Abstract
Rab5, a subfamily of Rab GTPases, regulates a variety of endosomal functions as a molecular switch. Arabidopsis thaliana has two different types of Rab5-member GTPases: conventional type, ARA7 and RHA1, and a plant-specific type, ARA6. We found that only one guanine nucleotide exchange factor (GEF), named VPS9a, can activate all Rab5 members to GTP-bound forms in vitro in spite of their diverged structures. In the vps9a-1 mutant, whose GEF activity is completely lost, embryogenesis was arrested at the torpedo stage. Green fluorescent protein (GFP)–ARA7 and ARA6-GFP were diffused in cytosol like GDP-fixed mutants of Rab5 in vps9a-1, indicating that both types of GTPase are regulated by VPS9a. In the leaky vps9a-2 mutant, elongation of the primary root was severely affected. Overexpression of the GTP-fixed form of ARA7 suppressed the vps9a-2 mutation, but overexpression of ARA6 had no apparent effects. These results indicate that the two types of plant Rab5 members are functionally differentiated, even though they are regulated by the same activator, VPS9a.
INTRODUCTION
Recent studies reveal that endocytosis is important for a variety of plant functions, including the establishment of cell polarity, polar transport of auxin, and cytokinesis, by regulating the localization and activity of plasma membrane proteins and extracellular components (Geldner et al., 2003; Russinova et al., 2004; Baluska et al., 2005; Dhonukshe et al., 2006; Robatzek et al., 2006). However, the underlying molecular mechanisms and the significance in plant development still remain largely unknown. To address these problems, we have been focusing our attention on Rab GTPases, one of the key regulators of vesicular traffic.
Rab GTPases are localized on organelle membranes in a specific manner and act as molecular switches by cycling between the GDP-bound and the GTP-bound states. Rab5, a member of Rab GTPases localizing on endosomes, has been shown in the mammalian system to organize many processes in the endocytic pathway, such as homotypic fusion between early endosomes, alteration of lipid composition of the endosomal membrane, and signal transduction through endosomes via specific interactions with effector proteins (Grosshans et al., 2006). To properly execute its wide spectrum of tasks, the GTPase cycle must be strictly controlled spatiotemporally, and several categories of regulator molecules have been identified to date (Zerial and McBride, 2001). Activation of Rab is mediated by the guanine nucleotide exchange factor (GEF), which stimulates the release of GDP, allowing GTP to bind. Inactivation of Rab through GTP hydrolysis is accelerated by the GTPase-activating protein. The inactivated (GDP-bound) Rab is detached from membranes and maintained in the cytosol by the Rab GDP dissociation inhibitor (Rab GDI) until the next round of the GTPase cycle begins. Among these regulatory processes, GEF is critical in determining when and where Rab is activated to induce downstream reactions.
In mammalian cells, the activation of Rab5 is regulated by several distinct GEF proteins: Rabex-5, RIN, Rme-6, ALS2, and ANKRD27. These Rab5 GEFs function at different steps in the endocytic pathway, which enables elaborate regulation of Rab5 activity (Carney et al., 2005). All of the Rab5 GEFs identified to date contain a highly conserved catalytic domain, the Vps9 domain, which was originally identified in yeast Vps9p (Burd et al., 1996; Hama et al., 1999).
In plants, a unique Rab5 system seems to be employed for the regulation of the endocytic pathway. The model plant Arabidopsis thaliana has three Rab5 members: ARA7, RHA1, and ARA6 (Ueda et al., 2001, 2004). ARA7 and RHA1 are regarded as the orthologs of mammalian Rab5 because they share very high similarity in structure. On the other hand, ARA6 is a plant-specific type of Rab5 member, which is well conserved among higher plants but not in other organisms. The most striking feature in its structure is that ARA6 lacks the conserved hypervariable region followed by the Cys motif at the C terminus, which is required for isoprenylation. This region is essential for membrane binding and other functions in conventional Rab GTPases. Instead, ARA6 harbors an N-terminal amino acid extension, wherein this protein is fatty-acylated to be anchored to membranes (Ueda et al., 2001). The sequence of the switch I region, which is known to be essential for interactions with some effectors, is also distinct between ARA6 and other conventional-type Rab5 members (see Supplemental Figure 4 online).
These two types of Rab5 members are localized on different populations of membrane-bound compartments with partial overlap, both of which should be regarded as endocytic organelles (endosomes collectively) because they are both stained by an endocytic tracer, FM4-64 (Ueda et al., 2001, 2004). ARA7 and RHA1 are mainly localized on the endosomes whose morphology is dependent on GNOM function in protoplasts (Geldner et al., 2003), whereas ARA6-residing endosomes are not affected by the gnom mutation. The endosomal property of ARA7-residing endosomes has also been confirmed by a study using a boron transporter, BOR1. Upon elevation of the boron concentration in the medium, the internalization of BOR1 from the plasma membrane is induced and the endocytosed BOR1 passes through the ARA7-positive compartment en route to the vacuole (Takano et al., 2005). On the other hand, ultrastructural analysis by immunoelectron microscopy recently demonstrated that both types of Rab5s are localized on multivesicular bodies (Haas et al., 2007). This observation strongly suggests that at least some portion of the Rab5-residing endosomes in Arabidopsis plants correspond to the organelles identified previously as the prevacuolar compartment. Colocalization studies by transient expression in Arabidopsis protoplasts and the heterologous expression approach using tobacco (Nicotiana tabacum) leaves are also consistent with the idea that these three Rab5 members are located on the prevacuolar compartment (Bolte et al., 2004; Kotzer et al., 2004; Lee et al., 2004; Ueda et al., 2004).
The precise functions of these Rab5 members in membrane traffic are still unclear at present, but several lines of evidence suggest that the conventional Rab5 members function both in the vacuolar transport pathway and in the endocytic pathway. The overexpression of dominant negative mutants of ARA7 and RHA1 perturbs the transport of soluble vacuolar proteins to the vacuole in tobacco leaf cells and Arabidopsis protoplasts, respectively (Sohn et al., 2003; Kotzer et al., 2004). On the other hand, dominant negative ARA7 has also been demonstrated to inhibit the internalization of FM4-64 into Arabidopsis root cells and tobacco BY-2 cells (Dhonukshe et al., 2006). Therefore, ARA7 and RHA1 may play roles at the point at which the endocytic and biosynthetic pathways are intimately associated or merged. For the ARA6-type Rab5 members, there have been inconsistent reports on their involvement in the vacuolar transport pathway. Whereas the overexpression of dominant negative ARA6 did not affect the transport of sporamin-GFP (for green fluorescent protein) to the vacuole (Sohn et al., 2003), dominant negative m-Rabmc, a close homolog of ARA6 in Mesembryanthemum crystallinum, had an inhibitory effect on the traffic of aleurain-GFP to the vacuole (Bolte et al., 2004). Since these results were obtained only by transient expression in protoplasts, it is obvious that further studies under more physiological conditions are needed to clarify the functions of these molecules.
In this study, we examined how the activation process of these structurally different Rab5 members is regulated. We have revealed that both types of Rab5 members in Arabidopsis, which are functionally differentiated, are activated by a single Rab5 GEF, VPS9a. We have also demonstrated that the activity of VPS9a is essential for various plant functions, such as cytokinesis, embryogenesis, and organ development.
