A, 2E, and 2I). At the uninucleate microspore stage, pollen wall of the transgenic plants was thinner than the wild type (Figures 2B, 2F, and 2J). In the ms1 mutant, we could not detect the pollen wall at all (Ito and Shinozaki, 2002). At the same time, tapetum vacuolation had occurred in the transgenic plants (Figures 2B, 2F, and 2J). At the bicellular pollen stage, pollen and tapetum were degenerating in the transgenic plants (Figures 2C, 2G, and 2K). Finally, less mature pollen had survived in the transformants (Figures 2D, 2H, and 2L). Both microspore and tapetum development were further examined by TEM. In early bicellular pollen stage, wild-type tapetum developed densely stained secretory organelle, elaioplast, and tapetosome (Figure 3A), but the ms1 tapetum did not (Figure 3C). In the ProMS1:MS1-SRDX transgenic plant, a few elaioplast and tapetosomes were observed (Figure 3B). We also observed tapetum vacuolation in the ms1 and the transgenic plant (arrowheads in Figures 3B and 3C). In microspores, vacuolation was more severe in ms1 than in the transgenic plant (arrows in Figures 3B and 3C). These results indicate that defects in the ProMS1:MS1-SRDX transgenic plants were similar to those in the ms1 mutant.
Figure 2.
Development of Anthers and Microspores in Col-0 and ProMS1:MS1-SRDX Transgenic Plants.
(A) to (D) Development of the Col-0 plant.
(E) to (H) Development of the ProMS1:MS1-SRDX transgenic T1 plant showing intermediate sterility.
(I) to (L) Development of the ProMS1:MS1-SRDX transgenic T1 plant showing strong sterility.
(A), (E), and (I) Tetrad stage. Bars = 50 μm.
(B), (F), and (J) Uninucleate microspore stage. Bars = 50 μm.
(C), (G), and (K) Bicellular pollen stage. Bars = 50 μm.
(D), (H), and (L) Mature pollen stage. Bars = 50 μm.
Figure 3.
TEM Analysis of the ms1-1 Mutant and the ProMS1:MS1-SRDX Transgenic Plant Showing Intermediate Sterility.
Microspore and tapetum development in the early bicellular stage is indicated. TA, tapetum; MI, microspore; Ba, bacula; Te, tectum; El, elaioplast (plastid); Ts, tapetosome (lipid body). Arrows and arrowheads indicate vacuoles in microspore and tapetum, respectively. Bars = 10 μm in (A) to (C), 2 μm in (D) to (F), and 1 μm in (G) to (I).
(A), (D), and (G) Ler.
(B), (E), and (H) ProMS1:MS1-SRDX transgenic plant (Col-0 background) showing intermediate sterility. Col-0 control images are not shown here, but the phenotypes are similar to those of Ler.
(C), (F), and (I) ms1-1 mutant (Ler background).
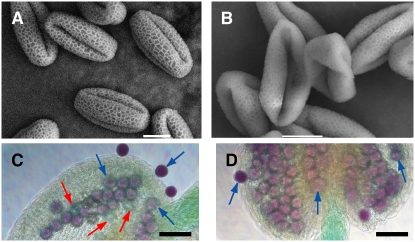
To address biological function of MS1 in exine formation, we observed exine abnormalities of the ms1 mutant and the less severe ProMS1:MS1-SRDX transgenic plant. Scanning electron microscopy analysis showed that surviving mature pollen grains of the transgenic plant showing intermediate sterility did not maintain their shapes and were collapsed along germination pores (Figures 4A and 4B). Nevertheless, these pollen grains were viable, as shown by the Alexander test (Figures 4C and 4D). Since the collapsed pollen structure suggested defective exine structure, we further examined exine development using TEM (Figure 3). TEM analysis showed that pollen of the ProMS1:MS1-SRDX transgenic plant retained sculptured exine structure, but bacula/tectum structure of the transgenic plant was smaller than that of the wild type (Figures 3D, 3E, 3G, and 3H). The smaller structure coincides with the thinner exine structure shown in Figures 2F, 2G, 3B, and 3E. On the other hand, in the ms1 mutant, there was no obvious bacula/tectum structure, which was seen in the wild type and the ProMS1:MS1-SRDX transgenic plant (Figures 3C, 3F, and 3I). However, the abnormal sporopollenin structure of the ms1 was anchored by the microspore surface (Figure 3I), suggesting that MS1 is important for the formation of sculptured bacula/tectum structure on a preformed scaffold. These results indicate that bacula/tectum structure of the transgenic plant was partially defective and that of the mutant completely defective. From these results, we propose that the MS1 protein transcriptionally regulates genes for pollen exine formation, pollen cytosolic content, and tapetum development.
Figure 4.
Scanning Electron Microscopy Analysis of Mature Pollens from the ProMS1:MS1-SRDX Transgenic Arabidopsis Plant Showing Intermediate Sterility.
(A) and (B) Scanning electron microscopy images. Bars = 10 μm.
(C) and (D) Alexander staining of pollen grains (Alexander, 1969). In the typical stain, aborted pollen is light green (red arrows) and nonaborted dark red or pink (blue arrows). Bars = 50 μm.
(A) and (C) Pollen grains from the Col-0 plant.
(B) and (D) Pollen grains from the ProMS1:MS1-SRDX transgenic plant showing intermediate sterility.
The PHD and Leu Zipper Regions in MS1 Are Required for Normal Function
To address the question whether the characteristic Leu zipper–like region and the PHD-finger motif are required for the MS1 function, we performed domain analysis. We introduced one of the constructs shown in Figure 5 into MS1/ms1-1 heterozygous plants and examined their ms1-1/ms1-1 descendants whether their mutant phenotype was complemented or not. As a control, we compared transgenic plants carrying the full-length MS1 cDNA fused to the CaMV 35S promoter with those carrying a fusion of the MS1 cDNA with its own promoter. In the Pro35S:MS1 plants, the mutant phenotype was not rescued (Figure 5, construct 1), indicating that the CaMV 35S promoter was inappropriate for this experiment. By contrast, the ProMS1:MS1 transgene was able to restore fertility to the ms1 mutant (Figure 5, construct 2). Therefore, we used the MS1 promoter to drive modified versions of MS1 cDNA. An internal deletion of MS1, including the Leu zipper–like region (Figure 5, construct 3) was not able to rescue the mutant phenotypes, indicating that this region is required for MS1 function. Next, we introduced base substitutions into the region encoding the PHD finger and found that a change in a single amino acid residue other than the conserved C or H did not affect the ability of the transgene to rescue (construct 4). On the other hand, substitutions of the conserved C or H residues caused partial loss of this ability (constructs 5 and 6). From these results, we conclude that the Leu zipper and the PHD regions are required for the function of the MS1 protein, although we cannot exclude a possibility that an unidentified motif other than Leu zipper in the deleted region is functional.
Figure 5.
Domain Analysis of the MS1 Protein.