RESULTS
Identification and Cloning of At VPS9a
Arabidopsis has two genes that encode proteins containing the Vps9 domain, VPS9a and VPS9b, in its genome (Figure 1A) (Vernoud et al., 2003). VPS9a is expressed in all tissues we examined by immunoblotting using an anti-VPS9a antibody (Figure 1B). This is also supported by published microarray data (Expression Atlas of Arabidopsis Development) (Schmid et al., 2005). We also found that mutations in VPS9a severely affect various stages of plant development, as will be described later. On the other hand, we have been unable to detect any expression of VPS9b by RT-PCR. There are no expression data such as EST clones deposited in the database, as far as we could determine. Furthermore, our analysis of T-DNA insertion mutants of VPS9b revealed no obvious phenotype (data not shown). These results strongly suggest that VPS9a functions predominantly throughout Arabidopsis development and that VPS9b has very limited roles, if any.
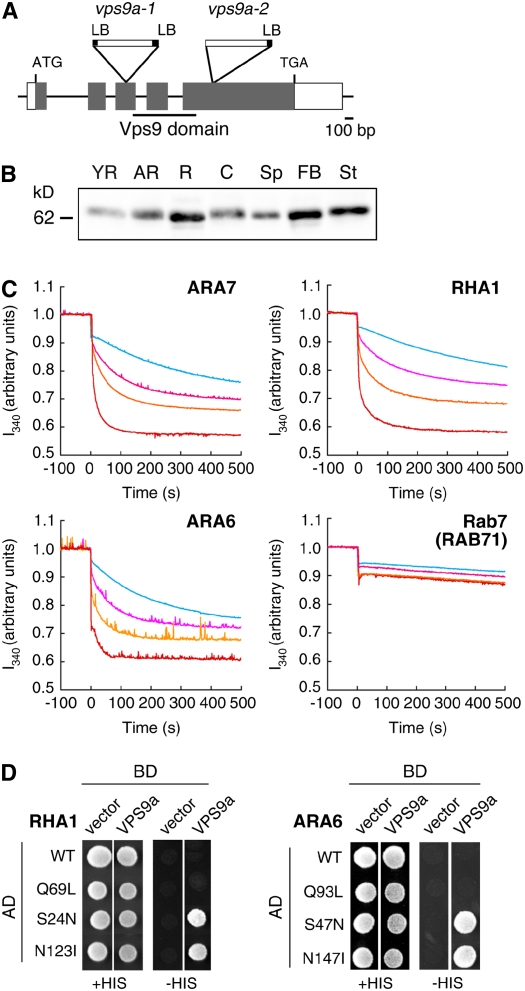
Figure 1.
Structure of the VPS9a Gene and in Vitro GEF Assay.
(A) Schematic structure of the VPS9a gene and positions of T-DNA insertions. The region encoding the Vps9 domain is underlined. LB, left border.
(B) Expression pattern of VPS9a in Arabidopsis organs. The protein samples prepared from young rosettes (YR), adult rosettes (AR), roots (R), cotyledons (C), seed pods (Sp), flower bud clusters (FB), and stems (St) were subjected to immunoblotting with an anti-VPS9a antibody.
(C) In vitro GEF assay of VPS9a. The conformational changes of Rab5s (ARA7, RHA1, and ARA6) and Rab7 (RAB71) upon GDP/GMP-PNP exchange were measured by monitoring Trp autofluorescence in the absence (light blue) or presence of 0.25 μM (pink), 0.5 μM (orange), or 1 μM (red) GST-VPS9a.
(D) Interactions between VPS9a and Rab5 members. VPS9a was expressed as a fusion protein with a DNA binding domain (BD), and Rab5s were expressed as fusions with a transcriptional activation domain (AD). Interactions between two proteins were tested using the HIS3 reporter gene.
VPS9a encodes a protein of 521 amino acid residues (VPS9a). Similarity analysis revealed that VPS9a and other Rab5 GEFs share similarity only within the Vps9 domain at the N-terminal half of the protein (Figure 1A). We did not find any known motif or similarity to other proteins for the C-terminal half of VPS9a.
VPS9a Activates All Arabidopsis Rab5 Members in Vitro
As there are two different types of Rab5s in Arabidopsis, it is an interesting question to ask whether VPS9a with the conserved Vps9 domain could activate all of these Rab5 members. To answer this question, we measured the GEF activity of VPS9a toward the three Rab5 members of Arabidopsis by monitoring the change in intrinsic Trp fluorescence upon nucleotide exchange. As we predicted, VPS9a stimulated the nucleotide exchange of conventional-type Rab5s (ARA7 and RHA1) in a dose-dependent manner (Figure 1C). Interestingly, VPS9a also activated the plant-specific ARA6 as efficiently as ARA7 and RHA1. These GEF activities are highly specific to Rab5 members: VPS9a showed no activity at all to other subclasses of Arabidopsis Rab GTPases (Figure 1C; see Supplemental Figure 1 online).
These specific interactions between VPS9a and Rab5 members were confirmed by the yeast two-hybrid assay and the in vitro pull-down assay. In these experiments, we examined the nucleotide dependence of the interactions. When wild-type and mutant versions of RHA1 were used as baits in the yeast two-hybrid assay, VPS9a showed positive interaction only with the GDP-fixed (S24N) and the nucleotide-free (N123I) forms but not with the wild-type and GTP-fixed (Q69L) forms (Figure 1D). ARA6 gave similar results. VPS9a interacted with GDP-fixed (S47N) and nucleotide-free (N147I) forms but not with wild-type and GTP-fixed (Q93L) forms of ARA6 (Figure 1D). The interactions of VPS9a with ARA7 and RHA1 were also detected in the pull-down assay (see Supplemental Figure 2 online). It should be noted here that wild-type ARA7 and RHA1 could form complexes with VPS9a in the pull-down assay, which may appear to contradict the results of the yeast two-hybrid assay. We assume that this is due to the difference of the nucleotide environment between these experimental systems; most purified Rab5s fused with glutathione S-transferase (GST) are presumably in the GDP-bound state, because they hydrolyze bound GTP to GDP during the purification procedure (Christoforidis and Zerial, 2000), while a considerable amount of wild-type Rab5s should be in the GTP-bound form, because of the high GTP:GDP ratio in yeast cells. We could not detect a stable interaction between VPS9a and ARA6 in the pull-down assay. This result may reflect the more transient association of these molecules. No physical interaction was observed for other subclasses of Rab GTPases, either in the yeast two-hybrid or the pull-down assay (see Supplemental Figure 2 online). From these data, we concluded that VPS9a acts as a specific GEF for all three members of the Arabidopsis Rab5 group in vitro.
Loss of VPS9a Function Causes Embryonic Lethality
Arabidopsis mutant lines, in which one of ARA7, RHA1, or ARA6 is knocked out, develop normally and do not show any significant phenotypes (K. Ebine and T. Ueda, unpublished data). In order to understand the importance of Rab5 activation in plant development, we characterized two mutant alleles of VPS9a, vps9a-1 and vps9a-2 (Figure 1A). The vps9a-1 mutant contains a T-DNA insertion in the third exon, which leads to the loss of the Vps9 domain. Thus, this mutant allele should have lost its GEF activity completely. PCR-based genotyping of the progeny derived from self-pollinated heterozygous mutant plants revealed that the homozygous vps9a-1 plant is inviable, whereas the heterozygous mutants were morphologically indistinguishable from wild-type plants. In the seed pods of the heterozygous mutant, ∼25% of seeds (normal:abnormal = 1010:329) were aborted, with small, brownish, and shriveled appearance (Figure 2A). These abnormal seeds contained small, rudimentary embryos whose development was arrested at the torpedo stage (Figure 2K). A genomic fragment containing the entire VPS9a gene rescued this embryonic-lethal phenotype (Figure 2A). Our results demonstrate that the Rab5 regulator VPS9a plays an essential role during Arabidopsis embryo development.