Lane 1 shows a construct in which MS1 cDNA was driven by the CaMV 35S promoter. Instead of CaMV 35S, MS1 promoter was used in the constructs in lanes 2 to 6. MS1(WT) is a construct expressing wild-type MS1 cDNA. MS1(Δ212-304) is an internal deletion mutant. The deleted region (amino acids 212 to 304) contains the Leu zipper–like region. MS1(E627K), MS1(C635A), and MS1(H640N, C643G) are amino acid residue–substituted mutants in the PHD region. NLS, LZ, and PHD shown by dark gray indicate nuclear localization signal, Leu zipper–like region, and PHD region, respectively. Fertile: fertilities of this column were equivalent to those of wild-type Ler plants. Semi-sterile: plants of this column were less fertile than wild-type Ler plants. Completely sterile: plants of this column produced no seeds.
Putative Downstream Genes Regulated by MS1
Our results strongly support the hypothesis that MS1 promotes pollen development by regulating gene expression. To further test this idea, we investigated MS1 function in regulating downstream genes. Yang et al. (2007) describe their previous differential expression profile analysis between wild-type (Landsberg erecta [Ler]) and ms1 mutant floral buds using the Affymetrix GeneChip (NASCArray at http://affymetrix.arabidopsis.info/; experiment title: Analysis of anther development by identifying downstream genes controlled by MS1). They compared wild-type and ms1 young and old buds. Because MS1 is expressed in young buds, we chose to focus on the genes that were expressed at higher levels in wild-type young buds than levels in ms1 young and old buds. From >22,000 genes on the ATH1 array, we identified 95 differentially expressed genes (Table 1), which included genes encoding enzymes for lipid and phenylpropanoid biosynthesis and metabolism, pollen coat proteins, cytochrome P450s, proteases, transcription factors, and prenyltransferases. Many of these genes encode predicted secretory proteins. All of the pollen coat proteins described by Mayfield et al. (2001) were included in the list. The complete list is shown in Supplemental Table 1 online.
Table 1.
Selected Genes Whose Expression Was Affected by the ms1 Mutation, from the Microarray Data of Yang et al. (2007)
| Transcript IDa | Probe Set IDa | DEX Inductionb | TargetP Predictionc | Gene Titlea | Possible Functiond | ms1 Younge | ms1 Olde | Ler Youngf | Ler Oldf |
|---|---|---|---|---|---|---|---|---|---|
| At1g06260 | 260788_at | Examined | S | Cys proteinase, putative | Protease | 28.1 ± 6.7 | 44.6 ± 13.8 | 1418.5 ± 349.8 | 222.8 ± 134.1 |
| At1g06990 | 256059_at | Examined | S | GDSL-motif lipase/hydrolase family protein | 4.6 ± 5.3 | 9.1 ± 9.0 | 152.3 ± 54.4 | 39.6 ± 23.5 | |
| At1g13140 | 262764_at | Examined | Cytochrome P450 family protein | Lipid, CYP86C3 | 24.2 ± 10.1 | 20.1 ± 8.4 | 647.0 ± 240.2 | 55.6 ± 42.9 | |
| At1g13150 | 262765_at | ND | S | Cytochrome P450, putative | Lipid, CYP86C4 | 13.0 ± 9.5 | 14.4 ± 10.6 | 301.4 ± 132.4 | 37.2 ± 19.1 |
| At1g23240 | 262987_at | ND | Caleosin-related family protein | PC | 9.5 ± 4.6 | 34.7 ± 38.0 | 404.4 ± 139.3 | 306.1 ± 205.4 | |
| At1g23250 | 263011_at | Examined | Caleosin-related | PC | 3.1 ± 3.2 | 4.5 ± 4.3 | 71.0 ± 49.1 | 34.3 ± 22.2 | |
| At1g28430 | 261499_at | ND | S | Cytochrome P450, putative | CYP705A24 | 7.9 ± 4.6 | 21.8 ± 11.1 | 300.3 ± 118.4 | 216.5 ± 184.2 |
| At1g61110 | 264882_at | Examined | No apical meristem (NAM) family protein | TF | 6.2 ± 2.0 | 10.5 ± 5.5 | 713.6 ± 262.8 | 69.4 ± 49.4 | |
| At1g67990 | 260001_at | ND | Caffeoyl-CoA 3-O-methyltransferase, putative | PP | 7.1 ± 0.4 | 12.8 ± 9.6 | 1501.5 ± 300.6 | 167.0 ± 195.1 | |
| At1g71160 | 259744_at | ND | S | β-Ketoacyl-CoA synthase family protein | Lipid | 25.9 ± 6.0 | 9.5 ± 2.7 | 225.2 ± 81.0 | 21.5 ± 21.2 |
| At1g74540 | 260228_at | Examined | Cytochrome P450, putative | PP, CYP98A8 | 5.5 ± 2.8 | 2.4 ± 0.1 | 498.8 ± 225.3 | 26.2 ± 31.8 | |
| At1g74550 | 260233_at | ND | Cytochrome P450, putative | PP, CYP98A9 | 40.7 ± 16.7 | 38.1 ± 5.2 | 353.3 ± 144.6 | 54.6 ± 15.6 | |
| At1g75910 | 262674_at | ND | S | Family II extracellular lipase 4 (EXL4) | PC | 20.2 ± 12.0 | 28.3 ± 19.1 | 1893.7 ± 591.5 | 275.1 ± 181.5 |
| At1g75920 | 262683_at | Examined | S | Family II extracellular lipase 5 (EXL5) | PC | 11.6 ± 6.1 | 17.9 ± 11.3 | 540.7 ± 215.7 | 89.1 ± 55.4 |
| At1g75930 | 262675_at | ND | S | Family II extracellular lipase 6 (EXL6) | PC | 3.3 ± 2.0 | 61.4 ± 56.9 | 1226.9 ± 351.7 | 564.7 ± 350.4 |
| At1g75940 | 262697_at | Examined | S | Glycosyl hydrolase family 1 protein/anther-specific protein ATA27 | 61.5 ± 11.7 | 56.7 ± 9.0 | 1830.9 ± 413.0 | 308.0 ± 243.5 | |
| At1g76470 | 259975_at | ND | Cinnamoyl-CoA reductase family | PP | 22.4 ± 6.9 | 19.6 ± 6.3 | 282.2 ± 129.8 | 32.5 ± 22.6 | |
| At2g03200 | 266708_at | Examined | Aspartyl protease family protein | Protease | 4.5 ± 4.3 | 24.8 ± 11.9 | 266.1 ± 76.9 | 38.4 ± 18.1 | |
| At2g19070 | 267440_at | Examined | Transferase family protein | PP | 8.3 ± 2.4 | 7.3 ± 2.1 | 382.0 ± 194.1 | 26.3 ± 18.3 | |
| At2g23800 | 267295_at | Examined | S | Geranylgeranyl pyrophosphate synthase (GGPS2) (GGPS5)/GGPP synthetase/farnesyltranstransferase | PT | 11.0 ± 14.9 | 36.4 ± 26.5 | 389.1 ± 193.9 | 246.6 ± 178.9 |
| At3g26125 | 257633_at | ND | Cytochrome P450, putative | Lipid, CYP86C2 | 64.6 ± 18.7 | 138.4 ± 45.1 | 849.0 ± 290.1 | 254.3 ± 166.7 | |
| At3g51590 | 252063_at | Examined | S | Lipid transfer protein, putative | Lipid | 22.1 ± 21.7 | 12.5 ± 8.8 | 1827.2 ± 484.9 | 225.4 ± 211.5 |
| At3g52160 | 252035_at | Examined | β-Ketoacyl-CoA synthase family protein | Lipid | 21.0 ± 7.4 | 13.5 ± 0.9 | 496.2 ± 223.9 | 45.3 ± 31.8 | |
| At3g57810 | 251558_at | ND | OTU-like Cys protease family protein | Protease | 108.8 ± 17.3 | 98.1 ± 11.4 | 487.3 ± 213.6 | 224.9 ± 92.1 | |
| At4g22080 | 254338_s_at | Examined | S | Pectate lyase family protein | 0.4 ± 0.4 | 0.1 ± 0.1 | 87.2 ± 36.5 | 7.0 ± 11.8 | |
| At4g22090 | |||||||||
| At4g23660 | 254230_at | ND | M | UbiA prenyltransferase family protein | PT | 49.2 ± 9.5 | 54.9 ± 3.5 | 297.8 ± 89.9 | 83.2 ± 23.0 |
| At4g28395 | 253772_at | Examined | Lipid transfer protein, putative | Lipid | 30.4 ± 6.4 | 11.6 ± 4..8 | 986.3 ± 249.3 | 69.8 ± 71.0 | |
| At5g07510 | 250635_at | ND | S | Gly-rich protein (GRP14) | PC | 7.4 ± 5.2 | 35.0 ± 7.5 | 422.6 ± 167.2 | 151.3 ± 81.5 |
| At5g07520 | 250636_at | ND | S | Gly-rich protein (GRP18) | PC | 9.9 ± 2.7 | 33.3 ± 8.3 | 491.3 ± 317.7 | 166.3 ± 105.5 |
| At5g07530 | 250637_at | Examined | Gly-rich protein (GRP17) | PC | 10.3 ± 0.4 | 119.5 ± 122.9 | 1344.0 ± 603.8 | 706.2 ± 455.9 | |
| At5g07540 | 250638_at | ND | S | Gly-rich protein (GRP16) | PC | 26.4 ± 12.1 | 41.6 ± 9.1 | 636.4 ± 322.5 | 232.2 ± 143.7 |
| At5g07550 | 250610_at | ND | S | Gly-rich protein (GRP19) | PC | 25.7 ± 20.3 | 337.8 ± 422.8 | 2450.7 ± 447.5 | 1502.9 ± 889.6 |
| At5g07560 | 250639_at | ND | Gly-rich protein (GRP20) | PC | 8.1 ± 6.6 | 85.3 ± 62.6 | 1856.4 ± 411.9 | 786.0 ± 519.3 | |
| At5g13380 | 250294_at | Examined | Auxin-responsive GH3 family protein | 9.0 ± 3.2 | 10.7 ± 7.6 | 403.1 ± 146.1 | 34.9 ± 34.5 | ||
| At5g49070 | 248638_at | ND | β-Ketoacyl-CoA synthase family protein | Lipid | 14.2 ± 9.8 | 9.3 ± 2.0 | 112.8 ± 49.0 | 15.1 ± 5.6 | |
| At5g60500 | 247639_s_at | Examined | Undecaprenyl pyrophosphate synthetase family protein/UPP synthetase family protein | PT | 1.3 ± 1.5 | 1.4 ± 0.1 | 154.8 ± 111.5 | 12.0 ± 12.9 | |
| At5g60510 | |||||||||
| At5g62320 | 247451_at | Examined | Myb family transcription factor (MYB99) | TF | 9.2 ± 2.1 | 9.6 ± 5.1 | 119.6 ± 41.2 | 16.4 ± 7.9 |
We used publicly available NASCArray data (experiment ID: NASCARRAYS-23) at http://affymetrix.Arabidopsis.info/. After eliminating genes with signal values <15.0 in Ler young, we selected genes with more than fivefold higher expression in Ler young than in ms1 old. Among them, we further selected genes with more than threefold higher expression in Ler young than in ms1 young. In total, we isolated 95 genes. Here, we show selected 39 genes among them. The complete list is shown in Supplemental Table 1 online.
These columns are the same as the original TIGR ATH1-121501 database.
Thirty-two genes out of 95 were examined for DEX inducibility using the DEX induction system as shown in Figure 6C and Supplemental Figure 1 online. See Supplemental Table 1 online for the complete list. ND, not determined.
Potential subcellular localization signal was predicted using the TargetP program (Emanuelsson et al., 2000; http://www.cbs.dtu.dk/services/TargetP/). Localization to the secretory pathway (S) and mitochondrion (M) with prediction reliability class 1 to 3 is indicated.
Possible function of a gene product involved in lipid biosynthesis and transfer (lipid), phenylpropanoid biosynthesis (PP), pollen coat protein (PC), cytochrome P450 family (CYP), protease, transcription factor (TF), or prenyltransferase (PT).
These columns show average of signal values (± sd) from three independent hybridization slides done by NASCArray.
These columns show average of signal values (± sd) from five independent hybridization slides done by NASCArray.
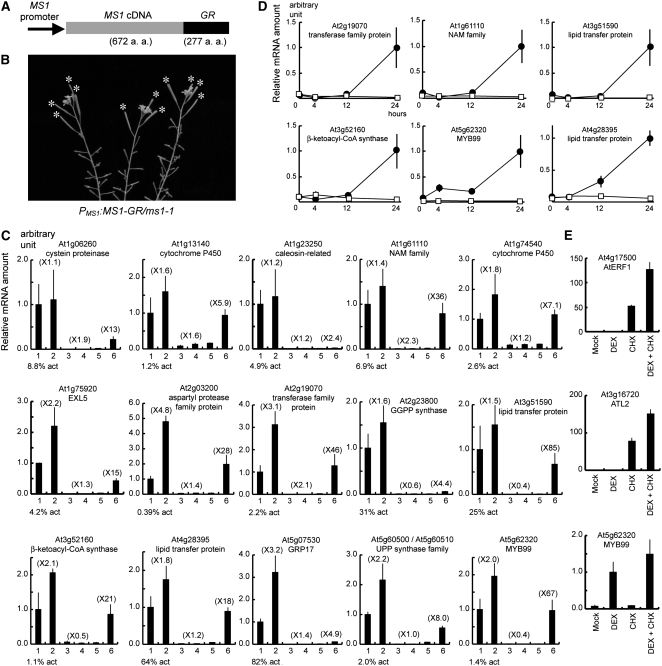
To further test whether these genes are indeed transcriptionally regulated by MS1, we examined whether they were induced immediately by the activation of the MS1 function. To this end, we adopted a glucocorticoid induction system, which was successful in conferring inducible activity to plant transcription factors (Lloyd et al., 1994; Aoyama et al., 1995; Sablowski and Meyerowitz, 1998; Wagner et al., 1999). We generated a transgenic plant homozygous for a construct (ProMS1:MS1-GR) of the MS1 promoter driving a fusion of the MS1 cDNA with the cDNA for the steroid binding domain of the rat glucocorticoid receptor (GR) (Figure 6A) in the ms1-1 mutant background. The transgenic plant was completely sterile under normal growth conditions; however, a single treatment with the synthetic steroid hormone dexamethasone (DEX) on the top of an inflorescence with a range of floral stages restored several fruitful seedpods (Figure 6B). This result is consistent with the observation that MS1 is expressed transiently in the anther (Ito and Shinozaki, 2002). In analogy with animal steroid receptors, DEX would lead to translocation of the MS1-GR protein from the cytosol to the nucleus, resulting in downstream events.