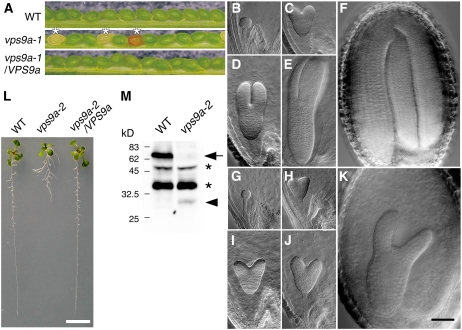
Figure 2.
Characterization of vps9a Mutants.
(A) The vps9a-1 mutation caused embryonic lethality. Seed pods collected from the wild type, heterozygous vps9a-1, and homozygous vps9a-1 harboring a genomic fragment containing VPS9a (vps9a-1/VPS9a) are shown. About 25% of seeds from the heterozygous vps9a-1 mutant showed small, brown, and shriveled appearance (asterisks).
(B) to (K) Embryogenesis of the vps9a-1 mutant was arrested at the torpedo stage. Embryos of the wild type or the heterozygous mutant ([B] to [F]) and the homozygous vps9a-1 mutant ([G] to [K]) are shown. (B) and (G), (C) and (H), (D) and (I), (E) and (J), and (F) and (K) are from the same seed pods. Bar = 50 μm.
(L) The vps9a-2 mutation affected root development. Nine-day-old seedlings of the wild type, vps9a-2, and vps9a-2 transformed with a genomic fragment containing VPS9a (vps9a-2/VPS9a) are shown. Bar = 5 mm.
(M) Accumulation of VPS9a protein in wild-type and vps9a-2 plants. Total protein lysates prepared from seedlings of the wild type and the vps9a-2 mutant were analyzed by immunoblotting using an anti-VPS9a antibody. The arrow and arrowhead represent the wild-type and the truncated VPS9a proteins, respectively. The asterisks indicate nonspecific bands.
To examine mutant embryogenesis in more detail, we observed the development of embryos in cleared seed pods from the heterozygous vps9a-1 mutants (Figures 2B to 2K). The development of the homozygous vps9a-1 mutant (Figures 2G to 2K) was delayed from the early globular stage, indicating that the function of VPS9a is required from very early stages of embryogenesis (Figures 2B and 2G).
vps9a-2 Is Defective in Root Development
To understand the role of VPS9a in postembryonic development, we next characterized a weaker mutant allele of VPS9a, vps9a-2. In this allele, T-DNA is inserted in the fifth exon, which is located behind the Vps9 domain–coding region (Figure 1A). Based on the analysis of transcripts in vps9a-2 by 3′ rapid amplification of cDNA ends, this mutant allele was expected to express an incomplete polypeptide with a molecular mass of 39 kD, which consists of the N-terminal 326 amino acid residues of VPS9a and an additional 15 residues translated from the vector sequence. By immunoblotting, however, only a faint band of 30 kD was detected in the vps9a-2 plant lysate (Figure 2M). Quantitative comparison of the amounts of accumulating products was not possible from this result, because we realized that the anti-VPS9a polyclonal antibody used in this experiment preferentially recognized the C-terminal region of VPS9a (data not shown), but it is likely that the VPS9a-2 protein is unstable and partially degraded in the mutant plants.
The vps9a-2 mutant apparently completed embryogenesis normally, but some postembryonic defects were observed in root development. The elongation of the primary root was severely repressed in vps9a-2, and the branching pattern was also affected, causing abnormal architecture of the root system (Figure 2L). The growth retardation of the primary root was observed at 3 d after germination (Figure 3B), and the average root length of 10-d-old mutant seedlings was 13.0 ± 5.2 mm (n = 20), which was approximately three times shorter than the length of wild-type roots (41.0 ± 2.9 mm [n = 20]) (Figure 3B). The mutant phenotypes of vps9a-2 were complemented by the genomic fragment containing the VPS9a gene (Figures 2L and 3A). These results further demonstrate that VPS9a function is required during postembryonic root growth.
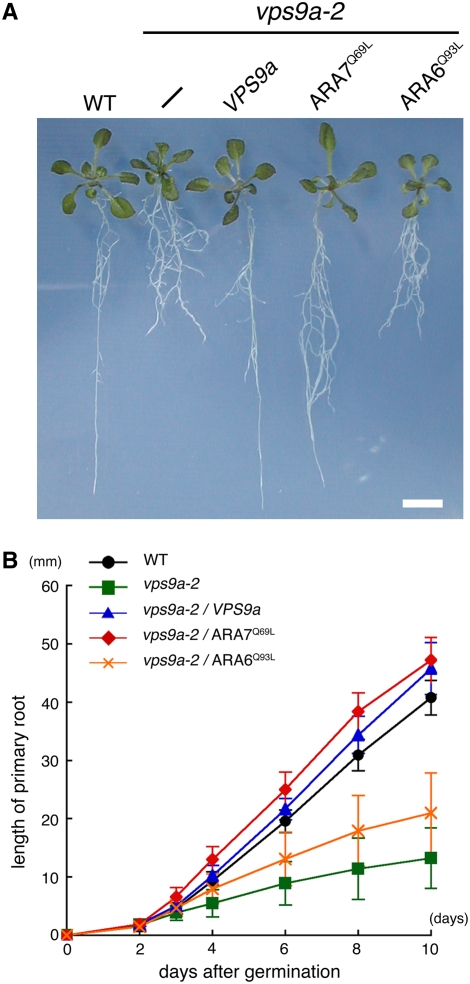
Figure 3.
GTP-Fixed ARA7 Suppressed the Defect in Root Elongation of vps9a-2.
(A) Constitutively active mutants of ARA7 and ARA6 are overexpressed in vps9a-2. Fourteen-day-old seedlings of the wild type, vps9a-2, vps9a-2 transformed with the VPS9a gene, and vps9a-2 expressing the constitutively active mutant of ARA7 or ARA6 are shown. Bar = 5 mm.