Figure 6.
Search for Immediate Downstream Genes of MS1 Using the DEX-Inducible ProMS1:MS1-GR/ms1-1 Transgenic Plant.
(A) ProMS1:MS1-GR construct. MS1-GR fusion gene was driven by the MS1 promoter.
(B) A 10-d-later ProMS1:MS1-GR/ms1-1 plant in a single addition of DEX on top of inflorescences. Fertility was recovered as shown by asterisks.
(C) Real-time RT-PCR analysis of 15 genes using RNA samples extracted from Ler (lanes 1 and 2), ms1-1 (lanes 3 and 4), or ProMS1:MS1-GR/ms1-1 (lanes 5 and 6) inflorescences 24 h after mock (lanes 1, 3, and 5) or DEX (lanes 2, 4, and 6) treatment. Arabidopsis Genome Initiative code and The Institute for Genomic Research (TIGR) annotation of each gene are indicated. Analyses of other 16 genes are shown in Supplemental Figure 1 online. Vertical axis indicates relative mRNA amount compared with the amount in lane 1. Parenthesis indicates the induction fold compared with the mock treatment. Expression level in lane 1 is indicated as percentage compared with actin mRNA amount (% act). Bars indicate sd.
(D) Time-course induction analysis of the six genes in (C) using real-time RT-PCR. RNA samples were extracted from ProMS1:MS1-GR/ms1-1 inflorescences 0, 4, 12, or 24 h after mock (squares) or DEX (circles) treatment. Vertical axis indicates relative mRNA amount compared with the amount of the plants 24 h after DEX treatment. Horizontal axis indicates hours after the treatment. Bars indicate sd.
(E) Real-time RT-PCR analysis of the At ERF1, ATL2, and MYB99 genes using the ProMS1:MS1-GR/ms1-1 samples 4 h after simultaneous treatment with DEX and CHX. Vertical axis indicates relative mRNA amount compared with the amount in lane 2. Mock (lane 1), DEX (lane 2), CHX (lane 3), and simultaneous treatment with DEX and CHX (lane 4). Bars indicate sd.
Using this system, we examined whether the selected putative MS1-regulated genes were induced after DEX treatment. We performed real-time RT-PCR analysis using RNA samples after 24-h induction and examined 32 genes out of the 95 in Table 1. Figure 6C and Supplemental Figure 1 online indicated that multiple genes were clearly induced within 24 h after DEX treatment. For example, the At1g61110 gene in Figure 6C was induced 36-fold (lanes 5 and 6), which was a much higher induction than those in positive (lanes 1 and 2) and negative (lanes 3 and 4) control plants, and the expression level was recovered up to that of the wild type. Here, if the induced expression reached the steady state level in the Ler plant within 24 h, we propose that the gene is transcriptionally regulated by MS1 directly or indirectly, whereas other genes are further downstream in a developmental regulatory hierarchy. The induced genes were further examined for their induction kinetics. Only the MYB99 (At5g62320) gene was induced as early as 4 h (Figure 6D). The others were not induced at 4 h after DEX treatment (Figure 6D; data not shown).
To test whether the MYB99 gene is a direct target of MS1, we performed a DEX induction experiment in the presence of a protein synthesis inhibitor, cycloheximide (CHX). To demonstrate that CHX was effective in the inflorescence of the ProMS1:MS1-GR plant, we monitored mRNA accumulation of At ERF1 (At4g17500) and an early elicitor-response gene (ATL2; At3g16720). These mRNAs are unstable (Gutierrez et al., 2002) and CHX inducible (Salinas-Mondragon et al., 1999; Fujimoto et al., 2000) in vegetative tissues, indicating that they can serve as reporters of CHX treatment. In the inflorescences of the ProMS1:MS1-GR plant, these genes were highly elevated after CHX treatment (Figure 6E), similar to previous observations (Salinas-Mondragon et al., 1999; Fujimoto et al., 2000). Therefore, CHX was effective in our system. We found that MYB99 was induced by DEX in the presence of CHX after 4 h (Figure 6E), strongly suggesting that induction of the MYB99 does not require de novo protein synthesis and that this gene is likely a direct target of MS1.
Phylogenetic Analysis of the MS1 Family and Phenotype of ProMS1:MS1-SRDX Transgenic Petunia
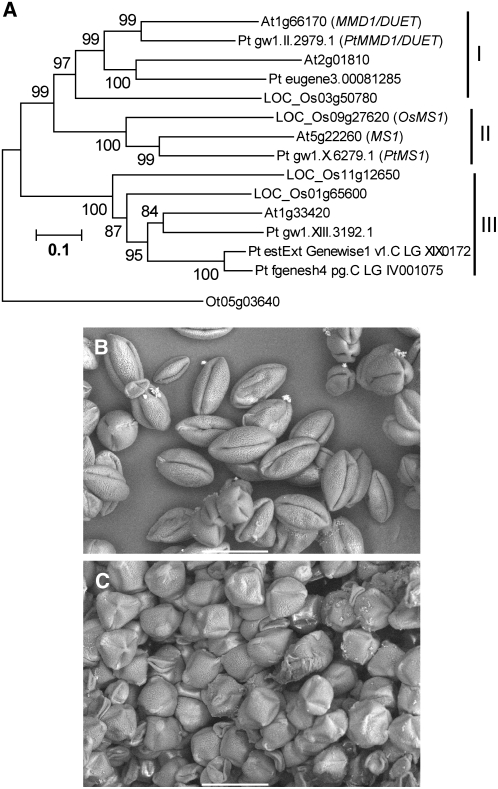
To test whether the regulatory system controlled by MS1 is conserved in other flowering plants, we performed phylogenetic analysis of MS1 and its homologs in Arabidopsis, rice (Oryza sativa), poplar (Populus trichocarpa), and the unicellular green algae Ostreococcus tauri (Prasinophyceae). In Arabidopsis, there are three homologous genes (At1g33420, At1g66170, and At2g01810) to MS1 (At5g22260). At1g66170 is the MALE MEIOCYTE DEATH1 (MMD1)/DUET gene and is thought to be a crucial regulator of meiosis (Reddy et al., 2003; Yang et al., 2003). The other two genes are not fully redundant with MS1 or MMD1/DUET, since ms1 or mmd1/duet single mutation causes severe pollen developmental defects. Rice has four MS1 homologs (LOC_Os01g65600, LOC_Os03g50780, LOC_Os09g27620, and LOC_Os11g12650), and in poplar, six MS1 homologs were detected. In a recently published genome sequence of O. tauri (Derelle et al., 2006), there is a single MS1 homolog (Ot05g03640). We also found other homologs in higher plant species whose genome analyses are underway (data not shown). We could not find any highly similar genes to MS1 in taxa other than Viridiplantae, although recognizably related genes were found in these species (data not shown). The phylogenetic analyses with the full-length sequence (Figure 7A) or with conserved regions (data not shown) yielded trees with very similar topologies. Our result indicates that MS1 (Arabidopsis), Pt MS1 (poplar), and Os MS1 (rice) form a separate clade within the entire family. The phylogenetic tree further suggests that the common ancestor of Arabidopsis, poplar, and rice had at least three PHD-finger genes in the MS1 family.