(B) Time course of the primary root elongation of seedlings. Each point represents the mean of 20 seedlings. Error bars indicate sd.
vps9a Mutations Affect Cell Plate Formation and Cell Wall Deposition
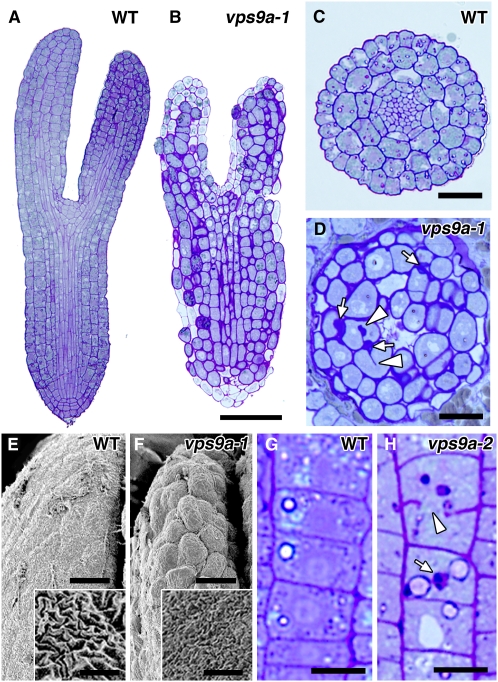
Next, we prepared plastic sections of the two mutants of VPS9a and observed them by light microscopy to examine the effects of mutations on tissue and cell structures. As shown in Figures 4A to 4D, the cell file alignment was disordered in both the cotyledon and the hypocotyl in vps9a-1 embryos at the torpedo stage, probably because the patterns of cell division were severely affected. Cells were significantly hypertrophied in such embryos (Figures 4B and 4D), and incomplete cell plates (arrowheads in Figure 4D) and abnormal cell wall depositions (arrows in Figure 4D) were frequently observed. Because similar phenotypes were also observed even in the 1.5-d-old root of the leaky mutant, vps9a-2 (Figures 4G and 4H), at which stage the root elongation defect was not yet apparent, the defects in cell plate formation and cell wall depositions could be direct consequences of the failure to activate Rab5s. These phenotypes were also confirmed by scanning electron microscopy (Figures 4E and 4F). The vps9a-1 embryos showed a bumpier surface, because of hypertrophied cells in disordered alignment. The texture of the cell wall was affected as well: the furrows observed on the cell wall were shallower on the mutant (insets in Figures 4E and 4F), which may be due to the impaired cell wall biogenesis or maintenance in the mutant.
Figure 4.
The Cell Plate and the Cell Wall Were Affected by vps9a Mutations.
(A) and (B) Longitudinal sections of wild-type (A) and vps9a-1 (B) embryos at the torpedo stage. Bar = 50 μm.
(C) and (D) Traverse sections of the hypocotyls of wild-type (C) and vps9a-1 (D) embryos at the torpedo stage. Incomplete cell plates (arrowheads) and abnormal cell wall depositions (arrows) were observed. Bars = 20 μm.
(E) and (F) Scanning electron micrographs of wild-type (E) and vps9a-1 (F) embryos at the torpedo stage. The cell wall texture was also affected by the vps9a-1 mutation (insets). Bars = 10 μm and 2 μm (insets).
(G) and (H) Longitudinal sections of the primary roots of 1.5-d-old wild-type (G) and vps9a-2 (H) plants. Incomplete cell plates (arrowhead) and aberrant cell walls (arrow) were observed in the vps9a-2 root. Bars = 10 μm.
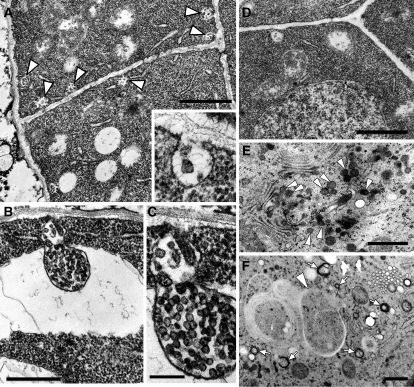
Abnormal Membrane Structures Accumulated in the vps9a-1 Mutant
To elucidate the effects of the vps9a mutation at the ultrastructural level, we next observed the vps9a-1 embryos at different developmental stages by transmission electron microscopy. The most conspicuous feature observed in the mutant embryos at the early globular stage, when only a little morphological alteration was detectable by light microscopy, was the accumulation of multivesiculated membrane structures associated with the plasma membrane (Figure 5A). These structures contained vesicular membrane structures between the plasma membrane and the cell wall (Figures 5A, inset, and 5C). Similar membranous structures were also occasionally seen in wild-type embryos, but the frequency was much lower than in the vps9a-1 mutant (0.018 ± 0.0044 paramural bodies [PMBs]/μm plasma membrane in wild-type cells [n = 50] versus 0.16 ± 0.020 PMBs/μm plasma membrane in vps9a-1 cells [n = 67]) (Figure 5D). Similar structures with a much larger size were also observed in mutant cells, which often appeared to be engulfed into vacuoles (Figure 5B). Structures with such characteristic morphology have been described in the literature as PMBs or plasmalemmasomes, whose biogenesis and function have been disputed for a long time (Marchant and Robards, 1968; Herman and Lamb, 1992; An et al., 2006b). Our observation in vps9a-1 embryos strongly suggests that the biogenesis of the PMBs should be regulated by Rab5 activity. Other organelles, such as the Golgi apparatus, vacuoles, and the endoplasmic reticulum, showed no significant difference in their structure from those in wild-type cells (data not shown).
Figure 5.
Accumulation of Aberrant Membrane Structures in the vps9a-1 Mutant.
Samples were fixed by the conventional ([A] to [D]) or high-pressure freezing ([E] and [F]) method.
(A) Abnormal accumulation of the PMBs (arrowheads) in the globular stage embryo of vps9a-1. The inset shows a high-magnification image of the PMBs. Bar = 1 μm.
(B) Large PMBs were sometimes engulfed by the vacuole. Bar = 500 nm.
(C) Magnified view of the PMBs shown in (B). Bar = 200 nm.
(D) Wild-type embryo at the globular stage, at which PMBs were not observed so frequently. Bar = 1 μm.
(E) and (F) Abnormal accumulation of dense vesicles (arrowheads in [E]), small cup- or ring-shaped structures (arrows in [F]), and autophagosome-like structures (arrowhead in [F]) observed in the torpedo stage embryo of vps9a-1. Bars = 500 nm.
We also observed the mutant embryos at the torpedo stage, whose development was arrested almost completely. We rarely found PMB-like structures in either mutant or wild-type cells at this stage. Instead, other abnormal membrane structures, such as dense vesicles around the Golgi apparatus (arrowheads in Figure 5E), small cup- or ring-shaped structures (arrows in Figure 5F), and autophagosome-like structures (arrowhead in Figure 5F), were accumulating. The protein storage vacuoles in the mutant cells showed lower electron density than those in the wild-type cells (data not shown). The accumulating Golgi-derived vesicles had a similar electron density to that of protein storage vacuoles observed in wild-type embryos; thus, these vesicles might be intermediates of the storage protein transport from the Golgi to vacuoles.
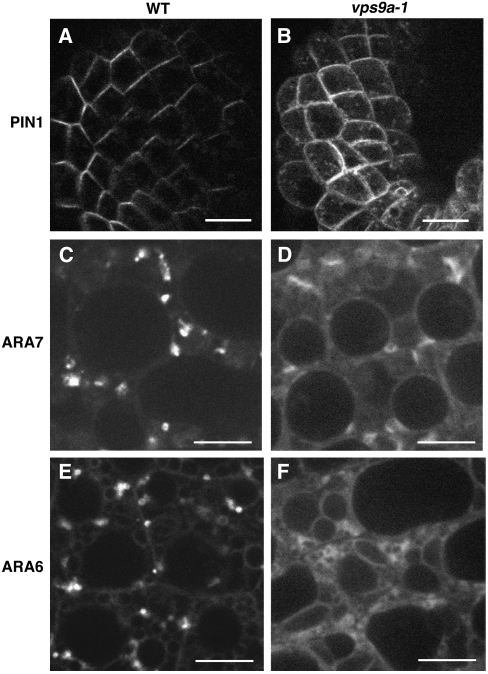
VPS9a Is Essential for the Establishment or Maintenance of Polar PIN1 Localization
The auxin efflux carrier PIN1 is polarly localized in various tissues and continuously cycles between endosomes and the plasma membrane (Steinmann et al., 1999; Geldner et al., 2003). This cycling is dependent on GNOM, a GEF of Arf GTPase, whose function is also essential for the correct localization of ARA7 and RHA1 (Geldner et al., 2003; Ueda et al., 2004). To examine whether the activation of Rab5s catalyzed by VPS9a plays some role in PIN1 trafficking, we examined the PIN1-GFP localization in the vps9a-1 embryos. In cotyledon epidermal cells of wild-type embryos at the heart stage, PIN1-GFP mainly localized on the apical region of the plasma membrane (Figure 6A) (Steinmann et al., 1999). On the other hand, PIN1-GFP lost the polarity and resided on the entire region of the plasma membrane in the cells of the mutant embryos (Figure 6B). In these cells, some punctate structures were also observed on or beneath the plasma membrane. These observations indicate that the activity of VPS9a is also essential for the establishment or maintenance of the polar localization of PIN1.
Figure 6.
Subcellular Localization of PIN1, ARA7, and ARA6 in the vps9a-1 Mutant.
(A) and (B) An auxin efflux carrier, PIN1, was mislocalized in the vps9a-1 embryo. Bars = 10 μm.
(C) to (F) In vps9a-1 cells, GFP-ARA7 ([C] and [D]) and ARA6-GFP ([E] and [F]) were dispersed in the cytosol, whereas they localized on endosomes in wild-type cells. Bars = 5 μm.
The vps9a Mutation Affects the Localization of ARA7 and ARA6
The conventional-type Rab GTPase cycles between cytosol and membranes depending on their nucleotide state. The GDP-bound Rab is held in cytosol by the Rab GDI, and when activated to the GTP-bound state by the GEF, Rab becomes associated with the membrane (Zerial and McBride, 2001; Seabra and Wasmeier, 2004). As for the plant-unique ARA6, which recycles between membrane and cytosol independently of Rab GDI (Ueda et al., 2001), the GDP-fixed mutant protein also disperses into cytosol when expressed in Arabidopsis plants (K. Ebine and T. Ueda, unpublished data). Thus, if VPS9a is the major activator of all Rab5 members in Arabidopsis, the defect in its GEF activity will cause the delocalization of all Rab5 members from the endosomal membranes to the cytosol. To test this possibility, we examined the subcellular localization of GFP-tagged Rab5 members in vps9a-1 embryos. To avoid possible mislocalization due to overexpression, we constructed translational fusions of their genomic sequences and GFP. As reported previously (Ueda et al., 2004), GFP-ARA7 and ARA6-GFP localized mainly on punctate organelles in wild-type embryos (Figures 6C and 6E). On the other hand, both GFP-ARA7 and ARA6-GFP showed diffuse localization in mutant embryos (Figures 6D and 6F). These results indicate that ARA6 and ARA7 were not activated properly in the vps9a-1 mutant, which strongly suggests that VPS9a is the direct and apparently the sole activator of both types of Rab5 members in vivo.
The GTP-Fixed Mutant of ARA7 Suppresses vps9a-2
vps9a-2 mutant plants show defects in primary root elongation and branching pattern. If these are caused by faulty Rab5 activation, the expression of the constitutively active (GTP-fixed) form of Rab5 is expected to alleviate the phenotypes. To verify this hypothesis, we overexpressed GTP-fixed mutants of ARA7 (Q69L) and ARA6 (Q93L) under the regulation of the cauliflower mosaic virus 35S promoter. As shown in Figure 3, ARA7Q69L clearly suppressed the defect in the primary root elongation of vps9a-2. By contrast, ARA6Q93L had no obvious effect on any vps9a-2 phenotypes, although ARA6Q93L was expressed at a comparable level to ARA7Q69L in suppressed lines (see Supplemental Figure 3 online). As the morphological alteration of endosomes caused by ARA6Q93L (Ueda et al., 2001) was also observed in Arabidopsis plants expressing it (K. Ebine and T. Ueda, unpublished data), ARA6Q93L should be active when expressed in Arabidopsis plants. Thus, our results strongly suggest that the defects in the primary root of vps9a-2 are attributed to the faulty activation of the conventional-type Rab5s. In other words, and more importantly, the physiological functions of the plant-unique ARA6 and the conventional-type ARA7 and RHA1 are clearly differentiated, even though they are all activated by the same GEF, VPS9a.
DISCUSSION
VPS9a Activates All Rab5 Members
In this study, we have isolated and characterized VPS9a, a specific activator of Rab5 members in Arabidopsis. As expected from its conserved Vps9 domain, VPS9a shows a clear GEF activity in vitro in the conventional-type Rab5s in Arabidopsis, ARA7 and RHA1. Interestingly, VPS9a also activates the plant-unique Rab5 homolog, ARA6, in spite of its unique structure. This GEF activity is specific to Rab5 members, and the selectivity seems to be conferred by a similar mechanism between animals and plants. Delprato et al. (2004) recently reported that a nonacidic residue (Ala/Ser) preceding the conserved Phe residue in the switch I region in mammalian Rab5 members (Rab5, Rab21, and Rab22) plays a crucial role in the interaction with the Vps9 domain, and an acidic amino acid residue is conserved at this position in other subfamilies of Rab GTPases. Of 57 Rab GTPases in Arabidopsis, only three Rab5 members, ARA6, ARA7, and RHA1, have a nonacidic residue at this position, whereas all other Rab GTPases conserve Asp or Glu at the corresponding site (see Supplemental Figure 4 online). This provides strong structural support that VPS9a specifically activates the three Rab5 members in Arabidopsis.
Several lines of evidence indicate that all Rab5 members are indeed activated by VPS9a in vivo. When expressed in the vps9a-1 mutant embryos, ARA6 and ARA7 tagged with GFP are both dispersed into cytosol, whereas these proteins are localized on punctate organelles in wild-type embryos (Figures 6C and 6D). As GDP-fixed mutants of Rab5 members show diffuse localization in cytosol (Sohn et al., 2003; Kotzer et al., 2004) (K. Ebine and T. Ueda, unpublished data), the above results support the notion that ARA6 and ARA7 stay at GDP-bound states in the mutant embryos because of the defect in VPS9a. Taking into account that we could not detect any expression of the VPS9b gene, VPS9a may represent the major active Rab5 GEF in Arabidopsis.
Genetic data further support the proposed function of VPS9a. Overexpression of the GTP-fixed ARA7 mutant rescues the root elongation phenotype of vps9a-2 (Figure 3). Similar genetic interactions between Rabs and cognate GEFs have also been reported in yeast studies. Duplication of the SEC4 gene, which encodes a Rab GTPase regulating the fusion between secretory vesicles and the plasma membrane, suppresses the defect of sec2, a mutant of the GEF for Sec4 (Nair et al., 1990). The overexpression of Ypt6 also suppresses the mutation of its GEF, Ric1 and Rgp1 (Siniossoglou et al., 2000). Furthermore, other classes of small GTPases, such as Ras, Sar/Arf, and Rho/Rac families, show similar genetic interactions (Broek et al., 1987; Nakano and Muramatsu, 1989; Ziman and Johnson, 1994). Another important finding is that the overexpression of GTP-fixed ARA6 does not suppress the root phenotype of vps9a-2 mutant plants. This result strongly suggests that the plant-unique ARA6 has different roles from conventional-type Rab5s, ARA7 and RHA1, in the development of Arabidopsis.