Figure 7.
Analyses of Conserved MS1 Function in Flowering Plants.
(A) A rooted neighbor-joining tree of Arabidopsis, rice, poplar, and Ostreococcus MS1 homologs. An Ostreococcus homolog was selected as an outgroup. Gene identifier numbers starting with At indicate genes from Arabidopsis; names of genes with functional information are given after the identifier numbers. LOC_Os indicates genes from rice (O. sativa), with names given according to gene identifier from TIGR. Pt indicates genes from poplar (P. trichocarpa), with temporary names given according to gene identifier from the Floral Genome Project. Ot indicates a gene from O. tauri. Os MS1, Pt MS1, and Pt MMD1/DUET after the identifier numbers are names of genes with deduced functional information. Bootstrap values are shown near the relevant nodes.
(B) A scanning electron microscopy image of petunia pollen grains transformed with a control vector pBE2113. Bar = 50 μm.
(C) A scanning electron microscopy image of petunia pollen grains transformed with the ProMS1:MS1-SRDX construct. Bar = 50 μm.
To test whether the MS1 function is conserved in higher plants, ProMS1:MS1-SRDX was introduced into heterologous garden petunia (Petunia hybrida), and pollen phenotype of transgenic plants was observed. Petunia is a member of the Asterids, a large group of eudicots that is distinct to the Rosids group that includes Arabidopsis. Compared with control pollens with rugby ball shape (Figure 7B), two out of 15 ProMS1:MS1-SRDX–independent transformants derived from separate calli exhibited obviously and uniformly mature pollen grains that were crushed along the longer axis (Figure 7C), although we could not obtain any transgenic petunias possessing a significantly reduced number of mature pollen grains, as observed in the transgenic Arabidopsis. The petunia pollen grains with abnormal shape were viable, as shown by the Alexander test (data not shown), suggesting a specific defect in its pollen wall. The crushed pollen phenotype in Figure 7C was similar to that of the transgenic Arabidopsis in Figure 4B. We confirmed the MS1-SRDX gene expression in the transgenic petunias exhibiting abnormal pollen shape using RT-PCR analysis (data not shown). These results indicate that the pollen wall of these transgenic petunias is abnormal and suggest the presence of an MS1-like regulatory system in petunia.
DISCUSSION
MS1 Regulates Pollen and Tapetum Development as a Transcriptional Activator
The MS1 protein contains motifs that suggest that it is a transcriptional regulator. We showed that the putative Leu zipper and PHD-finger domains were both required for the MS1 function, strengthening the idea that MS1 is a transcription factor. Leu zippers are found in bZIP and bHLH-ZIP transcription factors and mediate protein dimerization through a coiled-coil structure (Meshi and Iwabuchi, 1995). Therefore, the putative MS1 Leu zipper may facilitate protein–protein interaction. The PHD domain contains Cys4-His-Cys3 consensus and coordinates two zinc ions (Bienz, 2006). In yeast and animal, PHD-finger is found in histone methyltransferases, histone acetyltransferases, and chromatin binding and DNA binding proteins (Bienz, 2006). In plants, PHD-finger proteins constitute a large family, but their biochemical functions are unknown. Our results suggest that one possibility is that MS1 is a DNA binding transcription factor like the alfalfa (Medicago sativa) Alfin1 protein, which contains a PHD motif and binds to DNA in vitro (Bastola et al., 1998). Another possibility is that MS1 is a non-DNA binding transcription factor and functions via protein–protein interaction similar to some chromatin remodeling factors with PHD-fingers (Li et al., 2006; Pena et al., 2006; Shi et al., 2006; Wysocka et al., 2006).
To test whether MS1 is a positive transcriptional regulator, we used a transcriptional repression domain, SRDX, and showed that the chimeric MS1-SRDX construct conferred dominant ms1-like phenotype. This type of repression domain is found in plant DNA binding repressors, such as ERF and SUP, and was shown previously to cause repression when fused to DNA binding transcriptional activators, such as CUC1 and EIN3 (Hiratsu et al., 2003). Our results strongly support the hypothesis that MS1 is a transcriptional activator. This and our earlier observations that MS1 is localized to the nucleus and that MS1 mRNA was expressed specifically in tapetum and at anther stage 7 (Ito and Shinozaki, 2002) led us to propose that MS1 is important for transcriptional activation for postmeiotic pollen maturation.
Furthermore, the specific detection of the MS1 mRNA in the tapetum, but not in the microspore (Ito and Shinozaki, 2002), and the ms1 pollen defects suggest that the MS1 function in the tapetum is necessary for both microspore and tapetum development. It is plausible that some of the genes whose expression was affected by ms1 mutation are involved in this development. By transcriptome analysis, we have identified three putative protease genes as probable downstream targets of MS1 (Table 1). The DEX experiment indicated that an aspartyl protease gene (At2g03200) was fully induced after 24 h (Figure 6C). Since proteolytic enzymes are known to be associated with plant developmental events, such as biotic and abiotic stress responses, senescence, and programmed cell death (Beers et al., 2000; Simoes and Faro, 2004), this gene may play important roles in microspore and tapetum development.
We also observed that the ms1 mutant and the ProMS1:MS1-SRDX transgenic plants did not develop secretory organelle, elaioplast, and tapetosome in tapetum. This is in agreement with the results from a parallel study by Yang et al. (2007). Their microarray data (Yang et al., 2007) indicated that four genes relating to putative prenyltransferase were downstream of MS1 (Table 1). Three of the four genes were examined for DEX inducibility (Figure 6C), and two of them (At5g60500 and At5g60510), which are 92% identical at the nucleotide level, were induced after 24 h to a level close to that in the wild type. The predicted proteins of these two genes show 35% overall identity to a Saccharomyces cerevisiae cis-prenyltransferase, a key enzyme for the synthesis of dolichol, which is essential for protein glycosylation (Sato et al., 1999). A yeast mutation in this gene causes mislocalization of different types of endoplasmic reticulum membrane proteins (Sato et al., 1999), suggesting that the two Arabidopsis homologs may be involved in localization of membrane proteins in the tapetum after meiosis.
Putative Sporopollenin Biosynthetic Genes Regulated by MS1
We showed here that the ProMS1:MS1-SRDX transgenic plants and the ms1 mutant exhibit defects in exine formation and organization, suggesting that MS1 regulates exine development by organizing materials such as sporopollenin correctly on the microspore surface. Nevertheless, the finding that the abnormal sporopollenin structure was anchored near the microspore surface suggests that MS1 does not control the formation of a scaffold known as primexine, in agreement with that of Vizcay-Barrena and Wilson (2006), but not with the finding by Ariizumi et al. (2005) that sporopollenin in their ms1 allele (HKM/ms1-9) was randomly deposited onto the microspore plasma membrane. Therefore, it is unclear whether MS1 is needed for primexine formation on the microspore surface before exine elaboration.
Since phenolic compounds and lipids are important components for sporopollenin (Piffanelli et al., 1998), putative genes for phenylpropanoid and lipid syntheses in Table 1 may be involved in sporopollenin biosynthesis. Because the ms1 mutant anther formed lignin structure similar to that of wild-type anther (see Supplemental Figure 2 online), the genes affected by ms1 are likely important for the synthesis of lignin-like compounds, but not lignin. Therefore, we hypothesize that these putative phenylpropanoid synthetic gene products can catalyze novel reactions leading to sporopollenin monomers, such as p-coumaric acid and ferulic acid (Blokker et al., 2005). In addition, we have identified three groups of putative MS1 downstream genes with putative lipid biosynthetic functions (Table 1). The first group is homologous to a very-long-chain fatty acid elongase condensing enzyme, β-ketoacyl-CoA synthase (Lassner et al., 1996). The second (At1g13140, At1g13150, and At3g26125) are phylogenetically related to genes for fatty acid hydroxylases CYP86-A1 and A8 that catalyze ω-hydroxylation of fatty acids (Benveniste et al., 1998; Wellesen et al., 2001). The last group code for putative lipid transfer proteins.
Among the genes affected by the ms1 mutations, six genes (At1g13140, At1g74540, At2g19070, At3g51590, At3g52160, and At4g28395) were examined for their DEX inducibility and found be expressed up to wild-type mRNA levels after 24 h (Figures 6C and 6D). At4g28395 and At3g51590 are expressed at the free and uninucleate microspore stages, respectively (Rubinelli et al., 1998; Ariizumi et al., 2002), equivalent to anther stages 8 to 9 in which microspores generate an exine wall (Sanders et al., 1999). Independently, microarray data shows that the 12 genes for phenylpropanoid and lipid syntheses except At1g76470 in Table 1 are expressed specifically around stage 10/11 flower (Arabidopsis eFP Browser at http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). These findings suggest that MS1 directly or indirectly controls part of phenylpropanoid and fatty acid biosynthetic pathways leading to sporopollenin biosynthesis.
Most of the pollen coat proteins were isolated as downstream genes of MS1 (Table 1), but the glucocorticoid induction experiment indicated that genes encoding caleosin-related (At1g23250) and GRP17 (At5g07530) were not expressed, and a gene for EXL5 (At1g75920) was not expressed at a comparable level with the wild type after 24 h (Figure 6C). From these results, we conclude that these pollen coat protein genes are not transcriptionally regulated by MS1 but are further downstream in a developmental regulatory hierarchy. This conclusion is consistent with the observation that there is a temporal gap between the MS1 expression at tetrad stage (Ito and Shinozaki, 2002) and pollen coat formation at bicellular stage (Piffanelli et al., 1998).
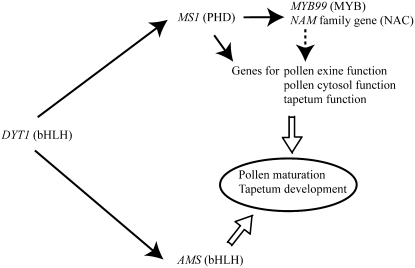
Genetic Control of Pollen and Tapetum Development after Meiosis
This and previous studies support a model for the genetic control of pollen maturation and tapetum development (Figure 8). Since MS1 and AMS1 genes are not expressed in the dysfunctional tapetum1 (dyt1) mutant, these genes are likely located downstream of DYT1, which is strongly expressed in tapetum at late stage 5 (Zhang et al., 2006). Although in situ mRNA hybridization analysis did not give a clear result, the AMS1 gene appears to function at similar stage as MS1, judging from the ams1 mutant phenotype (Sorensen et al., 2003). However, AMS1 does not seem to regulate exine formation since the ams1 mutant underwent initial exine deposition (Sorensen et al., 2003). These findings suggest that MS1 and AMS1 genes encoding transcription factors control different developmental/cellular processes at a similar stage.
Figure 8.
A Model for MS1 Function and Pollen and Tapetum Development after Meiosis.
The solid arrows indicate a positive genetic regulation. The dashed arrow indicates a putative genetic regulation. The open arrows represent a positive effect on the cellular process.
Two transcription factor genes encoding NAM family and MYB99 (At1g61110 and At5g62320, respectively) were found to be immediate downstream of MS1 (Figure 6C), but induction kinetics by DEX treatment was different (Figure 6D). The analysis using protein synthesis inhibitor suggested that the MYB99 gene was a direct target of MS1 (Figure 6E). These regulatory genes may regulate further downstream genes involved in pollen maturation and tapetum development. One current model is that MS1 directly regulates MYB99, and MYB99 regulates the other DEX-induced genes. Presence of consensus MYB binding site (CNGTTR) in promoter regions of the DEX-induced genes (data not shown) supports this model. MYB99 is also proposed to be downstream of MS1 by Yang et al. (2007).
Is the Regulation System by MS1 Conserved in Plant Species?
In Arabidopsis, MS1 plays a crucial role in pollen and tapetum development. Recent plant genome analyses revealed that there are multiple MS1 homologs in higher plant species. On the other hand, unicellular green algae O. tauri has a single MS1 homolog. O. tauri is a member of a lineage of green alga that is sister to a major clade that includes most other green algae and the land plants (Derelle et al., 2006). There are no MS1 homologs in species other than Viridiplantae (data not shown). These findings are consistent with the hypothesis that MS1 was generated by domain shuffling at or near the time of the endosymbiosis event. Phylogenetic analysis (Figure 7A) indicated that there have been two duplication events, one resulting in two clades, I/II and III, and the other producing clades I and II. Clade II contains MS1 and is distinct from other clades, supporting the genes in this clade as MS1 orthologs. The phenotype of the transgenic petunia (Figure 7C) indicated that the Arabidopsis MS1 can interfere with the putative petunia MS1 ortholog, suggesting that the biochemical activities of the two proteins are similar. It is plausible to propose that MS1 orthologs have conserved functions and similar cis regulatory elements in flowering plants.
METHODS
Plant Growth Conditions and Transformation Procedure
Arabidopsis thaliana was grown on soil at 22 to 24°C under a 16-h-light/8-h-dark cycle. Transformation was conducted by the Agrobacterium tumefaciens–mediated infiltration method (Clough and Bent, 1998). Petunia (Petunia hybrida cv Mitchel) was grown on soil at 25°C under a 16-h-light/8-h-dark cycle. Agrobacterium-mediated transformation was conducted as described by Yamada et al. (2005).
Domain Analysis of the MS1 Protein
For Pro35S:MS1 construction, MS1 cDNA was inserted into a SmaI site of the pBE2113 cassette (Mitsuhara et al., 1996). For ProMS1:MS1 construction, a genomic DNA fragment from −993 to −1 (the A residue of the MS1 translational start codon as +1) was inserted into HindIII-BamHI sites of pBI101.1 (Jefferson, 1987), and the β-glucuronidase gene was deleted from pBI101.1. This resulted in the pMS1P cassette. MS1 cDNA was inserted into a SmaI site of the pMS1P. For introduction of an internal deletion or a point mutation into MS1 cDNA, we performed a site-directed mutagenesis procedure.
To determine the MS1/MS1, MS1/ms1-1, and ms1-1/ms1-1 genotypes, we used cleaved amplified polymorphic sequence analysis. A PCR fragment was amplified with a primer set, 5′-GAGGATTGGAGTTTCTCGGTTAAG-3′ and 5′-ATGGTTTATACGGAACCTTGCAGG-3′, and was digested with an RsaI restriction enzyme, which recognizes the GTAC sequence. The latter primer is derived from the first intron sequence, and the primer set does not amplify introduced MS1 cDNA sequence. The 632-bp single fragment is ms1 specific and digested two fragments (252 and 380 bp) are MS1 specific.