These observations indicate that the three Rab5 members in Arabidopsis, ARA7, RHA1, and ARA6, are functionally and structurally differentiated, even though they are all activated by the same GEF, VPS9a.
Plants Seem to Have Evolved a Unique Rab5 System
There is a remarkable difference in the composition of endocytic Rab GTPases between animals and plants. In plants, we cannot find any homologs of Rab4 and Rab9, which regulate the endocytic pathway coordinately with Rab5 in animal cells (Rutherford and Moore, 2002; Ueda and Nakano, 2002). Instead, two distinct types of Rab5 members, ARA6 type and the conventional type, are present in higher plants (Ueda et al., 2001). This fact implies that plants and animals evolved the roles of Rabs in endocytosis in different ways. The regulation of Rab5 also seems to be differentiated between plants and animals. As we reported previously (Ueda et al., 2001), ARA6 is cycled between membranes and cytosol independently of Rab GDI, whereas the recycling of ARA7 and RHA1 is Rab GDI–dependent. In this study, we have further demonstrated that plants and animals employ significantly different systems for the activation of Rab5. Virtually only one Rab5 GEF regulates two different types of Rab5 members in Arabidopsis. On the other hand, there are several kinds of Rab5 GEFs in animals that activate Rab5 at different steps in the endocytic pathway. For example, at the beginning of endocytosis, Rme-6 regulates the formation of clathrin-coated vesicles on the plasma membrane (Sato et al., 2005). Rabex-5 accelerates the homotypic fusion between early endosomes (Horiuchi et al., 1997), and the RIN family appears to be involved in the external stimuli–induced endocytosis of specific cargos such as EGF receptors (Tall et al., 2001; Barbieri et al., 2003). It is also notable that animal Rab5 GEFs contain various domain structures in addition to the Vps9 domain (e.g., a zinc finger motif and a ubiquitin binding domain in Rabex-5, a Ras GTPase–activating protein domain in Rme-6, SH2 and Ras association domains in RIN family members, Ran GEF and PH domains in ALS2, and ankyrin repeats in ANKRD27) (Carney et al., 2005). Such diverse domain structures in Rab5 GEFs suggest that various signaling pathways are integrated at the activation step of Rab5 in animal cells. On the other hand, we could not find any known domain structure in VPS9a other than the Vps9 domain, which locates in the N-terminal half of protein. Plants have probably developed a different signaling mechanism via the endocytic pathway in which the C-terminal region of VPS9a might play a plant-specific role.
The Rab5 Regulator VPS9a Is Essential for Plant Development
In this study, we have demonstrated that the activation of Rab5s by VPS9a is essential in various plant functions. The Rab5-mediated endocytosis and/or vacuolar transport appears to be indispensable for embryogenesis, as shown by the embryonic lethality of vps9a-1. Postembryonic development also requires VPS9a activity; when it is impaired, organ development is severely affected (Figure 2). At the subcellular level, we frequently observe incomplete cell plates and abnormal cell wall depositions, which are seen even in the embryos and roots that show a normal macroscopic appearance (Figure 4). This result might support the direct involvement of the endocytic pathway in cell plate formation and cell wall biogenesis or maintenance, as was proposed recently. Baluska et al. (2005) demonstrated that cell wall pectins recycle between the cell wall and endosomes, from which they proposed that endocytosis should be involved in cell wall organization. Dhonukshe et al. (2006) reported that endocytosis contributes directly to cell plate formation by transporting materials for newly forming cell plates from preexisting cell walls. These functions of endocytosis are probably impaired in the vps9a mutant due to the faulty activation of Rab5 members, although we cannot rule out the possibility that the secretion of cell wall materials is affected by the vps9a mutations.
Some phenotypes observed in vps9a mutants may be explained in the context of polar auxin transport. It is well known that polar auxin transport plays crucial roles in various aspects of plant development, including embryogenesis and root development (Benková et al., 2003; Friml et al., 2003). As the localization of the PIN1 protein is disarrayed in vps9a-1 (Figures 6A and 6B), polar auxin transport is likely to be disorganized in this mutant. We have also reported that the endosomal localization of ARA7 and RHA1 in protoplasts depends on GNOM function (Geldner et al., 2003; Ueda et al., 2004). This raises the possibility that GNOM-dependent recycling of PIN1 at the plasma membrane (Geldner et al., 2003) may involve an ARA7- and RHA1-positive subpopulation of endosomes. These observations strongly suggest that the Rab5 members and VPS9a are involved in the regulation of PIN localization coordinately with the Arf/GNOM system, which serves for proper auxin transport. Further detailed studies on the auxin response and auxin accumulation in vps9a would be an interesting future project.
Subcellular Effects of the vps9a Mutation
The most notable ultrastructural phenotype of vps9a-1 observed at early stages of embryogenesis is the accumulation of PMBs. PMBs were discerned by transmission electron microscopy several decades ago, and subsequent morphological studies revealed that PMBs are frequently observed in plant tissues in which exocytosis and endocytosis occur actively (Marchant and Robards, 1968; Wang et al., 2005; An et al., 2006a, 2006b). Some studies also proposed that PMBs are involved in the internalization of cell wall components and plasma membrane proteins (Cox and Juniper, 1973; Herman and Lamb, 1992). However, the precise function and biogenetic process remained unknown for a long time. Although it is still unclear whether the PMBs are intermediates of endocytosis or the fusing of multivesicular bodies with plasma membranes, our result that PMBs accumulate in the vps9a-1 mutant strongly suggests that the activation of Rab5 members by VPS9a is required for the biogenesis of PMBs.
At the torpedo stage, vps9a-1 embryos also accumulate other membrane structures in the cells, such as Golgi-derived vesicles with high electron densities, cup- or ring-shaped membrane structures, and autophagosome-like structures. At this stage, embryogenesis is almost halted and it is not easy to distinguish the primary defect from secondary or tertiary effects, but phenotypes observed here still suggest the importance of VPS9a function. Electron densities of the contents in the accumulating vesicles are high, as in protein storage vacuoles in wild-type cells, implying that these vesicles are the intermediates in the biosynthetic pathway from the Golgi apparatus to vacuoles. This phenotype might be caused by the impairment of the proposed role of Rab5 members in the trafficking from the Golgi apparatus to vacuoles (Sohn et al., 2003; Lee et al., 2004), but it is also possible that this phenotype was caused by the improper recycling of receptors for storage proteins from the prevacuolar compartment/late endosomes, as is the case with the maigo1 mutant, which is defective in this recycling pathway (Shimada et al., 2006). vps9a-1 also accumulates autophagosome-like structures. A similar phenotype is reported for the vacuoleless1 (vcl1) mutant, which is defective in vacuolar biogenesis (Rojo et al., 2001). That vps9a-1 and vcl1 show embryonic lethality and the accumulation of autophagosome-like structures, although otherwise they look quite different, suggests that the membrane transport pathways that involve VPS9a and VCL1 partially overlap in the biosynthetic and/or endocytic pathways.
As we have demonstrated in this study, plants have developed a unique system to regulate Rab5 members. The next important question is how this unique system contributes to higher order plant functions. We are now investigating genetic interactions between rab5 and other mutations with a special focus on the endocytic pathway, and the search for effector proteins for Rab5 members is now under way. These approaches will lead us to a better understanding of the molecular mechanisms and the physiological significance of plant endocytosis.