Functional Analysis Using the Repression Domain
p35S-SRDXG containing the CaMV 35S promoter, TMV omega sequence, SRDX-coding sequence, and NOS terminator (Hiratsu et al., 2003) was digested with BamHI and EcoRI. The BamHI-EcoRI fragment containing TMV omega–SRDX-coding sequence–NOS terminator was inserted into BamHI-EcoRI sites of the pMS1P cassette, and this resulted in pMS1PSRDX. MS1 cDNA from the start ATG codon to the 672nd CCC codon without a stop codon was PCR amplified and inserted in frame into the SmaI site of pMS1PSRDX, and this resulted in pMS1PMS1SRDX (ProMS1:MS1-SRDX construct) shown in Figure 1A. This construct was transformed into Arabidopsis (Col-0 ecotype) and petunia.
Light and Electron Microscopy
Samples were fixed overnight at room temperature with FAA, which contained 1% (v/v) formaldehyde, 2.5% (v/v) acetic acid, and 45% (v/v) ethanol, and embedded in Technovit glycol methacrylate 7100 resin (Heraeus Kulzer). Sections (4 μm thick) were stained with 0.1% (w/v) toluidine blue O in 0.1 M phosphate buffer, pH 7.0. Specimens were photographed under bright-field illumination. Anthers and pollen grains of Arabidopsis and petunia were directly observed with a scanning electron microscope (JSM-5610LV; JEOL DATUM) at an accelerating voltage of 10 to 20 kV in a low vacuum mode. TEM analysis was performed as described by Motohashi et al. (2003).
The Glucocorticoid Induction System
We used a rat GR steroid binding domain (amino acids 519 to 795) as described by Aoyama et al. (1995). An in-frame MS1-GR fragment was inserted into a SmaI site of pMS1P cassette, and we obtained pMS1PMS1GR (ProMS1:MS1-GR construct) shown in Figure 6A. This construct was transformed into MS1/ms1-1 heterozygous plants. Both T-DNA and ms1-1 homozygous siblings (ProMS1:MS1-GR/ms1-1 plants) were used in the experiments. For DEX treatment, we applied 5 μM DEX in 0.015% (v/v) Silwet L-77 to inflorescences using a paintbrush. For both DEX and CHX treatment, 5 μM DEX and 10 μM CHX in 0.015% (v/v) Silwet L-77 was added. Stock solutions of 5 mM DEX in ethanol and 10 mM CHX in water were prepared before use. The same volume of ethanol was added to a negative control (mock) medium.
RNA Extraction, Reverse Transcription, and Real-Time PCR
Inflorescences of the Ler, ms1-1, and ProMS1:MS1-GR/ms1-1 plants were treated with mock or DEX solution, and the inflorescences were collected after 0, 4, 12, or 24 h as shown in Figure 6. In the experiment using protein synthesis inhibitor, the ProMS1:MS1-GR/ms1-1 plant was treated with mock, DEX, CHX, or DEX/CHX solution, and the inflorescences were collected after 4 h. RNA samples were extracted from the inflorescences containing stages 1 to 12 floral buds using the RNeasy plant mini kit (Qiagen). One microgram of total RNA was treated with RNase-free DNase I (Invitrogen) followed by inactivation of the DNase I. Reverse transcription was preformed using Supertranscriptase II (Invitrogen), and synthesized first-strand cDNA was used as a template for PCR.
Real-time PCR was performed using Brilliant SYBR Green QPCR Master Mix (Stratagene) and the ABI 7300 platform (Applied Biosystems). Each sample was analyzed in triplicate. The data were normalized using a reference actin transcript, and relative mRNA amount was calculated by comparative Ct method. ΔCt values of each experiment are shown in Supplemental Table 2 online. Primer sets were designed using Primer Express 2.0 software (Applied Biosystems). Primer sets used in this assay are indicated in Supplemental Table 3 online. The length of the PCR products is 151 bp. The PCR program is 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 57°C for 1 min, and 72°C for 1 min.
Phylogenetic Analysis of the MS1 Family
First, protein sequence of the MS1 was used to search for MS1 homologs in available genome sequences of all organisms (http://www.ncbi.nlm.nih.gov/BLAST/) and in the plant transcript database (http://plantta.tigr.org/) using both BLASTP and TBLASTN programs. Next, the same search was performed in the Arabidopsis (http://www.tigr.org/tdb/e2k1/ath1/), rice (Oryza sativa; http://www.tigr.org/tdb/e2k1/osa1/), poplar (Populus trichocarpa; http://www.floralgenome.org/tribe.php?action=blast), and Ostreococcus genomes (http://bioinformatics.psb.ugent.be/genomes/ostreococcus_tauri/) with a cutoff of 1e-50 (Arabidopsis, rice, and poplar) or 1e-30 (Ostreococcus). The multiple sequence alignment of protein sequence was performed using ClustalX (Plate-Forme de Bio-Informatique, Illkirch Cedex, France). The Pairwise alignment parameter was changed to GOP = 4.0 and GEP = 0.1, and multiple alignment parameter to GOP = 15.0 and GEP = 0.3. The whole regions were aligned (see Supplemental Figure 3 online) and used to perform neighbor joining analyses with the pairwise deletion option, Poisson correction model, and 1000 bootstrap replicates test using MEGA version 3.1 (Kumar et al., 2004; http://www.megasoftware.net/index.html). The single sequence of Ostreococcus was treated as an outgroup to the angiosperm sequences.
Microscopy Analysis of Anther Lignification
Whole inflorescences were fixed and bleached with ethanol overnight. For observation of lignin autofluorescence, the fixed inflorescences were cleared with chloral solution (5 g chloral hydrate, 0.5 g glycerol, and 1.25 mL water), and stage 13 anthers were photographed under UV illumination. For observation of lignin deposition by phloroglucinol, the fixed flowers were stained with 2% (w/v) phloroglucinol in 95% ethanol for 2 min. After washing with 5 n HCl, the flowers were photographed under bright-field illumination.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Real-Time RT-PCR Analysis of 16 Genes Other Than Those Shown in Figure 6C.
Supplemental Figure 2. Microscopy Analysis of Anther Lignification.
Supplemental Figure 3. Multiple Sequence Alignment of MS1 Homologs.
Supplemental Table 1. Complete List of Genes Whose Expression Was Affected by the ms1 Mutation, from the Microarray Data of Yang et al. (2007).
Supplemental Table 2. ΔCt Values in Figure 6 and Supplemental Figure 1.
Supplemental Table 3. Primers Used in the Real-Time PCR Experiment.
Supplemental Data Set 1. Text File of Multiple Sequence Alignment of MS1 Homologs (in ALN Format).
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Center and Z.A. Wilson for their microarray expression data and for sharing results before publication and TIGR and Affymetrix for information about gene annotations on the microarray. We also thank Y. Sun for his help with phylogenetic analysis, C.L.H. Hord for correcting English, and H. Kobayashi, S. Mizukado, D. Grobe, A. Omeis, M. Ooya, and Y. Ueda for their technical assistance. This work was supported in part by a grant for Genome Research from RIKEN (to K.S.), a grant from the U.S. Department of Energy (to H.M.), Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to T.I.) and from the Ministry of Education, Culture, Sports, Science, and Technology (to N.N.), a grant for Core Research for Evolutional Science and Technology from Japan Science and Technology (to M.O.-T. and K.S.), and a project grant of the Bio-oriented Technology Research Advancement Institution (to Y.Y.). T.I. is a recipient of a Research Fellowship from The Naito Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kazuo Shinozaki (sinozaki@rtc.riken.jp).
Online version contains Web-only data.
References
- Aarts, M.G.M., Hodge, R., Kalantidis, K., Florack, D., Wilson, Z.A., Mulligan, B., Stiekema, W.J., Scott, R., and Pereira, A. (1997). The Arabidopsis MALE STERILITY2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12 615–623. [DOI] [PubMed] [Google Scholar]
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., Dong, C.H., Wu, Y., Carabelli, M., Sessa, G., Ruberti, I., Morelli, G., and Chua, N.H. (1995). Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell 7 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi, T., Amagai, M., Shibata, D., Hatakeyama, K., Watanabe, M., and Toriyama, K. (2002). Comparative study of promoter activity of three anther-specific genes encoding lipid transfer protein, xyloglucan endotransglucosylase/hydrolase and polygalacturonase in transgenic Arabidopsis thaliana. Plant Cell Rep. 21 90–96. [Google Scholar]
- Ariizumi, T., Hatakeyama, K., Hinata, K., Inatsugi, R., Nishida, I., Sato, S., Kato, T., Tabata, S., and Toriyama, K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39 170–181. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., Hatakeyama, K., Hinata, K., Sato, S., Kato, T., Tabata, S., and Toriyama, K. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53 107–116. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., Hatakeyama, K., Hinata, K., Sato, S., Kato, T., Tabata, S., and Toriyama, K. (2005). The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sex. Plant Reprod. 18 1–7. [Google Scholar]
- Bastola, D.R., Pethe, V.V., and Winicov, I. (1998). Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol. 38 1123–1135. [DOI] [PubMed] [Google Scholar]
- Beers, E.P., Woffenden, B.J., and Zhao, C. (2000). Plant proteolytic enzymes: Possible roles during programmed cell death. Plant Mol. Biol. 44 399–415. [DOI] [PubMed] [Google Scholar]
- Benveniste, I., Tijet, N., Adas, F., Philipps, G., Salaun, J.P., and Durst, F. (1998). CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem. Biophys. Res. Commun. 243 688–693. [DOI] [PubMed] [Google Scholar]
- Bienz, M. (2006). The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31 35–40. [DOI] [PubMed] [Google Scholar]
- Blokker, P., Yeloff, D., Boelen, P., Broekman, R.A., and Rozema, J. (2005). Development of a proxy for past surface UV-B irradiation: A thermally assisted hydrolysis and methylation py-GC/MS method for the analysis of pollen and spores. Anal. Chem. 77 6026–6031. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dawson, J., Wilson, Z.A., Aarts, M.G.M., Braithwaite, A.F., Briarty, L.G., and Mulligan, B.J. (1993). Microspore and pollen development in six male-sterile mutants of Arabidopsis thaliana. Can. J. Bot. 71 629–638. [Google Scholar]
- Derelle, E., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 103 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, R.A., Ewing, R.M., Cherry, J.M., and Green, P.J. (2002). Identification of unstable tanscripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginson, T., Li, S.F., and Parish, R.W. (2003). AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 35 177–192. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Ohta, M., Matsui, K., and Ohme-Takagi, M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514 351–354. [DOI] [PubMed] [Google Scholar]
- Ito, T., and Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43 1285–1292. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lassner, M.W., Lardizabal, K., and Metz, J.G. (1996). A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Ilin, S., Wang, W., Duncan, E.M., Wysocka, J., Allis, C.D., and Patel, D.J. (2006). Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A.M., Schena, M., Walbot, V., and Davis, R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266 436–439. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., Fiebig, A., Johnstone, S.E., and Preuss, D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292 2482–2485. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., and Preuss, D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2 128–130. [DOI] [PubMed] [Google Scholar]
- Meshi, T., and Iwabuchi, M. (1995). Plant transcription factors. Plant Cell Physiol. 36 1405–1420. [PubMed] [Google Scholar]
- Mitsuhara, I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37 49–59. [DOI] [PubMed] [Google Scholar]
- Motohashi, R., Ito, T., Kobayashi, M., Taji, T., Nagata, N., Asami, T., Yoshida, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34 719–731. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders, D.M., Dodrill, C.H., Owen, H.A., and Makaroff, C.A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127 1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Pena, P.V., Davrazou, F., Shi, X., Walter, K.L., Verkhusha, V.V., Gozani, O., Zhao, R., and Kutateladze, T.G. (2006). Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P., Ross, J.H.E., and Murphy, D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11 65–80. [Google Scholar]
- Reddy, T.V., Kaur, J., Agashe, B., Sundaresan, V., and Siddiqi, I. (2003). The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130 5975–5987. [DOI] [PubMed] [Google Scholar]
- Rubinelli, P., Hu, Y., and Ma, H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37 607–619. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92 93–103. [DOI] [PubMed] [Google Scholar]
- Salinas-Mondragon, R.E., Garciduenas-Pina, C., and Guzman, P. (1999). Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol. Biol. 40 579–590. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11 297–322. [Google Scholar]
- Sato, M., Sato, K., Nishikawa, S., Hirano, A., Kato, J., and Nakano, A. (1999). The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., et al. (2006). ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes, I., and Faro, C. (2004). Structure and function of plant aspartic proteinases. Eur. J. Biochem. 271 2067–2075. [DOI] [PubMed] [Google Scholar]
- Sorensen, A.M., Krober, S., Unte, U.S., Huijser, P., Dekker, K., and Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33 413–423. [DOI] [PubMed] [Google Scholar]
- Vizcay-Barrena, G., and Wilson, Z.A. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57 2709–2717. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Sablowski, R.W.M., and Meyerowitz, E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584. [DOI] [PubMed] [Google Scholar]
- Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., Steiner-Lange, S., Saedler, H., and Yephremov, A. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc. Natl. Acad. Sci. USA 98 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28 27–39. [DOI] [PubMed] [Google Scholar]
- Wysocka, J., Swigut, T., Xiao, H., Milne, T.A., Kwon, S.Y., Landry, J., Kauer, M., Tackett, A.J., Chait, B.T., Badenhorst, P., Wu, C., and Allis, C.D. (2006). A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodeling. Nature 442 86–90. [DOI] [PubMed] [Google Scholar]
- Yamada, M., Morishita, H., Urano, K., Shiozaki, N., Yamaguchi-Shinozaki, K., Shinozaki, K., and Yoshiba, Y. (2005). Effects of free proline accumulation in petunias under drought stress. J. Exp. Bot. 56 1975–1981. [DOI] [PubMed] [Google Scholar]
- Yang, C., Vizcay-Barrena, G., Conner, K., and Wilson, Z.A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19 3530–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Makaroff, C.A., and Ma, H. (2003). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Sun, Y., Timofejeva, L., Chen, C., Grossniklaus, U., and Ma, H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133 3085–3095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.