METHODS
Plant Materials and Plasmids
vps9a-1 (SALK_018174) and vps9a-2 (GABI_557C02) were obtained from the ABRC (Alonso et al., 2003) and GABI-KAT (Rosso et al., 2003), respectively, and backcrossed at least three times with wild-type Arabidopsis thaliana (Columbia). For the complementation analysis, a 6.1-kb genomic fragment subcloned into pGWB1 (a kind gift from T. Nakagawa), which contains 2.8-kb upstream and 1.1-kb downstream sequences of VPS9a, was used to transform vps9a mutants. The translational fusions between GFP and Rab5 members were generated using the technique of fluorescence tagging of full-length proteins (Tian et al., 2004). The chimeric genes were then subcloned into pGWB1 and introduced into wild-type and mutant plants. For overexpression of ARA6Q93L and ARA7Q69L, we employed pGWB2, which contains a cauliflower mosaic virus 35S promoter and a Nos terminator. The transformation of Arabidopsis plants was performed via floral dipping using Agrobacterium tumefaciens (strain GV3101∷pMP90) (Clough and Bent, 1998).
Expression and Purification of GST Fusion Proteins
VPS9a and Rab proteins, Rab5s (ARA7, RHA1, and ARA6), Rab1 (ARA5), Rab2 (RAB2A), Rab7s (RAB71 and RAB75), Rab11 (ARA4), and Rab18 (RAB18-1), which were fused to GST, were expressed in Escherichia coli BL21 (DE3) using pGEX 4T-1 vector (GE Healthcare). The cells expressing fusion proteins were collected and resuspended in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% mercaptoethanol, and protease inhibitor cocktail [GE Healthcare]), sonicated, and centrifuged at 10,000g for 30 min. Supernatants were loaded onto a glutathione–Sepharose 4B column (GE Healthcare) and washed with 10 column volumes of washing buffer (50 mM Tris, pH 8.0, 500 mM NaCl, and 0.1% mercaptoethanol), and then fusion proteins were eluted with elution buffer (20 mM reduced glutathione, 50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% mercaptoethanol).
Antibody Preparation
GST-tagged VPS9a was used as an antigen to raise the anti-VPS9a polyclonal antibody. The obtained antibody was purified by protein G affinity column chromatography (GE Healthcare). Anti-RHA1 antibody was raised in a rabbit using a synthetic peptide (LPNGPGATAV) and affinity-purified using the same peptide. Anti-ARA6 (Ueda et al., 2001), anti-ARA4 (Anai et al., 1994), and anti-ARA7 (Haas et al., 2007) antibodies have been described elsewhere.
Nucleotide-Exchange Assay
Nucleotide exchange in the purified GST-tagged Rabs was measured by monitoring the change of autofluorescence from intrinsic Trp accompanying the structural conversion from the inactive to the active state (Pan et al., 1995; Antonny et al., 2001). Each purified Rab protein was preloaded with GDP and incubated with or without GST-VPS9a in reaction buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.5 mM MgCl2) for 100 s at 25°C. Then, GMP-PNP was added to 0.1 mM to start the nucleotide-exchange reaction. The fluorescence shift was detected with a fluorescence spectrophotometer (model F-2500; Hitachi High Technologies) at an excitation wavelength of 298 nm and an emission wavelength of 340 nm. The assay was repeated at least three times for each Rab GTPase.
In Vitro Pull-Down Assay
Each Rab GTPase was expressed in yeast strain YPH414 (MATa Δpep4:TRP1 ura3 lys2 ade2 trp1 his3 leu2) under the control of the constitutive TDH3 promoter. Collected cells were collapsed by vortexing with glass beads in PBS plus protease inhibitor cocktail (GE Healthcare) and 1% Triton X-100, and collected lysates were coincubated with GST-VPS9a prebound to glutathione–Sepharose 4B resin (GE Healthcare) for 60 min at room temperature. The protein complexes bound to resins were washed three times and subjected to immunoblot analysis. We confirmed that the results presented here were reproducible by independent assays repeated at least three times.
Yeast Two-Hybrid Assay
The cDNAs for wild-type Rab and mutant versions of Rab were subcloned into pAD-GAL4-2.1 (Stratagene). The open reading frame of VPS9a was subcloned into pBD-GAL4-GWRFC (from T. Demura at RIKEN). Plasmids containing each Rab and VPS9a were introduced into AH109 strain (Clontech). Empty vectors were used for negative controls. Transformation was performed at least twice independently, and at least three colonies were checked for interaction for each transformation.
Microscopy
For whole-mount visualization of embryos, ovules were cleared as described previously (Aida et al., 1997). For the observation of GFP fluorescence, embryos were mounted in 5% glycerol and observed with a fluorescence microscope (model BX51; Olympus) equipped with a confocal scanner unit (model CSU10; Yokogawa Electric) and a cooled CCD camera (model ORCA-AG; Hamamatsu Photonics). Images were processed with IPLab software (BD Biosciences).
For preparing sectioned specimens, samples were processed by a conventional fixation method (Karnovsky, 1965) or a high-pressure freezing and freeze substitution method (Ichikawa and Ichikawa, 1987; Otegui and Staehelin, 2000). Samples were embedded in Epon 812 or Spurr's resin and sectioned at 60- to 70-nm and 350- to 700-nm thickness for transmission electron microscopy and light microscopy, respectively. Specimens were stained with uranyl acetate and lead citrate (Reynolds, 1963) for transmission electron microscopy (model JEM-2000FX II; JEOL) and with 0.5% toluidine blue O (Sigma-Aldrich) for light microscopy. For scanning electron microscopy, conventionally fixed samples were dehydrated and then dried with a critical point dryer (model HCP-2; Hitachi High Technologies). Samples were coated with platinum/palladium using an ion coater (model IB-3; EIKO Engineering) and observed with a FE-SEM scanning electron microscope (model JSM-6330F; JEOL).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At3g19770 (VPS9a), At5g09320 (VPS9b), At3g54840 (ARA6/RABF1), At5g45130 (RHA1/RABF2a), At4g19640 (ARA7/RABF2b), At3g18820 (RAB71/RABG3f), At1g22740 (RAB75/RABG3b), At2g43130 (ARA4/RABA5c), At1g02130 (ARA5/RABD2a), At4g17170 (RAB2A/RABB1c), and At1g43890 (RAB18-1/RABC1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. VPS9a Does Not Activate Rab1, Rab2, Rab11, and Rab18 in Vitro.
Supplemental Figure 2. VPS9a Does Not Interact with Rab7 and Rab11.
Supplemental Figure 3. Constitutively Active ARA7 (ARA7Q69L), but Not ARA6 (ARA6Q93L), Suppresses the vps9a-2 Mutation.
Supplemental Figure 4. Alignment of the Switch I Region of Plant Rab GTPases.
Supplementary Material
Acknowledgments
We thank the Salk Institute and the Max Planck Institute for providing T-DNA insertion mutants of Arabidopsis. We also thank T. Demura (RIKEN) for pBD-GAL4-GWRFC, T. Nakagawa (Shimane University) for pGWB vectors, and J. Friml (University of Tübingen) for seeds of PIN1:GFP. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a fund from the Bioarchitect Project of RIKEN. T.G. was the recipient of a Junior Research Associate fellowship from RIKEN.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Takashi Ueda (tueda@biol.s.u-tokyo.ac.jp).
Online version contains Web-only data.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- An, Q., Ehlers, K., Kogel, K.H., van Bel, A.J., and Huckelhoven, R. (2006. a). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172 563–576. [DOI] [PubMed] [Google Scholar]
- An, Q., Huckelhoven, R., Kogel, K.H., and van Bel, A.J. (2006. b). Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 8 1009–1019. [DOI] [PubMed] [Google Scholar]
- Anai, T., Matsui, M., Nomura, N., Ishizaki, R., and Uchimiya, H. (1994). In vitro mutation analysis of Arabidopsis thaliana small GTP-binding proteins and detection of GAP-like activities in plant cells. FEBS Lett. 346 175–180. [DOI] [PubMed] [Google Scholar]
- Antonny, B., Madden, D., Hamamoto, S., Orci, L., and Schekman, R. (2001). Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 3 531–537. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Liners, F., Hlavacka, A., Schlicht, M., Van Cutsem, P., McCurdy, D.W., and Menzel, D. (2005). Cell wall pectins and xyloglucans are internalized into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 225 141–155. [DOI] [PubMed] [Google Scholar]
- Barbieri, M.A., Kong, C., Chen, P.I., Horazdovsky, B.F., and Stahl, P.D. (2003). The SRC homology 2 domain of Rin1 mediates its binding to the epidermal growth factor receptor and regulates receptor endocytosis. J. Biol. Chem. 278 32027–32036. [DOI] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Bolte, S., Brown, S., and Satiat-Jeunemaitre, B. (2004). The N-myristoylated Rab-GTPase m-Rabmc is involved in post-Golgi trafficking events to the lytic vacuole in plant cells. J. Cell Sci. 117 943–954. [DOI] [PubMed] [Google Scholar]
- Broek, D., Toda, T., Michaeli, T., Levin, L., Birchmeier, C., Zoller, M., Powers, S., and Wigler, M. (1987). The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell 48 789–799. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., Mustol, P.A., Schu, P.V., and Emr, S.D. (1996). A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol. Cell. Biol. 16 2369–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney, D.S., Davies, B.A., and Horazdovsky, B.F. (2005). Vps9 domain-containing proteins: Activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16 27–35. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., and Zerial, M. (2000). Purification and identification of novel Rab effectors using affinity chromatography. Methods 20 403–410. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cox, G.C., and Juniper, B.E. (1973). Autoradiographic evidence for paramural-body function. Nat. New Biol. 243 116–117. [Google Scholar]
- Delprato, A., Merithew, E., and Lambright, D.G. (2004). Structure, exchange determinants, and family-wide Rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 118 607–617. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., Baluska, F., Schlicht, M., Hlavacka, A., Samaj, J., Friml, J., and Gadella, T. W., Jr. (2006). Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev. Cell 10 137–150. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Grosshans, B.L., Ortiz, D., and Novick, P. (2006). Rabs and their effectors: Achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, T.J., Sliwinski, M.K., Martinez, D.E., Preuss, M., Ebine, K., Ueda, T., Nielsen, E., Odorizzi, G., and Otegui, M.S. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LIP5. Plant Cell 19 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama, H., Tall, G.G., and Horazdovsky, B.F. (1999). Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 274 15284–15291. [DOI] [PubMed] [Google Scholar]
- Herman, E.M., and Lamb, C.J. (1992). Arabinogalactan-rich glycoproteins are localized on the cell surface and in intravacuolar multivesicular bodies. Plant Physiol. 98 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi, H., Lippe, R., McBride, H.M., Rubino, M., Woodman, P., Stenmark, H., Rybin, V., Wilm, M., Ashman, K., Mann, M., and Zerial, M. (1997). A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90 1149–1159. [DOI] [PubMed] [Google Scholar]
- Ichikawa, M., and Ichikawa, A. (1987). The fine structure of sublingual gland acinar cells of the Mongolian gerbil, Meriones unguiculatus, processed by rapid freezing followed by freeze-substitution fixation. Cell Tissue Res. 250 305–314. [DOI] [PubMed] [Google Scholar]
- Karnovsky, M. (1965). A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27 137A–138A. [Google Scholar]
- Kotzer, A.M., Brandizzi, F., Neumann, U., Paris, N., Moore, I., and Hawes, C. (2004). AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117 6377–6389. [DOI] [PubMed] [Google Scholar]
- Lee, G.J., Sohn, E.J., Lee, M.H., and Hwang, I. (2004). The Arabidopsis Rab5 homologs Rha1 and Ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45 1211–1220. [DOI] [PubMed] [Google Scholar]
- Marchant, R., and Robards, A.W. (1968). Membrane systems associated with the plasmalemma of plant cells. Ann. Bot. (Lond.) 32 457–471. [Google Scholar]
- Nair, J., Muller, H., Peterson, M., and Novick, P. (1990). Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol. 110 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, A., and Muramatsu, M. (1989). A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 109 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M., and Staehelin, L.A. (2000). Syncytial-type cell plates: A novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J.Y., Sanford, J.C., and Wessling-Resnick, M. (1995). Effect of guanine nucleotide binding on the intrinsic tryptophan fluorescence properties of Rab5. J. Biol. Chem. 270 24204–24208. [DOI] [PubMed] [Google Scholar]
- Reynolds, E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Gillmor, C.S., Kovaleva, V., Somerville, C.R., and Raikhel, N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1 303–310. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Russinova, E., Borst, J.W., Kwaaitaal, M., Caño-Delgado, A., Yin, Y., Chory, J., and de Vries, S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5 518–528. [DOI] [PubMed] [Google Scholar]
- Sato, M., Sato, K., Fonarev, P., Huang, C., Liou, W., and Grant, B. (2005). Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat. Cell Biol. 7 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Seabra, M.C., and Wasmeier, C. (2004). Controlling the location and activation of Rab GTPases. Curr. Opin. Cell Biol. 16 451–457. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Koumoto, Y., Li, L., Yamazaki, M., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47 1187–1194. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., Peak-Chew, S.Y., and Pelham, H.R. (2000). Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19 4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, E.J., Kim, E.S., Zhao, M., Kim, S.J., Kim, H., Kim, Y.W., Lee, Y.J., Hillmer, S., Sohn, U., Jiang, L., and Hwang, I. (2003). Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15 1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Takano, J., Miwa, K., Yuan, L., von Wiren, N., and Fujiwara, T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall, G.G., Barbieri, M.A., Stahl, P.D., and Horazdovsky, B.F. (2001). Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1 73–82. [DOI] [PubMed] [Google Scholar]
- Tian, G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T., and Nakano, A. (2002). Vesicular traffic: An integral part of plant life. Curr. Opin. Plant Biol. 5 513–517. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Uemura, T., Sato, M.H., and Nakano, A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40 783–789. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A.C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Kong, L., Hao, H., Wang, X., Lin, J., Samaj, J., and Baluska, F. (2005). Effects of brefeldin A on pollen germination and tube growth. Antagonistic effects on endocytosis and secretion. Plant Physiol. 139 1692–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2 107–117. [DOI] [PubMed] [Google Scholar]
- Ziman, M., and Johnson, D.I. (1994). Genetic evidence for a functional interaction between Saccharomyces cerevisiae CDC24 and CDC42. Yeast 10 463–474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.